Abstract
In recent years, genome-wide association studies have identified 58 independent risk loci for coronary artery disease (CAD) on the autosome. However, due to the sex-specific data structure of the X chromosome, it has been excluded from most of these analyses. While females have 2 copies of chromosome X, males have only one. Also, one of the female X chromosomes may be inactivated. Therefore, special test statistics and quality control procedures are required. Thus, little is known about the role of X-chromosomal variants in CAD. To fill this gap, we conducted a comprehensive X-chromosome-wide meta-analysis including more than 43,000 CAD cases and 58,000 controls from 35 international study cohorts. For quality control, sex-specific filters were used to adequately take the special structure of X-chromosomal data into account. For single study analyses, several logistic regression models were calculated allowing for inactivation of one female X-chromosome, adjusting for sex and investigating interactions between sex and genetic variants. Then, meta-analyses including all 35 studies were conducted using random effects models. None of the investigated models revealed genome-wide significant associations for any variant. Although we analyzed the largest-to-date sample, currently available methods were not able to detect any associations of X-chromosomal variants with CAD.
In the last years, genome-wide association studies (GWAS) have uncovered numerous chromosomal risk loci for various complex diseases. Specifically, for coronary artery disease (CAD), 58 independent risk loci have been identified and verified in independent replication datasets1. However, a large part of the estimated heritability of CAD is not yet explained. This could be due partly to the fact that the X chromosome has routinely been excluded from GWAS. One reason for this is that the data has a different, sex-specific structure and, therefore, requires special analytical tools including special quality control and test statistics2. Thus, despite the profound effects of gender on the manifestation of CAD, no systematic association analyses of X-chromosomal variants with CAD have been reported so far. Therefore, an analysis of the X-chromosome from GWAS data might help to narrow the gap of missing heritability and help to yield new insights into the genetics of CAD.
X-chromosomal variants might be expected to play a role in the pathophysiology, since sex-specific features are known for CAD. Specifically, the risk to develop CAD varies between males and females independent from other risk factors. The symptoms of myocardial infarction (MI) as well as the prognosis after MI differ between males and females. Males are more likely than females to manifest CAD at young age, but females are more likely than males to die of a first MI. Furthermore, heart disease is the most common cause of death for females3. Thus, the analysis of X-chromosomal variants could help to explain the sex differences in CAD.
To comprehensively investigate the association of variants on chromosome X and CAD, we collected data from 35 world-wide study cohorts. All participating studies were part of the CARDIoGRAM + C4D consortium1. At each study site, quality control on subject level was performed, data were imputed on the basis of the 1000 genomes reference panel, and X chromosome-adapted association tests were calculated. After this, data were analyzed centrally at the University of Lübeck, where further quality control and the meta-analysis of all 35 studies were conducted. In the following, we will present the results of the association analysis of about 200,000 X-chromosomal single nucleotide polymorphisms (SNPs) with CAD on a sample of more than 100,000 subjects including more than 43,000 cases and 58,000 controls.
Results
Details on the investigated studies are summarized in Table 1. For each of the 35 studies, logistic regression models with additive scoring for the SNP were used. To account for the sex-specific structure of X-chromosomal data, sex was always included as a covariate. In addition, interactions between SNP and sex were investigated. Where appropriate, further covariates could be included. Since one of the two female X-chromosomes may or may not be inactivated at a specific locus, models were calculated that assumed inactivation as well as not assuming inactivation.
Table 1. Cohort descriptives of the 35 studies participating in the 1000G coronary artery disease meta-analysis of the X-chromosome.
| Study | Ancestry | Cases (% females) | Controls (% females) | % MI* (in cases) |
|---|---|---|---|---|
| ADVANCE | White European | 275 (59.9) | 311 (60.7) | 100.0 |
| BioMe_AfrAm | African American | 362 (66.5) | 2778 (70.9) | 36.0 |
| BioMe_EurAm | European American | 487 (35.0) | 1382 (61.6) | 30.4 |
| BioMe_HisAm | Hispanic American | 758 (55.1) | 3338 (71.3) | 36.7 |
| Cardiogenics | White European | 391 (13.7) | 410 (60.8) | 12.5 |
| CATHGEN | White European | 1191 (31.4) | 646 (58.9) | 48.1 |
| CCGB_2 | White European | 1547 (21.8) | 344 (45.0) | 60.3 |
| EGCUT | White European | 658 (49.6) | 5841 (56.1) | 19.6 |
| FGENTCARD | Libanese | 1802 (25.0) | 466 (50.8) | 16.0 |
| FINCAVAS | Finnish European | 774 (21.2) | 647 (44.8) | 12.4 |
| FINRISK/PredictCVD | Finnish European | 677 (30.9) | 1200 (42.7) | 40.0 |
| GerMIFSI | White European | 637 (33.9) | 1644 (51.2) | 100.0 |
| GerMIFSII | White European | 1222 (21.0) | 1298 (48.5) | 100.0 |
| GerMIFSIII | White European | 1096 (20.5) | 1509 (52.2) | 100.0 |
| GerMIFSIV | White European | 1002 (36.0) | 1147 (61.9) | 100.0 |
| GerMIFSV | White European | 2459 (24.5) | 1611 (53.0) | 100.0 |
| HPS | White European | 2700 (23.9) | 2748 (72.5) | 65.0 |
| HSDS | White European | 206 (0.0) | 258 (0.0) | 45.6 |
| ITH | White European | 402 (29.4) | 449 (32.9) | 100.0 |
| LIFE-Heart | White European | 1531 (24.8) | 768 (50.3) | 44.0 |
| LOLIPOP | Indian Asian | 2791 (18.5) | 3757 (14.1) | 43.9 |
| LURIC | White European | 2347 (25.5) | 621 (48.0) | 62.9 |
| MEDSTAR | White European | 875 (34.4) | 447 (55.9) | 100.0 |
| MIGEN | White European | 2905 (24.8) | 2998 (26.9) | 100.0 |
| OHGS_A2 | White European | 921 (24.9) | 1001 (50.5) | 64.3 |
| OHGS_B2 | White European | 1183 (22.4) | 1391 (49.5) | 55.6 |
| OHGS_C2 | White European | 833 (6.4) | 317 (66.7) | 44.3 |
| PENNCATH | White European | 933 (29.2) | 468 (58.8) | 100.0 |
| PIVUS | White European | 119 (24.3) | 830 (54.7) | 77.3 |
| PROCARDIS | White European | 5719 (29.2) | 6526 (62.5) | 80.0 |
| SDS | Indian Asian | 176 (29.5) | 1421 (49.0) | 100.0 |
| THISEAS | White European | 423 (21.3) | 593 (56.3) | 60.1 |
| TWINGENE | White European | 814 (29.1) | 5999 (55.8) | 70.5 |
| ULSAM | White European | 322 (0.0) | 857 (0.0) | 84.2 |
| WTCCC | White European | 1922 (20.9) | 2930 (51.2) | 71.5 |
| Total | — | 43120 (28.2) | 58291 (50.0) | 66.7 |
*MI = myocardial infarction.
The study-wise numbers of SNPs excluded due to quality control are given in Supplementary Tables S1 and S2. The inspection of inflation factors4 and Q-Q-plots (see also Supplementary Fig. S1) did not reveal any systematic inflation of specific studies. Thus, all studies were included in the meta-analysis.
None of the statistical models used for the meta-analysis revealed a genome-wide significant association with CAD for any SNP. Association results for the model without inactivation assumption and without SNP*sex interaction are presented in Fig. 1. Results of the other models are comparable and presented in Supplementary Figs S2–S6.
Figure 1. Chromosome-wide association results.
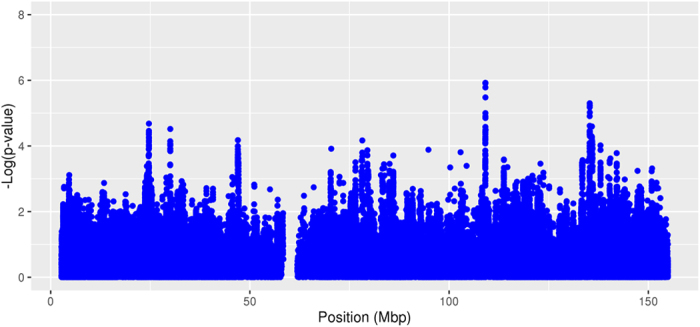
The statistical model assumes no inactivation and no SNP*sex interaction. Shown are logarithmized random effects p-values of all 184,673 quality controlled SNPs in order of physical position in mega base pairs (mbp).
Subgroup and sensitivity analyses
To investigate possible sex-specific effects, we conducted subgroup analyses of males and females separately. Association plots of these models are presented in Supplementary Figs S7 and S8. Again, no genome-wide significant associations were to be observed. To exclude a possible bias introduced by including study cohorts of non-European ancestry, we performed a subgroup analysis including only the 31 studies with European background. Excluding non-European studies did not show additional associations either (Supplementary Figs S9–S14). Finally, to eliminate possible influences of the quality control parameters, we varied our criteria on missing frequencies and imputation quality. Performing stricter quality control reduced the number of SNPs available for meta-analysis to about 90,000 (depending on model, between 90,658 and 96,502). Results of these analyses were comparable to the results from the primary analyses. Thus, none of the sensitivity analyses did reveal novel associations, supporting the null findings of the main analyses.
Power
We estimated the power of our study overall as well as in the smallest subgroup analyzed, i.e. the subgroup of females, as functions of the odds ratio and the effect allele frequency (EAF) (Fig. 2). In the entire sample of males and females, odds ratios of 1.1 or 1.11 would have reliably been detected with an EAF of 0.1 or higher. Even in the female subgroup, odds ratios of 1.15 and higher would have been detectable with a sufficiently large probability.
Figure 2. Estimated power.
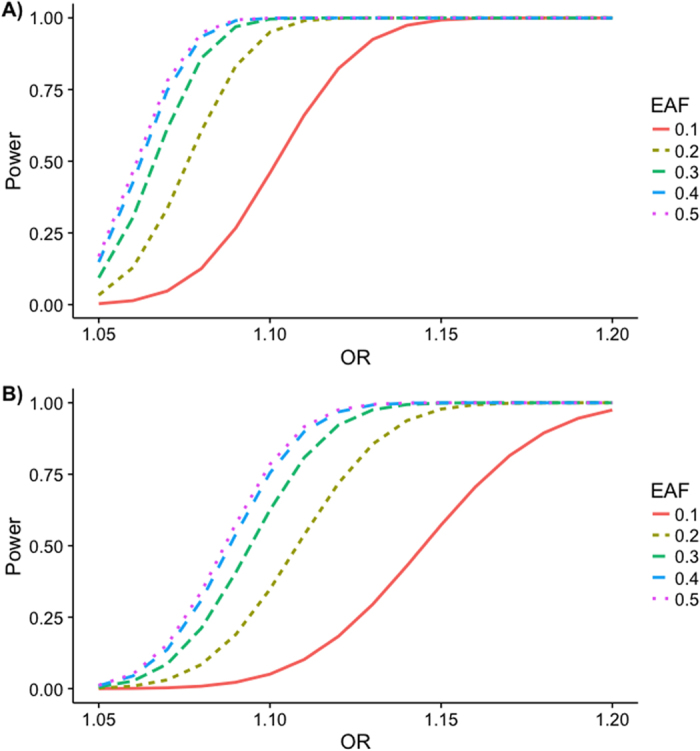
The power to detect an effect was estimated in dependence of the odds ratio (OR) and the effect allele frequency (EAF) using software Quanto (version 1.2.4 from May 2009). Parameters used for simulation: Binary (disease) phenotype, significance level α = 5·10−8, disease prevalence kP = 0.1, log-additive genetic model, no gene-environment interaction. (A) Effective Ncases = 27,640, 1.5817 effective controls per effective case (corresponding to 43,718 effective controls), (B) Female Ncases = 12,160, 2.3968 female controls per female case (corresponding to 29,145 female controls).
Discussion
Although the presented meta-analyses included more than 100,000 subjects (the largest to date), with 43,120 CAD cases (28.2% women) and 58,291 controls (50% women), no genome-wide significant associations could be detected. This negative finding was independent of the model chosen for analysis. There were no sex-specific associations for CAD. Stricter quality control or excluding non-European studies did not reveal any different findings.
The NHGRI GWAS catalog5,6 reports 52 genome-wide significant associations of variants on chromosome X with more than 600 traits. All of the reported studies are of much smaller sample size (less than 50,000 samples) and used fewer methods than our analyses. Yet, they successfully discovered associations. However, no genome-wide significant associations with CAD or any other correlated phenotype have been reported on the X chromosome. The only gene reported for CAD on chromosome X is CHRDL1 with a p-value of 9 · 10−7 for rs59430577, but this did not replicate in our analyses (p = 0.0172 for the model without inactivation assumption and without SNP*sex interaction). As our power estimates indicated (Fig. 2), in such a large dataset, the statistical power to detect medium to large effects is high, and only small effects are likely to have been missed. Therefore, the most natural explanation to the negative finding of this meta-analysis is that there are no substantial associations of X-chromosomal variants with CAD.
However, since the progression and the symptoms of CAD, as well as the prognosis after MI, are sex-specific, it may be that the genetics of chromosome X are more complex than previously assumed. For example, the inactivation patterns are not yet understood completely, and it has been shown that inactivation of the female X-chromosome can be cell-specific. The silenced X-chromosome is not necessarily chosen randomly, and silenced regions can differ between females8,9. Although we evaluated models with and without the assumption of inactivation, different inactivation patterns between females at one locus were not taken into account. Nor was non-random inactivation incorporated into the model. This could affect the power of the test statistics that were used and might result in an analysis that is less powerful than estimated. Further, it might be argued that the use of other statistical methods could have yielded significant findings. Specifically, most other studies (e.g. refs 10 and 11) are based on a separate analysis of males and females with a subsequent summary by a classical or sex-specific12 meta-analytic method. In contrast, we followed the approach to directly compute joint test statistics for males and females, taking the different structure of chromosome X in the sexes into account. However, examples for both methodological approaches have been compared in simulations13,14 with the overall result that the joint tests have greater power unless there are relevant differences in the effect sizes between males and females, which is not to be expected given our sex-specific subgroup analyses. Another potential limitation could be the lower coverage of chromosome X than autosomal chromosomes2,15. Specifically, depending on the specific genotyping chip, the distance between two known variations16 averages roughly 700 to 10,000 bp on all chromosomes but about 1400 to 22,500 bp on chromosome X. Accordingly, the median distance in our studies is 11,300 bp, so that the coverage is less than optimal but comparable to the use of an older genotyping array in general. More generally, the use of these technologies is restricted to finding associations with SNPs only; the effect of other structural variants or even having XX instead of XY cannot thus be detected. Another explanation for the lack of significant associations could be problems with the imputation. Using the IMPUTE2 algorithm, as most of the study sites did, a mixture of two populations, males and females, is imputed together. Perhaps this leads to a bias that has not been taken adequately into account. From a clinical perspective, coronary disease and its manifestations differ in women, including a larger proportion of younger women with myocardial infarction having the distinct pathophysiology of coronary dissection, which could complicate the dissection of genetic influence according to sex.
Although we have analyzed the largest sample to date, we were not able to detect genome-wide significant associations between chromosome X variants and CAD with currently available methods. Due to this lack of significant associations, the sex-specific differences in CAD are still unexplained. The genetics of chromosome X may be more complex than has been assumed, so that more sophisticated test statistics which allow for these complex biological processes would be required to detect associations of variants on the X-chromosome with CAD.
Materials and Methods
Sex-specific structure of chromosome X
One reason why chromosome X is usually excluded from GWAS is that the data has a different, sex-specific structure and, therefore, requires special analytical tools15,17. While there are two copies of each autosomal chromosome, males carry only one copy of the X chromosome whereas females, again, carry two copies. Therefore, at each SNP, females can carry one of three possible genotypes; that is, they can have 0, 1 or 2 copies of a specific allele. In contrast to this, there are only two possible genotypes for males, corresponding to 0 or 1 copies of a specific allele. Only for the so-called pseudo-autosomal regions, there exist homologous loci on the Y chromosome, and males can have up to 2 copies of a specific allele. In addition, one of the two female X chromosomes might be inactivated. In each cell, one of the two female X chromosomes is randomly selected to be silenced18. This means that the expression levels of this chromosome are much lower than for the second chromosome in the cell. This mechanism of dose compensation should result in comparable expression levels for males and females despite the different number of chromosome copies. However, this inactivation is incomplete: while some genes or regions will be completely inactivated, some genes might show expression levels that are reduced only slightly or not at all8. Therefore, to analyze X-chromosomal data, special quality control and test statistics are required2. Most of the quality control needs to be done separately for males and females, and test statistics for chromosome X should take into account the different data structure for males and females, for example by including sex as a covariate into the model. The choice of the best statistical test depends on the underlying genetic model and the inactivation patterns at a specific locus13.
Study cohorts
The meta-analysis includes data from 43,120 cases with CAD and 58,291 controls from 35 studies with 28.2% female cases and 50.0% female controls (for details, see Table 1). A subject was regarded as a CAD case if he/she had an inclusive CAD diagnosis, e.g. MI, acute coronary syndrome, chronic stable angina, or coronary stenosis >50%. More detailed information on the study cohorts can be found in the meta-analysis of autosomal variants of the CARDIoGRAMplusC4D Consortium1 which included most of the samples presented here. Thirty-one of the 35 studies consist of subjects with European ancestry, two study cohorts are of Asian ancestry, one of Hispanic and one of African ancestry.
Genotyping and imputation to 1000G data
Details on the genotyping arrays used for each study cohort have been published before1. At each study site, untyped SNPs were imputed on the basis of the 1000 genomes phase 1 version 3 reference panel19. Although this reference panel includes insertion and deletion variants (indels), these were excluded from further analyses. Prior to any quality control, there were 1,193,934 SNPs available for the non-pseudo-autosomal region of chromosome X.
Quality control
Quality control at subject level and at variant level was performed at each study site prior to imputation and association analysis as described previously1. In addition to quality criteria typically used for analyses of autosomal SNPs20,21, all subjects for whom the genotypic and reported sex could not be assigned unambiguously were excluded. Post-imputation quality control at SNP level was done centrally and in the same manner for all contributing studies. Here, SNPs were excluded if one of the following criteria was fulfilled: (1) ≥25% missing genotypes in either female or male cases or controls, (2) deviation from Hardy-Weinberg equilibrium in female controls with p < 0.0001, (3) minor allele frequency <1% in either males or females, and (4) imputation quality score (INFO for IMPUTE222,23,24,25 and r2 for Minimac22,26) <0.5. For sensitivity analysis, we additionally applied stricter criteria for imputation quality (INFO >0.7) and missing genotypes (<2% in either female or male cases or controls).
Study-wise association analysis
Study-wise association analyses were calculated at each study site. Logistic regression models with additive scoring for the SNP were used. The sex-specific structure of X-chromosomal data implies different variances in male and female sub-samples. To account for this, sex needs to be included as a covariate in the model. In addition, interactions between SNP and sex were investigated. Where appropriate, additional covariates to adjust for population stratification have been included in the model, for example in the form of variables calculated from a principal component analysis or variables describing the ethnic background of the subjects.
Since one of the two female X-chromosomes may or may not be inactivated at a specific locus, models were calculated that assumed inactivation as well as not assuming inactivation. If inactivation is present at a locus, two risk alleles of a female subject should show similar expression levels as compared to one risk allele in a male individual. Therefore, while the female genotypes for such a SNP are coded 0, 1 or 2, according to 0, 1 or 2 alleles, the genotypes for males should be coded 0 or 2 according to 0 or 1 alleles. If no inactivation occurs, the expression levels of one allele in females should be the same as one allele in males. Therefore, while the coding of female genotypes is unchanged, male genotypes should now be coded 0 or 1, according to 0 or 1 alleles. As an alternative to assuming complete or no inactivation approach, Wang et al.27 proposed likelihood ratio tests for the situation of non-random or skewed inactivation, which can be more powerful in the case of non-random inactivation. However, given that the gain in power is small and that these tests are available in Matlab28 only, which was not available for many of the participating study sites, we refrained from using this particular approach. These considerations resulted in four models being investigated (Table 2).
Table 2. Association models for chromosome X.
| Model | Mathematical descriptiona | SNP*sex interaction | Coding of SNP | Inactivation |
|---|---|---|---|---|
| I | Logit(CAD) = β0 + β1 · SNP + β2 · sex | No | Females: 0, 1, or 2; Males: 0 or 1 | No |
| II | Logit(CAD) = β0 + β1 · SNP + β2 · sex | No | Females: 0, 1, or 2; Males: 0 or 2 | Yes |
| III | Logit(CAD) = β0 + β1 · SNP + β2 · sex + β3 · SNP*sex | Yes | Females: 0, 1, or 2; Males: 0 or 1 | No |
| IV | Logit(CAD) = β0 + β1 · SNP + β2 · sex + β3 · SNP*sex | Yes | Females: 0, 1, or 2; Males: 0 or 2 | Yes |
aFor each study, further covariates to adjust for population stratification have been added to the model where appropriate.
Post-hoc quality control
After calculation of the association analyses for each single study, post-hoc quality control was applied for all studies. To control for population stratification or other sources of inflation of p-values, inflation factors according to Devlin and Roeder4 were calculated, and plots of expected versus observed test statistics were inspected visually. In addition, for each SNP, mean EAFs over all studies were calculated and compared to the study-wise EAFs. SNPs with extreme deviations (>0.1, corresponding to more than 4 standard deviations) from the mean EAF were excluded from further analyses. Finally, only SNPs for which at least half of the studies were available were included in the meta-analyses.
Meta-analyses
For the meta-analyses of the 35 studies, random effect models were calculated for each of the four models defined in Table 2. In the same way, meta-analyses of the effect estimates for the SNP*sex interaction were performed. In all of these analyses, outlier analyses according to Preuß et al.20 were performed to exclude studies with extremely inflated effect estimates.
Sensitivity and subgroup analyses
We conducted the following sensitivity and subgroup analyses: (1) Subgroup analyses of males and females separately with subsequent meta-analyses to gain further insight into sex-specific effects; (2) Subgroup meta-analysis including only the 31 studies with European background; (3) Meta-analysis of SNPs fulfilling stricter quality criteria as described above.
Power estimation
Using the software Quanto, version 1.2.429, we estimated the power of our analyses in two ways. Firstly, to take into account our entire sample of males and females, a simple combination of the data is not possible due to the sex-specific structure of X chromosomal variants. We therefore followed Clayton30 in assuming that the variance in males has twice the size of that in females in the additive model assuming inactivation. Therefore, we assumed that the effective sample size in males is halved, and this was added to the female sample size. Power was then estimated as a function of the odds ratio and the EAF at a significance level of α = 5·10−8, a disease prevalence kP = 0.1, and a log-additive genetic model. Secondly, we estimated the power for the smallest subgroup analyzed, i.e. the subgroup of females. The parameters in Quanto were set to the same values as before.
Additional Information
How to cite this article: Loley, C. et al. No Association of Coronary Artery Disease with X-Chromosomal Variants in Comprehensive International Meta-Analysis. Sci. Rep. 6, 35278; doi: 10.1038/srep35278 (2016).
Supplementary Material
Acknowledgments
This work was supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant 01ZX1313A-2014). The ADVANCE study was supported by a grant from the Reynold's Foundation and NHLBI grant HL087647. Sample collection in the Cardiogenics Consortium (http://www.cardiogenics.eu/web/) was funded by the 6th Framework Program of the European Union (LSHM-CT-2006-037593). We thank all the participants and clinicians involved in the recruitment process at Cambridge and Leicester (UK), Luebeck and Regensburg (Germany), and Paris (France). CATHGEN was supported by NIH grants HL095987 and HL101621. The Cleveland Clinic Gene Bank study was funded by P01HL076491 (to S.L.H). EGCUT was supported by Estonian Research Council grant no. IUT20-60 and Research Roadmap grant no. 3.2.0304.11-0312 and by University Tartu grant no. ARENG SP1GV. The FGENTCARD-Functional Genomic diagnostic tools for coronary artery disease project was funded by an EU FP6 award. We thank the patients for agreeing to participate in the study. We thank Sonia Youhanna, Nour Moukalled and Bariaa Khalil for their help with subject recruitment and data collection. The work of FINCAVAS was supported by the Competitive Research Funding of the Tampere University Hospital (Grant 9M048 and 9N035), the Finnish Cultural Foundation, the Finnish Foundation for Cardiovascular Research, the Emil Aaltonen Foundation, Finland, and the Tampere Tuberculosis Foundation. The authors thank the staff of the Department of Clinical Physiology for collecting the exercise test data. The GerMIFS studies were supported by grants from the German Federal Ministry of Education and Research (BMBF) within the framework of NGFN and NGFN-plus (Atherogenomics) and e:Med research and funding concept (e:AtheroSysMed, grant 01ZX1313A-2014), the Fondation Leducq (CADgenomics: Understanding CAD Genes, 12CVD02), and the European Union Sixth Framework Programme FP6 (under grant agreement FP6-LIFESCIHEALTH (Cardiogenics)) and the Seventh Framework Programme FP7/2007-2013 under grant agreement n° HEALTH-F2-2013-601456 (CVgenes-at-target). The Heart Protection Study (HPS) (ISRCTN48489393) was supported by the UK Medical Research Council (MRC), British Heart Foundation, Merck and Co (manufacturers of simvastatin), and Roche Vitamins Ltd (manufacturers of vitamins). Genotyping was supported by a grant to Oxford University and CNG from Merck and Co. Jemma C. Hopewell acknowledges support from the British Heart Foundation (FS/14/55/30806). HPS acknowledges the National Blood Service (NBS) donors and UK Twin study for using as population controls. A full list of the investigators who contributed to the generation of the NBS data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 07611. The UK Twin study was funded by the Wellcome Trust; European Community‟s Seventh Framework Programme (FP7/2007–2013). The Helsinki Sudden Death Study (HSDS) was financially supported by EU’s 7th Framework Programme (grant no. 201668 for AtheroRemo), the Tampere University Foundation, the Tampere University Hospital Medical Funds (grants X51001, 9M048 and 9N035 for Terho Lehtimäki, the Emil Aaltonen Foundation (Terho Lehtimäki, the Finnish Foundation of Cardiovascular Research (Terho Lehtimäki, Pekka J. Karhunen), the Pirkanmaa Regional Fund of the Finnish Cultural Foundation, the Yrjö Jahnsson Foundation, and the Tampere Tuberculosis Foundation (Terho Lehtimäki). LIFE-Heart is a part of the LIFE – Leipzig Research Center for Civilization Diseases, Universität Leipzig. LIFE is funded by means of the European Union, by the European Regional Development Fund (ERDF) and by means of the Free State of Saxony within the framework of the excellence initiative. The LOLIPOP study is supported by the National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centre Imperial College Healthcare NHS Trust, the British Heart Foundation (SP/04/002), the Medical Research Council (G0601966, G0700931), the Wellcome Trust (084723/Z/08/Z), the NIHR (RP-PG-0407-10371), European Union FP7 (EpiMigrant, 279143) and Action on Hearing (G51). We thank the participants and research staff who made the study possible. LURIC was supported by the 7th Framework Program (integrated project AtheroRemo, grant agreement number 201668 and RiskyCAD, grant agreement number 305739) of the European Union and by the INTERREG IV Oberrhein Program (Project A28, Genetic mechanisms of cardiovascular diseases) with support from the European Regional Development Fund (ERDF) and the Wissenschaftsoffensive TMO. We extend our appreciation to the participants of the LURIC study and thank the LURIC study team who were either temporarily or permanently involved in patient recruitment as well as sample and data handling, in addition to the laboratory staff at the Ludwigshafen General Hospital and the Universities of Freiburg and Ulm, Germany. The MIGen study was funded by R01HL087676 from the US National Heart, Lung, and Blood Institute. The Mount Sinai IPM Biobank Program is supported by The Andrea and Charles Bronfman Philanthropies. It was in part supported by NHGRI U01HG007417. OHGS_A2, OHGS_B2, and OHGS_C2 were funded by Canadian Institutes of Health Research (# MOP-2380941 to R.M.), (#MOP82810, MOP77682 to R.R., A.F.S. & R.M.); Canada Foundation for Innovation (#11966 to R.R., R.M. & A.F.S.; Heart & Stroke Foundation of Canada (#NA6001, #NA6650 to R.M). PIVUS was supported by Knut and Alice Wallenberg Foundation (Wallenberg Academy Fellow), European Research Council (ERC Starting Grant), Swedish Diabetes Foundation (grant no. 2013-024), Swedish Research Council (grant no. 2012-1397), and Swedish Heart-Lung Foundation (20120197). We thank the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se) for excellent genotyping. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project b2011036. PROCARDIS was supported by the European Community Sixth Framework Program (LSHM-CT- 2007-037273), AstraZeneca, the British Heart Foundation, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the Swedish Heart-Lung Foundation, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular Program of Karolinska Institutet and Stockholm County Council, the Foundation for Strategic Research and the Stockholm County Council (560283). Research in SDS was partly supported by NIH grants -R01DK082766 funded by the National Institute of Diabetes and Digestive and Kidney Diseases and NOT-HG-11-009 funded by National Genome Research Institute, and VPR Bridge grant from University of Oklahoma Health Sciences Center, Oklahoma City, USA. Recruitment for THISEAS was partially funded by a research grant (PENED 2003) from the Greek General Secretary of Research and Technology; we thank all the dieticians and clinicians for their contribution to the project. TwinGene was supported by grants from the Ministry for Higher Education, the Swedish Research Council (M-2005-1112 and 2009-2298), GenomEUtwin (EU/QLRT-2001-01254; QLG2-CT-2002-01254), NIH grant DK U01-066134, Knut and Alice Wallenberg Foundation (Wallenberg Academy Fellow), European Research Council (ERC Starting Grant), Swedish Diabetes Foundation (grant no. 2013-024), Swedish Research Council (grant no. 2012-1397), and Swedish Heart-Lung Foundation (20120197). We thank the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se) for excellent genotyping. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project b2011036. ULSAM was supported by Knut and Alice Wallenberg Foundation (Wallenberg Academy Fellow), European Research Council (ERC Starting Grant), Swedish Diabetes Foundation (grant no. 2013-024), Swedish Research Council (grant no. 2012-1397), and Swedish Heart-Lung Foundation (20120197). We thank the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se) for excellent genotyping. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project b2011036. Recruitment for the WTCCC study was funded by the British Heart Foundation and genotyping by the Wellcome Trust. Themistocles L. Assimes was supported by an NIDDK career development award DK088942. Panos Deloukas’s work forms part of the research themes contributing to the translational research portfolio of Barts Cardiovascular Biomedical Research Unit which is supported and funded by the National Institute for Health Research. Analysis was partly supported by BHF grant (to Panos Deloukas) RG/14/5/30893. Martin Farrall and Hugh Watkins acknowledge the support of the Wellcome Trust core award (090532/Z/09/Z) and Martin Farrall, Hugh Watkins and Theodosios Kyriakou, the BHF Centre of Research Excellence. Anuj Goel, Hugh Watkins and Theodosios Kyriakou acknowledge European Union Seventh Framework Programme FP7/2007-2013 under grant agreement no. HEALTH-F2-2013-601456 (CVGenes@Target) & and Anuj Goel, the Wellcome Trust Institutional strategic support fund. The UK Twin study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013). PoBI samples from the Wellcome Trust funded People of the British Isles project. Sekar Kathiresan is supported by the Donovan Family Foundation, Fondation Leducq, MGH Research Scholar Award, and R01 HL107816. Andrew P. Morris is a Wellcome Trust Senior Fellow in Basic Biomedical Science, funded under grant WT098017. Christopher P. Nelson and Nilesh J. Samani are funded by the British Heart Foundation and Nilesh J. Samani is a UK NIUHR Senior Investigator. Christopher P. Nelson and Nilesh J. Samani are funded by the British Heart Foundation and Nilesh J. Samani is a UK NIUHR Senior Investigator. Samuli Ripatti was supported by the Academy of Finland Center of Excellence in Complex Disease Genetics (Grant No. 213506 and 129680), Academy of Finland (Grant No. 251217 and 285380), the Finnish foundation for Cardiovascular Research, the Sigrid Juselius Foundation and the European Community’s Seventh Framework Programme (FP7/2007-2013) through the BioSHaRE-EU (Biobank Standardisation and Harmonisation for Research Excellence in the European Union) project, grant agreement 261433. Alexandre F. R. Stewart is supported by operating grants from the Canadian Institute of Health Research and Natural Sciences and Engineering Research Council of Canada. Hong-Hee Won is supported by a postdoctoral award from the American Heart Association (15POST23280019).
Footnotes
Author Contributions C.L., I.R.K., J.E. and H.S. conceived the idea and the design of the study. I.R.K. and J.E. recruited the participating studies for the meta-analysis. C.L., M.A., T.L.A., A.B., A.G., S.G., J.H., J.C.H., S.K., M.E.K., K.W.L., Y.L., L.P.L., C.P.N., M.N., L.Q., E.S., M.S., T.T., C.W., H.H.W., L.Z., W.Z., S.S.A., F.B., E.P.B., R.C., G.D., R.D., T.E., M.E., M.F., D.G., V.G., C.B.G., A.S.H., A.H., S.L.H., J.H., M.K., T.K., P.R.L., L.L., C.L., P.K.E.M., E.M., E.M., A.P.M., K.N., N.P., L.R., V.S., S.H.S., A.F.R.S., J.R.T., P.A.Z., J.C.C., R.C., E.I., C.I., P.J.K., J.S.K., T.L., R.J.F.L., W.M., R.M., A.M., M.P.R., S.R., D.K.S., J.T., H.W., P.D., S.K., N.J.S., H.S., J.E. and I.R.K. contributed to the data analysis on single study level. C.L. checked all single study data for validity and did all calculations and programming for the meta-analysis including quality control. C.L. and I.R.K. wrote the manuscript. C.L., M.A., T.L.A., A.B., A.G., S.G., J.H., J.C.H., S.K., M.E.K., K.W.L., Y.L., L.P.L., C.P.N., M.N., L.Q., E.S., M.S., T.T., C.W., H.H.W., L.Z., W.Z., S.S.A., F.B., E.P.B., R.C., G.D., R.D., T.E., M.E., M.F., D.G., V.G., C.B.G., A.S.H., A.H., S.L.H., J.H., M.K., T.K., P.R.L., L.L., C.L., P.K.E.M., E.M., E.M., A.P.M., K.N., N.P., L.R., V.S., S.H.S., A.F.R.S., J.R.T., P.A.Z., J.C.C., R.C., E.I., C.I., P.J.K., J.S.K., T.L., R.J.F.L., W.M., R.M., A.M., M.P.R., S.R., D.K.S., J.T., H.W., P.D., S.K., N.J.S., H.S., J.E. and I.R.K. contributed to the interpretation of the results of the meta-analysis and reviewed the manuscript.
References
- CARDIoGRAMplusC4D-Consortium. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- König I. R., Loley C., Erdmann J. & Ziegler A. How to include chromosome X in your genome-wide association study. Genet. Epidemiol. 38, 97–103 (2014). [DOI] [PubMed] [Google Scholar]
- Solimene M. C. Coronary heart disease in women: a challenge for the 21st century. Clinics (Sao Paulo) 65, 99–106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B. & Roeder K. Genomic control for association studies. Biometrics 55, 997–1004 (1999). [DOI] [PubMed] [Google Scholar]
- Hindorff L. et al. A Catalog of Published Genome-Wide Association Studies www.genome.gov/gwastudies (2014).
- Welter D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronary Artery Disease Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 43, 339–344 (2011). [DOI] [PubMed] [Google Scholar]
- Carrel L. & Willard H. F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005). [DOI] [PubMed] [Google Scholar]
- Wong C. C. et al. A longitudinal twin study of skewed X chromosome-inactivation. PLoS One 6, e17873 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukiainen T. et al. Chromosome X-wide association study identifies Loci for fasting insulin and height and evidence for incomplete dosage compensation. PLoS Genet. 10, e1004127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries P. S. et al. A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Hum. Mol. Genet. 25, 358–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mägi R., Lindgren C. M. & Morris A. P. Meta-analysis of sex-specific genome-wide association studies. Genet. Epidemiol. 34, 846–853 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loley C., Ziegler A. & König I. R. Association tests for X-chromosomal markers–a comparison of different test statistics. Hum. Hered. 71, 23–36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. et al. Accounting for eXentricities: analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS One 9, e113684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise A. L., Gyi L. & Manolio T. A. eXclusion: toward integrating the X chromosome in genome-wide association analyses. Am. J. Hum. Genet. 92, 643–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winham S. J., de Andrade M. & Miller V. M. Genetics of cardiovascular disease: Importance of sex and ethnicity. Atherosclerosis 241, 219–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J. C., Yen Z., Ziesche S. M. & Brown C. J. Silencing of the mammalian X chromosome. Annu. Rev. Genomics Hum. Genet. 6, 69–92 (2005). [DOI] [PubMed] [Google Scholar]
- Howie B. & Marchini J. 1,000 Genomes haplotypes – Phase I integrated variant set release (v3) in NCBI build 37 (hg19) coordinates https://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html (2012).
- Preuss M. et al. Design of the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: A Genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ. Cardiovasc. Genet. 3, 475–483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A. Genome-wide association studies: quality control and population-based measures. Genet. Epidemiol. 33 Suppl 1, S45–S50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B., Fuchsberger C., Stephens M., Marchini J. & Abecasis G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B. & Marchini J. IMPUTE2 https://mathgen.stats.ox.ac.uk/impute/impute_v2.html (2014).
- Howie B., Marchini J. & Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 1, 457–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J. & Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010). [DOI] [PubMed] [Google Scholar]
- Fuchsberger C., Abecasis G. R. & Hinds D. A. minimac2: faster genotype imputation. Bioinformatics 31, 782–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yu R. & Shete S. X-chromosome genetic association test accounting for X-inactivation, skewed X-inactivation, and escape from X-inactivation. Genet. Epidemiol. 38, 483–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathworks. Matlab http://www.mathworks.com/products/matlab/ (2016).
- Morris J. & Gauderman J. Software by USC Biostats http://biostats.usc.edu/software (2016).
- Clayton D. Testing for association on the X chromosome. Biostatistics 9, 593–600 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


