Abstract
C57BL/6 mice were tested in order to investigate the effects of dietary chitosan (COS) supplements on intestinal microflora and resistance to Citrobacter rodentium infection. The findings reveal that, after consuming a 300 mg/kg COS diet for 14 days, microflora became more diverse as a result of the supplement. Mice receiving COS exhibited an increase in the percentage of Bacteroidetes phylum and a decrease in the percentage of Firmicutes phylum. After Citrobacter rodentium infection, the histopathology scores indicated that COS feeding resulted in less severe colitis. IL-6 and TNF-α were significantly lower in colon from COS-feeding mice than those in the control group. Furthermore, mice in COS group were also found to experience inhibited activation of nuclear factor-kappa B (NF-κB) in the colonic tissue. Overall, the findings revealed that adding 300 mg/kg COS to the diet changed the composition of the intestinal microflora of mice, resulting in suppressed NF-κB activation and less production of TNF-α and IL-6; and these changes led to better control of inflammation and resolution of infection with C. rodentium.
1. Introduction
Citrobacter rodentium, a mucosal pathogen found in mice, and enteropathogenic Escherichia coli (EPEC) and enterohaemorrhagic E. coli (EHEC), enteric pathogens found in humans, create attaching and effacing (A/E) lesions that result in the colonization of mucosa within the intestinal tract [1, 2]. The intestinal epithelium becomes infiltrated by bacterial attachments, the brush border microvilli become depleted, and pedestal-shaped structures begin to form beneath the adherent bacterium [3]. Colonic tissue changes comparable to EHEC and EPEC infection, along with a rise in the production of inflammatory cytokine and leukocyte infiltration, are prompted by A/E pathogens following the colonization of the colonic epithelium [4]. C. rodentium amongst mice populations has been used by many researchers to represent intestinal E. coli since intestinal EPEC or EHEC does not infect mice.
A number of significant intestinal disorders found in humans are also frequently modelled by C. rodentium in mice, such as colon tumorigenesis, ulcerative colitis, and Crohn's disease [5]. Colitis develops in mice that have been infected with C. rodentium, which results in the bacteria becoming overabundant and the mice's natural microbiota reducing in variety and quantity [6]. When infected, 1–3% of the mice's intestinal microbiota becomes C. rodentium [7], whilst the colon comes under attack by 109 colony forming units (CFUs) per g [8]. The genetic makeup of the mouse impacts the formation of C. rodentium-induced colitis, with mice such as C3H/HeJ and FVB/N being prone to develop colitis as a result of the infection and mice such as CD-1 and C57BL/6 being considered amongst those that are not prone to develop colitis and are only prone to subclinical symptoms [9]. Vulnerability to contracting C. rodentium, as well as tendency to show a certain immune response, has been found to be highly related to intestinal microbiota composition [10].
The human intestinal tract houses 500–1,000 species of microbiota at a total quantity of almost 100 trillion [11]. Various factors, including location and diet, have a significant impact on the metagenome, although these factors do not have the same effect on a single organism's genome. Pathogen displacement, the extraction of energy such as SCFAs from nondigestible dietary substrates, and the development of the immune system all depend on intestinal microbiota. Significant changes in the natural microbiota homeostasis have been linked to various illnesses in humans [12].
COS is among the most plentiful polysaccharides found in insects, fungi, squid, oysters, krill, clams, and shellfish. It is a natural N-deacetylated derivative of chitin [13]. Many researchers have explored the ways in which the expression of Th1 and Th2 cytokines is impacted by COS [14]. In pigs [14], mice [15], rats [16], and fishes [17], COS serves as a regulator of the immune response. However, little is known about how intestinal microbiota is impacted by dietary COS despite a small number of studies attempting to study this in pigs and chickens [18]. It would be useful to gain insight into whether it is via the intestinal microbiota that COS is able to perform its advantageous biological functions, since it is already known that a number of biological functions are impacted by the intestinal microbiota [19, 20]. Nonetheless, at present, few findings exist on the topic of C. rodentium, intestinal microbiota, and the effects of COS supplements on them. Thus, the objective of this study is to evaluate the effects of COS supplementation on microbial flora composition changes, proinflammatory molecule reduction (and/or anti-inflammatory molecule increase), and C. rodentium infection.
2. Materials and Methods
2.1. Mice and Diet
This study used male C57BL/6 mice aged 6–8 weeks old, bred and reared at Hunan Agricultural University by breeders from China's Changsha-based SLAC Laboratory Animal Center. The reason for using only male mice was that sex and maternal factors have been shown to impact microflora composition. Hunan Agricultural University's Animal Care and Use Committees provided approval for the experiments. The mice were provided plentiful water and a normal diet [21], and they were kept individually in animal colonies that were free from pathogens. The colonies were kept at a temperature of 25°C, relative humidity of 53%, and an equal balance of 12 hours of dark/light. The mice were assigned at random to the COS and control groups after being housed in the colonies for three days, with 20 mice in each group. A normal rodent diet [21] was given to the control group. 300 mg/kg COS was added to the standard diet given to the COS group, which was consumed for 14 consecutive days. COS has a 6-sugar unit of N-acetyl glucosamine with β-(1–4)-linkages [8], an average molecular weight of less than 1,000 Da, and a degree of deacetylation over 95% and is free of endotoxins. The COS provided to the mice in this study was provided by the Chinese Academy of Sciences' Dalian Chemical and Physical Institute. The findings of an earlier research paper [14] were used as a guideline for the duration and amount of COS given. Throughout the experiment, the mice's weight gain, water consumption, and food consumption were recorded. Until being ready for analysis, samples were kept at −80°C.
2.2. C. rodentium Infection and Monitoring
After giving either the basal or COS diet for a period of 14 days, 10 mice in each group were infected with C. rodentium. CO2 asphyxiation was administered to all mice on D7 after infection. Samples of the mice's feces, tissue, and colonic content were gathered. Bacteria were grown in Luria Bertani broth (0.05 g/L nalidixic acid/mL) overnight. Centrifugation was used on the cultures and the pellet produced was resuspended in sterile phosphate-buffered saline (PBS). This resulted in a concentration of 5 × 109 CFU/mL. Mice were then challenged at 9 am and 5 × 109 CFU C. rodentium were orally administered to the mice using the gavage process. In order to prevent shedding mice from spreading infection, all mice were kept separately when infected with C. rodentium. The mice were fed with the same basal or COS diet for 7 days, respectively. Feces collection was conducted on D7 after infection, at which point they were also weighed and suspended in PBS. Serial dilutions were plated onto plates containing nalidixic acid. After a period of 24 hours, bacterial colonies were counted. Each dilution was grown at 37°C overnight using the pour plate method in MacConkey agar supplemented with the antibiotic kanamycin (40 μg/mL) in order to compute only C. rodentium.
2.3. Colon Tissue: Histological Analysis
The colon of each mouse was removed and fixed in 10% formalin. Hematoxylin and eosin were used to stain and prepare paraffin-embedded sections. Six criteria (edema, ulcers, erosion, inflammation, goblet cell hyperplasia, and cryptitis) were outlined in order to histologically grade the colitis. A scale of 0–4 was used to score the lesions as follows: no epithelial thickening/colitis (0); minor epithelial cell hyperplasia/greater quantity of mucosa leukocytes (1); inflammation at numerous loci, submucosa, and mucosa infiltrated by leukocytes and/or significant epithelial cell hyperplasia (with 2-3 times' rise in crypts) (2); greater epithelial cell hyperplasia (3–10 times higher crypt count), reduction of goblet cells secreting mucin, ulceration, and/or significant leukocytic infiltration of the submucosa and mucosa (3); extremely high epithelial cell hyperplasia (10 or more times higher crypt count), crypt abscesses, and/or serious infiltration of transmural leukocytes (4).
2.4. 16S rDNA and Illumina MiSeq Sequencing
After giving either the basal or COS diet for a period of 14 days, feces were collected and 10 mice in each group were killed to collect colon contents. Then, feces and colon contents were used for 16S rDNA sequencing. As per the guidelines for DNA isolation, the QiagenQIAamp DNA Stool Mini Kit was used to extract DNA from luminal colon and feces contents. In order to create a baseline sample for each sample type, equal quantities of DNA were gathered from six individual mice. Primers 515F 5′-GTGCCAGCMGCCGCGG-3′ and 907R 5′-CCGTCAATTCMTTTRAGTTT-3′ (the barcode being an eight-base sequence individual to each specific sample) were used to amplify the V4-V5 region of the bacteria 16S ribosomal RNA gene by PCR. Biotree, a commercial firm based in Shanghai, carried out the general data analyses and Illumina MiSeq sequencing. Previous findings were used to guide the MiSeq PE Libraries, MiSeq sequencing, and additional analysis [22]. The corresponding authors can be contacted to request further information on references, raw data, and the sequencing run.
2.5. Analysis of Colonic mRNA
IL-6 and TNF-α expression were incorporated in the analyses, which occurred after the collection and weighing of the proximal colon. TRIZOL regent (Invitrogen, USA) was used for the extraction of mRNA. Reverse transcription of the cDNA was conducted, and an ABI 7500 Fast RT-PCR machine (Applied Biosystems), Superscript II reverse transcriptase (Invitrogen), and oligo (dT) 20 were used to carry out real-time PCR.
2.6. Cytokine Measurement
A 50 mM Tris-HCl with 10 μg/mL protease inhibitor solution (Sigma-Aldrich Co., USA) was used to homogenise the colonic samples over ice. Centrifugation of 20 minutes was used on the homogenates at 30,000 ×g (4°C). Following centrifugation, a Sandwich ELISA Kit (ELISA Ready-SET-GO, eBioscience, CA, USA) was used to test the supernatants for TNF-α and IL-6. Normalization of cytokine was carried out to match the colonic samples' protein levels.
2.7. NF-κB (p65) Immunoblotting
The NF-κB (p65) transcription factor assay kit (Cayman Chemical Company, MI, USA) was used to measure NF-κB (p65) binding activity in the nuclear extracts. The proteins extracted from nuclear or cytoplasmic fractions were taken in equal quantities and then (1) divided using SDS-PAGE, (2) relocated to PVDF membranes (Millipore, MA, USA), and obstructed using 5% nonfat milk in a Tris-Tween buffered saline buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween-20) over a 3-hour period. Prior to conducting the analysis with Alpha Imager 2200 (Alpha Innotech Corporation, CA, USA), overnight incubation at 4°C was carried out with the primary antibodies, whilst 60-minute incubation at room temperature was carried out with the HRP-conjugated secondary antibodies. Finally, electronic quantification and normalization of signal intensity to Lamin B protein abundance were performed.
2.8. Statistical Data Analysis
SPSS 22.0 (Chicago, IL, USA) was used to carry out all statistical analysis. The data illustrated in this section are the means ± the standard error of the mean (SEM). Student's t-test was used to analyze the data between two groups. In this study, statistical significance is shown in values of P < 0.05.
3. Results
According to the weight measurements, there was no postinfection change in the weight of each mouse in either of the two groups. Additionally, as shown in Figure 1, at D7 after infection, no change was noted in the quantity of C. rodentium in the feces or colon contents of each group. However, as shown in Figure 2, a significant increase in colitis severity was observed at D7 after infection amongst the control group of mice compared to the mice provided with COS.
Figure 1.
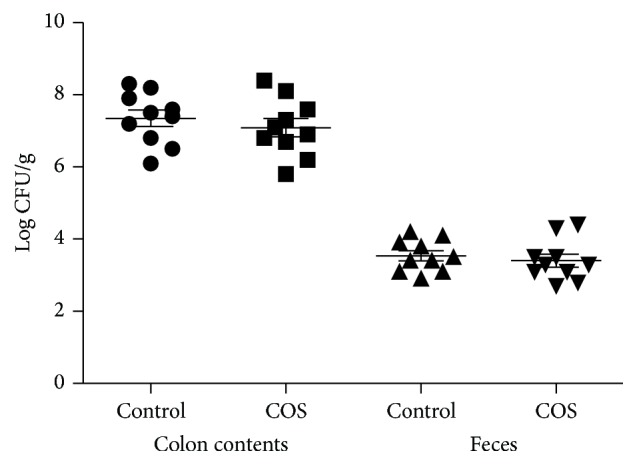
D7 postinfection C. rodentium levels in colon contents and feces of infected C57BL/6 mice. Feces and colon contents were gathered before undergoing homogenisation and being plated in serial dilution on LB agar (n = 10).
Figure 2.
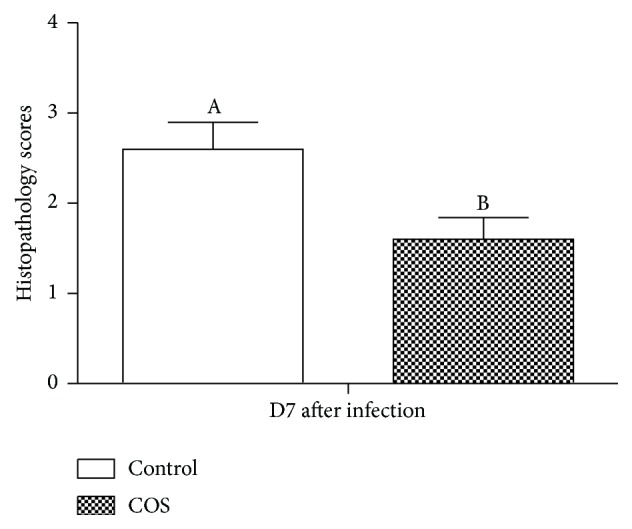
D7 postinfection histopathology scores of each mouse (n = 10).
As shown in Table 1, 16S rDNA sequencing was used to analyze the intestinal microbiota once the experiment was completed. The results of 16S rDNA sequencing were used to investigate the effects of COS supplements on histopathology scores. It is clear that the COS-fed mice showed reduced microbiota diversity (as per their feces samples), according to the Shannon and Simpson indices, compared to the control group of mice. The indices also showed that the COS-fed mice showed higher microbiota diversity than the control group of mice based on the colon analysis. Interestingly, it was indicated by the Ace and Chao richness indices that both groups of mice showed similar microbiota community richness in their faecal and colonic samples (Table 1).
Table 1.
Comparative results of 16S rRNA gene libraries' phylotype coverage and diversity at 97% similarity based on pyrosequencing analysis (n = 6).
| Number of reads | Number of OUT | Coverage | Richness estimator | Diversity index | |||
|---|---|---|---|---|---|---|---|
| Ace (95% Cl) | Chao (95% Cl) | Shannon (95% Cl) | Simpson (95% Cl) | ||||
| Colon | |||||||
| Control | 12,568 | 37 | 99.88% | 53 (43–86) | 58 (43–112) | 0.72 (0.69–0.74) | 0.68 (0.66–0.70) |
| COS | 12,794 | 46 | 99.88% | 62 (52–87) | 56 (50–81) | 0.88 (0.85–0.90) | 0.59 (0.58–0.60) |
| Feces | |||||||
| Control | 11,521 | 314 | 99.90% | 324 (312–344) | 329 (314–357) | 4.17 (4.23–4.39) | 0.025 (0.024–0.026) |
| COS | 10,956 | 271 | 99.43% | 304 (289–333) | 302 (281–337) | 4.05 (4.01–4.08) | 0.033 (0.032–0.034) |
The RDP classifier was used to perform a taxon-dependent analysis in order to identify the intestinal microbiota's taxonomy. For both groups of mice, the faecal microbiota showed nine phyla, including one candidate division (TM7), whilst the colon microbiota showed seven phyla, including one candidate division (TM7). Additionally, six phyla were found in each of the mice groups. As illustrated in Figure 3, in colon contents, Firmicutes (96.8%) and Proteobacteria (1.3%) were the phyla with the highest percentages in the colon microbiota of the COS-fed mice, whilst in feces Firmicutes (96.1%) and Proteobacteria (1.8%) were the phyla with the highest percentages in the control mice. The percentages of Bacteroidetes (61.4%), Firmicutes (27.1%), and Proteobacteria (4.5%) were the three most abundant in the faecal microbiota of the COS-fed mice, whilst the percentages of Bacteroidetes (53.8%), Firmicutes (39.6%), and Proteobacteria (3.5%) were the most abundant in the control mice.
Figure 3.
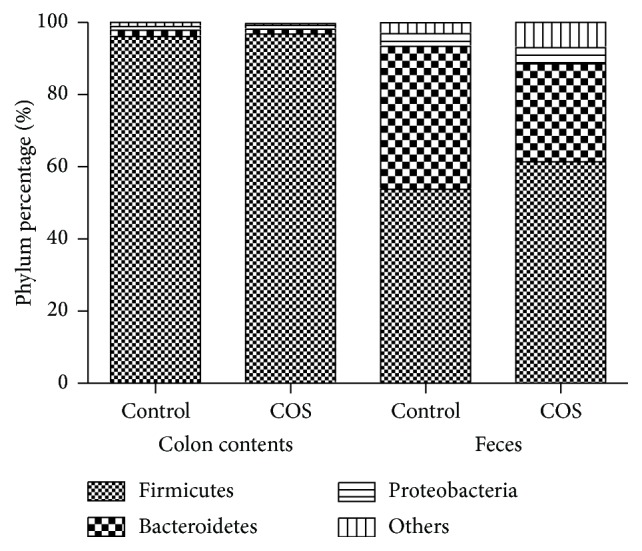
D7 postinfection composition of the intestinal microbiota. Microbial composition in the colon and feces of both groups (n = 6). Control mice were given standard drinking water and a basal rodent diet, whilst COS mice were given the same water and diet with the addition of COS at 300 mg/kg.
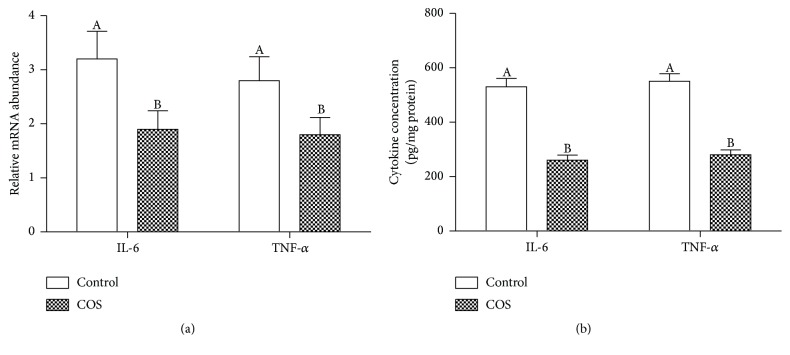
As shown in Figure 4(a), colon analysis of control mice showed significantly higher IL-6 and TNF-α mRNA than the COS-fed mice at D7 after infection. This finding is supported through the ELISA results (Figure 4(b)).
Figure 4.
Control and COS group (n = 6) mucosal inflammatory responses. (a) RT-PRC evaluation of IL-6 and TNF-α mRNA level. (b) ELISA evaluation of IL-6 and TNF-α protein level.
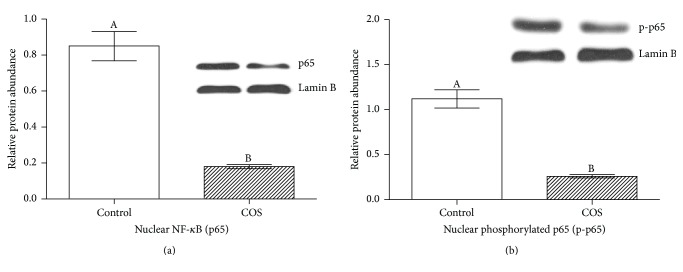
Nuclear NF-κB (p65) measurements were taken to identify the activation of NF-κB in the colons of the mice. The purpose of this was to identify whether the beneficial outcomes of COS (i.e., in reducing infection) are supported by the suppression of NF-κB activation pathways. The results showed that the control group of mice had significantly higher nuclear NF-κB (p65) in their colonic samples than the COS-fed group of mice, which suggests that COS suppresses NF-κB activation (see Figure 5).
Figure 5.
Immunoblotting of (a) nuclear NF-κB (p65) and (b) phosphorylated nuclear NF-κB (p65) in control and COS group (n = 6).
4. Discussion
C. rodentium, which is similar to the human enteropathogenic E. coli infection, is an extracellular enteric pathogen that infects mice. It has also been noted that inflammatory bowel disease (IBD) is often modelled using C. rodentium in mice. IBD results in intestinal inflammation. Thus, intestinal pathology is regulated by inflammatory regulators such as inducible TNFα and IL-6 [6]. Intestinal bacterial communities influence mice's susceptibility and ability to overcome the C. rodentium infection, as does the immune response of the mice [23]. E. coli is commonly modelled by the infection of mice with C. rodentium since mice are not affected by EPEC or EHEC. C. rodentium has been found in all mice strains but it is rare in human disease. Depending on the strain, an infected mouse can suffer subclinical disease or death after infection [9]. For instance, strains believed to be resistant to developing colitis as a result of C. rodentium infection include CD-1 and C57BL/6 mice. On the other hand, it is believed that susceptibility is present amongst mice strains C3H/HeJ and FVB/N [9]. Based on this information, human E. coli infection was modelled in this study by infecting C57BL/6 mice with C. rodentium.
The results showed that, after infecting the mice with C. rodentium, inflammation was resolved more quickly amongst mice receiving 300 mg/kg COS than the control mice. The results noted no impact on the quantity of C. rodentium taken from faecal samples, which suggests that the regulating impact of COS is not due to swift pathogenic eradication but rather due to an improvement in microflora diversity. Various biological functions are said to be influenced by such microbiota [19, 20], and it has been associated with cancer [24, 25], cirrhosis of the liver [26], and other diseases. Since Firmicutes has the ability to offer extra energy to the host by fermenting plant polysaccharide to SCFA, this is the reason for obesity being linked to higher concentrations of Firmicutes: Bacteroidetes [27]. At present, the reason behind COS's minimising impact on Firmicutes quantities is unclear. Some studies have also shown that a reduction in intestinal Firmicutes can be achieved through acidic oligosaccharides, a galactooligosaccharides/long-chain fructan solution (GOS/lcF, 9/1), fructooligosaccharides, and other oligosaccharides [28]. It may be that the N-acetyl glucosamine of COS may function as a binding agent that allows COS to influence the attachment of bacteria to the intestine. Given this, research on pigs and chickens indicates that intestinal microbial communities can be impacted by the introduction of COS supplements [18]. Furthermore, intestinal microbiota may change as a result of COS's role as a fermentable substrate for certain bacteria, as this could lower the pH level of the gut and activate natural acid production [26].
Various studies have indicated in vitro and in vivo anti-inflammatory effects of COS. In the current study, it has been found that IL-6 and TNF-α expression in the colon are reduced by COS, whilst microbial flora is impacted by the inflammation [23]. Thus, microbiome changes may be influenced by increased inflammation. Since the faecal samples showed no relationship between C. rodentium quantities and the introduction of COS, this finding is contradictory. Consequently, the speed at which C. rodentium resolution occurs is related to microflora changes and not inflammation [29]. The results of this study indicate that the effects of COS on the intestines only occur in specific areas of the gut and only take on a specific form. Mice fed with COS were found to have a lower expression of inflammatory cytokines, which is linked to bacterial flora shifts that lead to faster infection recovery.
COS is a d-glucosamine oligomer that cannot be eroded by enzymes in the gut. Thus, COS directly protects the host's intestinal epithelial cells (IEC). Research indicates that IEC expresses various chemokine and cytokine receptors such as TLR4 and other Toll-like receptors [30]. In the IEC, NF-κB-regulated proinflammatory cytokine production is promoted by the interaction between TNF-α and their respective receptors, TLR4, TNFR1, and TNFR2 [31]. Studies also show that dysfunction of the intestinal epithelial barrier can occur as a result of pronounced IEC-regulated mucosal inflammatory responses, which are associated with greater TLR4 and TNFR expression [32]. In the current study, it was revealed that a significant reduction in the activation of NF-κB along with colon production of TNF-α and IL-6 was achieved through the administration of COS through the basal diet. Since recent studies have found that mice can develop disease sharing similar characteristics to Crohn's as a result of IEC-derived TNF-α [32], this indicates that the anti-inflammatory impact of COS could be the result of COS' suppressive impact on the activation of NF-κB and production of proinflammatory cytokines in the IEC.
Overall, it is clear that in vivo C. rodentium infection can be attenuated by the introduction of COS into mice's diet. The findings suggest that this is due to the prevention of IL-6 and TNF-α expression and NF-κB activation as well as a shift in intestinal microflora. Innovative and successful methods for protecting hosts from C. rodentium infection could therefore be achieved through further investigation of the organic carbohydrate oligomer COS.
Acknowledgments
This study was in part supported by National Key Research and Development Program of China (2016YFD0500504), the National Natural Science Foundation of China (31402092), the Open Foundation of Key Laboratory of Agroecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences (ISA2015303), the Hunan Provincial Science and Technology Department (13JJ2034, 2013FJ3011, 2014NK3048, 2014NK4134, and 2014WK2032), and Open Project Program of State Key Laboratory of Food Science and Technology, Nanchang University (SKLF-KF-201416). The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding of this research through the Research Group Project no. RGP-213.
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this article.
References
- 1.Schauer D. B., Falkow S. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infection and Immunity. 1993;61(6):2486–2492. doi: 10.1128/iai.61.6.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luperchio S. A., Newman J. V., Dangler C. A., et al. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. Journal of Clinical Microbiology. 2000;38(12):4343–4350. doi: 10.1128/jcm.38.12.4343-4350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frankel G., Phillips A. D. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cellular Microbiology. 2008;10(3):549–556. doi: 10.1111/j.1462-5822.2007.01103.x. [DOI] [PubMed] [Google Scholar]
- 4.Lapointe T. K., O'Connor P. M., Buret A. G. The role of epithelial malfunction in the pathogenesis of enteropathogenic E. coli-induced diarrhea. Laboratory Investigation. 2009;89(9):964–970. doi: 10.1038/labinvest.2009.69. [DOI] [PubMed] [Google Scholar]
- 5.Chandrakesan P., Roy B., Jakkula L. U. M. R., et al. Utility of a bacterial infection model to study epithelial-mesenchymal transition, mesenchymal-epithelial transition or tumorigenesis. Oncogene. 2014;33(20):2639–2654. doi: 10.1038/onc.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mundy R., MacDonald T. T., Dougan G., Frankel G., Wiles S. Citrobacter rodentium of mice and man. Cellular Microbiology. 2005;7(12):1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 7.Lupp C., Robertson M. L., Wickham M. E., et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae . Cell Host and Microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Wiles S., Clare S., Harker J., et al. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium . Cellular Microbiology. 2004;6(10):963–972. doi: 10.1111/j.1462-5822.2004.00414.x. [DOI] [PubMed] [Google Scholar]
- 9.Vallance B. A., Deng W., Jacobson K., Finlay B. B. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium . Infection and Immunity. 2003;71(6):3443–3453. doi: 10.1128/iai.71.6.3443-3453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willing B. P., Vacharaksa A., Croxen M., Thanachayanont T., Finlay B. B. Altering host resistance to infections through microbial transplantation. PLoS ONE. 2011;6(10) doi: 10.1371/journal.pone.0026988.e26988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 12.Carmody R. N., Turnbaugh P. J. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. The Journal of Clinical Investigation. 2014;124(10):4173–4181. doi: 10.1172/jci72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cárdenas G., Orlando P., Edelio T. Synthesis and applications of chitosan mercaptanes as heavy metal retention agent. International Journal of Biological Macromolecules. 2001;28(2):167–174. doi: 10.1016/s0141-8130(00)00156-2. [DOI] [PubMed] [Google Scholar]
- 14.Xiao D., Wang Y., Liu G., et al. Effects of chitosan on intestinal inflammation in weaned pigs challenged by enterotoxigenic Escherichia coli . PLoS ONE. 2014;9(8) doi: 10.1371/journal.pone.0104192.e104192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi E. H., Yang H. P., Chun H. S. Chitooligosaccharide ameliorates diet-induced obesity in mice and affects adipose gene expression involved in adipogenesis and inflammation. Nutrition Research. 2012;32(3):218–228. doi: 10.1016/j.nutres.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Fang I.-M., Yang C.-M., Yang C.-H. Chitosan oligosaccharides prevented retinal ischemia and reperfusion injury via reduced oxidative stress and inflammation in rats. Experimental Eye Research. 2015;130:38–50. doi: 10.1016/j.exer.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Qin C., Zhang Y., Liu W., Xu L., Yang Y., Zhou Z. Effects of chito-oligosaccharides supplementation on growth performance, intestinal cytokine expression, autochthonous gut bacteria and disease resistance in hybrid tilapia Oreochromis niloticus ♀ × Oreochromis aureus ♂. Fish and Shellfish Immunology. 2014;40(1):267–274. doi: 10.1016/j.fsi.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Yang C. M., Ferket P. R., Hong Q. H., et al. Effect of chito-oligosaccharide on growth performance, intestinal barrier function, intestinal morphology and cecal microflora in weaned pigs. Journal of Animal Science. 2012;90(8):2671–2676. doi: 10.2527/jas.2011-4699. [DOI] [PubMed] [Google Scholar]
- 19.Lee W.-J., Hase K. Gut microbiota-generated metabolites in animal health and disease. Nature Chemical Biology. 2014;10(6):416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian S., Huq S., Yatsunenko T., et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren W., Chen S., Yin J., et al. Dietary arginine supplementation of mice alters the microbial population and activates intestinal innate immunity. Journal of Nutrition. 2014;144(6):988–995. doi: 10.3945/jn.114.192120. [DOI] [PubMed] [Google Scholar]
- 22.Gibson J., Shokralla S., Porter T. M., et al. Simultaneous assessment of the macrobiome and microbiome in a bulk sample of tropical arthropods through DNA metasystematics. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(22):8007–8012. doi: 10.1073/pnas.1406468111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann C., Hill D. A., Minkah N., et al. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infection and Immunity. 2009;77(10):4668–4678. doi: 10.1128/iai.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anhê F. F., Roy D., Pilon G., et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. 2015;64(6):872–883. doi: 10.1136/gutjnl-2014-307142. [DOI] [PubMed] [Google Scholar]
- 25.Louis P., Hold G. L., Flint H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nature Reviews Microbiology. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 26.Qin N., Yang F., Li A., et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 27.Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 28.Morel F. B., Oozeer R., Piloquet H., et al. Preweaning modulation of intestinal microbiota by oligosaccharides or amoxicillin can contribute to programming of adult microbiota in rats. Nutrition. 2015;31(3):515–522. doi: 10.1016/j.nut.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Kamada N., Kim Y.-G., Sham H. P., et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336(6086):1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cario E. Innate immune signalling at intestinal mucosal surfaces: a fine line between host protection and destruction. Current Opinion in Gastroenterology. 2008;24(6):725–732. doi: 10.1097/mog.0b013e32830c4341. [DOI] [PubMed] [Google Scholar]
- 31.Onizawa M., Nagaishi T., Kanai T., et al. Signaling pathway via TNF-α/NF-κB in intestinal epithelial cells may be directly involved in colitis-associated carcinogenesis. American Journal of Physiology—Gastrointestinal and Liver Physiology. 2009;296(4):G850–G859. doi: 10.1152/ajpgi.00071.2008. [DOI] [PubMed] [Google Scholar]
- 32.Roulis M., Armaka M., Manoloukos M., Apostolaki M., Kollias G. Intestinal epithelial cells as producers but not targets of chronic TNF suffice to cause murine Crohn-like pathology. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(13):5396–5401. doi: 10.1073/pnas.1007811108. [DOI] [PMC free article] [PubMed] [Google Scholar]