Abstract
Knowledge regarding the 3D structure of a protein provides useful information about the protein’s functional properties. Particularly, structural similarity between proteins can be used as a good predictor of functional similarity. One method that uses the 3D geometrical structure of proteins in order to compare them is the similarity value (SV). In this paper, we introduce a new definition of the SV measure for comparing two proteins. To this end, we consider the mass of the protein’s atoms and concentrate on the number of protein’s atoms to be compared. This defines a new measure, called the weighted similarity value (WSV), adding physical properties to geometrical properties. We also show that our results are in good agreement with the results obtained by TM-SCORE and DALILITE. WSV can be of use in protein classification and in drug discovery.
Keywords: Protein, Weighted similarity value, RMSD, Wigner-D functions
Introduction
In a quantitative manner, comparing two protein tertiary structures to evaluate their similarity is a major challenge. A successful comparison can provide answers to some important questions in structural biology, cell biology, and biochemistry [1]. In particular, it is believed that functional similarity can be predicted from the structural similarity between proteins. The 3D structure of a protein is obtained by various experimental methods such as X-ray or electron crystallography and sometimes NMR [2]. If there is no crystallographic structure of a protein, computational structure prediction methods exist that use sequence similarity. In sequence similarity, a technique called homology modeling is used based on the structure of a known protein as a template to predict the structure of an unknown protein [3]. If structural information of the protein exists, there are methods that have been developed to compare the structures [4–13].
Examples of methods based on numerical techniques to predict structural information are: SCOP (Structural Classification of Proteins) [9, 10], CATH (Class, Architecture, Topology and Homologous superfamily) [13], TM-SCORE [14, 15], STRUCTAL software [16], FSSP (Families of Structurally Similar Proteins) [17] and DALILITE [18].
Recently, in Ref. [19], the similarity value (SV), as a geometrical-based structural property, was introduced as a new protein similarity measure. The SV is an alternative measure to the root-mean-square deviation (RMSD). ‘SV’ is defined as a normalized RMSD of the protein distances in reciprocal space so that the protein’s atomic coordinates are mapped into the corresponding Fourier space. There are theorems in mathematics that allow us to perform this task by using Wigner-D functions and then by using crystallography concepts to arrive at the structure factor [19]. The advantage of defining SV in the reciprocal space is that it solves the known problem of different sizes of two compared proteins. Thus, there is no need to use partial or local similarity tests. An example of using a method that relies on the partial RMSD for computing the similarity value is STRUCTAL software [16]. SV is also sensitive to protein topology (for a brief explanation regarding the differences between SV and other methods see [19]).
In this paper, we propose an improved SV definition, called the ‘weighted similarity value’ (WSV) in order to add some important physical properties required to adequately compare any two proteins. We define WSV by adding a lower limit on the reciprocal space dimension for the two proteins that are being compared. This constraint ensures that we do not lose any information in mapping from the protein’s spatial space to the reciprocal space. We also consider the masses of the atoms as a physical property in the protein shape function. The importance of adding mass to the shape function comes from the structure factors in X-ray scattering data [20]. Thus, adding the atomic masses to the shape function provides more reliable computed structure factors as we show later in this paper.
We compare the results regarding protein similarity obtained by WSV with NRMSD, DALILITE, and TM-SCORE, and we show that our results are in good agreement with these methods. DALILITE is a multiple alignment method, which is based on the alignment of the amino acid sequences and the secondary structure states (helix, sheet, coil) of the two proteins being compared [18]. Since DALILITE is a multiple alignment method, the results given by DALILITE have multi-valued z-scores and corresponding similarity values between two proteins [18].
The template modeling score (TM-SCORE) is a global fold similarity measure between two protein structures with different tertiary structures and it is independent of proteins sizes. The TM-SCORE is a normalized measure and has a value in the [0,1] range; when it is equal to 1, the two proteins are similar [21].
Methods
RMSD is defined as a dissimilarity parameter between two proteins as follows:
| 1 |
where N is the number of proteins’ atoms and d ij is defined as the elements of the distance matrix between the atoms’ positions of a given protein, as is the case for d 'ij. Here, we assumed that the two proteins in question have the same number of atoms. If the numbers of atoms of the two proteins are not equal, we should use a partial RMSD definition. RMSD is a semi-bounded parameter (between zero and infinity). We now define ‘normalized RMSD’ (NRMSD) as a bounded similarity parameter between two proteins. First, we introduce the following auxiliary parameter:
| 2 |
and define:
| 3 |
where d 2 = 2∑Ni <j∑Nj = 1 d 2ij is the vector length (sum of the squares of arrays), as is the case for d ' 2. If the two proteins are not correlated, we have D 2 = d 2 + d ' 2 and NRMSD = 0. If we have a maximum correlation between these two proteins (two proteins are the same), i.e., D 2 = 0, then, NRMSD = 1/2. In the next step we define WSV.
The SV was defined by using the Wigner-D function in conjunction with a series expansion of the protein’s shape functions [19]. The Wigner-D functions [22] describe the surface of a 4-sphere and they are an extension of spherical harmonic oscillators (SHO). The surface of a 4-sphere is a three-dimensional manifold, which can be explored by using a set of three angles, defined as Euler angles. On the other hand, Euler angles describe a motion in three-dimensional Euclidean space. Thus, we can project a three-dimensional Euclidean space onto the three-dimensional manifold (4-sphere surface). This means we project a body onto the surface of a 4-sphere. Adding atomic masses, M atom to point coordinates gives gravitational attraction for a given projected point. Thus, we define the protein shape function as:
| 4 |
where i, j, k = 1, 2, …, N (N is the number of protein’s atoms) and M atom is the molar atomic mass in the atomic mass unit (in the definition of SV for all atoms we have M atom = 1). Here, (α i, β j, γ k) are three Euler angles corresponding to the position of this atom in the corresponding (x i, y j, z k) PDB (Protein Data Bank) entry. We now expand a protein shape function in terms of the Wigner-D functions, D lmn(α, β, γ), which span a basis set as follows:
| 5 |
where C lmn s are the coefficients of the series expansion and they are unique for a given function, f(α, β, γ). Some theorems in mathematics allow us to use the coefficients of expansion of a function by the Wigner-D function as a three-dimensional Fourier transform of this function [23, 24]. Thus, in the above expansion, the C lmn s corresponds to elements of the three-dimensional Fourier transform of f(α, β, γ). From crystallography considerations, it is readily recognized that these are the coefficients of the crystal shape function as a structure factor [25]. Thus, C lmn s are the protein structure factors. Now, we can see why adding the masses of atoms is so important because in X-ray scattering the atomic masses play an important role in determining the corresponding structure factors [26]. C lmn s can be obtained by the following relation:
| 6 |
where we have used the orthogonality relation between the Wigner-D functions as follows:
| 7 |
Now, in the reciprocal space, the two shapes (proteins) are described with the same dimensions [19], however, they have different numbers of atoms. This is due to the use of Wigner-D functions. The dimension of reciprocal space, N R, is given by:
| 8 |
where L max is an arbitrary maximum value chosen in the computation of C lmn.
The coefficients C lmn s belong to the complex space and we can embed them in the (N R × 2) -dimensional Euclidean space such that S≡(Real(C lmn), Imaginary(C lmn)) where S≡{S ij}, (i = 1, 2, ⋯, N R and j = 1, 2) is a matrix of structure factors. In this step, we can define an (N R × N R) -distance matrix for S and then, we define the SD parameter between two proteins as follows:
| 9 |
where sd ij and sd 'ij are the elements of the distance matrix in the reciprocal space of each of the two proteins that is defined by:
| 10 |
Here, we add a constraint on the definition of WSV by always making sure that N R ≥ max(N 1, N 2) where N 1 and N 2 are the numbers of atoms of the two compared proteins. Then, we define where ⌊.⌋ indicates the integer part of the number in the brackets. Now, we introduce a direct measure to characterize the similarity between two proteins, which depends on their geometries and physical properties (masses and positions of their atoms). Thus we define the weighted similarity values, WSV, as:
| 11 |
where , is the vector length (sum of the squares of arrays), as is the case for sd ' 2. If the two proteins are not correlated, we have SD 2 = sd 2 + sd ' 2 and then, WSV = 0. If we have a maximum correlation between these two proteins (two proteins are the same), i.e., SD 2 = 0, then, WSV = 1/2.
The range of atomic masses for the proteins is given in the following. The heaviest atom’s weight in a protein can be a sulfur atom, with a mass about 32.065 a.m.u. and the lightest atom mass is for hydrogen with a mass about 1.00794 a.m.u. We have also considered the atomic mass of some metal atoms in the liganded proteins.
We also wish to compare WSV with the other measures of protein similarity, namely (NRMSD, TM-SCORE, and DALILITE). We use these methods separately as targets and observe that WSV predicts the similarity or dissimilarity in close agreement with their predictions. To analyze it in this way, we compute ‘sensitivity’ (or the probability of prediction similarity between two proteins), ‘specificity’ (or the probability of prediction dissimilarity between two proteins), ‘accuracy’ (probability that the WSV measure is true or what it is supposed to measure), ‘precision’ (probability that if a test is repeated, it gives the same result), and ‘F-score’ (probability of giving a positive (similarity) prediction, or performance of higher sensitivity) [27] as explained below.
To compute sensitivity, specificity, etc., we first normalize TM-SCORE and DALILITE (referred to as M-score) to 0.5. Thus, when M = 0.5, the two proteins are completely similar and when it is 0, the two proteins are completely dissimilar. Then, we assume that a measure that predicts similarity between two proteins does so with any value greater than 0.251 and dissimilar proteins with a value less than 0.25. We have a true positive (TP) result when both measures predict similarity, true negative (TN) when both methods predict dissimilarity, false positive (FP) when WSV predicts similarity, and M-score predicts dissimilarity and false negative (FN) when WSV predicts dissimilarity and M-score predicts similarity. The definitions of sensitivity, specificity, etc., are given in Table 4.
Table 4.
The computation of sensitivity, specificity, accuracy, precision, and F-score for the 48 and 86 datasets
| Sensitivity1 | Specificity2 | Accuracy3 | Precision4 | F-Score5 | ||
|---|---|---|---|---|---|---|
| NRMSD | 48 dataset(i) | 0.800 | 0.375 | 0.729 | 0.865 | 0.831 |
| 86 dataset(ii) | 0.857 | 0.625 | 0.814 | 0.909 | 0.882 | |
| TM-SCORE* | 48 dataset(iii) | 0.791 | 0.500 | 0.766 | 0.944 | 0.861 |
| 86 dataset(iv) | 0.771 | 0 | 0.762 | 0.985 | 0.845 | |
| DALILITE** | 48 dataset(v) | 0.756 | 0 | 0.723 | 0.944 | 0.839 |
| 86 dataset(vi) | 0.765 | 0.250 | 0.741 | 0.954 | 0.849 |
(i) TP (true positive) = 32 TN (true negative) = 3 FP (false positive) = 5 FN (false negative)= 8
(ii) TP = 60, TN = 10, FP = 6, FN = 10
(iii) TP = 34, TN = 2, FP = 2, FN = 9
(iv) TP = 64 TN = 0 FP = 1 FN = 19
(v) TP = 34 TN = 0 FP = 2 FN = 11
(vi) TP = 62 TN = 1 FP = 3 FN = 19
*Because there are no TM-SCORE values for some pairs we have compared 47 and 84 pairs of the 48 and 86 datasets, respectively
**Maximum similarity values in DALILITE results are compared with WSV. Also, there are no DALILITE values for some pairs we have compared 47 and 85 pairs of the 48 and 86 datasets, respectively
1 Sensitivity = TP/(TP+FN)
2 Specificity = TN/(TN+FP)
3 Accuracy = (TP+TN)/(Num. of total population)
4 Precision = TP/(TP+FP)
5 F-Score = 2TP/(2TP+FP+FN)
We also compare WSV with the other scores by introducing a relative difference between WSV and M-score as:
| 12 |
When dif = 0, the WSV and M-score have the same prediction values and when dif = 1, this means the WSV and M-score have totally different prediction values. In other words, one predicts that the two proteins are similar and the other predicts that they are totally dissimilar.
Results and discussion
In this paper, we defined WSV as a development of SV by including some physical properties of proteins in its definition and a constraint on the dimension of the reciprocal space. In Tables 1 and 2, we show a comparison of the WSV with SV [19], RMSD, NRMSD, TM-SCORE [14, 15] and DALILITE [18] values for 48 and 86 datasets, respectively, where both liganded and unliganded proteins are listed in the supplementary material of Li et al. [1] (these sets are reported in http://dragon.bio.purdue.edu/visgrid_suppl). We reported only minimum and maximum similarity values between two proteins predicted by DALILITE. The data acquisition for the TM-SCORE [14, 15] was obtained by the Zhang Lab’s server http://zhanglab.ccmb.med.umich.edu/TM-SCORE/ for 48 and 86 datasets (only 84 data of the 86 dataset and 47 data of the 48 dataset were used; because there are no TM-SCORE values) and for DALILITE [18] it was obtained by the Holm’s Lab’s server: http://ekhidna.biocenter.helsinki.fi/dali_lite/start) for 48 and 86 datasets (only 85 data of the 86 dataset and 47 data of the 48 dataset were used because there are no DALILITE values).
Table 1.
A set of 48 protein structures with WSV, SV [19], and RMSD from Li et al. [1], NRMSD, TM-SCORE [14, 15], and DALILITE [18]
| First protein’s PDB ID | Second protein’s PDB ID | SV [8] | RMSD [1] | NRMSD | WSV | TM-SCORE [16, 17]* | DALILITE [18]* | L max |
|---|---|---|---|---|---|---|---|---|
| 1A6W | 1A6U | 0.198 | 0.34 | 0.459 | 0.438 | 0.9958 | 100 | 11 |
| 1MRG | 1AHC | 0.401 | 0.43 | 0.325 | 0.416 | 0.9958 | 99 | 12 |
| 1RNE | 1BBS | 0.316 | 0.61 | 0.0706 | 0.284 | 0.9567 | 100-14 | 16 |
| 1RBP | 1BRQ | 0.108 | 0.62 | 0.49 | 0.0460 | 0.9865 | 100 | 10 |
| 1BYB | 1BYA | 0.499 | 0.43 | 0.499 | 0.497 | 0.9923 | 100 | 14 |
| 1HFC | 1CGE | 0.399 | 0.37 | 0.335 | 0.49 | 0.9636 | 100-5 | 10 |
| 3GCH | 1CHG | 0.070 | 1.10 | 0.427 | 0.328 | 0.0439 | 100-7 | 11 |
| 1BLH | 1DJB | 0.497 | 0.23 | 0.478 | 0.49 | 0.9984 | 100 | 11 |
| 1INC | 1ESA | 0.397 | 0.21 | 0.499 | 0.224 | 0.2633 | 100-4 | 11 |
| 1GCA | 1GCG | 0.499 | 0.32 | 0.498 | 0.499 | 0.9975 | 100-8 | 13 |
| 1HEW | 1HEL | 0.498 | 0.21 | 0.499 | 0.49 | 0.9975 | 100 | 9 |
| 1IDA | 1HSI | 0.083 | 1.07 | 0.497 | 0.449 | 0.9360 | 100 | 10 |
| 1DWD | 1HXF | 0.150 | 0.27 | 0.461 | 0.212 | 0.8795 | 100-2 | 12 |
| 2IFB | 1IFB | 0.382 | 0.37 | 0.499 | 0.468 | 0.9927 | 100-9 | 9 |
| 1IMB | 1IME | 0.498 | 0.22 | 0.499 | 0.499 | 0.9987 | 100 | 15 |
| 2PK4 | 1KRN | 0.445 | 0.39 | 0.409 | 0.432 | 0.0802 | 100 | 7 |
| 2TMN | 1L3F | 0.266 | 0.62 | 0.497 | 0.09 | 0.9911 | 100 | 12 |
| 1IVD | 1NNA | 0.426 | 1.23 | 0.108 | 0.286 | 0.6688 | 49-4 | 18 |
| 1HYT | 1NPC | 0.332 | 0.88 | 0.470 | 0.107 | 0.9825 chain_1; 0.9795 chain_2 |
73 | 12 |
| 1PDZ | 1PDY | 0.499 | 0.66 | 0.499 | 0.49 | 0.9933 | 100-8 | 13 |
| 1PHD | 1PHC | 0.499 | 0.17 | 0.499 | 0.499 | 0.9994 | 100 | 13 |
| 1PSO | 1PSN | 0.499 | 0.33 | 0.49 | 0.499 | 0.9976 | 100-17 | 12 |
| 1SRF | 1PTS | 0.498 | 0.26 | 0.480 | 0.499 | 0.9694 | 100-1 | 11 |
| 1ACJ | 1QIF | 0.497 | 0.31 | 0.459 | 0.499 | 0.9908 | 100 | 15 |
| 1SNC | 1STN | 0.495 | 0.70 | 0.480 | 0.49 | 0.9741 | 100 | 9 |
| 1STP | 1SWB | 0.145 | 0.33 | 0.0284 | 0.362 | 0.9770 | 100-4 | 14 |
| 1ULB | 1ULA | 0.474 | 0.79 | 0.498 | 0.498 | 0.9674 | 94 | 12 |
| 2YPI | 1YPI | 0.165 | 1.27 | 0.499 | 0.0840 | 0.9727 | 100-5 | 14 |
| 2H4N | 2CBA | 0.498 | 0.20 | 0.465 | 0.499 | 0.9950 | 100 | 11 |
| 2CTC | 2CTB | 0.499 | 0.15 | 0.495 | 0.499 | 0.9995 | 100 | 12 |
| 5CNA | 2CTV | 0.034 | 0.40 | 0.0219 | 0.0063 | 0.9952 | 100 | 18 |
| 1FBP | 2FBP | 0.494 | 1.06 | 0.499 | 0.467 | 0.9600 | 100-5 | 17 |
| 2SIM | 2SIL | 0.499 | 0.14 | 0.499 | 0.499 | 0.9969 | 100-6 | 13 |
| 1MTW | 2TGA | 0.159 | 0.42 | 0.49 | 0.403 | 0.9837 | 100-5 | 10 |
| 1APU | 3APP | 0.498 | 0.40 | 0.498 | 0.499 | 0.9969 | 100-23 | 12 |
| 1QPE | 3LCK | 0.465 | 0.28 | 0.434 | 0.496 | 0.9984 | 2-4 | 12 |
| 5P2P | 3P2P | 0.480 | 0.42 | 0.49 | 0.457 | 0.9893 | 98 | 11 |
| 4PHV | 3PHV | 0.045 | 1.23 | 0.13 | 0.111 | 0.9268 | 99 | 10 |
| 3PTB | 3PTN | 0.122 | 0.26 | 0.499 | 0.292 | 0.9979 | 100 | 10 |
| 1BID | 3TMS | 0.499 | 0.24 | 0.499 | 0.499 | 0.9984 | 100 | 11 |
| 1OKM | 4CA2 | 0.472 | 0.22 | 0.482 | 0.498 | 0.9986 | 100 | 11 |
| 4DFR | 5DFR | 0.496 | 0.82 | 0.0887 | 0.348 | 0.9725 | 99 | 12 |
| 3MTH | 6INS | 0.381 | 1.09 | 0.437 | 0.479 | -- | -- | 8 |
| 6RSA | 7RAT | 0.440 | 0.18 | 0.158 | 0.463 | 0.9981 | 100 | 11 |
| 1CDO | 8ADH | 0.403 | 1.34 | 0.101 | 0.161 | 0.8304 | 55-8 | 16 |
| 7CPA | 5CPA | 0.132 | 0.40 | 0.499 | 0.128 | 0.9956 | 100 | 12 |
| 1ROB | 8RAT | 0.469 | 0.28 | 0.490 | 0.491 | 0.9955 | 100 | 8 |
| 1IGJ | 1A4J | 0.411 | 0.80 | 0.47 | 0.0707 | 0.2793 | 78-4 | 17 |
*For comparison, we have used TM-SCORE server at: http://zhanglab.ccmb.med.umich.edu/TM-SCORE/
**The numbers reported here are the maximum and minimum sequence identity of aligned positions in percent. The data are given from the Dali server: http://ekhidna.biocenter.helsinki.fi/dali_lite/start/ .
-- No value reported by the site
Table 2.
A set of 86 protein structures with WSV, SV [8], and RMSD from Li et al. [1], NRMSD, TM-SCORE [16, 17], and DALILITE [18]
| First protein’s PDB ID | Second protein’s PDB ID | SV [8] | RMSD [1] | NRMSD | WSV | TM-SCORE [16, 17]* | DALILITE [18]** | L max |
|---|---|---|---|---|---|---|---|---|
| 1AD4 | 1AD1 | 0.499 | 0.50 | 0.463 | 0.498 | 0.9445 | 100-8 | 14 |
| 1AHX | 1AHG | 0.499 | 0.24 | 0.494 | 0.500 | 0.9989 | 100 | 17 |
| 1AUR | 1AUO | 0.499 | 0.20 | 0.499 | 0.499 | 0.9987 | 100-9 | 13 |
| 1AXZ | 1AXY | 0.498 | 0.12 | 0.499 | 0.500 | 0.9995 | 100 | 11 |
| 1GN8 | 1B6T | 0.491 | 0.51 | 0.472 | 0.493 | 0.9904 | 100 | 12 |
| 1B9Z | 1B90 | 0.494 | 0.54 | 0.499 | 0.500 | 0.9929 | 100-3 | 15 |
| 1LRI | 1BEO | 0.498 | 1.05 | 0.471 | 0.495 | 0.9317 | 99 | 8 |
| 1BUL | 1BUE | 0.499 | 0.18 | 0.482 | 0.500 | 0.9991 | 100 | 11 |
| 1BYD | 1BYA | 0.499 | 0.43 | 0.499 | 0.498 | 0.9923 | 100-5 | 14 |
| 1C3R | 1C3P | 0.202 | 0.39 | 0.11 | 0.141 | 0.9970 | 99 | 17 |
| 1C5I | 1C5H | 0.494 | 0.13 | 0.499 | 0.500 | 0.9993 | 100 | 10 |
| 1QJW | 1CB2 | 0.498 | 0.63 | 0.495 | 0.495 | 0.9898 | 100-3 | 16 |
| 1CTE | 1CPJ | 0.499 | 0.29 | 0.492 | 0.500 | 0.9977 | 100 | 14 |
| 1SZJ | 1CRW | 0.499 | 0.33 | 0.499 | 0.498 | 0.9975 | 100 | 16 |
| 1ESW | 1CWY | 0.498 | 0.38 | 0.499 | 0.499 | 0.9978 | 100-4 | 14 |
| 1CY7 | 1CY0 | 0.155 | 1.12 | 0.45 | 0.085 | 0.9664 | 99-11 | 15 |
| 1DED | 1D7F | 0.481 | 0.26 | 0.364 | 0.499 | 0.9992 | 100-4 | 22 |
| 1P7T | 1D8C | 0.406 | 0.66 | 0.088 | 0.060 | 0.9827 | 97-4 | 21 |
| 1DMY | 1DMX | 0.499 | 0.19 | 0.499 | 0.500 | 0.9988 | 100 | 14 |
| 1DQY | 1DQZ | 0.052 | 0.75 | 0.0962 | 0.306 | 0.9494 | 99 | 15 |
| 1LP6 | 1DV7 | 0.471 | 0.56 | 0.126 | 0.443 | 0.9447 | 100-6 | 13 |
| 1E2S | 1E1Z | 0.499 | 0.13 | 0.492 | 0.500 | 0.9997 | 100 | 14 |
| 1ESE | 1ESC | 0.499 | 0.19 | 0.499 | 0.500 | 0.9991 | 100-8 | 12 |
| 6ALD | 1EWD | 0.477 | 0.44 | 0.434 | 0.435 | 0.9411 | 95-4 | 21 |
| 1NLM | 1F0K | 0.163 | 1.66 | 0.418 | 0.146 | 0.9454 | 15-6 | 16 |
| 1F4X | 1F4W | 0.488 | 0.25 | 0.499 | 0.500 | 0.9981 | 100-6 | 13 |
| 1JBW | 1FGS | 0.430 | 1.48 | 0.451 | 0.292 | 0.9286 | 100-3 | 13 |
| 1FR8 | 1FGX | 0.498 | 0.54 | 0.490 | 0.493 | 0.9935 | 100 | 15 |
| 1LD8 | 1FT1 | 0.416 | 0.92 | 0.480 | 0.048 | 0.9852 | 96-6 | 17 |
| 1HVC | 1G6L | 0.345 | 0.46 | 0.12 | 0.428 | -- | 97-96 | 13 |
| 1LSP | 1GBS | 0.360 | 0.26 | 0.499 | 0.483 | 0.9973 | 100 | 10 |
| 1LC3 | 1GCU | 0.458 | 0.77 | 0.4 | 0.405 | 0.9774 | 13-7 | 12 |
| 1GJW | 1GJU | 0.499 | 0.29 | 0.499 | 0.500 | 0.9989 | 100-7 | 16 |
| 1N75 | 1GLN | 0.422 | 1.47 | 0.49 | 0.095 | 0.9665 | 99 | 14 |
| 1GOY | 1GOU | 0.476 | 0.47 | 0.464 | 0.160 | 0.9727 | 100 | 11 |
| 1H46 | 1GPI | 0.193 | 0.15 | 0.499 | 0.253 | 0.9996 | 100 | 13 |
| 1GUZ | 1GV1 | 0.383 | 0.62 | 0.483 | 0.327 | 0.9908 | 13-7 | 19 |
| 1YDD | 1HEA | 0.491 | 0.18 | 0.499 | 0.500 | 0.9991 | 100 | 11 |
| 1YDA | 1HEB | 0.498 | 0.20 | 0.499 | 0.500 | 0.9989 | 100 | 11 |
| 1KIC | 1HOZ | 0.420 | 0.35 | 0.486 | 0.246 | 0.9911 | 100 | 16 |
| 1A80 | 1HW6 | 0.466 | 0.93 | 0.413 | 0.484 | 0.9540 | 100-11 | 11 |
| 1I3A | 1I39 | 0.498 | 0.40 | 0.499 | 0.500 | 0.9946 | 100 | 10 |
| 4AIG | 1IAG | 0.494 | 0.26 | 0.494 | 0.500 | 0.9946 | -- | 10 |
| 1JZS | 1ILE | 0.497 | 0.69 | 0.499 | 0.492 | 0.9952 | 100-7 | 17 |
| 1JQ3 | 1INL | 0.493 | 0.35 | 0.480 | 0.500 | 0.9996 | 100-99 | 20 |
| 1JAY | 1JAX | 0.435 | 0.60 | 0.493 | 0.397 | 0.9884 | 100 | 13 |
| 1UEH | 1JP3 | 0.499 | 0.67 | 0.451 | 0.499 | 0.9800 | 99 | 14 |
| 1JSO | 1JSM | 0.499 | 0.10 | 0 | 0.500 | 0.9998 | 100 | 14 |
| 1JYL | 1JYK | 0.208 | 0.94 | 0.0164 | 0.184 | 0.9724 | 99 | 18 |
| 1JVS | 1K5H | 0.351 | 1.16 | 0.253 | 0.067 | 0.7102 | 95-7 | 19 |
| 1K70 | 1K6W | 0.497 | 1.08 | 0.499 | 0.499 | 0.9838 | 100-8 | 13 |
| 1M6P | 1KEO | 0.136 | 1.05 | 0.450 | 0.126 | 0.8779 | 100 | 12 |
| 3KIV | 1KIV | 0.467 | 0.30 | 0.458 | 0.478 | 0.9908 | 99 | 7 |
| 1KMP | 1KMO | 0.498 | 0.64 | 0.439 | 0.492 | 0.9393 | 97-1 | 16 |
| 2NGR | 1KZ7 | 0.467 | 1.61 | 0.0536 | 0.233 | -- | 97 | 19 |
| 2MIN | 1L5H | 0.084 | 0.55 | 0.103 | 0.234 | 0.9546 | 99-5 | 24 |
| 1LL2 | 1LL3 | 0.496 | 0.37 | 0.480 | 0.500 | 0.7234 | 100 | 11 |
| 1LMC | 1LMN | 0.499 | 0.10 | 0.49 | 0.500 | 0.9994 | 100 | 8 |
| 1EYN | 1NAW | 0.208 | 1.02 | 0.112 | 0.221 | 0.9855 | 100-8 | 17 |
| 1BHT | 1NK1 | 0.033 | 0.58 | 0.465 | 0.044 | 0.9879 | 100 | 13 |
| 1PBO | 1OBP | 0.143 | 0.38 | 0.343 | 0.277 | 0.9062 | 99 | 13 |
| 1OPB | 1OPA | 0.295 | 0.68 | 0.129 | 0.471 | 0.9808 | 100 | 15 |
| 1I75 | 1PAM | 0.499 | 0.13 | 0.499 | 0.499 | 0.9998 | 100 | 20 |
| 1NME | 1PAU | 0.499 | 0.29 | 0.45 | 0.500 | 0.0643 | 100 | 11 |
| 1KEV | 1PED | 0.281 | 0.81 | 0.499 | 0.374 | 0.9864 | 100-10 | 20 |
| 1PIG | 1PIF | 0.495 | 0.32 | 0.499 | 0.499 | 0.9985 | 100-8 | 14 |
| 1PJC | 1PJB | 0.498 | 0.61 | 0.499 | 0.499 | 0.9942 | 100-9 | 12 |
| 1KLT | 1PJP | 0.168 | 0.97 | 0.358 | 0.409 | 0.9555 | 98-3 | 11 |
| 1QHG | 1PJR | 0.499 | 0.23 | 0.473 | 0.500 | 0.9974 | 100-6 | 16 |
| 1CEB | 1PKR | 0.041 | 0.58 | 0.0925 | 0.183 | 0.6458 | 100 | 9 |
| 2PK4 | 1PMK | 0.417 | 0.71 | 0.101 | 0.139 | 0.9566 | 100 | 9 |
| 1BK9 | 1PSJ | 0.494 | 0.24 | 0.321 | 0.500 | 0.9967 | 100 | 9 |
| 1QBB | 1QBA | 0.497 | 0.11 | 0.499 | 0.489 | 0.9999 | 100-5 | 22 |
| 1PYY | 1QME | 0.157 | 0.59 | 0.38 | 0.271 | 0.9775 | 99-5 | 15 |
| 1OSS | 1SGT | 0.367 | 0.27 | 0.488 | 0.408 | 0.9976 | 99-0 | 10 |
| 1SWN | 1SWL | 0.497 | 0.31 | 0.467 | 0.492 | 0.9941 | 100-1 | 14 |
| 1LBT | 1TCA | 0.440 | 0.24 | 0.0796 | 0.356 | 0.9987 | 100 | 15 |
| 1WBL | 1WBF | 0.371 | 0.39 | 0.122 | 0.197 | 0.9955 | 100 | 18 |
| 1YDB | 1YDC | 0.491 | 0.12 | 0.499 | 0.498 | 0.9996 | 100 | 11 |
| 1H0S | 2DHQ | 0.499 | 0.26 | 0.485 | 0.500 | 0.9856 | 10-5 | 9 |
| 1LLO | 2HVM | 0.498 | 0.12 | 0.499 | 0.500 | 0.9996 | 100-5 | 11 |
| 43CA | 43C9 | 0.491 | 0.23 | 0.495 | 0.499 | 0.9965 | 100-22 | 18 |
| 5BIR | 4BIR | 0.487 | 0.61 | 0.0932 | 0.193 | 0.9714 | 100 | 10 |
| 5EUG | 4EUG | 0.498 | 0.21 | 0.499 | 0.500 | 0.9986 | 100 | 11 |
| 5EAU | 5EAS | 0.064 | 0.40 | 0.38 | 0.019 | 0.9976 | 99-3 | 16 |
| 7TAA | 6TAA | 0.499 | 0.24 | 0.499 | 0.500 | 0.9990 | 100-7 | 14 |
*For comparison we have used TM-SCORE server at: http://zhanglab.ccmb.med.umich.edu/TM-SCORE/
** The numbers reported here are the maximum and minimum sequence identity of aligned positions in percent. The data are given from Dali server: http://ekhidna.biocenter.helsinki.fi/dali_lite/start/ .
-- No value reported by the site
A way to see how the mass of atoms and restriction on the space dimension perform the similarity criterion between two proteins is to compute the WSV and SV correlation with RMSD. The correlation between WSV and RMSD for the 48 dataset is 0.45 and for the 86 dataset is 0.55, which are better than the correlation between SV and RMSD for these two datasets, i.e., 0.32 and 0.36, respectively. In Ref. [19], a complete discussion is given to explain why we do not expect to see a high correlation between RMSD and SV (WSV). This is why we defined NRMSD for comparison with WSV. NRMSD is a bounded parameter that removes the inconvenience of semi-bounded RMSD. Also, both the parameters WSV and NRMSD are similarity criteria.
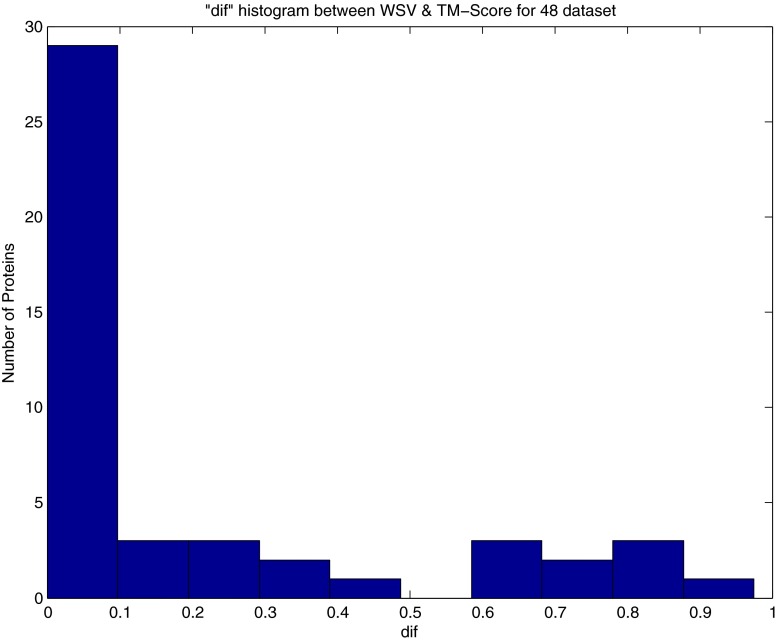
Figures 1 and 2 show the histogram of dif between WSV and NRMSD for the 48 and 86 datasets. We see that 60% of WSV and NRMSD prediction for the 86 dataset have less than 10% differences and 70% of their prediction values have less than 20% differences. The results for the 48 dataset also show a 60% agreement between WSV and NRMSD with less than 10% differences and 67% for 20% difference of prediction values. The disagreement between WSV and NRMSD by a 80% difference of prediction values for the 48 dataset is equal to 4.5% and for the 86 dataset is equal to 5.8% (a summary of these results is given in Table 3). Figures 3, 4, 5, and 6 show the histogram of dif computed between WSV and TM-SCORE and also DALILITE. These figures and also Table 3 show good agreement between WSV and these methods.
Fig. 1.
dif histogram between WSV and NRMSD for 48 dataset
Fig. 2.
dif histogram between WSV and NRMSD for 86 dataset
Table 3.
Differences between WSV and the other methods by using dif
| NRMSD | TM-SCORE | DALILITE | ||||
|---|---|---|---|---|---|---|
| 48 dataset (48)* | 86 dataset (86)* | 48 dataset (47)* | 86 dataset (84)* | 48 dataset (47)* | 86 dataset (85)* | |
| Less than 10% differences | 60.5% | 59.3% | 63.9% | 63.1% | 61.7% | 63.5% |
| Less than 20% differences | 66.7% | 69.7% | 68.1% | 69.1% | 68.1% | 67.1% |
| More than 80% differences | 4.1% | 5.8% | 8.5% | 4.8% | 6.4% | 4.7% |
| More than 90% differences | 0% | 2.3% | 2.1% | 1.2% | 4.3% | 1.1% |
*Number of proteins pairs compared. TM-SCORE and DALILITE have not given values for all protein pair comparisons
Fig. 3.
dif histogram between WSV and TM-SCORE [14, 15] for 48 dataset
Fig. 4.
dif histogram between WSV and TM-SCORE [14, 15] for 86 dataset
Fig. 5.
dif histogram between WSV and DALILITE [18] for 48 dataset
Fig. 6.
dif histogram between WSV and DALILITE [18] for 86 dataset
Table 4 shows the sensitivity, specificity, accuracy, and precision of WSV compared with the other scores (NRMSD, DALILITE, TM-SCORE) as targets. In summary, as Table 4 shows, comparing WSV with NRMSD, the ‘sensitivity’ or the probability that two proteins are determined to be similar by WSV is about 85.7% (80.0%) for the 86(48) dataset and the ‘specificity’ or the probability that the two proteins are determined to be dissimilar by WSV is equal to 62.5% (37.5%). The accuracy of the method (WSV) for the 86(48) dataset is 81.4% (72.9%) and the precision is 90.9% (86.5%), which indicates that both measures show good agreement between WSV and NRMSD predictions. The F-score for the 86(48) dataset shows that the performance to give similarity prediction is about 88.2% (83.1%), which is an expected result because the two datasets for the proteins examined here are closely similar. These results show that WSV could be a good alternative parameter for RMSD (or NRMSD); it does not involve the protein size issue and provides a normalized similarity criterion between any two proteins. The results of the comparison between WSV and TM-SCORE [14, 15] and DALILITE [18], in the same manner as for WSV and NRMSD and reported in Table 4, show that a good agreement exists between WSV and these methods’ predictions and also good precision of WSV.
All of the above results show that WSV appears to be a reliable alternative parameter for RMSD (or NRMSD). WSV is a geometrical criterion while it also includes physical properties. Moreover, it does not suffer from the protein size problem and it provides a similarity criterion between two proteins as well as other criteria.
For computing WSV, we used an i7 laptop with 8 GB RAM. The time required to complete this computation depends on the proteins’ sizes and on average it takes about 3 min (for small proteins it takes about 1 min and for large proteins the computation takes about 6 min). In Tables 1 and 2, we also show the L max used for each pair of proteins.
Conclusions
In this paper, we introduced WSV, which displays two major differences compared to SV. First, we weighted the shape function by atomic masses, which stresses the importance of the individual atoms in the computation. Second, we extended the dimensions of the reciprocal space at least up to the largest compared proteins’ sizes (measured by the number of atoms). This condition ensures that we do not lose any information about the proteins when we map them onto the reciprocal space. As discussed in the Results and discussion section, these two changes in SV improve the correlation between WSV with RMSD relative to SV. We compared WSV with NRMSD, TM-SCORE, and DALILITE by using statistical concepts such as sensitivity, specificity, etc. The results show good accuracy and precision for WSV. Also, we computed a relative difference (dif) between WSV and other methods, which also shows good agreement between WSV predictions and other scores. Our results confirm the reliability and usefulness of our method and show that WSV can be used alternatively with RMSD in helping to find protein similarity in various areas of protein science and in drug discovery.
WSV is now defined as a geometrical structural score. To develop this work in the future, it is suggested to define a score on both the WSV- and domain-based structural methods. Also, we wish to emphasize that WSV is a geometric-based method, sensitive to the protein's atoms positions and their masses. Thus, if one of these parameters changes, WSV will also change. Apparently, for two structurally similar proteins with dissimilar sequences, WSV does not give structural homologues as a result. This hypothesis will be examined in our future research and if it is indeed verified, this could present an advantage of WSV relative to SV or RMSD.
Acknowledgments
I thank Dr. Jack A. Tuszynski (University of Alberta) for his helpful comments.
Footnotes
When the TM-SCORE is less than 0.2 it corresponds to randomly chosen unrelated proteins whereas with a score higher than 0.5 we generally assume the same fold in SCOP/CATH [21]. Here we normalized the TM-SCORE to 0.5 (i.e., we divided it by 2).
References
- 1.Li, B., Turuvekere, S., Agrawal, M., La, D., Ramani, K.,Kihara, D.: Characterization of local geometry of protein surfaces with the visibility criterion. Proteins 71, 670 (2008) [DOI] [PubMed]
- 2.Rupp B, Wang J. Predictive models for protein crystallization. Methods. 2004;34:390. doi: 10.1016/j.ymeth.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 3.Arnold K, Bordoli L, Kopp J, Schwedem T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinf. (Oxford, England) 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 4.Kolodn R, Petrey D, Honig B. Protein structure comparison: implications for the nature of “fold space”, and structure and function prediction. Curr. Opin. Struct. Biol. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Carugo O. Recent progress in measuring structural similarity between proteins. Curr. Protein Peptide Sci. 2007;8:219–241. doi: 10.2174/138920307780831839. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9:101186. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betancourt MR, Skolnick J. Universal similarity measure for comparing protein structures. Biopolymers. 2001;59:305–309. doi: 10.1002/1097-0282(20011015)59:5<305::AID-BIP1027>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Kihara D, Sael L, Chikhi R, Esquivel-Rodriguez J. Molecular surface representation using 3D Zernike descriptors for protein shape comparison and docking. Curr. Protein Pept. Sci. 2011;12(6):520–530. doi: 10.2174/138920311796957612. [DOI] [PubMed] [Google Scholar]
- 9.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 10.Andreeva, A., Howorth, D., Chandonia, J.-M., Brenner, S.E., Hubbard, T.J.P., Chothia, C., Murzin, A.G.: Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 36, D419–425 (2008). [DOI] [PMC free article] [PubMed]
- 11.Maiorov VN, Crippen GM. Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. J. Mol. Biol. 1994;235(2):625–634. doi: 10.1006/jmbi.1994.1017. [DOI] [PubMed] [Google Scholar]
- 12.Carugo O, Pongor S. A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Sci.: Publ. Protein Soc. 2001;10(7):1470–1473. doi: 10.1110/ps.690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orengo, C.A., Michie, A.D., Jones, S., Jones, D.T., Swindells, M.B., Thornton, J.M.: CATH—a hierarchic classification of protein domain structures. Structure (London, England: 1993) 5, 1093–1108 (1997) [DOI] [PubMed]
- 14.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Zhang Y. How significant is a protein structure similarity with TM-SCORE = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levitt, M., Gerstein, M.: STRUCTAL. A structural alignment program. Stanford University (2005). Available from: http://csb.stanford.edu/levitt/Structal.
- 17.Holm L, Ouzounis C, Sander C, Tuparev G, Vriend G. A database of protein structure families with common folding motifs. Protein Sci. 1992;1:1691–1698. doi: 10.1002/pro.5560011217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Saberi Fathi SM, White DT, Tuszynski JA. Geometrical comparison of two protein structures using Wigner-D functions. Proteins. 2014;82:2756–2769. doi: 10.1002/prot.24640. [DOI] [PubMed] [Google Scholar]
- 20.Daniel KP, et al. Reconstruction of SAXS Profiles from Protein Structures. Comput. Struct. Biotechnol. J. 2013;8(11) doi: 10.5936/csbj.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-SCORE. Nucleic Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wigner EP. Gruppentheorie und ihre Anwendungen auf die Quantenmechanik der Atomspektren. Braunschweig: Vieweg Verlag; 1931. [Google Scholar]
- 23.Potts D, Prestin J, Vollrath A. A fast algorithm for nonequispaced Fourier transforms on the rotation group. Numer. Algorithms. 2009;52:355–384. doi: 10.1007/s11075-009-9277-0. [DOI] [Google Scholar]
- 24.Hielscher R, Potts D, Prestin J, Schaeben H, Schmalz M. The Radon transform on SO(3): a Fourier slice theorem and numerical inversion. Inverse Prob. 2008;24:025011. doi: 10.1088/0266-5611/24/2/025011. [DOI] [Google Scholar]
- 25.Lipson, H., Taylor, C.A.: Fourier Transforms and X-ray Diffraction. Bell, London (1958)
- 26.McKie, D., McKie, C., Essentials of Crystallography, Blackwell Scientific Publications, (1992). ISBN 0-632-01574-8.
- 27.Sokolova, M., Japkowicz, N., Szpakowicz, S.: Beyond Accuracy, F-score and ROC: a Family of Discriminant Measures for Performance Evaluation, American Association for Artificial Intelligence, (2006). https://www.aaai.org/Papers/Workshops/2006/WS-06-06/WS06-06-006.pdf.