An autophagy-related process is crucial for the mutualistic interaction of Phaseolus vulgaris with beneficial microorganisms such as Rhizobium tropici and Rhizophagus irregularis.
Abstract
Eukaryotes contain three types of lipid kinases that belong to the phosphatidylinositol 3-kinase (PI3K) family. In plants and Saccharomyces cerevisiae, only PI3K class III family members have been identified. These enzymes regulate the innate immune response, intracellular trafficking, autophagy, and senescence. Here, we report that RNAi-mediated downregulation of common bean (Phaseolus vulgaris) PI3K severely impaired symbiosis in composite P. vulgaris plants with endosymbionts such as Rhizobium tropici and Rhizophagus irregularis. Downregulation of Pv-PI3K was associated with a marked decrease in root hair growth and curling. Additionally, infection thread growth, root-nodule number, and symbiosome formation in root nodule cells were severely affected. Interestingly, root colonization by AM fungi and the formation of arbuscules were also abolished in PI3K loss-of-function plants. Furthermore, the transcript accumulation of genes encoding proteins known to interact with PI3K to form protein complexes involved in autophagy was drastically reduced in these transgenic roots. RNAi-mediated downregulation of one of these genes, Beclin1/Atg6, resulted in a similar phenotype as observed for transgenic roots in which Pv-PI3K had been downregulated. Our findings show that an autophagy-related process is crucial for the mutualistic interactions of P. vulgaris with beneficial microorganisms.
INTRODUCTION
To facilitate the acquisition of nutrients that are not readily available in the soil, most plants establish symbiotic interactions with soil microorganisms such as arbuscular mycorrhizal (AM) fungi of the monophyletic fungal lineage Glomeromycota and/or a wide range of gram-negative bacteria, collectively referred to as rhizobia (Bonfante and Genre, 2008; Oldroyd and Downie, 2008). Under stress conditions, the interaction between vascular plants and AM fungi results in an increase in the acquisition of mineral nutrients, particularly phosphate, from the soil by plant roots. By contrast, nitrogen fixation occurs as a consequence of the symbiosis between the roots of legumes and rhizobia. Both of these symbiotic interactions result in the development of de novo structures on the plant roots, namely, arbuscules (intracellular ramified branching hyphae) (Bonfante and Genre, 2008; Parniske, 2008) in the case of mycorrhization and nitrogen-fixing nodules in the case of rhizobial interactions (Oldroyd and Downie, 2008). These mutualistic associations share key evolutionary and developmental features, including a common set of essential plant genes, which are part of the common symbiotic pathway. AM invasion involves the formation of an infection peg from the hyphopodium (specialized fungal cell involved in attachment to the plant host), which mediates fungal hyphal growth into the epidermal cell, and then a prepenetration apparatus, which anticipates the direction of hyphal penetration in the plant cell. Finally, arbuscules arise from the intercellular hyphae in the inner root cortical cells. During nodulation, rhizobia trigger root hair cell curling and infection thread (IT) formation, which allows bacteria to be internalized in the root cortical cells. After reinitiating plant cell division, nodule primordia form. Finally, the IT reaches nodule primordia cells and rhizobia are released and taken up by these cells, which later mature into a functional nitrogen-fixing nodule. Nodulation and mycorrhization rely on Nod and Myc factor signaling, respectively.
The signaling network that underlies these mutualistic interactions has been well studied and is known to involve ionic and cytoskeletal changes (Cárdenas et al., 2000; Oldroyd, 2013). Phosphoinositides have been implicated in Nod factor-mediated signaling. However, little is known about the role of phosphoinositides in the modulation of either of these symbiotic interactions (Pingret et al., 1998; Engstrom et al., 2002; Charron et al., 2004; Peleg-Grossman et al., 2007; Xue et al., 2009).
Phosphatidylinositol 3-phosphate (PI3P) is a phosphoinositide that is present at very low levels in plant cells (Brearley and Hanke, 1992). Eukaryotes contain three types of lipid kinases, which belong to the phosphatidylinositol 3-kinase (PI3K) family. In plants and Saccharomyces cerevisiae, only PI3K class III family members have been identified. PI3P biosynthesis is catalyzed by PI3K, which is encoded by the only PI3K gene reported in Arabidopsis thaliana, Vacuolar protein sorting (Vps34). PI3P and PI3K/Vps34 are fundamental for normal plant growth and development (Welters et al., 1994; Liu et al., 2005; Lee et al., 2008a, 2008b) and have been implicated in various physiological functions, such as the innate immune response, intracellular trafficking, autophagy, senescence, and the symbiosis of legumes with rhizobia. In soybean (Glycine max), two isoforms of PI3K are differentially expressed in roots and nodules. The nodule isoform is highly expressed during nodule organogenesis and has been associated with membrane proliferation (Hong and Verma, 1994). In addition, PI3P mediates the entry of pathogen effector proteins that suppress host defenses and enable disease development during root oomycete infection (Kale et al., 2010). Therefore, PIP3 is an important regulator of the interaction between plants and microbes. Recently, PI3K was shown to have an essential role in autophagy (Klionsky and Ohsumi, 1999; Levine and Klionsky, 2004). PI3K/VPS34 interacts with VPS30/BECLIN1 and VPS15 to form a multiprotein complex that functions in autophagy (Kametaka et al., 1998). A novel protein, SH3P2 (SH3 domain protein 2), binds to PI3P downstream of the PI3K complex, which is involved in Arabidopsis autophagosome biogenesis (Zhuang et al., 2013). Autophagy is a highly regulated process in which cytoplasmic materials become enclosed in a double membrane vesicle that is then targeted to the vacuole or lysosome, where the contents are degraded and recycled (Xie et al., 2008). It also plays a central role in processes underlying several physiological responses, including plant development, the innate immune response, and nutrient recycling during starvation and senescence (Hanaoka et al., 2002; Yoshimoto et al., 2004; Liu et al., 2005; Thompson and Vierstra, 2005).
Here, we assessed the functional role of a Phaseolus vulgaris PI3K gene during the symbiosis with AM fungi or rhizobia using reverse genetics. Loss of function of Pv-PI3K or titration of the PI3P product by overexpression of the FYVE domain drastically inhibited root hair and IT growth and nodule formation, indicating that PI3P is essential for both mycorrhization and nodulation. Furthermore, arbuscule formation with Rhizophagus irregularis (formerly known as Glomus intraradices) was abrogated upon loss of function of Pv-PI3K. Downregulation of Pv-PI3K also reduced the transcript levels of autophagy-related genes. RNAi-mediated knocking down of an autophagy-related gene (Beclin1/Atg6) revealed that autophagy is indeed required for the symbiosis between P. vulgaris roots and the soil microorganisms, rhizobia, and AM fungi.
RESULTS
Identification of a P. vulgaris PI3K Gene
Using RT-PCR and Glycine max PI3K-specific primers (Supplemental Table 1), we amplified a cDNA fragment (847 bp) of P. vulgaris PI3K cDNA from total nodule RNA and then used this cDNA fragment as a probe to screen the P. vulgaris genomic library. A 13-kb genomic fragment, encoding a PI3K protein of the class III subfamily, was isolated. A BLAST survey of the Phytozome v11 database (http://www.phytozome.net) confirmed that Pv-PI3K (ID: Phvul.002G070100.1) is a single-copy gene localized on chromosome 2 and organized in 17 exons and 16 introns. A comparative analysis of PI3K genes from monocots and dicots revealed a well conserved PI3K gene organization in land plants. The Pv-PI3K transcription unit includes 69 and 288 bp of 5′- and 3′-untranslated region, respectively, and an open reading frame encoding a protein of 811 amino acids. In agreement with previously reported PI3K structures, Pv-PI3K contains the C2 (Pfam: PF00792) structural domain, which targets proteins to cell membranes; PIK (Pfam: PF00613), also referred to as the HR2 and PI3K accessory domain; and the catalytic domain (Pfam: PF00454), a PI3_PI4 kinase or HR1 domain (Vanhaesebroeck et al., 2010) containing two highly conserved motifs [GDD(I/L)RQD and LGVGDRH] (Herman and Emr, 1990; Hiles et al., 1992; Walker et al., 1999). In pairwise alignments, the Pv-PI3K amino acid sequence exhibited high levels of identity to that of PI3K from Arabidopsis (85%), Medicago truncatula (94%), and the root and nodule of G. max PI3K isoforms (95%). The phylogenetic tree based on reported PI3K class III amino acid sequences from P. vulgaris and other eukaryotes (Supplemental Figure 1; Supplemental Data Set 1) is consistent with the taxonomic order of the organisms included.
Pv-PI3K Transcript Levels during the Symbiosis between P. vulgaris and Rhizobium tropici
In Arabidopsis, PI3K is an essential gene that is expressed in almost all tissues, including pollen and root hairs (Welters et al., 1994; Lee et al., 2008a, 2008b). In the roots of 2-d-old P. vulgaris seedlings, the relative accumulation of PI3K transcript was higher (5.4-fold) in the root hairs, the site of rhizobial entry into the root, and in the root apex (4.1-fold) than in the stripped root (depleted of root hairs) (Figure 1A). To determine the role of Pv-PI3K during the P. vulgaris symbiosis with bacteria, we evaluated Pv-PI3K transcript levels in common bean roots soon after inoculation with R. tropici CIAT 899 and in uninoculated roots. No significant differences were observed between samples of R. tropici-inoculated and uninoculated roots harvested at 6, 12, 24, 48, or 72 h after inoculation (hai). However, the level of Pv-PI3K transcript in roots inoculated with rhizobia was higher at 96 hai (1.41 ± 0.24, sd) than in uninoculated roots (0.49 ± 0.073) and was 2.5-fold higher at 144 hai (3.99 ± 0.56 versus 1.57 ± 0.23) (Figure 1B) than in uninoculated roots. ENOD40 is an early nodulin gene that is upregulated at the onset of nodulation and therefore considered as a marker of nodule development (Crespi et al., 1994; Kumagai et al., 2006; Blanco et al., 2009; Montiel et al., 2012; Okazaki et al., 2013). A 10.4-fold increase in the transcript levels of Pv-ENOD40 in the inoculated roots collected at 72 hai (Figure 1C) confirmed that the molecular mechanisms underlying nodulation had been activated. In nodules collected from roots at 10 to 30 d after inoculation (dai), Pv-PI3K transcript levels increased during nodule development and peaked at 22 dai, corresponding to the stage at which nitrogen fixation activity in the nodule is highest (Mylona et al., 1995), and then decreased as the nodule senesced (30 dai; Supplemental Figure 2A). To evaluate the spatiotemporal expression of Pv-PI3K during nodule organogenesis, we cloned the Pv-PI3K promoter (region ∼1 kb directly upstream of the initiation codon) in a transcriptional fusion with the chimeric reporter GFP-GUS to yield PvPI3Kpro:GFP-GUS. Pv-PI3K promoter activity was therefore monitored as GUS activity or GFP fluorescence in P. vulgaris transgenic hairy roots generated by Agrobacterium rhizogenes K599-mediated transformation (Estrada-Navarrete et al., 2007). Control transgenic roots harbored a cassette with no promoter sequences upstream of the GFP-GUS sequences. In P. vulgaris transgenic roots harboring PvPI3Kpro:GFP-GUS, the activity of the Pv-PI3K promoter was detected at the tip and in the vascular tissue of the root and in the basal cells of the lateral root primordia (Figures 1D to 1F). PvPI3Kpro:GFP-GUS transgenic roots inoculated with R. tropici showed a strong GUS signal in the tips, but barely any signal in the vascular tissue adjacent to the infection site (Figures 1G and 1H). The Pv-PI3K promoter is highly active in infected root hairs and in cells of the nodule primordia (Figures 1H and 1I).
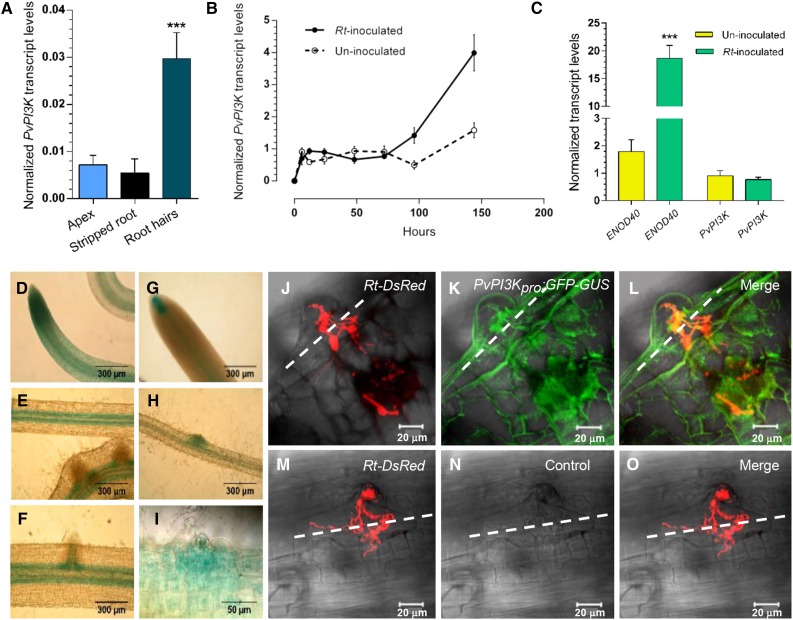
Figure 1.
Pv-PI3K Transcript Levels and Pv-PI3K Promoter Activity during the Early Stages of the Symbiosis between P. vulgaris and R. tropici CIAT 899.
(A) to (C) Quantification of transcript levels by RT-qPCR analysis.
(A) PI3K transcript levels were higher in root hairs than in the root apex and stripped roots collected from 2-d-old P. vulgaris seedlings. Bars represent the mean and sd (±sd) of two experiments (n = 2 from pool of 150 root tissues of seedlings treated to separate the apex and stripped roots).
(B) The level of Pv-PI3K transcript increases in wild-type P. vulgaris roots at 96 and 144 hai with R. tropici CIAT 899. At 6, 12, 24, 48, and 72 hai, PvPI3K transcript levels in inoculated roots were comparable to those in uninoculated roots. Black line, roots inoculated with R. tropici CIAT 899; dotted line, uninoculated roots. Bars represent mean ± sd of three experiments (n = 3 from pool of 10 roots).
(C) An increase in Pv-ENOD40 transcript level in rhizobia-inoculated roots (72 hai) confirmed the activation of nodulation. No significant difference was found in Pv-PI3K transcript levels following inoculation. Bars represent mean (±sd) of three experiments (n = 3 from pool of 10 roots).
In (A) to (C), transcript levels were quantified by reverse transcription and real-time PCR (RT-qPCR) and calculated using levels of Elongation Factor 1α as reference, as described in Methods. Each RNA sample was assessed in triplicate. The number of biological replicates (n) is indicated. Error bars indicate mean and sd (±sd). For (A) and (B), statistically significant differences were determined by one-way ANOVA followed by Tukey´s test (***P < 0.0001). For (C), statistically significant differences were confirmed by an unpaired two-tailed Student’s t test (***P < 0.001).
In (D) to (I), PvPI3Kpro:GFP-GUS activity in transgenic roots was detected at the root tip ([D] and [G]), the vascular tissue (E), and in the cells at the base of lateral root primordia (F), as assessed by GUS staining. When inoculated with R. tropici CIAT899, the Pv-PI3K promoter was highly active in the infected root hair (I) and in the cells adjacent to the infection site (H). Bars = 300 µm in (D) to (H) and 50 µm in (I).
(J) to (O) Hairy roots transformed with PvPI3Kpro:GFP-GUS were inoculated with R. tropici CIAT899 expressing the fluorescent marker DsRed (R. tropici-DsRed). Activity, detected as GFP fluorescence, was observed at the infection site (K) and merge in (L). Infection thread progression and ramification were traced by the fluorescence of DsRed (Rt-DsRed; red in [J] and [M]; merge in [L]). No green fluorescence was detected in P. vulgaris transgenic roots harboring a promoterless construct (control vector; [N]). White dashed lines indicate the border between the root epidermis and the adjacent cortical cell layer. Images were acquired in vivo using a Zeiss LSM 510 Meta Confocal Microscope; 21 optical sections each of 0.89 μm were acquired for PvPI3Kpro:GFP-GUS roots, and 16 optical sections each of 1.89 μm for the control roots. Transgenic roots were analyzed at 15 dai. Bars = 20 μm.
We next examined Pv-PI3K promoter-driven GFP-GUS expression during the early stages of infection in hairy roots inoculated with R. tropici expressing the fluorescent marker DsRed using laser scanning confocal microscopy. A representative image of a typical IT (harboring DsRed-expressing rhizobia), growing from the root hair through the GFP-expressing cells of a nodule primordium is shown in Figures 1J to 1L. In the mature nodule (22 dai), the Pv-PI3K promoter was mainly active in the central tissue (Supplemental Figure 2B), whereas at 30 dai, the promoter activity was restricted to the nodule vascular bundles (Supplemental Figure 2C). Neither rhizobia-inoculated nor uninoculated control transgenic roots bearing the promoter-less GFP-GUS construct exhibited GUS activity (Figures 1M to 1O; Supplemental Figures 2D to 2K).
Downregulation of PI3K Affects Root Hair Growth and Curling in Response to Rhizobia Inoculation
To gain insight into the role of Pv-PI3K during nodule organogenesis and AM symbiosis, we used RNAi to knock down the expression of Pv-PI3K in P. vulgaris composite plants, in which only the hairy roots were transgenic. A specific PvPI3K-RNAi construct (537 bp) was cloned downstream of the 35S promoter sequence in the Gateway-based hairpin vector pTdT-DC-RNAi (Valdés-López et al., 2008) to generate pTdT-PvPI3K-RNAi. A previously described construct bearing a truncated and irrelevant sequence from Arabidopsis pre-mir159 cloned in pTdT-DC-RNAi (Montiel et al., 2012) was used as a control vector and is herein referred to as pTdT-Sac-RNAi. Transgenic hairy roots were selected based on the expression of red fluorescent protein tdTomato (tandem dimer Tomato; Shaner et al., 2004) encoded by the vector. Nonfluorescent roots were discarded. In individual transgenic roots expressing the Pv-PI3K-specific RNAi, the levels of Pv-PI3K transcript were reduced by ∼60% with respect to the levels found in individual control transgenic roots (Figure 2A; Supplemental Figure 3A).
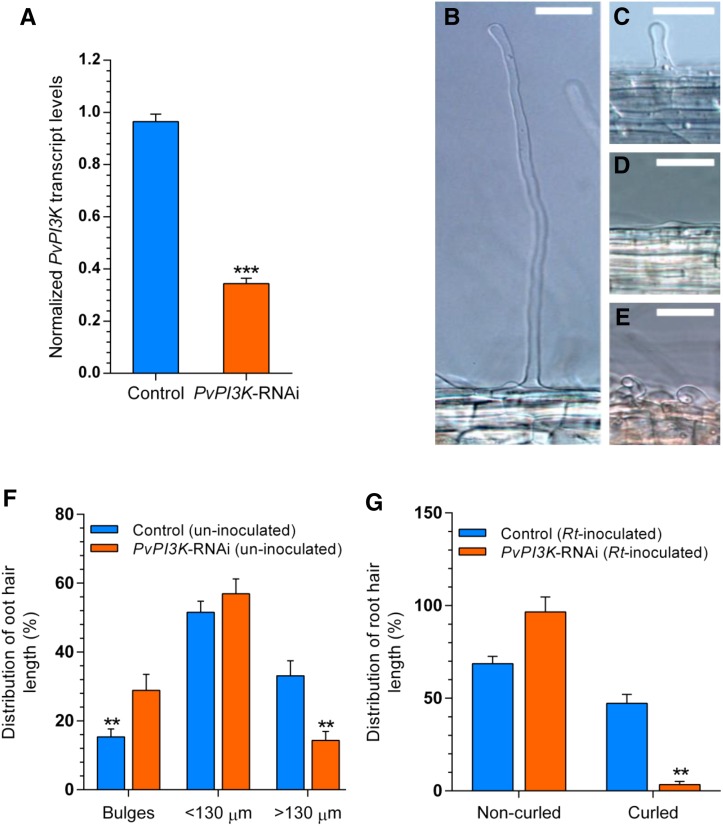
Figure 2.
Loss of Function of PvPI3K Inhibits Root Hair Growth and Root Hair Curling after R. tropici Inoculation.
(A) The extent of PvPI3K downregulation was determined by RT-qPCR analysis in individual red-fluorescent transgenic roots expressing PvPI3K-specific RNAi. A comparison between normalized PvPI3K transcript levels and control transgenic roots expressing TdT-Sac-RNAi. Normalized transcript levels were quantified by RT-qPCR using transcript levels of Elongation Factor 1α as reference. Bars are the mean ± sd of four (n = 4) independent transgenic roots (biological replicates), and each sample was analyzed in triplicate. Statistical significance was confirmed by an unpaired two-tailed Student’s t test (***P < 0.004).
(B) to (D) Representative images of P. vulgaris root hairs of different length, as described in this work: long, >130 µm (B); short, <130 µm (C); and emerging root hair or bulge (D). Bars = 50 µm.
(E) Representative image of a P. vulgaris curled root hair. Curling is typically observed after root inoculation with R. tropici CIAT 899. Bar = 50 µm.
(F) Distribution of root hair length in 12-d-old control (blue bars) and PvPI3K-RNAi (orange bars) transgenic roots, presented as percentage (%), where 100% is the sum of all root hairs counted.
(G) Distribution of noncurled and curled root hairs in control (blue bars) and PvPI3K-RNAi (orange bars) transgenic roots inoculated with R. tropici CIAT 866 and collected at 72 hai. Presented as percentage, where 100% is the sum of curled and noncurled root hairs.
Roots were cleared and root sections of ∼3 cm in length were mounted on glass microscope slides as described in Methods. Bars are the mean value (%) of 15 or 16 independent transgenic roots (n = 15 to 16). Bars refer to mean ± se. Statistically significant differences were confirmed by an unpaired two-tailed Student’s t test with Welch’s correction (**P < 0.005).
Loss of function of Pv-PI3K did not affect root hair density but had a striking effect on root hair length, possibly indicating that PvPI3K-RNAi transgenic roots had a lower proportion of long root hairs at 12 d. This indicates that PI3K has a direct effect on root hair length. However, the maximum length of root hairs in P. vulgaris roots is unknown. Data reported by Blanco et al. (2009) showed that root hairs in P. vulgaris transgenic roots expressing an irrelevant GUS-RNAi were ∼250 ± 80 µm in length (Blanco et al., 2009). To determine the relative length of root hairs in PvPI3K-RNAi and control transgenic roots, a series of digital images, representative of root zones presenting a similar density of root hairs, were analyzed (Supplemental Figure 3B). Although it cannot be established whether the root hairs measured were still growing or had ceased to grow, the length of the longest root hairs in control transgenic roots was 402 ± 60 µm, and we used this length as a parameter for length measurements. According to Dolan et al. (1993), Arabidopsis root hairs grow in three discrete, successive phases. After the appearance of small bulges, tip-growing root hairs first grow slowly and then grow more rapidly (Dolan et al., 1993). Based on these phases, P. vulgaris transgenic root hair cells were distributed in three groups: bulges or emerging root hairs; short root hairs, having a length equivalent to one-third or less of 402 ± 60 µm (i.e., <130 µm), which may include cells with a slow tip-growing phase that are transitioning to an increasing rate of tip growth; and long root hairs, having a length of >130 µm (Figures 2B to 2D). While no differences were found when comparing the percentage of short root hairs observed in PvPI3K-RNAi and control transgenic roots, more bulges were found in PvPI3K-RNAi (28.86% ± 4.69%) than in the control (15.37% ± 2.31%) and a decrease of ∼2-fold was found in the percentage of long root hairs (14.36% ± 2.60%) in PvPI3K loss-of-function transgenic roots with respect to control transgenic roots (33.12% ± 4.36%) (Figure 2F; Supplemental Table 2). Root hair curling, a typical response to the species-specific interaction between the root hair and the rhizobia, was affected by the downregulation of Pv-PI3K expression. Only 3.39% ± 1.70% of root hairs in PvPI3K-RNAi transgenic roots exhibited curling, compared with 31.30% (±4.20) in transgenic inoculated control roots (72 hai), as shown in Figures 2E and 2G.
Loss of Function of Pv-PI3K Impairs Nodule Organogenesis in Transgenic Roots
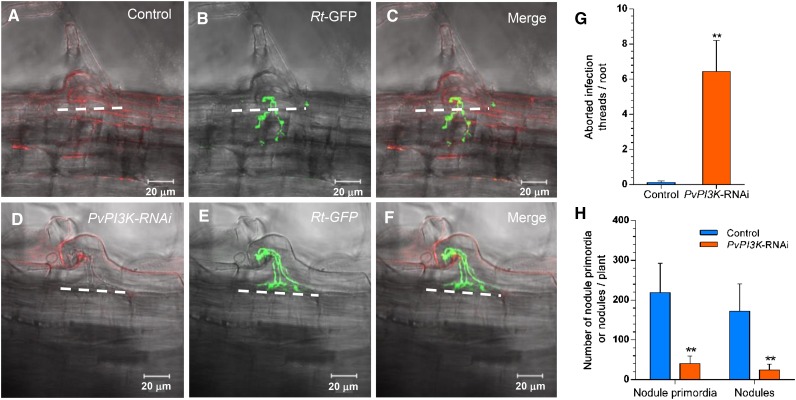
To assess the effects of the loss of function of Pv-PI3K on nodule organogenesis in P. vulgaris transgenic roots, we analyzed the events of infection and nodulation after inoculation with R. tropici expressing GFP by laser scanning confocal microscopy. At 12 dai, typical ITs formed and ramified in the root hair and further penetrated the dividing cells beneath the infection site in control transgenic roots (Figures 3A, 3B, and 3C). In PvPI3K-RNAi transgenic roots, IT growth was arrested at the base of the infected root hair, impairing its progress toward the first dividing cell layer beneath the infection site (Figures 3D to 3F). We then determined how many arrested ITs (AITs) were present in the transgenic roots. An average of 6.43 ± 0.71 AITs/root were found on PvPI3K-RNAi transgenic roots, whereas AITs on control transgenic roots were scarce (0.125 ± 0.03 AITs/root; Figure 3G). Loss of function of Pv-PI3K did not seem to affect reinitiation of the cell cycle in the outer cortical cells at the infection site (Figure 3F). This cell activity is a characteristic feature of the determinate type of nodulation, such as that exhibited by P. vulgaris (van Spronsen et al., 2001). The roots of control transgenic plants (Figure 3H; Supplemental Figures 4A to 4C) had 218.90 ± 73.40 nodule primordia and 172.40 ± 68.50 nodules per plant. In transgenic roots expressing PvPI3K-RNAi, the number of nodule primordia and nodules per plant was drastically reduced (40.66 ± 18.34 and 24.50 ± 14.39 per plant, respectively). Additionally, PvPI3K-RNAi nodules were smaller than those produced on control transgenic roots expressing TdT-Sac-RNAi (Supplemental Figures 4A and 4B). These data indicate that downregulation of Pv-PI3K led to a drastic reduction in the number of nodule primordia (5-fold) and nodules (7-fold) with respect to control transgenic roots. Another phenotypic feature affected by Pv-PI3K loss of function involved infection of cells in the central tissue of the nodule. Almost all cells in the central tissue of nodules generated in control transgenic roots were infected with rhizobia expressing GFP (Supplemental Figure 4D). In the case of PvPI3K-RNAi transgenic roots inoculated with R. tropici-GFP, the vast majority of the nodules did not contain cells infected with rhizobia-GFP (Supplemental Figure 4E), although ITs exhibiting R. tropici-GFP fluorescence were frequently observed at the periphery of the empty nodules, suggesting that the IT remained arrested in the root hair (Supplemental Figure 4F). As expected, the acetylene reduction assessment indicates that nitrogen fixation was barely detected in nodules harvested from PvPI3K-RNAi transgenic roots (10.28 ± 2.37 μmol ethylene/h/g of nodule dry weight) when compared with those collected from control transgenic roots (67.20 ± 10.39 μmol ethylene/h/g of nodule dry weight). Therefore, Pv-PI3K loss of function not only has an effect on nodule organogenesis, but also impairs nodule infection.
Figure 3.
Loss of Function of Pv-PI3K Impairs Infection Thread Growth, Ramification, and Nodule Development in P. vulgaris Transgenic Roots.
(A) to (F) Representative in vivo images of ITs formed in P. vulgaris control ([A] to [C]) and PvPI3K-RNAi transgenic roots ([D] to [F]) inoculated with R. tropici CIAT899 expressing GFP (Rt-GFP; green) in (B) and (E). Control (TdT-Sac-RNAi) and PvPI3K-RNAi roots expressed the red fluorescent tdTomato reporter marker in (A) and (D). Infection thread progression and ramification were traced based on GFP fluorescence. White dashed lines indicate the border between the root epidermis and the adjacent cortical cell layer. In PvPI3K-RNAi transgenic roots, IT growth was arrested at the base of the root hair, before entering the dividing cell layers adjacent to the infection site. This IT phenotype is referred to as an arrested or abortive IT (AIT). Images were acquired in vivo using a Zeiss LSM 510 Meta Confocal Microscope; 34 optical sections (1.0 μm) were acquired for the control and 27 optical sections (1.0 μm) for the PvPI3K-RNAi roots. Bars = 20 µm.
(G) More AITs were formed on the PvPI3K-RNAi transgenic roots than on the control (TdT-Sac-RNAi) transgenic roots. Bars represent the mean ± sd of AITs/root of independent transgenic roots (n = 16). Asterisks indicate statistically significant difference (unpaired two-tailed Student’s t test, **P < 0.05).
(H) The numbers of nodule primordia and nodules were reduced in Pv-PI3K-downregulated transgenic roots with respect to control transgenic roots. Bars represent mean ± sd (n = 10 transgenic roots from composite plants; 22 dai). Asterisks indicate statistically significant difference (unpaired two-tailed Student’s t test, **P < 0.003 and **P < 0.05, respectively).
Expression of the FYVE Domain Phenocopies Nodulation Impairment in Loss of Function of Pv-PI3K Transgenic Roots
To investigate whether PI3P, the product of Pv-PI3K kinase activity, is required for nodule organogenesis, a fluorescent biosensor (YFP-2xFYVE; Vermeer et al., 2006) was transgenically expressed in roots to specifically visualize PI3P in living cells. FYVE zinc finger domains present in four cysteine-rich proteins, i.e., Fab1, YOTB, Vac1, and EEA1 (Stenmark et al., 1996; Gaullier et al., 1998; Patki et al., 1998), were identified as having the ability to direct the highly specific binding of biosensor molecules, such as GFP or YFP, to PI3P in animal cells. A similar strategy was applied to study cowpea (Vigna unguiculata) protoplasts, tobacco (Nicotiana tabacum) BY-2 cells, and Arabidopsis root epidermal cells and stomata (Vermeer et al., 2006).
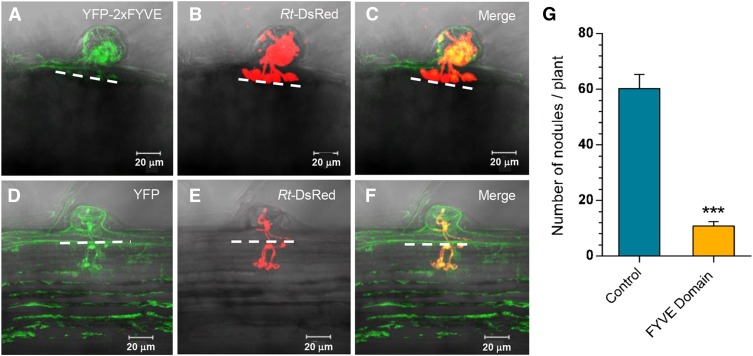
P. vulgaris transgenic roots harboring a 2x35S:YFP-2xFYVE construct and inoculated with DsRed-expressing R. tropici were phenotypically similar to transgenic roots in which Pv-PI3K expression was downregulated. Although rhizobia interacted with root hairs and ITs formed in transgenic roots expressing YFP-2xFYVE, IT progression was arrested in the root hair, but activation of the cell cycle in cortical cells adjacent to the infection site was not affected. Multiple AITs were often observed within the same root hair (Figures 4A to 4C). Neither IT elongation toward the dividing cortical cells (Figures 4D to 4F) nor the nodule number was affected in control transgenic roots bearing a 35S-YFP construct (Figure 4G). A reduction in the number of nodules of ∼5-fold was associated with the expression of the biosensor YFP-2xFYVE (Figure 4G). In summary, these data indicate that the expression of the PI3P binding biosensor YFP-2xFYVE mimics the loss of function of Pv-PI3K and strongly suggest that the catalytic activity of Pv-PI3K is instrumental for nodulation in P. vulgaris.
Figure 4.
Expression of the FYVE Domain Phenocopies the Loss of Function of Pv-PI3K in P. vulgaris Transgenic Roots after R. tropici Inoculation.
(A) to (F) Transgenic roots expressing YPF-2xFYVE or YFP (control) were inoculated with R. tropici-DsRed and fluorescence was observed in vivo at 15 dai by confocal microscopy. Representative images of an infection site in YPF-2xFYVE expressing roots revealed that IT progression was arrested (A) and ITs fail to reach the first cell layer of the outer cortex ([B] and [C]). A typical IT progression was observed in control transgenic roots ([D] to [F]). Images were acquired in vivo using a Zeiss LSM 510 Meta Confocal Microscope, and 23 optical sections each of 0.89 μm were acquired for YFP-2xFYVE and 25 optical sections each of 1.0 μm for the control roots. Bars = 20 µm.
(G) Fewer nodules were produced by transgenic roots expressing YPF-2xFYVE than by control transgenic roots (expressing YFP). Bars represent mean ± sd (n = 10 composite plants; 21 dai). Asterisks indicate statistically significant differences (unpaired two-tailed Student’s t test, ***P < 0.006).
Autophagy Is Downregulated in Pv-PI3K Loss of Function P. vulgaris Roots
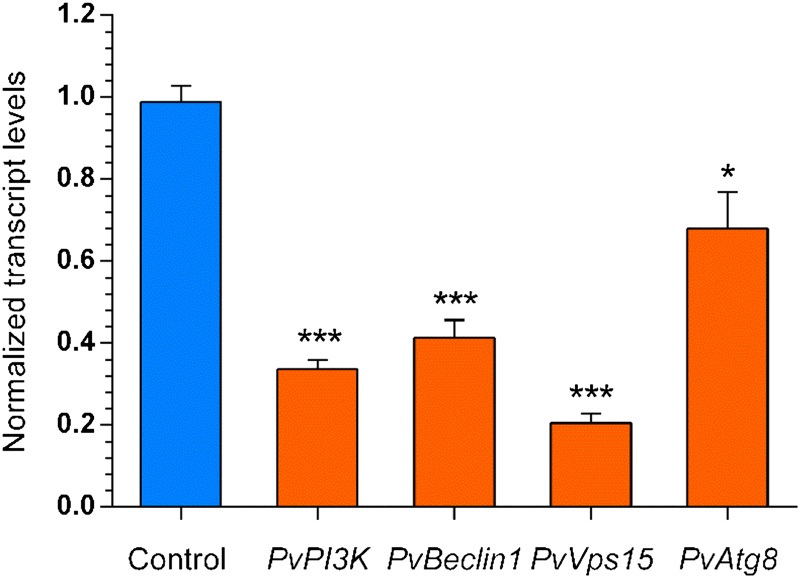
VPS34/PI3K, VPS15 (the regulatory subunit) and the accessory proteins VPS30/BECLIN1 and ATG14 form a protein multicomplex that is required for autophagy in yeast (Stack and Emr, 1993; Stack et al., 1995; Kametaka et al., 1998; Kihara et al., 2001). Whereas no Atg14 ortholog was found in plants (Levine and Klionsky, 2004), Vps15 and Beclin1 of this complex had orthologs in plants. To gain insight into the expression of Pv-Vps15, Pv-Beclin1, and the autophagy universal marker Pv-Atg8 in P. vulgaris, we performed a comparative analysis of the transcript accumulation of these genes in roots expressing PvPI3K-RNAi. The transcript level of PI3K in roots expressing PvPI3K-RNAi was decreased by 66.3% ± 2.3% relative to control (TdT-Sac-RNAi) roots. A 79.4% ± 2.28% decrease in the abundance of Pv-Vps15 transcript was found in Pv-PI3K loss-of-function transgenic roots. A statistically significant reduction was also found in the transcript accumulation patterns of Pv-Beclin1 (59% ± 3.4%) and Pv-Atg8 (32.1% ± 9.2%), as shown in Figure 5.
Figure 5.
Transcript Accumulation Analysis Showed That the Autophagy Genes Pv-Beclin1, Pv-Vps15, and Pv-Atg8 Were Downregulated upon Pv-PI3K Loss of Function.
RNA samples from P. vulgaris PvPI3K-RNAi transgenic roots with a reduction in Pv-PI3K transcript levels (∼66%) were further assessed by RT-qPCR analysis to quantify the transcript levels of Pv-Beclin1, Pv-Vps15, and Pv-Atg8. Calculations of normalized transcript level were made using Elongation Factor 1α as reference gene, as described in Methods. Bars refer to mean ± sd of two experiments (n = 2 from pools of 8 transgenic roots, biological replicates; each RNA sample was analyzed in triplicate). Statistically significant differences were determined using an unpaired two-tailed Student’s t test (*P < 0.05 and ***P < 0.001).
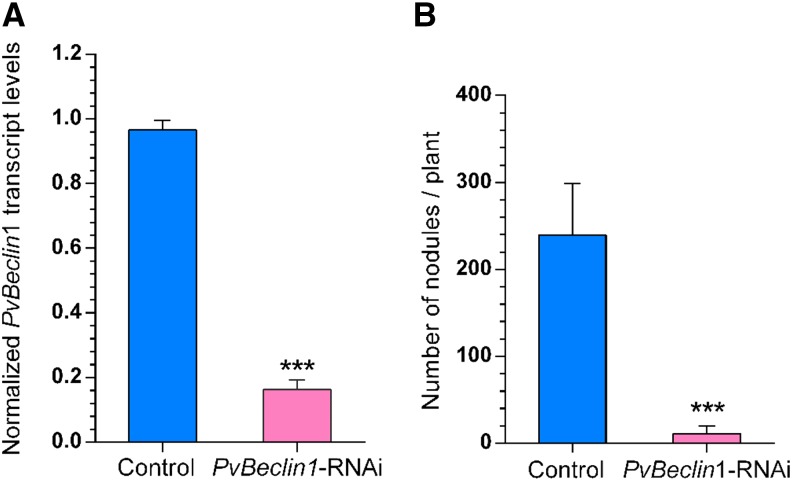
An Arabidopsis Beclin1-deficient mutant (beclin1) exhibits pollen germination defects, dwarfism, and early senescence. Genes involved in phosphatidylinositol metabolism and signaling and the glycosyl phosphatidyl inositol) anchor system are also significantly disrupted in bcl1 plants (Qin et al., 2007). Nicotiana benthamiana Beclin1 is essential for restriction of the hypersensitive response and programmed cell death (Liu et al., 2005). In an attempt to evaluate the functional relationship between Pv-PI3K and Pv-Beclin1 during nodule organogenesis, we generated a PvBeclin1-RNAi construct. Pv-Beclin1 transcript levels were reduced by 78.68% ± 2.74% in roots expressing PvBeclin1-RNAi compared with control transgenic roots (Figure 6A). The roots of plants in which Pv-Beclin1 expression was downregulated had far fewer nodules (11.2 ± 3.9) than did control roots expressing the TdT-Sac-RNAi (239.3 ± 34.2) at 21 dai with R. tropici (Figure 6B).
Figure 6.
Pv-Beclin1 Downregulation Impairs Nodulation in P. vulgaris Transgenic Roots.
(A) The levels of Pv-Beclin1 transcript were reduced in transgenic roots expressing PvBeclin1-RNAi, compared with control transgenic roots (expressing TdT-Sac-RNAi). Transcript levels were normalized using Elongation Factor 1α as reference gene, and calculations were made as described in Methods. Bars refer to mean ± sd of four experiments (n = 4 from pools of 10 hairy roots). Transcript levels were quantified by RT-qPCR at 14 dpi with A. rhizogenes K599. Statistically significant difference is indicated by asterisks (unpaired two-tailed Student’s t test; ***P < 0.001).
(B) Pv-Beclin1 downregulation led to a decrease in the number of nodules generated at 21 dai with R. tropici CIAT 899. Bars are the mean ± sd (n = 12 composite plants). Statistically significant differences were confirmed by an unpaired two-tailed Student’s t test (***P < 0.001).
Pv-PI3K Transcript Levels and Promoter Activity in P. vulgaris Roots after Inoculation with R. irregularis
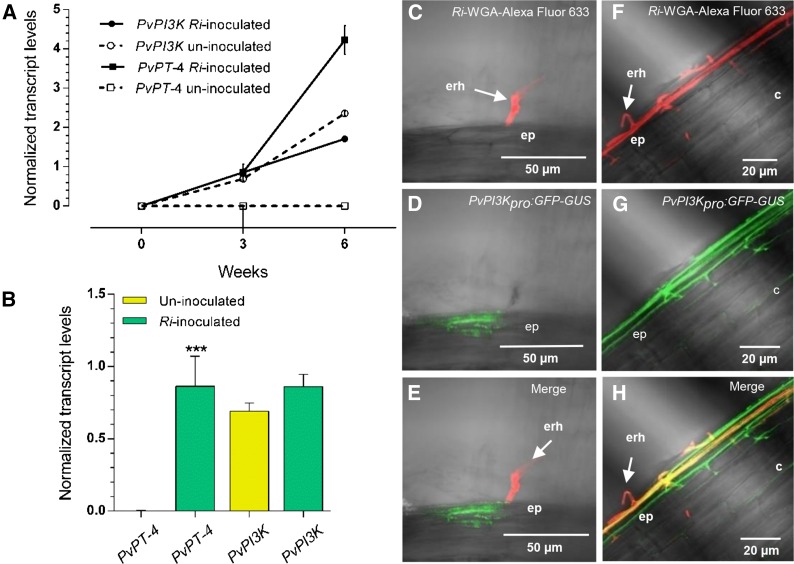
To address the participation of Pv-PI3K in the AM symbiosis in common bean roots, 5-d-old wild-type P. vulgaris seedlings were inoculated with R. irregularis and examined at 3 and 6 weeks after inoculation (wai). Uninoculated plants grown under identical conditions were used as controls. Fungal colonization was confirmed by Trypan blue staining and monitored by RT-qPCR measurement of P. vulgaris phosphate-transporter T4 (PvPT-4) transcripts, an indicator of progressive colonization (McGonigle et al., 1990; Arthikala et al., 2013). As expected, the relative levels of PvPT-4 transcript increased in response to R. irregularis inoculation, whereas no significant changes were observed in Pv-PI3K transcript levels between roots colonized with AM fungi and control uninoculated roots (Figures 7A and 7B).
Figure 7.
Pv-PI3K and Pv-PT-4 mRNA Levels and Pv-PI3K Promoter Activity in P. vulgaris Roots after Inoculation with R. irregularis.
(A) and (B) Pv-PI3K and Pv-PT-4 mRNA levels were quantified by RT-qPCR in wild-type P. vulgaris roots inoculated or not with R. irregularis and collected at 3 and 6 wai. Transcript levels were normalized to the reference gene EF1α, and values were quantified as described in Methods. Normalized Pv-PT-4 transcript levels are upregulated during the AM symbiosis (A). No significant differences in Pv-PI3K transcript level were observed between uninoculated roots and those inoculated with R. irregularis (B). Bars indicate mean ± sd from two independent biological replicates (n = 2 from a pool of 10 roots). Statistical significance was confirmed by an unpaired two-tailed Student’s t test (***P < 0.001).
(C) to (H) PvPI3Kpro:GFP-GUS activity, as evaluated by green fluorescence, was detected in epidermal cells neighboring the contact point of the fungal hyphae ([C] to [E]) and in those beneath the extending extraradical hyphae ([F] to [H]; merge). AM fungi were stained with WGA-Alexa Fluor 633 conjugate (Ri-WGA-Alexa Fluor 633, red fluorescence). Representative images of transgenic roots inoculated with R. irregularis and collected at 3 wai. Images were acquired in vivo using a Zeiss LSM 510 Meta Confocal Microscope. ep, epidermis; erh, extraradical hyphae; c, cortex.
Although the AM symbiosis in P. vulgaris has not been characterized in the same level of detail as in M. truncatula (Genre et al., 2008), a previous report described the key stages of AM infection in P. vulgaris roots (Arthikala et al., 2013). To survey the spatiotemporal Pv-PI3K promoter activity upon R. irregularis inoculation of transgenic roots harboring the construct PvPI3Kpro:GFP-GUS, we harvested the roots at 3 wai. R. irregularis was stained with an Alexa Fluor 633 conjugated to wheat germ agglutinin (WGA), which exhibits far-red fluorescence (Arthikala et al., 2013). Pv-PI3K promoter activity, revealed as GFP fluorescence, was restricted to epidermal cells (Figure 7D) neighboring the contact point of the fungal hyphae (Figures 7C and 7E, merge) and to those beneath the expanding extraradical hyphae (Figures 7F to 7H, merge). However, no PPA-like structures were detected, suggesting that the Pv-PI3K promoter was active prior to the formation of the PPA. The later stages of AM infection in cortical cell layers could not be analyzed due to the thickness of P. vulgaris transgenic roots beyond 3 wai.
Pv-PI3K Loss of Function Impairs AM Symbiosis
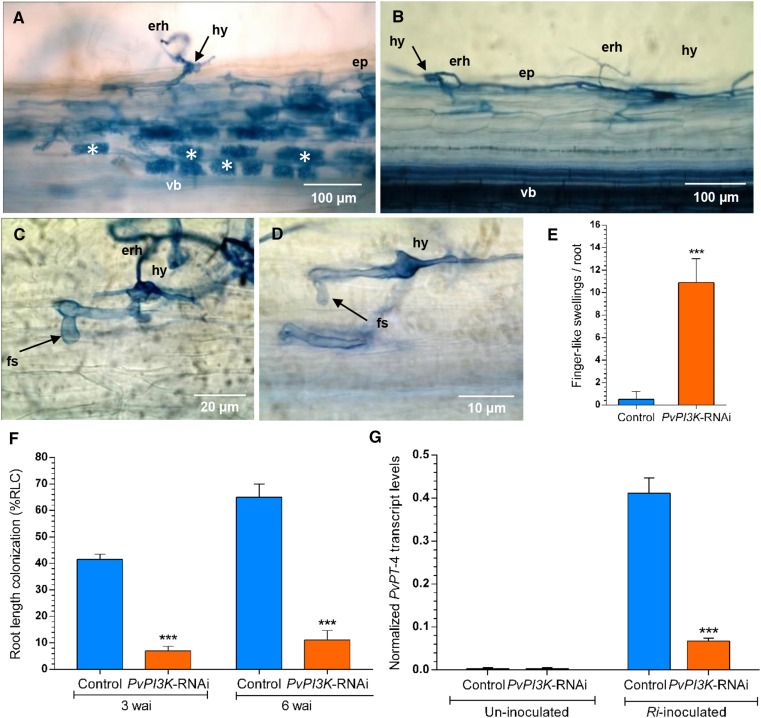
We next examined whether Pv-PI3K is required for the AM symbiosis in PvPI3K-RNAi transgenic roots (3 wai) stained with Trypan blue. As observed in Figure 8A, control transgenic roots showed a typical AM colonization pattern, which includes epidermal penetration by the fungal partner, formation of intracellular apoplastic compartments, ramification, and fungal colonization of both the outer and inner cortex with abundant arbuscules. All fungal structures were identified, namely, extraradical and intraradical hyphae, hyphopodium, and arbuscules with trunks and branches (Figure 8A). By contrast, typical mycorrhizal infection was drastically impaired in PvPI3K-RNAi transgenic roots. An increased number of extraradical hyphae and hyphopodia that seem to be arrested at the fungal contact site were observed with respect to control transgenic roots expressing TdT-Sac-RNAi (Figure 8B). Furthermore, finger-like outgrowths (10.90 ± 1.13/root) were often observed in extracellular hyphae in the downregulated transgenic roots (Figures 8C to 8E; Supplemental Figure 5A), whereas such structures were barely detected in control transgenic roots (less than one/root; Figure 8A; Supplemental Figure 5B). Occasionally, intraradical hyphae were found to penetrate the outer cortex (Figure 8B), but intracellular infection and arbuscule formation in the epidermis and outer cortex were never observed. To estimate the effect of Pv-PI3K downregulation on the extent of AM fungal colonization, we quantified the overall percentage of root length colonization (%RLC) (McGonigle et al., 1990) at 3 and 6 wai in control and PvPI3K-RNAi transgenic roots inoculated with R. irregularis. In PvPI3K-RNAi transgenic roots, an AM fungal colonization percentage of 7 ± 1.2 and 11.16 ± 3.5%RLC was observed at 3 and 6 wai, respectively, which represents about a 5.25-fold reduction with respect to control transgenic roots (i.e., 41.50 ± 2.0 and 65.0 ± 5.0%RLC at 3 and 6 wai, respectively; Figure 8F). Transcriptional activation of Pv-PT-4, a molecular tracker of AM colonization, was also impaired in PvPI3K-RNAi transgenic roots inoculated with R. irregularis. As shown in Figure 8G, after 3 wai, Pv-PT-4 transcript levels were lower (0.081 ± 0.009) in Pv-PI3K downregulated transgenic roots than in control transgenic roots (0.41 ± 0.036). Together, these results indicate that the AM symbiosis is drastically impaired in Pv-PI3K loss-of-function transgenic roots.
Figure 8.
Loss of Function of Pv-PI3K Impairs AM Symbiosis in P. vulgaris Roots.
(A) to (D) AM colonization in PvPI3K-RNAi and control TdT-Sac-RNAi transgenic roots (3 wai) was visualized using trypan blue staining. Bars = 100 μm in (A) and (B), 20 μm in (C), and 10 μm in (D).
(A) Control transgenic roots showing typical AM colonization with abundant arbuscules.
(B) PvPI3K-RNAi transgenic roots exhibited sparse hyphae entering and branching inside epidermal cells.
(C) and (D) Extraradical hyphae forming branched swellings known as hyphopodia were found. No arbuscules were observed. erh, extraradical hyphae; ep, epidermis; fs, finger-like swelling or outgrowth; hy, hyphopodium; vb, vascular bundle; asterisks, arbuscules.
(E) The number of finger-like outgrowths was higher in PvPI3K-RNAi transgenic roots than in control (TdT-Sac-RNAi) roots inoculated with R. irregularis and collected at 3 wai. Data are means ± sd from three independent experiments (n = 33). Statistical significance was determined using an unpaired two-tailed Student’s t test (***P < 0.001).
(F) AM colonization was impaired by the loss of function of Pv-PI3K. The root length colonization (%RLC) in PvPI3K-RNAi transgenic roots was lower than in control (TdT-Sac-RNAi) transgenic roots. The data represent means ± sd of three independent experiments (n = 39 roots). Statistical significance was determined using an unpaired two-tailed Student’s t test (***P < 0.001).
(G) Transcript level of PvPT-4, the molecular tracker of AM colonization. The expression of PvPT-4 was lower in PvPI3K-RNAi roots inoculated with R. irregularis than in inoculated control (TdT-Sac-RNAi) transgenic roots, as determined by RT-qPCR. Transcript levels were normalized to EF1α as reference gene. Calculations were made as described in Methods. The bars represent mean ± sd of two experiments (n = 2 from a pool of 12 transgenic roots). Asterisks represent statistically significant differences (unpaired two-tailed Student’s t test, **P < 0.004).
DISCUSSION
Leguminous plants, such as common bean, establish mutualistic relationships with soil microorganisms, such as AM fungi, to assimilate phosphate, and with rhizobia, to fix nitrogen symbiotically. In this study, we showed that PI3K has a previously undescribed role in mutualistic interactions. During the P. vulgaris-Rhizobium symbiosis, PI3K activity is essential for root hair growth, curling, and IT migration, since these responses were dramatically impaired in Pv-PI3K loss-of-function transgenic roots (PvPI3K-RNAi hairy roots). These results imply that Pv-PI3K not only plays a key role in supporting polar growth in root hair cells, as has been previously demonstrated (Figure 2F; Lee et al., 2008a), but also during IT growth from cell to cell, since the IT is arrested in the root hair cells and is unable to reach the cortical cells. Surprisingly, cortical cell division was not inhibited in PvPI3K-RNAi hairy roots and nodules developed. However, the few nodules that developed were severely impaired and lacked bacteria, since IT growth and penetration were largely arrested within the root hairs. Since the Pv-PI3K promoter is active in the root hair forming the IT and in the dividing cortical cells forming the primordium and thereafter the nodule, we anticipate that Pv-PI3K has a distinct role during the infection process and nodule development. This notion is supported by the observation that the few nodules that we did observe were smaller and lacked rhizobia, with some bacteria in the surface at the root hair forming the IT. It is possible that the IT is more sensitive to the absence of PI3P than is cell division, since this latter process was still observed. By the onset of nodule senescence, the Pv-PI3K promoter was only active in the nodule vascular bundles, suggesting that Pv-PI3K also functions in nutrient mobilization from the former sink organ.
It is surprising that R. irregularis hyphae did not enter the epidermal and cortical cells of PvPI3K loss-of-function bean roots (PvPI3K-RNAi), which resembles the phenotype observed for the rhizobia colonization using the same construction. Consequently, typical arbuscules did not form, and tubular protrusions (hyphopodium-like) were observed. These structures were unable to enter epidermal cells. Thus, we have presented compelling data that PI3K plays a key role in both the rhizobial and AM symbiosis in P. vulgaris roots. The finding that expressing a domain that specifically binds to PI3P, the catalytic product of PI3K, mimics the loss-of-function phenotype of Pv-PI3K in transgenic roots of common bean (Figure 4C), supports the notion that PI3K has a key role in P. vulgaris symbiotic interactions.
Root hairs are specialized plant cells, which, like pollen tubes, exhibit highly polarized, tip-localized expansion. Complex signaling networks, involving ion influxes, reactive oxygen species-localized gradients, actin filament reorganization, and enhanced membrane trafficking, operate at the tips of root hair cells (Fu et al., 2001; Hepler et al., 2001; Sieberer et al., 2005; Cárdenas et al., 2008; Oldroyd, 2013). Drugs that inhibit PI3K activity in pollen tubes and root hairs impair proper growth and development (Helling et al., 2006; Lee et al., 2008a, 2008b). The PI3K inhibitors LY294002 and Wortmannin also inhibit the actin filament reorganization induced by abscisic acid in Arabidopsis stomata (Choi et al., 2008). The mechanism by which IT growth is arrested inside root hairs remains unknown, but the mechanism that allows the migration of the IT from one cell to the next seems to be altered in the absence of PI3K. However, we know little about the processes that regulate the passage of the IT from one cell to the next. The nucleus migrates to the base of the root hair to guide IT formation in a cytoskeleton-dependent manner (Lloyd et al., 1897), but how this guidance is regulated is unknown. Our results suggest that PI3K is a key regulator of the polar growth of the IT.
Yeast contains two multiprotein VPS34 complexes that are evolutionarily conserved in autophagy and endocytic sorting. Both of these complexes contain VPS15 and VPS30/ATG6/BECLIN1. One of these complexes also contains ATG14, a regulator of autophagy, and the other contains a second VPS38, which regulates endocytic sorting (Kametaka et al., 1998; Kihara et al., 2001; Kim et al., 2013; Rostislavleva et al., 2015). Interestingly, the complexes are functionally distinct; deletion of Vps38 inhibits sorting of the lysosomal hydrolase (carboxypeptidase Y) but has no effect on autophagy-induced deprivation, while deletion of Atg14 and Atg6/Beclin1 inhibits autophagy and does not affect carboxypeptidase Y sorting. Deletion of Vps34 or Vps15 results in complete loss of autophagy activity, as described in other mutants defective in autophagy (aug mutants) (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Kihara et al., 2001). The main function of the VPS34 complex is to generate PI3P at the phagophore assembly site and recruit PI3P binding proteins. In Arabidopsis, SH3P2 binds to PI3P and ATG8 participates in autophagosome formation (Zhuang et al., 2013). In addition, deletion of Atg6/Beclin1 blocks recruitment of the ATGs required for the phagophore assembly site structure (Suzuki et al., 2001).
In plants, Vps34/PI3K, Beclin1, Atg3, and Atg7 restrict the hypersensitive response-programmed cell death response to tobacco mosaic virus infection (Liu et al., 2005). These findings are consistent with the observation that downregulating Pv-PI3K transcript levels with siRNA negatively affected the accumulation of transcripts encoding proteins involved in the formation of the PI3K multiprotein complex and also Atg8, which participates in autophagy. Pv-Beclin1 transcripts were among those that showed reduced accumulation. Fewer nodules formed on the roots of Pv-Beclin1 loss-of-function plants than on those of control plants. We found that Pv-Beclin1 loss-of function in PvPI3K-RNAi hairy roots and overexpression of the FYVE domain in common bean composite plants result in a similar phenotype, suggesting that autophagy is essential for mutualistic interactions between P. vulgaris and symbiotic microorganisms.
PI3P levels in plants are relatively low, typically comprising ∼2 to 15% of phosphoinositide pools, and they have a rapid turnover rate (Meijer et al., 2001). In Arabidopsis, the role of PI3K and PI3P in root hair growth has been examined using PI3K inhibitors or by expressing the 2xFYVE construct under a strong promoter, since homozygous knockout plants (Vps34 / Vps34) are not viable (Gillooly et al., 2000; Vermeer et al., 2006; Lee et al., 2008a). Reducing PI3P levels by expressing the Phosphatase and Tensin homolog deleted on chromosome 10 (PTEN) or by treatment with the WM PI3K inhibitor in tobacco pollen tubes also leads to accumulation of ATG8 in vacuolar autophagic bodies (Zhang et al., 2011). These findings suggest that PI3P levels regulate autophagy genes during the apical growth of root hairs and pollen tubes and that autophagy sustains the rapid growth rate of these cells.
We found that PI3K expression was upregulated 4 d after R. tropici inoculation (Figure 1B). However, recently published expression studies suggest that PI3K is not induced during rhizobial infection (Breakspear et al., 2014). The root hair infectome of M. truncatula was obtained by monitoring gene expression during rhizobial infection under selected growth conditions. However, no autophagy transcripts were found in the root hair infectome, probably due to the short half-life of autophagy-related transcripts and proteins. Roux et al. (2014) reported that nodule Mt-PI3K is highly expressed in the N-fixation zone (Roux et al., 2014), similar to our data, where Pv-PI3K transcript levels and PvPI3Kpro:GFP-GUS activity also coincided with maximum nitrogen fixation levels in nodules, as depicted in Supplemental Figure 2B.
PI3P binds to proteins through their FYVE domains, which are highly evolutionarily conserved. GFP fusions of these domains have been widely used as probes to monitor the subcellular localization of PI3P (Gillooly et al., 2000; Vermeer et al., 2006). The fact that expressing YFP-2xFYVE in bean hairy roots phenocopies the loss of function of Pv-PI3K strongly suggests that PI3P, the catalytic product of PI3K, is essential for the mutualistic interactions of P. vulgaris with AM fungi and rhizobia.
Our results suggest that autophagy provides precursors for root hair and IT growth, but apparently not for cortical cell divisions during common bean nodule organogenesis. The root epidermis penetration sites by hyphae (hyphopodia) and arbuscule formation of AM fungi also appear to require precursors derived from the autophagic process. Recently, Mi et al. (2015) reported that actin polymerization into short filaments and PI3P levels are crucial for both processes, since the shape and expansion of phagophores (the precursors of autophagosomes) depend on both of these processes to initiate autophagy, indicating that actin polymerization is regulated by the omegasome (i.e., the endoplasmic reticulum membrane protrusion enriched in the PI3P pool; Mi et al., 2015). This indicates that actin functions during the early stages of autophagosome formation. Specifically, dynamic actin polymerization and branching form a three-dimensional structure that supports the assembly of the omegasome and isolation membrane. Together, these data indicate that autophagy is a key player in mutualistic interactions between plants and symbiotic microorganisms.
METHODS
Bean Hairy Root Transformation
Common bean seeds (Phaseolus vulgaris cv Negro Jamapa) were used for Agrobacterium rhizogenes K599-mediated transformation to generate hairy roots in composite plants using a previously described method (Estrada-Navarrete et al., 2007). Usually at the second week after inoculation, hairy roots emerging from A. rhizogenes K599-induced calli are ∼3 cm long. When indicated, hairy roots were observed under an epifluorescence stereomicroscope (Olympus SZX7) and red fluorescent transgenic roots were selected on the basis of the expression of red fluorescent protein tdTomato (tandem dimer Tomato; Shaner et al., 2004) encoded by pTDT-DC-RNAi-derived constructs. Nonfluorescent hairy roots were excised with a scalpel from the site of emergence and discarded. Hairy roots were either frozen in liquid nitrogen and stored at −80°C or subjected to phenotype analysis.
Identification of the PI3K Gene in P. vulgaris
To identify P. vulgaris genomic sequences encoding a PI3K ortholog, the oligonucleotides PI3K_F and PI3K_R (Supplemental Table 1), based on the Glycine max PI3K cDNA sequence (Hong and Verma, 1994), were used in an RT-PCR reaction containing RNA extracted from common bean nodules (at 20 dai). The amplified fragment (847 bp) was further used as a probe to screen P. vulgaris genomic and cDNA libraries. A BLASTX sequence analysis of the isolated cDNA (2793 bp) and genomic clones (13.07 kb) isolated showed that both contain an open reading frame of 2436 bp encoding the same protein sequence (811 amino acids), which was 84% identical to Arabidopsis thaliana VPS34 (PI3K). The sequence of Pv-PI3K, originally reported in the GenBank database under accession number DQ118424.2, is identical to the sequence with ID number Phvul.002G070100.1 available in the Phytozome v11 database (http://www.phytozome.net).
Vector Construction
To generate the PvPI3Kpro:GFP-GUS construct, a 1050-bp fragment upstream of the Pv-PI3K initiation codon was amplified by PCR using P. vulgaris cv Negro Jamapa genomic DNA as template and the gene-specific primers PvPI3Kpro_F and PvPI3Kpro_R (Supplemental Table 1). The PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen), confirmed by sequencing, and recombined into the destination binary vector pBGWFS7,0 (Karimi et al., 2002) using Gateway LR Clonase II Enzyme Mix (Invitrogen). To create the PvPI3K-RNAi construct, a 537-bp fragment corresponding to the 3′-coding region of Pv-PI3K was amplified by PCR using the primers PvPI3K-RNAi_F and PvPI3K-RNAi_R (Supplemental Table 1) and cDNA from P. vulgaris nodules (20 dai). After cloning this fragment into pENTR/D-TOPO (Invitrogen) and confirming the presence of the insert by sequencing, the PCR product was recombined into the compatible recombination sites of the Gateway-based hairpin pTDT-DC-RNAi vector (Valdés-López et al., 2008). The correct orientation of the recombined fragments was confirmed by sequencing and by PCR using the primers Wrky-3_F or Wrky-5_R with PvPI3K-RNAi_F (Supplemental Table 1), as described (Valdés-López et al., 2008). The resulting construct (pTDT-PvPI3K-RNAi) drives the transcription of a hairpin loop PvPI3K-RNAi under control of the 35S promoter. The pTDT-DC-RNAi vector also harbors the NOSpro:tdT cassette, which mediates the hairy root expression of the molecular fluorescent marker tdTomato (Shaner et al., 2004) and allows identification of transformed roots. The PvBeclin1-RNAi construct was generated by recombining a Pv-Beclin1 fragment (400 bp of 5′-untranslated region) amplified by PCR using the primers PvBeclin1-RNAi_F and PvBeclin1_RNAi_R (Supplemental Table 1). To overexpress the FYVE domain, the pGreen35S:YFP-2xFYVE plasmid, kindly provided by Teun Munnik (Vermeer et al., 2006), was used. All constructed plasmids were introduced by electroporation into A. rhizogenes strain K599.
Nodulation Assays
Rhizobium tropici CIAT899 bacteria were grown in 250-mL flasks containing 100 mL of PY broth supplemented with 7 mM CaCl2, 50 µg/mL rifampicin, and 20 µg/mL nalidixic acid, in a shaking incubator (250 rpm) at 30°C until the suspension reached an OD600 of 0.8. For nodulation assays, transgenic composite plants were transplanted to pots (10 × 10 cm) containing sterile vermiculite, inoculated with 1 mL of a suspension of R. tropici CIAT899 diluted to an OD600 of 0.05 in 10 mM MgSO4, and grown in a controlled environmental chamber (16 h light/8 h darkness, at 26°C). Plants were watered three to four times per week with Broughton and Dilworth (B&D; Broughton and Dilworth, 1971) solution containing 2 mM KNO3. At the indicated time points after inoculation, transgenic roots were frozen in liquid nitrogen and stored at −80°C or analyzed to detect a phenotype.
Quantification of Transcript Levels by RT-qPCR Analysis
Total RNA was extracted from frozen tissues using TRIzol reagent (Life Technologies) following the manufacturer’s instructions. To eliminate contaminating genomic DNA, total RNA samples (1 µg in 20 µL) were treated with 1 unit of DNaseI (RNase-free; Invitrogen) at 37°C for 30 min and then at 65°C for 10 min. Two-step RT-qPCR was performed using a Maxima First Strand cDNA Synthesis Kit for RT-qPCR (ThermoFisher Scientific) and Maxima SYBR Green qPCR Master Mix (2X; ThermoFisher Scientific), respectively, following the manufacturer’s instructions. Each reaction was set up using 100 ng of cDNA as template in a 20 μL final volume. Gene-specific primers used in RT-qPCR reactions are listed in Supplemental Table 1. qPCRs were performed in a LightCycler 480 real-time PCR system (Roche). Relative transcript abundance was calculated using one the following formulae (Schmittgen and Livak, 2008):
 |
or
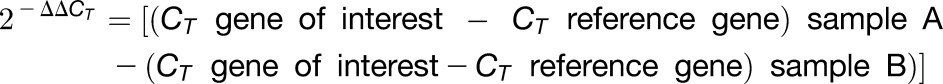 |
P. vulgaris Elongation Factor 1α (Pv-EF1α) was used as a reference gene, as previously described. RT-qPCR data are averages of three or four independent experiments or biological replicates, and each sample was assessed in triplicate.
Root Hair and Root Tip Isolation
Root hairs were isolated according to a protocol posted on the Root Hair Systems Biology website at Gary Stacey Laboratory, University of Missouri-Columbia, MO (http://www.soyroothair.org/index.php/education/4-research/protocols/12-root-hair-isolation-protocol). Briefly, P. vulgaris seedlings (48 h after germination) were held with tweezers and the root tips (∼0.5 mm) were cut from the seedlings with scissors. A second cut, at the base of the root, released the remaining segment of root. The freshly harvested root tips and root segments were dropped into separate beakers containing liquid nitrogen and stirred vigorously to separate the root hairs from the roots. Root hairs were isolated by pouring the liquid nitrogen mixture through a metal strainer. After nitrogen evaporation, root hairs, root tips, and stripped roots (which were retained in the strainer) were collected and stored at −80°C as separate frozen fractions until use.
Mycorrhizal Spore Inoculation and Mycorrhization
Composite bean plants (PvPI3K-RNAi and control) were transferred into pots (20-cm diameter) with sterile Metro-Mix and inoculated with 800 spores of Rhizophagus irregularis (kindly provided by Ignacio M. Mendoza, Instituto Politécnico Nacional, Mexico) per plant. Inoculated plants were irrigated twice weekly with half-strength B&D solution containing a low concentration of potassium phosphate (10 μM) to favor AM colonization (Smith et al., 2003). Infected roots were collected from plants at 21 and 42 dai, and AM fungi were stained either with Trypan blue (according to a modified staining procedure in Koske and Gemma, 1989) or with WGA conjugated to Alexa Fluor 633 (WGA-Alexa Fluor 633; Molecular Probes) as described before (Javot et al., 2007). AM fungal colonization status was determined by light and confocal microscopy, as indicated. The %RLC, including all fungal structures, was estimated by the magnified intersection method described by McGonigle et al. (1990).
Microscopy and Image Analysis
Transgenic roots were cleared and mounted in a solution containing 4.2 M NaI and 8 mM Na2S2O3 in 65% glycerol and 2% DMSO, as described (Dubrovsky et al., 2009). Prior to observation, root sections of ∼3 cm in length were mounted in rows on a glass microscope slide. The number of root hairs having a length of >130 or <130 μm per millimeter of mounted root was determined based on images captured using a NA40 ×/1 water immersion objective lens on a Zeiss Axiovert 200M microscope equipped with Nomarski optics and coupled to a Photometrics CoolSNAPcf Color Camera (Valley International). ITs and hand-sectioned nodules were analyzed in transgenic roots collected at 12 to 17 dai and observed under a confocal microscope. To visualize AM fungal structures, the roots were stained with Trypan blue and observed using a Zeiss Axioskop microscope with 10×, 20×, and 63× lenses. Infected roots were stained with WGA conjugated to Alexa Fluor 633 and observed under a confocal microscope. Transgenic roots harboring PvPI3Kpro:GFP-GUS were subjected to histochemical analysis of GUS activity, as described previously (Jefferson et al., 1987). Sample transgenic roots were observed under a Zeiss Axioskop microscope using 10×, 20×, and 63× lenses or by confocal microscopy. When indicated, the confocal microscopy analysis was performed at the Laboratorio Nacional de Microscopía Avanzada (LNMA-IBT UNAM) facility, using a Zeiss LSM 510 Meta Confocal Microscope. GFP fluorescence was excited with an argon/2 ion laser (488 nm), and emitted fluorescence was collected using a band-pass 500- to 530-nm IR filter. TdT and DsRed fluorescence were excited at 543 nm by a laser, and emission was filtered using a long-pass 560-nm filter. WGA-Alexa Fluor 633 (red channel) was excited by a HeNe2 laser at 633 nm, and the emitted fluorescence was collected using a band-pass 507-IR filter.
Acetylene Reduction
Nitrogen fixation was assessed using the acetylene reduction method (Vessey, 1994). Transgenic nodulated roots (20 dai; six individual roots per experiment) were placed in a 160-mL vial closed with a serum cap. Air (2 mL) was immediately withdrawn from the closed vial and replaced with an equal volume of acetylene gas. Samples were incubated for 60 min at room temperature, and ethylene production was assayed by gas chromatography in a Variant model 3300 chromatograph. Immediately after gas chromatography, nodules were separated from the roots and allowed to dry at 60°C for 2 d. Specific activity is expressed as mmol of ethylene/h/g of nodule dry weight.
Phylogenetic Analysis
Amino acid sequences of PI3K class III proteins were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/), EMBL-EBI (http://www.ebi.ac.uk/), Phytozome c11 (http://www.phytozome.net), and PLAZA (http://bioinformatics.psb.ugent.be/plaza/). Multiple sequence alignment was performed using the ClustalW algorithm, and the phylogenetic tree was generated using the neighbor-joining method with 1000 bootstrap trials, and both procedures were implemented in MEGA7 (Thompson et al., 1997). The phylogenetic tree was generated using MEGA7 (http://www.megasoftware.net/; Tamura et al., 2011).
Statistical Analysis
Data processing and statistical analysis were performed using GraphPad Prism version 6.00 for Windows (GraphPad Software). An unpaired two-tailed Student’s t test was used to determine whether data from two different groups were statistically significantly different or one-way ANOVA was performed for multiple comparisons. Single, double, and triple asterisks above the columns indicate differences that are statistically significant (P < 0.05 or P < 0.01) or very significant (P < 0.001), respectively.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers for PI3K class III proteins: Glycine max root isoform (NP_001236955.1), G. max nodule isoform (NP_001242315.1), P. vulgaris (ABA03136.1), Medicago truncatula (CAD56881.1), Populus trichocarpa (XP002318628.1), Vitis vinifera root isoform (XP_002267769.2), Nicotiana tabacum (AAW80628.1), Solanum lycopersicum (XP_004236633.1), Arabidopsis thaliana (AEE33693.1; AT1G60490.1), Brassica napus (AAN62481.1), Physcomitrella patens (XP_001762959.1), Zea mays (AFW62054.1), Hordeum vulgare (BAJ91813.1), Triticum urartu (EMS55477), Oryza sativa japonica 2 (NP_001054810.1), O. sativa japonica 1 (NP_001061506.1), Amborella trichopoda (ERN10666.1), Micromonas sp (XP_002506109.1), Chlamydomonas reinhardtii (XP_001689631.1), Dictyostelium discoideum (AAA85726.1), Saccharomyces cerevisiae (EDV08569.1), Candida albicans (Q92213), Schizosaccharomyces pombe (AAC49133.1), Trametes versicolor (EIW57772.1), Talaromyces marneffei (XP_002147483.1), Aspergillus oryzae (XP_001824747.1), Neosartorya fischeri (XP_001259818.1), Drosophila melanogaster (NP_477133.1), Homo sapiens (NP_002638.2), and Mus musculus (AAH24675.1).
Supplemental Data
Supplemental Figure 1. Phylogenetic tree of PI3K class III proteins.
Supplemental Figure 2. Pv-PI3K transcript levels during nodule development and tissue-specific activity of the Pv-PI3K promoter in mature and senescent nodules.
Supplemental Figure 3. Quantification of Pv-PI3K transcript levels in individual PvPI3K-RNAi transgenic roots.
Supplemental Figure 4. Loss of function of Pv-PI3K led to a reduction of Phaseolus vulgaris nodule number and impaired root nodule development.
Supplemental Figure 5. Extraradical hyphae growth and finger-like swellings are increased after Rhizophagus irregularis inoculation in PvPI3K loss of function of P. vulgaris roots.
Supplemental Table 1. Oligonucleotides used in this study.
Supplemental Table 2. Quantification of root hairs and curling in loss of function of PvPI3K and control P. vulgaris roots.
Supplemental Data Set 1. Text file of the alignment used to generate the phylogenetic tree of PI3K class III proteins shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Rosana Sánchez for her valuable contribution to the manuscript edition in its final version. We also thank members of F.S.’s laboratory for their helpful comments and discussions. We thank Teun Munnik (Section of Plant Physiology, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, The Netherlands) for providing the vector pGreen35S:YFP-2xFYVE. We thank Nelson Avonce for help and support with the phylogenetic analysis. We thank Olivia Santana, Chandrasekar B. Rajeswari, Carlos A. González Chávez, and Noreide Nava (Instituto de Biotecnología, Universidad Nacional Autónoma de México [UNAM]) for technical support. We thank Eugenio López and Arturo Yañez (Instituto de Biotecnología, UNAM) for synthesis of oligonucleotides. We thank Guillermo Krotzsch (Instituto de Física, UNAM) for helpful support with the images and figures. We thank Alfonso Leija for technical support with the acetylene assays (CCG-UNAM). This study was funded by CONACYT 177744 and DGAPA-IN206415.
AUTHOR CONTRIBUTIONS
F.S., G.E.-N., and R.L. conceived the study. F.S., G.E.-N., C.Q., and M.-K.A. designed the experiments. G.E.-N., N.C.-M., and R.L. produced the vector constructs. A.H.-B., L.C., J.E.O., and G.E.-N. performed quantitative analysis of root hairs. M.-K.A. and G.E.-N. performed phenotypic analyses of mycorrhizal transgenic roots. F.S., X.A.-A., G.E.-N., and N.C.-M. performed the confocal microscopy image analysis. G.E.-N., N.C.-M., and A.B. analyzed the experimental data, produced the graphs, and conducted the RT-qPCR analysis. F.S., L.C., C.Q., and G.E.-N. wrote the article.
Glossary
- AM
arbuscular mycorrhizal
- IT
infection thread
- hai
h after inoculation
- dai
d after inoculation
- AIT
arrested IT
- wai
weeks after inoculation
- WGA
wheat germ agglutinin
- %RLC
percentage of root length colonization
Footnotes
Articles can be viewed without a subscription.
References
- Arthikala M.K., Montiel J., Nava N., Santana O., Sánchez-López R., Cárdenas L., Quinto C. (2013). PvRbohB negatively regulates Rhizophagus irregularis colonization in Phaseolus vulgaris. Plant Cell Physiol. 54: 1391–1402. [DOI] [PubMed] [Google Scholar]
- Blanco F.A., Meschini E.P., Zanetti M.E., Aguilar O.M. (2009). A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association. Plant Cell 21: 2797–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P., Genre A. (2008). Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends Plant Sci. 13: 492–498. [DOI] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., Morieri G., Mysore K.S., Wen J., Oldroyd G.E., Downie J.A., Murray J.D. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brearley C.A., Hanke D.E. (1992). 3- and 4-Phosphorylated phosphatidylinositols in the aquatic plant Spirodela polyrhiza L. Biochem. J. 283: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton W.J., Dilworth M. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas L., Lovy-Wheeler A., Kunkel J.G., Hepler P.K. (2008). Pollen tube growth oscillations and intracellular calcium levels are reversibly modulated by actin polymerization. Plant Physiol. 146: 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas L., Holdaway-Clarke T.L., Sánchez F., Quinto C., Feijo J.A., Kunkel J.G., Hepler P.K. (2000). Ion changes in legume root hairs responding to Nod factors. Plant Physiol. 123: 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M.D., Jurkevitch E., Poiret M., d’Aubenton-Carafa Y., Petrovics G., Kondorosi E., Kondorosi A. (1994). enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 13: 5099–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron D., Pingret J.L., Chabaud M., Journet E.P., Barker D.G. (2004). Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 136: 3582–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Lee Y., Jeon B.W., Staiger C.J., Lee Y. (2008). Phosphatidylinositol 3- and 4-phosphate modulate actin filament reorganization in guard cells of day flower. Plant Cell Environ. 31: 366–377. [DOI] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Soukup A., Napsucialy-Mendivil S., Jeknic Z., Ivanchenko M.G. (2009). The lateral root initiation index: an integrative measure of primordium formation. Ann. Bot. (Lond.) 103: 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom E.M., Ehrhardt D.W., Mitra R.M., Long S.R. (2002). Pharmacological analysis of nod factor-induced calcium spiking in Medicago truncatula. Evidence for the requirement of type IIA calcium pumps and phosphoinositide signaling. Plant Physiol. 128: 1390–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Navarrete G., Alvarado-Affantranger X., Olivares J.E., Guillén G., Díaz-Camino C., Campos F., Quinto C., Gresshoff P.M., Sánchez F. (2007). Fast, efficient and reproducible genetic transformation of Phaseolus spp. by Agrobacterium rhizogenes. Nat. Protoc. 2: 1819–1824. [DOI] [PubMed] [Google Scholar]
- Fu Y., Wu G., Yang Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152: 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier J.M., Simonsen A., D’Arrigo A., Bremnes B., Stenmark H., Aasland R. (1998). FYVE fingers bind PtdIns(3)P. Nature 394: 432–433. [DOI] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Faccio A., Barker D.G., Bonfante P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19: 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling D., Possart A., Cottier S., Klahre U., Kost B. (2006). Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18: 3519–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187. [DOI] [PubMed] [Google Scholar]
- Herman P.K., Emr S.D. (1990). Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 6742–6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiles I.D., et al. (1992). Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell 70: 419–429. [DOI] [PubMed] [Google Scholar]
- Hong Z., Verma D.P. (1994). A phosphatidylinositol 3-kinase is induced during soybean nodule organogenesis and is associated with membrane proliferation. Proc. Natl. Acad. Sci. USA 91: 9617–9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H., Penmetsa R.V., Terzaghi N., Cook D.R., Harrison M.J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 104: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: b-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale S.D., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142: 284–295. [DOI] [PubMed] [Google Scholar]
- Kametaka S., Okano T., Ohsumi M., Ohsumi Y. (1998). Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 273: 22284–22291. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. (2001). Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2: 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim Y.C., Fang C., Russell R.C., Kim J.H., Fan W., Liu R., Zhong Q., Guan K.L. (2013). Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Ohsumi Y. (1999). Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15: 1–32. [DOI] [PubMed] [Google Scholar]
- Koske R.E., Gemma J.N. (1989). A modified procedure for staining roots to detect V-A mycorrhizas. Mycol. Res. 92: 486–488. [Google Scholar]
- Kumagai H., Kinoshita E., Ridge R.W., Kouchi H. (2006). RNAi knock-down of ENOD40s leads to significant suppression of nodule formation in Lotus japonicus. Plant Cell Physiol. 47: 1102–1111. [DOI] [PubMed] [Google Scholar]
- Lee Y., Bak G., Choi Y., Chuang W.I., Cho H.T., Lee Y. (2008a). Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol. 147: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kim E.S., Choi Y., Hwang I., Staiger C.J., Chung Y.Y., Lee Y. (2008b). The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol. 147: 1886–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6: 463–477. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Talloczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577. [DOI] [PubMed] [Google Scholar]
- Lloyd C.W., Pearce K.J., Rawlins D.J., Ridge R.W., Shaw P.J. (1897). Endoplasmic microtubules connect the advancing nucleus to the tip of legume root hairs, but F-actin is involved in basipetal migration. Cell Motil. Cytoskeleton 8: 27–36. [Google Scholar]
- McGonigle T.P., Millers M.H., Evans D.G., Fairchild G.L., Swan J.A. (1990). A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115: 495–501. [DOI] [PubMed] [Google Scholar]
- Meijer H.J., Berrie C.P., Iurisci C., Divecha N., Musgrave A., Munnik T. (2001). Identification of a new polyphosphoinositide in plants, phosphatidylinositol 5-monophosphate (PtdIns5P), and its accumulation upon osmotic stress. Biochem. J. 360: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi N., et al. (2015). CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat. Cell Biol. 17: 1112–1123. [DOI] [PubMed] [Google Scholar]
- Montiel J., Nava N., Cardenas L., Sanchez-Lopez R., Arthikala M.K., Santana O., Sanchez F., Quinto C. (2012). A Phaseolus vulgaris NADPH oxidase gene is required for root infection by Rhizobia. Plant Cell Physiol. 53: 1751–1767. [DOI] [PubMed] [Google Scholar]
- Mylona P., Pawlowski K., Bisseling T. (1995). Symbiotic nitrogen fixation. Plant Cell 7: 869–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S., Kaneko T., Sato S., Saeki K. (2013). Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA 110: 17131–17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol.11: 252–263. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546. [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6: 763–775. [DOI] [PubMed] [Google Scholar]
- Patki V., Lawe D.C., Corvera S., Virbasius J.V., Chawla A. (1998). A functional PtdIns(3)P-binding motif. Nature 394: 433–434. [DOI] [PubMed] [Google Scholar]
- Peleg-Grossman S., Volpin H., Levine A. (2007). Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J. Exp. Bot. 58: 1637–1649. [DOI] [PubMed] [Google Scholar]
- Pingret J.L., Journet E.P., Barker D.G. (1998). Rhizobium nod factor signaling. Evidence for a g protein-mediated transduction mechanism. Plant Cell 10: 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Ma Z., Zhang L., Xing S., Hou X., Deng J., Liu J., Chen Z., Qu L.J., Gu H. (2007). Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17: 249–263. [DOI] [PubMed] [Google Scholar]
- Rostislavleva K., Soler N., Ohashi Y., Zhang L., Pardon E., Burke J.E., Masson G.R., Johnson C., Steyaert J., Ktistakis N.T., Williams R.L (2015). Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350: aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77: 817–837. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Ketelaar T., Esseling J.J., Emons A.M. (2005). Microtubules guide root hair tip growth. New Phytol. 167: 711–719. [DOI] [PubMed] [Google Scholar]
- Smith S.E., Smith F.A., Jakobsen I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack J.H., Emr S.D. (1993). Genetic and biochemical studies of protein sorting to the yeast vacuole. Curr. Opin. Cell Biol. 5: 641–646. [DOI] [PubMed] [Google Scholar]
- Stack J.H., DeWald D.B., Takegawa K., Emr S.D. (1995). Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Aasland R., Toh B.H., D’Arrigo A. (1996). Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 271: 24048–24054. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20: 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using likelihood, distance, and parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Vierstra R.D. (2005). Autophagic recycling: lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 8: 165–173. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D.H. (1994). Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349: 275–280. [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333: 169–174. [DOI] [PubMed] [Google Scholar]
- Valdés-López O., Arenas-Huertero C., Ramírez M., Girard L., Sanchez F., Vance C.P., Luis Reyes J., Hernández G. (2008). Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 31: 1834–1843. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. (2010). The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11: 329–341. [DOI] [PubMed] [Google Scholar]
- van Spronsen P.C., Gronlund M., Pacios Bras C., Spaink H.P., Kijne J.W. (2001). Cell biological changes of outer cortical root cells in early determinate nodulation. Mol. Plant Microbe Interact. 14: 839–847. [DOI] [PubMed] [Google Scholar]
- Vermeer J.E., van Leeuwen W., Tobena-Santamaria R., Laxalt A.M., Jones D.R., Divecha N., Gadella T.W. Jr., Munnik T. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47: 687–700. [DOI] [PubMed] [Google Scholar]
- Vessey J.K. (1994). Measurement of nitrogenase activity in legume root nodules: in defence of the acetylene reduction assay. Plant Soil 158: 151–162. [Google Scholar]
- Walker E.H., Perisic O., Ried C., Stephens L., Williams R.L. (1999). Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 402: 313–320. [DOI] [PubMed] [Google Scholar]
- Welters P., Takegawa K., Emr S.D., Chrispeels M.J. (1994). AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc. Natl. Acad. Sci. USA 91: 11398–11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D.J. (2008). Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19: 3290–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H.W., Chen X., Mei Y. (2009). Function and regulation of phospholipid signalling in plants. Biochem. J. 421: 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li S., Zhou L.Z., Fox E., Pao J., Sun W., Zhou C., McCormick S. (2011). Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 68: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Zhuang X., Wang H., Lam S.K., Gao C., Wang X., Cai Y., Jiang L. (2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.