Abstract
To explore whether the roles of IL-1 family single nucleotide polymorphisms (SNPs) of the microRNA binding sites (miR-SNPs) in the 3′ untranslated region (3′-UTR) of their target genes in the progression of gastric cancer (GC) and verify the relationship between miR-197 with chronic inflammatory gene-IL1-F5 by microRNA target prediction, a case-control study which consisted of 500 cases and 500 frequency-matched healthy controls was conducted. Single nucleotide polymorphisms were analyzed by polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) or allele-specific PCR (AS-PCR). Association between SNPs and GC risk was evaluated by adjusted odds ratios (ORs) and 95% confidence intervals (CIs) in unconditional logistic regression analyses. Quantitative real-time (qRT) PCR assay and Western Blot analyses were performed to analyze the miR-197 expression and the IL1-F5 expression. The variant homozygote and heterozygote genotype of rs9005 in IL-1RN were significantly associated with increased risks of GC (ORadjusted [95%CI]: 1.71[1.04–2.81] and ORadjusted[95%CI]: 1.36 [1.04–1.78]). Compared with the wild heterozygote genotype, the variant heterozygote genotype of rs2472188 and rs2515401 in IL-1F5 polymorphisms were significantly associated with increased GC risks (ORadjusted [95%CI]: 1.51[1.15–1.99] and ORadjusted[95%CI]: 1.36[1.04–1.76]), but no significant differences existed in other 7 IL-1 family SNPs (rs2856836 in IL-1A, rs3732131 in IL-1R1, rs1135354 and rs3771157 in IL-18RA, rs3180235, rs957201 and rs2515402 in IL-1F5) with GC. The recombinant plasmid-pGenesil-1-miR-197 could upregulate the expression of miR-197 and downregulate the expression of IL-1F5 in human gastric cancer cell lines SGC-7901 and BGC-823 cells after transfection, and the miR-197 inhibitor could facilitate the expression of IL1-F5 after transfecting the same cell lines. These results suggested that SNPs in the IL-1 family genes play important roles in the development of GC and the IL-1F5 might be the target gene of miR-197, and miR-197 might negatively regulate its expression.
INTRODUCTION
Epidemiological studies were indicated that chronic inflammatory responses, cytokines, and immune mediators participated in the progressions of several cancers including gastric cancer (GC).1 It is widely recognized that one of the main causes of chronic inflammation of the stomach is infection with Helicobacter pylori (Hp). The infection of Hp can strongly involve the cytokine response of the immune system, particularly the interleukin family. Increasing evidence had demonstrated that the involvement of inflammatory cytokines such as interleukin-1 (IL-1), interleukin-1 receptor 1 (IL-1R1), and tumor necrosis factor (TNF)-α could influence the susceptibility of GC.2,3
MiRNA combines the 3′-UTR of its target gene mRNA in the form of base pairing, then regulate gene expression through inhibiting the translation process. 4 The SNPs of pathway genes that regulate the maturation of miRNAs are able to influence transcription, processing, transportation, and recognition of target genes by affecting the expression or function of the pathway genes; therefore SNPs could change the regulatory function of the miRNA.5 Studies have found that the overexpression of some miRNAs can negatively regulate antioncogenes or take part in the process of cell proliferation and apoptosis, so as to act as an oncogene.6 The functional SNPs in 3′ UTR of several genes have been reported to be associated with GC by impacting the expression of oncogenes.7
Recently, great progress about miRNA studies have been made in the field of cancer research, therefore, combining the certain function of miRNA with the SNP of its target sequence can better investigate the specific function of miRNAs in target gene regulation. To explore whether the IL-1 gene polymorphisms are involved in the gastric carcinogenesis, a case–control study was carried out to investigate the association between the SNPs and the risk for GC in Chinese-Han population and then qRT-PCR and Western Blot analyses were carried out to verify whether IL-1F5 is the target gene of miR-197.
MATERIALS AND METHODS
Study Subjects
According to the sample size calculation method of case-control study, totally 840 cases and controls were needed. In order to improve the statistical power, 500 GC cases and 500 healthy controls were included in this study. Newly diagnosed patients by histopathological examination at the First Affiliated Hospital of Zhengzhou University from May 1, 2012, to June 30, 2014, were eligible in this study. None of the cases received any treatment such as radiotherapy or chemotherapy before enrollment. Healthy controls were cancer-free subjects randomly selected from an epidemiological investigation in Henan Province during the corresponding period with case enrollment and were frequency-matched with cases by age (±5 years) and sex. The exclusion criteria for controls included: any kinship with GC patients, any sign or symptom of digestive disease previously or a history of other malignant tumors. For each individual, a 5 mL sample of venous blood was drawn and delivered to the laboratory for DNA extraction. Informed consents were obtained from all individuals and the protocol was approved by the Medical Ethics Committee of Zhengzhou University.
Data Collection
The information about demographic characteristics such as age, sex, tobacco use, alcohol intake, and family history of cancer was collected through a structured questionnaire. The definition of smokers is those who smoked more than 1 cigarette per day for >6 months, and subjects who consumed >100 g of alcoholic drinks once and more than once a week for at least 6 months were regarded as drinkers.
SNP Selection and Genotyping
Single nucleotide polymorphisms in IL-1A, IL-1R1, IL-1RN, IL-1F5, and IL-18RA were selected based on Chinese population data in the HapMap database (http://www.hapmap.org), then we used Haploview software, bioinformatics software packages such as TargetScan Human 6.2 (http://www.targetscan.org/), miRanda (http://microrna.org/), and Patrocles (http://www.patrocles.org/) to select SNPs of the microRNA-binding sites in 3′ UTR region of these genes. As a result, 10 SNPs in these genes were totally included for further genotyping with the criteria of a minor allele frequency (MAF) >5%. Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) was used to determine the genotypes of IL-1A, IL-1R1, IL-1RN, IL-18RA and IL-1F5 rs2472188, rs2515401, whereas IL-1F5 rs3180235, rs957201, and rs2515402 polymorphisms were determined by the allele-specific PCR (AS-PCR) method. Amplification was performed under the following conditions: first an initial denaturation for 5 min at 94 °C, then followed by 35 cycles of denaturation at 94 °C for 30 s, the annealing for 45 s, and extension at 72 °C for 45 s, followed by an extension at 72 °C for 5 min, stored at 4 °C lastly. The fragments after amplification of the first 7 SNPs were digested by restriction enzymes (MBI Ferments, St. Leon-Rot, Germany), 5 units for 12 h. The digestion products and AS-PCR products were visualized by electrophoresis on 3% agarose gel and the genotypes were inferred from the number of bands observed in the gel.
Cell Transfection
SGC-7901 and BGC-823 were transfected by pGenesil-1-miR-197, pGenesil-1, miR-197-inhibitor, and miRNA-inhibitor respectively together with lipofectamine 2000 transfection reagent in well culture plates. After cultivation of a certain period of time, total RNA and protein were extracted from each plate.
Quantitative Real-Time PCR analysis
By bioinformatics analysis, rs2472188 in IL-1F5 was found to exist the miRNA target binding site, which was most likely the miR-197 binding target site (ΔΔG = 6.33 kj/mol).8 The qRT-PCR assay was performed using the Eco Real-Time PCR System (Illumina) to analyze the miR-197 expression and the IL-1F5 expression. The real-time PCR amplification reactions were performed under the following reaction conditions: 40 cycles of PCR (95°C for 5 s and 60°C for 30 s) after an initial denaturation (95°C for 30 s). qRT-PCR data are presented as mean ± standard deviation (X ± S) and were normalized by the corresponding U6 and GADPH control cycle threshold values. Relative gene expression was calculated using the 2-ΔΔCt method.
Western Blot Analysis
Proteins were separated on 12% separation SDS polyacrylamide gels and 5% concentration SDS polyacrylamide gels, stained by Coomassie blue after electrophoresis. Then transferred to the nitrocellulose membrane followed Ponceau Stain followed by blocking the nitrocellulose membrane by using 5% skimmed milk. Immunoreactive bands were probed with primary antibody (Internal GAPDH antibody, Santa Cruz Biotechnology, Inc.) and secondary antibody (Goat Anti-rabbit IgG, Kang biological technology co., Ltd), then visualized via enhanced chemiluminescence. The Image J 1.48v software was used to analyze the gray stripe of the Western Blot band and corrected by the corresponding GAPDH internal.
Statistical Analysis
The SPSS 21.0 statistical software package (Analysis software, Shanghai, co., Ltd, 6761805c6989326cbf14) was used for statistical analyses. The differences in distribution of selected characteristics between cases and controls were tested by Pearson chi-square (c2) test for categorical variables and the Student t test for continuous variables. The Hardy–Weinberg equilibrium (HWE) was determined by using the goodness-of-fit c2 test. Online SHEsis (http://analysis.bio-x.cn/myAnalysis.php) was applied for haplotype prediction and analysis. Unconditional logistic regression was applied to estimate the associations between genetic polymorphisms and the risk of GC by computing the odds ratios (ORs) and its 95% confidence intervals (CIs). Data of qRT-PCR and Western Blot analysis are expressed as mean ± SD, and least significant difference (LSD) was made for multiple comparisons. A P-value of <0.05 was considered to be statistically significant and all the inspections were 2-way test.
RESULTS
Family History of GC and Smoke Could Increase GC Risk
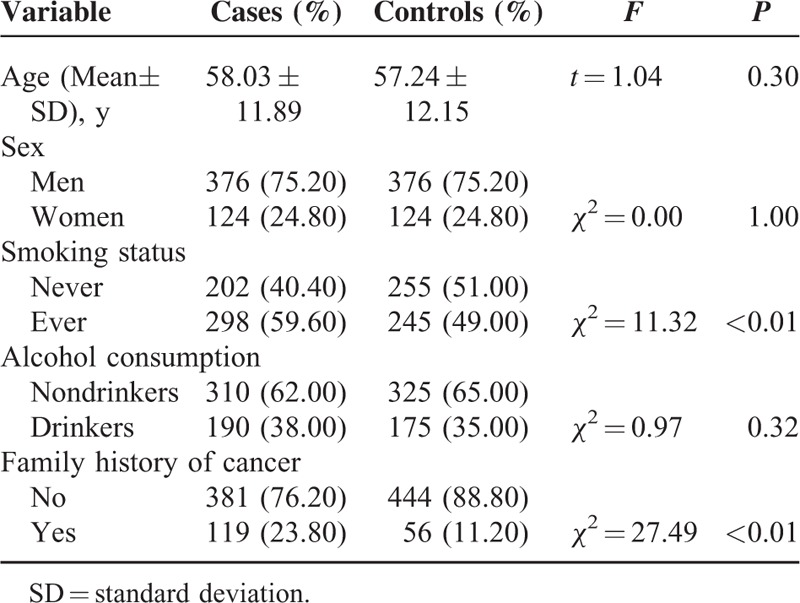
Table 1 reflected baseline characteristics of the samples, a total of 500 GC cases and 500 healthy controls were included in this study. The mean age of GC cases and controls were 58.03 ± 11.89 and 57.24 ± 12.15 respectively. The frequency matching case-control study on sex (χ2 = 0.00, P = 1.00) and age (t = 1.04, P = 0.30) was eligible since no statistically significant differences were found. Similarly, we found no statistically significant difference in alcohol consumption. However, the GC cases were more likely to have a family history of GC (χ2 = 11.32, P < 0.01) and smoke (χ2 = 27.49, P < 0.01).
TABLE 1.
Baseline Characteristics of Patients With Gastric Cancer and Controls
An Allele of rs9005 in IL-1RN, IL-1F5 rs2472188 GC, and IL-1F5 rs2515401 GC Could Increase GC Risk
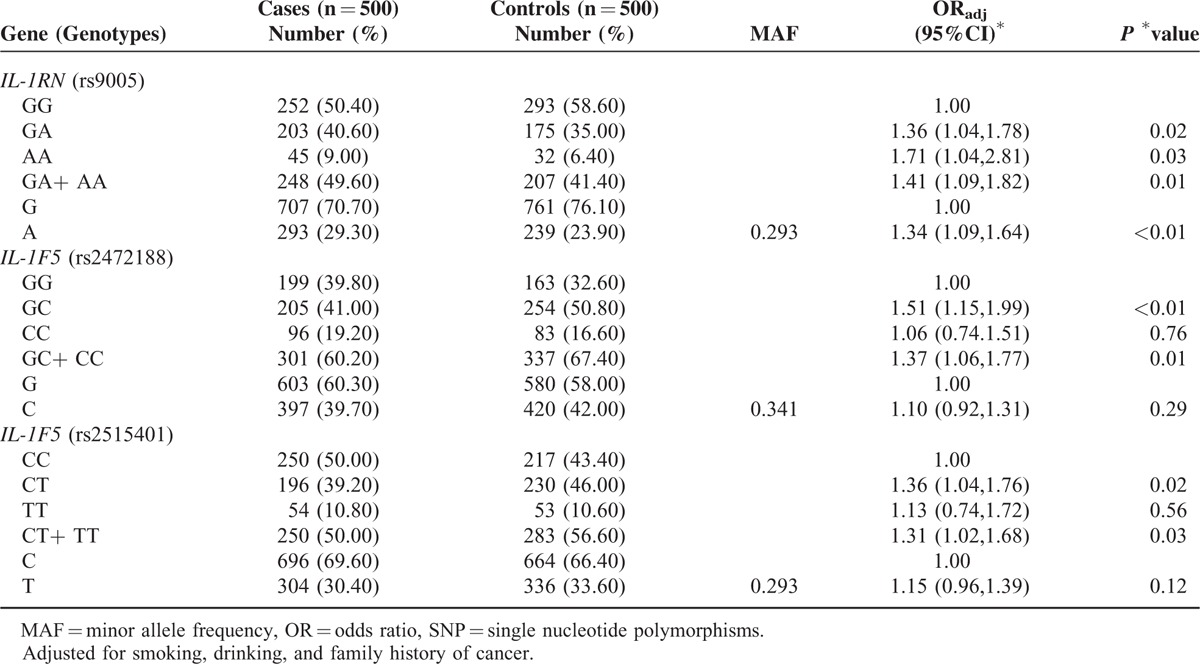
The healthy controls were representative, as 10 SNPs of the control group were all followed the Hardy–Weinberg equilibrium (P > 0.05). The meaningful genotype distributions and allele frequencies of the 10 selected SNPs between GC patients and healthy controls are shown in Table 2. The unconditional logistic regression analysis revealed that genotype and allele distributions of rs9005 exhibited significant differences between cases and controls. Subjects carrying GA (OR adjusted: 1.36; 95% CI: 1.04–1.78), AA (OR adjusted: 1.71; 95% CI: 1.04–2.81), and GA+AA (OR adjusted: 1.41; 95% CI: 1.09–1.82) genotypes in rs9005 showed significantly increase in risk for GC than individuals carrying GG genotype, and A allele also took an increased risk of GC compared with G allele (OR adjusted: 1.34; 95% CI: 1.09–1.64). Similarly, the genotyped distributions and allele frequencies of the SNPs also showed that the other 2 SNPs rs2472188 and rs2515401 in IL-1F5 had significant differences. As for rs2472188, individuals carrying the GC genotype (OR adjusted: 1.51; 95% CI: 1.15–1.99) and GC+CC genotype (OR adjusted: 1.37; 95% CI: 1.06–1.77) significantly associated with the risk of GC than GG genotype. And for rs2515401, compared with CC genotype, the individuals with CT (OR adjusted: 1.36; 95% CI: 1.04–1.76) and CT+TT (OR adjusted: 1.31; 95% CI: 1.02–1.68) genotypes showed increased risk to GC. However, no significant differences in either genotype or allelic analysis were observed between the other 7 SNPs (rs3732131 in IL-1R1, rs2856836 in IL-1A, rs1135354 and rs3771157 in IL-18RA, rs3180235, rs957201 and rs2515402 in IL-1F5) genetic variants and risk of GC.
TABLE 2.
Genotype and Allele Frequencies of 3 Meaningful SNPs and Their Associations With GC Risk
Haplotype IL-1F5 Associated With the Risk of GC
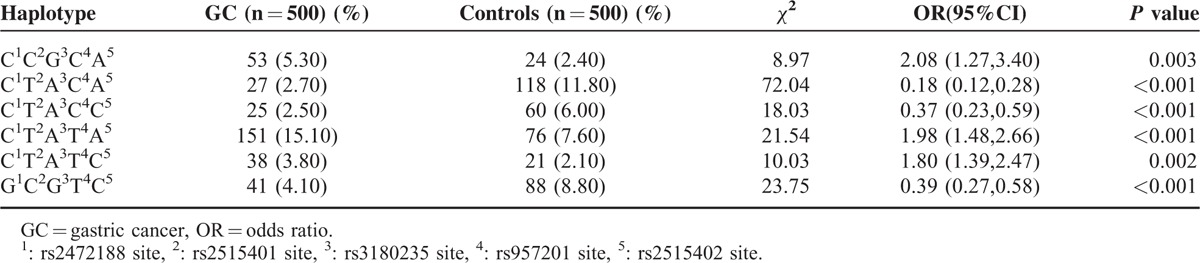
Haplotype analysis was performed with 2 SNPs (rs1135354 and rs3771157) across the IL-18RA gene and 5 SNPs (rs2472188, rs2515401, rs3180235, rs957201, and rs2515402) across the IL-1F5 gene. Both strong linkage disequilibrium was observed in IL-18RA (LD = 0.69) and IL-1F5 (LD = 0.61) gene SNPs. A total of 32 haplotypes were taken in the analysis across the IL-1F5 gene, and 4 haplotypes were taken across the IL-18RA gene. For each susceptibility analysis of a certain haplotype, all other haplotypes were taken as a reference. After abnegating those haplotypes frequency of cases and controls <3%, 9 haplotypes were analyzed. The meaningful haplotypes are shown in Table 3, haplotypes of C rs2472188C rs2515401G rs3180235C rs957201A rs2515402, C rs2472188T rs2515401A rs3180235T rs957201A rs2515402 and C rs2472188T rs2515401A rs3180235T rs957201C rs2515402 could increase the risk of GC (OR[95%CI]: 2.08[1.27–3.40], OR[95%CI]: 1.98[1.48–2.66] and OR[95%CI]: 1.80[1.39–2.47]). In contrast, haplotypes of C rs2472188Trs2515401A rs3180235C rs957201A rs2515402, C rs2472188T rs2515401A rs3180235C rs957201C rs2515402 and G rs2472188C rs2515401G rs3180235T rs957201Ars2515402 could decrease the risk of GC (OR[95%CI]: 0.18[0.12–0.28], OR[95%CI]: 0.37[0.23–0.59] and OR[95%CI]: 0.39[0.27–0.58]). However, none of the 2 haplotypes across the IL18-RA gene was associated with an increased or decreased GC risk (P > 0.05, data not shown).
TABLE 3.
The Meaningful Results of Haplotype Analysis of the IL-1F5 Gene in GC and Control Subjects
The pGenesil-1-miR-197 Enhanced the Expression of miR-197 in GC Cells
Extracting total RNA from 2 kinds of GC cells after transient transfection by SGC-7901 and BGC-823 GC cells, then detected the relation expression of miR-197 and internal gene-U6 by using the qRT-PCR method. MiR-197 relative expression after successfully transfected SGC-7901 and BGC-823 demonstrated that the pGenesil-1-miR-197 can significantly upregulated the expression of miR-197 in GC cells (Fig. 1). In the inhibitor group, miR-197 relative expression in the inhibitor transfection group was significantly lower than that in the control group (Fig. 1); it showed that miR-197 inhibitor transfected successfully and suppressed the miR-197 in the expression of SGC-7901 and BGC-823 cells.
FIGURE 1.
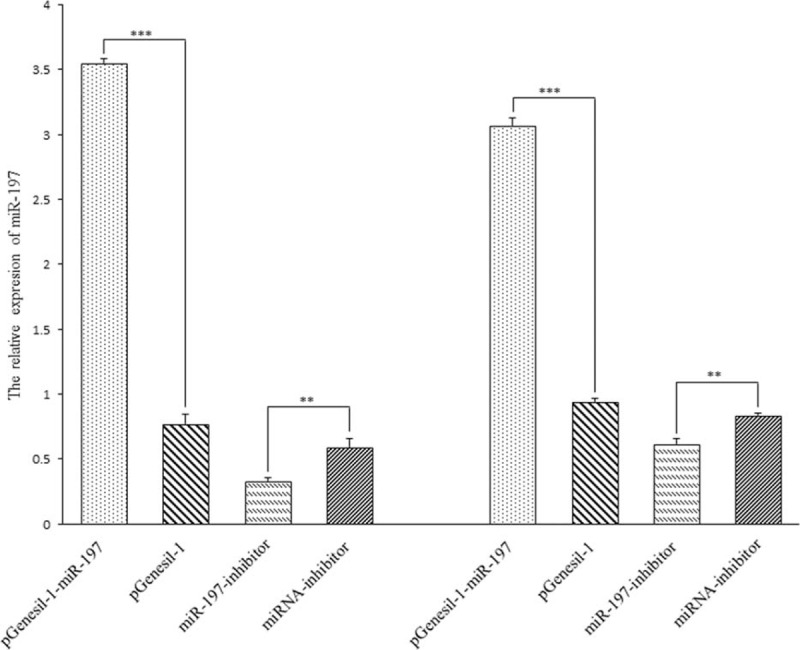
The relative expression of miR-197 after transfection in SGC-7901 and BGC-823Note: (∗∗: P < 0.01, ∗∗∗: P < 0.001). miR-197 expression was assessed by qRT-PCR in SGC-7901 (left) and BGC-823 (right) cell lines. Data was evaluated statistically using t-test and represent the mean ± SD from the experiments in triplicate.
The pGenesil-1-miR-197 Could Downregulated the Expression of IL-1F5 in GC Cells
We extracted total RNA from 2 kinds of GC cells (SGC-7901 and BGC-823 GC cells), then detected the expression of IL-1F5 and internal gene-GAPDH. The relative expression of IL-1F5 in pGenesil-1-miR-197 was significantly higher than that of the empty plasmid-pGenesil-1 (Fig. 2), indicated that pGenesil-1-miR-197 could downregulate the expression of IL-1F5 in both SGC-7901 and BGC-823 cells. In the inhibitor group, the relative expression of IL-1F5 in the inhibitor transfection group was significantly higher than that in the control group (Fig. 2); it showed that miR-197 inhibitor could upregulate the expression of IL-1F5 in SGC-7901 and BGC-823 cells after miR-197 inhibitor transfected successfully.
FIGURE 2.
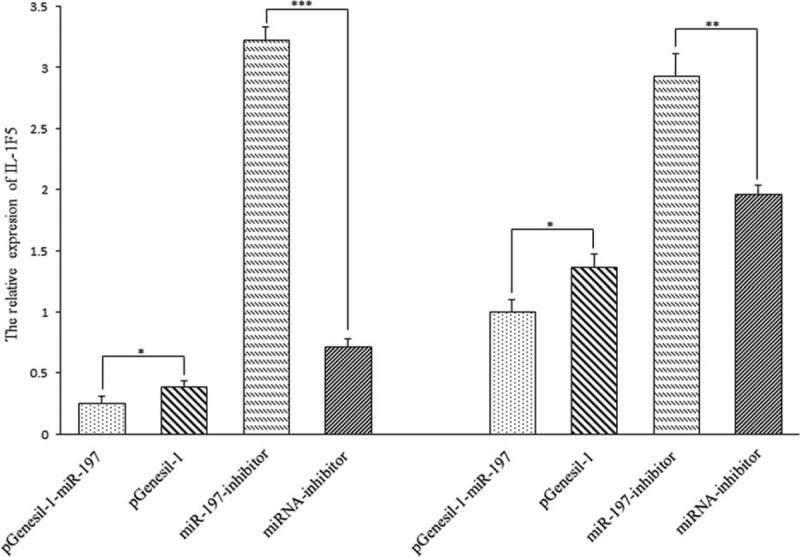
The relative expression of IL-1F5 after transfection in SGC-7901 and BGC-823 Note: (∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.001). IL-1F5 mRNA expression was assessed by qRT-PCR in SGC-7901 (left) and BGC-823 (right) cell lines. Data was evaluated statistically using t-test and represent the mean ± SD from the experiments in triplicate.
The Pgenesil-1-miR-197 Reduced Protein-IL-1F5 Translation in GC Cells
Total protein was extracted after transfected by SGC-7901 and BGC-823 GC cells and selected GAPDH as the internal gene, then tested the protein expression of IL-1F5 via Western Blot method in 2 kinds of cells. The relative expression of IL-1F5 in the pGenesil-1-miR-197 transfection group was significantly higher than that in the empty plasmid-pGenesil-1 group (Fig. 3), and it indicated that pGenesil-1-miR-197 could downregulate the expression of IL-1F5 in SGC-7901 and BGC-823 GC cells after successful transfection, whereas in the inhibitor group, the expression quantity of IL-1F5 in miR-197 inhibitor transfection group was significantly higher than in the control group (Fig. 3), with the result of upregulated expression of IL-1F5 in 2 GC cells.
FIGURE 3.
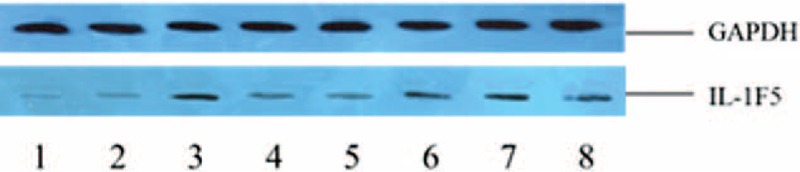
The Western Blot stripe of IL-1F5 and GAPDH after transfection in SGC-7901 and BGC-823Note: (1: pGenesil-1-miR-197 transfection group in SGC-7901; 2: pGenesil-1 transfection group in SGC-7901; 3: miR-197-inhibitor transfection group in SGC-7901; 4: miRNA-inhibitor transfection group in SGC-7901; 5: pGenesil-1-miR-197 transfection group in BGC-823; 6: pGenesil-1 transfection group in BGC-823; 7: pGenesil-1-miR-197 transfection group in BGC-823; 8: miRNA-inhibitor transfection group in BGC-823). The expressions of IL-1F5 protein in SGC-7901 and BGC-823 cell lines were analyzed by Western Blot. The protein profiles were normalized with GADPH antibody. The experiments were independently repeated three times.
DISCUSSION
Gastric cancer is an inflammation-related disease, which can induce a large amount of cytokines release, which includes IL-1, IL-6, IL-17, and TNF-α. The IL-1 gene cluster on chromosome 2q has been implicated in susceptibility to a large number of chronic inflammatory, autoimmune, and tumorous disorders. IL-1 can activate the vitality of both neutrophils and mononuclear phagocytes, thereby promoting the release of some inflammatory protein or medium from these immune cells. Related studies have shown that miRNAs have become the bridge which links the inflammation and tumor.9 In the present study, 10 SNPs of the microRNA-binding sites in 3′ UTR region of IL-1 family genes (rs2856836 in IL-1A, rs3732131 in IL-1R1, rs9005 in IL-1RN, rs1135354 and rs3771157 in IL-18RA, rs2472188, rs2515401, rs3180235, rs957201 and rs2515402 in IL-1F5) were selected to explore their relationship with GC, and future function verification tests were carried out to verify whether IL-1F5 is the target gene of miR-197.
The SNPs of Rs9005 in IL-1RN and Rs2472188, Rs2515401 of IL-1F5 Had Associations With the Risk of GC
The receptors of IL-1 gene cluster are TLR/IL-1R family, and it can initiate the transcription of pro-inflammatory factor, inducing the occurrence of inflammation.10 The inflammation-linked genes interleukin 1 (IL-1) A and IL-1RN are the most studied polymorphisms among the genetic susceptibility factors involved in GC risk.11 Recent studies have reported that associations of polymorphisms in IL-1 with risk of breast cancer (rs2856836, rs17561) 12 and GC (rs17042407).11 It was found that CT+CC genotype in IL-1A rs2856836 SNP might have a positive effect on breast cancer compared with TT genotype,10 but this association with the risk of GC was not observed in our subgroup analysis.
IL-1R1 acts as a bond to link IL-1A, IL-1B, and IL-1RN in healthy or disease conditions. The promoter region of IL-1R1 has been proven to be highly polymorphic, which can affect the expression level in a wide variety of organizations.13 IL-1R1 rs949963 was a significant genotypic predictor of class membership in the multivariable model as GA+AA genotype took a significantly increased risk in breast cancer.14 Few studies have reported the IL-1R1 SNP relationship with cancers, and none of them involved in rs3732131. In our subgroup analysis, we did not find that SNP rs3732131 of IL-1R1 was associated with the risk of GC.
Interleukin 18 (IL-18), a member of the IL-1 family of cytokines, is an important cytokine which can innate and acquire immune responses.15 The IL-18 receptor alpha chain (IL-18Ra) is the primary receptor responsible for specific binding of IL-18, with the function of inducing a variety of inflammatory cytokines. Up to now, we do not find any researches to study the relationship of IL-18RA SNPs with inflammatory diseases or tumors. Studies about IL-18 SNPs were reported to have significant associations with cancers.16,17 In the present study, 2 IL-18RA SNPs (rs1135354, rs3771157) were analyzed, but no significant associations were found between the 2 IL-18RA SNPs with GC.
As a member of IL-1 gene family, IL-1RN has been proven to act as an anti-inflammatory treatment in several chronic inflammatory diseases, which involve GC.18,19 A significant association was found between progression and several SNPs which include rs9005 and their haplotypes in the gene for IL-1RN, indicating that variants of this gene might be an important predictive marker of severity or progression in knee osteoarthritis.20,21 As far as we know, none of studies had researched the relationship between IL-1RN rs9005 SNP and tumors. In our present study, SNP rs9005 of IL-1RN was associated with the risk of GC, and in our subgroup analysis, compared with GG genotype, GA+ AA genotype could increase the risk of GC. Therefore, our findings supported the hypothesis that the functional SNP of IL-1RN might contribute to the etiology of GC.
IL-1F5 has been shown negatively to regulate TLR/IL-1 signaling pathway through the antagonism of IL-1R related proteins 2, hence inhibiting the activation of NF-κB.22 The relationship of IL-1F5 with GC and the SNPs of IL-1F5 gene in miRNA binding site with the susceptibility of GC was rarely reported. By analyzing 5 SNPs in IL-1F5, 2 SNPs (rs2472188 and rs2515401) were found to have a significant association with the risk of GC. Compared with the wild heterozygote genotype, the variant heterozygote genotypes of the 2 SNPs were significantly associated with increased GC risks.
Haplotype IL-1F5 Could Affect the Risk of GC
In this study, 2 haplotypes analyses were executed in 2 IL-1 family genes (IL-18RA and IL-1F5). The haplotype results of rs1135354 and rs3771157 in IL-18RA showed no relations with GC. The results of the haplotype analysis showed that haplotype of C rs2472188C rs2515401G rs3180235C rs957201A rs2515402, C rs2472188T rs2515401A rs3180235T rs957201A rs2515402 and C rs2472188T rs2515401A rs3180235T rs957201C rs2515402 could increase the risk of GC, whereas the haplotype of Crs2472188Trs2515401A rs3180235C rs957201A rs2515402, C rs2472188T rs2515401A rs3180235C rs957201C rs2515402 and G rs2472188C rs2515401G rs3180235T rs957201Ars2515402 could decrease the risk of GC. Taken together, the results indicated that the potentially functional SNPs of IL-1F5 might play a role in the etiology of GC.
IL-1F5 Might Be the Target Gene of miR-197 by Bioinformatics Prediction
The present study found rs2472188 SNP in IL-1F5 might have an association with GC; by further forecast analysis through MirSNP (http://cmbi.bjmu.edu.cn/mirsnp)23 and miRNASNP (http://www.bioguo.org/miRNASNP) 22 software, it was found that rs2472188 in IL-1F5 might exist the target binding site with miR-197 (ΔΔG = 6.33 kJ/mol). In this study, for further functional experiments, we built the eukaryotic expression vector pGenesil-1-miR-197 of miR-197 and transfected SGC-7901 and BGC-823.
The pGenesil-1-miR-197 Increased the Expression of miR-197 and Reduced the Expression of IL-1F5
By detecting the expressions of miRNA-197 and IL-1F5 after transfection using the qRT-PCR method, significant differences were found between the 2 expressions in both SGC-7901 and BGC-823 cells. The expressions of miR-197 in 2 GC cells were upregulated, and the IL-1F5 expression level was inhibited, which could be a preliminary estimation that IL-1F5 might be the target gene of miR-197. Meanwhile, similar results were observed in the miR-197 inhibitor group and the inhibitor control group, which indicated miR-197 can negatively regulate IL-1F5 from the reverse side and supported the finding that IL-1F5 might be the target gene of miR-197.
The pGenesil-1-miR-197 Reversed Protein-IL-1F5 Expression
In addition, in this study we also applied Western Blot analysis to test the IL-1F5 expression in SGC-7901 and BGC-823 cells after transfection. We found that the differences in the expression of IL-1F5 in the overexpression group and the inhibitor group in 2 GC cells were statistically significant. IL-1F5 might be the target gene of miR-197 with a further verification from the protein level.
By participating in biological processes, miR-197 plays an important role in the occurrence, development, and prognosis of a variety of tumors. The disordered expression of miR-197 associated with lung cancer.24 It was reported that the over expression of miR-197 had a close relationship with thyroid cancer.25 Furthermore, several studies indicated high expression of miR-197 was closely related to ovarian cancer,26 pancreatic cancer, 27 and esophageal cancer.28 Although miR-197 was reported to associate with many tumors, the researches of the relationship between miR-197 and GC were relatively few. Our study verified that IL-1F5 might be the target gene of miR-197 and miR-197 could downregulate the expression of IL-1F5. Therefore, miR-197 could affect the occurrence and development of GC via regulating IL-1F5.
In the present study, a strict quality control was conducted throughout the whole study. The investigators were trained strictly; for the data entry, parallel inputting by 2 keyboarders in different computers was used. Almost 10% of all the samples were randomly selected to assess the repeatability, and different PCR products of 10 genotypes in each SNP were sent to JiuShi Biotechnology Company for sequencing validation. Finally, qRT-PCR and Western Blot experiments were independently repeated 3 times, so it could effectively reduce the experimental error and increase the credibility of the experimental results. Meanwhile, some limitations in our study need to be noted. The overall population size was small, which might reduce the statistical power in the stratified analysis. Future prospective studies are needed to confirm the results of this topic.
CONCLUSIONS
The results suggested that rs9005 GA, AA and GA+AA genotype of IL1RN; rs2472188 GC and GC+CC genotype of IL1F5 and rs2515401 CT and CT+TT genotype of IL1F5 could increase the risk of GC. C rs2472188C rs2515401G rs3180235C rs957201A rs2515402, C rs2472188T rs2515401A rs3180235T rs957201A rs2515402 and C rs2472188T rs2515401A rs3180235T rs957201C rs2515402 haplotypes in IL-1F5 gene were risk factors for GC whereas C rs2472188Trs2515401A rs3180235C rs957201A rs2515402, C rs2472188T rs2515401A rs3180235C rs957201C rs2515402 and G rs2472188C rs2515401G rs3180235T rs957201Ars2515402 haplotypes were protective factors for GC. These results suggested that SNPs in the IL-1 family genes might play crucial roles in the development of GC. The expression of miR-197 in the GC cells was upregulated. The pGenesil-1-miR-197 could downregulate the expression of IL-1F5. The miR-197 inhibitor could upregulate the expression of IL-1F5.
Acknowledgments
We thank all the participants for their contributions and invaluable suggestions. We would also like to acknowledge our indebtedness to the respondents who provided blood sample.
Footnotes
Abbreviations: 3′UTR3′ = untranslated region, AS-PCR = allele specific-polymerase chain reaction restriction, CIs = confidence intervals, GAPDH = reduced glyceraldehyde-phosphate dehydrogenase, GC = gastric cancer, Hp = Helicobacter pylori, HWE = Hardy-Weinberg equilibrium, LD = linkage disequilibrium, LSD = least significant difference, MAF = minor allele frequency, miRNA = microRNA, NF-κB = nuclear factor-κb, ORs = odds ratios, PCR-RFLP = polymerase chain reaction restriction fragment length polymorphism, qRT-PCR = quantitative real-time polymerase chain reaction restriction, SNP = single nucleotide polymorphisms, TLR = toll-like receptor, TNF = tumor necrosis factor.
This research was supported by National Natural Science Foundation of China (81373097).
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Stubljar D, Jeverica S, Jukic T, et al. The influence of cytokine gene polymorphisms on the risk of developing gastric cancer in patients with Helicobacter pylori infection. Radiology and oncology Sep 2015; 49:256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panic N, Mastrostefano E, Leoncini E, et al. Susceptibility to Helicobacter pylori infection: results of an epidemiological investigation among gastric cancer patients. Mol Biol Rep 2014; 41:3637–3650. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe T, Takahashi A, Suzuki K, et al. Epithelial-mesenchymal transition in human gastric cancer cell lines induced by TNF-alpha-inducing protein of Helicobacter pylori. Int J Cancer 2014; 134:2373–2382. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell Jan 2009; 23: 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra PJ, Mishra PJ, Banerjee D, et al. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. Cell Cycle 2008; 7:853–858. [DOI] [PubMed] [Google Scholar]
- 6.Kong YW, Ferland-McCollough D, Jackson TJ, et al. microRNAs in cancer management. Lancet Oncol 2012; 13:e249–e258. [DOI] [PubMed] [Google Scholar]
- 7.Xuan Y, Yang H, Zhao L, et al. MicroRNAs in colorectal cancer: small molecules with big functions. Cancer Lett 2015; 360:89–105. [DOI] [PubMed] [Google Scholar]
- 8.Song CQ, Zhang JH, Shi JC, et al. Bioinformatic prediction of SNPs within miRNA binding sites of inflammatory genes associated with gastric cancer. Asian Pac J Cancer Prev 2014; 15:937–943. [DOI] [PubMed] [Google Scholar]
- 9.Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology 2012; 143:550–563. [DOI] [PubMed] [Google Scholar]
- 10.Barksby HE, Lea SR, Preshaw PM, et al. The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clin Exp Immunol 2007; 149:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duraes C, Munoz X, Bonet C, et al. Genetic variants in the IL1A gene region contribute to intestinal-type gastric carcinoma susceptibility in European populations. Int J Cancer 2014; 135:1343–1355. [DOI] [PubMed] [Google Scholar]
- 12.Han W, Kang SY, Kang D, et al. Multiplex genotyping of 1107 SNPs from 232 candidate genes identified an association between IL1A polymorphism and breast cancer risk. Oncol Rep 2010; 23:763–769. [PubMed] [Google Scholar]
- 13.McCann B, Miaskowski C, Koetters T, et al. Associations between pro- and anti-inflammatory cytokine genes and breast pain in women prior to breast cancer surgery. J Pain 2012; 13:425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merriman JD, Aouizerat BE, Cataldo JK, et al. Association between an interleukin 1 receptor, type I promoter polymorphism and self-reported attentional function in women with breast cancer. Cytokine 2014; 65:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinarello CA, Novick D, Kim S, et al. Interleukin-18 and IL-18 binding protein. Front Immunol 2013; 4:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh PK, Ahmad MK, Kumar V, et al. Effects of interleukin-18 promoter (C607A and G137C) gene polymorphisms and their association with oral squamous cell carcinoma (OSCC) in northern India. Tumour Biol 2014; 35:12275–12284. [DOI] [PubMed] [Google Scholar]
- 17.Bao J, Lu Y, Deng Y, et al. Association between IL-18 polymorphisms, serum levels, and HBV-related hepatocellular carcinoma in a Chinese population: a retrospective case-control study. Cancer Cell Int 2015; 15:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acosta JC, Banito A, Wuestefeld T, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol 2013; 15:978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neven B, Marvillet I, Terrada C, et al. Long-term efficacy of the interleukin-1 receptor antagonist anakinra in ten patients with neonatal-onset multisystem inflammatory disease/chronic infantile neurologic, cutaneous articular syndrome. Arthritis Rheum 2010; 62:258–267. [DOI] [PubMed] [Google Scholar]
- 20.Wu X, Kondragunta V, Kornman KS, et al. IL-1 receptor antagonist gene as a predictive biomarker of progression of knee osteoarthritis in a population cohort. Osteoarthritis Cartilage 2013; 21:930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Attur M, Wang HY, Kraus VB, et al. Radiographic severity of knee osteoarthritis is conditional on interleukin 1 receptor antagonist gene variations. Ann Rheum Dis 2010; 69:856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chustz RT, Nagarkar DR, Poposki JA, et al. Regulation and function of the IL-1 family cytokine IL-1F9 in human bronchial epithelial cells. Am J Respir Cell Mol Biol 2011; 45:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Zhang F, Li T, et al. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics 2012; 13:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujita Y, Yagishita S, Hagiwara K, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther 2015; 23:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokowy T, Eszlinger M, Swierniak M, et al. Analysis options for high-throughput sequencing in miRNA expression profiling. BMC research notes 2014; 7:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou D, Wang D, Li R, et al. MiR-197 induces Taxol resistance in human ovarian cancer cells by regulating NLK. Tumour Biol 2015. [DOI] [PubMed] [Google Scholar]
- 27.Hamada S, Satoh K, Miura S, et al. miR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. J Cell Physiol 2013; 228:1255–1263. [DOI] [PubMed] [Google Scholar]
- 28.Wang TY, Liu SG, Zhao BS, et al. Implications of microRNA-197 downregulated expression in esophageal cancer with poor prognosis. Genet Mol Res 2014; 13:5574–5581. [DOI] [PubMed] [Google Scholar]


