Brassica plants have altered transcript levels and coevolved neofunctionalization of duplicated genes to block developmental defects resulting from detrimental chimeric genes.
Abstract
Chimeric genes contribute to the evolution of diverse functions in plants and animals. However, new chimeric genes also increase the risk of developmental defects. Here, we show that the chimeric gene Brassica napus male sterile 4 (Bnams4b) is responsible for genic male sterility in the widely used canola line 7365A (Bnams3 ms3ms4bms4b). Bnams4b originated via exon shuffling ∼4.6 million years ago. It causes defects in the normal functions of plastids and induces aborted anther formation and/or albino leaves and buds. Evidence of the age of the mutation, its tissue expression pattern, and its sublocalization indicated that it coevolved with BnaC9.Tic40 (BnaMs3). In Arabidopsis thaliana, Bnams4b results in complete male sterility that can be rescued by BnaC9.Tic40, suggesting that BnaC9.Tic40 might restore fertility through effects on protein level. Another suppressor gene, Bnams4a, rescues sterility by reducing the level of transcription of Bnams4b. Our results suggest that Brassica plants have coevolved altered transcription patterns and neofunctionalization of duplicated genes that can block developmental defects resulting from detrimental chimeric genes.
INTRODUCTION
New genes can represent a major source of genetic innovation in genomes (Chen et al., 2010; reviewed in Chen et al., 2013; Cardoso-Moreira and Long, 2012) and may be involved in the establishment of novel biochemical pathways (Matsuno et al., 2009; Weng et al., 2012) as well as being integrated into existing gene networks (Capra et al., 2010; Chen et al., 2010, 2012b). They have been suggested to play essential roles in development (Chen et al., 2010; Ding et al., 2010), brain evolution (Chen et al., 2012a), and sexual dimorphism (Betrán et al., 2002; Ding et al., 2010). Natural chimeric genes arise from the fusion of existing sequence elements to create a new open reading frame (ORF) that contributes to novel molecular and cellular functions (Chen et al., 2013; Ding et al., 2012; Long and Langley, 1993). Most of the known chimeric genes were identified in insects and humans and were found to originate through RNA-based retrotransposition. For example, jingwei was generated by fusion of the N terminus of the yellow emperor gene with a retrotransposed Adh gene (Long and Langley, 1993). Although some chimeric genes induce early adaptive evolution and are finally fixed (Jones and Begun, 2005; Sayah et al., 2004; Wang et al., 2002), the majority are rapidly eliminated from the genome though pseudogenization. Only ∼1.4% of chimeric genes are preserved by natural selection (Rogers et al., 2009). Most of the information on the origin, evolution, and function of chimeric genes has come from RNA-based chimeric genes than from those derived by DNA-based exon shuffling. In Drosophila melanogaster, 8 of 14 DNA-based chimeric genes were found to be undergoing pseudogenization or were noncoding RNA genes (Zhan et al., 2011). Although chimeric genes arise frequently in plants, their use in agricultural breeding programs has seldom been reported (Wang et al., 2006). In addition, it is not clear how plants defend themselves against the potential detrimental effects of chimeric genes.
Canola (Brassica napus; allotetraploid, AACC, 2n = 38) is one of the most important sources of edible oil in the Brassicaceae and was first cultivated in Europe during the Middle Ages. It was formed by natural allopolyploidization between the Brassica oleracea ancestor (Mediterranean cabbage, CC, n = 9) and Brassica rapa ancestor (Asian cabbage or turnip, AA, n = 10) (Chalhoub et al., 2014; Nagaharu, 1935). According to the “triangle of U” hypothesis, this likely occurred by natural pairwise hybridization in which three diploid Brassica species, B. rapa (AA, n = 10), B. oleracea (CC, n = 9), and B. nigra (BB, n = 8), formed the allopolyploids B. napus (AACC, n = 19), B. juncea (AABB, n = 18), and B. carinata (BBCC, n = 17) (Nagaharu, 1935). Such hybridization and polyploidization constitute the major driving forces of genome evolution in B. napus (Marhold and Lihova, 2006; Nagaharu, 1935). The Brassica and Arabidopsis lineages diverged ∼20 million years ago (Mya) from a common ancestor (ancestral karyotype, n = 8) after the three polyploidization events (γ, β, and α) of angiosperms (Yang et al., 1999). Comparative genomic analyses of the 24 conserved chromosomal blocks (labeled A to X) suggested that the three basic diploid Brassica species descended from a common ancestor that was formed after whole-genome triplication (∼15.9 Mya) (Liu et al., 2014; Lysak et al., 2005; Schranz et al., 2006; Wang et al., 2011b).
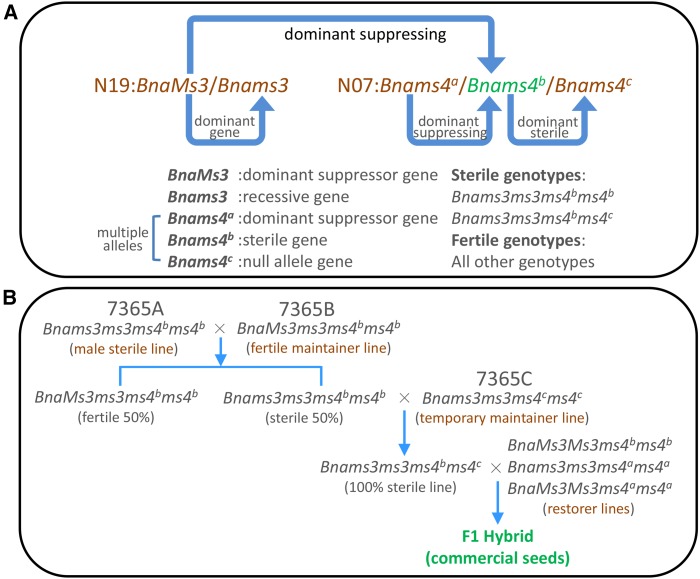
Male sterility is the most effective and commercially significant means of using heterosis. Cytoplasmic male sterility is widely used in canola hybrid breeding. However, the genic male sterile (GMS) breeding system 7365ABC offers further advantages such as stable sterility, a wide breadth of restorer lines, and no evident negative cytoplasmic effects. Thus, this system has considerable potential to be an important approach for future hybrid breeding (Huang et al., 2012; Xia et al., 2012). The 7365ABC GMS system was discovered in the 1990s and was regarded as a natural variation (Chen et al., 1993). Many commercial hybrids based on this GMS system have since been released (Chen et al., 2002, 2003; Fan et al., 2010; Sun et al., 2004; Wang et al., 2009); for example, the winter rapeseed varieties bred using this GMS system monopolized the European rapeseed market during the 2000s. 7365ABC is controlled by two independent loci, BnaMs3/Bnams3 (BnaC9.Tic40/ BnaC9.tic40) and Bnams4a/Bnams4b/Bnams4c (Figure 1A). Bnams4a/Bnams4b/Bnams4c is a multiallelic locus on chromosome N07, whereas BnaMs3/Bnams3 is located on N19. BnaMs3 and Bnams4a represent suppressor genes and Bnams4b is a male-sterile gene (Figure 1A). The dominance relationships of these alleles are BnaMs3>Bnams3 and Bnams4a>Bnams4b>Bnams4c (Figure 1A). The GMS system provides an excellent three-line breeding strategy for the production of commercial hybrids (Figure 1B). Thus, cloning and characterization of this multiallelic locus will be essential to understanding the biological and evolutionary mechanisms of this elite breeding system and to making better use of its potential capabilities.
Figure 1.
The Genetic Model and Three-Line Hybrid Breeding Procedure of Genic Male Sterile System 7365ABC in B. napus.
(A) The fertility of 7365ABC is controlled by two alleles of loci located in two different linkage groups: N07 and N19. The multigene locus (N07) consists of Bnams4a, Bnams4b, and Bnams4c. Bnams4b is a dominant male-sterile gene relative to the null allele Bnams4c; Bnams4b has two dominant suppressor genes BnaC9.Tic40 and Bnams4a. The locus N19 consists of BnaC9.Tic40 (BnaMs3) and BnaC9.tic40 (Bnams3). Curved blue arrows show the explicit-implicit relationships between these genes: BnaC9.Tic40>BnaC9.tic40 and Bnams4a>Bnams4b>Bnams4c. Therefore, only Bnams3ms3ms4bms4b and Bnams3ms3ms4bms4c are sterile for they have no suppressor gene, while Bnams3ms3ms4ams4b and BnaMs3ms3ms4bms4b are fertile as they each contain a suppressor gene. Bnams3ms3ms4cms4c is also fertile as it has no sterile gene Bnams4b, and this genotype can be used as temporary maintainer line.
(B) The genic male sterile breeding system 7365ABC consisted of male-sterile line 7365A (Bnams3ms3ms4bms4b), maintainer lines 7365B (BnaMs3ms3ms4bms4b) and 7365C (Bnams3ms3ms4cms4c), and restorer lines (BnaMs3Ms3ms4bms4b or Bnams3ms3ms4ams4a, etc.). First, male-sterile line 7365A is crossed with maintainer line 7365B to generate a 1:1 ratio of fertile and sterile plants; second, the sterile plants are crossed with temporary maintainer line 7365C to generate a 100% completely sterile population; this population makes manual removal of 50% fertile plants in field during hybrid seed production of large area unnecessary, compared with using other types of genic male sterile systems. Subsequently, the 100% sterile population was extensively tested with a variety of restorer lines to produce superior hybrid seed. This kind of genic male sterile system has extensive restorer lines, compared with cytoplasmic male sterile lines, due to the wide distribution of suppressor genes (BnaC9.Tic40 and Bnams4a) in natural varieties or inbred lines.
The evolutionary history of this GMS system has long been recognized as an important issue for the molecular biology of reproductive development and use of heterosis in B. napus. Consequently, we cloned BnaC9.Tic40 via map-based cloning (Dun et al., 2011) and showed that it encodes a protein that is a member of the translocon at the inner envelope membrane of chloroplasts (TIC) complex. BnaC9.Tic40 coordinates the participation of Tic110 and Hsp93 proteins in the translocation of specific preproteins across the inner envelope membrane into tapetal plastids (Chou et al., 2006; Chou et al., 2003; Dun et al., 2011). All Tic40 copies from Brassica species can rescue the albino leaf phenotype of the attic40 mutant of Arabidopsis thaliana. Only BolC9.Tic40 from the genome C09 in B. oleracea and BnaC9.Tic40 from N19 in B. napus can specifically rescue sterility in 7365A (Dun et al., 2014). BnaC9.Tic40 is a gain-of-function mutation and may represent neofunctionalization among the duplicated Tic40 homologs in B. oleracea lineages and has been associated with the male sterility gene Bnams4b (Dun et al., 2014). However, the origin, evolution, and function of the sterility gene Bnams4b remain unclear. Furthermore, the evolutionary relationship between the neofunctionalized suppressor gene BnaC9.Tic40 and Bnams4b is also unknown.
Our aims here were to clone Bnams4b by map-based cloning and to characterize gene structure, expression pattern, protein function, and cell biological effects. We also examined the origin and evolutionary relationship of Bnams4b with both suppressor genes BnaC9.Tic40 and Bnams4a. We found that the plants had evolved transcriptional and neofunctional mechanisms to block the developmental defects from this chimeric gene in B. napus.
RESULTS
Localization of Bnams4b in a 61-kb Insertion Region
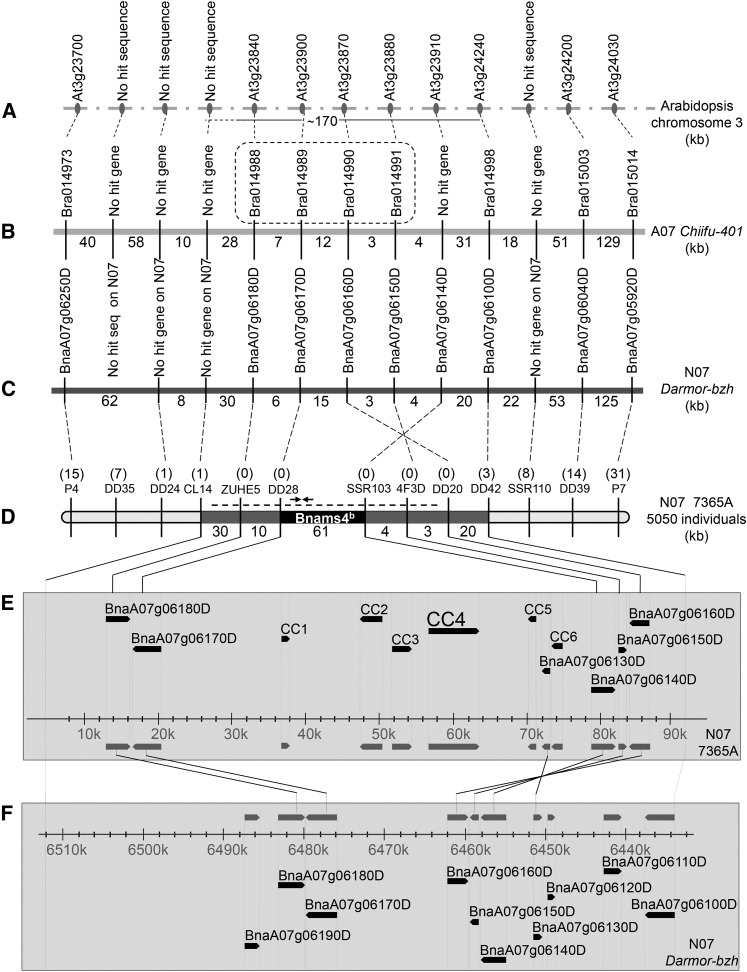
Fine mapping of Bnams4b was performed using a population of 5500 plants from the near-isogenic line (NIL) 7365AC (Bnams3ms3ms4bms4c/Bnams3ms3ms4cms4c). We used the published genome data of B. rapa to develop simple sequence repeat and sequence characterized amplified region markers. Marker polymorphisms were identified and genetic distances were determined to narrow down the candidate physical region. Finally, the markers CL14 and DD42 (Figure 2; Supplemental Table 1) were used to delimit Bnams4b within ∼170-kb (Figure 2A), 85-kb (Figure 2B), and 78-kb (Figure 2C) DNA fragments according to the published genome sequences of A. thaliana (Col-0) (Lamesch et al., 2012), B. rapa (Chiifu-401) (Wang et al., 2011b), and B. napus (Darmor-bzh) (Chalhoub et al., 2014), respectively. The 85-kb region of B. rapa contained four genes, Bra014988, Bra014989, Bra014990, and Bra014991 (Figure 2B); their corresponding A. thaliana orthologous genes were AT3G23840, AT3G23900, AT3G23870, and AT3G23880 (Figure 2A). Transgenic complementation experiments with A. thaliana and B. napus plants confirmed that the corresponding four genes in 7365A were not Bnams4b.
Figure 2.
Physical Mapping of the Predicted Candidate Gene of Bnams4b in B. napus.
(A) Corresponding A. thaliana chromosome 3 region homologous to the region in which Bnams4b resides. “No hit genes” means there is no homologous A. thaliana gene here. The small solid ellipses indicate corresponding genes of A. thaliana chromosome 3 or no hit genes.
(B) Corresponding B. rapa chromosome A07 region homologous to the region in which Bnams4b resides. The vertical bars indicate corresponding genes of B. rapa chromosome A07 or no hit genes. Dotted box represents the four predicted candidate genes for Bnams4b.
(C) Corresponding B. napus chromosome N07 region homologous to the region in which Bnams4b resides. The vertical bars indicate corresponding genes of B. napus chromosome N07 or no hit genes.
(D) Genetic and physical maps of Bnams4b. The numbers in parentheses above the molecular marker names indicate the number of recombinants of corresponding markers. Primer pair SD1R-SD2F and the BAC clone B107 are represented by the black arrows and the dotted line, respectively.
(E) Schematic view of genes in the candidate interval on 7365A chromosome N07. Six homologous Darmor-bzh genes are indicated. CC4 is the candidate gene for Bnams4b.
(F) Schematic view of genes in the corresponding interval on Darmor-bzh chromosome N07. Ten Darmor-bzh genes are indicated.
A mixed 7365AC genome BAC library was constructed to identify the location of Bnams4b via chromosome landing. After screening the BAC library using the cosegregated molecular markers SD1R-SD2F (Figure 2C; Supplemental Table 1), the BAC clone B107 was identified as containing Bnams4b (Figure 2C). We sequenced this clone using high-throughput sequencing. An additional insert fragment (∼61 kb) was also identified between the molecular markers DD28 and SSR103 (Figures 2D and 2E); however, this 61-kb fragment was not present in the Darmor-bzh and Chiifu-401 genome sequences (Figures 2B, 2C, and 2F). In this 61-kb region, the CC4 fragment (∼8.4-kb) contained a complete heat shock protein coding sequence. A protein homologous with this sequence has been shown to interact with the Hip (HSP70-interacting protein) domain of Tic40 (a homolog of BnaTic40) (Bédard et al., 2007). Therefore, CC4 was predicted to represent a Bnams4b candidate gene (Figure 2E). Other genes in this candidate region were excluded as they showed no annotation related to male reproduction, no obvious phenotype after transformation, and no information regarding associated interaction with BnaC9.Tic40 (Supplemental Table 2). Subsequently, CC4 was found to produce completely sterile anthers in all transgenic plants as well as variable frequencies of albino leaves in some plants when transformed into wild-type Col-0 A. thaliana (Figures 3A to 3D; Supplemental Figures 1A, 1B, and 1F). In addition, when CC4 was transformed into 7365C plants, six plants exhibited sterile anthers (Figure 3I), but all had some albino leaves and buds (Figure 3K; Supplemental Figures 1C, 1E, and 1G). Dead buds, which occasionally appeared in 7365A sterile plants, were also observed in these transgenic plants (Figure 3K). Thus, these transgenic complementation experiments indicated that the CC4 fragment contained the Bnams4b gene. In addition, transgenic A. thaliana and B. napus showed higher Bnams4b expression levels in small buds, siliques, and anthers than nontransgenic plants (Supplemental Figure 2). These male-sterile phenotypes support the conclusion that the complete sequence of CC4 contained the Bnams4b gene.
Figure 3.
Phenotype of Transformed Wild-Type A. thaliana and Transformed 7365C with Candidate Sequence CC4.
(A) and (B) Open flowers of wild-type and CC4 transgenic A. thaliana.
(C) and (D) Buds with removed perianth of wild-type (left) and CC4 transgenic A. thaliana (right).
(E) BnaC9.Tic40 can restore the male sterility caused by Bnams4b in A. thaliana. Complete fertile anther appeared in the F1 plant both contained BnaC9.Tic40 and Bnams4b.
(F) Sterile anther appeared in the F2 segregated plants which only contained Bnams4b.
(G) Wild-type A. thaliana grown under the same conditions.
(H) and (I) Buds with removed periapts of 7365C (left) and CC4 transgenic B. napus (right).
(J) and (K) Inflorescence of 7365C and CC4 transgenic B. napus.
RT-PCR and 5′- and 3′-rapid amplification of cDNA ends were used to confirm the whole gene structure of Bnams4b. The Bnams4b cDNA contains an 85-bp 5′ untranslated region, a 4161-bp ORF with nine exons, and a 113-bp 3′ untranslated region (Supplemental Figure 3; see Methods for gene structure confirmation). We transformed this ORF, driven by two different promoters, into A. thaliana and found that sterile anthers and siliques were present in transformed plants (Supplemental Figure 4), confirming that the ORF present in CC4 was Bnams4b.
Bnams4b Encodes a Deleterious Chimeric Protein That Localizes to Plastids
Sequence analysis of the Bnams4b ORF revealed a fusion of three segments (AA1, AA2, and AA3) from three independent B. napus genes (Figure 4A). AA1, AA2, and AA3 exhibited 93.6, 93.0, and 94.1% nucleotide identities with the corresponding portions of BnaA01g00420D, BnaA01g10280D, and BnaA01g00190D, respectively. In contrast, another segment between AA2 and AA3 did not demonstrate homology with any reference genome sequence (Figure 4A). BnaA01g00420D encodes a ribonuclease III family protein located in chloroplasts; BnaA01g10280D encodes pre-mRNA processing 8 (PRP8), which forms the conserved central component of the spliceosome (Grainger and Beggs, 2005); and BnaA01g00190D encodes mitochondrial heat shock protein 70-1 (mtHsc70-1). None of these three genes is associated with male sterility according to the current literature. These results indicated that Bnams4b was a newly formed functional chimeric gene composed of three segments from different genes.
Figure 4.
Gene Structure, Expression Pattern, and Subcellular Localization of Bnams4b.
(A) The structure of Bnams4b consists of four parts: portions of three genes and a segment (black dashed line) with no homologous sequences in four reference genome sequences.
(B) Gradient expression of Bnams4b in leaves with varying degrees of albino phenotypes in Bnams4b transgenic B. napus. Albino leaves had the highest expression level while green leaves almost had no expression of Bnams4b. Immunoblotting of Bnams4b showed the same tendency as transcript levels. Error bars indicate sd. The Duncan’s multiple-range test (P < 0.05, SPSS version 19.0) was used to investigate the significant differences of average relative expressions of the five samples. Different numbers of asterisks between two random samples indicated they have significant difference with means of relative expressions; same numbers mean no significant difference between them.
(C) RT-qPCR analysis of Bnams4b in sterile plants of NIL 7365AC. RL, rosette leaves; CL, cauline leaves; ST, stems; FL, flowers; Bud0, bud of 0 to 1 mm; Bud1, bud of 1 to 2 mm; Bud3, bud of 3 to 4 mm; Bud4, bud of 4 to 5 mm; SD5, siliques 5 d after pollination; SD15, siliques 15 d after pollination; SD30, siliques 30 d after pollination. Error bars indicate sd. The Duncan's multiple-range test (P < 0.05, SPSS version 19.0) was also used to investigate the significant differences between various tissues.
(D) GUS analysis of Bnams4b in transgenic wild-type A. thaliana.
(E) Subcellular localization of Bnams4b protein in A. thaliana leaf protoplasts. For the five images above, from left to right, the GFP signal of AA1-GFP fusion construct, blue fluorescence which was transformed from the chlorophyll red autofluorescent signal, red fluorescent signal of endoplasmic reticulum marker, the images in bright field, and merged images of the above all. Five images below indicated the sublocalization of (AA1+AA2)-GFP fusion construct with the same order of notes as Bnams4b AA1-GFP fusion construct. Bars =10 μm.
In addition to male sterility, an abnormal albino or yellow leaf morphology was observed in the plants with high expression levels of Bnams4b (Figure 4B; Supplemental Figures 1C and 1E to 1G). RT-qPCR and promoter GUS analyses revealed that Bnams4b was predominantly expressed in rosette leaves, cauline leaves, anthers, and siliques (Figures 4C and 4D). The variable frequencies of albino leaves resulted from different levels of Bnams4b transcription and translation in Bnams4b-transgenic B. napus plants (Figure 4B). In contrast, the mutant 7365A plants exhibited normal green leaves despite Bnams4b expression (Figure 4C), indicating that the leaves were subjected to a dose-dependent effect of Bnams4b expression (Figure 4B). The albino or yellow leaf phenotype was consistent with the chloroplast localization observed in an A. thaliana protoplast transformation assay (Figure 4E) and with Agrobacterium tumefaciens-mediated transient expression in tobacco leaf epidermal cells (Supplemental Figure 5). These data suggested that the Bnams4b protein has a deleterious effect in both anthers and leaves.
Bnams4b Affects the Normal Function of Plastid to Cause Aborted Anther and Albino Leaves
To investigate how Bnams4b caused aborted development of anthers, we used semi-thin sections and transmission electron microscopy (TEM) to investigate anther development in 7365A plants. Initiation of the abnormal phenotype occurred at the premeiosis stage and progressed to the tetrad stages (Supplemental Figures 6G and 6H). TEM revealed subtle differences in the vacuolar tapetum and microspores: defective biosynthesis of exine (Supplemental Figures 7F2 and 7H2); surrounded microspores throughout the development of the primary cell wall (Supplemental Figures 7H2 and 7J2); vacuolated tapetal cells and microspores (Supplemental Figures 7D1, 7D2, and 7F2); and reduced number of plastids, tapetosomes, and elaioplasts in tapetal cells (Supplemental Figures 7D3, 7F4 and 7J3). Ultimately, disintegration of the tapetal cells occurred in sterile anthers (Supplemental Figures 7H3, 7J1, and 7J3). The abnormal function of the tapetal plastids caused by Bnams4b in the premeiotic and tetrad stages thus led to anther abortion.
Overexpression of Bnams4b in wild-type Col-0 A. thaliana caused variable degrees of leaf albinism. Albino leaves exhibited few starch grains in their chloroplasts and insufficiently developed thylakoids (Figure 5A), indicating that Bnams4b overexpression caused aberrant chloroplast development. To further ascertain the effects of Bnams4b on chlorophyll synthesis, a leaf chlorophyll meter and photometric measurement of solvent-extracted chlorophyll were used to measure chlorophyll concentrations in albino A. thaliana leaves. Both methods indicated significantly lower absolute chlorophyll content in albino leaves than in wild-type green leaves (Figure 5B). Together, these results suggested that Bnams4b might block fatty acid synthesis in tapetal plastids as well as chlorophyll synthesis in leaf chloroplasts.
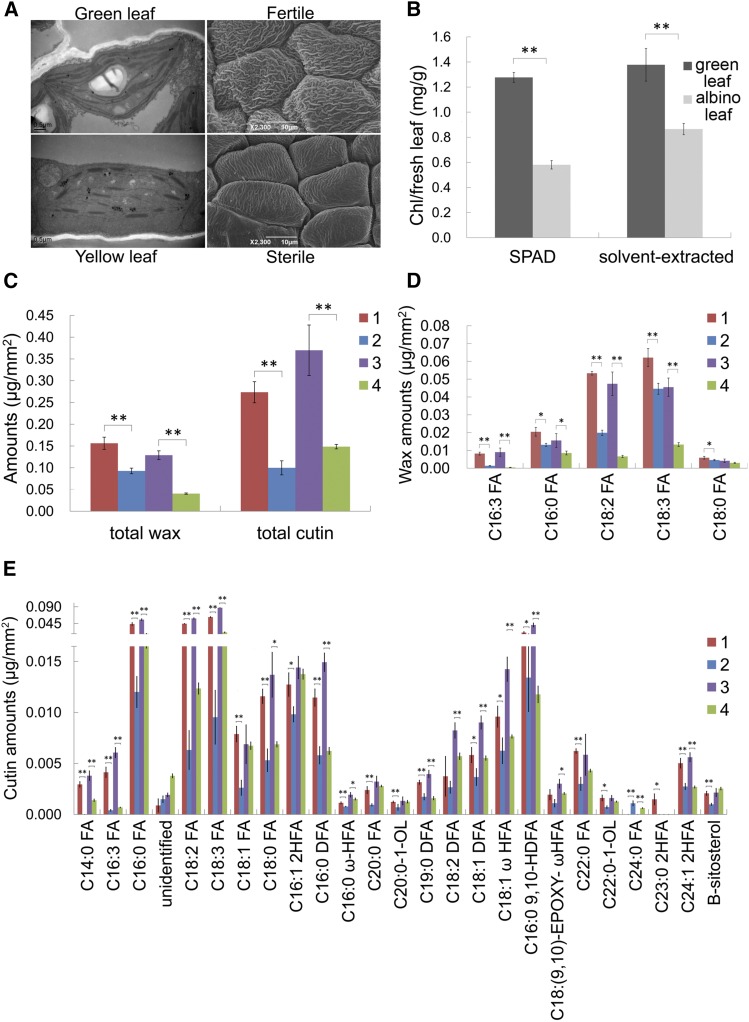
Figure 5.
Chlorophyll, Cutin, and Wax Measurements of Sterile and Fertile Anthers and Leaves.
(A) TEM observations of chloroplast in Bnams4b transgenic B. napus leaf and normal green leaf of 7365C (left). Scanning electron microscopy observations of fertile and sterile anther epidermis in NIL 7365AC (right).
(B) Chlorophyll content determination of albino leaves (Bnams4b transgenic) and green leaf (wild type) in A. thaliana. “SPAD” and “Solvent-extracted” represent two different methods of measurement.
(C) Comparison of total wax and cutin contents per unit surface area between fertile and sterile anther.
(D) Analysis of C16/C18 fatty acid wax components in the fertile and sterile anther epidermis.
(E) Analysis of cutin constituents in the fertile and sterile anther epidermis. Asterisks indicate components with significant difference between sterile and fertile anthers. 1, Anthers of Bnams3ms3ms4cms4c fertile plant with 2- to 3-mm buds; 2, anthers of Bnams3ms3ms4bms4c sterile plant with 2- to 3-mm buds; 3, anthers of Bnams3ms3ms4cms4c fertile plant with 4- to 5-mm buds; 4, anthers of Bnams3ms3ms4bms4c sterile plant with 4- to 5-mm buds. All data represent mean ± sd. *P < 0.05 and **P < 0.01. Data were subjected to one-way ANOVA F test for each parameter.
Scanning electron microscopy revealed that the sterile anthers of NIL 7365AC exhibited a relatively smooth epidermis (Figure 5A). Gas chromatography-mass spectrometry and gas chromatography-flame ionization detection (Bonaventure et al., 2004; Franke et al., 2005) revealed that the total wax and cutin contents decreased considerably in the sterile anthers compared with fertile anthers, regardless of the anther developmental stage (Figure 5C). Consistent with this, mid-chain fatty acids such as C16:0, C16:3, C18:2, and C18:3 (Figure 5D), which represent the main constituents of the total wax, were also reduced. Cutin monomer analysis of the sterile anther epidermis demonstrated significant reductions in C14:0, C16:0, C18:0, C16:3, C18:2, and C18:3 fatty acids in addition to a low abundance of components such as C16:0 DFA, C19:0 DFA, C18:1 DFA, C16:0 (9,10)-HDFA, C18:1-ω-HFA, and C18:1 (9,10)-EPOXY-ω-HFA (Figure 5E). Plastids are the most important subcellular organelles for C16 and C18 fatty acid synthesis. These results suggested that Bnams4b might affect lipid synthesis systems in the tapetal plastid.
Bnams4b Originated from Duplicated Genes by Exon Shuffling
We next attempted to compare Bnams4b with its orthologous genes in Capsella rubella, Arabidopsis lyrata, and A. thaliana to investigate the possible mechanism by which Bnams4b originated. C. rubella (n = 8) and A. lyrata (n = 8) were included in this analysis to increase the representation of Brassicaceae ancestral karyotypes (n = 8) (Figure 6A) (Koch and Kiefer, 2005; Lysak et al., 2006; Yogeeswaran et al., 2005). The results indicated that Bnams4b arose via the fusion of segments of three independent C. rubella, A. lyrata, and/or A. thaliana genes: Carubv10004547m/Alyrata490868/At4g37510.1, Carubv10003969m/Alyrata1477251/At1g80070.1, and Carubv10006443m/Alyrata490823/At4g37910.1 (Figure 4A). At least two exon-shuffling events occurred during the process of Bnams4b formation. Additionally, the sixth exon of Bnams4b consisted of an unidentified sequence that might have been recruited by the AA3 segment (Figure 4A). These data suggested that Bnams4b had a complex origin and likely arose from multiple exon shuffling events.
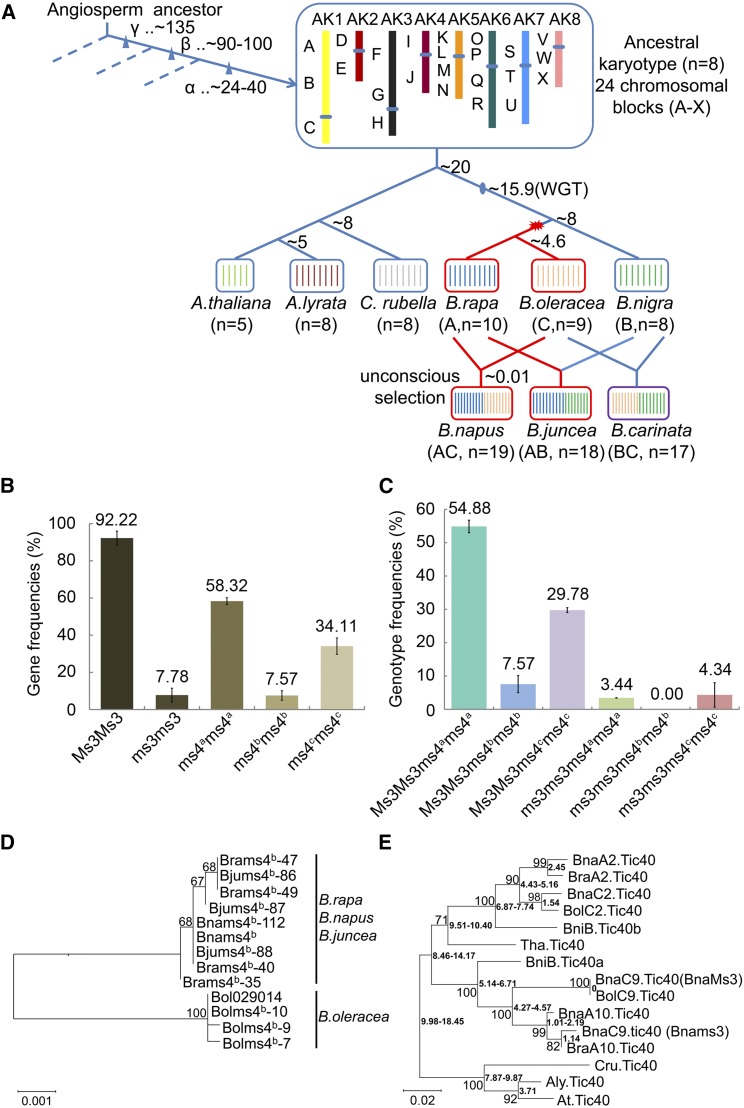
Figure 6.
The Age, Genes Frequencies, and Phylogenetic Relationship Analysis of Bnams4b and Divergence Time Estimations of Tic40 Homologies.
(A) Phylogeny and species differentiation times of certain Brassica and Arabidopsis lineages. Light-blue triangles represent the evolution of angiosperms three polyploidization events. Light-blue oval indicated the whole genome triplication (WGT) of Brassica. The vertical lines in the boxes represent the chromosome numbers of the haplotype. Red breaking symbol indicated the estimated time of Bnams4b generation. Red boxes and lines indicated evolutionary path of the Bnams4b gene. The six boxes in the lower right corner showed U’s triangle theory, which indicated B. napus was formed by natural hybridization of B. rapa and B. oleracea. Time unit is millions of years ago.
(B) Gene frequencies of two gene loci (Bnams4a/Bnams4b/Bnams4c and BnaMs3/Bnams3) in two populations of B. napus. Different numbers of asterisks indicated significant differences between different gene frequencies (Duncan’s multiple-range test, P < 0.05, SPSS version 19.0). Data represent mean ± sd.
(C) Genotype frequencies in two populations of B. napus with same analysis as in (A).
(D) The phylogenetic tree for Bnams4b homologs in the Brassicaceae. The full-length ORF sequences of Bnams4b homologs were used for phylogenetic analysis using MEGA4.0. Bna, B. napus; Bol, B. oleracea; Bra, B. rapa; Bni, B. nigra. The numbers on nodes are bootstrap values.
(E) Divergence time estimations of Tic40 homologies by Ks value, which indicated that the divergence time of BnaC9.Tic40 and BnaC9.tic40 is between 4.27 and 4.57 Mya, which is consistent with the age of Bnams4b (∼4.6 Mya). The estimated divergence time between other genes was also consistent with the previous studies (see Results).
Phylogenetic analysis of AA1, AA2, and AA3 showed that Bnams4b had high sequence similarity with Brams4b-40 (Supplemental Figure 8) and Bol029014 (from Bolbase, http://www.ocri-genomics.org/bolbase/) (Supplemental Figure 9). Amino acid sequence alignment suggested that Bol029014, Brams4b-40, and Bnams4b represent highly similar orthologs that exhibited different transcript structures (Supplemental Figure 9 and Supplemental Table 3). In addition, an ∼490-amino acid deletion was identified in Bol029014 (Supplemental Figure 9). AA1, AA2, and AA3 all exhibited higher sequence similarity to Brams4b-40 than to Bol029014 in B. oleracea (Supplemental Figure 8), suggesting that Bnams4b was directly derived from its orthologous gene in B. rapa rather than from its orthologous gene in B. oleracea. Additionally, AA1, AA2, and AA3 were shown to have corresponding paralogous gene fragments in B. rapa and B. oleracea (AA1, Bra011770 and Bol028906; AA2, Bra021701, Bra013431, Bol004508, and Bol000723; AA3, Bra010620, Bra011794, Bol032741, and Bol028880). This indicated that Bnams4b was generated from the DNA fragments of duplicated genes in Brassica.
To further confirm that Bnams4b was derived from B. rapa, the full-length cDNAs and genomic sequences of the identified ms4b genes of Brassica species were isolated. A phylogenetic tree was constructed using the full-length ORFs of 13 Bnams4b homologs (Supplemental Data Set 1). The 13 ORFs were distributed into two lineage groups: a B. oleracea group and a group that included B. rapa, B. napus, and B. juncea (Figure 6D). Bnams4b exhibited the highest sequence similarity to Bnams4b-112, Bjums4b-88, and Brams4b-40. The other four homologs in B. oleracea, all of which lacked the 490 amino acid region, were clustered into the B. oleracea group (Figure 6D). This analysis suggested that the Bnams4b homologs in B. napus and B. juncea might have been derived from B. rapa.
Bnams4b Is a Relatively Young Gene and Not Completely Fixed in B. napus
According to the U-triangle theory, B. rapa (AA; n = 10) and B. oleracea (CC; n = 9) were the parental species for the derivative allotetraploid species, B. napus (AACC; n = 19) (Nagaharu, 1935). Therefore, we investigated whether these species contained the intact sequence or partially combined fragments of Bnams4b. Three pairs of degenerate primers were used to amplify two of the three segments of Bnams4b: AA1, AA2, and AA3 (Figure 4A; Supplemental Table 1). For example, the degenerate primer pair HY1F and HY1R was used to specifically amplify the chimeric structure between AA1 and AA2 (based on the presence/absence of specific PCR fragment bands). The degenerate primer pairs were used to screen 30 lines of B. oleracea, 23 lines of B. rapa, 7 lines of B. nigra, 200 lines of B. napus, 10 lines of B. juncea, and 20 lines of B. carinata (Supplemental Data Set 1). However, we found no partially combined fragments (e.g., AA1+AA2, AA2+AA3, or AA1+AA3) in these lines. The intact sequence of Bnams4b was only present in B. oleracea (16/30), B. rapa (14/23), B. napus (123/200), and B. juncea (8/10), whereas it did not appear in B. nigra (0/7) or B. carinata (0/20) (Figure 6A). These findings indicated that Bnams4b might have originated after the divergence of B. nigra and B. oleracea, which has been suggested to occur <8 Mya (Figure 6A) (Liu et al., 2014; Lysak et al., 2005). Therefore, Bnams4b represents a relatively young chimeric gene.
Bnams4b was not found in all B. napus lines, suggesting that it had not been completely fixed in this species. Therefore, the gene frequencies of two loci (BnaMs3/Bnams3 and Bnams4a/Bnams4b/Bnams4c) and the genotype frequencies were investigated using linked molecular markers in the 200 lines from the above analysis plus an additional 148 inbred lines. The genotype frequencies of the suppressor genes BnaMs3Ms3 (92.22%) and Bnams4ams4a (58.32%) were higher than for the sterility gene Bnams4bms4b (7.57%) (Figure 6B). The genotype frequencies of BnaMs3Ms3ms4ams4a and BnaMs3Ms3ms4cms4c were also significantly higher than other genotypes whereas the sterile genotype had zero frequency (Figure 6C). These results indicated that the sterility gene Bnams4b was present at a low frequency while the two suppressor genes BnaC9.Tic40 and Bnams4a showed higher gene frequencies due to their stronger adaptive properties in B. napus. Therefore, we speculate that Bnams4b is a detrimental chimeric gene whose existence depends on the widespread suppressor genes BnaC9.Tic40 and Bnams4a.
Bnams4b Coevolved with the Suppressor Gene BnaC9.Tic40
The neofunctionalization of BnaC9.Tic40 (BnaMs3) is restricted to B. oleracea and its derivative species (such as B. montana, B. villosa, B. carinata, and B. napus) (Dun et al., 2014). Thus, neofunctionalization likely occurred after the divergence of the Brassica A, B, and C genomes (Dun et al., 2014). Bnams4b was generated between the time of B. nigra divergence from the Brassica lineage and the divergence of B. rapa and B. oleracea (Figure 6A). These data suggested that the sterility gene Bnams4b and the suppressor gene BnaC9.Tic40 probably coevolved in B. oleracea and its derivatives. Tic40 is an ancient conserved molecular chaperone that transfers nuclear-encoded proteins into the chloroplast, and its orthologs exist in nearly every green plant species. We attempted to estimate the age of BnaC9.Tic40 based on the synonymous (Ks) substitution rate. Using the DnaSP program, the age of BnaC9.Tic40 was estimated at 4.27 to 4.57 Mya (Figure 6E; Supplemental Data Set 2). The estimated divergence times of Brassica species as determined by Ks corresponded with those reported in previous studies. For example, the divergence of the Arabidopsis and Brassica lineages in the current study was 9.98 to 18.45 Mya versus the reported 14.5 to 20.4 Mya (Yang et al., 1999), that of B. nigra and B. oleracea was 6.87 to 7.74 Mya versus the reported 7.9 Mya (Lysak et al., 2005), that of B. oleracea and B. rapa was 4.43 to 5.16 Mya versus the reported 4.6 Mya (Liu et al., 2014), that of A. thaliana and A. lyrata was 3.71 Mya versus the reported 5 Mya (Koch and Matschinger, 2007), and that of A. thaliana and C. rubella was 7.87 to 9.87 Mya versus the reported 8 Mya (Franzke et al., 2009; German et al., 2009) (Figure 6E). We also calibrated the molecular clock to estimate the divergence time (functional differentiation time) between BnaC9.Tic40 and BnaC9.tic40. The estimated divergence time of BnaC9.Tic40 using MEGA package software was 4.45 Mya (Supplemental Figure 10). In conclusion, the estimated age of BnaC9.Tic40 (4.27 to 4.57 Mya or 4.45 Mya) essentially coincided with the time at which Bnams4b originated. Therefore, we speculate that the detrimental gene Bnams4b might have coevolved with the suppressor gene BnaC9.Tic40.
BnaC9.Tic40 Rescues the Sterility Caused by Bnams4b in A. thaliana
The presence of the Bnams4b gene caused complete male sterility in A. thaliana (Figures 3B and 3D). We performed a genetic transformation experiment to investigate whether BnaC9.Tic40 was able to restore male fertility. First, BnaC9.Tic40 and Bnams4b were individually transformed into wild-type Col-0 A. thaliana. Subsequently, sterile plants expressing Bnams4b were used as the female parent for hybridization with BnaC9.Tic40-transgenic plants. F1 plants containing both BnaC9.Tic40 and Bnams4b had completely fertile anthers (Figure 3E; Supplemental Table 4), indicating that BnaC9.Tic40 completely rescued the sterility caused by Bnams4b in A. thaliana. Fertility segregation was analyzed in an F2 population derived from a few F1 plants; most plants had a male fertile phenotype, and only a few were sterile due to the presence of multiple copy numbers of the transgene or the negative effect of Bnams4b (Figure 3F; Supplemental Table 4). Therefore, the pollination control system of 7365AB was successfully reconstructed in A. thaliana, indicating that the BnaC9.Tic40 protein in plastids could block the detrimental defect of the chimeric Bnams4b protein during anther development. It is possible that Bnams4b and BnaC9.Tic40 could potentially be used for utilization of heterosis in other species.
Bnams4a Rescues Fertility by Reducing the Transcriptional Level of the Chimeric Gene
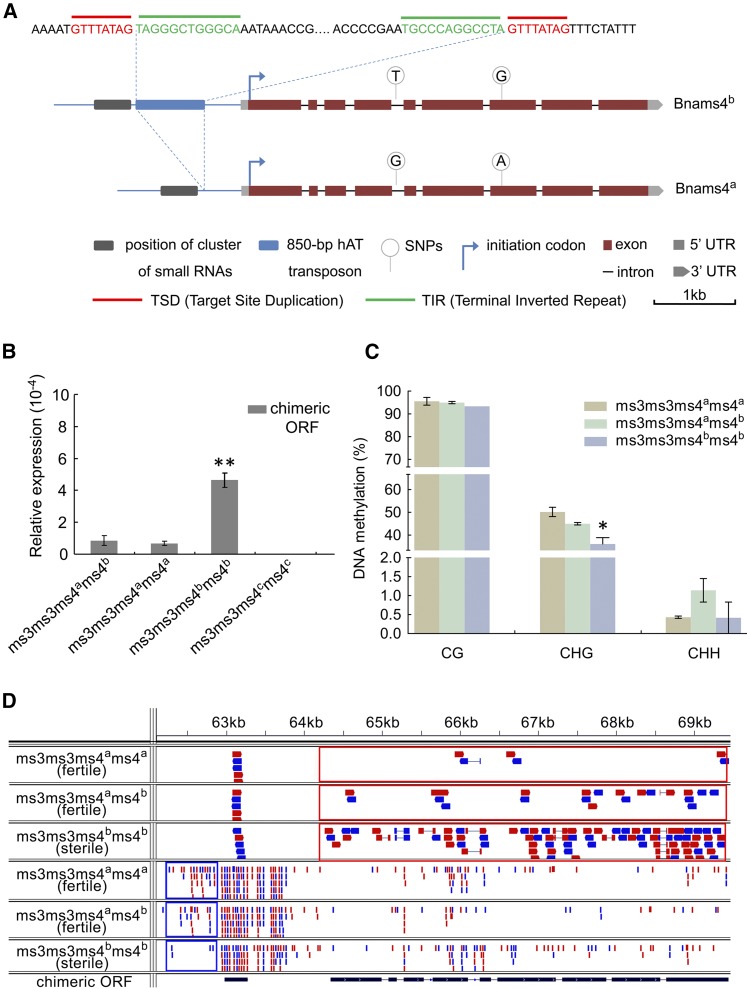
Comparative sequencing showed that the core promoter, nine exons, and eight introns showed no critical differences between Bnams4b and the chimeric gene in the Bnams4ams4a genotype. The chimeric gene sequence corresponding to the Bnams4ams4a genotype contained two single nucleotide polymorphisms (SNPs) compared with the ORF of Bnams4b (one in the fourth intron and another synonymous SNP in the seventh exon) (Figure 7A). However, these two SNP differences in the Bnams4ams4a genotype resulted in consistent anther sterility with the Bnams4b gene in A. thaliana (Supplemental Figure 11). As there was no hAT-type miniature inverted repeat transposable element (hAT-MITE) in the Bnams4ams4a genotype (Figure 7A), we explored transcript levels of the Bnams4ams4a chimeric ORF and the Bnams4b gene in three genotypes, namely, Bnams3ms3ms4ams4a, Bnams3ms3ms4ams4b, and Bnams3ms3ms4bms4b, that were generated by self-crossing of heterozygous individuals (Bnams3ms3ms4ams4b) in NIL 736512AB (Xia et al., 2012). The transcript levels were significantly higher in the sterile anthers than in the fertile anthers of two genotypes (Figures 7B and 7D), indicating that expression of Bnams4b and Bnams4ams4a-chimeric-ORF was reduced by Bnams4a. Furthermore, small RNA sequencing revealed that Bnams4a was able to produce a cluster of small RNAs (21 to 24 nucleotides) in the regulatory region of the Bnams4ams4a-chimeric-ORF (Figure 7D). We speculated that these small RNAs might block transcription of Bnams4b and Bnams4ams4a-chimeric-ORF by RNA-directed DNA methylation (Figure 7C; Supplemental Figure 12). In addition, the candidate region for Bnams4c in 7365C did not contain the 61-kb segment, indicating that Bnams4c was a null allele (Supplemental Figure 13).
Figure 7.
Bnams4a Probably Rescued the Sterility by Suppressing Expression of the Chimeric ORF.
(A) Structure comparison of the chimeric genes in Bnams3ms3ms4ams4a (Bnams4ams4a-chimeric-ORF) and Bnams3ms3ms4bms4b (Bnams4b) from NIL 736512AB. It shows an insertion of 850-bp MITE in the Bnams4b regulatory region but not in regulatory region of the Bnams4ams4a-chimeric-ORF. Although there are two SNPs between Bnams4ams4a-chimeric-ORF and Bnams4b, both of them can lead to male sterility in A. thaliana. This suggests both the chimeric ORF in Bnams3ms3ms4ams4a and Bnams3ms3ms4bms4b are functional in causing sterility.
(B) The transcript level of the chimeric ORF in three kinds of genotypes from self-crossing of fertile individuals in NIL 736512AB. By self-crossing, the fertile individuals in NIL 736512AB produced three genotypes: Bnams3ms3ms4ams4a (fertile), Bnams3ms3ms4ams4b (fertile), and Bnams3ms3ms4bms4b (sterile). The anthers from about the tetrad stage (buds of 1.5 to 2 mm) were used for analysis. 7365C (Bnams3ms3ms4cms4c) which did not contain Bnams4b was used as negative control.
(C) DNA methylation analysis of the small RNA regions of Bnams4ams4a-chimeric-ORF and Bnams4b by bisulphite-based method. It shows methylation level of CHG type is higher in the small RNAs region of Bnams4ams4a-chimeric-ORF than the sterile genotype: Bnams3ms3ms4bms4b. All data represent mean ± sd. *P < 0.05.
(D) Integrative Genomics Viewer screen capture of RNA-seq reads mapping to chimeric ORF. The three panels above showed transcriptome sequencing reads (∼100 bp), while three panels below represented microRNA sequencing reads (∼21 to 24 bp). These results suggested that a cluster of microRNA (blue box) in promoter region might repress the expression of chimeric ORF (red box). Both sense (represented in red) and antisense (represented in blue) strand reads are shown.
DISCUSSION
In this study, Bnams4b was found to be a relatively young chimeric gene (∼4.6 to 8 Mya) formed by the fusion of partially duplicated segments (AA1, AA2, and AA3) of three independent genes through exon shuffling events (Figure 4A). Overexpression of Bnams4b caused yellow or albino leaves that were regarded as representing various degrees of lethal effect (Figure 4B). This dominant function is in contrast to the general functioning of newly evolved genes observed in animals. Many new genes in Drosophila rapidly become essential for organ formation and patterning, as demonstrated by the high probability of lethality in the pupal and larval stages following their knockdown (Chen et al., 2010). In mammals, the newly evolved genes FGF4 (Parker et al., 2009), SRGAP2C (Dennis et al., 2012), CYPA (Sayah et al., 2004), and POLDI (Heinen et al., 2009) also contribute to essential functions in development and disease resistance providing them with an adaptive evolutionary advantage and enabling their subsequent fixation. In contrast, Bnams4b is not fixed and is of relatively recent origin. Consistent with this, Bnams4b has almost no intraspecific nucleotide variation (Supplemental Table 3) and is thus not suitable for rigorous statistical analysis since the few observable sequence changes are not sufficient to support powerful statistical testing (Supplemental Table 5). Nevertheless, a Ka/Ks ratio for Bnams4b is smaller than 1 (0.57752), which was calculated using the CODEML program of the Phylogenetic Analysis by the Maximum Likelihood statistical package (Goldman and Yang, 1994; Zhang et al., 2011) (to test the possibility of functional constraints; Supplemental Table 6), supporting its functionality. As an artificially selected gene for breeding, it can be maintained in the population at detectable levels despite its slight detrimental effects. This differs from naturally existing new genes whose existence depends on the positive selection of advantageous functions. The identification of a functional constraint is interpreted to mean that the gene product has certain functions valuable for the wild-type species that permits its retention over a substantial period prior to its identification and utilization.
Bnams4b encodes a novel chimeric protein that causes anther abortion and albino leaves, which are detrimental to reproduction and vegetative growth. However, plants evolved two patterns to prevent these unfavorable effects. The suppressor gene Bnams4a altered transcription levels of Bnams4b while BnaC9.Tic40 showed a coevolutionary protein-level epistatic control of Bnams4b. The neofunctionalization of BnaC9.Tic40 within B. oleracea enabled the new function of restoring fertility (Dun et al., 2014). BnaC9.Tic40 constitutively expressed primarily in seedlings, leaves, anthers, siliques, and seeds and localized in the chloroplast (Dun et al., 2011). This pattern of expression is similar to that of Bnams4b (Figures 4C to 4E). Additionally, the identified age of BnaC9.Tic40 is also consistent with the estimated age of Bnams4b (Figures 6A and 6E). These consistent results indicate that coevolution probably enabled the persistence of Bnams4b, based on a complex protein interaction network (but not a direct protein interaction; see Supplemental Figure 14) involving BnaC9.Tic40 that rescues sterility by a coevolutionary protein-level pattern. By contrast, Bnams4a was shown to act by reducing the level of Bnams4b expression. According to those further results (Figure 7), we speculated that the hAT-MITE close to the small RNAs site is not present in the regulatory region, resulting in the normal synthesis of small RNAs in Bnams3ms3ms4ams4a plants. These small RNAs might induce RNA-directed DNA methylation to suppress the transcription of the chimeric ORF and thereby rescue sterility. Anyway, Bnams4a rescues the sterility by a transcriptional pattern. The capacitor theory indicated that some genes can serve as buffering capacitor for genetic variation (Rutherford and Lindquist, 1998), and it has been applied to A. thaliana (Queitsch et al., 2002), Drosophila (Sollars et al., 2003), yeast (Jarosz and Lindquist, 2010), and cavefish (Rohner et al., 2013). BnaC9.Tic40 and Bnams4a can buffer the detrimental genetic variation (male sterility). Under the protection of the buffer genes, the genetic change behaves like neutral variant. Once BnaC9.Tic40 is changed to BnaC9.tic40 or Bnams4a disappears, the detrimental effect (male sterility) was then exposed. Our results may suggest that BnaC9.Tic40 and Bnams4a act as standing capacitors in an evolutionary time scale for the genetic variation through genetic and epigenetic mechanisms, respectively.
By destroying tapetal plastids and chloroplasts, Bnams4b caused a reduction in unsaturated C16/C18 fatty acids and induced abnormal chlorophyll synthesis (Figures 5B to 5E). However, this gene differs from the previously reported genes implicated in cutin- and sporopollenin-defective mutants (Chen et al., 2011; de Azevedo Souza et al., 2009; Li et al., 2010; Shi et al., 2011), which all encode lipid biosynthesis-related enzymes. Instead, Bnams4b mainly consists of three segments (AA1, AA2, and AA3) that exhibit high similarity with three A. thaliana genes: AT4G37510, a ribonuclease III family protein; AT1G80070, a component of the spliceosome; and AT4G37910, known as mtHsc70-1. AA1 appeared to encode an N-terminal chloroplast transit peptide, whereas AA2 was homologous to the partial sequence of AT1G80070, whose null mutations were shown to lead to embryonic lethality and abnormal suspensor development (Schwartz et al., 1994). AA3 contained an intact HSP70 functional domain. None of these segments has been individually implicated in lipid biosynthesis or male sterility. Therefore, we speculate that the AA2 and AA3 protein had been mistransferred into the plastid to cause defective development. Additionally, both cpHsp70 and mtHsp70 are essential for the translocation of important proteins across the inner membranes of chloroplasts and mitochondria in vivo (D’Silva et al., 2004; Su and Li, 2010). Therefore, Bnams4b might partially impair such membrane transport systems, probably resulting in the failed transport of specific chlorophyll and fatty acid synthesis-related proteins into the tapetal plastid stroma and the chloroplast and leading to the malfunction of tapetal plastids and male sterility or to the variable frequencies of albino leaves and buds.
The ability of Bnams4b to function in other species will likely make it a valuable research tool with multiple potential prospects for practical applications. In A. thaliana, Bnams4b causes stable, complete sterility. Plants with normal leaves but sterile anthers could be isolated among such transgenic plants, suggesting that a stable male-sterile line might be derived by the inclusion of the dominant sterile gene Bnams4b in Brassicaceae species. Notably, the sterility in A. thaliana could be reversed by BnaC9.Tic40. This indicated that restorer lines could potentially be derived using BnaC9.Tic40 in A. thaliana. Therefore, it might be possible to introduce ideal agronomic traits into male sterile or restorer lines to rapidly cultivate superior hybrids. We are in the process of determining whether this system might potentially be available in other species such as the dicotyledons tobacco (Nicotiana tabacum) and cotton (Gossypium hirsutum) and in the monocotyledon rice (Oryza sativa). Overall, the fertility conversion system described herein, which consists of two co-evolved genes, shows considerable potential value for future transgenic breeding.
METHODS
Plant Materials and Growth Conditions
The NIL 7365AC (Bnams3ms3ms4bms4c/Bnams3ms3ms4cms4c) was obtained by repeated backcrossing for map-based cloning of Bnams4b, construction of a BAC library, genetic transformation, RT-PCR, transcriptome sequencing, small RNA sequencing, cuticle analysis, and cytological observations. Two hundred varieties of Brassica napus were provided by Yongming Zhou in Huazhong Agricultural University. A total of 148 inbred lines of B. napus were conserved in our laboratory. These two populations of B. napus, collected in 2009 and 2014, respectively, were representative inbred lines or natural pure-lines used for association mapping with various genetic backgrounds. Wild-type Col-0 Arabidopsis thaliana was used for genetic transformation, GUS analysis, and subcellular localization analysis. Diverse basic species were collected from the Centre for Genetic Resources, Plant Genetic Resources, The Netherlands (CGN-PGR) (Supplemental Data Set 1). Information on other experimental materials can be found in the following method. Transgenic B. napus and A. thaliana plants were grown in the greenhouse under conditions of 20 to 22°C and 16/8 h night/day). Nontransgenic B. napus plants were grown under normal farming practices in the field of Huazhong Agricultural University, Hubei, China.
Physical Fine Mapping of Bnams4b
The sterile gene Bnams4b was defined in a physical region 304 kb in size according to the published genome sequence of B. rapa (Wang et al., 2011b; Xia et al., 2012). The candidate region of Bnams4b (namely, BnRf) had favorable genomic synteny with Scaffold000017 of the B. rapa genome, but limited synteny with the A. thaliana genome (Xia et al., 2012). Therefore, a series of sequence characterized amplified regions and simple sequence repeat markers were designed according to the nucleotide sequence of Scaffold000017 to narrow down the physical region of Bnams4b (Xia et al., 2012). Mixed pools of DNA from sterile and fertile plants were used to screen polymorphic markers linked to Bnams4b as described (Dun et al., 2011). The genetic distances of the polymorphic markers were determined using 5500 NIL 7365AC individuals. The nearest flanking markers, CL14 and DD42, located Bnams4b to 85-kb (Figure 2B) and 78-kb (Figure 2C) DNA fragments according to the Chiifu-401 and Darmor-bzh genome sequences, respectively. This smaller physical region contained four candidate genes. Although these four genes have not been previously characterized or predicted to be involved in anther development, the respective complete sequences of the four genes in the Bnams4b candidate region were cloned. These represented four candidate genes for Bnams4b. Each of the four candidate genes for Bnams4b was transformed into fertile 7365C (Bnams3ms3ms4cms4c) plants and wild-type Col-0 A. thaliana. None of the transgenic plants had a sterile phenotype.
We were unable to amplify the intergenic sequence between two of the four candidate genes. Therefore, thermal asymmetric interlaced-PCR was repeatedly performed to clone the intergenic sequence in the Bnams4b candidate region as described (Li et al., 2013). Finally, ∼15 kb was obtained, from which markers were developed and screened. Using a population of 5500 individuals, we further identified several cosegregated markers to screen the BAC clone library of 7365AC (SD2F-SD1R). All PCR products were separated on 6% polyacrylamide denaturing gels. Bands were visualized with a silver-staining method (Yi et al., 2006) or separated on 0.8 to 1% agarose gels and visualized with an agarose gel imaging system (Bio-Rad). The primer sequences used for mapping Bnams4b are listed in Supplemental Table 1.
Construction of a BAC Library and the Screening, Sequencing, and Assembling of the Target Monoclone
The BAC clone library was constructed according to a previously described method (Luo and Wing, 2003). We cooperated with COSETE Technology Company to complete the construction of the BAC library. After the seeds were sown, seedlings with very small leaves were grown under weak light for ∼10 d. To prepare high molecular weight genomic DNA fragments (average length of ∼700 kb), ∼150 g of fresh pale green leaves was ground in liquid N2 in a mortar until small tissue chunks could be seen. This process was followed by gentle mixing of the incubated nuclear suspensions at 45°C for 5 min with 1% low-melting-temperature agarose prepared in NIB and preincubated at 45°C. The mixture was added to plug molds and placed on ice for ∼40 min to form plugs. After the preparation of genomic DNA plugs, each half DNA plug (50 µL) was chopped into fine pieces and transferred to a microcentrifuge tube containing 45 μL of restriction enzyme buffer and was incubated on ice for 40 min. Gradient dilutions of restriction enzyme were used. The microcentrifuge tubes were then incubated in a 37°C water bath for 40 min. CHEF agarose gels were used to separate the digested product. DNA fragments of 90 to 110 kb were selected to perform DNA ligation. CopyControl pCC1BAC (HindIII Cloning-Ready) Vector (cat. no. CBAC311H) was used for ligation with selected DNA fragments. The ligation product was transformed into thawed ElectroMax DH10B T1 phage-resistant competent cells in prechilled electroporation cuvettes. Gradient volume transformation solutions of each culture were plated on 150-mm-diameter Petri dish agar plates. The number of recombinant clones was adjusted to 800 to 1000 per Petri dish. We calculated the average length of the insertion to be ∼93 kb (Supplemental Figure 15). The colonies were collected into a 2-mL centrifuge tube to obtain mixed BAC clones pools. Ultimately, we obtained 100 mixed pools, and the genome size contained in this BAC library was 93 kb × 900 clones × 100 pools (∼7.982 Gb).
Plasmids from each mixed pool were extracted using the protocol described by Sambrook and Russell (2001). The primer pairs 4F3DF-4F3DR and SD2F-SD1R were used to screen the target monoclone containing the Bnams4b locus. Serial numbers of clone pools identified by 4F3DF-4F3DR were 5, 6, 7, 17, 33, 37, 49, and 58. Those identified by SD2F-SD1R were 5, 35, 46, 58, 80, and 100. We sequenced the amplified fragments and confirmed number 58 to be a candidate BAC pool, which we used to screen the final monoclone containing Bnams4b. Gradient dilutions of 5-, 10-, and 50-fold after a 10,000-fold dilution of mixed bacteria pools of the number 58 were plated on 150-mm-diameter Petri dish agar plates; then the monoclones (nearly 800 to 1000 per dish) were picked using a toothpick-dipping method to identify those containing Bnams4b (Schenk, 2014). B107 was the final monoclone that contained Bnams4b. Plasmid extraction from B107 was performed with a Qiagen Large-Construct Kit (cat. no. 12462). The protocol used to construct the sequencing library for the BAC plasmid was provided by the high-throughput sequencing facility (HiSeq 2000 platform) at the State Key Laboratory of Crop Genetic Improvement. Paired-end sequencing was performed with ∼1 Gb per monoclone. SOAP (BGI Technology) was used for sequence assembly.
Plasmid Construction and Complementation Transformation
Vector construction for the complementation test and B. napus transformation was performed as described (Dun et al., 2011). All fragments for vector construction were amplified by Phusion DNA Polymerase (F-530S; Thermo Scientific). The four candidate genes showing sequence similarity with Bra014988, Bra014989, Bra014990, and Bra014991 were cloned into the vector pCAMBIA2300 using the traditional digestion and ligation method with restriction endonuclease and T4 ligase, respectively. Each construct contained a native promoter (∼2 kb), exons, introns, and 3′ sequence (∼1 kb). The whole sequence of CC4 (containing a 1906-bp promoter, nine exons, eight introns, and an 1185-bp 3′ sequence) was cloned into the vector pCAMBIA2300. For Bnams4a, the same method was adopted as used for CC4. To further confirm the function of the Bnams4b ORF, two other vectors were constructed with the A9 (At5g07230) promoter (1199 bp) and RBCS (At1g67090) promoter (1061 bp), followed by the Bnams4b ORF using pMDC83 binary vector. The primers used for promoter-GUS analysis were GUSF and GUSR (Supplemental Table 1). GUSR was designed close to the translation initiation site with no base gap. The length of GUSFR amplification was 2084 bp. This fragment was cloned into the PBI101 promoter-GUS analysis vector using a suitable restriction enzyme. To reconstruct the pollination control system in A. thaliana, the whole sequence of BnaC9.Tic40 (containing a 1951-bp promoter, 14 exons, 13 introns, and a 1227-bp 3ʹ sequence) was cloned into the vector pCAMBIA2300 to recover sterility by Bnams4b (CC4). Detailed procedures of B. napus Agrobacterium tumefaciens-mediated transformation using the hypocotyls infection method followed those previously described (Yi et al., 2010). Agrobacterium-mediated transformation of A. thaliana was performed using the floral-dip method (Clough and Bent, 1998).
Subcellular Localization of Bnams4b
To determine the subcellular localization of the Bnams4b protein, an A. thaliana protoplast transformation assay and Agrobacterium-mediated transient expression of tobacco leaf epidermal cells were employed. These two subcellular localization assays were performed according to published methods, with some modifications (Sparkes et al., 2006; Yoo et al., 2007). For the A. thaliana protoplast transformation assay, two segmental Bnams4b ORFs (AA1 and AA1+AA2, 657 and 1557 bp, respectively; Figure 4A) starting from the translation initiation site were inserted individually into a GFP-fusion expression vector pM999-35S-GFP. PEG-calcium-mediated transfection was used to deliver DNA into the A. thaliana protoplast, followed by a 5- to 24-h incubation to for gene expression (Yoo et al., 2007). RFP fusion with the chaperone binding protein BIP was used as an additional endoplasmic reticulum marker protein (Kim et al., 2001). Chloroplast autofluorescence was artificially changed to a blue color. For Agrobacterium-mediated transient expression by Nicotiana tabacum, the two segments were individually cloned into another GFP-fusion expression vector, pMDC83. The mGFP6 sequence in this pMDC83 vector was replaced with a pM999-GFP sequence using KpnI and SacI. Expression of these two fusion genes in greenhouse-grown 6-week-old tobacco leaves was achieved by agroinfiltration (∼0.6 OD) with GV3101 harboring pMDC83-AA1 and pMDC83-(AA1+AA2). Fluorescence of GFP and RFP were visualized using a FluoView FV1000 laser scanning confocal microscope (Olympus). Conditions for imaging were set at 488-nm excitation, collecting bandwidth at 500 to 520 nm for GFP and 559-nm excitation, collecting bandwidth at 570 to 583 nm for RFP.
5′, 3′ RACE and Confirmation of Bnams4b Gene Structure
RACE was performed using the GeneRacer Kit (for full-length, RNA ligase-mediated rapid amplification of 5′ and 3′ cDNA ends; catalog no. L1502-01). Total RNA from sterile buds of Bnams4b transgenic A. thaliana and buds of sterile plants in 7365AC was reverse transcribed into cDNA. After amplifying the cDNA ends of each sample, nested PCR was performed to obtain the terminal sequences. The full-length cDNAs of Bnams4b were submitted to NCBI BLAST-N to confirm the exact sequences of eight introns according to conservative exon sequences of related species in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=WebandPAGE_TYPE=BlastHome). The rules of eukaryotic intron initiation and termination sequences were used to examine the authenticity of start and stop bases of Bnams4b introns. Meanwhile, deep transcriptome sequencing data of the 7365A anther (the stage of abortion with 11 to 2-mm buds) was also used to check the authenticity of eight intron sequences (Figure 7D).
RT-qPCR and Immunoblotting
Total RNAs from albino leaves of varying degrees from transgenic B. napus, various tissues from NIL 7365AC sterile plants, and anthers of three genotypes from self-crossing of fertile individuals in NIL 736512AB were isolated using TRIzol reagent (Invitrogen) according to the user manual. One or two micrograms of RNA from each sample was reverse transcribed to cDNA using the SuperScript III reverse transcriptase (Invitrogen) or RevertAid First Strand cDNA Synthesis Kit (#K1622; Thermo Fisher Scientific). To guarantee the specification of quantitative primers, we first designed consensus primers to amplify the transcribed cDNA of buds, then sequenced the bands and compared them to the sequence of Bnams4b cDNA. Next, specific quantitative primers of Bnams4b were designed to perform RT-PCR and RT-qPCR. RT-PCR procedures were performed as described (Zhou et al., 2012). qRT-PCR was performed on an CFX96TM real-time system (C1000 thermal cycler; Bio-Rad) using SYBR Green Realtime PCR Master Mix (QPK-201; TOYOBO) and Hard-Shell PCR plates (catalog: HSP9655; Bio-Rad). For each sample, the experiment was performed with four technical replicates or three biological replicates. Bnaactin3 and A. thaliana ACTIN2 were used as the reference genes for normalization in B. napus and Arabidopsis samples, respectively. For RT-PCR of microsamples, buds (0.3 to 0.6 mm, tetrad stage) and siliques (4 d after pollination) from wild-type and transgenic A. thaliana were collected for total RNA extraction using the SV Total RNA Isolation and Purification System (Z3100; Promega). RNA isolation from transgenic and 7365AC anthers (1- to 2-mm buds) were performed with the same kits. Reverse transcription of these RNAs was performed by RevertAid First Strand cDNA Synthesis Kit (#K1622; Thermo Fisher Scientific). RT-PCR primers are listed in Supplemental Table 1.
For immunoblotting, the Bnams4b fragment was amplified from the cDNA of sterile buds using the primers WB-F and WB-R. The pGEX-4T-3 vector (GE Healthcare Life Sciences) was used to construct pGEX-4T-3-Bnams4b. The purified fusion protein was used as the antigen. The antibody was prepared by ABclonal Technology. Immunoblot experiments were performed as previously described (Jing et al., 2012). Total protein (0.1 g) of five albino cauline leaves from independent transgenic B. napus lines was extracted using extraction buffer (0.1 mmol/L Tris-HCl, 0.1 mmol/L EDTA, 0.15 mol/L NaCl, and 0.5% butanol, pH 7.5 to 8.0). β-Actin (for plants, AC009) was used as an endogenous standard.
Bisulfite Sequencing and McrBC PCR
DNA extraction (anthers from the tetrad period of Bnams3ms3ms4ams4a and Bnams3ms3ms4bms4b genotypes, ∼1.5 mm) was performed following the Qiagen DNeasy Plant Mini Kit (cat. no. 69106) handbook. A sample of 500 ng genomic DNA was treated for sodium bisulfite conversion using the Qiagen EpiTect Bisulfite kit (cat. no. 59104). Control primers NADFR were used to check the samples were treated adequately (Supplemental Table 1). DNA purification was performed using the Qiagen PCR purification kit (cat. no. 28106). Primers for bisulfate sequencing PCR were designed using Kismeth (https://phytozome.jgi.doe.gov/pz/portal.html#!search?show=BLAST; Supplemental Table 1). Amplification, cloning, and sequencing were performed as previously described (Wang et al., 2011a). DNA cytosine methylation in the CG, CHG, and CHH sites were analyzed on the above website.
McrBC-PCR experiments were performed as previously described (Ding et al., 2007; Rabinowicz et al., 2003). Digestion was performed by McrBC enzyme following the instruction (M0272S; New England Biolabs). PCR amplifications were performed using equal amounts of McrBC-digested and nondigested DNA of the anthers from the three genotypes of plants. Real-time McrBC-PCR was performed to quantitatively measure the methylation level. The primers for these experiments can be found in Supplemental Table 1.
Analysis of Anther Wax and Cutin
Wax and cutin from four types of anther epidermis, derived from three genotypes and two developmental stages, were analyzed as previously described (Jung et al., 2006; Li et al., 2010). Six different univariate linear regression equations were generated to estimate the ratios of dried anther weight-to-surface area (Supplemental Figure 16). Area was determined from pixel numbers in microscopy images, assuming the B. napus anthers to possess a cylindrical body. To extract wax, 6 to 10 mg of each sample of freeze-dried anther material was submersed in 1 mL chloroform for 1 min. Ten milligrams of tetracosane (Fluka) was used as an internal standard and was added to the resulting chloroform extracts and transferred to new vials. After evaporation under a gentle stream of nitrogen gas, free hydroxyl and carboxyl groups were converted to their trimethylsilyl ethers and esters by adding 20 mL of BSTFA (N,N-bis-trimet hylsilyltrifluoroacite amide; Macherey-Nagel) and 20 mL of pyridine to the extracts, which were then incubated for 40 min at 70°C. The remaining delipidated anthers were dried over silica gels and used to analyze the monomer composition of cutin polyester as described (Franke et al., 2005). The final data were subjected to one-way ANOVA F test for each comparison (Supplemental Figure 17).
Measurement of Chlorophyll
An SPAD-502 meter (Konica-Minolta) and a spectrophotometer (Mapada; UV-1800) were used to measure the chlorophyll concentration in Bnams4b transgenic and wild-type A. thaliana leaves. Two wild-type individuals and two albino individuals were measured during flowering. The SPAD values were converted to absolute units of chlorophyll concentration (total chlorophyll per unit fresh weight; mg/g, R2 = 0.9809) according to calibration equations (y = 0.0007x2 + 0.0230x + 0.0544) (Ling et al., 2011). Photometric measurements of solvent-extracted chlorophyll were performed according to a method previously described (Lichtenthaler, 1987). Five technical replicates of each experiment were performed.
Characterization of Anther Phenotypes
TEM analysis was performed as described (Li et al., 2006). Anthers from buds of various sizes from sterile and fertile plants in 7365AB and cut leaves (2 mm) were prefixed and vacuumized in 2.5% (w/v) glutaraldehyde with 0.1 M phosphate buffer (pH 7.4). After the anthers sank, the samples were kept in the refrigerator at 4°C. Samples were fixed in 2% OsO4, dehydrated, embedded, and used to generate ultrathin sections as previously described (Li et al., 2006). Scanning electron microscopy and the generation of semithin sections were performed as previously described (Yi et al., 2010; Zhu et al., 2010). The stages of anther development in this study were distinguished according to the methods described in a previous study (Sanders et al., 1999).
Phylogenetic Analysis and Sequence Divergence Estimation
The amino acid sequence of Bnams4b was divided into three sections (AA1, AA2, and AA3) and then used for BLAST searches against the phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#!search?show=BLAST) with relative species. Homologous sequences in B. napus (http://www.genoscope.cns.fr/brassicanapus/), B. rapa (Brams4b), and B. oleracea (http://www.ocri-genomics.org/bolbase/) were added manually. Multiple homologous sequences were derived and aligned with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Three maximum likelihood phylogenetic trees were obtained by aligning homologous protein sequences of the three sections with those in related species using the MEGA package version 6.0.6 (Tamura et al., 2013), and the LG+G model with all gaps deleted. Bootstrap values were obtained from 1000 replicates. The same method was used to perform phylogenetic analysis of whole sequences of Bnams4b homologs in the four species after removing the corresponding nucleotide sequences of the 490 amino acids in Bol029014. The divergence times of Tic40s among Brassica species, A. thaliana, Arabidopsis lyrata, and Capsella rubella were estimated by MEGA 6.0. Nucleotide substitution rate was set as 1.5 × 10−8 (Koch et al., 2000). Four times were used as molecular clock calibration: calibration name = 'B. rapa and B. oleracea', MinTime = 3.60000000 and MaxTime = 5.60000000 (Liu et al., 2014); calibration name = 'A. thaliana and A. lyrata', MinTime = 3.00000000 and MaxTime = 5.80000000 (Koch and Matschinger, 2007); calibration name = 'C. rubella and A thaliana', MinTime = 5.00000000 and MaxTime = 9.00000000 (Franzke et al., 2009; German et al., 2009); calibration name = 'A thaliana and B. rapa', MinTime = 14.00000000 and MaxTime = 24.00000000 (Yang et al., 1999). All sequence data used to analyze phylogeny relationships or divergence times were provided with a FASTA format in a sequence compressed file (Supplemental Files 1 to 5).
Anther mRNA Transcriptome Sequencing and Small RNA Sequencing
Total RNA samples of three kinds of anthers from self-crossing progeny of fertile individuals in NIL 736512AB were used for mRNA transcriptome and small RNA sequencing. Anthers from the tetrad period (buds 1.5 to 2 mm) were used for analysis. mRNA sequencing was performed on the HiSeq 2000 platform at the State Key Laboratory of Crop Genetic Improvement. Three libraries corresponding to three samples were sequenced producing 30-Gb clean data (∼10 Gb per library). Small RNA sequencing was performed by Novogene in Beijing; three libraries corresponding to three samples were sequenced producing 5.60 Gb clean data (1.94, 1.70, and 1.87 Gb). The reference sequence in IGV was the B107 BAC sequence. Parameters of the reads matched those of the reference sequence by 100%. Small RNA reads with <19 or >24 nucleotides were excluded for IGV mapping.
Yeast Two-Hybrid Assays
Gal4-based yeast two-hybrid assay was performed using the Matchmaker Gold Yeast Two-Hybrid System from Clontech according to the manufacturer’s instructions. Full-lengths CDS of BnaC9.Tic40, BnaC9.tic40, and Bnams4b were cloned into the PGBKT7 vector (GAL4-BD domain) and the PGADT7 vector (GAL4-AD domain) (primers are listed in Supplemental Table 1), respectively. The pairs of constructs were cotransformed into AH109 yeast cells and dropped on synthetic dropout nutrient medium (SD/-Trp-Leu-His-Ade) or (SD/-Trp-Leu, as positive control) agar plates.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Bnams4a, KX756983; Bnams4b, KX756984; Bol029014, KX756985; Bnaactin3, LOC106418315; and ACTIN2, AT3G18780.
Supplemental Data
Supplemental Figure 1. The sterility and albino phenotypes of transformed wild-type A. thaliana and 7365C by CC4.
Supplemental Figure 2. Relative expression levels of Bnams4b in both transgenic A. thaliana and transgenic B. napus tissues.
Supplemental Figure 3. The nucleotide sequence of Bnams4b.
Supplemental Figure 4. Sterile phenotypes of Bnams4b-transgenic A. thaliana driven by the A. thaliana A9 promoter.
Supplemental Figure 5. Chloroplast localization of Bnams4b protein by Agrobacterium-mediated transient expression in tobacco leaf epidermal cells.
Supplemental Figure 6. Semithin sections of fertile and sterile anthers in NIL 7365AC.
Supplemental Figure 7. TEM examination of fertile and sterile anthers in NIL 7365AC.
Supplemental Figure 8. Maximum likelihood phylogenetic trees summarizing the evolutionary relationships among the Bnams4b-related proteins (AA1, AA2, and AA3).
Supplemental Figure 9. Amino acid alignment of Brams4b-40, Bnams4b, and Bol029014.
Supplemental Figure 10. The estimated divergence times of Tic40s among Brassica species, A. thaliana, A. lyrata, and C. rubella by MEGA 6.0.
Supplemental Figure 11. Wild-type A. thaliana flower and transgenic flowers carrying the Bnams4ams4a allele that differs by two SNPs.
Supplemental Figure 12. Small RNA regions of all three genotypes showed high-level DNA methylation by McrBC-PCR.
Supplemental Figure 13. Comparison of sequence collinearity between partial BAC sequence of Bnams4b and sequence corresponding to Bnams4cms4c by Unipro UGENE.
Supplemental Figure 14. Bnams4b could not directly interact with BnaC9.Tic40 and BnaC9.tic40.
Supplemental Figure 15. Average insert size assay of the BAC library of 7365AC.
Supplemental Figure 16. Linear regression equations of anther dry weight and anther surface area with sterile and fertile anthers of different genotypes and developmental stages.
Supplemental Figure 17. Simple ANOVA tables in this study.
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. Homology and annotations of genes in candidate region.
Supplemental Table 3. Amino acid and nucleotide identities between Bnams4b and its intact homologous genes in B. rapa, B. oleracea, B. napus, and B. juncea
Supplemental Table 4. Number of individuals containing Bnams4b or BnaMs3 in F1 and F2 transgenic A. thaliana.
Supplemental Table 5. Neutral selection tests of Bnams4b by Mcdonald-Kreitman1, Tajima’s D2, and Fu and Li3.
Supplemental Table 6. Detection of positive selection of seven ms4b homologous genes in B. napus, B. rapa, B. oleracea, and B. juncea.
Supplemental File 1. Fifteen homologous sequences of AA1 in Brassicaceae.
Supplemental File 2. Thirty-one homologous sequences of AA2 in Brassicaceae.
Supplemental File 3. Thirty-eight homologous sequences of AA3 in Brassicaceae.
Supplemental File 4. Thirteen homologous sequences of Bnams4b in Brassicaceae.
Supplemental File 5. tic40 homologous sequences in Brassicaceae.
Supplemental Data Set 1. The information for diverse Brassicaceae accessions.
Supplemental Data Set 2. Ka/Ks value and divergence time of Tic homologous genes estimated by DnaSP.
Supplementary Material
Acknowledgments
We thank Hanhui Kuang and Yidan OuYang (Huazhong Agricultural University) for helpful discussions. This research was supported by funds from the National Key R&D Program for Crop Breeding (2016YFD0100305), the National Natural Science Foundation of China (31130040), National Natural Science Funds of China (31501374), China postdoctoral Science Foundation (2015M570646), and the Natural Science Foundation of Hubei province Key program (2014CFA008).
AUTHOR CONTRIBUTIONS
S.X. wrote the initial draft of the manuscript and performed all of the experiments involving the map-based cloning, cytological observation, expression, and biochemical analysis. Z.W. participated in the genetic transformation work. H.Z. participated in the genotype frequencies investigation and statistical analysis and contributed to partial genetic transformation. K.H. analyzed the sequence data and performed the phylogenetic and dN/dS ratio analyses. M.Q. performed the immunoblotting. Z.Z. participated in subcellular localization. X.D. supplied the sequence information on BnaC9.Tic40 orthologs. B.Y., J.W., C.M., J.S., and T.F. supervised the study. J.T. conceived and supervised the writing.
Glossary
- ORF
open reading frame
- Mya
million years ago
- GMS
genic male sterile
- NIL
near-isogenic line
- TEM
transmission electron microscopy
- SNP
single nucleotide polymorphism
References
- Bédard J., Kubis S., Bimanadham S., Jarvis P. (2007). Functional similarity between the chloroplast translocon component, Tic40, and the human co-chaperone, Hsp70-interacting protein (Hip). J. Biol. Chem. 282: 21404–21414. [DOI] [PubMed] [Google Scholar]
- Betrán E., Thornton K., Long M. (2002). Retroposed new genes out of the X in Drosophila. Genome Res. 12: 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G., Beisson F., Ohlrogge J., Pollard M. (2004). Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 40: 920–930. [DOI] [PubMed] [Google Scholar]
- Capra J.A., Pollard K.S., Singh M. (2010). Novel genes exhibit distinct patterns of function acquisition and network integration. Genome Biol. 11: R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M., Long M. (2012). The origin and evolution of new genes. Methods Mol. Biol. 856: 161–186. [DOI] [PubMed] [Google Scholar]
- Chalhoub B., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953. [DOI] [PubMed] [Google Scholar]
- Chen F.X., Hu B.C., Li Q.S. (1993). Discovery and Study of Genic Male Sterility(GMS) Material 9012 A in Brassica napus L. Acta Agric. Univ. Pekinensis 19: 57–61. [Google Scholar]
- Chen F.X., Hu B.C., Li Q.S., Hou S.M., Wu X.J., Fei W.X. (2002). The breeding of the recessive epistatically genic male sterile hybrid rapeseed-Wanyou18 with double low. Journal of Anhui Agricultural Sciences 30: 535–537. [Google Scholar]
- Chen F.X., Hu B.C., Li Q.S., Hou S.M., Wu X.J., Fei W.X., Li C., Chen W.S. (2003). Breeding of Wanyou 14, a recessive epistatically genic male sterile hybrid rapeseed (Brassica napus) with double low. Chinese Journal of Oil Crop Sciences 25: 63–65. [Google Scholar]
- Chen S., Zhang Y.E., Long M. (2010). New genes in Drosophila quickly become essential. Science 330: 1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Krinsky B.H., Long M. (2013). New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 14: 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Spletter M., Ni X., White K.P., Luo L., Long M. (2012a). Frequent recent origination of brain genes shaped the evolution of foraging behavior in Drosophila. Cell Reports 1: 118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Ni X., Krinsky B.H., Zhang Y.E., Vibranovski M.D., White K.P., Long M. (2012b). Reshaping of global gene expression networks and sex-biased gene expression by integration of a young gene. EMBO J. 31: 2798–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Yu X.H., Zhang K., Shi J., De Oliveira S., Schreiber L., Shanklin J., Zhang D. (2011). Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 157: 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.L., Chu C.C., Chen L.J., Akita M., Li H.M. (2006). Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175: 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.L., Fitzpatrick L.M., Tu S.L., Budziszewski G., Potter-Lewis S., Akita M., Levin J.Z., Keegstra K., Li H.M. (2003). Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22: 2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., Haughn G.W., Kombrink E., Douglas C.J. (2009). A novel fatty Acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M.Y., et al. (2012). Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149: 912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Zhou Q., Wang W. (2012). Origins of new genes and evolution of their novel functions. Annu. Rev. Ecol. Evol. Syst. 43: 345–363. [Google Scholar]
- Ding Y., Wang X., Su L., Zhai J., Cao S., Zhang D., Liu C., Bi Y., Qian Q., Cheng Z., Chu C., Cao X. (2007). SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Zhao L., Yang S., Jiang Y., Chen Y., Zhao R., Zhang Y., Zhang G., Dong Y., Yu H., Zhou Q., Wang W. (2010). A young Drosophila duplicate gene plays essential roles in spermatogenesis by regulating several Y-linked male fertility genes. PLoS Genet. 6: e1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Silva P., Liu Q., Walter W., Craig E.A. (2004). Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat. Struct. Mol. Biol. 11: 1084–1091. [DOI] [PubMed] [Google Scholar]
- Dun X., Zhou Z., Xia S., Wen J., Yi B., Shen J., Ma C., Tu J., Fu T. (2011). BnaC.Tic40, a plastid inner membrane translocon originating from Brassica oleracea, is essential for tapetal function and microspore development in Brassica napus. Plant J. 68: 532–545. [DOI] [PubMed] [Google Scholar]
- Dun X., Shen W., Hu K., Zhou Z., Xia S., Wen J., Yi B., Shen J., Ma C., Tu J., Fu T., Lagercrantz U. (2014). Neofunctionalization of duplicated Tic40 genes caused a gain-of-function variation related to male fertility in Brassica oleracea lineages. Plant Physiol. 166: 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q.X., Meng D.Q., Tang T.Z., Li Z.F., Xu L., Liu N. (2010). Effect of fertilizer and density on the yield of Mianyou 63. Anhui Agric. Sci. Bull. 16: 93–94. [Google Scholar]
- Franke R., Briesen I., Wojciechowski T., Faust A., Yephremov A., Nawrath C., Schreiber L. (2005). Apoplastic polyesters in Arabidopsis surface tissues--a typical suberin and a particular cutin. Phytochemistry 66: 2643–2658. [DOI] [PubMed] [Google Scholar]
- Franzke A., German D., Al-Shehbaz I.A., Mummenhoff K. (2009). Arabidopsis family ties: molecular phylogeny and age estimates in Brassicaceae. Taxon 58: 425–437. [Google Scholar]
- German D.A., Friesen N., Neuffer B., Al-Shehbaz I.A., Hurka H. (2009). Contribution to ITS phylogeny of the Brassicaceae, with special reference to some Asian taxa. Plant Syst. Evol. 283: 33–56. [Google Scholar]
- Goldman N., Yang Z. (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Grainger R.J., Beggs J.D. (2005). Prp8 protein: at the heart of the spliceosome. RNA 11: 533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen T.J.A.J., Staubach F., Häming D., Tautz D. (2009). Emergence of a new gene from an intergenic region. Curr. Biol. 19: 1527–1531. [DOI] [PubMed] [Google Scholar]
- Huang Z., Xiao L., Dun X.L., Xia S.Q., Yi B., Wen J., Shen J.X., Ma C.Z., Tu J.X., Meng J.L., Fu T.D. (2012). Improvement of the recessive genic male sterile lines with a subgenomic background in Brassica napus by molecular marker-assisted selection. Mol. Breed. 29: 181–187. [Google Scholar]
- Jarosz D.F., Lindquist S. (2010). Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 330: 1820–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing B., Heng S., Tong D., Wan Z., Fu T., Tu J., Ma C., Yi B., Wen J., Shen J. (2012). A male sterility-associated cytotoxic protein ORF288 in Brassica juncea causes aborted pollen development. J. Exp. Bot. 63: 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.D., Begun D.J. (2005). Parallel evolution of chimeric fusion genes. Proc. Natl. Acad. Sci. USA 102: 11373–11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K.H., Han M.J., Lee D.Y., Lee Y.S., Schreiber L., Franke R., Faust A., Yephremov A., Saedler H., Kim Y.W., Hwang I., An G. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Eu Y.J., Yoo C.M., Kim Y.W., Pih K.T., Jin J.B., Kim S.J., Stenmark H., Hwang I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Kiefer M. (2005). Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species--Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana. Am. J. Bot. 92: 761–767. [DOI] [PubMed] [Google Scholar]
- Koch M.A., Matschinger M. (2007). Evolution and genetic differentiation among relatives of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 6272–6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498. [DOI] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Yan L., Lai J., Ma C., Gautam M., Fu T. (2013). Molecular cloning and mRNA expression profile of sucrose transporter gene BnSUT1C from Brassica napus L. Indian J. Exp. Biol. 51: 1130–1136. [PubMed] [Google Scholar]
- Li H., Pinot F., Sauveplane V., Werck-Reichhart D., Diehl P., Schreiber L., Franke R., Zhang P., Chen L., Gao Y., Liang W., Zhang D. (2010). Cytochrome P450 family member CYP704B2 catalyzes the omega-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids:pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382. [Google Scholar]
- Ling Q., Huang W., Jarvis P. (2011). Use of a SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynth. Res. 107: 209–214. [DOI] [PubMed] [Google Scholar]
- Liu S., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5: 3930–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M., Langley C.H. (1993). Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260: 91–95. [DOI] [PubMed] [Google Scholar]
- Luo M., Wing R.A. (2003). An improved method for plant BAC library construction. Methods Mol. Biol. 236: 3–20. [DOI] [PubMed] [Google Scholar]
- Lysak M.A., Koch M.A., Pecinka A., Schubert I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Berr A., Pecinka A., Schmidt R., McBreen K., Schubert I. (2006). Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103: 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold K., Lihova J. (2006). Polyploidy, hybridization and reticulate evolution: lessons from the Brassicaceae. Plant Syst. Evol. 259: 143–174. [Google Scholar]
- Matsuno M., et al. (2009). Evolution of a novel phenolic pathway for pollen development. Science 325: 1688–1692. [DOI] [PubMed] [Google Scholar]
- Nagaharu U. (1935). Genomic anlaysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7: 389–452. [Google Scholar]
- Parker H.G., et al. (2009). An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325: 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C., Sangster T.A., Lindquist S. (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624. [DOI] [PubMed] [Google Scholar]
- Rabinowicz P.D., Palmer L.E., May B.P., Hemann M.T., Lowe S.W., McCombie W.R., Martienssen R.A. (2003). Genes and transposons are differentially methylated in plants, but not in mammals. Genome Res. 13: 2658–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers R.L., Bedford T., Hartl D.L. (2009). Formation and longevity of chimeric and duplicate genes in Drosophila melanogaster. Genetics 181: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner N., Jarosz D.F., Kowalko J.E., Yoshizawa M., Jeffery W.R., Borowsky R.L., Lindquist S., Tabin C.J. (2013). Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science 342: 1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S.L., Lindquist S. (1998). Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanders P.M., Bui A.Q., Weterings K., McIntire K., Hsu Y.-C., Lee P.Y., Truong M.T., Beals T., Goldberg R. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322. [Google Scholar]
- Sayah D.M., Sokolskaja E., Berthoux L., Luban J. (2004). Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430: 569–573. [DOI] [PubMed] [Google Scholar]
- Schenk P.M. (2014). Rapid cloning of genes and promoters for functional analyses. Methods Mol. Biol. 1099: 123–132. [DOI] [PubMed] [Google Scholar]
- Schranz M.E., Lysak M.A., Mitchell-Olds T. (2006). The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11: 535–542. [DOI] [PubMed] [Google Scholar]
- Schwartz B.W., Yeung E.C., Meinke D.W. (1994). Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235–3245. [DOI] [PubMed] [Google Scholar]
- Shi J., et al. (2011). Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollars V., Lu X., Xiao L., Wang X., Garfinkel M.D., Ruden D.M. (2003). Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat. Genet. 33: 70–74. [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Su P.H., Li H.M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.C., Zhao H., Wang W.R., Li Y.L., Qian X.F., Fang G.H. (2004). Breeding of a recessive genic male sterile (GMS) hybrid variety HuyouzaNo.1 with double low in Brassica napus L. Zhongguo You Liao Zuo Wu Xue Bao 26: 63–65. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang C., Long Y., Hopkins C., Kurup S., Liu K., King G.J., Meng J. (2011a). Universal endogenous gene controls for bisulphite conversion in analysis of plant DNA methylation. Plant Methods 7: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Brunet F.G., Nevo E., Long M. (2002). Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99: 4448–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2006). High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18: 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.R., Sun C.C., Li Y.L., Zhou X.R., Zhuang J., Yang L.Y., Jiang M.Y., Jin Q.L. (2009). Breeding of the Brassica napus cultivar Huyouza 4, a recessive genic male sterile (GMS) hybrid with double low content in rapeseed. Shanghai Nong Ye Xue Bao 25: 69–71. [Google Scholar]
- Wang X., et al. ; Brassica rapa Genome Sequencing Project Consortium (2011b). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Weng J.K., Li Y., Mo H., Chapple C. (2012). Assembly of an evolutionarily new pathway for α-pyrone biosynthesis in Arabidopsis. Science 337: 960–964. [DOI] [PubMed] [Google Scholar]
- Xia S., Cheng L., Zu F., Dun X., Zhou Z., Yi B., Wen J., Ma C., Shen J., Tu J., Fu T. (2012). Mapping of BnMs4 and BnRf to a common microsyntenic region of Arabidopsis thaliana chromosome 3 using intron polymorphism markers. Theor. Appl. Genet. 124: 1193–1200. [DOI] [PubMed] [Google Scholar]
- Yang Y.W., Lai K.N., Tai P.Y., Li W.H. (1999). Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48: 597–604. [DOI] [PubMed] [Google Scholar]
- Yi B., Chen Y., Lei S., Tu J., Fu T. (2006). Fine mapping of the recessive genic male-sterile gene (Bnms1) in Brassica napus L. Theor. Appl. Genet. 113: 643–650. [DOI] [PubMed] [Google Scholar]
- Yi B., Zeng F., Lei S., Chen Y., Yao X., Zhu Y., Wen J., Shen J., Ma C., Tu J., Fu T. (2010). Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. Plant J. 63: 925–938. [DOI] [PubMed] [Google Scholar]
- Yogeeswaran K., Frary A., York T.L., Amenta A., Lesser A.H., Nasrallah J.B., Tanksley S.D., Nasrallah M.E. (2005). Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhan Z.B., Zhang Y., Zhao R.P., Wang W. (2011). [Evolutionary fate and expression patterns of chimeric new genes in Drosophila melanogaster]. Zool. Res. 32: 585–595. [DOI] [PubMed] [Google Scholar]
- Zhang C., Wang J., Xie W., Zhou G., Long M., Zhang Q. (2011). Dynamic programming procedure for searching optimal models to estimate substitution rates based on the maximum-likelihood method. Proc. Natl. Acad. Sci. USA 108: 7860–7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Dun X., Xia S., Shi D., Qin M., Yi B., Wen J., Shen J., Ma C., Tu J., Fu T. (2012). BnMs3 is required for tapetal differentiation and degradation, microspore separation, and pollen-wall biosynthesis in Brassica napus. J. Exp. Bot. 63: 2041–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Dun X., Zhou Z., Xia S., Yi B., Wen J., Shen J., Ma C., Tu J., Fu T. (2010). A separation defect of tapetum cells and microspore mother cells results in male sterility in Brassica napus: the role of abscisic acid in early anther development. Plant Mol. Biol. 72: 111–123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.