ChIP-seq analyses of RBP LIF2 and its LHP1 partner in various backgrounds and stress conditions revealed target regions for the two proteins enriched in antagonistic marks.
Abstract
LHP1-INTERACTING FACTOR2 (LIF2), a heterogeneous nuclear ribonucleoprotein involved in Arabidopsis thaliana cell fate and stress responses, interacts with LIKE HETEROCHROMATIN PROTEIN1 (LHP1), a Polycomb Repressive Complex1 subunit. To investigate LIF2-LHP1 functional interplay, we mapped their genome-wide distributions in wild-type, lif2, and lhp1 backgrounds, under standard and stress conditions. Interestingly, LHP1-targeted regions form local clusters, suggesting an underlying functional organization of the plant genome. Regions targeted by both LIF2 and LHP1 were enriched in stress-responsive genes, the H2A.Z histone variant, and antagonistic histone marks. We identified specific motifs within the targeted regions, including a G-box-like motif, a GAGA motif, and a telo-box. LIF2 and LHP1 can operate both antagonistically and synergistically. In response to methyl jasmonate treatment, LIF2 was rapidly recruited to chromatin, where it mediated transcriptional gene activation. Thus, LIF2 and LHP1 participate in transcriptional switches in stress-response pathways.
INTRODUCTION
In eukaryotes, the control of gene expression is central to development and environmental adaptation. The establishment and maintenance of specific transcriptionally active and repressive chromatin states participate in this control. Polycomb repressive complexes (PRCs) and Trithorax complexes shape chromatin states and have general transcriptional repressor and activator activities, respectively (Simon and Kingston, 2013; Del Prete et al., 2015). Over the past few years, the regulatory function of PRCs has been challenged in both plants and animals (Tavares et al., 2012; Simon and Kingston, 2013; Calonje, 2014; Pu and Sung, 2015; Förderer et al., 2016). For instance, novel PRC1 complexes have been identified; the canonical model of PRC repression, in which PCR2-dependent H3K27 trimethylation is followed by PRC1-dependent H2A monoubiquitination, is no longer regarded as the unique mode of action (Tavares et al., 2012; Calonje, 2014), and a novel transcriptional activation function has been reported for PRC1 (Gil and O’Loghlen, 2014).
However, the mechanism underlying the transition from active to repressed chromatin states remains poorly understood. Documented recruitment of Polycomb group proteins (PcG) to chromatin identified thousands of target regions in eukaryotic genomes. In plants, the PRC1 subunit LHP1 is distributed throughout the genome and colocalizes with the H3K27me3 repressive histone mark (Turck et al., 2007; Zhang et al., 2007), as observed for animal PcG proteins. The distribution of FERTILIZATION INDEPENDENT ENDOSPERM (FIE), a plant PRC2 subunit, somewhat overlaps with H3K27me3 regions (Deng et al., 2013). The chromatin context, which is determined by the combination of specific DNA motifs, histone marks, or other chromatin-associated proteins, largely determines PcG recruitment. For instance, Drosophila melanogaster PRCs contain sequence-specific DNA binding factors and are classically recruited at Polycomb/Trithorax response elements (PRE/TREs or PREs) in the genome, which are composed of a variable combination of short DNA motifs and participate in the maintenance of the transcriptional status (Bauer et al., 2015). Only a few PRE-like elements have been reported in mammals (Bauer et al., 2015). PRCs interact with various chromatin-associated proteins, such as histone modifying enzymes or transcription factors (TFs), which may also contribute to their targeting. In plants, several TFs, such as SCARECROW, ASYMMETRIC LEAVES1, and AS2, interact with PRC subunits (reviewed in Del Prete et al., 2015). Recently, the Arabidopsis thaliana GAGA binding factor BASIC PENTACYSTEINE6 (BPC6) was shown to recruit LHP1 to GAGA motifs (Hecker et al., 2015), reminiscent of the recruitment of PcG proteins to GAGA motifs present in animal PREs. Finally, whereas some long noncoding RNAs are involved in the scaffolding of chromatin modifying complexes associated with PRC function (Brockdorff, 2013) or mediate intrachromosomal interactions (Zhang et al., 2014), they also emerged as novel interacting partners of both PRC2 and PRC1 subunits (Del Prete et al., 2015) that participate in their genomic recruitment. In Arabidopsis, long noncoding RNAs were proposed to function in the transcriptional regulation of FLOWERING LOCUS C mediated by PcG proteins (Swiezewski et al., 2009; Heo and Sung, 2011; Csorba et al., 2014). Intriguingly, LIF2, a heterogeneous nuclear ribonucleoprotein Q (hnRNP-Q) with three RNA recognition motifs (RRMs), was identified as a partner of LHP1 (Latrasse et al., 2011), highlighting the diversity of plant proteins associated with PRC1 and suggesting that RNA binding proteins (RBPs) mediate interactions between plant PRC1 and RNA components.
To investigate the interplay between LIF2 and LHP1, we compared the genome-wide chromatin profiles of LIF2 and LHP1 in wild-type and mutant backgrounds. This is the first report of the genome-wide chromatin profile of a plant RBP. Our ChIP-seq data analyses revealed that LIF2 had a more restricted distribution than LHP1, being mainly present at stress-responsive genes. The spatial analysis of LHP1 distribution showed that LHP1 regions tend to aggregate locally, suggesting a role for LHP1 in genome topography. Specific and antagonistic histone marks were associated with each protein, as well as cis-regulatory DNA elements. We identified the GAGA motif and telo-box motifs in the LHP1 target genes. Also present in FIE binding sites (Deng et al., 2013), these two motifs may thus be part of a PRC targeting signature. Given the role of LIF2 in pathogen responses (Le Roux et al., 2014), we investigated the distribution of LIF2 in response to methyl jasmonate (MeJA), a key hormone in plant biotic and abiotic stress responses. We showed that LIF2 distribution was dynamic in response to MeJA treatment and that LIF2 was required for transcriptional gene activation. Thus, we highlighted a complex interplay between LIF2 and LHP1 in stress-response pathways.
RESULTS
LIF2 and LHP1 Target a Common Set of Chromatin Regions
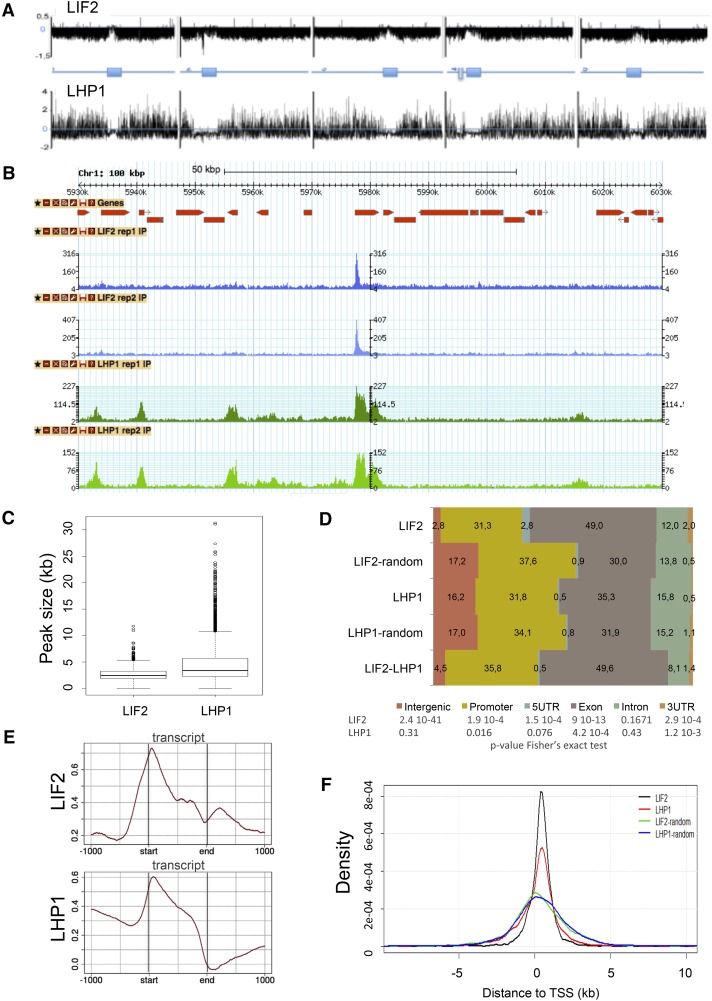
Prompted by the observation that hnRNP-Q LIF2 physically interacts with the chromatin-associated protein LHP1 (Latrasse et al., 2011), we performed ChIP-seq experiments to identify the chromatin regions enriched in LIF2 and LHP1 (enrichment regions [ERs]). For this purpose, we produced transgenic lines expressing 3xHA-tagged LIF2 (HA:LIF2) and 3xHA-tagged LHP1 (LHP1:HA) under the control of endogenous genomic regulatory sequences, in the lif2-1 and lhp1-4 genetic backgrounds, respectively. Two independent ChIP-seq libraries were sequenced for each protein ( TPC201600244RAR2 ure 1). We observed good overlaps between replicates, as well as high Pearson coefficients of the MACS peak fold-change correlations between replicates (0.93 [LIF2] and 0.81 [LHP1]; Supplemental Figure 1). We identified 1457 ERs present in both replicates for LIF2 and 4844 for LHP1 (at a false discovery rate [FDR] of < 0.05) and determined the summit (i.e., position with the maximum read number) in each ER. The comparison of the two genome-wide distributions allowed us to identify 488 genomic regions where LIF2 and LHP1 were detected, corresponding to the intersection of the two genomic distributions (named LIF2-LHP1 intersect regions [IRs]) (Figures 1A and 1B). We confirmed binding to 10 ERs by ChIP experiments followed by quantitative PCR (ChIP-qPCR) (Supplemental Figure 1). We established that 52.8% of the DamID-identified LHP1 target genes (Zhang et al., 2007) was present in our ChIP-seq data set (despite differences in tissue, developmental stages, and growth conditions). These data suggest that LIF2 has a more specialized function in the genome than does LHP1, with each protein having independent and specific functions. However, in agreement with their physical interactions, a subset of genomic regions was identified where the two proteins were located.
Figure 1.
Genome-Wide Distributions of LIF2 and LHP1.
(A) Chromosomal view of the peaks using model-based analysis.
(B) Screenshot of a 100-kb window with the distributions.
(C) Size distributions of the ERs defined as intersects of MACS peaks for the biological replicates.
(D) Distributions of ER-associated annotations (percentage). Regions with identical sizes were randomly shuffled in the genome and compared with the observed ERs, using a Fisher’s exact test.
(E) Distributions of IP enrichment (log2(# reads IP/# read input)) over the transcript structures.
(F) Distance to closest TSSs of LIF2 and LHP1 ERs, and the corresponding randomized regions.
LIF2 Is Present in Narrow Chromatin Regions in 5′ and 3′ Untranslated Regions
LIF2 and LHP1 exhibited different chromatin-associated profiles. Whereas the LIF2 profile had narrow, discrete peaks, LHP1 peak sizes were larger (Figure 1C). By analyzing the distribution of LIF2 and LHP1 over annotated genomic features and comparing this distribution with a random distribution over genome regions of similar size, we found that LIF2 had a preference for 5′ untranslated regions (UTRs; 2.52-fold compared with LIF2-random), exons (especially exon 1) (Supplemental Figure 2), and 3′ UTRs (4-fold compared with LIF2-random), whereas LHP1 had a more balanced distribution over all regions (Figure 1D). Interestingly, the distribution of the LIF2-LHP1 IRs was similar to that of the LIF2 ERs. Our analysis of the peak distributions over transcripts showed that LIF2 was enriched at transcription start sites (TSSs) and depleted at transcription termination sites (TTSs) (Figure 1E). The low but significant level of LIF2 downstream TTSs was not due to the proximity of another TSS. LHP1 was more prevalent at promoter regions and gene bodies, with a marked preference for TSSs, resulting in an asymmetry between upstream and downstream genic regions (Figure 1E). The presence of LIF2 and LHP1 ERs in regions close to the TSS was confirmed with the analysis of the distance of the ERs to the closest TSS compared with the randomly shuffled control regions (Figure 1F).
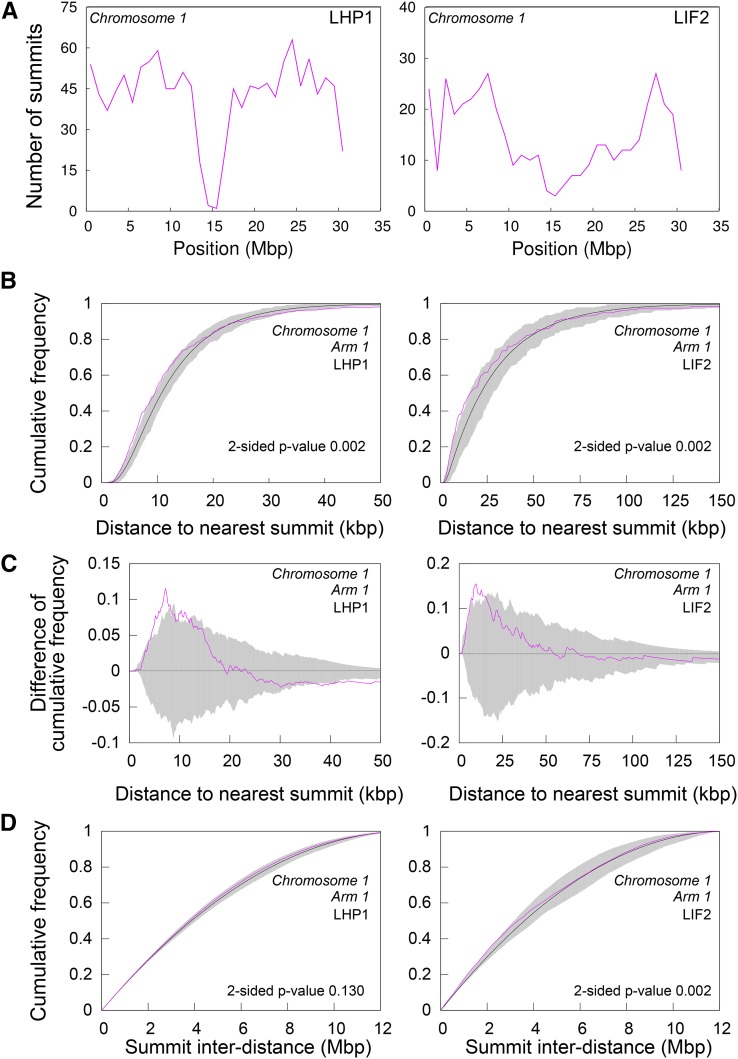
The Targeted Regions Tend to Form Clusters
Given the role of PcG proteins in structuring the genome in animal species (Del Prete et al., 2015), we analyzed the distribution of LHP1 and LIF2 along each chromosome. The distribution of the number of ER summits in 1-Mb windows revealed that portions of the genome were enriched for LHP1 and LIF2 (Figure 2A; Supplemental Figure 3). To analyze the distribution of LHP1 ERs further and to test the existence of an underlying organization principle in this distribution, we compared the summit distribution of LHP1 ERs to a random distribution model, conditioned on the size of LHP1 ER regions. Observed and model-predicted distributions were compared using local-scale (i.e., cumulative distribution of the distance between each ER and its closest neighbor) and global-scale (i.e., cumulative distribution of the inter-distances between all ERs) spatial descriptors. A significant discrepancy with the completely random model was observed at the local scale, with the measured distances between ERs and their closest neighbors being significantly smaller than expected under a random distribution for all chromosome arms (Figure 2B; Supplemental Figure 4). Compared with the random distribution, the distance to the closest ER was enriched in the range of short values of up to ∼10 kb (Figure 2C). This range was constant across chromosome arms, suggesting the existence of common spatial constraints despite differences in arm length and ER density. Overall, no significant difference to the random distribution was observed (Figure 2D; Supplemental Figure 5) when comparing the distribution of all interdistances (Figure 2D; Supplemental Figure 4), consistent with the globally uniform distribution of LHP1 ERs in 1-Mb windows (Figure 2A; Supplemental Figure 3). LIF2 ERs exhibited spatial clustering that was similar to that of LHP1 ERs. Despite the lower density of LIF2 ERs, the range of distances between nearest neighbors was similar to that observed for LHP1 ERs (Figure 2C), suggesting that the distributions of the two proteins’ target regions were under shared constraints.
Figure 2.
Nonrandom Distributions of the LHP1 ERs and LIF2 ERs in the Arabidopsis Genome.
(A) Number of summits in 1-Mb windows along Chromosome 1.
(B) to (D) Observed (pink) and random model (black, average; gray, 95% envelope) distributions of distance to nearest ER ([B] and [C]) and of all ER interdistances (D) on the first arm of Chromosome 1. Similar results were obtained for all chromosome arms (Supplemental Figures 3 to 5).
To further investigate the clustering of LHP1 ERs, we analyzed the relationship between the LHP1 ER genome-wide distribution and the distribution of repeated genes in the Arabidopsis genome. Indeed, repeated genes may participate in this clustering tendency. It was previously shown using ChIP-chip experiments that out of the 679 tandemly repeated genes located on chromosome 4, 30% were targeted by LHP1 (Turck et al., 2007). In the whole Arabidopsis genome, 1564 tandem duplications (T-clusters) and 1680 segmental duplications (with a 1:1 duplication relation, S-clusters) were described (Haberer et al., 2004). The T-clusters contain from 2 to up to 21 repeated genes, with a mean value <3 genes. Using our ChIP-seq data, we observed that 20.6% of the T-cluster genes were targeted by LHP1, compared with 11.7% for the S-cluster genes. However, only 23.1% of the LHP1-targeted genes were located in T-clusters. On average, there was less than one LHP1 target gene per T-cluster and only 11% of T-clusters had two or more LHP1 target genes, accounting for only 9.6% of all LHP1 targets (Supplemental Table 1). These low figures suggest that LHP1 binding to T-cluster genes is not sufficient to explain the clustering tendency of LHP1-targeted regions observed at the local scale on the chromosome arms.
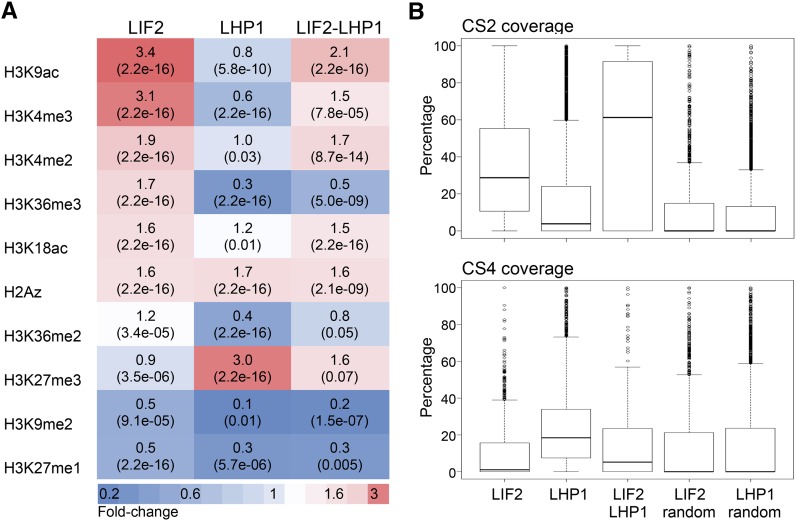
The Presence of Antagonistic Histone Marks and H2A.Z Characterize LIF2-LHP1 IRs
A limited number of chromatin states, which are based on histone posttranslational modifications or histone variants, have been reported for the Arabidopsis genome (Sequeira-Mendes et al., 2014). We thus examined whether specific epigenetic marks were preferentially associated with the identified ERs, using data sets for nine histone marks (Luo et al., 2013) and the H2A.Z histone variant (Zilberman et al., 2008; Coleman-Derr and Zilberman, 2012). We observed that LHP1 ERs were enriched in the repressive mark H3K27me3, confirming our previous genome-wide analysis (Zhang et al., 2007), and were depleted in active histone marks, such as H3K4me3 (Figure 3; Supplemental Figure 6). By contrast, LIF2 ERs were enriched in H3K4me3 and H3K9ac histone marks, which are hallmarks of active/open chromatin. Interestingly, a similar enrichment in H3K4me3 and H3K27me3 was observed in LIF2-LHP1 IRs, and this was associated with a noticeable depletion in the active mark H3K36me3 compared with LIF2 ERs. LIF2 and LHP1 ERs also had similar levels of H2A.Z, with LIF2-LHP1 IRs having slightly higher levels. In Arabidopsis, H2A.Z is enriched within the nucleosomes surrounding the TSSs of genes (Zilberman et al., 2008), but also across the bodies of genes with low transcription levels and high responsiveness (Coleman-Derr and Zilberman, 2012). Our data suggest that LIF2-LHP1 IRs may correspond to subdomains of chromatin state 2 (CS2), which is characterized by relatively high levels of both active H3K4me3 and inactive H3K27me3 histone marks and is mostly associated with bivalent regions and highly constrained gene expression (Sequeira-Mendes et al., 2014). We thus analyzed the coverage of CS2 in the distributions of LHP1 and LIF2 ERs and compared this coverage with CS4 coverage; CS4 has high levels of H3K27me3 but reduced levels of active marks. We confirmed enrichments in CS2 for both LIF2 IRs and LIF2-LHP1 IRs (Figure 3B). By comparing the lists of LIF2 and LHP1 target genes with the bivalent genes identified by sequential ChIP experiments (Luo et al., 2013), we observed that ∼14.92% of the LIF2-LHP1 IR genes have been annotated as bivalent (Luo et al., 2013), whereas only 4.97% and 5.97% of the LIF2 and LHP1 target genes have been annotated as bivalent, respectively. Thus, the genome-wide distributions of LIF2 and LHP1 contributed to the functional topographical organization of the Arabidopsis genome (Sequeira-Mendes et al., 2014).
Figure 3.
Posttranslational Histone Modifications and the H2A.Z Histone Variant in the LIF2 ERs, LHP1 ERs, and LIF2-LHP1 IRs.
(A) Heat map presenting the fold changes (P value paired t test) between targeted and randomized regions.
(B) Percentage of chromatin states 2 and 4 (CS2 and CS4; defined in Sequeira-Mendes et al., 2014) covering LHP1 ERs, LIF2 ERs, LIF2-LHP1 IRs, and randomized control regions.
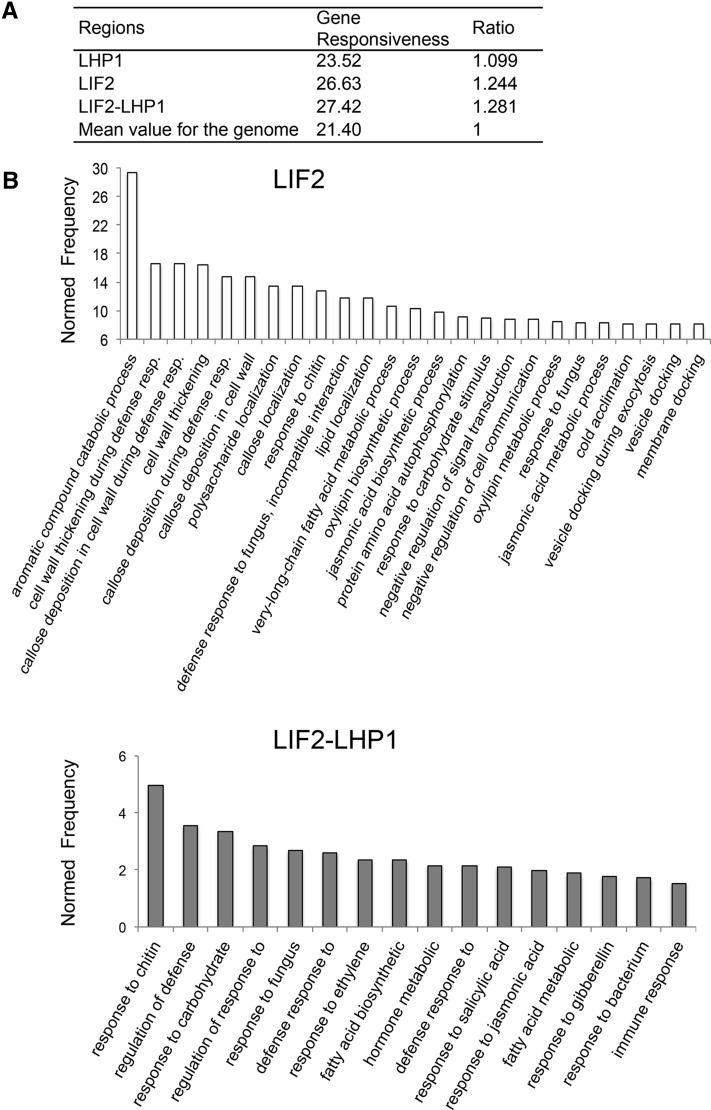
LIF2-LHP1 IRs Are Enriched in Stress-Responsive Genes
To predict the functions of genes of LIF2 ERs and LIF2-LHP1 IRs, we determined the gene responsiveness index of the binding regions based on the expression profiles of the genes located in the ERs (Aceituno et al., 2008; Coleman-Derr and Zilberman, 2012). We found that they were enriched in responsive genes (Figure 4A). Our analysis of the functional Gene Ontology (GO) terms revealed that LIF2 ERs and LIF2-LHP1 IRs were enriched in stress-responsive genes (Figure 4A; Supplemental Figures 7 and 8). The Bio-Array Resource for Plant Functional Genomics classification Superviewer program (Provart et al., 2003) showed that both LIF2 ERs and LIF2-LHP1 IRs were enriched in the GO term “response to abiotic or biotic stimulus” (normed frequency [NF]; LIF2, NF = 3, P value 1.399 10−78; LIF2-LHP1, NF = 2.7, P value 2.662 10−19) (Supplemental Table 2). A more detailed analysis using AgriGO revealed that the first two enriched GO terms for LIF2 ERs were “aromatic compound catabolic process” (NF = 29.35, FDR 9.4 10−5, P value 3.8 10−6) and “callose deposition in cell wall during defense response” (NF = 16.57, FDR 1.2 10−4, P value 5.1 10−6). For LIF2-LHP1 IRs, they were “response to chitin” (NF = 4.98, FDR 5.9 10−12, P value 1.7 10−14) and “regulation of defense response” (NF = 3.56, FDR 1.3 10−3, P value 9.5 10−5) (Supplemental Figure 7). These results were in agreement with those of our previous transcriptome analysis of the lif2 mutant (Latrasse et al., 2011) and the response of lif2 to pathogens (Le Roux et al., 2014), but also with the epigenetic marks present at LIF2-LHP1 IRs. Indeed, genes present in CS2 were shown to have constrained transcription profiles (Sequeira-Mendes et al., 2014).
Figure 4.
LIF2 Preferentially Binds Stress-Response Genes.
(A) Average gene responsiveness scores were calculated based on a published data set (Aceituno et al., 2008) and normalized to the genome-wide average.
(B) GO analysis of LIF2 ERs and LIF2-LHP1 IRs using the AgriGO toolkit. The biological process GO terms, with the 25 best NF and with NF ≥ 1.5 are presented for LIF2 ERs and LIF2-LHP1 IRs, respectively.
Further GO term analysis revealed an enrichment in “transcription factor activity” in the LIF2 ER and LIF2-LHP1 IR data sets (NF = 2.39, P value 3.730 10−18 and NF = 3.7, P value 2.739 10−17, respectively) (Supplemental Table 2). Using the Plant GeneSet Enrichment Analysis (PlantGSEA) (Yi et al., 2013) and Arabidopsis Gene Regulatory Information Server (AGRIS) (Yilmaz et al., 2011) toolkits, we observed that the main TFs targeted by LIF2 belonged to the AP2-EREBP and WRKY families, consistent with a role for LIF2 in the stress response, whereas TFs in LHP1-ERs belonged to a larger range of families (Supplemental Table 3). Interestingly, TFs present in the 488 LIF2-LHP1 IRs also belonged to the AP2-EREBP family. Some of the target genes were also targeted by other TFs, such as LONG HYPOCOTYL5 (HY5) (Lee et al., 2007; Zhang et al., 2011) and PHYTOCHROME-INTERACTING FACTOR1 (PIF1) (Chen et al., 2013) (Supplemental Table 4), suggesting a complex interplay between LIF2, LHP1, and TFs.
Identification of cis-Regulatory DNA Elements Associated with LIF2 and LHP1 Binding
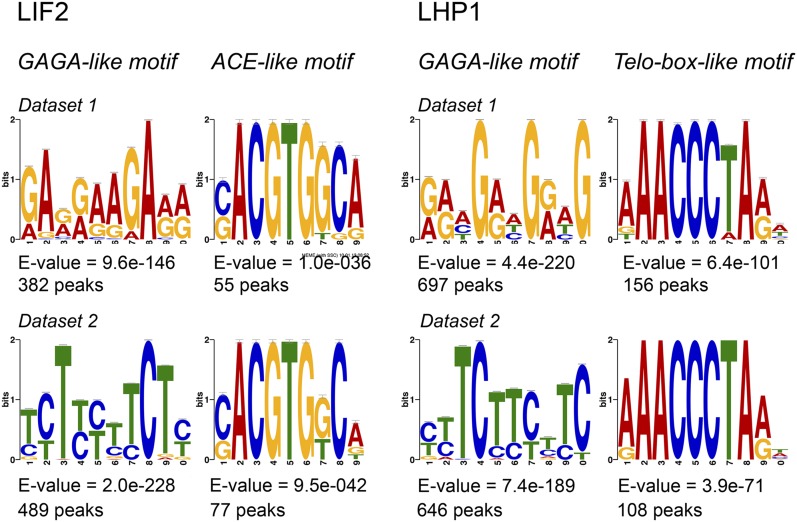
We next searched for putative DNA binding motifs around the summits. Using the MEME algorithm (Bailey and Elkan, 1995), two consensus motifs were discovered in the 51-bp regions centered on the LIF2 binding summits: a GAGA-like motif and a (C/G)ACGTG(G/T)C(A/G) consensus motif, which belongs to the ACGT-containing element (ACE) family (Figure 5). The ACGTGGCA word was present at moderate levels in the whole genome, mostly in the distal promoter regions of genes (region from −1000 bp to −3000 bp relative to the TSS) (Supplemental Table 5). Some of the ACE elements are recognized by TFs, among which HY5 and PIF1 (Song et al., 2008; Chen et al., 2013), previously identified as having common targets with LIF2 (Supplemental Table 4) and two physically interacting TFs involved in plant growth and, in particular, in the crosstalk between light and reactive oxygen species signaling. In the LHP1 data sets, we identified a GAGA-like motif as a putative recognition motif (Figure 5). In addition, we identified the (A/G/T)AACCCTA(A/G) motif. Despite being less represented among the LHP1 peaks, this putative and highly significant DNA motif (-log10(E-value) > 20) was discovered with both MEME and “peak-motif” algorithms (Bailey and Elkan, 1995; Thomas-Chollier et al., 2012b) (Figure 5B). This motif contains the AAACCCTA short interstitial telomere motif, also named the telo-box, which was originally described in the 5′ regions of genes encoding the translation elongation factor EF1α and ribosomal proteins (Regad et al., 1994; Gaspin et al., 2010). The AAACCCTA word/telo-box is mainly present in introns and 5′ UTRs (Supplemental Table 5). Interestingly, the (A/G/T)AACCCTA(A/G) motif recognized by LHP1 was present in a LHP1-target subset, which was enriched in the molecular function GO term “nucleic acid binding transcription factor activity” (GO:0001071, fold-enrichment 3.62, P value 2.67 10−03) and in the biological process GO term “carpel development” (GO:0048440, fold-enrichment >5, P value 4.04 10−02) (Panther classification system), suggesting the existence of a small and specialized subset of LHP1 targets containing the (A/G/T)AACCCTA(A/G) telo-box-like motif. Interestingly, TELOMERE REPEAT BINDING PROTEIN1 (TRB1) also binds to the AAACCCTA motif and it was proposed that TRB1 may act as a transcriptional repressor in the absence of LHP1 (Zhou et al., 2016).
Figure 5.
Identification of Putative cis-Regulatory DNA Motifs in LIF2 ERs and LHP1 ERs.
The regions centered on LIF2 and LHP1 summits were used to screen for putative targeting motifs. The E-value of MEME program is an estimate of the expected number of motifs with the given log likelihood ratio (or higher), and with the same width and number of occurrences, that one would find in a similarly sized set of random sequences.
LIF2 Has a Major Transcriptional Activation Activity on Its Targets
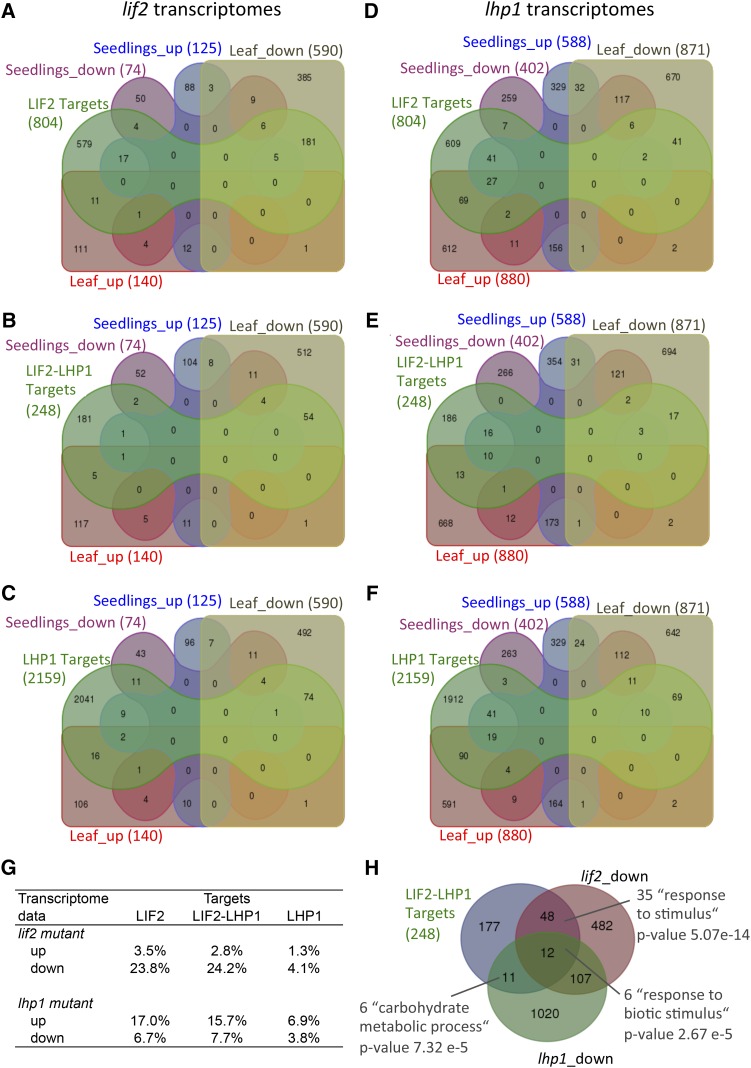
To better understand the mode of action of LIF2, we compared the binding profiles of LIF2 with our previous transcriptome data obtained from the seedlings and rosette leaves of lif2 and lhp1 mutants (Latrasse et al., 2011) (Figure 6). We observed a bias toward downregulated genes among LIF2 targets (23.8%), suggesting that LIF2 had a global transcriptional activator role on its own targets. The lif2 mutation had no significant impact on the transcription of LHP1 target genes, whereas LHP1 had a general repressor activity on LIF2 targets (25.5% of the LIF2 targets were deregulated in the lhp1 mutant). A proportion of genes located in the LIF2-LHP1 IRs were activated by LIF2 and repressed by LHP1, suggesting that LIF2 and LHP1 have general antagonistic transcriptional roles in activation and repression, respectively. Nevertheless, small sets of LIF2-LHP1 IR genes were downregulated in the mutants and enriched in stress response-associated GO terms (Figure 6G), suggesting that LIF2 and LHP1 can also act synergistically to activate specific genes.
Figure 6.
LIF2 Functions Mainly as a Transcriptional Activator on Its Targets.
(A) to (F) Venn diagrams between genes of LIF2 ERs ([A] and [D]), LIF2-LHP1 IRs ([B], [E], and [G]), and LHP1 ERs ([C] and [F]) and deregulated genes in vegetative tissues of the lif2 ([A] to [C]) and lhp1 ([D] to [F]) mutants. The analysis involved genes for which the binding was located in coding sequences or in UTRs.
(G) Comparisons between target genes and deregulated genes in lif2 and lhp1 mutants.
(H) Venn diagram and GO annotations of LHP1-LIF2 IR genes and genes activated by LHP1 and LIF2, respectively, revealed a small set of genes that requires a synergistic and activation function of both LIF2 and LHP1.
A Complex Interplay between LIF2 and LHP1 Recruitments
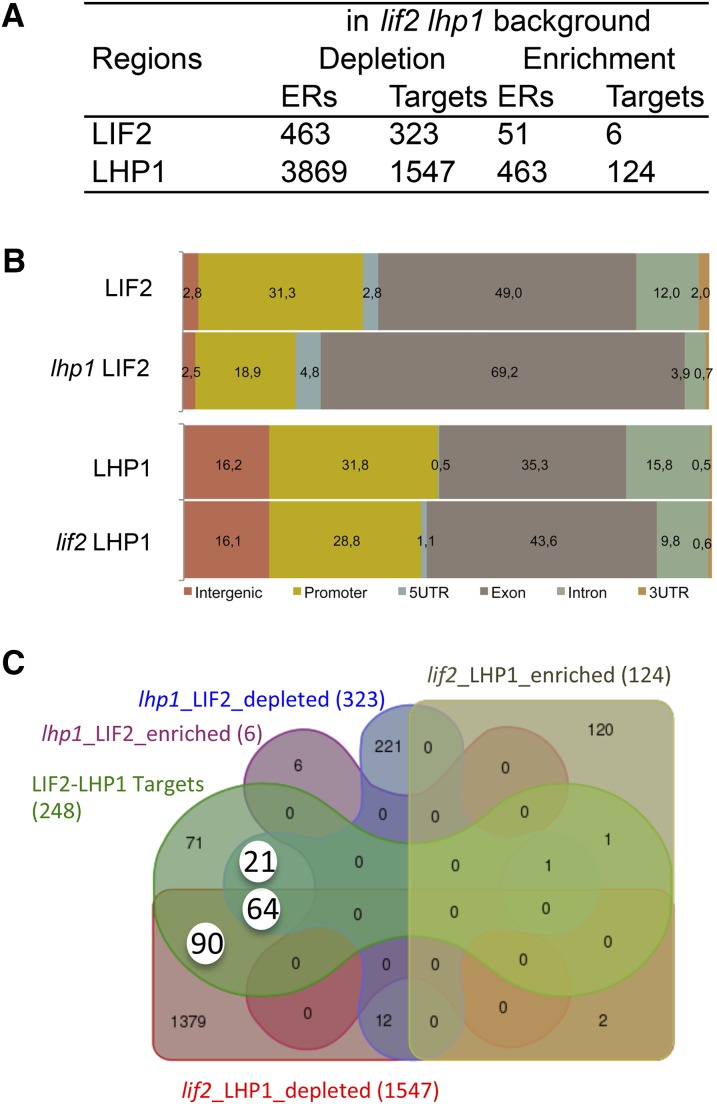
To investigate the impact of LIF2 and LHP1 on each other’s binding, we crossed the complemented mutant lines expressing tagged LIF2 or LHP1 with the lif2-1 lhp1-4 double mutant and selected transgenic lines in single mutant backgrounds (named lif2 LHP1 and lhp1 LIF2). We performed ChIP-seq experiments and identified regions that exhibited differences in the binding of the tagged proteins compared with the binding in the original complemented mutant lines (lif2 LIF2 and lhp1 LHP1). The analysis revealed a strong bias toward a depletion of any protein binding in the double mutant background (Figure 7A). In the absence of LHP1, LIF2 binding decreased strongly in regulatory regions (UTRs and promoters) and increased strongly in gene bodies (exons). Similar findings were observed for LHP1 (Figure 7B). The gene set depleted in LIF2 binding in the lhp1 background was enriched in stress-related genes and in the GO term “transcription repressor activity” (GO:0016564, NF = 17.1, P value 8.9 10−07; Supplemental Table 6). Since modifications of the binding of one protein at a precise locus in the mutant background could result from a direct loss of binding of the other or from indirect effects of its loss of function, we focused our analysis on the LIF2-LHP1 IR genes identified in the first part of this study (Figure 1). Among these LIF2-LHP1 IR genes, we identified three subsets that presented an alteration in LHP1 and LIF2 binding in the lif2 LHP1 and lhp1 LIF2 backgrounds (Figure 7C). The three sets were enriched in stress response-associated GO terms (Supplemental Figure 8). In Set-64 (64 genes), the presence of both proteins was mutually required for their binding, suggesting a synergistic mode of action, whereas for Set-90 and Set-21, LIF2 and LHP1, respectively, were necessary for the presence of the other one. Therefore, these data suggest a prominent role for LIF2 in LHP1 recruitment to chromatin and regulation in the LIF2-mediated stress response pathway. This role may be underestimated, as we only considered locations occupied by the two proteins under normal physiological conditions. Most of the genes of the three sets were not deregulated in our lif2 and lhp1 transcriptomes (Latrasse et al., 2011). This might be due to redundant mechanisms of gene regulation. Furthermore, transcriptome profiles, established in mutants under normal physiological conditions, may not highlight deregulation in responses to various cues. However, 45.3% of the Set-64 genes were downregulated in lif2, in agreement with the major transcriptional activity of LIF2 (Supplemental Figure 8D).
Figure 7.
Complex Interplay between LIF2 and LHP1 for Their Recruitment.
(A) LIF2 and LHP1 binding in the mutant backgrounds.
(B) Distribution of the annotations of the targeted regions.
(C) Venn diagram highlighting Set-21, Set-64, and Set-90 (white circles), which contain LIF2-LHP1 IR genes, depleted in one or the other protein, in the mutant backgrounds.
Rapid Recruitment of LIF2 in Response to MeJA
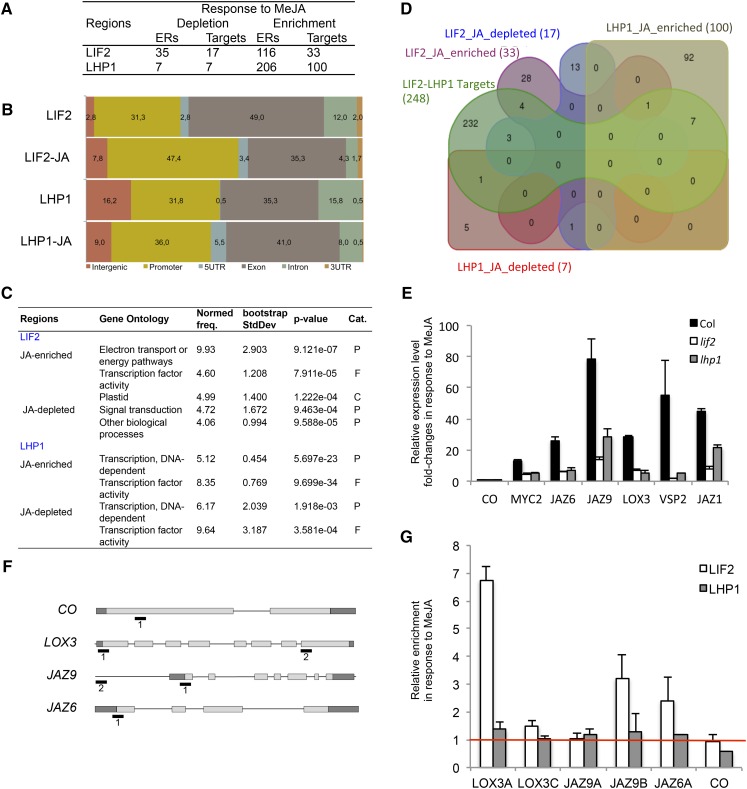
Due to the enrichment in stress GO terms, such as “JA-mediated signaling pathway,” in both lif2 transcriptomes (Le Roux et al., 2014) and LIF2 ERs (Supplemental Figure 7), we investigated whether JA treatment affects LIF2 recruitment to chromatin by comparing ChIP-seq data obtained from plants subjected or not to JA treatment. For the JA treatment, we used a short-term (1 h) oxylipin-derived MeJA treatment to avoid complex downstream regulatory events, as a 1-h treatment was sufficient to transcriptionally activate JA-inducible marker genes in wild-type plants (Supplemental Figure 9). For each protein, we identified a reduced number of regions with binding modifications in response to MeJA (JA-ERs), and observed a bias toward enrichments in LIF2 and LHP1 in response to MeJA (Figure 8A). Short-term MeJA treatment promoted LIF2 binding in promoter and intergenic regions and LHP1 binding at 5′ UTRs (Figure 8B). Interestingly, after MeJA treatment, the ERs that exhibited the greatest enrichment in LIF2 or LHP1 were enriched in “transcription factor activity” GO term and also in the “energy pathway” GO term for LIF2 ERs (Figure 8C). When the JA ERs were compared with LIF2-LHP1 IRs under normal conditions, only a limited number of loci were identified, suggesting that we had access to very early regulatory events, in agreement with the observed enrichment in TFs and/or that both proteins have independent functions in response to MeJA (Figure 8D). Alternatively, the use of a gene set in which both proteins might already be present before the treatment introduced a bias in the analysis.
Figure 8.
LIF2 and LHP1 Binding in Response to MeJA.
A 1-h MeJA treatment was performed on 2-week-old seedlings.
(A) Dynamics of LIF2 and LHP1 binding in response to MeJA.
(B) Distribution of the annotations of the binding regions.
(C) GO terms with NF > 4 (AgriGO toolkit). Cat., category; P, process; F, function; C, cellular component.
(D) Venn diagram with the genes of the LIF2-LHP1 IRs.
(E) Fold changes of the relative expression in response to MeJA in the mutant backgrounds of stress-related genes. Mean ± se. Three biological replicates were performed.
(F) and (G) Relative enrichments of LIF2 and LHP1 in response to MeJA. The targeted regions (i.e., 1 and 2) are indicated in the schematic representations (F). ChIP-QPCR experiments (G). Three biological replicates were performed.
To further characterize LIF2 binding in response to MeJA, we examined the expression of JA-inducible genes, MYC2, JASMONATE-ZIM DOMAIN (JAZ1, JAZ6, and JAZ9), VEGETATIVE STORAGE PROTEIN2 (VSP2), and LIPOXYGENASE3 (LOX3). LOX3 is among the bivalent genes identified by sequential ChIP (Luo et al., 2013). These genes were upregulated in wild-type, lif2-1, and lhp1-4 plants in response to MeJA treatment; however, the activation levels were higher in wild-type plants than in any of the mutants (Figure 8E). Under normal growth conditions, LIF2 and LHP1 were present on LOX3, a gene present in Set-64, whereas JAZ6 and JAZ9 were only targeted by LIF2 (our ChIP-seq data). These data suggested that the two proteins were cooperatively recruited to LOX3 (our ChIP-seq data). Upon MeJA treatment, LIF2 binding increased in the TSS regions of the three loci, whereas LHP1 binding was not significantly affected (Figure 8F). These data revealed that the early events of the transcriptional activation of the three JA-inducible genes require LIF2 recruitment. The presence of LHP1 on LOX3 seemed to be required to reach a full level of activation, as suggested by LOX3 expression in the lhp1 mutant, but its distribution on the locus was not significantly affected. Therefore, LHP1 seems to be required for the early transcriptional events of JA-dependent activation of LOX3 by LIF2. Whether long-term treatments would impact LHP1 binding requires further investigation.
DISCUSSION
Dynamic switches that mediate the transition between active and inactive chromatin states are crucial for the development and adaptation of organisms. PRC and TRX complexes, with their antagonistic effects on transcriptional gene regulation, play a crucial role in these chromatin-associated transitions. Chromatin may be regarded as a bistable system composed of two main antagonistic chromatin states and transitory intermediate chromatin states. The mechanism by which chromatin changes from one state to another remains poorly understood. To decipher this mechanism, we studied two interacting partners, LHP1, a plant PRC1 subunit, and the LIF2 hnRNP-Q protein. Our comparative analysis of their genome-wide binding profiles in wild-type and mutant backgrounds and under normal and stress conditions, and of their transcriptomes, revealed that these two proteins interact in a complex manner to control gene transcription.
Contrasting profiles were obtained for these interacting proteins: LHP1 was distributed over large genomic regions similar to histone marks, while LIF2 occurred in narrow binding regions, mainly located in promoters and in proximity to TSSs, which is reminiscent of TF binding at precise regulatory DNA elements. Furthermore, whereas LHP1 ERs were associated with the Polycomb H3K27me3 mark, as we previously reported using the DamID approach (Zhang et al., 2007), LIF2 was present in chromatin states characterized by the presence of H3K9ac and H3K4me3, which are usually associated with active/open chromatin.
The LIF2-LHP1 IRs were identified at the intersection of LIF2 and LHP1 protein distributions. However, to pursue and fully demonstrate that they are simultaneously binding to the exact same chromatin fiber, further analyses, such as sequential ChIP experiments, would be required. The LIF2-LHP1 IRs were associated with antagonistic marks, which may correspond to bivalent regions (Sequeira-Mendes et al., 2014) or to intermediate heterochromatin such as telomeric heterochromatin (Vrbsky et al., 2010; Vaquero-Sedas et al., 2012). Interestingly, 9.8% of the H3K9ac target genes in Arabidopsis are also marked by H3K27me3 (Zhou et al., 2010; Karmodiya et al., 2012) and H3K9ac is present in bivalent chromatin regions of mouse promoters of developmentally regulated genes (Karmodiya et al., 2012). LHP1 interacts with MSI1 (Derkacheva et al., 2013), which associates with histone deacetylase 19 (HDAC19) in the same in vivo complex, to maintain a low H3K9ac level at genes involved in the ABA signaling pathway (Mehdi et al., 2016). Furthermore, the LIF2-LHP1 IRs had a good coverage with Chromatin State 2 and were enriched in stress-responsive genes, demonstrating that the LIF2/LHP1 duo seems to have a specialized function in the stress response pathway and a putative role in maintaining or regulating a distinctive chromatin state at a specific gene set. Furthermore, the binding maps of each protein, established in the absence of its partner, revealed various scenarios that were highly dependent on the genomic contexts, with synergistic binding, as well as binding dependent on one or the other protein.
The identification of GAGA motifs in LHP1 ERs confirmed a recent discovery (Hecker et al., 2015). Indeed, the BPC6 GAGA binding factor interacts with LHP1 and recruits LHP1 at GAGA motif-containing DNA probes in vitro (Hecker et al., 2015). Interestingly, GAGA motifs were also present in FIE ERs (Deng et al., 2013), as was Motif 2, which is similar to a telo-like box. The presence of these two types of motifs in LHP1 and FIE ERs suggests the existence of common recruitment motifs between plant PRC1 and PRC2 subunits, but also between PRC1 and LIF2. Thus, these different DNA motifs may correspond to modules that participate to form putative plant PREs.
In addition to establishing the global rules governing LIF2 and LHP1 binding, we observed that the two proteins exhibited different recruitment dynamics in response to a short-term MeJA treatment. A rapid increase in LIF2 binding was observed, especially at the TSS of LOX3, JAZ6, and JAZ9, with an associated increase in gene expression. These data were in agreement with the global downregulation of LIF2 targets in lif2. At LOX3, the presence of LHP1 was not modulated by the MeJA treatment, but LHP1 was required for LIF2-mediated activation. Removal of LHP1 was not a prerequisite for the early transcriptional activation, suggesting that the two proteins may have different kinetics of action. Thus, one hypothesis would be that the RNA binding protein LIF2 functions in transcriptional activation, especially in JA-dependent activation, and may counteract gene repression via its interaction with LHP1. Further investigation is needed to understand this dynamic and complex interplay. For instance, it remains unclear how LIF2 specifically interacts with chromatin. Perhaps this interaction is mediated by RRMs. Indeed, RRMs are plastic protein domains and some RRMs also have DNA binding properties (Enokizono et al., 2005; Grinstein et al., 2007; Wan et al., 2007). Alternatively, RNA molecules interacting with RRMs may participate in RNA/DNA recognition and thus help target RBP via their interaction with RNA molecules. Since RNA molecules play diverse functions in modulating animal PRC activities, further investigation of putative interactions between LIF2 and RNA molecules will be of key importance.
Finally, we showed that LHP1 ERs had a significant and robust tendency to form clusters (in the ∼10 kb range), regardless of the chromosome arm identity. Due to the large number of LHP1 ERs in the genome, the distribution of LHP1 clusters may not be neutral and may influence the functional organization of the genome. Indeed, proteins in the HP1 family have dimerization properties and SWI6 even has an oligomerization property, which contributes to heterochromatin formation (Canzio et al., 2011). Thus, a clustering of the LHP1 ERs may have 3D consequences on genome organization. In animals, PcG proteins contribute to the modular organization of the linear epigenome, but also to the 3D genome organization (Cavalli, 2014; Del Prete et al., 2015). In Arabidopsis, recent HiC studies highlighted long-range genome interactions, but the absence of large chromatin modules as observed in animal genomes (Feng et al., 2014; Grob et al., 2014), possibly due to resolution limitations. Although restricted to one dimension, our approach in this study, in which spatial statistics are applied to genome-wide data, represents a complementary tool for deciphering eukaryotic genome organization. It allowed us to evaluate distribution patterns of chromatin-associated proteins at different scales and highlighted the existence of short-range clusters on the linear organization of the Arabidopsis genome. It will be interesting to determine whether the linear proximity of LHP1 ERs contributes to the formation of LHP1 foci (Gaudin et al., 2001), promotes silent plant chromatin formation, or influences the 3D genome organization.
METHODS
Materials and Hormonal Treatment
All Arabidopsis thaliana lines used in this study are in the Col-0 background. The lif2-1 and lhp1-4 mutants were previously described (Latrasse et al., 2011). For all experiments, plants were grown in vitro for 14 d under controlled long-day conditions as previously described (Gaudin et al., 2001). For MeJA treatments, a filter paper was imbibed with 10 μL of 95% MeJA (Sigma-Aldrich) and placed in a Petri dish. Plates were hermetically sealed and placed for 1 h under identical growth conditions. MeJA-treated and mock seedlings were either directly harvested for gene expression analyses or fixed for ChIP assays after the 1-h treatment. All primers are listed in Supplemental Table 7.
Plasmid Constructs
For the 3xHA:LIF2 binary construct, the 3xHA tag was PCR amplified from the pGWB15 vector (Invitrogen) using the 3HA-1 and 3HA-2 primers bearing PstI and XbaI restriction sites, respectively. After digestion and purification, the 3xHA fragment was inserted into the pCambia1300 vector giving the pCa-HA vector. The Nos terminator, amplified from plasmid pUC-SPYNE (Walter et al., 2004) using the Nost-1 and Nost-2 primers (bearing KpnI and EcoRI sites, respectively) was digested, gel-purified, and inserted into the KpnI/EcoRI-digested pCa-HA vector. A 3-kb promoter region of LIF2 (including the first three codons) was amplified from the T18A10 BAC plasmid (ABRC DNA stock center) using primers AD379-28 and AD379-29 (bearing a PstI restriction site). The PstI-digested LIF2 promoter fragment was gel-purified and inserted into the pCa-HA-tNos vector at the PstI and blunt-made HindIII sites. Finally, the LIF2 genomic region was amplified from T18A10 using the primers AD379-30 and AD379-32, digested with XhoI, and inserted into the SalI/SmaI-digested pCa-ProLIF2:HA-tNos vector giving the pCa-ProLIF2:HA:LIF2-tNos vector (N-terminal HA-tagged gLIF2).
For the ProLHP1:LHP1:HA binary construct, a 3xHA fragment was PCR amplified from the pGWB15 vector (Invitrogen) using the 3HA-2 and 3HA-2 primers and digested with EcoRV and XhoI. The 3xHA fragment was inserted into the EcoRV restriction site of the vector bearing a 5569-bp genomic LHP1 fragment (Latrasse et al., 2011). Subsequently, the NcoI/BstEII fragment containing the LHP1:3xHA-tagged region was substituted to the wild-type genomic fragment of the pCaSSP vector giving the gLHP1:HA binary plasmid (C-terminal HA tagged gLHP1). All subcloning steps were confirmed by sequencing. Col-0 plants were transformed by floral dip. For each construct, homozygous transgenic lines with wild-type phenotypes were selected, in which the functional HA-tagged protein was detected.
RNA Extraction
Total RNA was isolated from 14-d-old in vitro-grown seedlings, subjected or not to MeJA treatment, using the RNeasy Plant Mini Kit (Qiagen) according to supplier’s instructions. Total RNA (1 to 2 μg) was treated with RNase-free DNaseI (Invitrogen) and reverse transcribed with Superscript II reverse transcriptase (Invitrogen).
Quantitative Real-Time PCR
Relative levels of cDNA (RT-qPCR) and immunoprecipitated DNA fragments (ChIP-qPCR) were analyzed by quantitative real-time PCR on an Eppendorf Mastercycler ep Realplex using SsoAdvanced SYBR (Bio-Rad). Immunoprecipitated DNA levels were normalized to input and to the internal reference gene EF1 (AT5G60390). The cDNA levels were normalized to EF1.
ChIP Library Construction and Sequencing
ChIP assays were performed on 5 g of 14-d-old in vitro seedlings from transgenic lines expressing LHP1-HA or LIF2-HA in the single or double mutant genetic backgrounds, using a previously published protocol (Latrasse et al., 2011), with the following minor modifications. Chromatin was immunoprecipitated overnight using high-affinity anti-HA antibody (Roche). Immunoprecipitated DNA enrichment was controlled by quantitative real-time PCR (qPCR). DNA quantity and quality were checked using a Qubit fluorometer (ThermoFisher Scientific) and an Agilent 2100 Bioanalyzer (Agilent Technologies,). Several independent experiences were pooled for library construction. Then, 10 to 15 ng of immunoprecipitated DNA was fragmented to a 100- to 500-bp range using the Covaris E210 instrument. Libraries were prepared according to the Illumina standard procedure using the NEBNext DNA Sample Preparation Reagent Set 1 (New England Biolabs) and homemade ligation adaptors. The ligated product was amplified by 12 cycles of PCR using Platinum Pfx DNA Polymerase (ThermoFisher Scientific). Amplified material was purified using Agencourt Ampure XP beads (Beckmann Coulter Genomics). Libraries were then quantified by qPCR and library profiles were evaluated using an Agilent 2100 Bioanalyzer. Two independent libraries for each protein were sequenced using 100 base-length read chemistry in a paired-end flow cell on the HiSeq 2000 (Illumina).
ChIP-Seq Data Analyses
After Illumina sequencing, Illumina read processing and quality filtering were performed. An in-house quality control process was applied to reads that passed the Illumina quality filters. Low quality nucleotides (Q < 20) were discarded from both ends of the reads. Next, Illumina adapter and primer sequences were removed from the reads. Then, reads shorter than 30 nucleotides after trimming were discarded. These trimming and removing steps were achieved using internal software based on the FastX package (FASTX-Toolkit; http://hannonlab.cshl.edu/fastx_toolkit/index.html). This processing yields high-quality data and improves subsequent analyses. The sequencing reads were uniquely mapped to the Arabidopsis genome (TAIR10; http://www.arabidopsis.org) using Bowtie 4.1.2 mapper (Langmead et al., 2009) with default mismatch parameters and retaining only reads mapping uniquely to the genome for further analysis. The main heterochromatic regions of the genome were thus excluded from our analysis.
To identify biologically relevant binding regions, peak prediction and normalization were performed using MACS1.4.1 (Zhang et al., 2008) and peak analysis was performed using S-MART (Langmead et al., 2009) or the “annotatePeaks.pl” software from Homer (http://homer.salk.edu/homer; Heinz et al., 2010). High-confidence target regions (i.e., ERs) were defined as strict overlap of the MACS peaks from the corresponding biological replicates.
By default, a TSS region was defined from −1 kb to +100 bp from TSS and the TTS region was defined from −100 bp to +1 kb from the TTS. The process of annotating peaks/regions was divided into two primary parts. The first determined the distance to the nearest TSS and assigned the peak to that gene. The second determined the genomic annotation of the region occupied by the center of the peak/region.
Bioinformatics Analyses
Motifs were predicted using the integrated online pipeline “peak-motifs” (http://plants.rsat.eu/; Thomas-Chollier et al., 2012a, 2012b). Briefly, 50 and 300 bp surrounding protein binding summits were scanned for a global overrepresentation of words (oligo-analyses) or spaced words (dyad-analyses). Then, 5000 random, artificial 300-bp long sequences were generated by the “RSAT-random sequence tool” (http://plants.rsat.eu/) and were used as background control for motif discovery. In parallel, sequences were analyzed by the motif prediction program “MEME” (Bailey and Elkan, 1995). The word occurrence was determined using the word frequency program in AtcisDB from AGRIS (http://arabidopsis.med.ohio-state.edu/AtcisDB/).
The functional annotation and classification of gene populations was performed using the online “AgriGO” gene Ontology tool (http://bioinfo.cau.edu.cn/agriGO/) using preset parameters. Venn diagrams were generated using the online tool provided by T. Hulsen (http://bioinformatics.psb.ugent.be/webtools/Venn/).
To analyze the histone mark enrichments over the ERs, ChIP-seq data presented by Luo et al. (2013) were used and available at SRA under IDs GSM701923-701931. Raw data were mapped onto the TAIR10 genome with the Bowtie mapper (Langmead et al., 2009) (unique hits, one mismatch at most). Mapped reads were processed using SAMtools (Li et al., 2009) and BEDtools (Quinlan and Hall, 2010). The number of reads per bp of the selected loci was counted and compared with that of randomized loci (using the shuffle BEDtool). Fold enrichment/depletion was calculated as the ratio between the mean read number in regions of interest versus randomized regions. Statistical significance was assessed by t tests. Box plots represent distribution of histone mark ChIP-seq reads within LIF2, LHP1, LIF2-LHP1, and the corresponding randomized regions.
Spatial Distributions of the Targeted Regions
The spatial distributions of LHP1 and LIF2 targeted regions were quantified and analyzed for each individual biological replicate, using the cumulative distribution functions of (1) the distance to the nearest neighbor of each targeted region and (2) the interdistance between every pair of targeted regions. Departure from randomness was assessed by adapting a Monte Carlo procedure developed for 3D data (Andrey et al., 2010). Observed distributions were compared with distributions obtained under complete randomization of targeted regions without overlap (999 randomizations for computing averages of distance functions under randomness; 999 further randomizations for computing envelopes around averages). The relative position of the empirical distance function within the range of variations under randomness was used to estimate P values (Andrey et al., 2010).
Accession Numbers
Sequencing data were deposited at NCBI under the Sequence Read Archive number SRP068984.
Supplemental Data
Supplemental Figure 1. ChIP-seq experiments.
Supplemental Figure 2. Exon distributions of LIF2 ERs.
Supplemental Figure 3. Distributions of the number of summits in 1-Mb windows.
Supplemental Figure 4. Cumulative distribution of the distance to the nearest LHP1 summit.
Supplemental Figure 5. Cumulative distribution of LHP1 summit interdistances.
Supplemental Figure 6. Posttranslational histone modifications and their distributions in LIF2 ERs, LHP1 ERs, and LIF2-LHP1 IRs.
Supplemental Figure 7. GO term analysis of the target loci of LIF2 and LHP1.
Supplemental Figure 8. Analyses of the LIF2-LHP1 IRs with binding alterations in the mutant backgrounds.
Supplemental Figure 9. Expression kinetics of JA-induced marker genes in response to MeJA treatment in wild-type plants.
Supplemental Table 1. Tandem duplications and LHP1 target genes.
Supplemental Table 2. GO term analysis of the genes present in LIF2 ERs and LIF2-LHP1 IRs using the Plant Functional Genomics (BAR) classification Superviewer program.
Supplemental Table 3. Enrichments of LIF2 and LHP1 targets in specific transcription factor families using the PlantGSEA resource.
Supplemental Table 4. LIF2 and LHP1 targets are also bound by specific transcription factors.
Supplemental Table 5. Occurrences of the two identified DNA words.
Supplemental Table 6. GO term analysis of LIF2 or LHP1 depleted regions in the mutant backgrounds (AgriGO).
Supplemental Table 7. List of primers.
Supplementary Material
Acknowledgments
We thank Bruno Letarnec and Hervé Ferry for plant care in the greenhouses, Georg Haberer for providing the S-cluster listing, and Crisanto Gutierrez for helpful exchange. We thank our colleagues, Franziska Turck and Dierk Wanke, for critical reading of the manuscript and suggestions. D.L., A.M.M., and M.H. were supported by fellowships from the ANR (ANR-08-BLAN-0200, Polycombara) from the French Research Ministry. S.D.P. was supported by a PhD fellowship provided by the European Commission Seventh Framework-People-2012-ITN Project EpiTRAITS (epigenetic regulation of economically important plant traits, no-316965). Génoscope supported the sequencing in the frame of a large-scale DNA sequencing project (N°11). The IJPB benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS).
AUTHOR CONTRIBUTIONS
A.M.M., D.L., M.Z., P.A., N.H.-H., M.H., C.B., S.D.P., A.A., and V.G. performed the experiments. A.M.M., M.Z., M.Z., P.A., M.H., H.Q., and V.G. analyzed the data. A.M.M., M.Z., P.A., and V.G. prepared the figures. A.M.M. and V.G. wrote the manuscript.
Glossary
- TF
transcription factor
- RRM
RNA recognition motif
- RBP
RNA binding protein
- MeJA
methyl jasmonate
- FDR
false discovery rate
- ER
enrichment region
- UTR
untranslated region
- TSS
transcription start site
- TTS
transcription termination site
- IR
intersect region
- NF
normed frequency
- GO
Gene Ontology
References
- Aceituno F.F., Moseyko N., Rhee S.Y., Gutiérrez R.A. (2008). The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics 9: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrey P., et al. (2010). Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLOS Comput. Biol. 6: e1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. (1995). The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3: 21–29. [PubMed] [Google Scholar]
- Bauer M., Trupke J., Ringrose L. (2015). The quest for mammalian Polycomb response elements: are we there yet? Chromosoma 125: 471–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. (2013). Noncoding RNA and Polycomb recruitment. RNA 19: 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonje M. (2014). PRC1 marks the difference in plant PcG repression. Mol. Plant 7: 459–471. [DOI] [PubMed] [Google Scholar]
- Canzio D., Chang E.Y., Shankar S., Kuchenbecker K.M., Simon M.D., Madhani H.D., Narlikar G.J., Al-Sady B. (2011). Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41: 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G. (2014). Chromosomes: now in 3D! Nat. Rev. Mol. Cell Biol. 15: 6. [DOI] [PubMed] [Google Scholar]
- Chen D., Xu G., Tang W., Jing Y., Ji Q., Fei Z., Lin R. (2013). Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D., Zilberman D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T., Questa J.I., Sun Q., Dean C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 111: 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete S., Mikulski P., Schubert D., Gaudin V. (2015). One, two, three: Polycomb proteins hit all dimensions of gene regulation. Genes (Basel) 6: 520–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Buzas D.M., Ying H., Robertson M., Taylor J., Peacock W.J., Dennis E.S., Helliwell C. (2013). Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genomics 14: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M., Steinbach Y., Wildhaber T., Mozgová I., Mahrez W., Nanni P., Bischof S., Gruissem W., Hennig L. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32: 2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enokizono Y., Konishi Y., Nagata K., Ouhashi K., Uesugi S., Ishikawa F., Katahira M. (2005). Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 280: 18862–18870. [DOI] [PubMed] [Google Scholar]
- Feng S., Cokus S.J., Schubert V., Zhai J., Pellegrini M., Jacobsen S.E. (2014). Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 55: 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förderer A., Zhou Y., Turck F. (2016). The age of multiplexity: recruitment and interactions of Polycomb complexes in plants. Curr. Opin. Plant Biol. 29: 169–178. [DOI] [PubMed] [Google Scholar]
- Gaspin C., Rami J.F., Lescure B. (2010). Distribution of short interstitial telomere motifs in two plant genomes: putative origin and function. BMC Plant Biol. 10: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin V., Libault M., Pouteau S., Juul T., Zhao G., Lefebvre D., Grandjean O. (2001). Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128: 4847–4858. [DOI] [PubMed] [Google Scholar]
- Gil J., O’Loghlen A. (2014). PRC1 complex diversity: where is it taking us? Trends Cell Biol. 24: 632–641. [DOI] [PubMed] [Google Scholar]
- Grinstein E., Du Y., Santourlidis S., Christ J., Uhrberg M., Wernet P. (2007). Nucleolin regulates gene expression in CD34-positive hematopoietic cells. J. Biol. Chem. 282: 12439–12449. [DOI] [PubMed] [Google Scholar]
- Grob S., Schmid M.W., Grossniklaus U. (2014). Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol. Cell 55: 678–693. [DOI] [PubMed] [Google Scholar]
- Haberer G., Hindemitt T., Meyers B.C., Mayer K.F. (2004). Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol. 136: 3009–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker A., Brand L.H., Peter S., Simoncello N., Kilian J., Harter K., Gaudin V., Wanke D. (2015). The Arabidopsis GAGA-binding factor BASIC PENTACYSTEINE6 recruits the POLYCOMB-REPRESSIVE COMPLEX1 component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA motifs. Plant Physiol. 168: 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., Tora L. (2012). H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D., et al. (2011). Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS One 6: e16592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C., Del Prete S., Boutet-Mercey S., Perreau F., Balagué C., Roby D., Fagard M., Gaudin V. (2014). The hnRNP-Q protein LIF2 participates in the plant immune response. PLoS One 9: e99343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Sidote D.J., Zhang Y., Kerstetter R.A., Michael T.P., Lam E. (2013). Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 73: 77–90. [DOI] [PubMed] [Google Scholar]
- Mehdi S., Derkacheva M., Ramström M., Kralemann L., Bergquist J., Hennig L. (2016). The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 28: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart N.J., Gil P., Chen W., Han B., Chang H.S., Wang X., Zhu T. (2003). Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiol. 132: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu L., Sung Z.R. (2015). PcG and trxG in plants - friends or foes. Trends Genet. 31: 252–262. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad F., Lebas M., Lescure B. (1994). Interstitial telomeric repeats within the Arabidopsis thaliana genome. J. Mol. Biol. 239: 163–169. [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J., Aragüez I., Peiró R., Mendez-Giraldez R., Zhang X., Jacobsen S.E., Bastolla U., Gutierrez C. (2014). The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. (2013). Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., et al. (2008). DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins. Plant Physiol. 146: 1862–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802. [DOI] [PubMed] [Google Scholar]
- Tavares L., et al. (2012). RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Herrmann C., Defrance M., Sand O., Thieffry D., van Helden J. (2012a). RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 40: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Darbo E., Herrmann C., Defrance M., Thieffry D., van Helden J. (2012b). A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat. Protoc. 7: 1551–1568. [DOI] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero-Sedas M.I., Luo C., Vega-Palas M.A. (2012). Analysis of the epigenetic status of telomeres by using ChIP-seq data. Nucleic Acids Res. 40: e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrbsky J., Akimcheva S., Watson J.M., Turner T.L., Daxinger L., Vyskot B., Aufsatz W., Riha K. (2010). siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 6: e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wan F., Anderson D.E., Barnitz R.A., Snow A., Bidere N., Zheng L., Hegde V., Lam L.T., Staudt L.M., Levens D., Deutsch W.A., Lenardo M.J. (2007). Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 131: 927–939. [DOI] [PubMed] [Google Scholar]
- Yi X., Du Z., Su Z. (2013). PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 41: W98–W103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A., Mejia-Guerra M.K., Kurz K., Liang X., Welch L., Grotewold E. (2011). AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res. 39: D1118–D1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang H., et al. (2014). Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J. Cell Biol. 204: 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Germann S., Blus B.J., Khorasanizadeh S., Gaudin V., Jacobsen S.E. (2007). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14: 869–871. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang X., He K., Charron J.B., Elling A.A., Deng X.W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72: 585–595. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Hartwig B., Velikkakam James G., Schneeberger K., Turck F. (2016). Complementary activities of TELOMERE REPEAT BINDING proteins and Polycomb Group complexes in transcriptional regulation of target genes. Plant Cell 28: 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Coleman-Derr D., Ballinger T., Henikoff S. (2008). Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.