MODD negatively regulates OsbZIP46 activity and stability through HDAC-related chromatin remodeling and PUB70-mediated ubiquitination, respectively, to fine-tune ABA signaling and drought resistance.
Abstract
Plants have evolved complicated protective mechanisms to survive adverse conditions. Previously, we reported that the transcription factor OsbZIP46 regulates abscisic acid (ABA) signaling-mediated drought tolerance in rice (Oryza sativa) by modulating stress-related genes. An intrinsic D domain represses OsbZIP46 activity, but the detailed mechanism for the repression of OsbZIP46 activation remains unknown. Here, we report an OsbZIP46-interacting protein, MODD (Mediator of OsbZIP46 deactivation and degradation), which is homologous to the Arabidopsis thaliana ABSCISIC ACID-INSENSITIVE5 binding protein AFP. MODD was induced by ABA and drought stress, but the induction was much slower than that of OsbZIP46. In contrast to OsbZIP46, MODD negatively regulates ABA signaling and drought tolerance, and inhibits the expression of OsbZIP46 target genes. We found that MODD negatively regulates OsbZIP46 activity and stability. MODD represses OsbZIP46 activity via interaction with the OsTPR3-HDA702 corepressor complex and downregulation of the histone acetylation level at OsbZIP46 target genes. MODD promotes OsbZIP46 degradation via interaction with the U-box type ubiquitin E3 ligase OsPUB70. Interestingly, the D domain is required for both deactivation and degradation of OsbZIP46 via its interaction with MODD. These findings show that plants fine-tune their drought responses by elaborate regulatory mechanisms, including the coordination of activity and stability of key transcription factors.
INTRODUCTION
Plants respond and adapt to stresses through a complex network of factors involved in stress signaling and regulation of gene expression (Hirayama and Shinozaki, 2010). The phytohormone abscisic acid (ABA), which is considered a stress hormone, plays pivotal roles in abiotic stress tolerance in plants (Zeevaart and Creelman, 1988; Cutler et al., 2010). Many components involved in ABA signaling have been identified, including protein phosphatases, protein kinases, and various transcription factors (Dai et al., 2013; Guo et al., 2002; Mustilli et al., 2002; Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). In particular, the identification of the PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS has established a core pathway for ABA signaling: The ABA-PYR complex inhibits Type 2C protein phosphatases (PP2Cs), leading to the activation of SNF1-related type 2 protein kinases (SnRK2s), which thereby phosphorylate specific targets, including transcription factors (Cutler et al., 2010; Raghavendra et al., 2010).
Plants undergo dramatic transcriptional regulation to efficiently coordinate stress responses. The transcriptional regulators, including various transcription factors and cofactors, function through ABA-dependent or ABA-independent pathways (Xiong et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). The bZIP (basic region and leucine zipper) family of transcription factors, especially members of the AREB/ABF/ABI5 subfamily, function in the ABA-dependent pathway and are major targets of SnRK2 protein kinases in ABA core signaling (Fujii and Zhu, 2009; Fujita et al., 2009).
Rapid signal transduction and transcriptional regulation facilitate the quick and effective stress response of plants. Plants have also evolved elaborate regulatory mechanisms to control the response and minimize metabolic cost. These mechanisms effectively retune the expression of stress-responsive genes to prestress levels when the stress is relieved and mostly involve the regulation of diverse transcription factors (Krogan and Long, 2009; Zhai et al., 2013). Transcription factors bind to their target promoters and thereby regulate the expression of target genes, and the posttranslational modification of transcription factors fine-tunes their function to effectively and precisely implement the stress response. In particular, the activities of many transcription factors, including the ABF subfamily members, can be modulated by posttranslational modifications such as phosphorylation and dephosphorylation (Kagaya et al., 2002; Choi et al., 2005; Furihata et al., 2006; País et al., 2009; Fujii et al., 2011). Posttranslational modification can also influence other processes, including conformation modification, cellular localization, and protein stability (Wang et al., 2014; Nelson and Millar, 2015).
Transcription factors function in transcriptional activation and repression; both of these functions are critical to biological processes (Mitsuda and Ohme-Takagi et al., 2009). Proteins that indirectly affect transcription, for example, by steric hindrance of activation-related components, are termed passive repressors. In contrast, active repressors possess an intrinsic repressive capacity conferred by conserved repression domains. Many of the active repressors identified so far in plants have the EAR (ethylene-responsive element binding factor-associated amphiphilic repression) motif, which is defined by the consensus sequence LxLxL or DLNxxP in the ERF transcriptional repressors and AUX/IAA repression domains (Cowell, 1994; Johnson, 1995; Ohta et al., 2001; Tiwari et al., 2004; Krogan and Long, 2009; Kagale and Rozwadowski, 2011).
The repression domains harbored by transcriptional repressors generally interact with non-DNA binding corepressors, which thereby recruit other regulators, including chromatin modifiers, to facilitate the repression of transcription (Shahbazian and Grunstein, 2007; Liu and Karmarkar, 2008; Kagale and Rozwadowski, 2011). Many processes involved in transcriptional repression have been found to be associated with histone deacetylase (HDAC) complexes, which can remove the acetyl groups from histone N-terminal lysine residues, and in turn promote the formation of a repressive chromatin state, leading to gene silencing. Groucho/Tup1-type corepressors in Arabidopsis thaliana were reported to function as global corepressors by recruiting HDACs (Keleher et al., 1992; Liu and Karmarkar, 2008). The TOPLESS/TOPLESS-Related (TPL/TPR) type corepressors have been intensively studied for their general function in multiple hormone pathways through the direct interaction with the EAR motifs in specific transcriptional repressors or adaptor proteins (Ohta et al., 2001; Weigel et al., 2005; Long et al., 2006; Szemenyei et al., 2008; Krogan and Long, 2009; Pauwels et al., 2010; Causier et al., 2012).
In addition to activation/deactivation, the stabilization and destabilization of transcription factors also posttranscriptionally fine-tune their functions. The turnover of most transcription factors in eukaryotes involves the ubiquitin-26S proteasome system (UPS). UPS requires the sequential action of the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin ligase (E3), which ultimately ligate one or more ubiquitin molecules to the substrates (Hare et al., 2003; Vierstra, 2003). Plants have four types of E3s: the single-polypeptide HECT, RING, and U-box domain containing proteins and the multisubunit cullin-RING ligases; these have essential functions in defining substrate specificity and function in a wide range of biological processes (Vierstra, 2009; Moon et al., 2004).
We previously reported that OsbZIP46, an ABF subfamily member, is a positive regulator of ABA signaling and drought stress tolerance in rice (Oryza sativa). The activity of native OsbZIP46 is blocked by a negative regulatory region designated domain D (Tang et al., 2012). OsbZIP46 has been reported under different names, including OsbZIP12, OsABF2, OsAREB1, ABL1, and OsAREB8 (Corrêa et al., 2008; Hossain et al., 2010; Jin et al., 2010; Yang et al., 2011; Yoshida et al., 2010). Here, we identify the OsbZIP46-interacting protein MODD (Mediator of OsbZIP46 deactivation and degradation, a homolog of AFP), which interacts with OsbZIP46 through the D domain and plays pivotal roles in the negative regulation of OsbZIP46 activity and stability by mediating HDAC-related chromatin remodeling and OsPUB70-related ubiquitination.
RESULTS
Repression of OsbZIP46 Activity via the D Domain
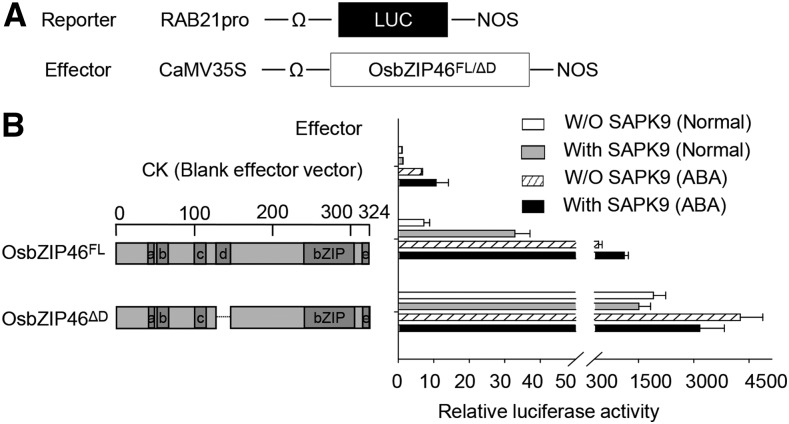
As the deletion of domain D of OsbZIP46 resulted in constitutive transactivation activity in yeast in our previous study (Tang et al., 2012), we further examined the repression function of domain D using an effector-reporter system in rice protoplasts based on a dual luciferase reporter assay. The native form of the full-length OsbZIP46 (designated as OsbZIP46FL) and the domain D-truncated mutated form (designated OsbZIP46ΔD) were transiently expressed in the rice leaf mesophyll protoplasts under the control of the constitutive CaMV 35S promoter. A reporter plasmid was constructed by fusing the promoter of the OsbZIP46 target gene RAB21 (Tang et al., 2012) to the firefly luciferase gene, and this was cotransfected into the protoplasts. Simultaneously, the Renilla luciferase gene driven by the 35S promoter was cotransfected as an internal control. Consistent with the previous results in a yeast system, the OsbZIP46ΔD-transfected protoplasts had a much higher relative luciferase activity compared with the blank vector control, but the OsbZIP46FL-transfected protoplasts had only weak relative luciferase activity (Figures 1A and 1B), indicating that the transactivation activity of OsbZIP46 could be dramatically suppressed by domain D in vivo.
Figure 1.
Analysis of the Transcriptional Activation Activity of OsbZIP46.
(A) Scheme of the constructs used in the cotransfection experiments. The reporter construct contained the promoter of RAB21, which has been previously reported as a putative target gene of OsbZIP46 (Tang et al., 2012), the firefly gene for luciferase (LUC), and a nopaline synthase (Nos) terminator. The effector construct contained the coding region of the tested genes driven by the CaMV 35S promoter; a translational enhancer sequence from tobacco mosaic virus (Ω) was located upstream of the site of the initiation of translation.
(B) Relative luciferase activities in rice protoplasts that had been cotransfected with a reporter plasmid and two different forms of OsbZIP46, together with or without (W/O) SAPK9 cotransfection in normal conditions or under ABA treatment. Schemes of the native full length (FL) and the domain D-missing mutated form (ΔD) of OsbZIP46 are shown on the left. All luciferase activities are expressed relative to values obtained with the cotransfection of a reporter construct and the blank effector vector (CK; set arbitrarily at 1). Error bars indicate the standard deviations based on six replicates.
A previous study indicated that the transcriptional activity of OsbZIP46 may be dependent on posttranslational regulation, such as ABA-dependent phosphorylation in vivo (Tang et al., 2012). We further examined the influence of domain D when OsbZIP46 was undergoing phosphorylation-mediated activation. We performed the transient expression analysis with exogenous ABA treatment or in leaves cotransfected with the ABA-activated kinase SAPK9. As expected, both the native OsbZIP46FL and truncated OsbZIP46ΔD showed significantly enhanced activity when treated with ABA, and OsbZIP46FL showed significantly enhanced activity when cotransfected with SAPK9. Nevertheless, the activity of OsbZIP46FL was still lower than that of OsbZIP46ΔD, but the difference was smaller compared with the difference under nonactivated conditions, indicating a decreased repressive effect of domain D under activation conditions (Figures 1A and 1B). Similarly, the full-length proteins of two other rice ABF members, OsbZIP23 and OsbZIP72, also showed significantly enhanced activity when treated with ABA (Supplemental Figure 1). These results demonstrated that ABA-dependent phosphorylation could activate the transcriptional activity of OsbZIP46 and meanwhile partially suppressed the negative regulation function of domain D.
OsbZIP46 Physically Interacts with MODD Depending on Domain D
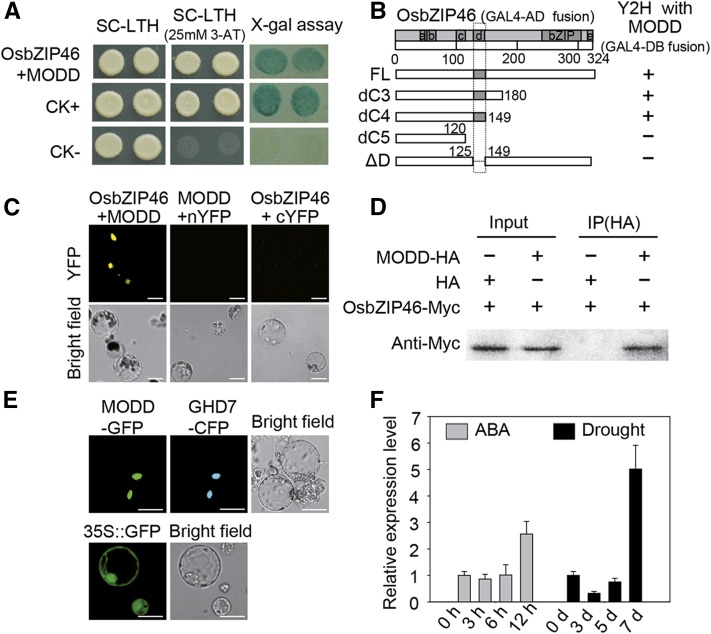
We proposed that the negative regulatory function of domain D might be attributed to an interacting partner of OsbZIP46. Therefore, a yeast two-hybrid (Y2H) screen was conducted to identify OsbZIP46-interacting proteins. Approximate 106 yeast transformants were screened, and three positive clones were identified to contain the same cDNA with its full-length sequence encoding a protein designated as MODD (Figure 2A). Sequence analysis indicated that MODD (LOC_Os03g11550), which is homologous to AFP in Arabidopsis, contains the unknown functional domain DUF1675 according to the RGAP annotation database (http://rice.plantbiology.msu.edu/; Supplemental Figures 2A and 2B). We further examined which region in OsbZIP46 is indispensable for the OsbZIP46-MODD interaction in a yeast system. Surprisingly, the domain D was necessary for the interaction of the two proteins (Figure 2B). Moreover, the interaction between OsbZIP46 and MODD was further confirmed by bimolecular fluorescence complementation (BiFC) and coimmunoprecipitation (Co-IP) in rice protoplasts (Figures 2C and 2D). Further experiments showed that two other ABF members, OsbZIP23 and OsbZIP72, could also interact with MODD (Supplemental Figure 3). In addition, according to the Plant Interactome Database (http://interactome.dfci.harvard.edu/A_thaliana), AFP, a MODD homolog in Arabidopsis, could interact with multiple ABFs including ABI5 (Supplemental Table 1; Garcia et al., 2008).
Figure 2.
Identification of the OsbZIP46 Interacting Protein MODD.
(A) Yeast two-hybrid assay of OsbZIP46 and MODD. CK+ and CK− indicate the positive and negative control, respectively. SC-LTH, synthetic complete-Leu-Trp-His medium; 3-AT, 3-amino-1,2,4-triazole.
(B) Detection of the interaction domain between OsbZIP46 and MODD by Y2H assays using serially truncated forms of OsbZIP46. The + indicates positive (interaction) result; − indicates negative (no interaction) result.
(C) Confirmation of the interaction of OsbZIP46 and MODD by BiFC in rice protoplasts as shown by a yellow fluorescence signal. nYFP, the construct for the YFP N-terminal fusion expression. cYFP, the construct for the YFP C-terminal fusion expression. Bars = 20 μm.
(D) Confirmation of the interaction of OsbZIP46 and MODD by Co-IP assay. Total proteins from the protoplasts expressed with OsbZIP46-Myc and MODD-HA or a vector control (HA) were used. Proteins before (input) and after IP were detected with the anti-Myc antibody.
(E) Subcellular localization of MODD. GHD7-CFP is a nuclear marker. 35S:MODD-GFP (MODD-GFP) and 35S:GHD7-CFP (GHD7-CFP) were cotransformed into etiolated shoot protoplasts of rice. 35S:GFP was transformed as a control. Bars = 20 μm.
(F) The expression of MODD is inducible by ABA and drought stress. The x axis is the time course for sampling (h, hours; d, days). Error bars indicate standard errors based on three replicates.
Since OsbZIP46 is a nuclear protein (Tang et al., 2012), we further investigated the subcellular localization of the MODD protein. The MODD protein with a GFP tag under the control of a constitutive cauliflower mosaic virus 35S promoter was transiently expressed in rice mesophyll protoplasts. The nuclear localization of MODD-GFP was confirmed by its colocalization with the CFP-fused nuclear protein GHD7 (Figure 2E; Supplemental Figure 4), indicating that MODD is also a nuclear protein.
According to the expression profiles in the RiceXPro database (http://ricexpro.dna.affrc.go.jp/), MODD and OsbZIP46 were coexpressed across various tissues and organs (Supplemental Figure 5). Both genes showed circadian rhythmic expression, but their peaks were different (Supplemental Figure 6). Moreover, considering that the expression of OsbZIP46 was induced by drought and ABA, we checked the expression of MODD based on RT-qPCR. The results showed that MODD expression was also induced by ABA and drought stress, but the induction appeared only at the late stage of the treatments (Figure 2F), which contrasts with the induction of OsbZIP46 at the early stage (Tang et al., 2012), implying that MODD may have different roles in ABA signaling and the drought stress response when compared with OsbZIP46.
MODD Negatively Regulates ABA Signaling and Drought Resistance
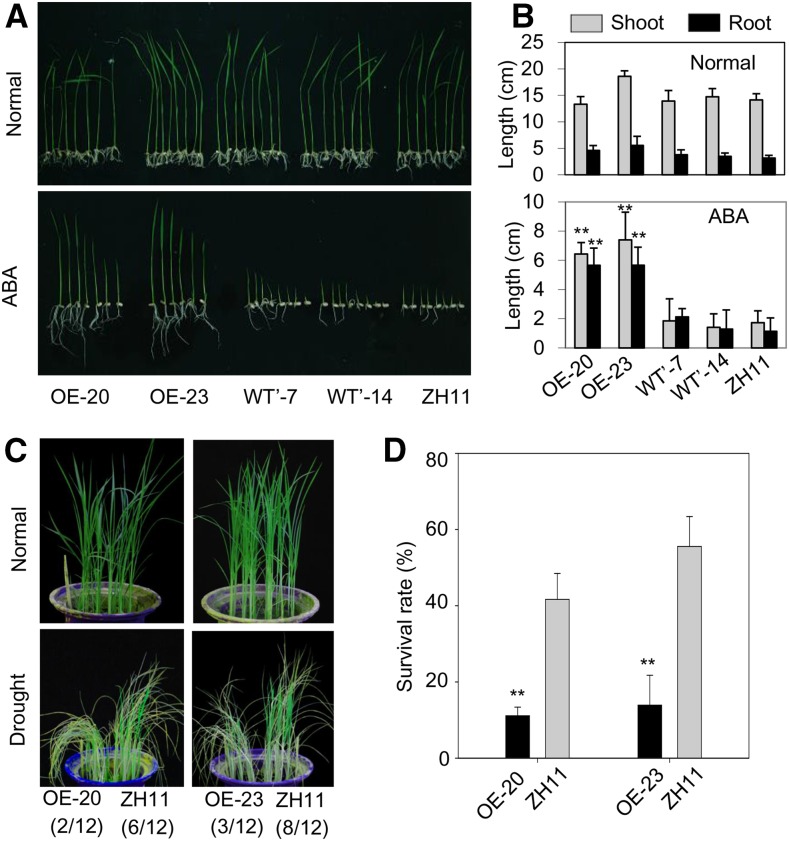
Since OsbZIP46 functions as a positive regulator of ABA signaling and drought stress response in rice, we investigated the possible roles of MODD in ABA signaling and the drought stress response. We generated transgenic rice overexpressing MODD (Supplemental Figure 7) and tested these plants for ABA sensitivity and drought resistance. The MODD overexpression plants exhibited significantly longer shoots and roots than those of the negative control transgenic lines and the wild type under ABA treatment, but no differences were observed when the plants were grown under normal growth conditions (Figures 3A and 3B). This indicates the decreased ABA sensitivity of the MODD overexpression rice plants, in contrast to the results of the OsbZIP46 overexpression rice plants (Tang et al., 2012). After drought stress treatment, the survival rates of the MODD overexpression lines were significantly lower than that of the wild type (Figures 3C and 3D), which was consistent with their decreased ABA sensitivity. These results suggest that MODD may have a negative role in ABA signaling and drought resistance.
Figure 3.
Phenotype of MODD Overexpression Plants under ABA Treatment and Drought Stress Treatment.
(A) and (B) Reduced ABA sensitivity of the MODD overexpression plants.
(A) Performance of two independent MODD-overexpression transgenic lines (OE-20 and OE-23), two negative transgenic lines (WT’-7 and WT’-14), and the wild type (ZH11) in MS medium containing 0 µM ABA (Normal) or 3 µM ABA (ABA treatment).
(B) The shoot and root lengths were measured at 10 d after germination. Error bars indicate standard deviations (n ≥ 9). The statistical significance between data in the OE and wild-type plants was determined using the Student's t test (**P < 0.01).
(C) and (D) Drought tolerance testing of MODD overexpression plants.
(C) Performance of two MODD overexpression lines (OE-20 and OE-23) and the wild type (ZH11) under normal or drought stress conditions. Values in parentheses indicate the number of survived plants out of the total number of plants presented in the pictures.
(D) Survival rates of the overexpression lines and the wild type after drought stress treatment. Error bars indicate the standard deviations based on three replicates (**P < 0.01, Student's t test).
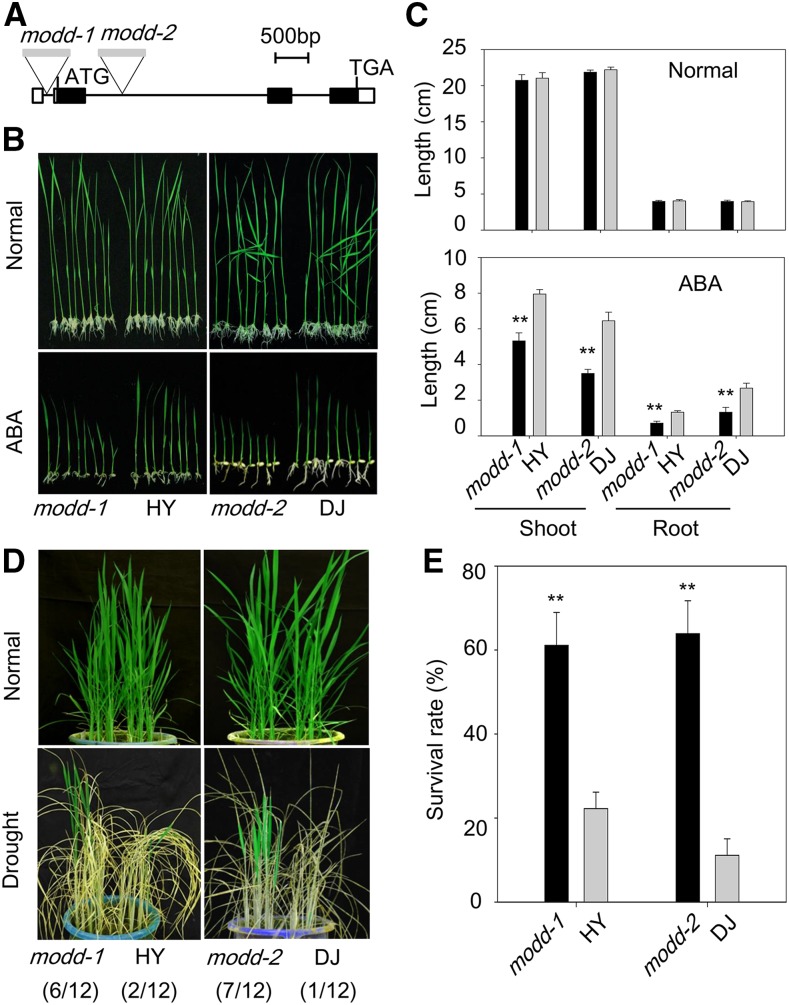
To confirm the negative role of MODD in ABA signaling and drought resistance, we collected two allelic rice T-DNA insertion mutants, modd-1 (1D-03345) and modd-2 (3A-14659), with insertions in the 5′-untranslated region and the first intron of the MODD gene, respectively. The T-DNA insertions and the lack of an intact MODD transcript in the modd mutants were confirmed using PCR and RT-qPCR, respectively (Supplemental Figure 8; the primers are listed in Supplemental Data Set 1). Then, the ABA sensitivity and drought stress tolerance of both mutants were further investigated by evaluating the growth performance under ABA treatment and the survival rates after drought stress treatment, respectively. Contrary to the phenotypes of the MODD overexpression lines, the mutant lines showed strikingly increased ABA sensitivity and drought resistance compared with the wild type (Figures 4A to 4E). Taken together, these results indicate that MODD functions as a negative regulator in ABA signaling and drought response in contrast to the positive regulator OsbZIP46.
Figure 4.
Phenotype of the modd Loss-of-Function Mutant Plants under ABA Treatment and Drought Stress Treatment.
(A) Characterization of two allelic modd T-DNA insertion mutants (modd-1 and modd-2). Schematic representation of the MODD gene and insertion position of the T-DNA. The rectangular blocks indicate exons.
(B) and (C) Increased ABA sensitivity of the modd T-DNA insertion mutants.
(B) Performance of two allelic mutants (modd-1 [HY background] and modd-2 [DJ background]) and the wild type (HY or DJ) under normal growth conditions or in MS medium containing 3 µM ABA.
(C) The shoot and root lengths were measured at 14 d after germination. The error bars indicate standard deviations (n ≥ 9). Statistical significance between data in the mutants and the wild type was determined using the Student's t test (**P < 0.01).
(D) and (E) Drought tolerance testing of the modd T-DNA insertion mutants.
(D) Performance of two allelic mutants and the wild type under normal or drought stress conditions. Values in parentheses indicate the number of survived plants out of the total number of plants presented in the pictures.
(E) Survival rates of the mutants and the wild type after drought stress treatment. Error bars indicate the standard deviations based on three replicates (**P < 0.01, Student's t test).
MODD Adversely Affected the Expression of OsbZIP46 Target Genes
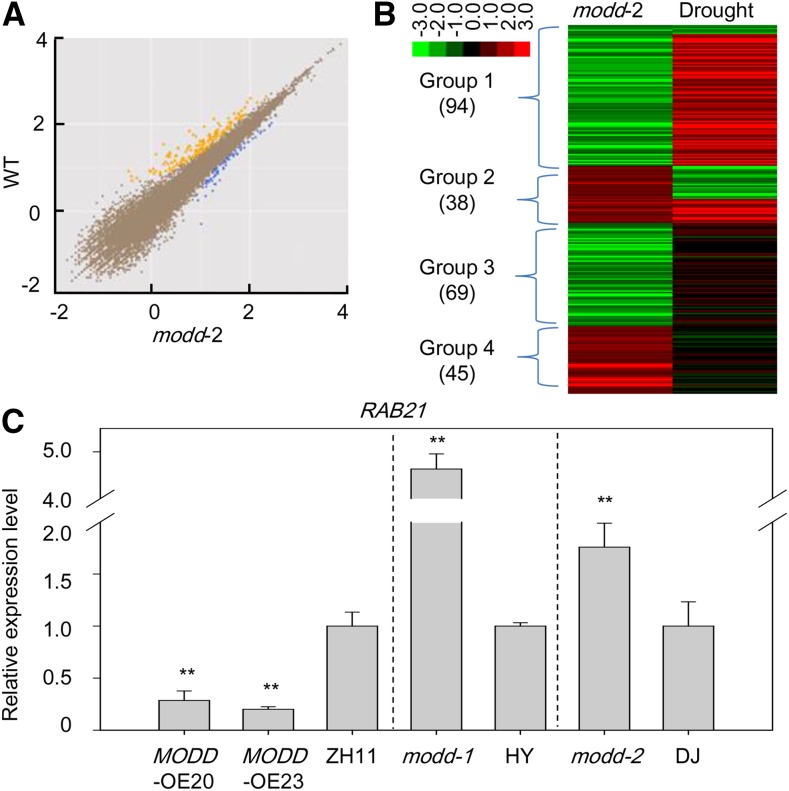
To elucidate the molecular function of MODD, we compared expression profile changes in the knockout mutant modd-2 using RNA-seq. Compared with the wild type, the transcriptome of the modd-2 mutant exhibited significant changes. With a threshold of 2-fold change (diverge probability ≥0.8), a total of 163 and 83 genes were down- and upregulated, respectively, in the modd-2 plants (Figure 5A; Supplemental Data Sets 2 and 3). In addition, we further classified all of these genes based on their change patterns in the mutant and their responsiveness to drought stress. Based on the published microarray results (Jain et al., 2007), more than half of these genes were regulated by exposure to drought stress treatment. Among the 246 expression-affected genes, 94 downregulated (group 1) and 32 upregulated genes (group 2) were drought responsive, suggesting the important function of MODD in the regulation of drought responsive gene expression (Figure 5B).
Figure 5.
Expression Profiles of Genes Regulated by MODD.
(A) Scatterplots of the whole-genome expression profiles of genes in the modd knockout (modd-2) compared with the wild type by RNA-seq. The x and y axes indicate the gene expression levels (log10 transformation) in the modd knockout and the wild type, respectively. The blue and yellow dots indicate the genes with mutant-to-wild type signal ratios of >2 and <0.5, respectively.
(B) Drought-responsive patterns of differentially expressed genes in the modd knockout and wild type. Group 1 genes are downregulated in modd-2 and are drought responsive; group 2 genes are upregulated in modd-2 and are drought responsive; group 3 and group 4 genes are regulated in modd-2 but are not drought responsive.
(C) The expression of the OsbZIP46-targeting genes including RAB21, which are positively regulated by OsbZIP46, were downregulated in the MODD overexpression lines OE-20 and OE-23, and upregulated in the modd loss-of-function mutants (modd-1 and modd-2); ZH11, HY, and DJ are the wild-type controls of the MODD overexpressors, modd-1 and modd-2, respectively. Error bars indicate the standard errors based on three replicates (**P < 0.01, Student's t test).
Considering that MODD physically interacts with OsbZIP46 and that OsbZIP46 functions as a transcription activator to regulate ABA and stress signaling by directly binding the promoters of its target genes and regulating their expression (Tang et al., 2012; Supplemental Figure 9), we speculated that MODD may affect the expression of the OsbZIP46 target genes. To verify this speculation, the expression levels of the OsbZIP46 target genes were checked in the MODD overexpression and mutant lines by RT-qPCR. The results showed that the transcript levels of RAB21, which is positively regulated by OsbZIP46 (Tang et al., 2012), were downregulated in the MODD overexpression lines and upregulated in the modd mutants respectively (Figure 5C). Most of the other OsbZIP46-regulated genes also showed reverse change patterns between the OsbZIP46-OE and MODD-OE plants or similar change patterns between the OsbZIP46-OE and modd-2 plants (Supplemental Figure 10). These results imply that MODD can antagonize the function of OsbZIP46 by negatively regulating the expression levels of OsbZIP46-targeted genes involved in ABA signaling and the drought stress response.
MODD Counteracts the Transcriptional Activation of OsbZIP46 via Domain D
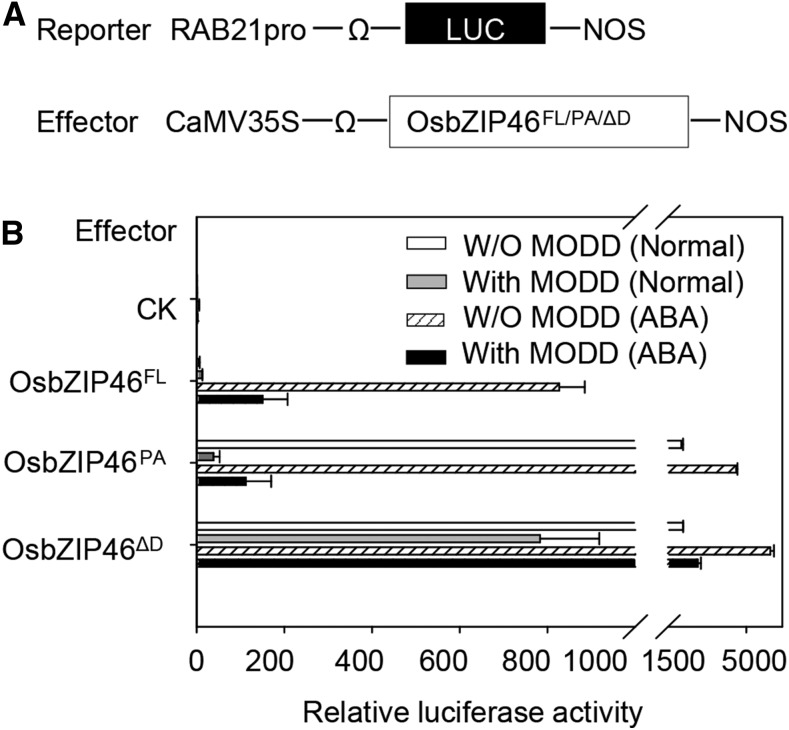
Given that domain D negatively regulates OsbZIP46 activity and that MODD interacts with domain D to antagonize OsbZIP46 function, we were curious about the possible influence of MODD on the transcriptional activation activity of OsbZIP46. By using the dual-luciferase reporter system, we analyzed the transcriptional activity of OsbZIP46FL, OsbZIP46PA (the phosphorylation active form with domain D; Tang et al., 2012), and OsbZIP46ΔD along with MODD as a coeffector (Figure 6A) under normal conditions and ABA treatment. The results showed that the protoplasts transfected with OsbZIP46FL (under ABA treatment) or OsbZIP46PA (under normal conditions or ABA treatment) without the cotransfection of MODD had very high relative luciferase activity, but their activities decreased sharply when cotransfected with MODD (Figure 6B). These results suggested that MODD could significantly inhibit the activity of OsbZIP46FL and OsbZIP46PA. Distinctively, the inhibition effect that resulted from MODD was decreased in the protoplasts transfected with OsbZIP46ΔD, which showed high relative luciferase activities under normal conditions or ABA treatment, even when cotransfected with MODD (Figure 6B). These results indicated that MODD can counteract the transcriptional activation activity of OsbZIP46 through the domain D.
Figure 6.
MODD Antagonizes the Transcriptional Activation Activity of OsbZIP46 Dependent on the Presence of Domain D.
The scheme of the constructs for the effector and reporters is shown in (A). The relative luciferase activities in rice protoplasts that had been cotransfected with a reporter plasmid and three different forms of OsbZIP46, together with or without (W/O) MODD cotransfection in normal conditions or under ABA treatment are shown in (B). OsbZIP46FL, the native full length; OsbZIP46PA, the phosphorylation active form with multiple amino acid substitutions (Ser/Thr to Asp), providing a negative charge to mimic the phosphorylated status, and domain D is contained (Tang et al., 2012); OsbZIP46ΔD, domain D-missing mutated form of OsbZIP46. The error bars indicate standard deviations (n = 3).
MODD Mediates Transcriptional Repression by Recruiting the OsTPR3-HDA702 Complex and Regulating Chromatin Remodeling
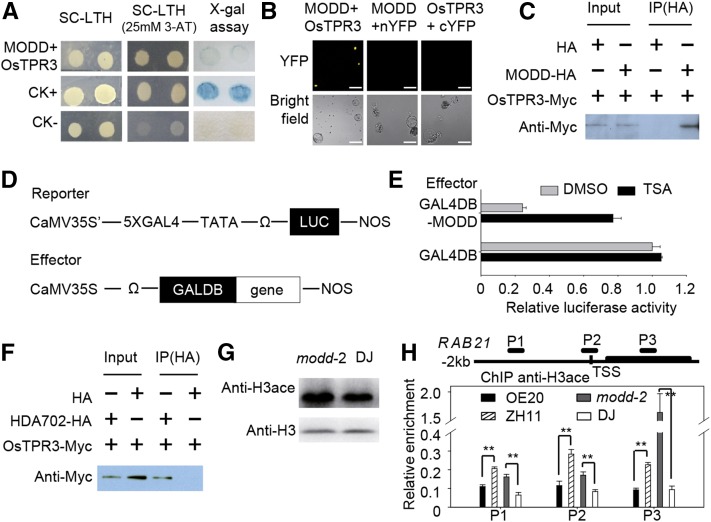
Since MODD could directly bind to OsbZIP46 to counteract its transcriptional activation activity, we attempted to clarify the mechanism of regulation by studying the interacting partners of MODD. Fine analysis of the MODD protein sequence revealed a remarkable EAR motif (LxLxL), a hallmark of transcriptional repressors (Kazan, 2006) in the N-terminal conserved domain (Supplemental Figures 2C and 2D). Considering that TPL/TPR-type transcriptional corepressors have been reported to function as direct interactors of EAR motif-containing proteins (Krogan and Long, 2009), we speculated that a direct interaction between the MODD and TPL/TPR corepressors may exist in rice. The results showed that OsTPR3, one of the three TPL/TPR corepressors in rice, indeed interacted with MODD in the Y2H assay, and the interaction was further confirmed by BiFC and Co-IP assays in rice protoplasts (Figures 7A to 7C).
Figure 7.
MODD Mediates Transcriptional Repression through Facilitating Chromatin Deacetylation.
(A) to (C) Identification of OsTPR3 as a MODD-interacting protein by a Y2H assay (A), BiFC (B), and Co-IP assays (C) in rice protoplasts. For the Co-IP assays, total proteins from the protoplasts expressed with OsTPR3-Myc and MODD-HA or a vector control (HA) were used. Proteins before (input) and after IP were detected with the anti-Myc antibody. Bars in (B) = 20 μm.
(D) Scheme of the constructs used in the cotransfection experiments. The GAL4-responsive reporter construct contained the CaMV35S enhancer (CaMV35S’), five copies of the GAL4 binding site in tandem, and a minimal TATA region of the CaMV 35S promoter, the firefly gene for luciferase (LUC), and a nopaline synthase (Nos) terminator. The effector construct contained a GAL4 DNA binding domain and the coding region of the tested genes driven by the CaMV 35S promoter; a translational enhancer sequence from tobacco mosaic virus (Ω) was located upstream of the translation initiation site.
(E) MODD acts as a transcriptional repressor in rice protoplasts, and the repression activity is abolished by TSA (histone deacetylation inhibitor) treatment. Rice leaf protoplasts were transfected with a MODD-GALDB derivative or GAL4DB-only effector plasmids plus the 35S:GAL4-LUC reporter plasmid and treated with 20 μM TSA (black bars) or DMSO (gray bars) for 6 h before fluorescence levels were determined. The data indicate mean values with standard deviations of three biological replicates.
(F) Identification the interaction between OsTPR3 and HDA702 by Co-IP in rice protoplasts. Total proteins from protoplasts expressed with OsTPR3-Myc and HDA702-HA or the vector control (HA) were used.
(G) Acetylated histone H3 levels in the knockout mutant (modd-2) and wild-type (DJ) plants were revealed by protein gel blots. The histone extraction was tested with anti-acetyl-Histone H3 antibody. H3 was detected as a loading control.
(H) ChIP combined with qPCR was used to measure histone acetylation levels at the OsbZIP46 target genes in the MODD overexpressor (OE20), modd-2 mutant, and wild type (ZH11 and DJ). ChIP was performed with anti-acetyl-histone H3 antibody, and signals from the ChIP assays are represented as enrichment values normalized to actin. The diagram of the RAB21 locus is shown in the upper panel. The transcribed region is represented by the thick line. The relative positions of the transcription start site (TSS) and the primer sets (P1 to P3) used in the ChIP experiments are indicated. The data indicate mean values plus standard errors of three biological replicates. The statistical significance between data in the mutant and wild type was determined using Student's t test (**P < 0.01).
Considering the association with OsTPR3, we proposed that MODD might have active transcriptional repression activity. To confirm this, we directly assessed the transcriptional repression capability of MODD using a dual-luciferase reporter system. Given that MODD has no predicted DNA binding domain, MODD fused with the GAL4 DNA binding domain (GAL4DB) at the N terminus was constructed as an effector. The firefly luciferase gene driven by the CaMV35S enhancer (CaMV35S’) with five copies of the GAL4 binding element in front of a TATA box was used as a reporter (Figure 7D), and the Renilla luciferase gene driven by a 35S promoter was used as the internal control. The results showed that the effector containing MODD had significantly lower relative luciferase activity compared with the GAL4DB control, and the repressive effect represented by the reduced activity ratio reached almost 70% (Figure 7E), indicating that MODD can actively mediate transcriptional repression.
Increasing evidence supports the idea that transcriptional repression may involve chromatin remodeling (Wolffe, 1996; Goodrich and Tweedie, 2002). Especially, the removal of histone acetylation modifications by HDACs is largely correlated with transcription repression (Shahbazian and Grunstein, 2007); therefore, we checked whether the repression function of MODD involved HDAC-dependent mechanisms. Using the dual-luciferase reporter system, we found that the transcriptional repression effect of MODD was reduced to 30% upon exposure to the HDAC inhibitor Trichostatin A (TSA) (Figure 7E), compared to when TSA was absent. This result suggested that the transcriptional repression effect of MODD was largely dependent on the activity of HDAC. Moreover, an interaction between OsTPR3 and HDA702, the closest rice homolog of the stress-related Arabidopsis HDAC AtHD1/HDA19 (Hu et al., 2009; Chen and Wu, 2010), was confirmed using Co-IP (Figure 7F), suggesting that OsTPR3 connects MODD with HDA702.
To further reveal whether deacetylation is implicated in the MODD-mediated transcriptional repression of OsbZIP46 target genes in vivo, we checked the histone acetylation level in the modd mutant and the wild type by immunoblot with the antibody against acetylated histone. The results showed that the acetylation level in the modd mutant was increased, compared with wild type (Figure 7G). Next, we examined the histone acetylation levels of the OsbZIP46 target genes, including RAB21, in the modd mutant, MODD overexpressor, and the wild type using ChIP-qPCR with an antibody against acetylated histone H3. The enrichment of acetylated histone at the target genes was compared among the modd mutant, MODD-OE, and wild type. Consistent with the elevated expression levels of RAB21 in the modd mutant, we found that the acetylated histone level at RAB21 was significantly increased in the modd mutant compared with that in the wild type (Figure 7H). Similarly, consistent with the reduced expression level of RAB21 in the MODD-OE plants, acetylated histone level at RAB21 in the MODD-OE was also decreased, compared with the wild type (Figure 7H). The gene expression and histone acetylation levels of the other two OsbZIP46-regulated genes were similarly consistent (Supplemental Figure 11), implying that MODD can affect the histone acetylation at the OsbZIP46 target genes to exert its transcriptional repression function.
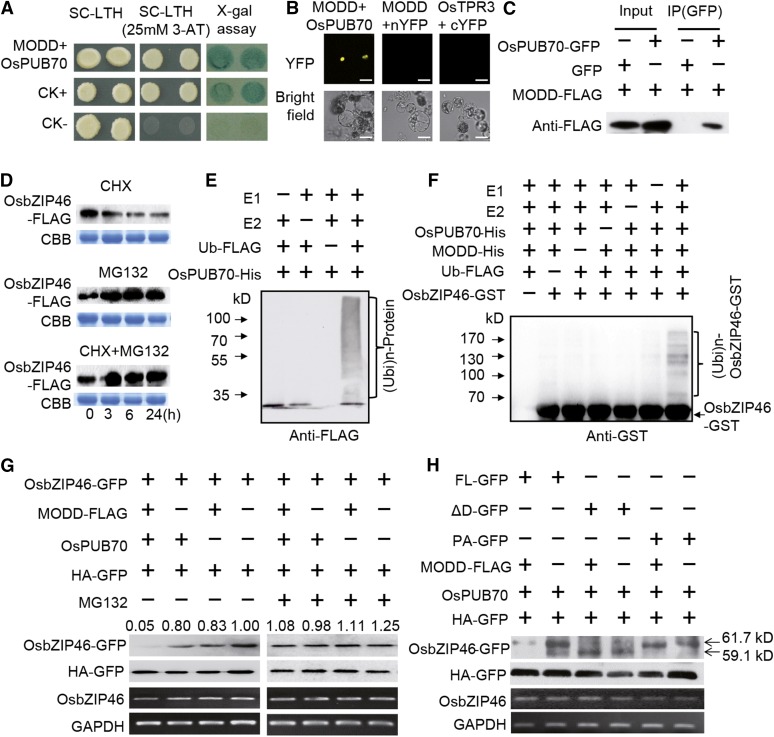
MODD Promotes OsbZIP46 Degradation by Interacting with the E3 Ubiquitin Ligase OsPUB70
Although our results clearly indicate that MODD can mediate transcriptional repression to negatively regulate the activity of OsbZIP46, MODD could also affect the abundance or stability of the OsbZIP46 protein. To further investigate the antagonistic role of MODD, a MODD-interacting protein OsPUB70, which has been predicted to be a potential U-box domain containing ubiquitin E3 ligase (Zeng et al., 2008), was identified from a Y2H screening (two positive clones acquired from screening 106 transformants). The interaction was validated by BiFC in rice protoplasts and Co-IP in tobacco leaves (Figures 8A to 8C). A further Y2H experiment showed that there is no direct interaction between OsbZIP46 and OsPUB70 (Supplemental Figure 12). Ubiquitin E3 ligases have been intensively documented for their essential functions in defining the substrate specificity of the UPS-mediated protein degradation (Vierstra, 2009). Immunoblot assays using protein extracts from OsbZIP46-FLAG seedlings pretreated with cycloheximide (CHX), an inhibitor of de novo protein synthesis, showed reduced OsbZIP46-FLAG levels, while the addition of MG132, a 26S proteasome inhibitor, led to an increased OsbZIP46-FLAG level (Figure 8D). Furthermore, cotreatment with MG132 and CHX largely suppressed the effect of CHX (Figure 8D). These results indicate that OsbZIP46 is subjected to UPS-mediated protein degradation. Therefore, it is intriguing to detect whether MODD could promote OsbZIP46 ubiquitination and degradation by recruiting OsPUB70.
Figure 8.
MODD Promotes OsbZIP46 Protein Degradation by Recruiting the E3 Ligase OsPUB70 Dependent on the Presence of Domain D.
(A) and (B) Identification of OsPUB70 as a MODD-interacting protein by Y2H screen (A) and BiFC (B). Bars in (B) = 20 μm.
(C) Co-IP assays. Total proteins from the protoplasts expressed with OsPUB70-GFP and MODD-FLAG or a vector control (GFP) were used. Proteins before (input) and after IP were detected with the anti-FLAG antibody.
(D) OsbZIP46 is subjected to proteasome-mediated degradation. Fourteen-day-old seedlings of OsbZIP46-FLAG were treated with 50 μM CHX and/or 20 μM MG132 for 0, 3, 6, and 24 h before total proteins were extracted for immunoblotting using an anti-FLAG antibody. CBB, Coomassie blue staining.
(E) E3 ubiquitin ligase activity of OsPUB70. OsPUB70-His fusion protein was tested for E3 ubiquitin ligase activity in the presence of E1, E2, and ubiquitin-Flag (Ub-FLAG). Ubiquitinated proteins [(Ubi)n-Protein] were detected using anti-FLAG antibody.
(F) OsPUB70 ubiquitinates OsbZIP46 in vitro in the presence of MODD. The full-length OsbZIP46 protein was fused with GST tag (OsbZIP46-GST) and used as the substrate for the assays, which were performed using OsPUB70-His, MODD-His, E1, E2, and Ub-FLAG. GST-tagged substrate protein was detected by immunoblot with anti-GST, and the ubiquitinated OsbZIP46 was indicated as (Ubi)n-OsbZIP46-GST.
(G) MODD-OsPUB70 promotes OsbZIP46 protein degradation dependent on the proteasome. Immunoblot analysis of protein extracts from tobacco leaves agroinfiltrated with the indicated plasmids in the presence or absence of MG132. OsbZIP46-GFP and HA-GFP were detected using anti-GFP antibody and anti-HA antibody, respectively. OsbZIP46-GFP (indicated as OsbZIP46) and GAPDH mRNA expression levels were analyzed by RT-PCR. Values indicate the relative band intensity of OsbZIP46-GFP, which was normalized to control (HA-GFP) and expressed relative to the fourth lane from the left (set as 1.00).
(H) MODD-OsPUB70 promotes degradation of the native full-length OsbZIP46 (FL-GFP, 61.7 kD), but not the domain D-missing mutated form OsbZIP46ΔD (ΔD-GFP, 59.1 kD) and the phosphorylation active form OsbZIP46PA (PA-GFP, 61.7 kD).
We performed in vitro ubiquitination experiments to confirm the function of OsPUB70 as an E3 ligase. OsPUB70-His fusion protein was incubated with E1 (activating enzyme), E2 (conjugating enzyme), and FLAG-tagged ubiquitin. Immunoblot using an anti-FLAG antibody revealed a polyubiquitination signal (Figure 8E, first lane from the right), suggesting that OsPUB70 has E3 ubiquitin ligase activity. To examine whether OsPUB70 could mediate OsbZIP46 protein ubiquitination, OsbZIP46-GST fusion protein was used as a substrate in the in vitro ubiquitination assay in the presence of OsPUB70, MODD, E1, and E2, and ubiquitin. Immunoblot analysis with anti-GST showed that the ubiquitinated OsbZIP46 protein, indicated by a series of shifted bands with higher molecular weight (Figure 8F, first lane from right), was detected only in the presence of all these components. These results suggest that MODD can recruit OsPUB70 to promote OsbZIP46 ubiquitination.
Furthermore, we conducted an in vivo protein degradation assay with an efficient agroinfiltration expression system established in Nicotiana benthamiana (Liu et al., 2010, 2012). GFP-tagged OsbZIP46 was transiently expressed via agroinfiltration, and the OsbZIP46-GFP protein was detected by immunoblot. OsbZIP46-GFP protein abundance rapidly decreased when coexpressed with MODD, but only in the presence of OsPUB70 (Figure 8G, first lane from the left). However, when the 26S proteasome inhibitor MG132 was infiltrated into the same region, the degradation of the OsbZIP46-GFP protein by MODD-OsPUB70 was inhibited (Figure 8G, fifth lane from the left). Moreover, in contrast to the native form of the full-length OsbZIP46 (OsbZIP46FL), two mutated forms of the OsbZIP46 protein, the D domain-missing form (OsbZIP46ΔD), and the phosphorylation active form (OsbZIP46PA) could not be degraded by MODD and OsPUB70 in the protein degradation assays (Figure 8H). These results suggest that MODD promotes the native OsbZIP46, but not the active mutated forms (OsbZIP46ΔD and OsbZIP46PA), for UPS-mediated protein degradation by recruiting OsPUB70, suggesting an antagonistic role of MODD on OsbZIP46 protein stability.
OsPUB70 Negatively Regulates ABA Signaling
To support the biochemical results above, transgenic rice overexpressing OsPUB70 were generated (Supplemental Figure 13). Interestingly, the ABA sensitivity of OsPUB70 overexpression plants was decreased compared with the wild type (Figures 9A and B), which is similar to the results of the MODD overexpression rice plants. We further identified the ospub70 knockout T-DNA insertion mutant (3A-08808) (Supplemental Figure 14). Contrary to the phenotype of the OsPUB70 overexpression lines, the mutant line showed increased ABA sensitivity compared with the wild type (Figures 9C and 9D). Taken together, these results indicate that OsPUB70 functions as a negative regulator of ABA signaling.
Figure 9.
Reduced ABA Sensitivity of the OsPUB70 Overexpression Plants and Increased ABA Sensitivity of the ospub70 T-DNA Insertion Mutant.
(A) and (B) Two independent OsPUB70 overexpression transgenic lines (OsPUB70-OE6 and OE-7) and the wild type (ZH11) were tested for their ABA sensitivity.
(A) Performance of an OsPUB70 overexpression transgenic line (OsPUB70-OE6) and the wild type in MS medium containing 0 µM (normal) or 3 µM ABA.
(B) The shoot and root lengths were measured at 10 d after germination. Error bars indicate the sd (n ≥ 9). The statistical significance between data in the overexpression line and wild type was determined using the Student's t test (*P < 0.05 and **P < 0.01).
(C) and (D) Increased ABA sensitivity of the ospub70 T-DNA insertion mutant.
(C) Performance of ospub70 (DJ background) and the wild type (DJ) under normal growth conditions or in MS medium containing 3 µM of ABA.
(D) The shoot and root lengths were measured at 14 d after germination. The error bars indicate the sd (n ≥ 9). Statistical significance between data in the mutants and wild type was determined using the Student's t test (*P < 0.05).
DISCUSSION
The Negative Regulatory Domain D Commonly Exists in the ABF Subfamily
In this report, we focused on dissecting the negative regulatory mechanism of OsbZIP46 transcriptional activity mediated by domain D. Fine regulation of transcriptional activity is important for the functions of ABF subfamily members, which have prominent roles in ABA signaling (Fujii and Zhu, 2009; Fujita et al., 2009). In accordance with that, OsbZIP46 activity was almost completely repressed under nonstressed conditions, and OsbZIP46 requires ABA for full activation, implying regulation of OsbZIP46 activity. Similarly, two other rice ABF members, OsbZIP23 and OsbZIP72, also require ABA for full activation in a protoplast assay (Supplemental Figure 1; Yoshida et al., 2010). Moreover, the ABF members in Arabidopsis, including AREB1, AREB2, and ABF3, have also been reported to require ABA for full activation (Yoshida et al., 2010). These results implied that the regulation of their activity was pivotal for most ABF subfamily members. In addition, the domain D, which corresponds to the C3 domain in Arabidopsis ABF members (Bensmihen et al., 2002), could also have a negative regulatory function in Arabidopsis, based on the observation of activity regulation of modified AREB1 and ABI5 (Fujita et al., 2005; Tezuka et al., 2013). A constitutively active form, AREB1∆QT, was produced with an internal deletion in which domain D is included (Fujita et al., 2005). Also, the abi5-9 allele, which encodes an intact ABI5 protein with one amino acid substitution (A214G) in domain D, showed a change in the transcriptional activation activity (Tezuka et al., 2013), suggesting the importance of the conserved Ala in domain D. Considering that the domain D sequence was quite conserved in most of the ABF subfamily bZIP members (Tang et al., 2012; Yoshida et al., 2010), domain D may function as a general negative regulator of the activities of plant ABFs.
The Function of Domain D Involves the Interacting Partner MODD
Since the domain D is important for regulation of transcriptional activity, it is tempting to dissect the negative regulatory mechanism. Domain D might be important for the conformational maintenance of the protein or for interacting with its partner(s). The activities of transcription factors could be regulated, sometimes even in a reverse manner, by interacting with different partners, which could be coactivators or corepressors. Increasing evidence suggests that the interacting protein largely determines whether a transcription factor acts as a transcriptional activator or repressor. For instance, flower development-related MADS box family members SEPALLATA3 and AGAMOUS-like 15 could function as either activators or repressors depending on their association with different interacting factors (Castillejo et al., 2005; Sridhar et al., 2006; Hill et al., 2008).
The OsbZIP46-MODD interaction appeared to be very intriguing since domain D is indispensable for mediating the interaction, which strongly implies that the stress-inducible interacting partner MODD could facilitate the function of domain D. This hypothesis was further validated by functional analysis of the MODD protein. Overexpression of MODD in rice resulted in decreased ABA sensitivity and drought resistance, whereas modd-1 and modd-2 mutants showed increased ABA sensitivity and drought resistance. MODD was identified to be a negative regulator of ABA signaling and the drought stress response, in contrast to the positive regulatory function of OsbZIP46. Moreover, the expression levels of most OsbZIP46 target genes decreased with increasing expression of MODD. More interestingly, a dual-luciferase reporter assay showed that MODD could negatively regulate the expression of OsbZIP46 target genes through suppressing the activity of OsbZIP46 harboring domain D, but not the activity of OsbZIP46ΔD, which was missing domain D, indicating that the repression effect on OsbZIP46 possessed by MODD is dependent on domain D. Meanwhile, the interaction of OsbZIP46 with MODD, mediated essentially by domain D, also supports the negative regulatory function of domain D.
In addition, the interaction with MODD was not exclusively confined to OsbZIP46 because we found that other ABF members in rice, including OsbZIP23 (Xiang et al., 2008) and OsbZIP72 (Lu et al., 2009), could also interact with MODD through domain D (Supplemental Figure 3). Consistent with this, AFP, a MODD homolog in Arabidopsis, was reported to interact with multiple ABFs including ABI5 (Supplemental Table 1; Garcia et al., 2008). According to the negative regulatory function of domain D in OsbZIP46 and the similar interaction mediated by domain D in the ABF subfamily, we propose that domain D plays an important role in regulating the activities of ABF transcription factors through interaction with other regulatory partners, including MODD or MODD-like proteins. Nevertheless, it cannot be excluded that domain D may contribute to a conformational change that affects posttranslational modifications such as phosphorylation.
MODD Is Involved in the Regulation of Both the Activity and Stability of OsbZIP46 through Recruiting Different Partners
Our data clearly demonstrated that MODD, which harbors a typical transcription repression motif LxLxL, acts as a general transcriptional repressor. Repression domains can usually mediate the interaction with corepressors, which may in turn recruit other chromatin remodeling regulators including HDACs to promote the formation of a repressive chromatin state (Shahbazian and Grunstein, 2007; Krogan and Long, 2009). Previous reports implicated TPL/TPR-type transcriptional corepressors in multiple hormone signaling pathways (Krogan and Long, 2009). The jasmonic acid signaling component NINJA, which has been reported to be an interaction mediator of TOPLESS and a JAZ transcriptional repressor (Pauwels et al., 2010), showed certain similarity to MODD on the basis of the protein sequence comparison (Supplemental Figure 2B and Supplemental Data Set 4), though the sequence similarity is relatively low. To date, no direct evidence is available to support the idea that the TPL/TPR related modules are involved in ABA or drought stress signaling. There are three homologs of TPL-type corepressors in rice (OsTPR1, 2, and 3), which function in the regulation of meristem fate in rice (Yoshida et al., 2012). Here, we showed that OsTPR3 could connect MODD with a histone deacetylase HDA702 by forming a MODD-OsTPR3-HDA702 interaction complex. Consistent with this, our results revealed that the repression activity of MODD was dependent on the activity of HDAC, and MODD significantly affected the histone acetylation levels of the OsbZIP46 target genes to regulate their expression, indicating that MODD represses OsbZIP46 activity through the regulation of chromatin remodeling, which may be attributed to the OsTPR3-HDA702 partners.
In addition, sequence analysis revealed that MODD is homologous to the Arabidopsis ABI5 binding protein AFP (Supplemental Figure 2B and Supplemental Data Set 4), which promotes the degradation of the ABI5 protein (Lopez-Molina et al., 2003), although the detailed molecular mechanism of AFP action is unclear. Therefore, it will be intriguing to test whether MODD has a similar function as AFP on OsbZIP46 protein stability. Interestingly, our results showed that MODD could recruit E3 ligases such as OsPUB70 and thereby promote the ubiquitination and UPS-mediated degradation of the OsbZIP46 protein, which is dependent on domain D. This suggests that domain D may also act as a degron, in addition to its prominent effect on transcriptional repression, and interact with MODD to regulate OsbZIP46 protein stability through recruiting OsPUB70. Such a dual-posttranslational regulation of OsbZIP46 activity and stability mediated by MODD and the interaction with domain D may be fulfilled through recruiting different interacting partners.
Coordination of MODD-Mediated Negative Regulation of OsbZIP46 in ABA Signaling and the Drought Stress Response
In contrast with the prominent role of OsbZIP46 in positively regulating ABA signaling and the drought stress response (Tang et al., 2012), MODD functions as a negative regulator of ABA signaling and the drought stress response through the regulation of OsbZIP46 activity and/or stability. Hence, the coordination of these two regulators with reverse roles in the same processes may be especially important for the regulation of ABA signaling and drought tolerance.
MODD and OsbZIP46 showed coexpression patterns across various tissues and organs (Supplemental Figure 5), and both genes showed circadian rhythmic expression, but their peaks of expression were shifted (Supplemental Figure 6), implying that the temporal actions of the two genes does not completely overlap. Of special note, MODD was also inducible by ABA and drought stress, but the induction lagged behind that of OsbZIP46 (Figure 2F), implying that MODD may be involved in the attenuation of ABA and stress signaling at the late stage of the stress time course.
We have shown that OsbZIP46FL had significantly enhanced activity when cotransfected with SAPK9 and/or under ABA treatment (Figure 1), suggesting that OsbZIP46 can be activated by SnRK2-type protein kinases in ABA signaling. Correspondingly, the phosphorylation-mimic form OsbZIP46PA showed significantly enhanced activity, like the domain D-truncated form OsbZIP46ΔD (Figure 6B), implying that these two forms may exhibit a similar conformation change that may affect the recruitment of interacting partners for degradation and/or deactivation of the active forms. Indeed, the stability of OsbZIP46PA was not affected by MODD, which is similar to the domain D-truncated form (Figure 8H). However, the activity of OsbZIP46PA could still be partially inhibited by MODD (Figure 6B). It is possible that the native phosphorylated OsbZIP46 may also possess a conformation change that disturbs the MODD-mediated recruitment of ubiquitination partners and the negative role of MODD in activation of OsbZIP46, but the phosphorylation mimic may not completely resemble the native fully phosphorylated OsbZIP46.
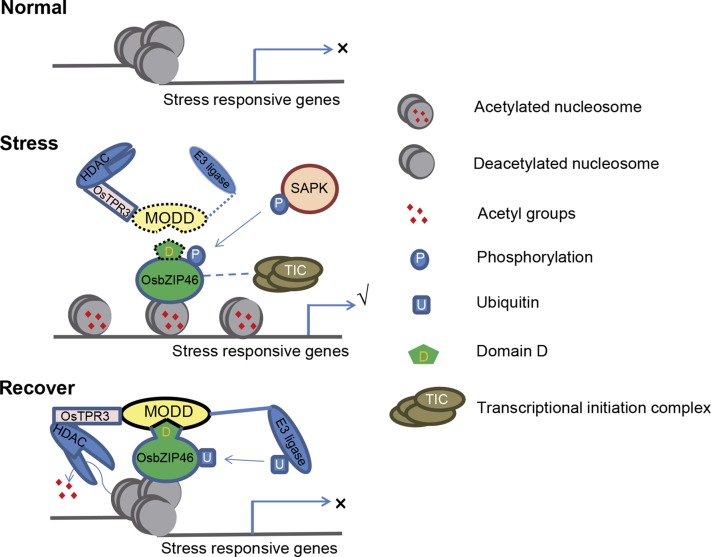
Taking these results together, we propose a model to describe the function of MODD-mediated negative regulation of OsbZIP46 in ABA signaling and the drought stress response (Figure 10). Under normal conditions, both OsbZIP46 and MODD have relatively low expression levels, and the stress-responsive genes may not be activated or have low expression levels. Upon exposure to drought stress conditions, OsbZIP46 is induced and activated by SnRK2-type protein kinases such as SAPK9, but MODD is not induced at the early stage of the stress, so that the function of activated OsbZIP46 is not significantly affected by low levels of the MODD protein. Meanwhile, it is also possible that the negative regulatory effect of MODD is weakened due to a disturbance in the physical interaction of MODD with the phosphorylated active OsbZIP46. In either case, the phosphorylated OsbZIP46 is dominant and antagonizes the negative regulatory effect from MODD at the early stage after ABA or drought stress signaling. When the stress continues for a certain length of time, MODD is also induced and MODD interacts with OsbZIP46 to attenuate the transcriptional activity and/or stability of OsbZIP46, resulting in attenuation of stress signaling. In other words, MODD acts like a brake to avoid the excessive acceleration of the stress signaling and to facilitate the timely decay of the stress signaling when the stress is removed, thus allowing plants to reach a reasonable balance in dealing with the stress responses and other biological processes, especially growth.
Figure 10.
Working Model for MODD in the Negative Regulation of Stress Responses.
Domain D mediates the interaction with MODD that plays pivotal roles in the activity attenuation and degradation of OsbZIP46. The dashed line for transcriptional initiation complex (TIC) means a predicted interaction. The dotted MODD represents a low expression level in the early stage of stress. The dotted domain D represents the possible conformation change of OsbZIP46 resulting from phosphorylation, which may partially disturb the domain D-mediated protein interaction complex. The dotted line to E3 ligase represents the potential disturbed recruitment of ubiquitination machinery resulting from the OsbZIP46 conformation change.
Considering that the dephosphorylation by several types of phosphatases has been reported to negatively regulate the activities of ABF members (Lynch et al., 2012; Dai et al., 2013; Hu et al., 2014), it will be intriguing to clarify the potential relationship between dephosphorylation and MODD-mediated deactivation in further studies.
In conclusion, this study revealed the mechanism of the negative regulatory function of domain D in OsbZIP46, an important ABF member participating in ABA signaling and the drought stress response, and found that MODD negatively regulates OsbZIP46 function by posttranslational regulation of both the activity and stability of OsbZIP46. The phosphorylation activation by a SnRK2 (SAPK)-type protein kinase and the activity and/or stability regulation mediated by MODD (through interaction with domain D) are coordinately integrated with OsbZIP46 to fine tune ABA signaling and the drought stress response.
METHODS
cDNA Clones
The full-length cDNAs of SAPK9 (AK069697), OsTPR3 (AK111518), HDA702 (AK068682), and OsPUB70 (AK069675) were obtained from KOME cDNA library (http://cdna01.dna.affrc.go.jp/), and the full-length cDNA of MODD (EI#84-J20) was obtained from a REDB cDNA library of Minghui 63 (Oryza sativa ssp indica; http://redb.ncpgr.cn/).
Rice Protoplast Preparation and Transformation
Rice protoplasts were prepared from 13-d-old seedlings of rice Zhonghua 11 (O. sativa ssp japonica) growing on half-strength Murashige and Skoog (1/2MS) media as described previously (Xie and Yang, 2013). The protoplasts were isolated by digesting rice sheath strips in digestion solution (10 mM MES, pH 5.7, 0.5 M mannitol, 1 mM CaCl2, 5 mM β-mercaptoethanol, 0.1% BSA, 1.5% Cellulase R10 [Yakult Honsha], and 0.75% Macerozyme R10 [Yakult Honsha]) for 4 h. Then, the protoplasts were collected and incubated in W5 solution (2 mM MES, pH 5.7, 154 mM NaCl, 5 mM KCl, and 125 mM CaCl2) at room temperature for 60 min. Next, the protoplasts were collected by centrifugation at 100g for 8 min and resuspended in MMG solution (4 mM MES, 0.6 M mannitol, and 15 mM MgCl2) to a final concentration of 0.5 to 1.0 × 107 mL–1. For transformation, 5 to 20 µg plasmid was gently mixed with 100 μL protoplasts and 110 μL PEG-CaCl2 solution (0.6 M mannitol, 100 mM CaCl2, and 40% PEG4000) and incubated at room temperature for 15 min. Two volumes of W5 solution was then added to stop the transformation, and the protoplasts were collected by centrifugation, resuspended in WI solution (4 mM MES, pH 5.7, 0.6 M mannitol, and 4 mM KCl), and cultured in 24-well culture plates for 20 h in the dark. Finally, the transformed protoplasts were collected by centrifugation at 200g for 5 min.
Transcriptional Regulation Activity Assay in Protoplasts
To determine the transcriptional activation activity of OsbZIP46, the reporter construct was produced by fusing a fragment of ∼1.7 kb of the promoter region (containing eight ABRE cis-elements) of RAB21, an OsbZIP46 target gene reported previously (Tang et al., 2012), to the firefly luciferase (LUC) gene. The sequences of primers used for cloning of RAB21 promoter are listed in Supplemental Data Set 1. For the effector constructs, different mutated forms of OsbZIP46 were cloned into the BamHI and KpnI sites of the effector vector to generate constructs driven by the CaMV 35S promoter. The effector and reporter constructs, together with the construct containing the Renilla luciferase gene driven by the Arabidopsis thaliana UBIQUITIN3 promoter (Hao et al., 2011) as an internal control, were cotransfected into rice protoplasts in a ratio of 6:12:1 (effector:reporter:reference). The cotransfected protoplast cells were cultured for 12 h at 24°C in the dark, and then the luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. The relative reporter gene expression levels were expressed as the ratio of firefly LUC to the Renilla luciferase. Moreover, to check the effect of SAPK9 or MODD on the transcriptional activation activity of OsbZIP46, CaMV35S:SAPK9 or CaMV35S:MODD constructs were cotransfected as coeffectors with an amount equal to the OsbZIP46 effector construct.
To determine the transcriptional repression activity of MODD, the GAL4-responsive reporter construct was produced by fusing the CaMV35S enhancer (CaMV35S’), five copies of the GAL4 binding site in tandem, and a minimal TATA region of the CaMV 35S promoter to the firefly LUC gene. For the effector constructs, the full-length MODD was cloned into the BamHI and KpnI sites of the effector vector that contained a GAL4 DNA binding domain (GAL4DB) to generate the GALDBD-MODD fusion construct driven by the CaMV35S promoter. The effector, reporter, and internal control constructs were cotransfected into rice protoplasts in a ratio of 6:6:1.
For ABA treatment of protoplasts, 100 μM ABA (Sigma-Aldrich; A1049) was added to the cultures of the transfected protoplasts. For the TSA (Sigma-Aldrich; T1952) treatment of protoplasts, transfected protoplasts were cultured for 6 h at 24°C in the dark, and then 20 μM TSA (in DMSO) or DMSO alone was added to the cultures for another 6 h. Then the luciferase activities were measured as described above.
Protein Interaction Assay
The Y2H assay was performed using the ProQuest two-hybrid system (Invitrogen). The coding regions of OsbZIP46 and MODD were cloned into the pDEST32 vector using the Gateway Technology (Invitrogen) to generate a bait vector with OsbZIP46 or MODD fused to the GAL4 DNA binding domain. The bait construct was further cotransformed into the yeast strain Mav203 with a prey cDNA library of rice, which was constructed by fusing cDNAs with the GAL4 activation domain in the pEXP-AD502 vector. The transformants were identified according to the manufacturer’s instructions (Invitrogen). Positive clones were isolated and their self-activation activities were checked by cotransformation with an empty pDEST32 vector. Those positive clones without self-activation activities were further identified by sequence. The detailed procedure conducted was referred to in the manufacturer’s manual (Invitrogen).
For the BiFC assay, OsbZIP46, OsTPR3, and OsPUB70 were respectively cloned into the pVYNE vector and fused to the N terminus of YFP, and MODD was cloned into the pVYCE vector and fused to the C terminus of YFP. Both constructs were coexpressed transiently in rice leaf protoplasts via PEG transformation. The fluorescence signal was detected by confocal microscopy (Leica).
For the Co-IP assays in rice protoplasts, total proteins were extracted from rice protoplasts expressed with two candidates, which were tagged with Myc and HA, in Co-IP buffer (50 mM Tris-HCl, pH 8.0, 150 mM KCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF, and 1× protease inhibitor cocktail tablets [Roche]). The protein extracts were mixed with HA agarose beads (Roche) and then incubated at 4°C for 2 h. After being washed at least five times, the agarose beads were recovered and mixed with the SDS sample buffer. The samples were detected by immunoblot using anti-Myc antibody (ABclonal). For Co-IP assays in tobacco, total protein was extracted from tobacco leaves that express the two candidate interacting proteins, which were tagged with FLAG and GFP, in Co-IP buffer as described above. After the immunoprecipitation using GFP beads (GFP-TRAP; Chromo Tec), the samples were detected with the anti-Flag antibody (Sigma-Aldrich).
Subcellular Localization
MODD was cloned into the pM999-33 vector to generate a MODD-GFP fusion construct driven by a CaMV35S promoter (35S:MODD-GFP), which was further cotransformed with the nuclear marker 35S:GHD7-CFP into rice protoplasts prepared from etiolated shoots via polyethylene glycol-mediated transformation. The florescence signal was checked by confocal microscopy (Leica) at 24 h after transformation.
Plant Materials and Growth Conditions
To construct the MODD overexpression vector, the sequence-confirmed clone containing the full-length cDNA of MODD was digested with KpnI and BamHI and cloned into the KpnI and BamHI sites of the binary expression vector pCAMBIA1301U (driven by a maize ubiquitin promoter). The construct was introduced into Zhonghua11 (O. sativa ssp japonica) by Agrobacterium tumefaciens-mediated transformation. For the loss-of-function mutants, two allelic T-DNA insertion mutants of modd were obtained from the RISD rice mutant library (http://cbi.khu.ac.kr/RISD_DB.html). Homozygous lines were further identified based on PCR with MODD-specific T-DNA border primers presented in Supplemental Data Set 1. For the overexpression of OsPUB70 in rice, the full-length cDNA of OsPUB70 (AK069675) was cloned into pCAMBIA1301U and introduced into rice Zhonghua11 by Agrobacterium-mediated transformation. The loss-of-function mutant ospub70 was obtained from the RISD rice mutant library.
The plants were grown in the greenhouse with a 12-h-light/12-h-dark cycle at 25 to 30°C, and 600-W sodium lamps were used for supplemental light.
Stress Treatments
The ABA sensitivity and drought tolerance of the overexpression plants and T-DNA insertion mutants were checked compared with the wild type as described previously (Tang et al., 2012). In brief, for ABA sensitivity, different genotype plants (12 plants each and 3 repeats) were germinated and then transplanted to normal 1/2MS medium or 1/2MS medium containing 3 µM ABA. The shoot and root growth was observed after about 2 weeks. For drought stress tolerance testing, plants were grown in a half-split manner (half transgenic half control, 12 plants each, and three repeats) in pots filled with sandy soil. At the 4-leaf stage, drought stress testing was conducted by withholding irrigation for 1 week followed by recovery for 1 week, and then the survival rates were calculated. The plants with green leaves and regenerating shoots were considered as survived plants.
To check the expression level of the MODD gene under drought stress or ABA treatment, the wild-type Zhonghua11 rice plants were grown in the soil (for drought) or hydroponic culture medium (for ABA treatment) for ∼3 weeks under normal growth conditions. The water supply was then removed for the drought stress treatments, and 100 μM ABA was sprayed on the leaves for the ABA treatment followed by sampling at the designated time.
Quantification of Gene Expression
The total RNAs of the rice leaves were extracted and reverse-transcribed using the TRIzol reagent (Invitrogen) and SuperScript reverse transcriptase (Invitrogen), respectively, according to the manufacturer’s instructions. RT-qPCR was performed on an optical 96-well plate with an ABI PRISM 7500 real-time PCR system (Applied Biosystems) using SYBR Premix Ex Taq (TaKaRa). The details of the qPCR were essentially the same as described previously with the rice Actin1 gene as the endogenous control (Tang et al., 2012). The primer sequences used in the qPCR are listed in Supplemental Data Set 1x.
RNA-Seq and Data Analysis
Total RNA was extracted from wild-type rice and modd-2 mutants at the 4-leaf stage under normal conditions with two biological replicates. The RNA samples were sequenced and analyzed by the Beijing Genomics Institute. We used the BWA software package (Li and Durbin, 2009) to map clean reads to the reference genome of rice and used Bowtie (Langmead et al., 2009) to reference genes. Gene expression levels were quantified by the software package RSEM (Li and Dewey, 2011) with FPKM calculation method. The Noiseq method (Tarazona et al., 2011) was used to screen for differentially expressed genes between the two groups according to the criteria fold change ≥2 and diverge probability ≥0.8.
Chromatin Immunoprecipitation
To measure histone acetylation levels at the OsbZIP46 target gene in the MODD overexpression and modd loss-of-function mutant and the wild type, ChIP (chromatin immunoprecipitation) combined with qPCR was conducted in the MODD-OE20, modd-2 mutant, and wild-type plants with the Anti-acetyl-Histone H3 antibody (Millipore). For validation of the direct binding of OsbZIP46 to the promoters of target genes, wild-type and OsbZIP46-FLAG transgenic rice plants were used for ChIP-qPCR analysis with the antibody against FLAG (Sigma-Aldrich).
The chromatin extraction and immunoprecipitation were conducted as described by Huang et al. (2007). Briefly, 2 g of mutant and wild-type seedlings were harvested and cross-linked in 1% formaldehyde in a vacuum. Chromatin was extracted and fragmented by sonication, and then the immunoprecipitations with the anti-acetyl-Histone H3 antibody or anti-FLAG were performed. The precipitated and input DNAs were further analyzed using qPCR with specific primers (Supplemental Data Set 1) corresponding to the OsbZIP46 target genes, including RAB21.
In Vivo Protein Degradation Assay
In vivo protein degradation assays were performed following the protocols described by Liu et al. (2010, 2012). GFP-tagged OsbZIP46 was coexpressed with OsPUB70, MODD-FLAG, and the HA-GFP internal control in tobacco leaves. The protein abundances of OsbZIP46 and HA-GFP were detected with anti-GFP antibody (Abcam) and anti-HA antibody (Roche), respectively.
In Vitro Ubiquitination Assays
The ubiquitination assays were performed following the protocols described by Zhao et al. (2012). For the E3 ubiquitin ligase activity assay, 6×His tag fused OsPUB70 protein was expressed in Escherichia coli and purified by Qiagen Ni-NTA agarose kit. Five micrograms of purified OsPUB70-His fusion protein was mixed with 50 ng E1 (Boston Biochem; E-302), 300 ng E2 (Boston Biochem; UbcH5b), and 5 µg Ub-FLAG (Boston Biochem) in buffer containing 50 mM Tris, pH 7.5, 3 mM DTT, 5 mM MgCl2, and 2 mM ATP. After 1.5 h incubation at 30°C, reactions were stopped by adding Laemmli sample buffer. A quarter of the mixture was subjected to 10% SDS-PAGE and immunoblotting using anti-FLAG antibody (Sigma-Aldrich). For the substrate ubiquitination assay, OsbZIP46-GST, MODD-His, and OsPUB70-His fusion proteins were expressed in Es. coli and purified. Two micrograms of OsbZIP46-GST, 1 µg MODD-His, and 0.5 µg OsPUB70-His were incubated together with the ubiquitination mixture including 5 ng E1, 30 ng E2, and 0.5 µg Ub-FLAG for 1.5 h. The mixture was then subjected to 10% SDS-PAGE and immunoblotting using anti-GST antibody (Abcam).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: MODD, LOC_Os03g11550; OsTPR3, LOC_Os03g14980; HDA702, LOC_Os06g38470; OsPUB70, LOC_Os06g06760; OsbZIP46, LOC_Os06g10880; OsbZIP23, LOC_Os02g52780; OsbZIP72, LOC_Os09g28310; RAB21, LOC_Os11g26790; OsLEA3, LOC_Os05g46480; OsLEA14, LOC_Os01g50910; RAB16B, LOC_11g26780; and SAPK9, LOC_Os12g39630. The RNA-seq data were deposited in the Gene Expression Omnibus under accession number GSE83075.
Supplemental Data
Supplemental Figure 1. The transcriptional regulation activity of OsbZIP23, OsbZIP72, and OsbZIP46 is activated by ABA.
Supplemental Figure 2. Protein sequence analysis of the MODD family members.
Supplemental Figure 3. OsbZIP23 and OsbZIP72 interact with MODD.
Supplemental Figure 4. Statistical information on the subcellular localization of MODD-GFP and the control (GFP).
Supplemental Figure 5. Similar expression patterns of MODD and OsbZIP46 across various tissues and organs.
Supplemental Figure 6. Circadian rhythmic expression profiles of MODD and OsbZIP46.
Supplemental Figure 7. Generating transgenic rice overexpressing MODD.
Supplemental Figure 8. Characterization of modd T-DNA insertion mutants.
Supplemental Figure 9. Validation of the direct binding of OsbZIP46 to the promoters of target genes.
Supplemental Figure 10. The expression of OsbZIP46-targeted genes in the MODD overexpression (OE-20) and modd loss-of-function mutant (modd-2) lines.
Supplemental Figure 11. Histone acetylation levels at the OsbZIP46 target genes in MODD overexpressor (OE20), modd-2 mutant, and wild type (ZH11 and DJ) using ChIP-qPCR.
Supplemental Figure 12. Yeast two-hybrid assay of OsbZIP46 and OsPUB70.
Supplemental Figure 13. Generation of OsPUB70 overexpression transgenic rice.
Supplemental Figure 14. Characterization of ospub70 T-DNA insertion mutants.
Supplemental Table 1. Interaction proteins of AFP obtained by searching the Plant Interactome Database.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. Genes downregulated in the modd-2 mutant.
Supplemental Data Set 3. Genes upregulated in the modd-2 mutant.
Supplemental Data Set 4. Alignment corresponding to the phylogenetic analysis in Supplemental Figure 2B.
Supplementary Material
Acknowledgments
We thank Masaru Ohme-Takagi (National Institute of Advanced Industrial Science and Technology, Japan) and Shouyi Chen (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the gift of vectors for the transcriptional regulation activity assay in protoplasts. This work was supported by grants from the National Program for Basic Research of China (2012CB114305) and the National Program on High Technology Development of China (2016YFD0100604 and 2014AA10A600).
AUTHOR CONTRIBUTIONS
N.T., S.M., W.Z., N.Y., Y.L., C.Y., Z.G., J.L., Xu.L., Y.X., H.S., J.X., and Xi.L. performed research. N.T., S.M., and L.X. analyzed data. N.T., S.M., and L.X. wrote the article.
Glossary
- ABA
abscisic acid
- UPS
ubiquitin-26S proteasome system
- Y2H
yeast two-hybrid
- Co-IP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- TSA
Trichostatin A
- CHX
cycloheximide
- 1/2MS
half-strength Murashige and Skoog
References
- Bensmihen S., Rippa S., Lambert G., Jublot D., Pautot V., Granier F., Giraudat J., Parcy F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C., Romera-Branchat M., Pelaz S. (2005). A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 43: 586–596. [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.T., Wu K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5: 1318–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.I., Park H.J., Park J.H., Kim S., Im M.Y., Seo H.H., Kim Y.W., Hwang I., Kim S.Y. (2005). Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 139: 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa L.G., Riaño-Pachón D.M., Schrago C.G., dos Santos R.V., Mueller-Roeber B., Vincentz M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One 3: e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell I.G. (1994). Repression versus activation in the control of gene transcription. Trends Biochem. Sci. 19: 38–42. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Dai M., Xue Q., Mccray T., Margavage K., Chen F., Lee J.H., Nezames C.D., Guo L., Terzaghi W., Wan J., Deng X.W., Wang H. (2013). The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25: 517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Wang M.L., Lynch T.J., Rao S., Goodman H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106: 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. (2011). Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA 108: 1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50: 2123–2132. [DOI] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103: 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.E., Lynch T., Peeters J., Snowden C., Finkelstein R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67: 643–658. [DOI] [PubMed] [Google Scholar]
- Giraudat J., Hauge B.M., Valon C., Smalle J., Parcy F., Goodman H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J., Tweedie S. (2002). Remembrance of things past: chromatin remodeling in plant development. Annu. Rev. Cell Dev. Biol. 18: 707–746. [DOI] [PubMed] [Google Scholar]
- Guo Y., Xiong L., Song C.P., Gong D., Halfter U., Zhu J.K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3: 233–244. [DOI] [PubMed] [Google Scholar]
- Hao Y.J., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68: 302–313. [DOI] [PubMed] [Google Scholar]
- Hare P.D., Seo H.S., Yang J.Y., Chua N.H. (2003). Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr. Opin. Plant Biol. 6: 453–462. [DOI] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S.E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53: 172–185. [DOI] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Hossain M.A., Cho J.I., Han M., Ahn C.H., Jeon J.S., An G., Park P.B. (2010). The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 167: 1512–1520. [DOI] [PubMed] [Google Scholar]
- Hu R., Zhu Y., Shen G., Zhang H. (2014). TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiol. 164: 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Qin F., Huang L., Sun Q., Li C., Zhao Y., Zhou D.X. (2009). Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388: 266–271. [DOI] [PubMed] [Google Scholar]
- Huang L., Sun Q., Qin F., Li C., Zhao Y., Zhou D.X. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144: 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Arora R., Agarwal P., Ray S., Sharma P., Kapoor S., Tyagi A.K., Khurana J.P. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143: 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X.F., Xiong A.S., Peng R.H., Liu J.G., Gao F., Chen J.M., Yao Q.H. (2010). OsAREB1, an ABRE-binding protein responding to ABA and glucose, has multiple functions in Arabidopsis. BMB Rep. 43: 34–39. [DOI] [PubMed] [Google Scholar]
- Johnson A.D. (1995). The price of repression. Cell 81: 655–658. [DOI] [PubMed] [Google Scholar]
- Kagale S., Rozwadowski K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Hobo T., Murata M., Ban A., Hattori T. (2002). Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14: 3177–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. (2006). Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11: 109–112. [DOI] [PubMed] [Google Scholar]
- Keleher C.A., Redd M.J., Schultz J., Carlson M., Johnson A.D. (1992). Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68: 709–719. [DOI] [PubMed] [Google Scholar]
- Krogan N.T., Long J.A. (2009). Why so repressed? Turning off transcription during plant growth and development. Curr. Opin. Plant Biol. 12: 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhao Q., Xie Q. (2012). In vivo ubiquitination assay by agroinfiltration. Methods Mol. Biol. 876: 153–162. [DOI] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Liu Z., Karmarkar V. (2008). Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13: 137–144. [DOI] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17: 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Gao C., Zheng X., Han B. (2009). Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229: 605–615. [DOI] [PubMed] [Google Scholar]
- Lynch T., Erickson B.J., Finkelstein R.R. (2012). Direct interactions of ABA-insensitive (ABI)-clade protein phosphatase(PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol. Biol. 80: 647–658. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Ohme-Takagi M. (2009). Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 50: 1232–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Parry G., Estelle M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.J., Millar A.H. (2015). Protein turnover in plant biology. Nat. Plants 1: 15017. [DOI] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- País S.M., Téllez-Iñón M.T., Capiati D.A. (2009). Serine/threonine protein phosphatases type 2A and their roles in stress signaling. Plant Signal. Behav. 4: 1013–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra A.S., Gonugunta V.K., Christmann A., Grill E. (2010). ABA perception and signalling. Trends Plant Sci. 15: 395–401. [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76: 75–100. [DOI] [PubMed] [Google Scholar]
- Sridhar V.V., Surendrarao A., Liu Z. (2006). APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166. [DOI] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tang N., Zhang H., Li X., Xiao J., Xiong L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158: 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S., García-Alcalde F., Dopazo J., Ferrer A., Conesa A. (2011). Differential expression in RNA-seq: a matter of depth. Genome Res. 21: 2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka K., Taji T., Hayashi T., Sakata Y. (2013). A novel abi5 allele reveals the importance of the conserved Ala in the C3 domain for regulation of downstream genes and salt tolerance during germination in Arabidopsis. Plant Signal. Behav. 8: e23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8: 135–142. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Wang Y.C., Peterson S.E., Loring J.F. (2014). Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 24: 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel R.R., Pfitzner U.M., Gatz C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P. (1996). Histone deacetylase: a regulator of transcription. Science 272: 371–372. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Tang N., Du H., Ye H., Xiong L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148: 1938–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Yang Y. (2013). RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant 6: 1975–1983. [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (Suppl): S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803. [DOI] [PubMed] [Google Scholar]
- Yang X., Yang Y.N., Xue L.J., Zou M.J., Liu J.Y., Chen F., Xue H.W. (2011). Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol. 156: 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Ohmori Y., Kitano H., Taguchi-Shiobara F., Hirano H.Y. (2012). Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 70: 327–339. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685. [DOI] [PubMed] [Google Scholar]
- Zeevaart J.A.D., Creelman R.A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39: 439–473. [Google Scholar]
- Zeng L.R., Park C.H., Venu R.C., Gough J., Wang G.L. (2008). Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant 1: 800–815. [DOI] [PubMed] [Google Scholar]
- Zhai Q., Yan L., Tan D., Chen R., Sun J., Gao L., Dong M.Q., Wang Y., Li C. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9: e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Liu L., Xie Q. (2012). In vitro protein ubiquitination assay. Methods Mol. Biol. 876: 163–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.