Abstract
The objective was to investigate porcine epidemic diarrhea (PED) outbreak that occurred in 2014 in Japan and its effects on herd-level productivity using a data recording system (PigINFO). The study herds were selected from farrow-to-finish herds (n=99) that entered in the PigINFO system between July 2013 and March 2015. From 1 April to 30 June 2014 (PED epidemic), any herds with clinical signs of PED and feces positive for porcine epidemic diarrhea virus (PEDV) on polymerase chain reaction analysis and/or immunohistochemical staining were defined as PED-positive (n=38). They were further classified into those with long PED periods (L-PED-positive; n=28) and those with short PED periods (S-PED-positive; n=10). Herds with no clinical signs of PED were classified as PED-negative (n=61). Herd-level production data, including preweaning mortality (%; PRWM), postweaning mortality (%; POWM), pigs weaned per litter (PWL), pigs born alive per litter, litters per mated female per year and pigs marketed per sow (MP), were calculated every 3 months during study period. During the PED epidemic, L-PED-positive herds had significantly higher PRWM and POWM than PED-negative herds, and L-PED-positive and S-PED-positive herds had significantly lower PWL. During October–December 2014, L-PED-positive herds had significantly fewer MP than PED-negative herds. The PED outbreak increased mortality and consequently reduced the numbers of marketed pigs. The rapid control of an outbreak is important for reducing the financial losses arising from PED infections.
Keywords: benchmarking, pigINFO, porcine epidemic diarrhea, swine
Porcine epidemic diarrhea (PED) was first reported in the United Kingdom in 1971 [16]. The clinical signs of PED are acute diarrhea, vomiting, dehydration and high mortality in neonatal piglets, resulting in significant economic losses [5]. Outbreaks of PED continued in Europe during the 1980s and 1990s [5]. In October 2010, a severe outbreak of PED occurred in >10 provinces in southern China, and >1,000,000 piglets died [13]. PED outbreaks were subsequently reported in other Asian countries, including Korea [6], Taiwan [7] and Thailand [11], and were then reported in European countries, including Germany in 2014 [4] and Belgium in 2015 [15]. PED was identified in the United States for the first time in May 2013 [12] and spread rapidly to approximately 50% of the US swine breeding herds from July 2013 to July 2014 [3]. The PED isolates collected in the United States were genetically similar to those obtained in Asian [2, 6, 7, 10] and European countries [4, 15], indicating potential transcontinental transmission.
In Japan, PED was first confirmed at one swine farm in 1982 [14]. In 1996, a large PED epidemic occurred in 102 affected herds, and 39,539 pigs died [10]. From 1997 to 2006, only sporadic cases of PED were reported (1–3 cases/year), and no cases were reported from 2007 to 2012 [10]. On 1 October 2013, a new case of PED was reported in a herd located in Okinawa Prefecture. From December 2013 to February 2014, the first PED epidemic (0–25 cases/week) was reported, primarily involving herds located on the southern island of Japan. From March to June 2014, a more severe PED epidemic (5–100 cases/week) was reported among herds throughout Japan [10]. By the end of August, these two PED epidemics had affected 817 herds, detected in 38 prefectures [9] and caused serious economic damage to the swine industry, as in the United States.
Although many case reports have described PED outbreaks and compared PED viruses genetically, very little information is available regarding the effects of PED outbreaks on swine productivity and economic factors. The objective of this study was to summarize the characteristics of the PED infections in the second epidemic in Japan using data from herds belonging to the clients of consulting veterinarians. We also investigated the effects of the herd-level PED status on productivity using a data recording system (PigINFO) recently developed for Japanese swine producers [17].
MATERIALS AND METHODS
Study population: The study herds were selected from among farrow-to-finish herds that had been entered in the PigINFO system (n=119) between July 2013 and March 2015 (study period). Data associated with the porcine epidemic diarrhea virus (PEDV) infections in each herd were investigated by 15 veterinarians belonging to the Japan Association of Swine Veterinarians (JASV). From 1 April to 30 June 2014 (PED epidemic), any herds with clinical signs of PED and feces that tested positive for PEDV with polymerase chain reaction and/or immunohistochemical staining at local livestock hygiene centers were defined as PED-positive. Herds that showed no clinical signs of PED or were negative for PEDV on the abovementioned laboratory tests were classified as negative. Herds for which there was no information on PED status, with missing data, or in which PED was confirmed before 31 March 2014 were excluded from the study. The final sample sizes in the PED-positive and PED-negative groups were 38 and 61 herds, respectively. For all PED-positive herds, the participating veterinarians recorded the types of barns in which the pigs showed clinical symptoms. In the PED-positive herds, the dates of the initial PED diagnosis and the disappearance of clinical signs from the herd were also recorded by the veterinarians. The PED period for each PED-positive herd was defined as the number of days between the initial PED diagnosis and the disappearance of clinical signs. The PED-positive herds were then further classified into those with a long PED period (≥30 days; L-PED-positive; n=28) and those with a short PED period (<30 days; S-PED-positive; n=10). This cutoff value (30 days) for the PED period was determined by visually inspecting the distribution of PED periods. Thus, the targeted herds were classified into three groups (PED-negative, L-PED-positive and S-PED-positive), and their productivity, characterized with the production parameters described below, was compared during the study period.
Production parameters: The herd-level production data were obtained every 3 months from the PigINFO system [17]. For each herd, the numbers of sows and mated gilts were counted at the end of each month, and the average female inventory (AFI) for each herd was defined as the average number of sows and mated gilts in 2014. The herd-level productivity data, such as preweaning mortality (%; PRWM), postweaning mortality (%; POWM), pigs weaned per litter (PWL), pigs born alive per litter (PBA), litters per mated female (LMFY) and numbers of marketed pigs (/sow/year; MP), were calculated based on the definitions as described previously [17]. To calculate MP, the data obtained from each 3-month period were converted to 1-year data by multiplying them by 4. Data handling was conducted based on a research contract between the National Institute of Animal Health of Japan and JASV.
Data analysis: AFI and all the productivity parameters were initially tested for the normality of their distributions by visually inspecting histograms of the data and with the Shapiro–Wilk test. Log-transformations were performed for variables that were not normally distributed, and these were then retested for normality. For variables, such as AFI, PRWM and POWM, the log-transformed data were normally distributed and were used for subsequent analyses. Group mean comparisons of the log-transformed AFI were made with Scheffe’s test. Analysis of variance, with repeated measures, was used to determine the differences in the productivity parameters among the three groups, the study periods and their interactions. If the interactions were significant for any of the parameters, analysis of covariance, with log-transformed AFI as the covariate, was performed among the three groups at different time points. Multiple comparisons were made to detect differences between the means of the groups at different time points using Bonferroni’s test. Statistical differences were defined as P<0.05. All statistical analyses were performed with SPSS version 21.0.0.0 for Windows and R version 2.13.0 (R Foundation for Statistical Computing, 2011).
RESULTS
In the 38 PED-positive herds, PED was diagnosed in 23 herds in April, in 14 herds in May and in one herd in June. The median PED period was 43.5 days (range, 13 to >90 days). During the PED epidemic, clinical signs were identified in farrowing barns (36 herds, 94.7%), nursery barns (25 herds, 65.8%), finishing barns (23 herds, 60.5%) and pregnant sow barns (29 herds, 76.3%). The means and confidence intervals (CI) for AFI in the PED-negative, S-PED-positive and L-PED-positive herds were 580 (95% CI, 378–782), 464 (95% CI, 234–695) and 1,259 (95% CI, 620–1897), respectively, and the L-PED-positive herds had significantly higher AFI than the PED-negative herds (P<0.05). In the repeated-measure analysis, the time factor and the interaction between time and group were significant for PRWM, POWM, PWL and MP. However, only the time factor was significant for PBA and LMFY. The expression as “time factor was significant” means average values of all 3 groups differ significantly among different time points. For example, the average preweaning mortality was significantly higher in April-June 2014 than at other time points. The mean values of all the parameters differed significantly at different time points, so the time factor was significantly associated with all the variables investigated in this study. The expression as “interaction between time and groups was significant” means some groups (S-PED-positive and L-PED-positive groups) had significantly lower or higher values for these variables at specific time points compared with the other group (PED-negative group). For example, the average preweaning mortality in April–June was significantly lower in the PED-negative group than in the L-PED-positive group, even though it did not differ among the groups at the other sampling times, which can be explained by the interaction between time and group. We observed significant time–group interactions for PRWM, POWM, PWL and MP, in that the values for these parameters were significantly lower in specific groups (S-PED-positive and L-PED-positive) than in the other group (PED-negative) at specific time points.
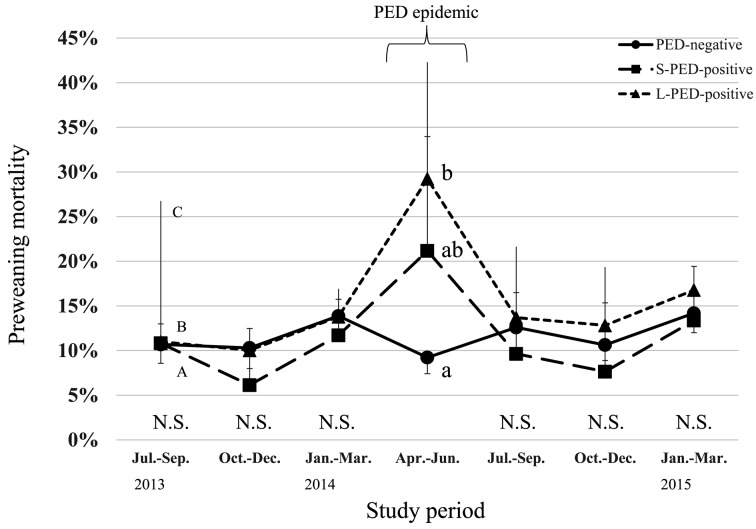
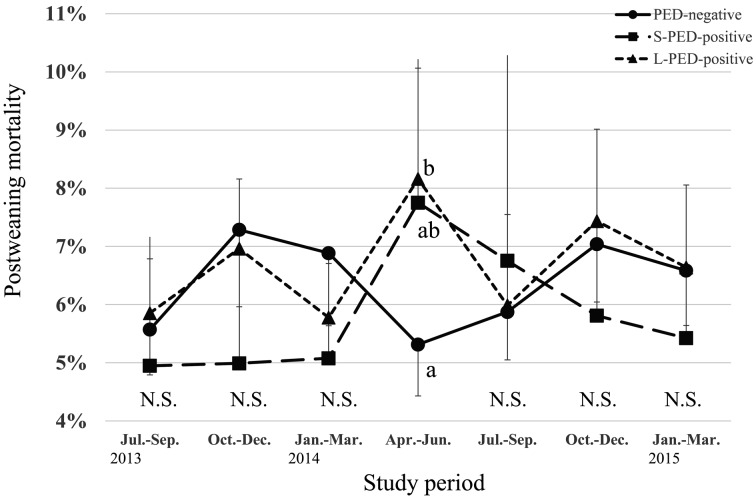
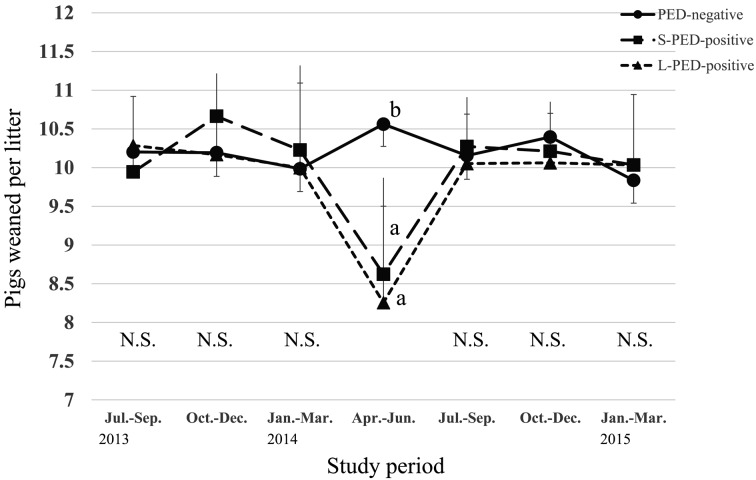
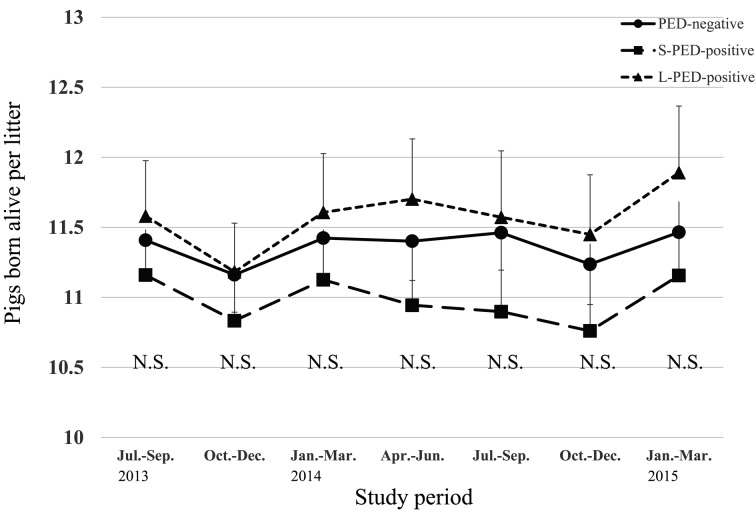
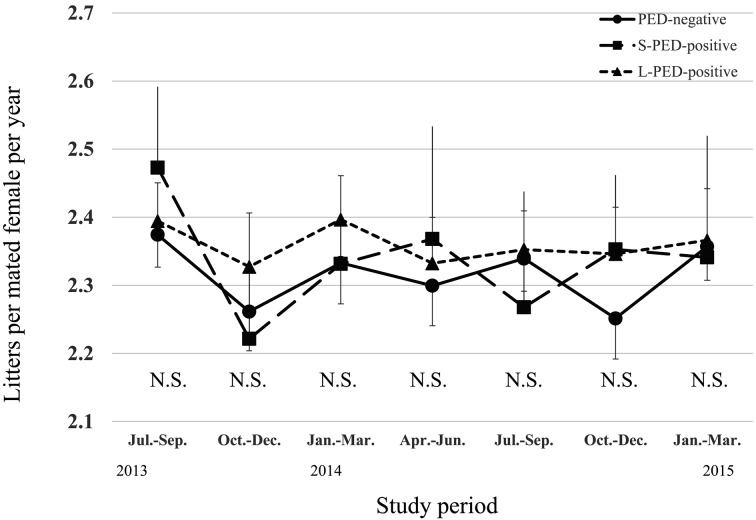
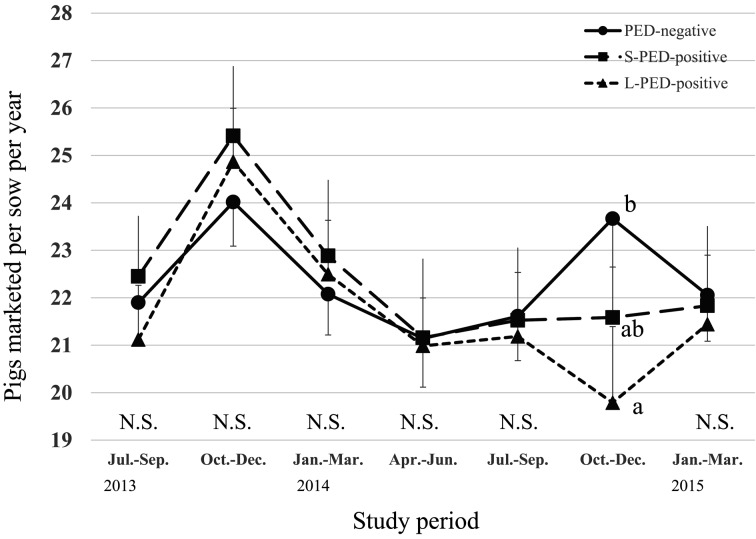
During the PED epidemic, L-PED-positive herds had significantly higher PRWM and POWM than the PED-negative herds, but no significant difference was observed between the S-PED group and the other groups (Figs. 1 and 2). The L-PED-positive and S-PED-positive herds had significantly lower PWL than the PED-negative herds during the PED epidemics (Fig. 3). No significant difference was observed in PBA or LMFY among the groups during the study period (Figs. 4 and 5). During October–December 2014, the L-PED-positive herds had significantly fewer MP than the PED-negative herds, whereas the S-PED-positive herds did not differ significantly from the other groups (Fig. 6).
Fig. 1.
Average preweaning mortality. a, b: points with different letters differ significantly (P<0.05). N.S.: not significantly different (P≥0.05). A (lower vertical line with capsule):lower value of 95% confidence interval for PED-negative group. B (higher vertical line with capsule):higher value of 95% confidence interval for L-PED-positive group. C (higher vertical line without capsule):higher value of 95% confidence interval for S-PED-positive group.
Fig. 2.
Average postweaning mortality.
Fig. 3.
Average pigs weaned per litter.
Fig. 4.
Average pigs born alive per litter.
Fig. 5.
Average litters per mated female per year.
Fig. 6.
Average pigs marketed per sow per year. Data obtained for 3 months were converted into annual data by multiplying them by 4.
DISCUSSION
This study demonstrates that the L-PED-positive herds had significantly reduced productivity, characterized by higher PRWM and POWM and lower PWL, during the PED epidemic. We observed similar result in the S-PED-positive herds, and the lack of significant differences between the S-PED-positive and other groups might be attributable to the small sample size (n=10) of this group. Among the PED-positive herds, the maximum PRWM was 45%, which was close to that of another study (21.7%) [8], but lower than those reported in other studies, in which piglets under 2 weeks of age displayed much higher mortality (>90%) [7, 12]. This discrepancy might be attributable to differences in the ages of the suckling pigs targeted for investigation, the methods used to calculate suckling pig mortality in the different studies, virulence between different strains and farm managements between different countries.
Reduced productivity, characterized by PRWM, POWM and PWL, was only observed during April-June 2014, which indicates that in many of the PED-positive herds, the outbreaks were controlled within a short period. This finding agrees with those reported in the United States, in which many PED-infected herds reached their baseline production levels within 12.6 weeks [3]. This may be because the herds rapidly developed immunity and therefore recovered from the severe POWM after the PED outbreak [3]. Alternatively, the owners of the herds targeted in the present study may have taken urgent preventive action, because they used consulting veterinary services.
In many of the PED-positive herds, the clinical signs of PED were observed in the nursery and finishing barns, which is consistent with the evidence of higher POWM in the PED-positive herds. This finding is also consistent with that of a previous report [8], but contradicts to that of another study [7], in which no mortality was observed in stages other than the suckling stage. These different results may be attributable to differences in the swine management systems used. For example, most of the herds targeted in the present study were farrow-to-finish systems, which are at higher risk of introducing pathogens into the postweaning stages than other production systems. The increased POWM in the PED-positive herds in the present study was similar to that reported in a retrospective cohort study in which pigs that survived PED infection showed a 10% increase in postweaning mortality [1].
The L-PED-positive herds had significantly higher AFI than the PED-negative herds. This may be because herds with larger size required more time and efforts to control PED. The L-PED-positive herds had higher PBA and LMFY than in the PED-negative herds, although these differences were not significant. This indicates that the L-PED-positive herds are more common among larger herds, which usually involve more intensive production systems and more rigorous biosecurity measures. Many participating veterinarians commented that the first PED cases in their regions appeared in herds with excellent biosecurity controls (personal communication), which is inconsistent with the standard theories regarding many other infectious swine diseases, and constitutes a mysterious aspect of the PED outbreaks in Japan.
In the present study, the levels of PBA and LMFY in the PED-positive herds did not decline during the PED epidemic. A previous observational study indicated that only pregnant sows and gilts infected with PED at 0–30 days of pregnancy had smaller litters, whereas those infected at other points during the gestation period were not affected by PED [11]. If litter sizes are only reduced by PEDV infection within a particular gestational stage, the calculation of herd-level PBA, as used in the present study, may be insufficient to demonstrate the actual effects of PED status on the subsequent litter size.
In Japanese pork markets, MP generally increased between October–December 2013 (Fig. 6), mainly because fattening pigs grow faster in this season. However, the L-PED-positive herds, and to a lesser extent the S-PED-positive herds, had fewer MP than the PED-negative herds during this period in 2014 (Fig. 6), approximately 180 days (fattening period) after the PED epidemic. This was a direct consequence of the reduced number of weaned pigs, caused by piglet death and slaughtering during the PED epidemic. The fact that the S-PED-positive herds had higher MP than the L-PED-positive herds indicates that the rapid control of an outbreak (<30 days) is economically justified because it increases the number of pigs available for marketing.
The difference in MP between the PED-negative and L-PED-positive herds in October–December 2014 was 3.88 pigs/sow/year, which was equivalent to 0.97 pigs/sow/3 months. During October–December 2014, the average price of marketed pigs in the L-positive-herds was $332/pig (data not shown). Therefore, the estimated average loss for an L-PED-positive herd caused by the PED outbreak, based only on the fewer pigs marketed, is estimated to be $314/sow ($322 × 0.97). Using the mean AFI (1,259) for the L-PED-positive herds, the estimated average loss from the reduced MP caused by the PED outbreaks per L-PED-positive herd was $395,326 ($314/sow ×1,259), which is a significant loss for pig producers. In addition to the losses mentioned above, the potential reduced productivity described by the reduced daily weight gain and feed conversion rate have been reported previously [1]. Other indirect losses, including the costs of vaccines and disinfectants, animal movement restrictions and additional labor cost for disease control, were also estimated.
To the best of our knowledge, this is the first report of the direct effects of a PED outbreak on the subsequent numbers of pigs marketed. We are continuously recording the production parameters of these herds with our data-recording system PigINFO and plan to investigate the potential risk factors for PED outbreaks. Sporadic PED continues to occur in 2015, and the strict biosecurity measures against PED must be strengthened because PED outbreaks cause serious economic losses.
Acknowledgments
This study was supported by collaborative research grants from the National Institute of Animal Health and the Japan Association of Swine Veterinarians. The authors thank the veterinarians and farmers who participated in this study.
REFERENCES
- 1.Alvarez J., Sarradell J., Morrison R., Perez A.2015. Impact of porcine epidemic diarrhea on performance of growing pigs. PLoS ONE 10: e0120532. doi: 10.1371/journal.pone.0120532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Q., Li G., Stasko J., Thomas J. T., Stensland W. R., Pillatzki A. E., Gauger P. C., Schwartz K. J., Madson D., Yoon K. J., Stevenson G. W., Burrough E. R., Harmon K. M., Main R. G., Zhang J.2014. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 52: 234–243. doi: 10.1128/JCM.02820-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goede D. M. R.2015. Production impact study update. Swine Health Monitoring Project 08/01/2014. University of Minnesota. Available: http://www.cvm.umn.edu/sdec/SwineDiseases/pedv/SHMP_14/index.htm. Accessed 5/7/2015.
- 4.Hanke D., Jenckel M., Petrov A., Ritzmann M., Stadler J., Akimkin V., Blome S., Pohlmann A., Schirrmeier H., Beer M., Höper D.2015. Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerg. Infect. Dis. 21: 493–496. doi: 10.3201/eid2103.141165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung K., Saif L. J.2015. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 204: 134–143. doi: 10.1016/j.tvjl.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S., Lee C.2014. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 20: 1223–1226. doi: 10.3201/eid2007.140294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C. N., Chung W. B., Chang S. W., Wen C. C., Liu H., Chien C. H., Chiou M. T.2014. US-like strain of porcine epidemic diarrhea virus outbreaks in Taiwan, 2013-2014. J. Vet. Med. Sci. 76: 1297–1299. doi: 10.1292/jvms.14-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martelli P., Lavazza A., Nigrelli A. D., Merialdi G., Alborali L. G., Pensaert M. B.2008. Epidemic of diarrhoea caused by porcine epidemic diarrhoea virus in Italy. Vet. Rec. 162: 307–310. doi: 10.1136/vr.162.10.307 [DOI] [PubMed] [Google Scholar]
- 9.Ministry of Agriculture Forestry and Fisheries2014. available from http://www.maff.go.jp/j/syouan/douei/ped/ped.html. Accessed 5/7/2015.
- 10.Miyazaki A., Suzuki T., Ohashi S., Shibahara T., Yamakawa M., Tsutsui T., Tsuda T.2014. Overview and current topics in porcine epidemic diarrhea. Proc. Jpn. Pig Vet. Soc. 64: 15–24(in Japanese). [Google Scholar]
- 11.Olanratmanee E. O., Kunavongkrit A., Tummaruk P.2010. Impact of porcine epidemic diarrhea virus infection at different periods of pregnancy on subsequent reproductive performance in gilts and sows. Anim. Reprod. Sci. 122: 42–51. doi: 10.1016/j.anireprosci.2010.07.004 [DOI] [PubMed] [Google Scholar]
- 12.Stevenson G. W., Hoang H., Schwartz K. J., Burrough E. R., Sun D., Madson D., Cooper V. L., Pillatzki A., Gauger P., Schmitt B. J., Koster L. G., Killian M. L., Yoon K. J.2013. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 25: 649–654. doi: 10.1177/1040638713501675 [DOI] [PubMed] [Google Scholar]
- 13.Sun R. Q., Cai R. J., Chen Y. Q., Liang P. S., Chen D. K., Song C. X.2012. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 18: 161–163. doi: 10.3201/eid1801.111259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K., Okada K., Ohshima K.1983. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Nippon Juigaku Zasshi 45: 829–832. doi: 10.1292/jvms1939.45.829 [DOI] [PubMed] [Google Scholar]
- 15.Theuns S., Conceição-Neto N., Christiaens I., Zeller M., Desmarets L. M. B., Roukaerts I. D. M., Acar D. D., Heylen E., Matthijnssens J., Nauwynck H. J.2015. Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in belgium, january 2015. Genome Announc. 3: e00506–e00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood E. N.1977. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 100: 243–244. doi: 10.1136/vr.100.12.243 [DOI] [PubMed] [Google Scholar]
- 17.Yamane I., Ishizeki S., Yamazaki H.2015. Aujeszky’s disease and the effects of infection on Japanese swine herd productivity: a cross-sectional study. J. Vet. Med. Sci. 77: 579–582. doi: 10.1292/jvms.14-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]