Using the maize Bbm and Wus2 genes enhances transformation efficiency in maize and other monocots, broadens the genotype range, and permits transformation of mature seed-derived embryos and leaf segments.
Abstract
While transformation of the major monocot crops is currently possible, the process typically remains confined to one or two genotypes per species, often with poor agronomics, and efficiencies that place these methods beyond the reach of most academic laboratories. Here, we report a transformation approach involving overexpression of the maize (Zea mays) Baby boom (Bbm) and maize Wuschel2 (Wus2) genes, which produced high transformation frequencies in numerous previously nontransformable maize inbred lines. For example, the Pioneer inbred PHH5G is recalcitrant to biolistic and Agrobacterium tumefaciens transformation. However, when Bbm and Wus2 were expressed, transgenic calli were recovered from over 40% of the starting explants, with most producing healthy, fertile plants. Another limitation for many monocots is the intensive labor and greenhouse space required to supply immature embryos for transformation. This problem could be alleviated using alternative target tissues that could be supplied consistently with automated preparation. As a major step toward this objective, we transformed Bbm and Wus2 directly into either embryo slices from mature seed or leaf segments from seedlings in a variety of Pioneer inbred lines, routinely recovering healthy, fertile T0 plants. Finally, we demonstrated that the maize Bbm and Wus2 genes stimulate transformation in sorghum (Sorghum bicolor) immature embryos, sugarcane (Saccharum officinarum) callus, and indica rice (Oryza sativa ssp indica) callus.
INTRODUCTION
Since the first successful genetic transformations of major crop species such as soybean (Glycine max), cotton (Gossypium hirsutum), maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum), sorghum (Sorghum bicolor), and sugarcane (Saccharum officinarum), steady progress has been made in all facets of this process. For monocots, there has been a progression from particle gun transformation to that mediated by Agrobacterium tumefaciens, as well as refinements in tissue culture protocols and selection strategies (Shrawat and Lörz, 2006). Since the earliest reports of maize protoplast transformation (Rhodes et al., 1988; Shillito et al., 1989; Golovkin et al., 1993), the preferred target cells for transformation have gone from maize cells in liquid suspension (Fromm et al., 1990; Gordon-Kamm et al., 1990; Frame et al., 1994), through embryogenic callus (Walters et al., 1992; Wan et al., 1995), and finally to the transformation of scutellar cells of freshly isolated immature embryos (Ishida et al., 1996; Songstad et al., 1996; Frame et al., 2002). For other monocots such as barley (Hordeum vulgare), wheat, rice, and sorghum, immature embryos remain the predominant transformation target, despite reports over the years of successfully initiating tissue culture responses from explants other than immature embryos. These alternative explants have included (1) leaf bases to initiate callus cultures in maize (Wenzler and Meins, 1986), rice (Ramesh et al., 2009), and wheat (Yu et al., 2012); (2) immature inflorescences to initiate cultures in sorghum (Brettell et al., 1980), wheat (Maddock et al., 1982; Ozias-Akins and Vasil, 1982), rice (Chen et al., 1985; Rout and Lucas, 1996), barley (Wen et al., 1991), tritordeum (Barcelo et al., 1994), and maize (Pareddy and Petolino, 1990); (3) multiple-shoot cultures from apical meristems in maize (Zhong et al., 1992); and (4) regenerable callus from mature seed-derived embryos in rice (Lee et al., 2002).
Thus, successful transformation in monocot species has been achieved based on various explants and culture responses. However, all previous reports describing explants other than immature embryos have used these explants to initiate callus or meristem cultures that were subsequently transformed. There have been no previous reports of directly transforming differentiated cells in explants such as mature maize embryos or leaf segments and directly stimulating dedifferentiation and subsequent callus formation. Among reports of successful transformation of callus, the explants from which the callus was first derived include (1) immature tassels, immature ears, or anthers in maize (Cheng et al., 2004); (2) leaf bases from maize (Sidorov et al., 2006; Ahmadabadi et al., 2007); and (3) scutellum from mature seeds in rice (Chen et al., 1998; Dai et al., 2001). Alternatively, explants used to initiate proliferating meristem cultures for subsequent transformation have included maize apical or nodal meristems (Zhong et al., 1996; Zhang et al., 2002) or mature seeds in species such as rice (Cho et al., 2004), oat (Avena sativa; Cho et al., 1999), orchardgrass (Dactylis glomerata; Cho et al., 2000a), Kentucky bluegrass (Poa pratensis; Ha et al., 2000), and fescue (Festuca sp; Cho et al., 2000b). Regardless of the culture type, all of these reports have relied on the manipulation of exogenous hormones in the culture media to produce either embryogenic callus or multiple meristems for use as the transformation target.
As an adjunct to altering hormone levels and nutrients in the culture medium, strategies have evolved to use expression of non-plant growth-stimulating transgenes to improve plant transformation (Ebinuma et al., 1997, 2005; Sugita et al., 2000; Endo et al., 2002; Gordon-Kamm et al., 2002). In addition, several reports have described the production of embryo-like structures or somatic embryos on various explants in response to overexpression of plant morphogenic genes such as LEAFY COTYLEDON1 (Lotan et al., 1998), Lec1 (Lowe et al., 2002), LEAFY COTYLEDON2 (Stone et al., 2001), WUSCHEL (WUS; Zuo et al., 2002), and BABY BOOM (BBM; Boutilier et al., 2002) as well as enhanced regeneration (Srinivasan et al., 2007; Deng et al., 2009). However, none of these studies on various dicotyledonous species using morphogenic genes reported an increase in transformation frequency.
Similar to the observations for WUS and BBM expression in dicots, we report here that overexpression of the maize Wus2 (Nardmann and Werr, 2006) and Bbm genes in monocots after Agrobacterium-mediated transformation of immature embryos resulted in a growth stimulation of embryogenic tissue. Importantly, and in contrast to the dicot literature, this embryogenic response enhanced the recovery of transgenic plants particularly in recalcitrant or marginally transformable maize, rice, sorghum, and sugarcane varieties. Furthermore, expression of Wus2 and Bbm enabled direct Agrobacterium-mediated transformation of mature seed-derived embryo axes or leaf segments, without an intervening callus or meristem culture step.
RESULTS
Early Growth Phenotypes Produced by Transient Expression of Bbm and Wus2
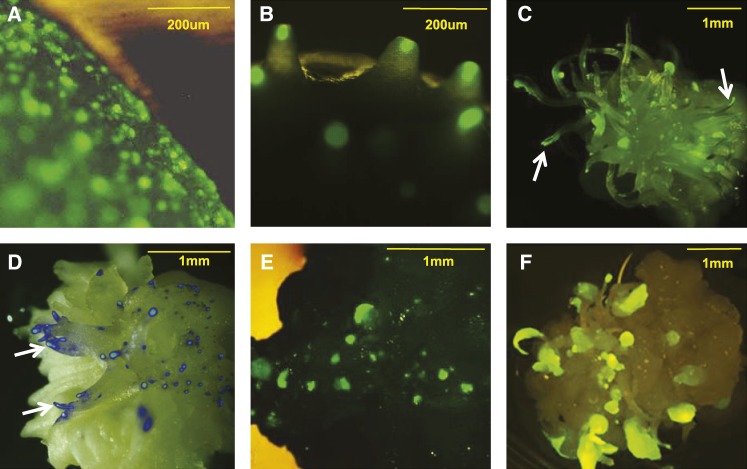
When scutellar cells of 18 d after pollination (DAP) embryos of inbred PH581 were particle bombarded with a fluorescent protein expression cassette, only single fluorescing cells were observed after 7 d (Figure 1A). Cobombardment of Wus2 and the fluorescent protein expression cassette resulted in foci of fluorescing cells that remained small and confined to the tips of protrusions. These foci appeared on the surface of the scutellum (Figure 1B) and continued to elongate over time, consistent with non-cell-autonomous activity of WUS protein (Figures 1C and 1D). Cobombardment for expression of Bbm and moGFP enhanced growth in a cell-autonomous manner, with only the cells receiving and expressing the transgenes being stimulated to grow (Figure 1E). When constructs for expression of Bbm, Wus2, and moGFP were cobombarded, growth stimulation was most pronounced. Numerous, rapidly growing cell clusters exhibiting a mixture of the Bbm and Wus2 growth phenotypes were observed on the surface of each scutellum (Figure 1F). When transgenic callus harboring Oleosinpro:Wus2 was allowed to grow, the callus exhibited a chimeric phenotype, with large sectors of nontransgenic callus growing between the transgenic sectors. In addition, continued expression of Wus2 behind this strong callus promoter often led to callus necrosis, and when regeneration was attempted, only nontransgenic plants were produced. For this reason, any further experimentation using the combination of Bbm and Wus2 was done using the strong maize Ubiquitin promoter (Ubi; Christensen et al., 1992) driving Bbm (which was tolerated in callus), while Wus2 was expressed using the weak Agrobacterium-derived nopaline synthase (nos) promoter (An, 1986).
Figure 1.
Early Growth Response at 7 d Showing Morphogenic Stimulation of Non-Cell-Autonomous Wus2 and Cell-Autonomous Bbm Gene Delivery into the Scutellum of 18-DAP Embryos.
(A) Introduction of Ubipro:moGFP:pinII alone (control). Single green fluorescent cells were observed on the scutellum surface.
(B) to (D) nospro:Wus2:pinII cobombardment with the moGFP cassette. Foci of fluorescing cells appeared to enlarge slightly but remained confined to the tips of elongating protrusion (B) or formed a file of fluorescing cells from the tip as the protrusions continued to elongate (arrows in [C] and [D]).
(D) A light micrograph and epifluorescence micrograph from the cyan filter set were superimposed onto each other with no other changes made to the data.
(E) Ubipro:Bbm:pinII cointroduced with moGFP. Green fluorescent multicellular clusters were observed.
(F) Cobombardment of the Wus2, Bbm, and moGFP expression cassettes. High growth stimulation was observed with a mixed phenotype exhibiting attributes from both Bbm and Wus2.
Immature Embryo Transformation Using Agrobacterium
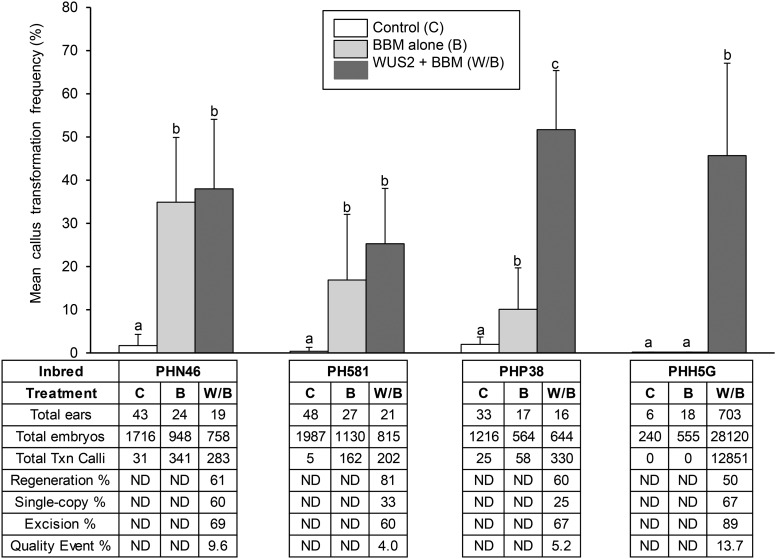
The effect of Bbm and Wus2 on transformation frequencies after Agrobacterium infection (transgenic plant recovery based on plants per starting embryo or explant) was evaluated in a number of Pioneer maize inbred lines, including both stiff-stalk and non-stiff-stalk lines. These inbred lines were chosen based on commercial importance rather than culture response. For each treatment, immature embryos were harvested from multiple ears (replicates) and infected with Agrobacterium strain LBA4404; transformation data were tabulated as the number of callus transformation events for the embryos from each ear, with means and standard deviations then being calculated (with total sample sizes ranging from a minimum of 240 to more than 28,000 embryos per treatment). Baseline transformation frequencies using a control vector were low or nonexistent depending on the line, ranging from 0% for inbred PHH5G to 2.0% for PHP38 (Figure 2). For inbred lines PHN46, PH581, and PHP38, transformation using nospro:Wus2 plus Ubipro:CYAN (with no Bbm) produced low frequencies of chimeric callus, which based on cyan fluorescence contained a mixture of transgenic and nontransgenic sectors that produced only nontransgenic plants upon attempted regeneration. Thus, for this treatment, event numbers and frequencies were not tabulated because no transgenic plants could be recovered.
Figure 2.
Ectopic Expression of Bbm and Wus2 Increased Transformation Frequencies in Four Maize Inbreds.
Immature embryos from inbreds PHN46, PH581, PHP38, and PHH5G were transformed using Agrobacterium in which the T-DNA contained no Bbm or Wus2 (pPHP24600), Bbm alone (pPHP24955), or Bbm and Wus2 (pPHP35648). For each inbred, significant differences between treatments are indicated by letter designations determined using penalized logistic regression analysis (P = 0.05). ND, not determined.
Each inbred line responded differently to either Ubipro:Bbm alone or Ubipro:Bbm plus nospro:Wus2 (Figure 2). When inbred PHN46 was transformed with Bbm alone, there was a substantial increase in callus transformation frequency from 1.7% in the control treatment to 34.9%, while addition of Wus2 resulted in a modest additional increase to 38.0%. In inbred PH581, Bbm alone elicited an increase in callus transformation as in PHN46, from 0.4% without Bbm or Wus2 to 16.9% with Bbm alone, and the combination of Bbm plus Wus2 increased transformation frequency further to 25.3%. Inbred PHP38 produced a different trend in response to these two treatments; Bbm alone resulted in the lowest mean transformation frequency of 10.1% (relative to the Bbm response observed in PHN46 and PH581), but when Bbm and Wus2 were used together, the transformation frequency increased to 51.7%. As all three of these inbred lines produce low levels of compact, Type I callus even in the absence of transformation, all of the above results were generated while applying our standard bialaphos selection (3 mg/L). The addition of Bbm and/or Wus2 increased overall callus transformation rates for PHN46, PH581, and PHP38, with the callus morphology retaining an embryogenic phenotype.
Inbred PHH5G produced a distinctly different response to Bbm and Wus2 relative to the other three inbred lines and required no chemical (i.e., bialaphos) selection. Using the control vector, PHH5G produced no transgenic events following a brief initial swelling of the tissue with no apparent cell divisions (Figure 2). In contrast to the other three inbred lines, the addition of the individual nospro:Wus2 or Ubipro: Bbm expression cassettes also produced no transformed callus. However, when Ubipro:Bbm plus nospro:Wus2 was used, transgenic callus was produced at a mean frequency of 45.7% (12,851 independent events from 28,120 immature embryos; Figure 2). Transgenic callus grew vigorously and exhibited a mixed morphology of type I and type II embryogenic callus, defined as having either compact or friable growth patterns (Armstrong and Green, 1985). For all four inbred lines, transformation frequencies were increased by the use of the morphogenic genes, but callus morphology and regeneration capacity were still dependent on hormones in the medium, such as the auxin 2,4-D. In the absence of hormones, regenerable callus was not recovered in response to expression of Bbm and Wus2.
To determine how these two morphogenic genes would affect transformation responses in a larger panel of inbred lines, an additional set of 50 commercially important Pioneer inbred lines (spanning heterotic groups) was screened for transformation response by Agrobacterium-mediated delivery of Ubipro:Bbm plus nospro:Wus2 into immature embryos and then scored for the percentage of embryos that produced transgenic calli. Of these 50 inbred lines, 17 inbred lines did not produce transgenic events, 16 produced transgenic calli at less than a 1% frequency, 12 produced transgenic calli at a frequency between 1% and 10%, and 5 produced transgenic calli at a frequency above 10%. Thus, using the same protocol and plasmid, with no adjustments in medium formulations, 33 of 50 inbred lines produced transgenic calli. In addition to the increased rate of transgenic callus initiation for all the above inbred lines, the calli grew more rapidly with Ubipro:Bbm plus nospro:Wus2 but again overall callus morphology was dictated by the response of each inbred to the hormones in the medium (i.e., callus morphology for all responding inbred lines was variable between type I and a mix of type I and type II).
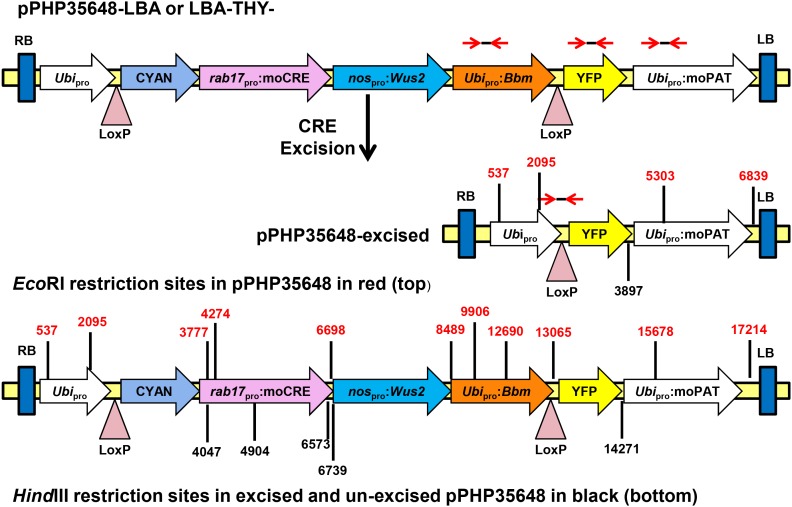
An important requirement for plant regeneration is the removal of the Bbm and Wus2 expression cassettes before attempts to regenerate plants. Constructs were designed with loxP sites (in the same orientation) flanking the entire sequence containing the Wus2, Bbm, and CRE expression cassettes. To drive excision, the rab17 promoter (Vilardell et al., 1990, 1991), a strong drought-inducible maize promoter, was used to drive expression of the CRE recombinase gene. Using desiccation to induce the rab17 promoter and CRE expression resulted in the removal of the sequences between the loxP sites (Figure 3). Failure to excise Bbm and Wus2 resulted in aberrant phenotypes, including thick, short roots in plantlets (a phenotype associated with ectopic Bbm expression) and if allowed to grow further, plants were stunted, twisted, and usually sterile at maturity. Thus, excision of the morphogenic genes was necessary to produce healthy, fertile T0 transgenic plants. As with other in vitro manipulations of maize inbred lines, differences were observed in the frequency of regeneration, the efficiency of CRE/Bbm/Wus2 excision, and the frequency of single-copy T0 plants (Figure 2). Following a 3-d desiccation treatment of calli on dry filter paper, regeneration frequencies for callus varied from 50% in PHH5G to 81% in PH581, single-copy frequency ranged from 25% in PHP38 to 67% in PHH5G, and excision frequency for single-copy T0 plants ranged from 60% in PH581 to 89% in PHH5G. While PCR analysis indicated that CRE-mediated excision occurred after the 3-d desiccation, this interpretation required validation through DNA gel blot hybridization analysis.
Figure 3.
nospro:Wus2:pinII Plus Ubipro:Bbm:pinII Containing T-DNA Used for DNA Gel Blot Analysis in the T1 Generation.
Red opposing arrows indicate approximate locations of qPCR primers for YFP and moPAT for copy number determinations and PCR primers to detect the absence of the Bbm cDNA sequence and the formation of the newly formed junction across the one remaining loxP site after excision. Locations of EcoRI (red numbers on top) and HindIII (black numbers on bottom) restriction sites used in DNA gel blot analysis are also shown.
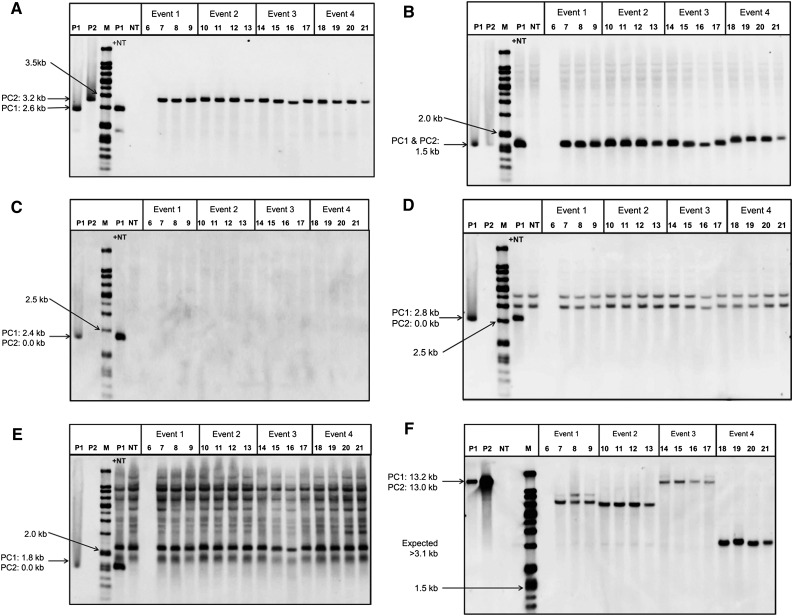
For DNA gel blot evaluation, leaf samples were taken from four T1 plants arising from each of 47 randomly chosen events (188 total plants) from inbred PHH5G that had been characterized as single copy for the remaining YFP and moPAT transgenes (after complete excision of the CRE, Bbm, and Wus2 transgenes; Figure 3), and the leaf samples were processed for DNA gel blot analysis. Results for 187/188 T1 plants tested confirmed the conclusions based on PCR analysis, and an example set of DNA gel blots for plants arising from four transformation events is shown in Figures 4A to 4F. EcoRI restriction digestion of plasmids pPHP35648 and pPHP35648-excised (pPHP35648 that had undergone in vitro CRE-mediated excision to produce the final excision product) produced insert integrity data for all of the T-DNA genes (CRE plus Bbm and Wus2 plus YFP plus moPAT), while excised HindIII digestion yielded the copy numbers of YFP (data not shown) and moPAT. Plasmids pPHP35648 (Figure 4, lane P1) and pPHP35648-excised (Figure 4, lane P2) were used as controls. Since the sites for EcoRI enzyme were known in the T-DNA fragment, a single specific band size corresponded to the intact insert, while the expected fragment size of genes upon HindIII digestion varied from event to event.
Figure 4.
Representative DNA Gel Blot Analysis of Four Transgenic PHH5G Inbred T1 Plants Derived from Each of Four Independent Transgenic Events.
For all blots, P1 = pPHP35648 plasmid, while P2 = pPHP35648-excised (pPHP35648 after in vitro CRE-mediated excision). M, molecular weight ladder; P1 + NT, wild-type PHH5G genomic DNA spiked with control plasmid; NT, genomic DNA from the nontransgenic, wild-type plant. PC1, positive control unexcised; PC2, positive control excised. For event 1, DNA was inadvertently left out of lane 6.
(A) T1 plant samples digested with EcoRI and probed for YFP; a single 3.2-kb band indicates intact gene sequence.
(B) T1 plant samples digested with EcoRI and probed for moPAT; a single 1.5-kb band indicates intact gene sequence.
(C) T1 plant samples digested with EcoRI and probed for CRE; the absence of a single 2.4-kb band indicates that CRE had been excised.
(D) T1 plant samples digested with EcoRI and probed for Bbm; two bands result from hybridization to the endogenous genomic Bbm sequence and were also present in the two nontransgenic control lanes (labeled “NT” and “P1 plus NT”). The absence of a single 2.8-kb band indicates that Bbm had been excised.
(E) T1 plant samples digested with EcoRI and probed for Wus2; numerous bands resulting from hybridization to the endogenous genomic Wus2 sequence were also present in the two nontransgenic control lanes (labeled “NT” and “P1 plus NT”). The absence of a single 1.8-kb band indicates that Wus2 had been excised.
(F) T1 plant samples digested with HindIII and probed for moPAT; single bands larger than 3.1 kb and differing in size between the lines derived from the four events indicate that all four events were independent and single copy.
As expected, a single band corresponding to 3.2 and 1.5 kb sizes, respectively, in blots probed for YFP and moPAT, indicated an intact gene sequence (Figures 4A and 4B, respectively). Diagnostic bands on the EcoRI-digested DNA gel blots probed for CRE (2.4 kb), Bbm (2.8 kb), and Wus2 (1.8 kb) would be indicative of nonexcision. The plasmid control pPHP35648 (Figure 4, lane P1) produced the appropriately sized single intact bands for CRE, Bbm, and Wus2, while pPHP35648 excised (Figure 4, lane P2) resulted in no bands for these genes after excision. As a negative control, nontransgenic DNA (Figure 4, lane NT) alone or spiked with plasmid DNA (Figure 4, lane P1+NT) was used in this study. As expected, no hybridizing band corresponding to CRE was observed after excision (Figure 4C). While bands from the endogenous Bbm or Wus2 family members hybridized at different sizes, the unique band corresponding to the cDNAs at 2.8 and 1.8 kb in the control lanes were clearly discernable (Figures 4D and 4E, respectively) and were not observed in the regenerated progeny plants, as expected after CRE-mediated excision of the morphogenic genes.
In vector pPHP35648-derived PHH5G transformants, a HindIII site between YFP and moPAT provided copy number information only for lines in which excision occurred. A single band of >3.9 kb with the YFP probe denoted a single copy of the gene, while for moPAT, a single band at >3.1 kb indicated a single copy. For all lines, the bands for both YFP (data not shown) and moPAT (Figure 4F) were consistent with single-copy integration.
Out of the 188 plants sampled for DNA gel blot analysis, there was the exception of one plant derived from one transformation event (Figure 4, lane 6 for Event 1) for which it appeared that the DNA from this plant may not have been loaded onto the gel, since there were no bands on any of the blots, including those probed for Bbm and Wus2 in which endogenous bands should have been visible. For the remaining three plants arising from this transformation event and for all four plants from the remaining 46 events tested, DNA gel blot results were consistent and showed that (1) the moPAT gene was present in all T0 plants tested and, as expected, the band size did not change after excision (compared with the control lanes; Figure 4B); (2) YFP was present in all plants tested and the band size was consistent with excision having occurred (data not shown); and (3) the CRE, Bbm, and Wus2 transgenes were no longer present in the T0 plants (Figures 4C to 4E, respectively). See Figure 3 for details regarding digestion patterns for EcoRI and HindIII used for this DNA gel blot analysis.
All of the above (initial transformation, regeneration, single copy, and excision) contribute to the overall efficiency of recovering only perfect single-copy, completely excised T0 plants (quality events), which were then used for further characterization. Taking all these criteria into consideration and based on the starting number of immature embryos used for transformation, the frequency of single-copy, completely excised transformants that generated fertile T0 plants in the greenhouse was 4.0, 5.2, 9.6, and 13.7% for inbred lines PH581, PHP38, PHN46, and PHH5G, respectively (Figure 2). In addition, Agrobacterium plasmid backbone (outside the T-DNA) was detected in 20% of these transformants and they were discarded, resulting in a final event frequency of 3.2, 4.2, 7.7, and 11.0% for these four respective inbred lines. For all four inbred lines, “single-copy and excised” plants in the greenhouse were healthy and both male and female fertile.
Transformation of Mature Seed Embryo Sections
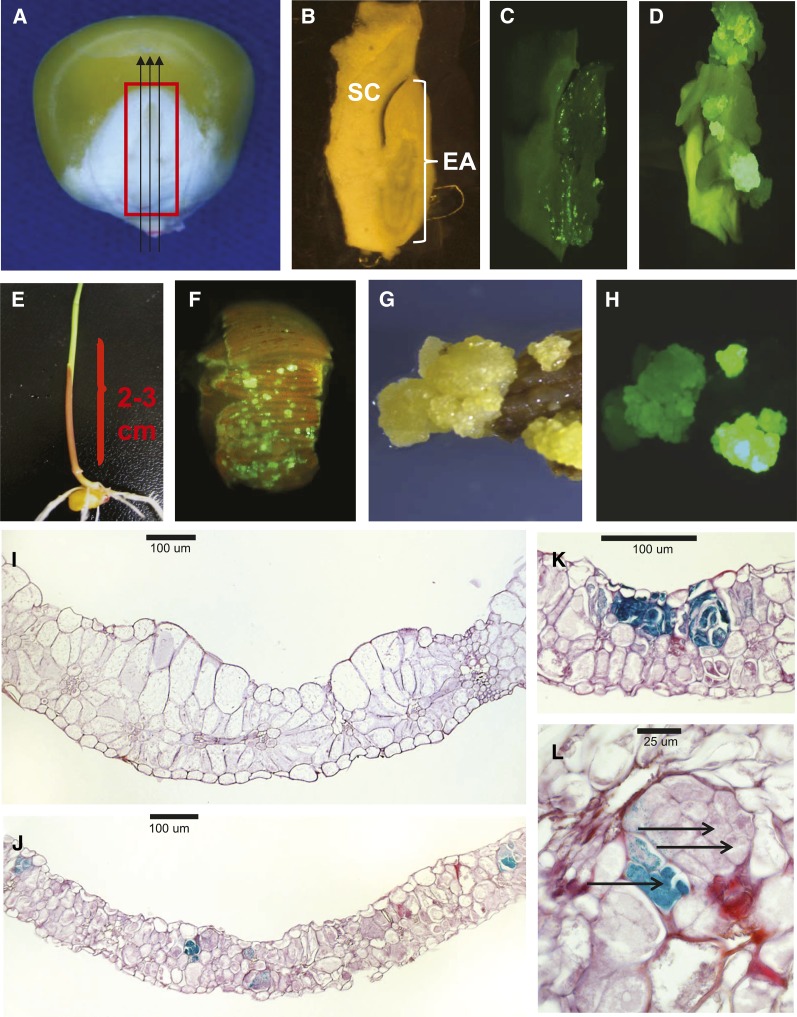
In addition to demonstrating that overexpression of Wus2 and Bbm enabled transformation of immature embryos, we evaluated similar vectors for their ability to transform alternative explants from maize. In one series of experiments, sections of embryo axes from mature maize seed (Figures 5A and 5B) were infected with Agrobacterium strain LBA4404 (THY-) containing pPHP54733 (Supplemental Figure 1E), which delivered T-DNA into embryo axis cells of inbred PH0AZ, resulting in a distribution of individual cells transiently expressing the ZS-GREEN1 fluorescent protein (Figure 5C). After 3 weeks in culture with no selection, multicellular fluorescent tissue was observed growing from what had originally been the embryo axis (Figure 5D) with multiple embryogenic events frequently observed from the same original mature embryo section.
Figure 5.
Transformation of Alternative Explants Derived from Maize Seed or Seedlings.
(A) to (D) To prepare mature embryo sliced for transformation, the mature embryo was cut out of the kernel (red box in [A]), and parallel cuts were then made to produce 300- to 400-μm-thick sections (black arrows) in which the scutellum (SC) and embryo axis (EA) could be discerned (B). After Agrobacterium-mediated transformation (strain LBA4404 with pPHP54733), transient ZS-GREEN1 expression was observed predominantly in the embryo axis (C), and after 2 to 3 weeks, callus could be observed growing from this region (D).
(E) to (H) To prepare leaf tissue for transformation, a segment of the leaf whorl was removed from individual PHH5G seedlings (E) and cut into ∼1-mm segments. After Agrobacterium transformation (strain AGL1 with pPHP54733) and culture for 7 to 14 d, a dispersed pattern of multicellular cell clusters was clearly observed emerging from the leaf surface (F), and with continued culture multiple independent events were often observed arising from the surface of a single leaf segment under light microscopy (G) or under epifluorescence illumination (H).
(I) to (L) Early observation of ectopic cell division patterns in the leaf were made by comparing cross sections of leaves from a nontransformed seedling leaf (I) with those of leaves transformed via Agrobacterium strain AGL1 delivering a T-DNA containing BSVpro:GUS:pinII + nospro:Wus2:pinII +Ubipro:Bbm:pinII (pPHP46344), which revealed numerous GUS-staining multicellular foci (J) 7 d after infection. Within this population of growing multicellular foci, there were two types of division patterns: with all dividing cells within the cluster transformed based on GUS staining (K) or with GUS-staining cells (arrow in [L]) appearing to stimulate ectopic cell division in neighboring cells (double arrow in [L]).
To assess the reproducibility of this method, 25 separate experiments were conducted using four to six mature embryo sections for each kernel, with each experiment initiated on different days using a fresh preparation of Agrobacterium and freshly prepared kernel cross sections (as shown in Figure 5B). From 773 kernels (and 3960 sections) across 25 experiments, 608 embryogenic calli were recovered 10 weeks after transformation initiation, with a mean callus transformation efficiency per section of 16.5% ± 8.2% on a per-experiment basis (or 87.5% ± 46.6% based on starting seed). Of these 608 calli, 590 were moved onto regeneration medium, with 240 producing one or more plants, for a regeneration frequency of 40.7%. Using all T0 plants regenerated from the 240 calli (including calli for which multiple plants were regenerated), 373 regenerated plants were sampled for PCR analysis, with 154 (41.3%) being single-copy for the T-DNA (with no Agrobacterium backbone) and with 17.4% showing complete excision of CRE, Bbm, and Wus2. T0 plant morphology was similar to that of nontransgenic PH0AZ plants grown in the greenhouse, and the T0 plants exhibited vigorous growth and good fertility. Among the single-copy, excised PH0AZ T0 plants generated in this set of experiments, a subset of 58 were evaluated for seed set, producing a mean of 277 (±159) kernels/ear after self-pollination (n = 35) and a mean of 209 (±189) kernels/ear when wild-type pollen was carried to the T0 ear (n = 23). These seed set values were comparable to those of T0 transgenic plants produced using immature embryos as the starting explant for this inbred.
Transformation of Seedling-Derived Leaf Segments
Agrobacterium-mediated transformation was used to assess the morphogenic plasticity of maize leaf cells in response to delivery of the Ubipro:Bbm and nospro:Wus2 expression cassettes. Agrobacterium strain AGL1 (a more virulent strain than LBA4404) containing pPHP54733 (pPHP54733-AGL1; Supplemental Figure 1E) delivered T-DNA into leaf cells from 15- to 16-d-old seedlings (Figure 5E) in Pioneer inbred PHH5G, producing a scattered distribution of individual dividing leaf cells expressing the ZS-GREEN1 fluorescent protein (Figure 5F). With continued culture, multiple embryogenic events were often observed from the same original explant (Figures 5F to 5H). In the example shown in Figures 5F to 5H, although there were clearly multiple transgenic multicellular clusters growing from the same explant, we made no attempt in these experiments to separate these independent colonies. Instead, we let them grow together and scored the callus mass growing from a single explant reflecting a single transgenic event (realizing that our calculations of transformation frequencies would substantially underestimate the actual frequency). In 28 separate experiments (on separate days), 334 seedlings were used for explant preparation (12.4 ± 5 seedlings/experiment) and Agrobacterium transformation. From these experiments, 151 embryogenic calli were recovered 10 weeks after transformation initiation, giving a transformation frequency (on a per seedling basis) of 45%. Of these calli, 46% regenerated to produce T0 plants, of which 33% were single copy. Of the single-copy T0 plants, 45% produced qPCR results that indicated complete excision of CRE and the morphogenic genes. Thus, from 334 seedlings used to prepare leaf segments for transformation, a final total of 10 single-copy, morphogenic gene-minus (excised) T0 plants were produced. These T0 plants still contained Ubipro:ZS-GREEN1:Sb-ACTIN 3′ sequence and Sb-Ubipro:PMI:Sb-Ubi 3′ sequence expression cassettes left behind after morphogenic gene excision. All were phenotypically normal, fertile, and produced transgenic T1 progeny at the expected Mendelian frequencies. As mentioned above, only one T0 plant was regenerated from each callus mass derived from a leaf section. This was done by simply regenerating the healthiest, fastest growing plantlet from a single mass of callus. With the realization that the callus arising from a single explant was likely a mixture of independent events, we could still potentially increase our transformation frequencies even further by separating independent events derived from a single section at an early stage in the experiment.
In separate experiments, we were interested in determining which cells in the leaf were being transformed and contributing to callus formation. To assess which leaf cells were beginning to divide after transformation, observations of transgenic cell(s) in leaf cross section were required, for which the GUS reporter gene was more suitable than fluorescent proteins. For these experiments, leaf segments were transformed with pPHP46344-AGL1, a vector containing the GUS gene driven by the Banana Streak Virus promoter (Supplemental Figure 1D). Seven days after starting the transformation process, no GUS-expressing cells or foci were observed in wild-type (nontransformed) leaf sections (Figure 5I). However, 7 d after transformation with T-DNA containing the Bbm, Wus2, and GUS expression cassettes, GUS-expressing multicellular foci were numerous and appeared to derive predominantly from mesophyll cells (Figure 5J). Interestingly, two distinct types of early growth response were observed. In the first, dividing mesophyll-derived cells within a cluster all appeared to express GUS (Figure 5K). In the second type of response, clusters of non-GUS-stained but clearly dividing cells were observed adjacent to GUS-staining cells (Figure 5L), indicating ectopic, non-cell-autonomous WUS2 activity.
Transformation of Other Monocot Crops
The combination of Ubipro:Bbm plus nospro:Wus2 expression cassettes was also tested in other monocot crops. Using immature embryos harvested from the same plant and transformed on the same day, the transformation frequency in sorghum cultivar Tx430 was increased from a control level of 1.9 to 18.3% when the morphogenic genes were present in the T-DNA (Table 1). The sorghum transformation frequencies reported here are low compared with those published by Wu et al. (2014), as this work was performed much earlier than that of Wu et al. (2014). These transgenic sorghum lines (produced using our desiccation-induced maize rab17pro:CRE excision method as described earlier) were characterized by PCR and verified to contain the marker genes and no longer to contain Bbm, Wus2, and the CRE recombinase gene.
Table 1. Use of Wus2 and Bbm to Enhance Transformation of Sorghum, Rice, and Sugarcane.
| Species | Vector | Developmental Gene Expression Cassettes | Marker Expression Cassettes | No. Explants Treated | No. Txn Events | Txn Freq. (%) |
|---|---|---|---|---|---|---|
| Sorghum bicolor TX430 | pPHP32371 | Ubipro:Bbm:pinII + nospro:Wus2:pinII | Ubipro:CYAN:pinII + Ubipro:moPAT:pinII | 393 immature embryos | 72 | 18 |
| Sorghum bicolor TX430 | pPHP32269 | None | Ubipro:moPAT-YFP: pinII + Ubipro:PMI:pinII | 376 immature embryos | 7 | 2 |
| Oryza sativa (ssp indica IRV95) | pPHP57324 | Ubipro:OsBbm:pinII + nospro:Wus2:pinII | CaMV 35Spro:YFP-HYG:pinII | 300 calli | 130 | 43 |
| Oryza sativa (ssp indica IRV95) | pPHP66801 | Ubipro:Bbm:pinII + nospro:Wus2:pinII | CaMV 35Spro:YFP-HYG:pinII | 300 calli | 80 | 27 |
| Oryza sativa (ssp indica IRV95) | pPHP48195 | None | CaMV 35Spro:HYG:nos3′ + Ubipro:CYAN:pinII | 300 calli | 8 | 3 |
| Oryza sativa (ssp indica IRV95) | pPHP56800 | Ubipro:Bbm:pinII + nospro:Wus2:pinII | Ubipro:CYAN:pinII + Ubipro:PMI:pinII | 300 calli | 45 | 15 |
| Oryza sativa (ssp indica IRV95) | pPHP49754 | None | Ubipro:CYAN:pinII + Ubipro:PMI:pinII | 300 calli | 0 | 0 |
| Saccharum officinarum (v. CP01-1372) | pPHP54561 | Ubipro:Bbm:pinII + nospro:Wus2:pinII | Ubipro:YFP:pinII + Ubipro:moPAT:pinII | 20 calli | 177 | 885 |
| Saccharum officinarum (v. CP01-1372) | pPHP35648 | Ubipro:Bbm:pinII + nospro:Wus2:pinII | Ubipro:CFP:pinII + Ubipro:moPAT:pinII | 40 calli | 109 | 273 |
| Saccharum officinarum (v. CP01-1372) | pPHP24600 | None | CaMV 35Spro:PAT:35S 3′ + Ubipro:DsRED:pinII | 48 calli | 1 | 2 |
Transformation of sorghum immature embryos, indica rice established callus explants, and sugarcane established callus explants using Agrobacterium strain AGL1 in sugarcane and LBA4404 in rice and sorghum that delivered either (1) T-DNAs containing Ubipro:Bbm and nospro:Wus2 or (2) T-DNAs with marker genes but no Bbm or Wus2 (control vectors). Sorghum and rice transformation frequencies based on phenotype (both resistance to a selectable marker and fluorescence) and qPCR, while sugarcane frequencies were based on fluorescence. For rice transformation experiments, one experiment was performed with the rice ZmBbm ortholog OsBbm + the Zea mays Wus2 (pPHP57324). For all other experiments, the Zea mays Bbm and Wus2 genes were used. Transformation is abbreviated Txn in the column labels.
For the rice indica variety IRV95, after transformation of callus with Agrobacterium strain LBA4404 delivering a T-DNA containing Ubiquitin-driven PMI and CYAN genes, no transgenic events were recovered. However, when the maize Bbm and Wus2 expression cassettes were also in the T-DNA, the transformation frequency was 15% (Table 1). Using a hygromycin resistance gene (HYG) alone for selection in IRV95 resulted in recovery of transgenic calli at a frequency of 3%, and the addition of Ubipro:Bbm plus nospro:Wus2 expression cassettes (containing either the rice or maize Bbm orthologs along with the maize Wus2) plus a HYG-YFP fusion cassette resulted in transformation frequencies of 43 and 27% in two experiments, respectively. The addition of morphogenic genes also improved Agrobacterium-mediated transformation of callus cells (callus clusters between 1 and 2 mm in diameter) in the sugarcane variety CP01-1372 (Table 1). In the control plasmid containing CaMV 35Spro:PAT and Ubipro:DsRED, the transformation frequency was around 2%, while using constructs containing the morphogenic genes in addition to PAT and a fluorescent marker, the transformation frequencies (based on bialaphos resistance and YFP fluorescence) increased to 273 and 885% in two separate treatments (these frequencies were derived by scoring all the individual calli recovered from the original number of calli treated with Agrobacterium, for example, 177 calli recovered from 20 starting calli exposed to Agrobacterium = 885%).
The putative transformed calli recovered from sorghum, rice, and sugarcane were not characterized by DNA gel blot analysis, so they could not be confirmed to be independent events. For all three of the above species, the results were based on the number of recovered calli exhibiting the transgenic phenotype (resistance and fluorescence) in each treatment. Transgenic calli from all three crops successfully went through excision of morphogenic genes and regeneration of transgenic plants, as measured by PCR analysis in T0 plants. For example, in sugarcane variety CP01-1372, a subset of 53 callus events transformed with pPHP54561-LBA were subjected to the desiccation-induced excision treatment, and of these, 36 (68%) were determined to have excised Wus2, Bbm, and CRE, based on PCR analysis at the T0 plant level.
DISCUSSION
The concept of using growth-stimulating genes, such as the ipt gene from Agrobacterium, to stimulate morphogenesis in plants has been in the literature since the late 1980s (Smigocki and Owens, 1988). The utility of using such growth-stimulating genes to aid in the recovery of transgenic T0 plants was elegantly demonstrated using genes that stimulate cytokinin production in dicots by Ebinuma et al. (1997). Unfortunately, because good venues for publishing negative results do not exist, we must assume that researchers attempting to repeat this strategy in monocots met with poor results. For example, there have only been a few reports of using growth-stimulating genes to enhance maize transformation (in High Type-II germplasm; see Armstrong and Green, 1985), using either the wheat dwarf virus RepA, which stimulates cell division (Gordon-Kamm et al., 2002), or Lec1, a gene involved in embryo development (Lowe et al., 2002). While both genes stimulated growth in transgenic callus sectors in Hi-II germplasm, there was minimal impact on the transformation of recalcitrant inbred lines. By contrast, in this study the influence of Bbm and Wus2 on transformation of a wide variety of inbred lines has been dramatic, and visualization of this growth stimulation can be observed after just a few days.
Since the first report of using maize immature embryos (harvested 9 to 12 DAP) for Agrobacterium transformation (Ishida et al., 1996), maize transformation approaches have focused on this explant. An important attribute of immature embryos contributing to transformation is that many maize genotypes produce either embryogenic or organogenic culture responses on hormone-containing media (O’Connor-Sánchez et al., 2002). However, maize embryos removed from the kernels at 18 DAP will simply germinate if placed on hormone-free medium (Weymann et al., 1993) and will not produce an organized callus response when placed on medium with hormones. Typically, maize embryos past 15 DAP will swell slightly and produce irregular patches of cells that most closely resemble root cells (large, elongated, and vacuolated) that do not continue to divide. Thus, when the Ubipro:moGFP plasmid (or Ubipro:CYAN) along with a second control plasmid (CaMV 35Spro:GUS) were cobombarded into the scutellum of 18 DAP embryos, numerous GFP-expressing single cells could be seen with no signs of cell division after 7 d on nonselective culture medium (Figure 1A). By contrast, ectopic expression of Bbm in scutellar cells of 18 DAP embryos produced multiple foci on the scutellum surface in which cell proliferation had been stimulated (Figure 1E), and it appeared that all the cells within each developing multicellular cluster were uniformly expressing the fluorescent protein. This same type of growth stimulation was observed in leaf mesophyll, where only the GUS-expressing cells appeared to divide (Figure 5K). Both observations are consistent with the Bbm transcription factor exerting its influence in a cell-autonomous manner.
Transient expression of Wus2 also stimulated growth but with a markedly different phenotype. Wus2 expression resulted in the development of numerous protrusions from the scutellar surface (Figures 1B to 1D), with the expression of the fluorescent protein (and by inference WUS2) confined to cells at the tip of the protrusion. Closer examination of the tips of these protrusions revealed that fluorescent protein was most likely being expressed in a few cells, and surrounding cells were being stimulated to divide. This is consistent with data from other groups demonstrating the WUS2 protein is diffusible and is a major contributor to apical meristem organization and maintenance (Mayer et al., 1998; Gallois et al., 2002; Yadav et al., 2011, 2013). As these protrusions continued to elongate from the scutellum of the embryo, a file of cells in the center of the protrusions was observed to contain the fluorescent protein, with the fluorescence gradually diminishing as the file progressed further from the tip of the protrusion. After transformation of leaf cells with Bbm and Wus2, it was also observed that cell division was being stimulated in cells neighboring the transgene-expressing cells (Figure 5L). These macroscopic and microscopic observations are consistent with WUS2 acting in a non-cell-autonomous manner. Also consistent with the transient phenotype, strong constitutive expression of Wus2 resulted in chimeric callus that upon regeneration produced only nontransformed plants. To potentiate this negative effect, the weakly expressing nos promoter was used to drive Wus2 expression, which when coexpressed with Bbm, eliminated the proliferation of chimeric nontransgenic sectors. Transient expression of both Bbm and Wus2 in the same cells often produced growth stimulation greater than that by Bbm or Wus2 alone, with a mixed phenotype in which apparently embryogenic and organogenic structures were intermingled (Figure 1F). In dicots, ectopic expression of BBM has been reported to produce somatic embryos in various species and tissue types, including Arabidopsis cotyledons (Boutilier et al., 2002), Arabidopsis leaves (Morcillo et al., 2007; Bandupriya et al., 2014), Brassica napus leaves (Boutilier et al., 2002), and callus from Populus tomentosum (Deng et al., 2009). However, none of these reports indicates any stimulation of transformation frequencies. By contrast, we have shown that expression of Bbm in maize results primarily in a stimulation of callus proliferation and overall transformation efficiency, with callus embryogenic quality and regeneration capacity determined by exogenously applied hormones. Similarly, ectopic expression of WUSCHEL has also been observed to result in de novo embryo formation in Arabidopsis (Zuo et al., 2002). With the constructs and monocot germplasm used in this study, we have not seen indications that Wus2 expression alone can lead to formation of complete de novo embryos, but instead ectopic expression resulted in formation of de novo meristem-like structures.
In some of our inbred lines, the addition of Wus2 provided no significant increase beyond using Bbm alone (for example, in PHN46 and PH581 in Figure 2). However, for the inbred PHP38, the addition of Wus2 produced a substantial increase in transformation relative to Bbm alone. The response of inbred PHH5G was even more dramatic, with transgenic plants being recovered only when both expression cassettes were present, stimulating overall callus growth but with the embryogenic morphology remaining dependent on the medium composition. For inbred PHH5G, a medium formulation that contained only the auxin 2,4-D produced the best embryogenic callus and subsequent regeneration, while for other inbred lines (such as PH0AZ), medium containing the cytokinin BAP and a lower 2,4-D concentration was required for optimal callus morphology and regeneration. This is similar to results reported in tobacco (Nicotiana tabacum), where ectopic expression of Bbm has been reported to result in organogenesis, but upon the addition of cytokinin produced somatic embryos (Srinivasan et al., 2007), but differs from what has been reported in Arabidopsis thaliana and B. napus, where ectopic expression of WUS (Zuo et al., 2002) and BBM (Boutilier et al., 2002) has been reported to directly produce somatic embryos.
Longer term expression of the morphogenic genes Bbm and Wus2 during the development of transgenic calli clearly aided in the recovery of transgenic plants, predominantly through the stimulation of growth rates. Improved transformation frequencies were observed in a number of inbred lines, extending our ability to transform some previously recalcitrant inbred lines (for example PHH5G) and increasing transformation frequencies in other inbred lines from marginally useful levels to levels that are practical for production transformation. However, as stated in the results, not all inbred lines responded to this combination of genes/promoters. Thus, while we have clearly broadened the range of inbred lines we can transform, a genotype-independent transformation method for maize remains elusive.
Although the addition of Bbm/Wus2 had a dramatic positive effect on the recovery of transgenic plants in maize, there was still a great deal of variability associated with this process, as evidenced by the large standard deviations in these treatments. There were two potential sources of variability associated with the source material for immature embryos. The first was that while a large number of immature embryos were used for the different treatments and tests in this study, these embryos were collected from the greenhouse during different seasons of the year, and the changing greenhouse environment may have influenced the phenotype and physiology of the source material. The second contributing factor that has been historically observed is simple ear-to-ear variability. Thus, even for plants from a given inbred grown side-by-side in the same greenhouse and harvested on the same day, tremendous variation in transformation results for immature embryos harvested from the two ears were commonly observed. While we can still only speculate on the cause of this variability, it is clear that even though using the morphogenic genes Bbm and Wus2 increased overall transformation frequencies, they had minimal effect on the large deviation observed between donor plants.
Two main strategies have been used to control expression of growth-stimulating genes during the transformation process. Such control is critical for optimal use of these genes, relying on either inducible excision (Ebinuma et al., 1997; Deng et al., 2009) or chemically induced expression of the gene (Zuo et al., 2002) to take advantage of the growth stimulation provided by the gene while eliminating later pleiotropic effects on plant growth and fertility. For maize, we found the desiccation-responsive promoter rab17 to provide highly efficient control of CRE excision, permitting high frequencies of transgenic callus to be recovered while also producing high frequencies of complete excision.
Producing a continuous supply of maize immature embryos for transformation is a costly process. Accordingly, explants from mature seed or seedlings are an attractive alternative. For maize, there have been reports on how readily the exposed tissues inside the kernel can be used to initiate either regenerable embryogenic callus or regenerable multiple shoot cultures (for example, see Al-Abed et al., 2006). However, to date, the closest researchers have come to directly using mature seed tissues for transformation in monocots has been a report in which a brief preculture period (1 to 5 d) was used before Agrobacterium-mediated DNA delivery in japonica rice (Toki et al., 2006). The remaining reports of “mature seed transformation” in monocots have all progressed through an intermediate culture step, using either embryogenic callus (Hiei et al., 1997; Chen et al., 1998; Dai et al., 2001; Sidorov et al., 2006; Ahmadabadi et al., 2007), organogenic callus (O’Connor-Sánchez et al., 2002) or proliferating meristem cultures (Zhong et al., 1996; Zhang et al., 2002), and finally using the proliferating cultured cells as the transformation target. Here, using the transcription factors Bbm and Wus2, we demonstrated that cells in the exposed embryo axis of mature maize seed can reproducibly produce transgenic callus and regenerate healthy, fertile T0 plants. In contrast to scutellum transformation typically observed for immature embryo transformation (Songstad et al., 1996), we never observed transgenic events originating from the mature scutellum of maize embryos. The simplest explanation for this difference is that the Agrobacterium strain used in our experiments for mature embryo transformation predominantly delivered DNA into cells of the embryo axis and not the scutellum (Figure 5C). However, it is equally plausible that morphogenic changes that occur during scutellar maturation in the maize embryo render these cells incapable of dedifferentiating and dividing.
Differentiated cells must dedifferentiate in order to respond to embryogenic signals is an underlying tenet in plant cell transformation. In this regard, with the exception of tobacco among the wide array of crops in which transformation work has been done, leaf cells are often seen as one of the most intractable. In the hope of finding a reasonable compromise, researchers in the grasses have moved down the leaf to develop callus culture methods that focus on the more meristematic, less differentiated region directly above the mesocotyl, at the base of the leaf (Wenzler and Meins, 1986; Conger et al., 1987; Ramesh et al., 2009; Yu et al., 2012). Despite the development of callus culture methods, the direct transformation of leaf-base cells in maize has never been demonstrated. When starting with leaf-base explants, successful reports of transformation in maize have all relied on an intermediate callus-culture step to produce the transformable explant. For example, leaf bases in germinating maize seedlings have been used to produce callus for later transformation (Sidorov et al., 2006; Ahmadabadi et al., 2007). Importantly, we demonstrated here that mesophyll cells from maize seedlings can be directly transformed using Agrobacterium-mediated delivery of Bbm/Wus2 and that those cells divide and appear to be the source of the embryogenic, regenerable callus produced in our experiments. In other experiments using Wus2 and Bbm, we were able to recover transgenic plants from mature maize leaves, such as the flag leaf immediately below the tassel of mature plants. Furthermore, transgenic plants produced from leaf-derived callus were healthy and fertile.
The ploidy of the transgenic plants was of interest, since there have been numerous observations that endoreduplication can occur in many plant organs including leaves (Barlow, 1985; Galbraith et al., 1991; Kondorosi et al., 2000; Aubry et al., 2014). We had two types of evidence indicating that the transgenic events derived from leaf transformation were diploid. First, although the number of samples analyzed was small, all 13 of the randomly tested leaf-derived transgenic calli produced typical diploid histograms in flow cytometry analyses similar to wild-type control plants. Second, we observed hundreds of transgenic regenerated plants that were derived using leaf transformation, and none exhibited the typical plant phenotypes associated with increased ploidy levels, such as thicker stem girth, wider leaves, and larger ears (Randolph, 1935; Randolph et al., 1944).
This is not the first report of direct leaf cell transformation in a monocot; transgenic events have been recovered after transformation of leaf cells in orchardgrass (Denchev et al., 1997). However, before performing these transformation experiments, these researchers selected accessions of orchardgrass that exhibited an atypically high level of somatic embryogenesis when leaf tissue was exposed to exogenous auxin (Conger et al., 1983). More recently, it has been reported that Agrobacterium-mediated transformation directly into young sugarcane leaf tissue has been accomplished (Eldessoky et al., 2011), but again in a species in which somatic embryogenesis from leaf cells in response to auxin has been well established (Lakshmanan et al., 2006). In this report, we demonstrated successful leaf cell transformation in maize inbred lines that are normally recalcitrant to hormone-mediated organogenesis or somatic embryogenesis from leaves. Likewise, we demonstrated direct transformation of mature maize seed tissues, and while we have still not attained the “holy grail” of a truly genotype-independent transformation method for maize, we have opened the doors much wider than with previous methods. Furthermore, we demonstrated that this combination of Bbm and Wus2 can be used to enhance transformation in sorghum, sugarcane, and indica rice. With further refinements and discoveries in this area, use of genes such as Bbm and Wus2 will undoubtedly continue to be improved upon to make transformation of difficult monocot crops, and routine transformation of new explants in monocots, a tractable reality for both academic and industrial research and product development.
METHODS
Cloning of the Maize Bbm and Wus2 Genes
Both the Bbm and Wus2 genes were cloned from the Pioneer/DuPont maize EST library (5′RACE was used to obtain a full-length maize (Zea mays) Wus2 cDNA). Identity was confirmed relative to other AP2-domain proteins by alignment of the encoded BBM protein to the published BBM protein sequences of Brassica napus and Arabidopsis thaliana (Boutilier et al., 2002). The identity of Wus2 was confirmed by alignment with Arabidopsis WUS family members (Laux et al., 1996; Gallois et al., 2002).
Plant Material
Five DuPont Pioneer inbred lines were used in these experiments, including two stiff-stalk inbred lines (PHH5G and PHP38) and three non-stiff-stalk inbred lines (PHN46, PH581, and PH0AZ). Two of the DuPont Pioneer inbred lines reported in this article (PHP38 and PHN46) are nonproprietary and publicly available from USDA-GRIN. The other three DuPont Pioneer inbred lines described in this research are proprietary (PHH5G, PH581, and PH0AZ). If a proprietary inbred is requested, Pioneer will provide at its discretion the most closely related nonproprietary Pioneer inbred or a nontransgenic Pioneer hybrid derived from the requested inbred.
Agrobacterium Strains and Vectors
Agrobacterium tumefaciens strains LBA4404, AGL1 (Lazo et al., 1991), and thymidine auxotrophic (THY-) versions of LBA4404 were used in this study. For clarity, the suffixes -LBA, -AGL1, and -THY are indicated following the pPHP numbers in Figure 3 and Supplemental Figure 1. Vectors pSB1 and pSB11 (Komari, 1990; Komari et al., 1996) were used to construct the superbinary vectors (with the T-DNA components shown in Figure 3 and Supplemental Figure 1). For a list of molecular components (promoters, genes, 3′ sequences, etc.), see Supplemental Table 1. All expression cassettes in all the plasmids used in this study contained the pinII 3′ regulatory sequence from the potato proteinase inhibitor II gene (An, 1986) unless otherwise specified. pPHP24600-LBA and pPHP32269-AGL1 or pPHP32269-LBA were used as control vectors in different experiments that did not contain the maize morphogenic genes Bbm and Wus2. pPHP24600-LBA (Supplemental Figure 1A) contained two expression cassettes in opposite orientations, a CaMV 35S promoter driving expression of a phosphinothricin acetyltransferase (PAT) gene followed by a CaMV 35S 3′ regulatory sequence (in the 3′ to 5′ orientation going toward the T-DNA right border) and a maize Ubiquitin (Ubi) promoter and Ubi intron driving expression of a DsRED coding sequence (Baird et al., 2000) with an introduced potato LS1 intron (Vancanneyt et al., 1990). pPHP32269-LBA (Supplemental Figure 1B) contained two expression cassettes, both in the 5′ to 3′ orientation; the first contained the Ubi promoter and intron driving expression of the phosphomannose isomerase (PMI) gene (Negrotto et al., 2000), and the second cassette contained the maize Ubi promoter and 3′ sequence driving expression of a maize codon-optimized version (moPAT; Jayne et al., 2000) of the gene encoding phosphinothricin acetyltransferase (White et al., 1990) fused to Zs-Yellow-N1 (Matz et al., 1999).
All other vectors included combinations of cassettes encoding BBM and WUS2. The plasmids containing Bbm and Wus2 all contained three expression cassettes, a recombinase (moCRE or moFLP; maize codon-optimized versions), Wus2 and Bbm in between recombination target sites (loxP or FRT sites, respectively). The T-DNA in pPHP32371 contained five complete expression cassettes plus a promoterless YFP coding sequence. Starting at the right border, the first expression cassette contained the maize Ubi promoter and intron driving expression of Am-Cyan1 (Matz et al., 1999), with a FLP-recombinase target site (FRT1) positioned between the Ubi intron and the Am-Cyan1 (CYAN) coding sequence. The next three expression cassettes contained (1) the maize rab17 promoter and 5′UTR (Vilardell et al., 1990, 1991) driving expression of moFLP (with a potato LS1 intron), (2) the nos promoter driving the Wus2 structural gene, and (3) the maize Ubi promoter and intron driving the maize Bbm gene. The last pinII sequence in this three-cassette series was followed by a second FRT site and a Zs-Yellow-N1 coding sequence (YFP). The final expression cassette in this T-DNA was a maize Ubi promoter and intron driving the maize-optimized PAT (moPAT). Vector pPHP35648 (Figure 3) was identical to pPHP32371 (Supplemental Figure 1C) with the following exceptions: pPHP35648 contained a maize codon-optimized CRE recombinase (instead of FLP) with an intervening potato LS1 intron behind the rab17 promoter and the FRT sites were replaced by loxP recombination target sites. Vector pPHP40710 (Supplemental Figure 1G) was an exact duplicate of pPHP35648 except that the Ubipro:Bbm expression cassette had been removed.
While the recombinase system used in the constructs differed, pPHP32371 and pPHP35648 both produced the same end result upon recombinase-mediated excision; removal of CYAN, the CRE, Wus2, and Bbm expression cassettes, and activation of Zs-Yellow expression, while leaving behind a single FRT or loxP target site after excision (Figure 3). pPHP40710 produced the same end result after excision, except only Wus2 and CRE were excised. pPHP35648-LBA-THY- is the exact replica of pPHP35648-LBA, except the plasmid was moved into a LBA4404 THY- strain. The T-DNA for pPHP54733-AGL1 (Supplemental Figure 1E) in Agrobacterium strain AGL1 contained loxP, rab17pro:CRE, nospro:Wus2, Ubipro:Bbm, loxP, Ubipro:GFP:SB-ACTIN 3′ sequence, and SB-Ubi pro:PMI:SB-Ubi 3′ sequence . The T-DNA for pPHP54561 (Supplemental Figure 1F) in LBA4404 contained a Ubi promoter and intron driving expression of moPAT with a loxP site in between the Ubi intron and the moPAT gene. This was followed by Ubipro:YFP, rab17pro:CRE, nospro:Wus2, Ubipro:Bbm, and then the second loxP target site. Immediately after this second loxP site, the GAT gene and a UBQ3 3′ sequence were positioned. Upon recombinase-mediated excision at loxP sites, moPAT, CRE, Wus2, and Bbm were removed and the GAT gene was activated.
The materials reported in this article contain selectable markers (PAT, moPAT, and PMI) owned by third parties. Pioneer Hi-Bred International (Pioneer) will provide materials to academic investigators for noncommercial research under an applicable material transfer agreement subject to proof of permission from any third-party owners of all or parts of the material and to governmental regulation considerations. Obtaining permission from third parties will be the responsibility of the requestor. Alternatively, Pioneer will provide replacement materials containing a maize HRA-selectable marker. This expression cassette contains a sorghum Acetolactate Synthase (ALS) promoter, an HRA gene, and a terminator. The HRA gene is a highly herbicide-resistant ALS allele with two mutations, W574L and P197A, that confers resistance to selective herbicides such as ethametsulfuron and imazapyr. This HRA expression cassette will replace the Ubipro:moPAT:pinII expression cassette (Figure 3; Supplemental Figures 1C, 1F, 1G, and 1H) or the CaMV 35Spro:PAT:35S 3′ expression cassette (Supplemental Figure 1A) or the Ubipro:PMI expression cassettes (Supplemental Figures 1B and 1E). In addition, the GAT gene in Supplemental Figure 1F will be replaced with NPTII, which confers resistance to antibiotics such as kanamycin and G418.
Particle Gun Transient Phenotype Observations
To assess the ability of Ubipro:Bbm and nospro:Wus2 to stimulate rapid growth, the two expression cassettes (on separate plasmids) were introduced into 18 DAP embryos of the Pioneer inbred PH581 using the Biolistics helium particle gun (Bio-Rad). Mixtures of plasmids were cobombarded for the various treatments described below. All treatments contained a plasmid with the expression cassette Ubipro:moGFP (moGFP indicates a maize-codon-optimized GFP gene, see US6486382). For the control treatment, a second plasmid containing CaMV 35Spro:GUS was mixed with the moGFP-containing plasmid to ensure that the total DNA content and the molar ratios of individual plasmids were consistent. The morphogenic gene plasmids contained either (1) Wus2 with a maize Oleosin promoter and the Agrobacterium nopaline synthase 3′ sequence or (2) Bbm with a maize Ubiquitin promoter and the pinII 3′ sequence. Treatments included (1) moGFP alone, (2) moGFP plus Wus2, (3) moGFP plus Bbm, or (4) moGFP plus both morphogenic genes. Gold particle preparation, attachment of the DNA mixture to the 0.6-µm gold particles, and bombardment parameters were described by Ananiev et al. (2009), with the following changes in the attachment protocol. The DNA was coated onto 0.6-µm (average diameter) gold particles using a water-soluble cationic lipid Tfx-50 (Promega) as follows: A DNA mixture was prepared by adding separate plasmids, containing 50 ng GFP plasmid plus 100 ng GUS plasmid (Trt. 1); 50 ng each of GFP, GUS, and Wus2 (Trt. 2) plasmids; 50 ng each of GFP, GUS, and Bbm plasmids (Trt. 3); or 50 ng each of GFP, Wus2, and Bbm plasmids (Trt. 4), for a total of 150 ng DNA in all treatments (roughly equimolar since plasmid sizes were similar) and adjusting to a final volume of 40 µL. To the DNA solution, 50 μL of a 0.01 mg/μL gold solution and 5 μL Tfx-50 were added, followed by mixing and shaking at room temperature for 10 min. The suspension was centrifuged at 10,000g for 1 min, the supernatant discarded, and the gold particles (with coated DNA/lipofectin) were resuspended in 120 μL ethanol by brief sonication. Ten microliters of the gold suspension in ethanol was pipetted onto carrier discs and then bombarded into immature embryos in a Bio-Rad PDS-1000/He Gun using a rupture disc of 400 p.s.i.
After particle bombardment, the embryos were transferred to our standard tissue culture medium 605J (with no selective agent; see Supplemental Table 2) and cultured in the dark at 26°C. Fourteen days after particle bombardment, embryos were observed using a Nikon SMZ1500 stereomicroscope with standard xenon epifluorescence illumination using a FITC HYQ filter set.
Immature Embryo Isolation, Agrobacterium-Mediated Transformation, and Tissue Culture
Immature embryos derived from greenhouse-grown Pioneer inbred lines PHN46, PH581, PHP38, and PHH5G were used for Agrobacterium-mediated transformation. All embryo donor and transgenic plants were grown in greenhouses (Johnston, IA) in pots containing Metro-Mix 838 (Sun Gro Horticulture Canada) with greenhouse day/night average temperatures of ∼29/20°C with a 12-h day photoperiod and supplemental lighting provided by a 3:1 ratio of metal halide (1000 W) and high pressure sodium (1000 W) lamps. Maize immature ear harvest, immature embryo isolation, Agrobacterium-mediated transformation, callus culture, and plant regeneration were essentially as described by Zhao et al. (2002), with some modifications to accommodate transformation of inbred germplasm (see Supplemental Table 2 for medium formulations). For inbred lines PHN46, PH581, PHP38, and PHH5G, immature embryos were removed from the ears at 9 to 12 d after pollination, producing embryos with an average length of 1.0 to 2.0 mm. Agrobacterium containing transformation vectors were grown for 1 d on YP medium (Ishida et al., 1996), and colonies were collected and suspended in 700 liquid medium. Immature embryos were mixed with Agrobacterium strain LBA4404 (OD = 0.7 at 550 nm) in 700 liquid medium for 5 min and were then removed from the liquid and placed scutellum side up on 710I solid medium for 3 d at 21°C in the dark. Embryos were then cultured on resting medium 605J with 100 mg/L carbenicillin for 1 week at 26°C in the dark, and transferred to 3 mg/L bialaphos callus selection medium (605K) for inbred lines PHN46, PH581, and PHP38 or onto medium containing no bialaphos selective agent (605T) in the case of inbred PHH5G. Callus was subcultured at 2- to 3-week intervals for 2 to 2.5 months.
Callus transformation frequency was defined as the number of treated immature embryos that produced either bialaphos-resistant calli (for inbred lines PHN46, PH581, and PHP38) or rapidly growing, embryogenic calli for PHH5G after 2 to 2.5 months of culture (with no chemical selection) relative to the total number of immature embryos inoculated with Agrobacterium. Transgenic sectors growing on 605K or 605T medium were confirmed by visualizing fluorescence (DsRED for pPHP24600, Zs-Yellow1-N1 for pPHP32269, and Am-Cyan1 for pPHP35648 and pPHP32371) under a Nikon SMZ1500 stereomicroscope with standard xenon epifluorescence illumination and Nikon filter sets for DsRED, YFP, and Cyan (8345M TRITC, FITC HYQ, and 83456M CYGFP, respectively). After 2 to 2.5 months of growth, samples representing transgenic callus events were desiccated on a stack of three dry Whatman 60-mm filter papers in a 60-mm Petri dish for 3 d at 26°C in the dark; alternatively, the Petri dishes were placed in a commercially available food dehydrator (Nesco) at the lowest setting for 3 h. This desiccation treatment activated the late-embryogenic rab17 promoter, driving expression of the recombinase and stimulating high levels of excision.
For pPHP32371 and pPHP35648, expression of Am-CYAN1 was lost as a result of the excision process and expression of ZS-Yellow N1 was activated. In this fashion, excision could be monitored under the dissecting microscope with epifluorescence and the appropriate filters. After transgenic calli had grown for 2 to 2.5 months on 605K (for lines produced with pPHP24600, pPHP32371, pPHP35648, or pPHP24944) or on 605T (for lines produced using pPHP38333), calli were moved onto regeneration medium (289O, 272V, and 13158) at 28°C for 2 to 3 weeks to initiate shoots. Once shoots and roots had been established, plantlets were transferred to pots in a greenhouse.
Transformation of sorghum immature embryos was performed in a similar fashion to maize immature embryos, following the detailed methods and medium formulations described by Wu et al. (2014).
Preparation of Mature Seed-Derived Explants for Transformation
Dry seeds of maize inbred PH0AZ were surface sterilized with 80% ethanol for 3 min followed by 50% commercial bleach (2.6% sodium hypochlorite) with 0.1% Tween 20 for 20 min, then washed three times with sterile water. Surface sterilized seeds were soaked overnight (18 to 24 h) in sterile water at room temperature and mature embryos were dissected out from the softened seeds. Longitudinal slices (Figure 5A) of ∼300 to 400 um in thickness were prepared by hand-sectioning the embryo. Each slice contained exposed leaf primordia, mesocotyl, and root primordium regions (Figure 5B). Fresh slices were immediately transferred into liquid medium 700 before Agrobacterium infection. These regions on the embryo slice were the target area for T-DNA delivery during Agrobacterium-mediated transformation and contained cells that were culture responsive.
Preparation of Leaf Segments for Transformation
Dry seeds of inbred line PHH5G were surface sterilized as described above. Sterilized seeds were placed onto solid medium 13158 (Supplemental Table 2) for direct germination at 26°C in the dark for 15 to 16 d, and shoot segments 2 to 3 cm long above the first leaf base node of the seedling were used to prepare transformation explants (Figure 5E). The coleoptile was removed and the leaf fragment was split longitudinally first, then cross-dissected into 1- to 2-mm leaf pieces. Leaf pieces were immediately transferred into liquid medium 700 before Agrobacterium infection.
Infection of Alternative Explants with Agrobacterium
For mature embryo slice infection, Agrobacterium strain LBA4404 THY- was grown on solid culture medium containing yeast extract with 50 mg/L spectinomycin medium and incubated at 26°C in the dark for 1 d. The Agrobacterium suspension was adjusted to OD 0.7 at 550 nm using liquid medium 700 containing 200 µM acetosyringone and 0.04% Silwet L-77. For leaf segment infection, Agrobacterium strain AGL1 was grown on solid medium (5 g/L yeast extract, 10 g/L peptone, 5 g/L NaCl, 15 g/L Bacto-Agar, and 100 mg/L spectinomycin) and incubated at 26°C in the dark for 1 d. The Agrobacterium suspension was adjusted to an OD of 0.4 at 550 nm using 10 mM MgSO4 solution containing 200 µM acetosyringone and 0.02% Silwet L-77.
Liquid medium bathing either the embryo sections or leaf segments was drawn off and replaced with freshly prepared Agrobacterium suspension. The culture plates were then transferred into a plastic vacuum desiccator. The desiccator was connected to house vacuum (24 inches Hg) and kept on a shaker platform with a speed of 100 rpm for 15 to 30 min. After infection, the Agrobacterium suspension was drawn off from the culture plates and the embryo slices or leaf pieces were blotted dry with sterile filter paper before being transferred onto solid medium 710I and incubated at 21°C in the dark for 3 d.
After 3 d cocultivation on medium 710I, embryo slices or leaf pieces were transferred onto culture medium 605J or 605T containing antibiotics to kill Agrobacterium. After 6 to 8 weeks of culture, fast-growing embryogenic calli could be identified. These fast growing calli were confirmed as transformed by expression of the fluorescent marker. Embryogenic calli were transferred onto fresh 605J or 605T culture medium for further proliferation before plant regeneration. Transformed callus tissues were transferred into an empty Petri dish containing a piece of autoclaved glass filter paper (VWR Glass Microfiber Filter 691) and covered with the lid but not sealed. Petri dishes with callus tissues were placed into a culture box with a loose cover and incubated at 26°C in the dark for 3 d. This 3-d, gradual desiccation treatment induced rab17:CRE expression and excision of the morphogenic genes. After the desiccation treatment, calli were transferred to maturation medium 289O for 2 to 3 weeks in the dark to induce shoot formation. Calli with 1- to 2-cm shoots were then transferred to hormone-free medium 272V for further development of shoots and roots under low light (10 to 30 mE m−2 s−1). Plantlets with well developed shoots and roots were sampled for comprehensive molecular analysis before being transferred to the greenhouse for seed production. Detailed PCR analyses were performed to determine copy number of transgenes as well as to confirm the excision of morphogenic genes from the final transgenic plants.
Transformation Methods for Callus in Sugarcane and Rice
Two morphogenic gene vectors (pPHP35648-LBA or pPHP54561-LBA) were compared with a control vector containing Ubipro:PAT plus Ubipro:DsRED in the US sugarcane cultivar CP01-1372 in order to evaluate the effect of these maize transcription factors on sugarcane transformation frequency (see Supplemental Table 3 for medium formulations). pPHP35648 contained Ubipro:loxP:CYAN plus rab17pro:CRE plus nospro:Wus2 plus Ubipro:Bbm:loxP:YFP plus Ubipro:moPAT. pPHP54561 contained Ubipro:loxP:moPAT plus Ubipro:YFP plus rab17 pro:CRE plus nospro:Wus2 plus Ubipro:Bbm:loxP:GAT. Callus tissue was initiated from young leaf cylinders on DBC3 medium using in vitro-cultured plantlets and maintained on the same medium. Callus transformation methods and selection conditions were similar for both sugarcane and indica rice. Callus pieces 4 to 5 mm in diameter were infected with Agrobacterium in liquid 10 mM MgSO4 plus 100 µM acetosyringone and were then dissected into smaller segments and cocultivated on filter paper saturated with liquid DBC3(M5G) medium plus 100 µM acetosyringone in Petri dishes at 21°C in the dark. After 3 d of cocultivation, the tissue was transferred to solid DBC3 medium containing 100 mg/L cefotaxime and 150 mg/L timentin, and incubated at 26°C (±1°C) in the dark or dim light for 3 to 7 d. Afterwards, the tissue was transferred to the same medium plus 5 mg/L bialaphos. After 2 to 3 weeks, the tissue was transferred onto DBC6 medium containing antibiotics and 5 mg/L bialaphos. When a sufficient mass of callus was obtained, tissue was moved onto MSA medium for maturation and to MSB for rooting. Two months after the initiation of the experiment, transformation frequency was calculated based on the number of independent calli exhibiting either CYAN or YFP expression relative to the number of explants initially infected by Agrobacterium.
Molecular Analysis
qPCR (Wu et al., 2014) was used to estimate the copy number of the transgenes to determine if T-DNA integrations were intact or truncated, if recombinase-mediated excision had occurred, and to screen for the presence of Agrobacterium binary vector backbone integration. Genomic DNA was extracted from a single piece (200 ng) of fresh leaf tissue from each plant (Truett et al., 2000). Nontransgenic maize inbred lines PHN46, PH581, PHP38, PHH5G, and PH0AZ were used as the negative controls. Quantification was based on detection of amplification of gene sequences using gene-specific forward and reverse primers along with the corresponding gene-specific FAM/Vic-based MGB fluorogenic probes (Applied Biosystems). The 2−ΔΔCT method (Livak and Schmittgen 2001; ABI’s user bulletin #2, www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf) was used to estimate the copy number. For plants transformed with pPHP24600, two regions of the T-DNA (PAT and Ds-Red; see Supplemental Figure 1A) were screened. Plants with single-copy scores for both sequences were classified as having intact single-copy T-DNA integrations. Plants that had single-copy scores for only one of the two sequences were classified as truncated single-copy T0 plants. Plants with multiple-copy scores for either of the two sequences were classified as having multiple-copy integrations, which could either be intact or truncated. For plants derived from pPHP32269, primer sets were used that amplified segments of either PMI or YFP (Supplemental Figure 1B). For plants derived from excision vectors, qPCR was used to detect both the presence and copy number of genes outside the excision fragment, screening for ZS-YELLOW1 N1 and moPAT in pPHP32371- and pPHP35648-derived plants, and screening for GAT in pPHP54561-derived plants. In addition, T0 plants were screened by qPCR for the Bbm cDNA that should have been excised by the recombinase (Figure 3) using pPHP35648 as an example, with qPCR screening assays for excision vectors. For all maize transgenic plants, detection of Agrobacterium vector backbone integration was based on screening for sequences from five regions outside of the T-DNA (5′ of the RB, virG, SPC, and Tet, and 3′ of the LB). Plants with negative qPCR signals for all five regions were considered to be backbone-negative. Otherwise, they were classified as backbone-positive. Plants with intact single-copy T-DNA integrations without vector backbone were defined as quality events.
DNA Gel Blot Analysis
For DNA gel blot analysis, the Pioneer inbred PHH5G was transformed with the plasmid pPHP35648 and 47 PHH5G single-copy T0 plants were identified by PCR. A control nontransgenic inbred line was included to verify background hybridization of probe to the maize genome. The plasmid pPHP35648 contained the original T-DNA introduced into PHH5G, with the CRE, Wus2, and Bbm expression cassettes all between flanking loxP sites, with a promoterless YFP and the moPAT expression cassettes downstream of the 3′ loxP site (Figure 3). The plasmid pPHP35648-excised was created using CRE protein to excise the loxP-flanked rab17pro:CRE plus nospro:Wus2 plus Ubipro:Bbm from pPHP35648 and then retransforming the plasmid back into Escherichia coli, and after picking colonies and growing in liquid, performing a miniprep to purify the resultant plasmid containing Ubipro:loxP:YFP plus Ubipro:moPAT (as an excision control).
Total genomic DNA of 188 independent PHH5G T1 plants expressing moPAT and YFP (pPHP35648-excised) from 47 T0 plants and the PHH5G nontransgenic inbred line (negative control) was prepared from 50 mg lyophilized leaf powder (Paint Shaker-SK450; AGS Transact Technology). Genomic DNA was extracted using a high salt extraction buffer (2.0 M NaCl, 100 mM Tris-HCl, pH 8.0, 50 mM EDTA sodium salt, and 100 mM sodium metabisulphite) and sequentially precipitated using one-tenth volume of 5 M potassium acetate and isopropyl alcohol followed by two 70% ethanol washes. DNA pellets were air dried and dissolved in 1× TE. Genomic DNA was then treated with ribonuclease (RNase treatment) and purified. Following the extraction, DNA was quantified on a SPECTRAmax M2 plate reader using Pico Green reagent (Molecular Probes, Invitrogen) and visualized on 1% agarose gel to check the quality of the isolated DNA both qualitatively and quantitatively.
Genomic DNA of PHH5G transgenic lines was restriction digested with HindIII (NEB) for copy number analysis of the marker genes, YFP, and moPAT in the recombinant and nonrecombinant plants of pPHP35648. EcoRI enzyme (NEB) was used for integrity information of the genes within the loxP cassette (CRE, Wus2, and Bbm) along with YFP and moPAT. The plasmid controls and genomic DNA of transgenic plants digested with suitable enzymes were purified and electrophoresed on 1% agarose gels. The gels were documented under UV illumination (Bio-Rad Gel Doc, XR+). Following agarose gel electrophoresis, the separated DNA fragments were depurinated (0.25 M HCl for 10 to 15 min), denatured (0.5 M NaOH and 1.5 M NaCl for 30 min), neutralized in situ (1.5 M NaCl and 1.0 M Tris-HCl, pH 8.0 for 30 min), and transferred overnight to a N+ nylon membrane (GE healthcare) in 20× SSC buffer by the capillary method. Following transfer, the DNA was bound to the membrane by UV cross-linking (UV Stratalinker, UVP) at 120,000 μJ/cm3.
Two probes homologous to YFP and moPAT genes on pPHP35648 were used for hybridization to confirm the recombination status and copy number of these genes, while the presence of YFP, moPAT CRE, Bbm, and Wus2 genes were confirmed by hybridizing to their respective homologous probes. The probes were labeled by PCR reaction incorporating a digoxigenin (DIG)-labeled nucleotide, [DIG-11]-dUTP, according to the procedures of the PCR DIG Probe Synthesis Kit (Roche). The labeled probes were hybridized to the target DNA on the blots for detection of specific fragments following the DIG Easy Hyb Solution (Roche). DNA molecular weight markers III and VII DIG labeled (Roche) were used for visualization of the size standards.
Prehybridization was performed using 10 mL of prewarmed DIG Easy Hyb buffer in a 4.5 × 15-cm hybridization bottle at 50°C, 60 rpm in the hybridization incubator for a minimum of 30 min to block nonspecific binding sites on the membrane(s) in order to lower the background signal. After discarding the prehybridization solution, the membranes were incubated in heated probe-Hyb mix overnight at 50°C at 60 rpm in the hybridization incubator. The membranes were washed twice at room temperature for 5 min each in 2× SSC, 0.1% SDS solution. The membranes were then washed twice for 15 min each at 65°C using 0.1× SSC, 0.1% SDS solution on an orbital shaker at 60 rpm. After stringent washes, the blots were visualized using the CDP-Star chemiluminescent nucleic acid detection system with DIG Wash and Block Buffer Set (Roche) in a Chemiluminiscent reader (Syngene). Membranes were equilibrated with 1× washing buffer and blocked with 1× blocking buffer at room temperature for 30 min with constant shaking on an orbital shaker. The membranes were then incubated in anti-DIG antibody:blocking buffer (1:10,000) mix at room temperature for 30 min. The membranes were further rinsed twice with 1× washing buffer at room temperature for 15 min, with constant shaking and equilibrated for 5 min in detection buffer. The substrate CDP star solution was applied to the membrane and incubated for 5 min at room temperature. The chemiluminescence on the membranes was digitally captured using Syngene G-Box ChemiXT16 Lumi-Imager. Following hybridization and detection, membranes were stripped of DIG-labeled probe to prepare blots for subsequent rehybridization to a different probe. Membranes were rinsed briefly in distilled deionized water and then stripped in a solution of 0.2 M NaOH and 0.1% SDS at 37°C with constant shaking. The membranes were then rinsed in 2× SSC and either used directly for subsequent hybridizations or stored for later use. The alkali-based stripping procedure effectively removed probes labeled with alkali-labile DIG used in these experiments.
Fluorescence Microscopy and Histology, GUS Activity, Paraffin Sectioning, and Microscopy
Leaves were cut into 5-mm pieces and immediately placed in fresh fixative containing 2% paraformaldehyde and 2% glutaraldehyde (Electron Microscopy Sciences) in PBS, pH 7.2, on ice. Material in fixative was placed under vacuum (15 p.s.i.) for 2 min and then removed from vacuum and further fixed for 30 min at room temperature. Tissue was washed in five changes of PBS with 2 to 3 min/change prior to the GUS assay. GUS assay was performed according to Jefferson (1987) with some modifications. Material was placed in GUS-staining solution, vacuum infiltrated 15 min, and then incubated at 37°C overnight. The reaction was stopped by replacing substrate with PBS for 5 min, followed by a quick rinse in PBS. Tissue was refixed in 2% glutaraldehyde and 2% paraformaldehyde in PBS overnight at 4°C. Tissue was washed in three changes of PBS, 15 min each change. Material used for paraffin sectioning was dehydrated in a graduated ethanol series, changed to xylene, and then infiltrated at 60°C and embedded in Paraplast Plus (McCormick Scientific). Paraffin sectioning was done with a Leica RM 2165 microtome at 10-µm thickness and mounted on glass slides with Haupt’s adhesive (Ruzin, 1999). Paraffin sections were counterstained with safranin (Sigma-Aldrich) according to Johansen (1940). Observations and images were captured with a Leica DMRXA microscope and DC500 camera. Final image manipulations (contrast and hue adjustments) were applied uniformly using Adobe Systems Photoshop.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Zm-Bbm cDNA (CS155772), Zm-BBM protein (CAJ29869), Zm-Wus2 cDNA (EA275154), and Zm-WUS2 protein (ABW43772). The Os-Bbm gene can be found at OSA1 Release 7 (LOC_Os1g67410.1).
Supplemental Data
Supplemental Figure 1. T-DNAs used in monocot transformation experiments.
Supplemental Table 1. List of components and abbreviations used in vector maps.
Supplemental Table 2. Medium formulations used for maize and rice transformation and tissue culture.
Supplemental Table 3. Medium formulations used for sugarcane transformation and tissue culture.
Supplementary Material
Acknowledgments
We thank Hua Mo and Lu Liu for performing the statistical analysis and Scott Betts for critical review and editorial suggestions.
AUTHOR CONTRIBUTIONS
B.G.-K., K.L., E.W., N.W., M.-J.C., C.H., and G.H. designed the experiments. C.S., W.B., and L.N. cloned Wus2 while B.S. and J.Z. cloned Bbm. K.L., G.H., E.W., N.W., C.S., L.R., J.C.-Y., W.H., M.Y., T.K., and J.B. performed the transformation and tissue culture experiments. C.S., J.C., C.H., L.W., and Z.B. constructed vectors. B.L. designed PCR assays and performed all PCR analysis. K.B., E.I., B.R., and P.S. designed and performed DNA gel blots. Microscopy was done by B.G.-K., M.C., E.W., and N.W. B.G.-K., K.L., E.W., M.-J.,C., C.H., D.B., C.F., J.R., Z.-Y.Z., D.Z., and T.J. were involved in strategic planning and new vector design. K.L., M.-J.C., B.L., B.R., J.R., T.J., and B.G.-K. wrote the article.
Glossary
- DAP
days after pollination
Footnotes
Articles can be viewed without a subscription.
References
- Ahmadabadi M., Ruf S., Bock R. (2007). A leaf-based regeneration and transformation system for maize (Zea mays L.). Transgenic Res. 16: 437–448. [DOI] [PubMed] [Google Scholar]
- Al-Abed D., Rudrabhatla S., Talla R., Goldman S. (2006). Split-seed: a new tool for maize researchers. Planta 223: 1355–1360. [DOI] [PubMed] [Google Scholar]
- An G. (1986). Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiol. 81: 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev E.V., Wu C., Chamberlin M.A., Svitashev S., Schwartz C., Gordon-Kamm W., Tingey S. (2009). Artificial chromosome formation in maize (Zea mays L.). Chromosoma 118: 157–177. [DOI] [PubMed] [Google Scholar]
- Armstrong C.L., Green C.E. (1985). Establishment and maintenance of friable, embryogenic maize callus and the involvement of L-proline. Planta 164: 207–214. [DOI] [PubMed] [Google Scholar]
- Aubry S., Kneřová J., Hibberd J.M. (2014). Endoreduplication is not involved in bundle-sheath formation in the C4 species Cleome gynandra. J. Exp. Bot. 65: 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird G.S., Zacharias D.A., Tsien R.Y. (2000). Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97: 11984–11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandupriya H.D., Gibbings J., Dunwell J.M. (2014). Overexpression of coconut INTEGUMENTA-like gene, CnANT, promotes in vitro regeneration in transgenic Arabidopsis. Plant Cell Tissue Organ Cult. 116: 67–79. [Google Scholar]
- Barcelo P., Hagel C., Becker D., Martin A., Lörz H. (1994). Transgenic cereal (tritordeum) plants obtained at high efficiency by microprojectile bombardment of inflorescence tissue. Plant J. 5: 583–592. [DOI] [PubMed] [Google Scholar]
- Barlow P.W. (1985). The nuclear endoreduplication cycle in metaxylem cells of primary roots of Zea mays L. Ann. Bot. (Lond.) 55: 445–457. [Google Scholar]
- Boutilier K., Offringa R., Sharma V.K., Kieft H., Ouellet T., Zhang L., Hattori J., Liu C.-M., van Lammeren A.A.M., Miki B.L.A., Custers J.B.M., van Lookeren Campagne M.M. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettell R.I.S., Wernicke W., Thomas E. (1980). Embryogenesis from cultured immature inflorescences of Sorghum bicolor. Protoplasma 104: 141–148. [Google Scholar]
- Chen L., Zhang S., Beachy R.N., Fauquet C.M. (1998). A protocol for consistent, large-scale production of fertile transgenic rice plants. Plant Cell Rep. 18: 25–31. [Google Scholar]
- Chen T.H., Lam L., Chen S.C. (1985). Somatic embryogenesis and plant regeneration from cultured young inflorescences of Oryza sativa L. (rice). Plant Cell Tissue Organ Cult. 4: 51–54. [Google Scholar]
- Cheng M., Lowe B.A., Spencer T.M., Ye X., Armstrong C.L. (2004). Invited review: factors influencing Agrobacterium-mediated transformation of monocoteldonous species. In Vitro Cell. Dev. Biol. Plant 40: 31–45. [Google Scholar]
- Cho M.-J., Choi H.W., Lemaux P.G. (2000a). Transgenic Orchardgrass (Dactylis glomerata L.) plants produced from highly regenerable tissues derived from mature seed. Plant Cell Rep. 20: 318–324. [Google Scholar]
- Cho M.-J., Ha C.D., Lemaux P.G. (2000b). Production of transgenic tall fescue and red fescue plants by particle bombardment of seed-derived highly regenerable tissues. Plant Cell Rep. 19: 1084–1089. [DOI] [PubMed] [Google Scholar]
- Cho M.-J., Jiang W., Lemaux P.G. (1999). High-frequency transformation of oat via microparticle bombardment of seed-derived highly regenerable cultures. Plant Sci. 148: 9–17. [Google Scholar]
- Cho M.-J., Yano H., Okamoto D., Kim H.-K., Jung H.-R., Newcomb K., Le V.K., Yoo H.S., Langham R., Buchanan B.B., Lemaux P.G. (2004). Stable transformation of rice (Oryza sativa L.) via microprojectile bombardment of highly regenerative, green tissues derived from mature seed. Plant Cell Rep. 22: 483–489. [DOI] [PubMed] [Google Scholar]
- Christensen A.H., Sharrock R.A., Quail P.H. (1992). Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18: 675–689. [DOI] [PubMed] [Google Scholar]
- Conger B.V., Hanning G.E., Gray D.J., McDaniel J.K. (1983). Direct embryogenesis from mesophyll cells of orchardgrass. Science 221: 850–851. [DOI] [PubMed] [Google Scholar]
- Conger B.V., Novak F.J., Afza R., Erdelsky K. (1987). Somatic embryogenesis from cultured leaf segments of Zea mays. Plant Cell Rep. 6: 345–347. [DOI] [PubMed] [Google Scholar]
- Dai S., Zheng P., Marmey P., Zhang S., Tian W., Chen S., Beachy R.N., Fauquet C. (2001). Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol. Breed. 7: 25–33. [Google Scholar]
- Denchev P.D., Songstad D.D., McDaniel J.K., Conger B.V. (1997). Transgenic orchardgrass (Dactylis glomerata) plants by direct embryogenesis from microprojecticle bombarded leaf cells. Plant Cell Rep. 16: 813–819. [DOI] [PubMed] [Google Scholar]
- Deng W., Luo K., Li Z., Yang Y. (2009). A novel method for induction of plant regeneration via somatic embryogenesis. Plant Sci. 177: 43–48. [Google Scholar]
- Ebinuma H., Sugita K., Matsunaga E., Yamakado M. (1997). Selection of marker-free transgenic plants using the isopentenyl transferase gene. Proc. Natl. Acad. Sci. USA 94: 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinuma H., Sugita K., Endo S., Matsunaga E., Yamada K. (2005). Elimination of marker genes from transgenic plants using MAT vector systems. In Methods in Molecular Biology, Vol. 286: Plants: Methods and Protocols, Pena L., ed (Totowa, NJ: Human Press), pp. 237–253. [DOI] [PubMed] [Google Scholar]
- Eldessoky D.S., Ismail R.M., Abdel-Hadi A.-H.A., Abdallah N. (2011). Establishment of regeneration and transformation system of sugarcane cultivar GT54-9 (C9). GM Crops 2: 126–134. [DOI] [PubMed] [Google Scholar]
- Endo S., Sugita K., Sakai M., Tanaka H., Ebinuma H. (2002). Single-step transformation for generating marker-free transgenic rice using the ipt-type MAT vector system. Plant J. 30: 115–122. [DOI] [PubMed] [Google Scholar]
- Frame B.R., Drayton P.R., Bagnell S.V., Lewnau C.J., Bullock W.P., Wilson H.M., Dunwell J.M., Thompson J.A., Wang K. (1994). Production of fertile transgenic maize plants by silicon carbide whisker-mediated transformation. Plant J. 6: 941–948. [Google Scholar]
- Frame B.R., Shou H., Chikwamba R.K., Zhang Z., Xiang C., Fonger T.M., Pegg S.E.K., Li B., Nettleton D.S., Pei D., Wang K. (2002). Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M.E., Morrish F., Armstrong C., Williams R., Thomas J., Klein T.M. (1990). Inheritance and expression of chimeric genes in the progeny of transgenic maize plants. Biotechnology (NY) 8: 833–839. [DOI] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Knapp S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois J.-L., Woodward C., Reddy G.V., Sablowski R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129: 3207–3217. [DOI] [PubMed] [Google Scholar]
- Gordon-Kamm W.J., et al. (1990). Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell 2: 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Kamm W., et al. (2002). Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc. Natl. Acad. Sci. USA 99: 11975–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M.V., Ábrahám M., Mórocz S., Bottka S., Fehér A., Dudits D. (1993). Production of transgenic maize plants by direct DNA uptake into embryogenic protoplasts. Plant Sci. 90: 41–52. [Google Scholar]
- Ha C.D., Lemaux P.G., Cho M.-J. (2000). Stable transformation of a recalcitrant Kentucky bluegrass (Poa pratensis L.) cultivar using highly-regenerable tissues. In Vitro Cell. Dev. Biol. 37: 6–11. [Google Scholar]
- Hiei Y., Komari T., Kubo T. (1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35: 205–218. [PubMed] [Google Scholar]
- Ishida Y., Saito H., Ohta S., Hiei Y., Komari T., Kumashiro T. (1996). High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14: 745–750. [DOI] [PubMed] [Google Scholar]
- Jayne S., Barbour E., Meyer T. (2000). Methods for improving transformation efficiency. Patent No. US6096947.
- Jefferson R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Report. 5: 387–405. [Google Scholar]
- Johansen D.A. (1940). Plant Microtechnique. (New York: McGraw-Hill; ). [Google Scholar]
- Komari T. (1990). Transformation of cultured cells of Chenopodium quinoa by binary vectors that carry a fragment of DNA from the virulence region of pTiBo542. Plant Cell Rep. 9: 303–306. [DOI] [PubMed] [Google Scholar]
- Komari T., Hiei Y., Saito Y., Murai N., Kumashiro T. (1996). Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 10: 165–174. [DOI] [PubMed] [Google Scholar]
- Kondorosi E., Roudier F., Gendreau E. (2000). Plant cell-size control: growing by ploidy? Curr. Opin. Plant Biol. 3: 488–492. [DOI] [PubMed] [Google Scholar]
- Lakshmanan P., Geijskes R.J., Wang L., Elliott A., Grof C.P., Berding N., Smith G.R. (2006). Developmental and hormonal regulation of direct shoot organogenesis and somatic embryogenesis in sugarcane (Saccharum spp. interspecific hybrids) leaf culture. Plant Cell Rep. 25: 1007–1015. [DOI] [PubMed] [Google Scholar]
- Laux T., Mayer K.F., Berger J., Jürgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96. [DOI] [PubMed] [Google Scholar]
- Lazo G.R., Stein P.A., Ludwig R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (NY) 9: 963–967. [DOI] [PubMed] [Google Scholar]
- Lee K., Jeon H., Kim M. (2002). Optimization of a mature embryo-based in vitro culture system for high-frequency somatic embryogenic callus induction and plant regeneration from japonica rice cultivars. Plant Cell Tissue Organ Cult. 71: 237–244. [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A.L., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lowe K., Hoerster G., Sun X., Rasco-Gaunt S., Lazerri P., Abbitt S., Glassman K., Gordon-Kamm B. (2002). Maize LEC1 improves transformation in both maize and wheat. In Plant Biotechnology 2002 and Beyond, Vasil I.K., ed (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 255–258. [Google Scholar]
- Maddock S.E., Lancaster V.A., Risiott R., Franklin J. (1982). Plant regeneration from cultured immature embryos and inflorescences of 25 cultivars of wheat (Triticum aestivum). J. Exp. Bot. 34: 915–926. [Google Scholar]
- Matz M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., Lukyanov S.A. (1999). Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17: 969–973. [DOI] [PubMed] [Google Scholar]
- Mayer K.F.X., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- Morcillo F., Gallard A., Pillot M., Jouannic S., Aberlenc-Bertossi F., Collin M., Verdeil J.L., Tregear J.W. (2007). EgAP2-1, an AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm. Planta 226: 1353–1362. [DOI] [PubMed] [Google Scholar]
- Nardmann J., Werr W. (2006). The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 23: 2492–2504. [DOI] [PubMed] [Google Scholar]
- Negrotto D., Jolley M., Beer S., Wenck A.R., Hansen G. (2000). The use of phosphomannose–isomerase as a selectable marker to recover transgenic maize plants (Zea mays L.) via Agrobacterium transformation. Plant Cell Rep. 19: 798–803. [DOI] [PubMed] [Google Scholar]
- O’Connor-Sánchez A., Cabrera-Ponce J.L., Valdez-Melara M., Téllez-Rodríguez P., Pons-Hernández J.L., Herrera-Estrella L. (2002). Transgenic maize plants of tropical and subtropical genotypes obtained from calluses containing organogenic and embryogenic-like structures derived from shoot tips. Plant Cell Rep. 21: 302–312. [Google Scholar]
- Ozias-Akins P., Vasil I.K. (1982). Plant regeneration from cultured immature embryos and inflorescences of Triticum aestivum L. (wheat): evidence for somatic embryogenesis. Protoplasma 110: 95–105. [Google Scholar]
- Pareddy D.R., Petolino J.F. (1990). Somatic embryogenesis and plant regeneration from immature inflorescences of several elite inbred lines of maize. Plant Cell Rep. 7: 673–676. [Google Scholar]
- Ramesh M., Murugiah V., Gupta A.K. (2009). Efficient in vitro plant regeneration via leaf base segments of indica rice (Oryza sativa L.). Indian J. Exp. Biol. 47: 68–74. [PubMed] [Google Scholar]
- Randolph L.F. (1935). Cytogenetics of tetraploid maize. J. Ag. Res. 50: 591–605. [Google Scholar]
- Randolph L.F., Abbe E.C., Einset J. (1944). Comparison of shoot apex and leaf development and structure in diploid and tetraploid maize. J. Ag. Res. 69: 47–76. [Google Scholar]
- Rhodes C.A., Lowe K.S., Ruby K.L. (1988). Plant regeneration from protoplasts isolated from embryogenic maize cell cultures. Biotechnology (NY) 6: 56–60. [Google Scholar]
- Rout J.R., Lucas W.J. (1996). Characterization and manipulation of embryogenic response from in-vitro-cultured immature inflorescences of rice (Oryza sativa L.). Planta 198: 127–138. [Google Scholar]
- Ruzin S.E. (1999). Plant Microtechnique and Microscopy. (Oxford, UK: Oxford University Press; ). [Google Scholar]
- Shillito R.D., Carwell G.K., Johnson C.M., Dimaiom J.J., Harms C.T. (1989). Regeneration of fertile plants from protoplasts of elite inbred maize. Biotechnology (NY) 7: 581–587. [Google Scholar]
- Shrawat A.K., Lörz H. (2006). Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol. J. 4: 575–603. [DOI] [PubMed] [Google Scholar]
- Sidorov V., Gilbertson L., Addae P., Duncan D. (2006). Agrobacterium-mediated transformation of seedling-derived maize callus. Plant Cell Rep. 25: 320–328. [DOI] [PubMed] [Google Scholar]
- Smigocki A.C., Owens L.D. (1988). Cytokinin gene fused with a strong promoter enhances shoot organogenesis and zeatin levels in transformed plant cells. Proc. Natl. Acad. Sci. USA 85: 5131–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songstad D.D., Armstrong C.L., Petersen W.L., Hairston B., Hinchee M.A.W. (1996). Production of transgenic maize plants and progeny by bombardment of Hi-II immature embryos. In Vitro Cell. Dev. Biol. Plant 32: 179–183. [Google Scholar]
- Srinivasan C., Liu Z., Heidmann I., Supena E.D.J., Fukuoka H., Joosen R., Lambalk J., Angenent G., Scorza R., Custers J.B.M., Boutilier K. (2007). Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta 225: 341–351. [DOI] [PubMed] [Google Scholar]
- Stone S.L., Kwong L.W., Yee K.M., Pelletier J., Lepiniec L., Fischer R.L., Goldberg R.B., Harada J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98: 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita K., Kasahara T., Matsunaga E., Ebinuma H. (2000). A transformation vector for the production of marker-free transgenic plants containing a single copy transgene at high frequency. Plant J. 22: 461–469. [DOI] [PubMed] [Google Scholar]
- Toki S., Hara N., Ono K., Onodera H., Tagiri A., Oka S., Tanaka H. (2006). Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47: 969–976. [DOI] [PubMed] [Google Scholar]
- Truett G.E., Heeger P., Mynatt R.L., Truett A.A., Walker J.A., Warman M.L. (2000). Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29: 52–54, 54. [DOI] [PubMed] [Google Scholar]
- Vancanneyt G., Schmidt R., O’Connor-Sanchez A., Willmitzer L., Rocha-Sosa M. (1990). Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol. Gen. Genet. 220: 245–250. [DOI] [PubMed] [Google Scholar]
- Vilardell J., Goday A., Freire M.A., Torrent M., Martínez M.C., Torné J.M., Pagès M. (1990). Gene sequence, developmental expression, and protein phosphorylation of RAB-17 in maize. Plant Mol. Biol. 14: 423–432. [DOI] [PubMed] [Google Scholar]
- Vilardell J., Mundy J., Stilling B., Leroux B., Pla M., Freyssinet G., Pagès M. (1991). Regulation of the maize rab17 gene promoter in transgenic heterologous systems. Plant Mol. Biol. 17: 985–993. [DOI] [PubMed] [Google Scholar]
- Walters D.A., Vetsch C.S., Potts D.E., Lundquist R.C. (1992). Transformation and inheritance of a hygromycin phosphotransferase gene in maize plants. Plant Mol. Biol. 18: 189–200. [DOI] [PubMed] [Google Scholar]
- Wan Y., Widholm J.M., Lemaux P.G. (1995). Type I callus as a bombardment target for generating fertile transgenic maize (Zea mays L.). Planta 196: 7–14. [Google Scholar]
- Wen F.S., Sorensen E.L., Barnett F.L., Liang G.H. (1991). Callus induction and plant regeneration from anther and inflorescence culture of sorghum. Euphytica 52: 177–181. [Google Scholar]
- Wenzler H., Meins F. Jr (1986). Mapping regions of the maize leaf capable of proliferation in culture. Protoplasma 131: 103–105. [Google Scholar]
- Weymann K., Urban K., Ellis D.M., Novitzky M.E.R., Dunder E., Jayne S., Pace G. (1993). Isolation of transgenic progeny of maize by embryo rescue under selective conditions. In Vitro Cell. Dev. Biol. 29P: 33–37. [Google Scholar]
- White J., Chang S.Y., Bibb M.J., Bibb M.J. (1990). A cassette containing the bar gene of Streptomyces hygroscopicus: a selectable marker for plant transformation. Nucleic Acids Res. 18: 1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E., Lenderts B., Glassman K., Berezowska-Kaniewska M., Christensen H., Asmus T., Zhen S., Chu U., Cho M.-J., Zhao Z.-Y. (2014). Optimized Agrobacterium-mediated sorghum transformation protocol and molecular data of transgenic sorghum plants. In Vitro Cell. Dev. Biol. Plant 50: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G.V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25: 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jönsson H., Reddy G.V. (2013). Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Wang W., Wang Y., Hou B. (2012). High frequency wheat regeneration from leaf tissue explants of regenerated plantlets. Adv. Biosci. Biotechnol. 3: 46–50. [Google Scholar]
- Zhang S., Williams-Carrier R., Lemaux P.G. (2002). Transformation of recalcitrant maize elite inbred lines using in vitro shoot meristematic cultures induced from germinated seedlings. Plant Cell Rep. 21: 263–270. [Google Scholar]
- Zhao Z.-Y., Glassman K., Sewalt V., Wang N., Miller M., Chang S., Thompson T., Catron S., Wu E., Bidney D., Jung R. (2002). Nutritionally improved transgenic sorghum. In Plant Biotechnology 2002 and Beyond, Vasil I.K., ed (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 413–416. [Google Scholar]
- Zhong H., Srinivasan C., Sticklen M.B. (1992). In-vitro morphogenesis of corn (Zea mays L.) : I. Differentiation of multiple shoot clumps and somatic embryos from shoot tips. Planta 187: 483–489. [DOI] [PubMed] [Google Scholar]
- Zhong H., Sun B., Warkentin D., Zhang S., Wu R., Wu T., Sticklen M.B. (1996). The competence of maize shoot meristems for integrative transformation and inherited expression of transgenes. Plant Physiol. 110: 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.-W., Frugis G., Chua N.-H. (2002). The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 30: 349–359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.