Abstract
Antibiotics resistance in Helicobacter pylori (H. pylori) is the major factor for eradication failure. Molecular tests including fluorescence in situ hybridization, PCR-restriction fragment length polymorphism, and dual priming oligonucleotide-PCR (DPO-PCR) play critical roles in the detection of antibiotic susceptibility; however, limited knowledge is known about application of multiple genetic analysis system (MGAS) in the area of H. pylori identification and antibiotics resistance detection.
The aim of this study is to determine the antibiotics resistance using different molecular tests and evaluate the treatment outcomes of E-test-based genotypic resistance.
A total of 297 patients with dyspepsia complaint were recruited for gastroscopies. Ninety patients with H. pylori culture positive were randomly divided into 2 groups (test group and control group). E-test, general PCR, and MGAS assay were performed in test group. Patients in control group were treated with empirical therapy (rabeprazole + bismuth potassium citrate + amoxicillin [AMX] + clarithromycin [CLR]), whereas patients in test group received quadruple therapy based on E-test results twice daily for 14 consecutive days. The eradication effect of H. pylori was confirmed by 13C-urea breath test after at least 4 weeks when treatment was finished.
Rapid urease test showed 46.5% (128/297) patients with H. pylori infection, whereas 30.3% (90/297) patients were H. pylori culture positive. E-test showed that H. pylori primary resistance rate to CLR, AMX, metronidazole, tetracycline, and levofloxacin (LVX) was 40.0% (18/45), 4.4% (2/45), 53.3% (24/45), 0% (0/45), and 55.6% (25/45), respectively. In addition, there are many multidrug resistant (MDR) phenotypes, and the MDR strains have higher minimum inhibitory concentration than their single-drug resistant counterparts. Considering E-test as the reference test, the sensitivities of general PCR and MGAS in detecting CLR resistance were 83.3% (15/18) and 94.4% (17/18), whereas in detecting LVX resistance were 100% (25/25) and 83.3% (15/18), respectively. Finally, the eradication rate in test group was significantly higher than that in control group as demonstrated by intention-to-treat analysis and per-protocol analysis.
MGAS is a promising assay for H. pylori identification and antibiotic susceptibility testing. Phenotypic resistance-guided quadruple therapy showed a high efficacy in treating patients with H. pylori infection.
INTRODUCTION
Helicobacter pylori (H. pylori) infection is closely associated with upper gastrointestinal diseases, including gastritis, peptic ulcer, and malignancies (gastric cancer and mucosa-associated lymphoid tissue lymphoma).1,2 Therefore, H. pylori were identified as a Class 1 carcinogen to human beings by the International Agency for Research on Cancer monograph committee in 1994.3H. pylori infection is a worldwide issue involving half of the world's population. In China, the prevalence of H. pylori infection ranges from 41.35% to 72.3%, and this rate varies with the population studied and the geographic area.4 Owing to the increasingly serious antibiotic resistance, the eradication rate is, however, now unacceptable in most areas of the world.5
Currently, many tests are available to detect antibiotic susceptibility.6 The conventional drug sensitive test is time-consuming and difficult to popularize. Other common molecular tests, such as dual priming oligonucleotide-PCR (DPO-PCR), PCR-restriction fragment length polymorphism, fluorescence in situ hybridization, and real-time PCR, have been used to identify H. pylori and detect clarithromycin (CLR) resistance on gastric specimens and even on stool samples.7–10 However, these methods failed to detect multiple antibiotic resistances at the same time and have not been used clinically. Therefore, an assay for rapid and accurate diagnosing H. pylori infection and determining multiple antibiotic susceptibilities simultaneously is urgently needed. Previously, we have reported a multiple genetic analysis system (MGAS), which is suitable for H. pylori detection and screening.11
In this study, we first investigated antibiotic susceptibility of H. pylori in Shanghai population. Then, we identified H. pylori infection and detected antibiotic susceptibility of CLR, levofloxacin (LVX), and furazolidone based on MGAS. A highly conserved and stably expressed gene of H. pylori, 16S rRNA, was chosen as the identification gene.12 Resistance to CLR, LVX, and furazolidone is conferred by specific point mutations in 23S rRNA gene (A2143G), gyrA gene (C261A/G), and porD gene, respectively. The sensitivity, specificity, and accuracy of this assay were assessed. Meanwhile, the efficiency of phenotypic resistance-guided quadruple therapy was evaluated.
MATERIALS AND METHODS
Ethics Statement
This study was approved by the Ethics Committee of Huadong Hospital, Shanghai Medical College, Fudan University. The informed consent to participate was signed by all the patients.
Clinical Specimens
The inclusion criteria in this study were as follows: those permanently living in Shanghai and >18 years old; those with dyspepsia syndrome, including epigastric pain and postprandial discomfort syndrome. And the exclusion criteria were as follows: endoscopy confirmed with active bleeding; accepted the H. pylori eradication therapy; taken bismuth agent, proton pump inhibitor, H2 receptor blockers, and antibiotics in the recent 1 month; be allergic to the drugs used for the treatment or patients whose skin test are positive (especially amoxicillin [AMX]); patients in pregnancy and lactation; residual stomach; those with severe basic diseases, such as severe liver and kidney insufficiency, cerebrovascular disease, and malignant tumor; and long-term use of corticosteroids and nonsteroidal anti-inflammatory drugs. A total of 297 gastric biopsy samples were collected from patients undergoing gastroenterology because of dyspepsia symptoms between July 2014 and December 2014 in Huadong Hospital, Shanghai Medical College, Fudan University. In the course of the gastroscopy, 2 biopsy specimens from gastric antrum were collected from each patient for rapid urease test (RUT) and H. pylori culture. A consort flow diagram of this study was shown in Figure 1.
FIGURE 1.

A consort flow diagram of this study. H. pylori eradication program adopted quadruple therapy (rabeprazole + bismuth potassium citrate + 2 kinds of antibiotics). Antibiotics include amoxicillin, clarithromycin, metronidazole, levofloxacin, and furazolidone. Patients in control group received empirical therapy (rabeprazole + bismuth potassium citrate + amoxicillin + clarithromycin, twice daily for 14 days), whereas patients in test group received individualized eradication program (rabeprazole + bismuth potassium citrate + 2 kinds of relatively sensitive antibiotics, twice daily for except for levofloxacin once daily for 14 days) based on the result of E-test. All the 90 patients accepted 13C-urea breath test after 4 to 6 weeks when treatment finished, negative results means successful eradication.
H. pylori Culture
Gastric biopsy specimens were homogenized in different tubes and cultured on selective Columbia agar supplemented with 5% sheep blood in a 37°C incubator under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) for 3 to 5 days. H. pylori were successfully cultured from 90 gastric biopsy samples and were confirmed by Gram stain and oxidase, catalase, and urease test.
Antimicrobial Susceptibility Testing
Susceptibility to CLR, AMX, metronidazole (MTZ), tetracycline (TCY), and LVX was determined by the E-test according to the manufacturer's recommendations (BioMerienx, France). Briefly, H. pylori inoculum was prepared from a 72-hour-old culture and spread on a nonselective Columbia agar plate before the placement of E-test strip. Three days later, the minimal inhibitory concentration (MIC) was determined. An isolate was considered resistant as follows: ≥1 μg/mL for CLR and LVX; ≥8 μg/mL for MTZ; ≥0.5 μg/mL for AMX; and ≥4 μg/mL for TCY.
DNA Extraction, General PCR Analysis, and Sequencing
Genomic DNA from the cultivated H. pylori was isolated using a bacterial DNA extraction kit (Tiangen Biotech Co., Ltd., Beijing, China). The concentration of each sample was determined using the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Inc, Waltharm, MA). DNA was aliquoted and stored at −20°C until required for analysis. The primers for general PCR of target genes are as follows. 16S rRNA: forward, TCGATGACACTCAGACATCCGTAAGGAGGAGGAAGGTG, reverse, GTACGACTCACTATAGGGATGAAGATTGGCTCCACTTCG; 23S rRNA, forward, GAGCGACCGCCCCAGTCAAAC, reverse, CTGCATGAATGGCGTAACGAG; gyrA, forward, AGCTTATTCCATGAGCGTGA, reverse, TCAGGCCCTTTGACAAATTC; porD, forward, GCAAGAAGTCATTGACGC, reverse, GGGGTGATAGGATAGGCT; oorD, forward, TTTAGCACAAAGGAGAATG, reverse, AACTTGGCGTAATAGGAT. The amplification was conducted using a PCR Master-Mix (Tiangen Biotech Co., Ltd.) and performed in an automatic thermal cycler (Applied Biosystems, Rockville, MD). Reaction conditions are available if needed. The PCR products were run in 2% agarose gel electrophoresis, observed under UV gel imaging system (Gel Doc XR+, Bio-Rad Ltd., Hercules, CA), and purified by a Gel and PCR Clean-Up System (Tiangen Biotech Co., Ltd.). Sequencing was performed by Sangni Biotech Co., Shanghai, China. The sequencing results were aligned and coordinated to H. pylori ATCC 26696 (Accession: NC_000915.1). Comparative sequence analysis between resistant and the standard strains was performed using the Vector NTI suite 7.1 software.
Multiple Genetic Analysis System and Capillary Electrophoresis
The multiplex PCR was performed to simultaneously determine the mutant gene (23S rRNA gene, gyrA, and porD) and identification gene (16S rRNA) in a reaction system. The mix for this reaction (10 μL) was carried out using 2 μL MgCl2 solution, 3.65 μL distilled H2O, 0.35 μL Taq DNA polymerase (Thermo Fisher Scientific Inc), 1 μL 10 × PCR buffer (Beckman Coulter, Ningbo HEALTH Gene Technologies Co., Ltd, Ningbo, China), 1 μL primers, 1 μL fluorescent markers and dNTP mixture, and 1 μL DNA template. The primers for targeted genes were shown in Table 1. The amplification was conducted under the following conditions: 105°C for pretreatment, 95°C for 2 minutes, followed by 33 cycles of 94°C for 30 seconds, 58°C for 30 seconds, 70°C for 1 minute, with a final extension at 72°C for 2 minutes. The product was kept at 4°C for storage. Capillary electrophoresis was performed as previously described.11 Briefly, the Beckman Coulter GeXP Genetic Analysis system was used to detect the multiple PCR products. A total of 40μL mixture contained 1 μL PCR product, 38.5 μL sample loading solution, and 0.5 μl DNA Size Standard 400 (Beckman Coulter, Inc) were added to the 96-well GeXP electrophoresis plate, followed by capillary electrophoresis and fragment separation using the GeXP Genetic Analysis System.
TABLE 1.
Primers for MGAS

Statistical Analysis
The SPSS software program was used for statistical analysis (SPSS Inc, Chicago, IL). Age was presented as the means ± standard deviation. Sensitivity and specificity of the tests were calculated with the formulas: a/(a + c) × 100% and d/(b + d) × 100%, respectively, where a = true positive, b = false positive, c = false negative, d = true negative. And test accuracy was calculated as follows: (a + d)/(a + b + c + d) × 100%. Per-protocol (PP) and intention-to-treat (ITT) analyses were used to evaluate the eradication rates of H. pylori between the 2 groups and calculated with 95% confidence intervals. The Fisher exact test was used to compare the difference in eradication rate between the 2 regimens. All P values are 2-sided and <0.05 was considered statistically significant.
RESULTS
Prevalence of H. pylori Infection Among Patients Studied
Among all 297 patients, 46.5% (128/297) were positive for RUT and 30.3% (90/297) were H. pylori culture positive. The consistent rate of them was 85.2%. Among these culture positive patients (54 men and 36 women), aged 20 to 82 years (mean age, 50.4 ± 12.8), 46 cases were diagnosed with chronic gastritis and 44 cases with peptic ulcer.
Detection of Antibiotic Susceptibility by E-Test
The E-test showed that CLR, AMX, MTZ, TCY, and LVX resistance were observed in 40.0% (18/45), 4.4% (2/45), 53.3% (24/45), 0% (0/45), and 55.6% (25/45) specimens detected, respectively (Table 2). Among these resistance strains, 44.4% (20/45) were found in the presence of multidrug resistance (MDR). In those MDR strains, as shown in Table 3, resistant to MTZ + LVX or CLR + MTZ + LVX were most frequently observed, followed by CLR + MTZ and CLR + LVX. Only 2.2% (1/45) was resistant to CLR + AMX. And one strain was resistant to all the antibiotics except for TCY. Meanwhile, the average MIC value of MDR was demonstrated to be significantly higher than that of single-drug resistant in CLR, MTZ, and LVX (Figure 2).
TABLE 2.
Detection of Antibiotic Susceptibility by E-Test
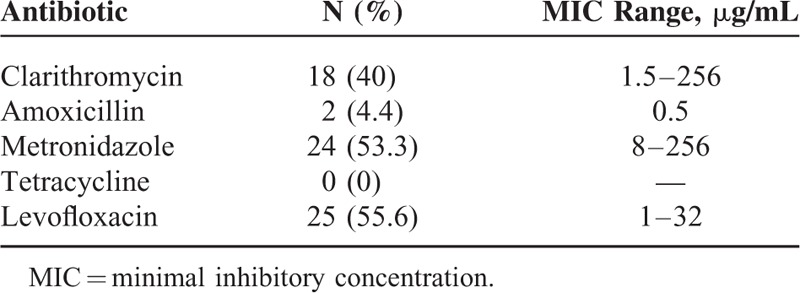
TABLE 3.
MDR Pattern in 45 Strains H. pylori
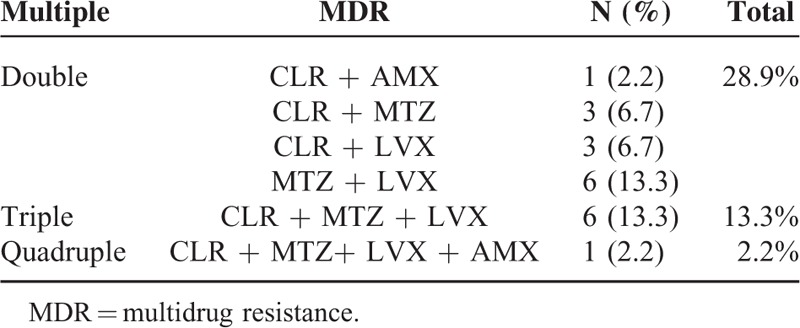
FIGURE 2.
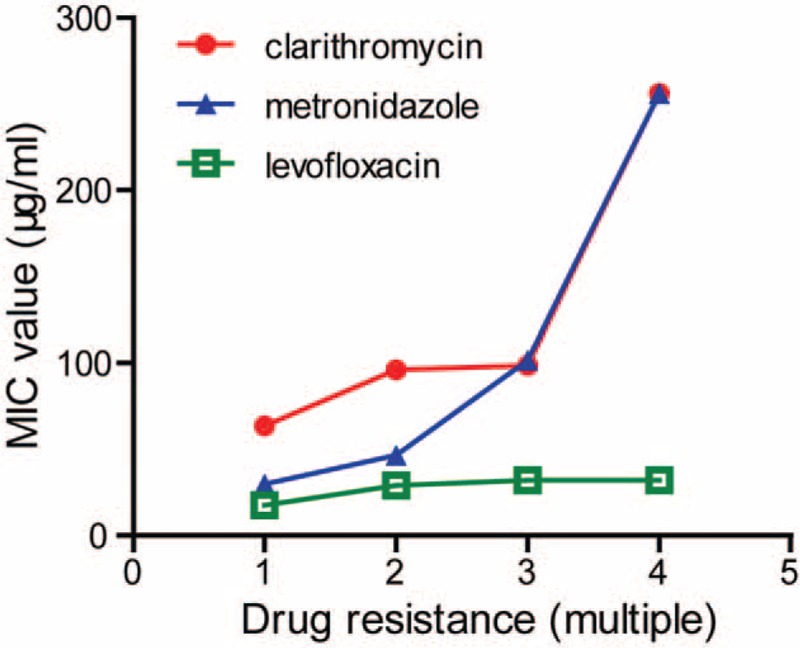
MIC values of clarithromycin, metronidazole, and levofloxacin in the presence of different drug resistance. MIC = minimal inhibitory concentration.
Genetic Variations of 23S rRNA, gyrA, and porD in H. pylori Strains
To further evaluate the antibiotic susceptibility in test group, we performed DNA sequencing analysis and detected mutations to CLR and LVX resistance. By analysis of 23S rRNA sequence, we observed 15 strains with A2143G mutation and 2 strains without A2143G mutation in the 18 resistant strains as demonstrated by E-test (Table 4). Sensitivity, specificity and test accuracy were 83.3%, 92.6%, and 88.9% respectively. Analysis in the gyrA gene showed 25 strains with mutations, including C261A (11/25), C261G (7/25), and G271T (7/25) (Table 4). This result was completely consistent with previous findings in E-test (Table 2 and Table 6). Meanwhile, mutations involved in furazolidone resistance also determined (Table 5). Among the mutations detected, C165T mutation was the most common (43/45), followed by C156T (29/45) and a double mutation A112G + A335G (10/45).
TABLE 4.
Sequencing Results in 23S rRNA and gyrA Mutaion

TABLE 6.
Comparison Between E-Test, General PCR, and MGAS Conferring Resistance to Clarithromycin and Levofloxacin

TABLE 5.
Mutations Involved in Furazolidone Resistance in 45 Strains H. pylori

Multiple Genetic Analysis System Mediated Molecular Detection on Resistant Strains
Next, we determined the genetic variation of 23S rRNA (A2143G), gyrA (C261A/G), porD (C347A/T/G), and the H. pylori identification gene 16S rRNA simultaneously by MGAS (Figure 3). The identification gene was detected in all of the 45 specimens by MGAS. Furthermore, as demonstrated in Table 6, 17 of the 18 specimens with CLR resistance were found to be resistant to CLR and 25 out of 27 specimens sensitive to CLR were found to be susceptible by MGAS. Sensitivity, specificity, and test accuracy for CLR resistance detected by MGAS were 94.4%, 92.6%, and 93.3%, respectively. However, the sensitivity, specificity, and test accuracy for LVX resistance detected by MGAS were 93.8%, 86.4%, and 89.5%, respectively, indicating a reduced diagnostic value in detection of LVX resistance by MGAS compared with general PCR. And consistent with the data in general PCR, the presence and frequency of C347T/G in porD gene were also observed by MGAS.
FIGURE 3.
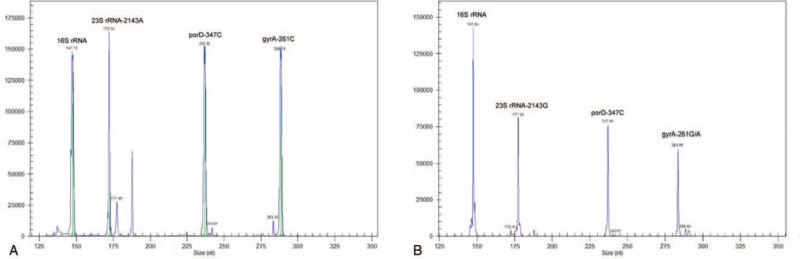
Capillary electrophoresis figures of MGAS. (A) The MGAS result obtained by DNA template from the standard ATCC 26695 strains. (B) Representative pictures of the MGAS result obtained by clinical samples. Different genes detected were marked at the top of corresponding specific peak. MGAS = multiple genetic analysis system.
Eradication Rate of H. pylori Between Test Group and Control Group
Based on E-test results, the eradication program in test group was shown in Table 7, whereas patients in control group received empirical therapy (Figure 1). The empirical therapy is rabeprazole 10 mg bid, Bismuth potassium citrate 600 mg bid, AMX 1.0 g bid, and CLR 0.5 g bid, for 14 consecutive days. And no significant difference was found in sex (P = 0.197) and age (P = 0.279) between test and control group (Table 8). Two patients in test group and 3 patients in control group lost to follow-up. The success eradication was confirmed by 13C-urea breath test at 4-6 weeks after treatment. In ITT analysis, the eradication rate in test group (91.1%, 41/45) was significantly higher than that in control group (73.3%, 33/45). And in PP analysis, the individualized therapy in test group (95.3%, 41/43) was also demonstrated to be more effective compared with empirical therapy in control group (78.6%, 33/42). Besides, adverse effects including dizziness, headache, abdominal pain, oral malodor, and rash were recorded (Table 8). These symptoms did not affect life quality severely and disappeared when the therapy ended.
TABLE 7.
Therapeutic Regimen in Test Group
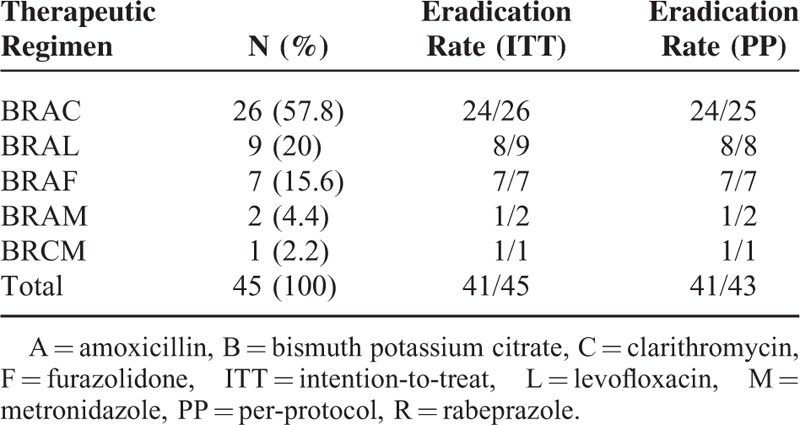
TABLE 8.
Comparison of Eradication Rates Between Test Group and Control Group

DISCUSSION
The ideal eradication rate of H. pylori should be >80%.5 However, the rate is gradually decreasing due to increasing antibiotic resistance rate. Therefore, antimicrobial susceptibility testing seems quite essential before the therapy. In the southeast coastal region of China, Su et al13 reported that H. pylori prevalence of antibiotic resistance was: CLR 21.5%, MTZ 95.4%, LVX 20.6%, AMX 0.1%, gentamicin 0.1%, and furazolidone 0.1%, respectively. In Shanghai, Sun et al14 revealed increased rates of H. pylori resistance to CLR (from 8.6% to 20.7%) and LVX (from 10.3% to 32.5%) during the year 2000 to 2009; the resistant rate of H. pylori to MTZ remained relatively stable (40%–50%); all H. pylori strains were sensitive to AMX and furazolidone; and only one strain of H. pylori isolated in 2005 was resistant to TCY. In our study, the resistance rate of H. pylori to CLR, MTZ, AMX, and TCY is close to the reported, but its resistant rate to LVX is much higher than the recorded. This observation may be explained by different methods used in detecting antimicrobial susceptibility and antibiotics usage habits.15
As a key antibiotic in quadruple therapy, macrolide is crucial to H. pylori eradication. The Maastricht IV consensus put forward that if H. pylori resistance to CLR exceeding 15% to 20%, CLR should be abandoned for the first-line therapy without drug sensitive test guidance.16,17 And in this study, we observed the resistant rate is 40%, so the use of CLR in Shanghai population had better refer to the antibiotic susceptibility test. Quinolones, as the second-line drugs in H. pylori eradication, is now attracting extensive attention. In this study, however, H. pylori resistance to LVX is as high as 55.6%, much higher than other domestic areas, likely due to the widely clinical use of quinolones causing secondary resistance.18 Based on overall analysis of the H. pylori antibiotic resistance, we found that the percentage of MDR has reached as high as 44%, which will greatly reduce H. pylori eradication rate. And consistent with the previous reports, MDR strains usually have higher MIC values.19,20 Therefore, clinicians should take corresponding preventive measures, such as referring to drug sensitive test and monitoring local drug resistance before prescription.
According to the Maastricht IV guidance, molecular technologies can be used as an alternative for bacterial culture and the conventional susceptibility test for H. pylori identification and antibiotic resistance detection.17 By general PCR, we found the main mutation site in 23S rRNA of CLR resistant strains is A2143G in Shanghai. Meanwhile, several new mutation sites, such as A2223G and C2244T, were also observed. However, whether these mutations confer H. pylori with CLR resistance needs further study. The main mutation sites in gyrA of LVX resistant strains are C261A, C261G, and G271T, whereas other common mutations at positions 260 and 272 were not found.21 It has been reported that mutations (G353A, A356G, and C357T) occurred in porD and another 3 mutations, A041G, A122G, and C349A(G), occurred in oorD genes contribute to furazolidone resistance.22 Inconsistent with this, we detected A78G, A112G, A335G, C156T, and C165T mutations in oorD gene, and C347T, C347G, and C346A mutations in porD gene. Because furazolidone susceptibility was not evaluated by E-test, so whether these mutations correlate with furazolidone resistance remains further investigation.13,14
As shown by capillary electrophoresis, an identification gene and 3 diagnostic genes of H. pylori were detected independently by MGAS. And through optimizing primer design, MGAS will confirm H. pylori infection and detect multiple antibiotic susceptibilities of H. pylori simultaneously, which is of great clinical significance and shows an advantage toward DPO multiplex PCR.23,24 Given the results of MGAS detection are intuitive and show a good application prospect, we will try to directly extract DNA from gastric mucosa biopsies and perform MGAS analysis in subsequent experiments. However, not all the mutations were detected by MGAS. This may be explained by the mutations detected by MGAS are specific and the underlying existence of mutated site in DNA sequence before the detecting site, which will contribute to the termination of PCR assay.
Based on phenotypic resistance-guided quadruple therapy, the eradication rate of H. pylori in test group is much higher than that of the control group. Therefore, developing individualized programs according to the drug resistance situation can obviously improve the H. pylori eradication rate. However, there are 2 main limitations in our study. For one thing, a multicenter and large-scale study is needed to make the data more convincing. For another, without access to the E-test strips containing furazolidone, we failed to get the resistance information about furazolidone. Furthermore, we will use agar diffusion or disc diffusion method to test it. These results would be more convincing if the limits were eradicated.
CONCLUSION
We reveal a high prevalence of H. pylori infection in patients with dyspepsia and detected antibiotic susceptibility in Shanghai Population. Our data indicate that MGAS is a fast, accurate technology for the simultaneous detection of H. pylori infection and antibiotic resistance. It is promising to be popularized in clinical testing for H. pylori.
Footnotes
Abbreviations: AMX = amoxicillin, CLR = clarithromycin, DPO-PCR = dual priming oligonucleotide-PCR, ITT = intention-to-treat, LVX = levofloxacin, MDR = multidrug resistance, MGAS = multiple genetic analysis system, MTZ = metronidazole, PP = per-protocol, RUT = rapid urease test, TCY = tetracycline.
FD, DJ, and RH are co-first authors.
ZB conceived and designed the experiments, and contributed to final approval of the version to be published. FD, FZ, MK, LN, XZ, and YW contributed to perform experiments, analysis, and interpretation of the data, drafting the article. DJ, RH, and PX contributed to collection of clinical specimens, analysis, and interpretation of the data. YH contributed to technical support.
This work was supported by grant from Science and Technology Commission of Shanghai, China (No. 134119a1700), Shanghai Health System (No. 2013SY049), and Shanghai Key Laboratory of Clinical Geriatric Medicine (13dz2260700).
The authors declare no conflict of interest.
REFERENCES
- 1.Ishaq S, Nunn L. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench 2015; 8 suppl 1:S6–S14. [PMC free article] [PubMed] [Google Scholar]
- 2.Asano N, Iijima K, Koike T, et al. Helicobacter pylori-negative gastric mucosa-associated lymphoid tissue lymphomas: a review. World J Gastroenterol 2015; 21:8014–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohammadi M, Kashani SS, Garoosi YT, et al. In vivo measurement of Helicobacter pylori infection. Methods Mol Biol 2012; 921:239–256. [DOI] [PubMed] [Google Scholar]
- 4.Xie C, Lu NH. Review: clinical management of Helicobacter pylori infection in China. Helicobacter 2015; 20:1–10. [DOI] [PubMed] [Google Scholar]
- 5.Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 2010; 59:1143–1153. [DOI] [PubMed] [Google Scholar]
- 6.Papastergiou V, Georgopoulos SD, Karatapanis S. Treatment of Helicobacter pylori infection: past, present and future. World J Gastrointest Pathophysiol 2014; 5:392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scaletsky IC, Aranda KR, Garcia GT, et al. Application of real-time PCR stool assay for Helicobacter pylori detection and clarithromycin susceptibility testing in Brazilian children. Helicobacter 2011; 16:311–315. [DOI] [PubMed] [Google Scholar]
- 8.Ciftci IH, Ugras M, Acarturk G, et al. Comparison of FISH, RFLP and agar dilution methods for testing clarithromycin resistance of Helicobacter pylori. Turk J Gastroenterol 2014; 25 suppl 1:75–80. [DOI] [PubMed] [Google Scholar]
- 9.Lehours P, Siffre E, Megraud F. DPO multiplex PCR as an alternative to culture and susceptibility testing to detect Helicobacter pylori and its resistance to clarithromycin. BMC Gastroenterol 2011; 11: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerqueira L, Fernandes RM, Ferreira RM, et al. Validation of a fluorescence in situ hybridization method using peptide nucleic acid probes for detection of Helicobacter pylori clarithromycin resistance in gastric biopsy specimens. J Clin Microbiol 2013; 51:1887–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L, Zhao F, Hu B, et al. A creative Helicobacter pylori diagnosis scheme based on multiple genetic analysis system: qualification and quantitation. Helicobacter 2015; 20:343–352. [DOI] [PubMed] [Google Scholar]
- 12.Mesquita B, Goncalves MJ, Pacheco P, et al. Helicobacter pylori identification: a diagnostic/confirmatory method for evaluation. Curr Microbiol 2014; 69:245–251. [DOI] [PubMed] [Google Scholar]
- 13.Su P, Li Y, Li H, et al. Antibiotic resistance of Helicobacter pylori isolated in the Southeast Coastal Region of China. Helicobacter 2013; 18:274–279. [DOI] [PubMed] [Google Scholar]
- 14.Sun QJ, Liang X, Zheng Q, et al. Resistance of Helicobacter pylori to antibiotics from 2000 to 2009 in Shanghai. World J Gastroenterol 2010; 16:5118–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanks AM, El-Omar EM. Helicobacter pylori infection, host genetics and gastric cancer. J Dig Dis 2009; 10:157–164. [DOI] [PubMed] [Google Scholar]
- 16.Malfertheiner P, Megraud F, O’Morain C, et al. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut 2007; 56:772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malfertheiner P, Megraud F, O’Morain CA, et al. Management of Helicobacter pylori infection—the Maastricht IV/Florence Consensus Report. Gut 2012; 61:646–664. [DOI] [PubMed] [Google Scholar]
- 18.Boltin D, Ben-Zvi H, Perets TT, et al. Trends in secondary antibiotic resistance of Helicobacter pylori from 2007 to 2014: has the tide turned? J Clin Microbiol 2015; 53:522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teh X, Khosravi Y, Lee WC, et al. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PloS One 2014; 9:e101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan TN, Santona A, Tran VH, et al. High rate of levofloxacin resistance in a background of clarithromycin- and metronidazole-resistant Helicobacter pylori in Vietnam. Int J Antimicrob Agents 2015; 45:244–248. [DOI] [PubMed] [Google Scholar]
- 21.Trespalacios AA, Rimbara E, Otero W, et al. Improved allele-specific PCR assays for detection of clarithromycin and fluoroquinolone resistant of Helicobacter pylori in gastric biopsies: identification of N87I mutation in GyrA. Diagn Microbiol Infect Dis 2015; 81:251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Z, Xu H, Zhang C, et al. Mutations in Helicobacter pylori porD and oorD genes may contribute to furazolidone resistance. Croat Med J 2006; 47:410–415. [PMC free article] [PubMed] [Google Scholar]
- 23.Chung WC, Jung SH, Oh JH, et al. Dual-priming oligonucleotide-based multiplex PCR using tissue samples in rapid urease test in the detection of Helicobacter pylori infection. World J Gastroenterol 2014; 20:6547–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo HY, Park DI, Park H, et al. Dual-priming oligonucleotide-based multiplex PCR for the detection of Helicobacter pylori and determination of clarithromycin resistance with gastric biopsy specimens. Helicobacter 2009; 14:22–28. [DOI] [PubMed] [Google Scholar]


