KNOTTED1-like Homeobox (KNOX) gene expression reorients both tissue cell and regional polarity in Hooded barley, generating altered patterns of identity, anisotropic growth, and cell differentiation.
Abstract
Plant development involves two polarity types: tissue cell (asymmetries within cells are coordinated across tissues) and regional (identities vary spatially across tissues) polarity. Both appear altered in the barley (Hordeum vulgare) Hooded mutant, in which ectopic expression of the KNOTTED1-like Homeobox (KNOX) gene, BKn3, causes inverted polarity of differentiated hairs and ectopic flowers, in addition to wing-shaped outgrowths. These lemma-specific effects allow the spatiotemporal analysis of events following ectopic BKn3 expression, determining the relationship between KNOXs, polarity, and shape. We show that tissue cell polarity, based on localization of the auxin transporter SISTER OF PINFORMED1 (SoPIN1), dynamically reorients as ectopic BKn3 expression increases. Concurrently, ectopic expression of the auxin importer LIKE AUX1 and boundary gene NO APICAL MERISTEM is activated. The polarity of hairs reflects SoPIN1 patterns, suggesting that tissue cell polarity underpins oriented cell differentiation. Wing cell files reveal an anisotropic growth pattern, and computational modeling shows how polarity guiding growth can account for this pattern and wing emergence. The inverted ectopic flower orientation does not correlate with SoPIN1, suggesting that this form of regional polarity is not controlled by tissue cell polarity. Overall, the results suggest that KNOXs trigger different morphogenetic effects through interplay between tissue cell polarity, identity, and growth.
INTRODUCTION
Class 1 KNOTTED1-like Homeobox (KNOX) genes are central to shoot meristem function (Barton and Poethig, 1993; Kerstetter et al., 1997). Loss-of-function mutants typically lack shoot meristems or have reduced organ complexity (Barton and Poethig, 1993; Long et al., 1996; Kerstetter et al., 1997; Vollbrecht et al., 2000), suggesting that class 1 KNOX genes contribute to meristem identity. When overexpressed, class 1 KNOX genes have a diverse range of developmental effects: enhanced leaf lobing, leaflet formation, ectopic meristems, knotted leaves, forked leaves, and petal spurs (Vollbrecht et al., 1991; Smith et al., 1992; Sinha et al., 1993; Lincoln et al., 1994; Müller et al., 1995; Chuck et al., 1996; Williams-Carrier et al., 1997; Janssen et al., 1998; Golz et al., 2002; Hake et al., 2004; Ramirez et al., 2009; Shani et al., 2009). This raises the question of how a gene controlling meristem identity generates such a diverse range of morphological effects when ectopically expressed. One hypothesis is that KNOX genes have additional roles in organ outgrowth. This may be indicated by the localization of KNOTTED1 protein in the base of developing maize (Zea mays) leaves (Jackson, 2002) and in the involvement of KNOX genes during evolution for leaf margin elaboration (Bharathan et al., 2002; Hay and Tsiantis, 2006; Shani et al., 2009; Piazza et al., 2010).
Previous studies have suggested that class 1 KNOX genes influence organ development by acting as regulators of cell fate determination (Smith et al., 1992; Sinha et al., 1993; Lincoln et al., 1994; Janssen et al., 1998; Shani et al., 2009), involving the modulation of the cytokinin to gibberellic acid ratio (Jasinski et al., 2005; Yanai et al., 2005). There is also evidence that class 1 KNOX genes influence cell division (Smith et al., 1992; Sinha et al., 1993; Schneeberger et al., 1995). For example, the barley (Hordeum vulgare) Hooded (Hd) mutant, which ectopically expresses the class 1 KNOX gene BKn3 in the lemma/awn boundary (Müller et al., 1995), has altered cell division patterns (Stebbins and Yagil, 1966). The Hd mutant also develops wing-like outgrowths in the lemma margin, indicating that class 1 KNOX genes also influence growth patterns (Bonnett, 1938; Stebbins and Yagil, 1966; Williams-Carrier et al., 1997). Modulation of growth is also indicated in studies where leaf margins are modified (Hay and Tsiantis, 2010). The effect of class 1 KNOX genes on growth is further supported by their effect on the cytokinin: gibberellic acid ratio, influencing the distribution of cell division and cell elongation in a tissue (Sakamoto et al., 2001; Jasinski et al., 2005; Yanai et al., 2005; Bolduc and Hake, 2009; Bolduc et al., 2012).
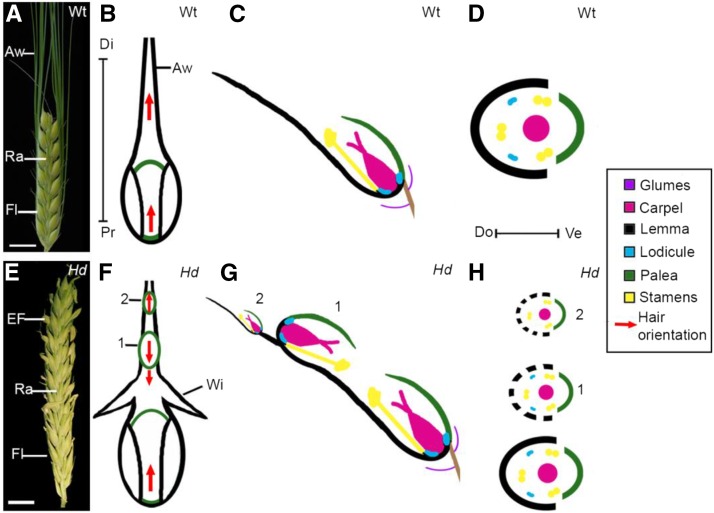
Some of these developmental effects of class 1 KNOX genes may also reflect their influence on polarity. We may distinguish between two types of polarity: regional polarity and tissue cell polarity. Regional polarity reflects spatial variation in regional identities. For example, the maize leaf has abaxial-adaxial, proximodistal, and mediolateral polarities, which refer to the difference between the upper and lower leaf zones, the basal sheath and upper blade, and the midvein and lateral margins, respectively. Mathematically, regional polarity can be described as a field of values (e.g., gene expression levels) associated with positions in space (a scalar field; Lawrence et al., 2007). Analysis of class 1 KNOX gene overexpression mutants has led to the suggestion that morphological changes arise due to modulation of regional polarity (Golz et al., 2002; Ramirez et al., 2009). For example, the maize Knotted1 mutant produces proximal sheath-like outgrowths on the distal blade margins (Ramirez et al., 2009), suggesting a change in proximodistal polarity. The barley Hd mutation also affects regional polarity. Wild-type barley (Figure 1) has a protective bract-like floral organ, the lemma, which has a distal extension called the awn (Figures 1A to 1D). Instead of an awn, the Hd mutant (Figure 1) develops ectopic flowers on the lemma (Figures 1E to 1H). These ectopic flowers have the same whorled structure of wild-type flowers (Figure 1D versus 1H), but inverted regional polarity, evidenced by the positions of the palea (Figures 1F and 1G) (Harlan, 1931; Bonnett, 1938; Stebbins and Yagil, 1966; Müller et al., 1995; Williams-Carrier et al., 1997). It has been proposed that the inversion of regional polarity in the Hd lemma arises due to ectopic BKn3 expression in the lemma, inducing a new “polarizing gradient centre” based on hormone concentrations (Stebbins and Yagil, 1966) or a new inflorescence meristem unit (Williams-Carrier et al., 1997), generating inverted ectopic flowers.
Figure 1.
The Hd Mutant Exhibits Polarity Reversals.
(A) Photograph of wild-type barley (Bowman, 2-row) inflorescence, illustrating the arrangement of florets (Fl) along the rachis (Ra). Clusters of three spikelets are initiated in a distichous pattern along the rachis (only the central spikelet produces a mature floret). Each floret has an outer protective organ, the lemma, with a distal awn (Aw). Bars = 1 cm.
(B) to (D) Floral diagrams of the wild type.
(B) Adaxial wild-type morphology. The lemma and palea are visible and hairs on the lemma point distally (hair orientation, red arrows).
(C) Longitudinal cross section through a wild-type spikelet. The floret has a dorsal lemma and ventral palea.
(D) Transverse cross section illustrating the organ whorls in the wild-type floret.
(E) The Hd inflorescence phenotype resembles the wild type, although the lemma develops ectopic flowers (EF) instead of an awn. Bars = 1 cm.
(F) to (H) Floral diagrams of Hd.
(F) One or more flowers can form on the Hd lemma’s adaxial surface. Analyses of hair and organ orientations (Harlan, 1931; Bonnett, 1938; Stebbins and Yagil, 1966; Müller et al., 1995; Williams-Carrier et al., 1997) have shown that the first ectopic flower (1) is inverted, pointing toward the lemma base, whereas the second ectopic flower (2) points toward the lemma tip. The Hd lemma develops wing-like (Wi) margin outgrowths below the first ectopic flower.
(G) Longitudinal cross section through an Hd spikelet.
(H) Floral diagram of each Hd flower, showing that each ectopic flower retains the same whorled arrangement of organs. Each ectopic flower is thought to use the existing lemma as its own (dashed black line). Pr-Di, proximal-distal axis; Do-Ve, dorsal-ventral axis.
Tissue cell polarity refers to asymmetries across individual cells (cell polarity) and their coordination across a tissue. Mathematically, tissue cell polarity corresponds to a field of vectors associated with positions in space (a vector field). Tissue cell polarity (a vector field) is therefore a distinct notion from regional polarity (a scalar field) (Lawrence et al., 2007), although the two may interact. Cellular localization of the auxin transporter PIN-FORMED1 (PIN1) can be used as a marker of tissue cell polarity and may also play a role in its establishment or coordination. Several models have been proposed for how PIN1 may be involved in the establishment of tissue cell polarity: (1) Up-the-gradient(Jönsson et al., 2006; Smith et al., 2006), (2) With-the-flux model (Sachs, 1969, 1981; Mitchison, 1980; Mitchison et al., 1981; Stoma et al., 2008), and (3) Indirect cell-cell coupling (Abley et al., 2013). With all of these models, coordination of cell polarity across a tissue can be achieved through modulating auxin dynamics (Abley et al., 2016). Analysis of PIN1 localization and class 1 KNOX gene expression in meristems and compound leaves has led to the suggestion that KNOX expression confers competency for PIN1 convergence point formation (Barkoulas et al., 2008; Hay and Tsiantis, 2010), indicating that class 1 KNOX genes and tissue cell polarity may interact. Tissue cell polarity has also been proposed to provide the axial information from which growth is oriented (Abley et al., 2013). In Drosophila melanogaster, epidermal wing hairs point distally on the wing and are used as a readout of planar cell polarity (Adler, 2002). Similarly, hairs on the adaxial surface of the wild-type barley lemma point distally toward the lemma tip (Figure 1B). In the Hd mutant, this uniform orientation of hairs is disrupted, with hairs pointing proximally below the first ectopic flower (Figure 1F) (Bonnett, 1938; Stebbins and Yagil, 1966; Williams-Carrier et al., 1997). Taken together, these results suggest that some of the effects of ectopic expression of BKn3 in the developing lemma may be mediated through early changes in tissue cell polarity.
The Hd mutant is a good model for exploring the relationship between class 1 KNOX genes, polarity, and shape, as all three aspects are affected. Ectopic expression of BKn3 in Hd is specific to the lemma/awn boundary (Müller et al., 1995; Williams-Carrier et al., 1997), so the subsequent effect on lemma development can be analyzed in a specific spatiotemporal manner. Such spatiotemporal analysis is more difficult with other KNOX overexpression systems, like maize Kn1 (Ramirez et al., 2009), where ectopic expression patterns are more variable. The morphology of the Hd mutant is not unique to ectopic expression of BKn3, as the Hd phenotype can be replicated by overexpressing maize KN1 in barley (Williams-Carrier et al., 1997). This suggests that understanding the role of BKn3 in Hd development could shed light on class 1 KNOX gene functions in general. Although the study of overexpression mutants generally identifies roles of genes outside of their wild-type context, they can also highlight wild-type functions not previously observed in the corresponding null mutants. This can be particularly important for a gene, like KNOTTED1, for which the null mutant exhibits complete loss of an organ or meristem, masking gene functions that are additional to those required for organ or meristem formation.
Here, we characterize changes in Hd lemma morphology using 3D imaging and analyze the spatial and temporal effects of the Hd mutation on growth, tissue cell polarity, regional polarity, identity, and differentiation. We show that at the time that ectopic BKn3 expression is activated, there is a change in tissue cell polarity, as marked by SISTER OF PIN-FORMED1 (SoPIN1) cellular localization. Concurrent with these changes in tissue cell polarity, changes in identity occur, illustrated by the upregulation of BKn3 and ectopic activation of barley homologs of LIKE AUX1 (HvLAX1) and NO APICAL MERISTEM (HvNAM). These changes in tissue cell polarity and identity precede the later changes in regional polarity (inverted ectopic flowers), cell differentiation (inverted hair orientation), and growth (wing development). Experimental analysis and modeling of these developmental events suggest that interactions between identity and tissue cell polarity play a key role in regulating cell differentiation and growth. Ectopic flower polarity (regional polarity) does not directly reflect tissue cell polarity patterns, suggesting that this aspect of regional polarity is regulated via a separable mechanism. These studies on ectopic BKn3 expression thus show how class 1 KNOX genes may regulate development through interactions between identity, tissue cell polarity, and regional polarity.
RESULTS
Two Separable Morphological Changes Are Observed for the Hd Lemma
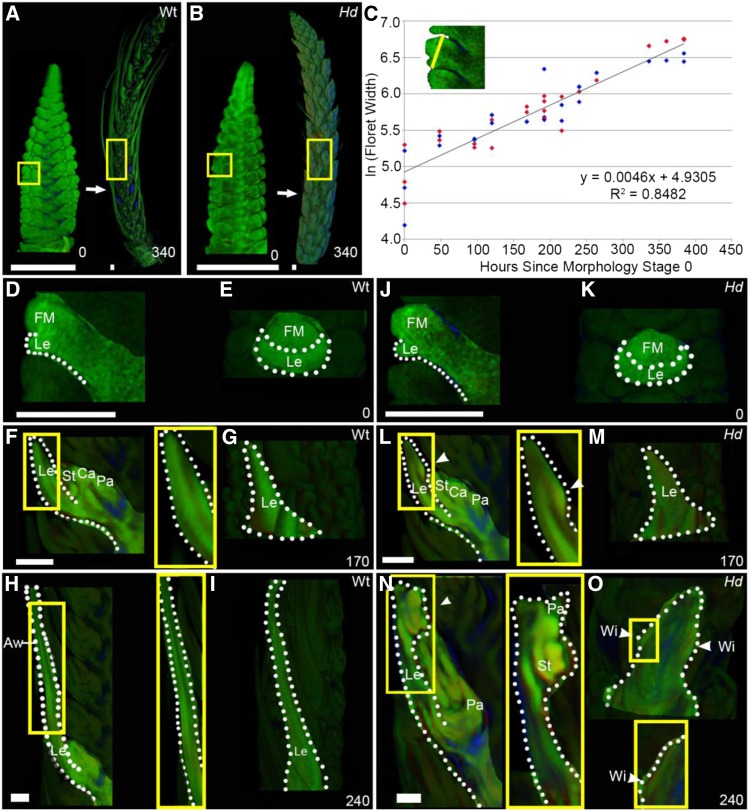
To determine the downstream effects of the Hd mutation, we first established the timing of both cellular and tissue level events in wild-type and Hd inflorescence development (Figure 2). We used optical projection tomography (OPT) (Sharpe et al., 2002; Lee et al., 2006) to image in 3D fixed inflorescences harvested at intervals over a 380-h period of development (Figures 2A and 2B). Using digital slices through the images, the width of floret 5 was measured for each time-course sample and used to generate a growth curve (Figure 2C). This showed that the natural logarithm of floret width increased linearly at a similar rate in both the wild type and Hd, corresponding to a growth rate of 0.46% h−1. Using the equation of this graph, we could assign a standard developmental time, in hours, based on floret width. Zero hour (illustrated in Figures 2A, 2B, 2D, and 2J) corresponded to a floret width of 140 µm when the lemma primordium was just visible (Figures 2D, 2E, 2J, and 2K). At this stage, the wild type and Hd were morphologically indistinguishable. Morphological divergence between the wild type and Hd was first clearly detected at 170 h, when the wild-type lemma had a smooth adaxial surface (Figure 2F), while an ectopic meristem was just visible as a cushion on the adaxial surface of the Hd lemma (Figure 2L). By 240 h, an extended awn was visible at the distal end of the wild-type lemma (Figures 2H and 2I) but absent from the Hd lemma (Figures 2N and 2O). At this stage, the margins of the wild-type lemma were curving (Figure 2I), while those of the Hd lemma showed small bulges that eventually developed into the wings (Figure 2O). Thus, two separable morphological changes are observed during Hd lemma development: formation of the ectopic meristem on the adaxial surface at 170 h and outgrowth of the wings from the lemma margins at 240 h.
Figure 2.
The Hd Phenotype Emerges Early in Development.
OPT of fixed time-course samples of wild-type ([A] and [D] to [I]) and Hd ([B] and [J] to [O]) Bowman barley inflorescences.
(A) to (C) Examples of inflorescence morphology at 0 and 340 h. Yellow boxes indicate the position of floret 5. The natural logarithm (ln) of floret 5 width (yellow line in the inset in [C]) was used to generate a combined log-linear growth curve (C) for the wild type (dark blue) and Hd (red) (n = 2 to 4 for each time point). The equation of the line of best fit (y = 0.0046x + 4.9305) (R2 value of 0.8482) was used to stage all further experimental data. All times are hours since the initial morphology stage (0 h, illustrated in [A] and [B]).
(D) to (O) Lateral cross sections through the center of floret 5 ([D], [F], [H], [J], [L], and [N]) and images of the abaxial surface of the lemma ([E], [G], [I], [K], [M], and [O]) for different time points identified key morphology changes in the Hd lemma ([J] to [O]) versus the wild type ([D] to [I]). At 0 h ([D], [E], [J], and [K]), Hd and the wild type shared the same morphology: by 170 h ([F], [G], [L], and [M]), the ectopic meristem started to form on the adaxial surface of the Hd lemma (yellow box, white arrowhead), while the wild-type lemma elongated to form the awn ([F] to [I]). By 240 h, the wing outgrowths started to form in the margins below the ectopic flower in the Hd lemma ([N] and [O]). Zoomed-in images (yellow boxes) of the ectopic meristem and wings in Hd and the corresponding positions in the wild type are shown. Floral meristem (FM), lemma (Le), stamen (St), carpel (Ca), palea (Pa), wings (Wi), and awns (Aw). White dotted lines, shape of the lemma; white arrowheads, position of the ectopic meristem and wings.
Bars = 500 μm in (A) and (B) and 200 μm in (D) to (O).
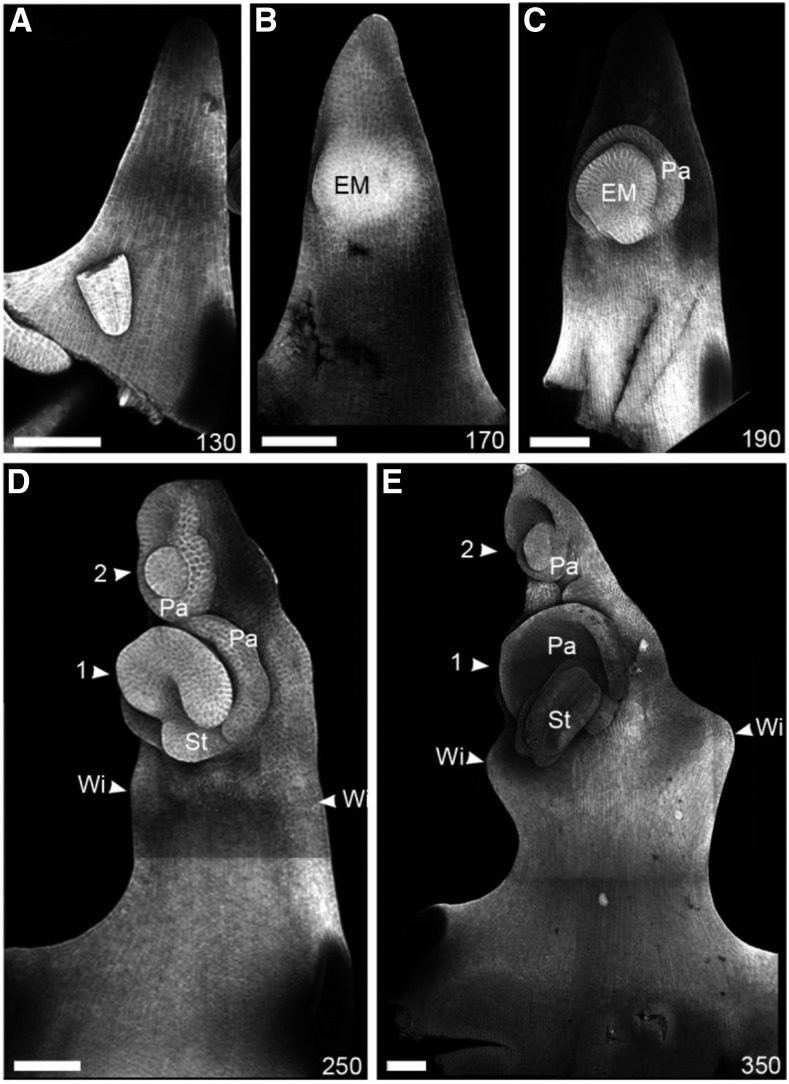
To visualize these changes at higher resolution, we performed confocal imaging of the adaxial surface of fixed Hd lemmas at different developmental stages (Figure 3). At 130 h, when the wild-type and Hd lemmas were morphologically indistinguishable, the Hd lemma had a smooth adaxial surface (Figure 3A). By 170 h, the Hd lemma had a distinct adaxial cushion (Figure 3B). This cushion corresponds to the ectopic meristem identified in the Hd OPT images. The ectopic meristem continued to develop, forming distinct organ primordia. By 190 h, a distal palea primordium was visible (Figure 3C) and by 250 h other floral organ primordia started to emerge (Figure 3D, 1). A second ectopic meristem can also form on the Hd lemma distal to the first and was clearly visible by 250 h (Figure 3D, 2). At 250 h, small bulges were also visible in the margin of the Hd lemma (Figure 3D, Wi), which developed into more pronounced wing outgrowths by 350 h (Figure 3E, Wi).
Figure 3.
Ectopic Flowers Emerge on the Adaxial Surface of the Hd Lemma.
Confocal microscope images of the adaxial surface of Hd lemmas (cell walls highlighted by calcofluor staining) illustrating ectopic meristem and wing development. The lemma initially had a smooth surface and triangular shape ([A]; 130 h). At 170 h (B), the first ectopic meristem (EM) had started to develop. By 190 h (C), organ primordia were initiated, first forming a distal palea (Pa). If a second ectopic meristem formed ([D], 2), the palea was proximally positioned ([D], 2, Pa). By 250 h (D), the wings (Wi) started to develop from the margins below the first ectopic meristem. The wings formed distinct rounded outgrowths by 350 h (E). n > 4 for each time point. Bars = 100 μm.
Ectopic BKn3 Expression Is Activated by 110 h and Is Strongly Expressed by 170 h
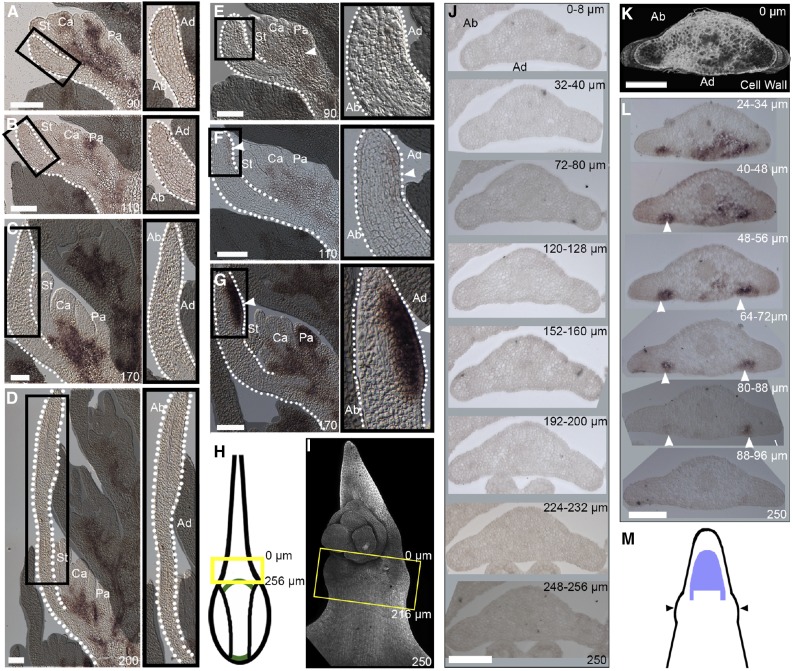
To establish when and where ectopic expression of BKn3 is activated during Hd lemma development, RNA in situ hybridization of BKn3 was performed on barley inflorescences (Figure 4). Throughout wild-type development, BKn3 mRNA was completely excluded from the developing lemma and was restricted to the meristematic region at the base of the flower (Figures 4A to 4D, 4H, and 4J). Similarly, in the Hd lemma at 90 h, BKn3 mRNA was localized to the meristematic region in the base of the flower (Figure 4E, arrowhead) and was excluded from the developing organs, including the lemma. However, in contrast to the wild type, by 110 h, faint BKn3 mRNA expression was detected in the distal adaxial region of the developing Hd lemma (Figure 4F). By 170 h (Figure 4G) when the ectopic meristem had started to form, BKn3 mRNA expression was strong within this meristem. This distribution of BKn3 mRNA localization in wild-type and Hd lemmas contrasts with previous observations of mRNA localization by Müller et al. (1995), who reported BKn3 mRNA throughout the lemma at early stages of development. Our observations are consistent with the protein localization reported by Williams-Carrier et al. (1997). However, both of these earlier studies did not establish timings, so direct comparisons cannot be made.
Figure 4.
In Hd, Ectopic BKn3 Expression Is Activated around 110 h, Is Strong by 170 h, and Extends beyond the Ectopic Meristem Region by 250 h.
(A) to (G) BKn3 mRNA in situ hybridization of longitudinal midsections through wild-type florets ([A] to [D]) and Hd florets ([E] to [G]) (zoomed-in lemma images, black boxes) at 90 h ([A] and [E]), 110 h ([B] and [F]), 170 h ([C] and [G]), and 200 h ([D]; wild-type only). At 90 h ([A] and [E]), BKn3 mRNA (dark precipitate) was localized to the base of the flower (white arrowhead) and excluded from developing organs, including the lemma, in both the wild type and Hd. Throughout wild-type development, this pattern is maintained, with BKn3 mRNA excluded from the lemma ([B] to [D]). In Hd at 110 h (F), there was faint BKn3 mRNA localization in the middle of the lemma on the adaxial side (zoomed in image, white arrowhead). By 170 h in Hd (G), there was strong BKn3 mRNA localization in the adaxial half of the lemma (zoomed in image, white arrowhead), where the ectopic meristem formed.
(H) Diagram illustrating mature wild-type morphology; the yellow box illustrates the approximate position of the sections in (J). In Hd, wings start to develop by 250 h (I). Confocal image of a calcofluor-stained Hd lemma (I) at a similar developmental time as the sectioned lemma in (K) and (L); the yellow box indicates the approximate position of the sections.
(J) to (L) Transverse sections through a lemma at 250 h for the wild type (J) and Hd ([K] and [L]).
(J) In the wild type, BKn3 was not expressed in the lemma.
(K) and (L) Transverse sections through the Hd lemma.
(K) 3D reconstruction of calcofluor-stained slices, viewed from the top, showing the curve of the wing.
(L) In situ hybridization of transverse slices throughout the wing region indicated that BKn3 was expressed adaxially in two proximodistal stripes on either side of the lemma midline (the position of each slice within the 3D reconstructed region is indicated in micrometers).
(M) Diagram depicting the predicted BKn3 expression pattern on the adaxial surface of the lemma (BKn3 expression, purple; lemma outline, black) in relation to the position of the wings (black arrows). Stamen (St), carpel (Ca), palea (Pa), abaxial (Ab), adaxial (Ad). Dotted white lines: lemma outline.
n > 4 for each time point in (A) to (G); n = 3 in (J) to (L). Bars = 100 μm.
At later stages in Hd development, around 250 h, when the wings started to form, transverse cross sections through the lemma indicated that BKn3 expression extended below the ectopic meristem region (Figures 4I and 4K to 4M). BKn3 was expressed in two short stripes (∼40 to 50 µm long) either side of the lemma midline, specific to the adaxial cell layers (Figure 4L, arrowheads). This region was just above where the tissue starts to deform toward the wing outgrowths (Figures 4I and 4K to 4M) and may suggest a role of BKn3 in wing development. (No BKn3 expression was seen in transverse sections of the wild type; Figures 4H and 4J.) The timing of the initial expression of the BKn3 in the Hd lemma, combined with the staging of ectopic flower and wing formation, is consistent with BKn3 modulating the development of the adaxial surface of the Hd lemma after 110 h.
Ectopic BKn3 Expression Leads to Reorientation of Tissue Cell Polarity
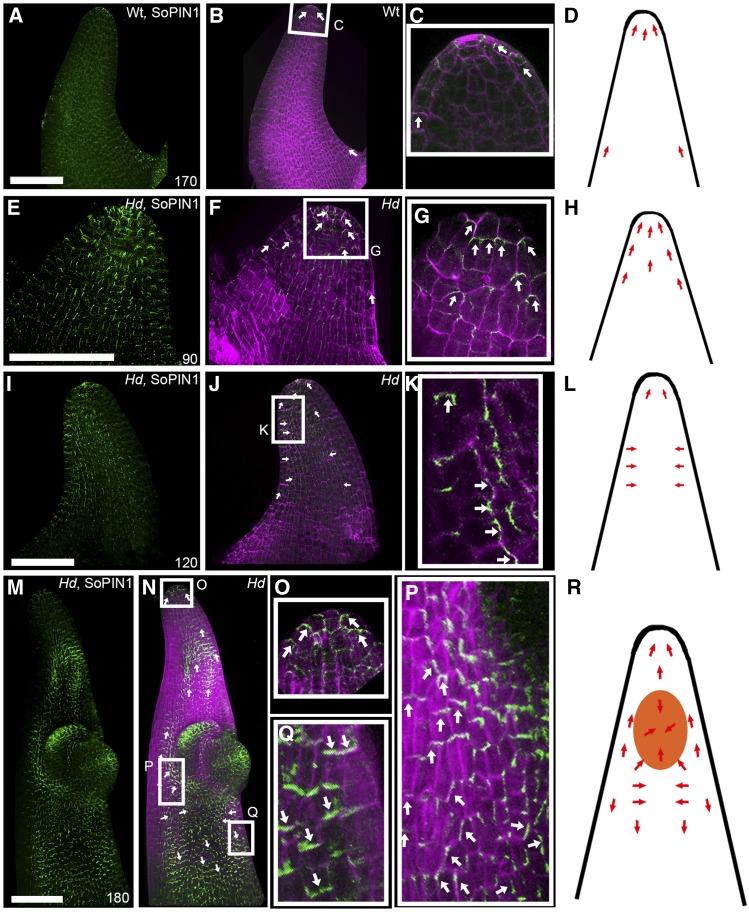
Previous studies based on floral organ orientation have shown that regional polarity is inverted in the Hd mutant at late stages of development (Bonnett, 1938; Harlan, 1931; Müller et al., 1995; Williams-Carrier et al., 1997; Stebbins and Yagil, 1966). In addition, lemma hair orientation suggests that tissue cell polarity may also be altered in Hd. To assess when and how BKn3 first influences tissue cell polarity, we determined the distribution of the cell polarity marker, SoPIN1 (O’Connor et al., 2014), in the adaxial surface of the lemma using whole-mount immunolocalization (Figure 5). At early stages in wild-type and Hd lemma development, SoPIN1 (green signal, wild-type 170 h, Figures 5A to 5D; Hd 90 h, Figures 5E to 5H) was localized at the distal end of epidermal cells (Figures 5C and 5G, white arrows), indicating that tissue cell polarity was coordinately oriented toward the lemma tip (Figures 5D and 5H, red arrows). As wild-type development progressed, SoPIN1 signal was lost.
Figure 5.
SoPIN1 Localization Reorients When Ectopic BKn3 Expression Is Activated.
3D projections of whole-mount immunolocalization of SoPIN1 (green) and calcofluor-stained cell walls (magenta) in whole wild-type lemmas at 170 h ([A] to [D]) and Hd lemmas at 90 h ([E] to [H]), 120 h ([I] to [L]), and 180 h ([M] to [R]). Orientation of SoPIN1 is based on cellular SoPIN1 localization in the epidermal layer only. Each panel illustrates SoPIN1 localization alone ([A], [E], [I], and [M]), both SoPIN1 and calcofluor combined ([B], [F], [J], and [N]), and a zoomed-in image of the boxed regions ([C], [G], [K], [O], [P], and [Q]), alongside a cartoon illustrating the inferred relationship between the ectopic meristem (orange) and SoPIN1 localization (red arrows) ([D], [H], [L], and [R]). Overlapping SoPIN1 and calcofluor signals appear white. In wild-type lemmas, SoPIN1 pointed distally toward the lemma tip throughout development ([A] to [D]), although SoPIN1 signal in the adaxial surface was lost over developmental time. In Hd lemmas, SoPIN1 signal was not lost over developmental time ([E] to [R]). Before BKn3 was ectopically expressed in the Hd lemma, SoPIN1 was localized distally, toward the tip of the developing lemma ([E] to [G], white arrows; [H], red arrows). Around the time that ectopic BKn3 expression was activated in the lemma (110 h), SoPIN1 localization reoriented (with respect to earlier stages in development) in the adaxial surface. Initially, SoPIN1 oriented laterally toward the center of the adaxial surface ([I] to [K], white arrows; [L], red arrows). After the ectopic meristem had formed ([M] to [R], 180 h), near the distal tip, cellular SoPIN1 was oriented distally toward the tip of the lemma (O), below the ectopic meristem SoPIN1 pointed toward the center of the adaxial surface (P), and below this SoPIN1 was oriented proximally ([Q], white arrows; [R], red arrows). n = 4 for each time point. Bars = 100 μm.
In contrast to the wild type, in Hd, epidermal SoPIN1 signal was not lost at later stages of development. At 120 h, after BKn3 expression was activated in the adaxial half of the Hd lemma, SoPIN1 localization became oriented toward the center of the region of ectopic BKn3 expression (Figures 5J and 5K, white arrows; Figure 5L, red arrows). At 180 h, after the ectopic meristem had clearly formed and BKn3 expression was strong in the Hd lemma, the reorientation of SoPIN1 in the adaxial surface, relative to its orientation at earlier stages, was more clearly seen (Figures 5M to 5R). At the distal end of the lemma, SoPIN1 pointed toward the lemma tip (Figures 5N and 5O, white arrows). Near the ectopic meristem, SoPIN1 was oriented toward the lemma midline (Figures 5N and 5P, white arrows). This midline-oriented SoPIN1 may have been focused toward the center of the BKn3 expression region and the ectopic meristem (Figure 5R). Below the ectopic meristem, SoPIN1 pointed proximally (Figures 5N and 5Q, white arrows), the opposite orientation to that observed at 90 h. This suggests that BKn3 is able to modulate tissue cell polarity early in lemma development.
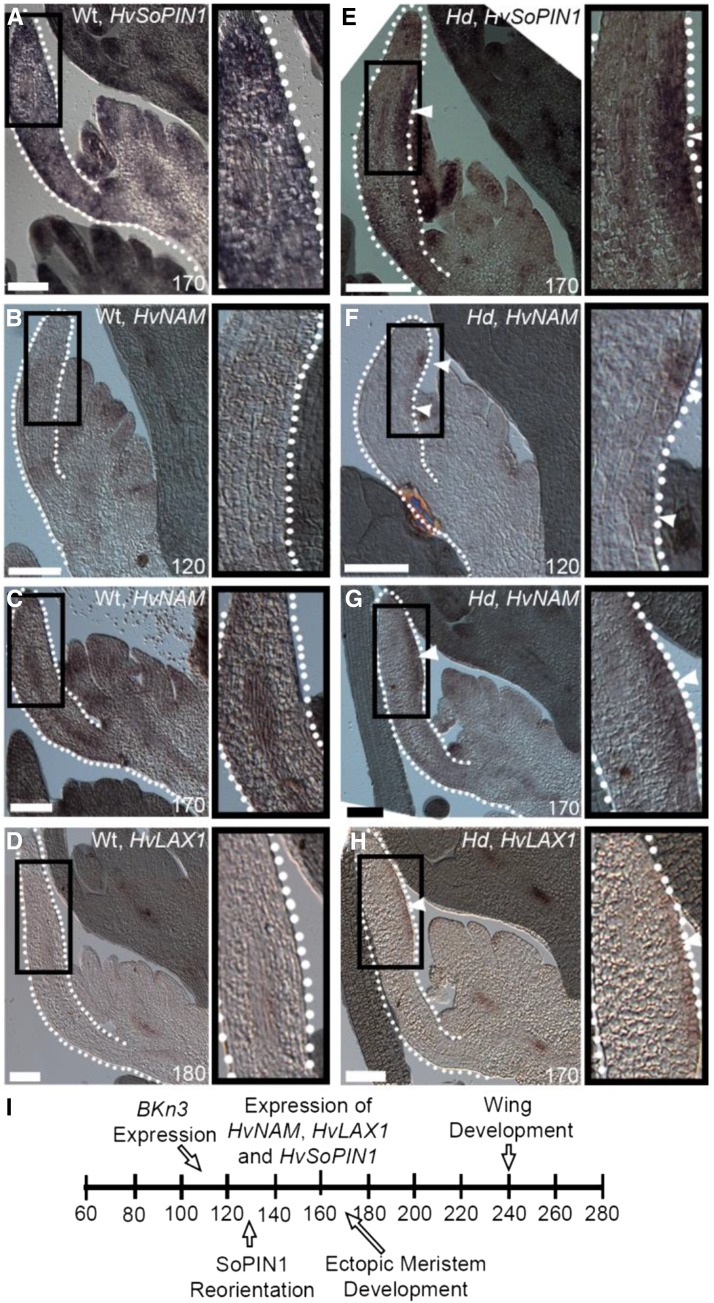
To test whether the higher levels of SoPIN1 in the Hd lemma were due to transcriptional activation of HvSoPIN1 by BKn3, we used RNA in situ hybridization (Figure 6). At 170 h in wild-type florets, HvSoPIN1 expression was largely excluded from the developing lemma (Figure 6A). By contrast, in the Hd lemma at 170 h, there was high expression of HvSoPIN1 in the region of the ectopic meristem (Figure 6E, white arrowhead). This HvSoPIN1 expression pattern supports the immunolocalization observation that SoPIN1 protein levels are not detected in the adaxial surface of the wild type, in contrast to what is observed for Hd at later stages of development. Combined, the mRNA localization patterns and the protein localization suggest that BKn3 induces expression of HvSoPIN1 in the Hd lemma.
Figure 6.
Ectopic Expression of BKn3 Leads to the Expression of HvSoPIN1, HvNAM, and HvLAX1.
(A) to (H) RNA in situ hybridization of HvSoPIN1 ([A] and [E]), HvNAM ([B], [C], [F], and [G]), and HvLAX1 ([D] and [H]). In the wild type, HvSoPIN1, HvNAM, and HvLAX1 were excluded from the lemma, except in some vascular regions ([A] to [D]). At 120 h, HvNAM was expressed flanking the region of BKn3 expression ([F], white arrowheads). By 170 h, when BKn3 expression was high, all three genes (HvSoPIN1, HvNAM, and HvLAX1) were strongly ectopically expressed in the adaxial half of the Hd lemma ([E], [G], and [H], white arrowheads). Black box, zoomed-in image of the lemma; white dotted line, lemma outline; white arrowheads, ectopic expression. n > 4 for each. Bars = 100 μm.
(I) Approximate times (h) of different events during Hd development.
BKn3 Activates Candidate Polarity Organizers
It has previously been proposed that tissue cell polarity may be anchored by regions called organizers, which could influence auxin dynamics (Abley et al., 2013). Candidate organizers include the boundary gene NAM (Souer et al., 1996) and the auxin importer LAX1 (Péret et al., 2012). To test whether BKn3 may influence tissue cell polarity through the modulation of organizers, we performed RNA in situ hybridization (Figure 6) of barley homologs of candidate organizer genes: HvNAM and HvLAX1. RNA in situ hybridization of these components in wild-type florets showed that HvNAM and HvLAX1 mRNA were largely excluded from developing organs, including the lemma (Figures 6B to 6D). By contrast, after 110 h (when BKn3 is expressed in the Hd lemma), ectopic expression of HvNAM and HvLAX1 was detected in the Hd lemma. At 120 h, ectopic HvNAM expression was seen in the adaxial half of the developing lemma, flanking the region where BKn3 mRNA would be expected to be localized (Figure 6F, white arrowheads). By 170 h, ectopic expression of HvNAM had spread throughout the meristem cushion region (Figure 6G, white arrowheads). At this stage, HvLAX1 mRNA was localized in the epidermis of the ectopic meristem (Figure 6H, white arrowheads). This modulation of the expression patterns of the potential organizer components by BKn3 may contribute to the observed reorientation of tissue cell polarity.
The sequence of changes in identity, growth, tissue cell polarity, and morphology is summarized in Figure 6I.
Formation of the Wings Involves a Reorientation of Growth in the Lemma Margin
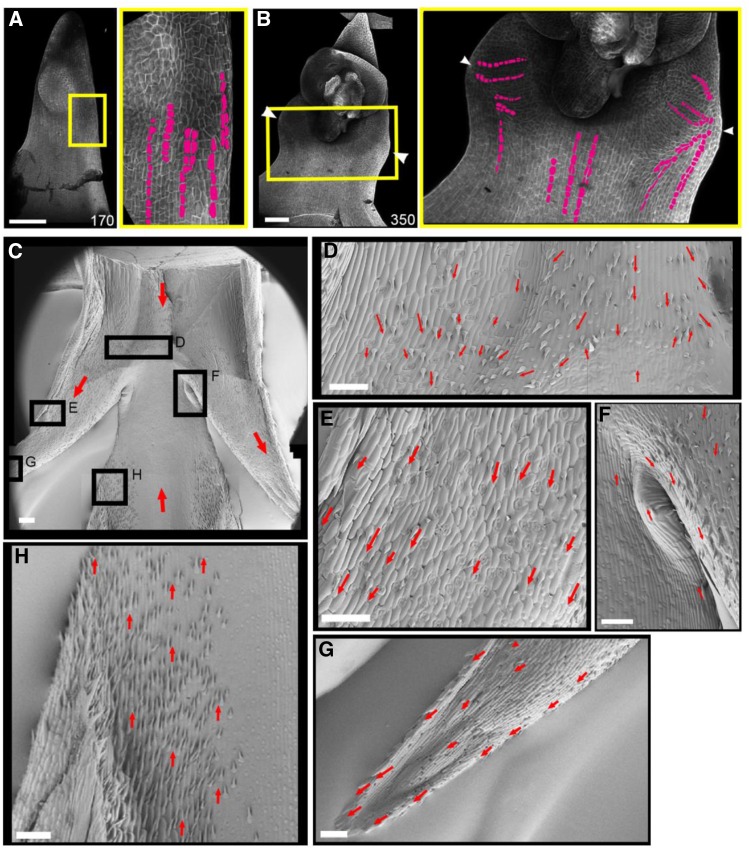
In addition to floral meristem activation, ectopic expression of BKn3 also leads to the initiation of wings at around 240 h. To determine how these wings form, we analyzed the pattern of cell files in the margin during Hd lemma development (Figure 7). Confocal images of calcofluor-stained Hd lemmas (Figures 7A and 7B) showed that at 170 h, when the ectopic meristem had formed but before the wings had emerged, cell files were largely oriented proximodistally in the lemma margin (Figure 7A, magenta cell files). At 350 h, when the wings had formed (Figure 7B), cell files in the center of the lemma body retained the proximodistal orientation, whereas in the margins where the wings were developing, the cell files were oriented toward the tip of the wing (Figure 7B, magenta). This suggests that there is a change in growth orientation in the margins of the developing lemma, which leads to the formation of wings.
Figure 7.
Reorientation of Growth and Tissue Cell Polarity in the Margins of the Hd Lemma.
Confocal images of the adaxial surface of the Hd lemma, stained with calcofluor ([A] and [B]). Zoomed-in images of the region where wings form (yellow boxes) are also shown. Before wing formation (A), cell files (magenta) in the margin of the Hd lemma were oriented proximodistally. After wing formation was initiated (B), the cell files in the margins became aligned with the outgrowing tip of the wing (n = 6). Scanning electron microscopy images of mature Hd wings (zoomed-in images of the boxed regions in [C] shown in [D] to [G]) show the orientation of hairs on the adaxial surface of the Hd lemma. Below the ectopic flower (removed), the hairs pointed basally ([D], red arrows). There is also a ridge where different cell patterns and hairs seem to collide; this may correspond to the convergence of two different polarity fields, as in the lemma body (H), the hairs pointed distally. In the region of the wing outgrowth, the hairs pointed toward the wing tip ([E] and [G]). In the proximal axil of the wing (F), the hairs in the main lemma body orient distally, and those above in the wing orient proximally; near the node, the hairs appear to flow toward the wing (n = 2). Bars = 100 μm.
To evaluate whether the change in growth orientation may reflect a change in tissue cell polarity in the lemma margin, we assessed the orientation of mature hairs on the adaxial surface of the Hd lemma using scanning electron microscopy (Figure 7C). Below the wing outgrowths in the body of the lemma, hairs pointed distally (Figure 7H). In the middle of the lemma body, below the ectopic flower and above the emergence point of the wings, hairs pointed proximally (Figure 7D, red arrows). Closer to the proximal axil of the wing (the bend point of the tissue; Figure 7F), hairs from the body of the lemma pointed distally, whereas those from below the ectopic meristem pointed proximally, and along the edge of the wing they pointed toward the wing tip, suggesting a flow of margin hair orientation toward the wing tip. Hairs along the wings pointed toward the wing tip (Figures 7E and 7G, red arrows), indicating that the two polarity fields converged at the tips of the wings. This pattern of hair orientation suggests that during wing development, tissue cell polarity in the lemma margin may become oriented toward the wing tip.
Interaction between Tissue Cell Polarity and Growth Could Account for Wing Formation
The observed cell file and hair orientations suggest that wings may form due to anisotropic growth, possibly oriented by the changes in tissue cell polarity in the lemma margin after the formation of the ectopic meristem. To evaluate this hypothesis, we used the growing polarized tissue (GPT) framework to model the growth of the wings (Kennaway et al., 2011) (Figure 8; Supplemental Figure 1). This framework approximates plant tissues as a connected continuous sheet of material with two surfaces, termed the canvas. Anisotropic growth is oriented by a polarity field that propagates through the canvas, which may correspond to the tissue cell polarity field at the cellular level. Regional factors determine local specified growth rates parallel (Kpar) and perpendicular (Kper) to the polarity field. The growth pattern is determined by three interacting networks: (1) the gene regulatory network, which defines the regional factors in the model; (2) the growth regulatory network, which defines the specified growth rates; and (3) the polarity regulatory network (PRN), which defines the distribution of a diffusible factor called POLARISER (POL), the gradient of which determines the polarity field. POL distribution is determined by the location of organizer regions that generate POL (plus organizers) and remove POL (minus organizers).
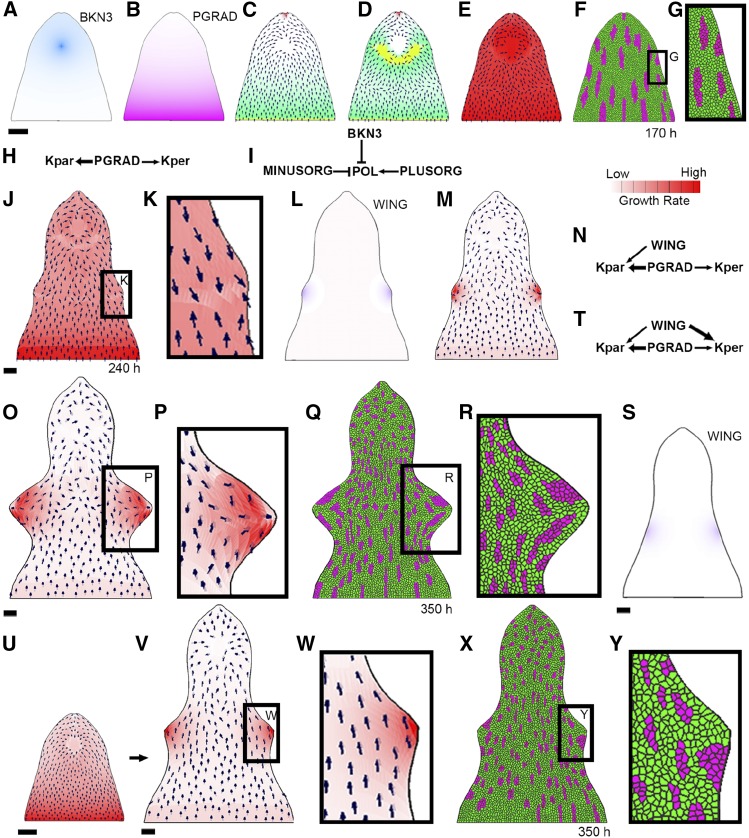
Figure 8.
Changes in Growth and Polarity Can Account for the Formation of Wings.
GPT framework models exploring wing development hypotheses. Initially at 170 h, there is a distal region of BKN3 identity (dark blue; [A]) and a proximodistal gradient of the identity factor PGRAD (pink; [B]). To start with, there is a proximodistal polarity field, which converges toward BKN3 ([C], small black arrows), defined by the local gradient of POL (green). POL is produced at PLUSORG (yellow) and degraded at MINUSORG (red) and BKN3 (PRN in [I]). A reversal in polarity is introduced at 170 h (by adding PLUSORG to the lower boundary of BKN3 [D]). Initially, growth is promoted by PGRAD (KRN in [H], growth rate in [E]). If a layer of virtual dividing cells is added to the surface of the canvas at 0 h and a random selection of cells are shocked, clonal sectors by 170 h are proximodistally elongated ([F] and [G]). When this model is run to later stages, it forms small outgrowths in the margins (240 h in [J]) where the polarity fields converge ([K], black arrows); however, these outgrowths remain small in size. To enhance the size of the outgrowths, both the inversion in polarity and a change in growth rate pattern can be combined ([L] to [R]). A diffusible growth factor, WING ([L], purple) is produced at the wing tips at 240 h. WING promotes Kpar where the concentration of WING is <65% of its maximum value ([M] and [N]). This model generates larger outgrowths by 350 h ([O] and [P]). Clonal sectors induced at 170 h in the margin of the canvas become deformed toward the wing tip by 350 h ([Q] and [R]). Alternatively, enhanced growth rates in the margins could generate the outgrowths ([S] to [Y]). This model has the same setup ([A] and [B]), initial KRN (H), PRN (I), and proximodistal polarity field (C) as the previous model, but no inversion of polarity occurs (the polarity and growth rate patterns for this model at 170 h are summarized in (U). WING is promoted in the margins at 240 h (S) and WING promotes Kper more than Kpar (T). By 350 h, outgrowths form in the margins where WING is promoting growth ([V] and [W]). Clonal sectors induced at 170 h are mostly proximodistal and do not deform toward the wing tip by 350 h ([X] and [Y]). Black arrows illustrate the orientation of the polarity field defined by the local concentration of POL. Bars = 100 μm.
The model has two phases: (1) setup and (2) wing. The setup phase is the same for all models and initially grows a small semicircular canvas (approximately the size of a lemma primordium at 0 h in our time course) to the shape of a lemma at 120 h (Supplemental Figures 1A to 1C). During this initial stage, the canvas has a proximodistal gradient in specified growth rates and has higher Kpar than Kper (determined by the identity factor PGRAD, pink, which declines distally; Supplemental Figure 1A), consistent with higher growth rates in the base of monocot organs (Poethig, 1984; Sylvester et al., 1990; Fiorani and Beemster, 2006; Nelissen et al., 2012) and a proximodistal polarity field (determined by the gradient of POL, green, which is produced at PLUSORG, yellow, and degraded at MINUSORG, red; Supplemental Figure 1B), consistent with SoPIN1 localization in early Hd and wild-type lemmas. If this simulation is run past 120 h, the canvas develops an elongated triangle shape with smooth margins (Supplemental Figure 1D); this shape is similar to that of a wild-type lemma. In this model, polarity is assumed to be proximodistal throughout the lemma, even though the marker of tissue cell polarity we use, SoPIN1 signal, is absent from much of the wild-type lemma in later stages. The assumed proximodistal polarity pattern correlates with hair orientations in the mature wild-type lemma.
After 120 h, distal BKN3 is added to the canvas. BKN3 promotes specified growth rates (both Kpar and Kper) simulating the formation of a new meristematic cushion. BKN3 also acts as a new minus organizer, and polarity is reset to orient toward the center of the canvas surface and the tip, as we observe a reorientation of SoPIN1 localization toward the center of the lemma’s adaxial surface soon after ectopic BKn3 expression. This generates the shape of a Hd lemma at 170 h (Figures 8A to 8F). After 170 h is reached, the specified growth effect of BKN3 is removed but the polarity effect remains (Figure 8C). This is the starting canvas shape for all of the wing simulations (wing phase).
During the wing phase, hypotheses for wing development in the lemma margins are tested. At the start of the wing phase (170 h), the canvas has a distal region of BKN3 identity (Figure 8A, dark blue). The polarity field is proximodistal with a reorientation toward the BKN3 region (Figures 8C and 8I). PGRAD (Figure 8B) promotes both Kpar and Kper, promoting Kpar more than Kper (Figures 8E and 8H). Clonal sectors in the margin of canvas at 170 h are elongated proximodistally (Figures 8F and 8G), similar to the marginal cell files in Figure 7A.
The wings could form as a result of the inversion of tissue cell polarity induced by BKn3. To test this hypothesis, inversion of the model polarity field (consistent with the SoPIN1 patterns in later stage Hd lemmas) is introduced by adding a PLUSORG region in the boundary of BKN3 (Figure 8D, yellow), perhaps corresponding to the activation of HvNAM, and then resetting the polarity. This simulation (Figure 8E) results in small outgrowths forming in the margins at 240 h, where the polarity field converges (Figures 8J and 8K). However, the outgrowths remain much smaller than those observed at 350 h in the confocal lemma images (compared to Figure 3E). These results suggest that a convergent polarity field may be involved in generating the wing outgrowths, but additional factors are also needed to account for their size.
One possible explanation for the size of the wings is that specified growth rates are enhanced in the margin where the polarity field converges, possibly due to the accumulation of auxin. To evaluate this hypothesis, we produced WING at the same place as the polarity convergence points at 240 h (this could be a proxy for the convergence points activating WING; Figure 8L, purple). WING promotes Kpar but also inhibits Kpar where the concentration of WING is more than 65% of its maximum value (Figures 8M and 8N; without this inhibition of growth at the wing tip, the wings become sharp and triangular; Supplemental Figure 1E). The inhibition of growth at the wing tip could result from a reduction in the concentration of growth promoting factor. For example, if WING corresponds to auxin, epidermal auxin at convergence points can be internalized through the formation of vascular traces. Or, it could be achieved through threshold responses; for example, different AUX-IAA (AUXIN/INDOLE-3-ACETIC ACID protein)/ARFs (AUXIN RESPONSE FACTORs) combinations are proposed to respond to different concentrations of auxin (Calderón Villalobos et al., 2012). By 350 h, the shape of the outgrowths (Figures 8O and 8P) broadly matches the shape of the Hd lemma wings at 350 h (Figure 3E). The clonal sectors in the wing region of the canvas also become deformed toward the wing tip (Figures 8Q and 8R), broadly matching the flow pattern of the cell files observed in the confocal data (Figure 7B). The position and shape of the new PLUSORG in the BKN3 boundary can influence the position and shape of the outgrowths (Supplemental Figures 1F to 1I). Robustness in wing shape formation may result from activation of an additional factor, such as new MINUSORG at the wing tips possibly due to an accumulation of auxin.
It may be that the promotion of growth in the lemma margin alone can generate wing outgrowths. To evaluate this hypothesis, we do not invert the polarity field (same polarity field as Figure 8C), and we specify the production of WING in the margin (Figure 8S, purple) at 240 h, which is the approximate time when the wing outgrowths are first observed. WING promotes both Kpar and Kper, but it promotes Kper more than Kpar (Figure 8T). This simulation results in the formation of outgrowths (Figures 8V and 8W) that have clonal sectors that do not seem to flow toward the tip of the outgrowth (compare Figures 8X to 8Y with Figures 3E and 7B). This suggests that an increase in growth rate in the margin can generate outgrowths but is insufficient alone to account for the orientations of growth observed in wing formation.
The computational modeling suggests that the wing outgrowth in the margin may arise through the convergence of tissue cell polarity fields, combined with enhanced growth in the lemma margin.
DISCUSSION
Class 1 KNOX genes are central to meristem function, and when overexpressed, they have profound effects on morphology. Understanding how these effects are mediated may shed light on KNOX gene functions. We show that ectopic expression of the class 1 KNOX gene, BKn3, in the barley lemma leads to a defined series of temporal and spatial changes in tissue cell polarity (SoPIN1 localization) and identity (ectopic expression of HvSoPIN1, HvNAM, and HvLAX1). These events precede changes in growth (wing formation), regional polarity (inverted ectopic flowers), and oriented cell differentiation (hair development).
Ectopic BKn3 expression is activated between 90 and 110 h in the distal region of the lemma. By 120 h, SoPIN1 in the lemma is upregulated and relocalizes to point centrally, indicating a change in tissue cell polarity. Several hypotheses might account for how BKn3 alters SoPIN1 localization. BKn3 could initially stimulate the formation of a minus organizer, causing the reorientation of polarity toward it. The possible molecular basis of the organizer would depend on the mechanism for coordinating tissue cell polarity (Abley et al., 2016). If auxin is a component of tissue cell polarity, BKn3 could generate a localized increase in intracellular auxin, for example, through enhancing auxin biosynthesis, as suggested by Bolduc et al. (2012). SoPIN1 could then relocalize to point centrally if an up-the-gradient model applies (Jönsson et al., 2006; Smith et al., 2006). Alternatively, BKn3 could trigger a localized reduction in extracellular auxin, for example, by promoting auxin import or vein formation. The observed upregulation of HvLAX1 expression in the epidermis of the ectopic meristem (Figure 6H) supports the hypothesis that BKn3 can promote local auxin import. SoPIN1 could perhaps then reorient toward the region of low extracellular auxin if the with-the-flux or indirect coupling models apply (Sachs, 1969, 1981; Mitchison, 1980; Mitchison et al., 1981; Stoma et al., 2008; Abley et al., 2013). Whatever the mechanism, our results show that KNOX gene activity is intimately connected with the orientation of tissue cell polarity.
After strong BKn3 expression has been established and SoPIN1 has reoriented toward the BKn3 expression region (170 h), SoPIN1 below the ectopic meristem reorients to point proximally. This may be a result of the activation a new plus organizer in the boundary of the BKn3 expression region. For example, the HvNAM boundary gene is initially expressed in the margin of the BKn3 domain (120 h) and later throughout the BKn3 region. This activation of HvNAM could be a direct effect of BKn3, as KNOXs are able to bind upstream of NAM genes (Bolduc et al., 2012). Boundary identity regions themselves may act as plus organizers of SoPIN1 localization (Abley et al., 2013). Thus, BKn3 modulates tissue cell polarity during organ development, possibly through altering auxin dynamics and/or boundary identities. This may be a common class 1 KNOX function, as other overexpression phenotypes, like the altered vein patterns in maize Kn1 (Ramirez et al., 2009), could involve changes in underlying tissue cell polarity.
In both wild-type and Hd lemmas, the orientation of epidermal hairs correlates with the pattern of SoPIN1 localization. This suggests that differentiating cells have an inherent polarity based upon patterns of tissue cell polarity established earlier in development. Therefore, by modulating the local patterns of tissue cell polarity in the Hd lemma, BKn3 is able to influence oriented cell differentiation patterns at later stages.
Ectopic BKn3 expression also leads to changes in growth, as illustrated by the development of wings in the lemma margin below the ectopic meristem at 240 h. BKn3 expression extends a short distance below the ectopic flower into the wings but does not occur throughout the wings. Epidermal hair orientations and cell file patterns suggest that tissue cell polarity converges toward the tips of the developing wings. These observations suggest that the wings reflect effects of tissue cell polarity on orienting growth and may be a nonautonomous effect of BKn3 expression. Computational modeling results are consistent with this hypothesis. Models in which a polarity field (i.e., a vector field) orients specified growth, and in which growth rate is enhanced where polarity converges at the margin, lead to the formation of wing-shaped outgrowths. The position of the polarity convergence points may be reinforced by the formation of new minus organizers at the wing tips, possibly induced by the accumulation of auxin. The increase in growth rate could be a consequence of an accumulation of a growth promoting factor in the margin; this factor would need to be activated outside of the BKn3 expression domain, as wing development is a non-cell-autonomous effect. Auxin could be a strong candidate for this growth promoting factor, as it is known to promote growth (Teale et al., 2006), and it could accumulate in the margin due to the reversal of the SoPIN1 polarity field. The small stripe of BKn3 expression near the axil of the wing may also influence margin growth rates through influencing SoPIN1 localization, auxin dynamics, or other factors involved in growth (Sakamoto et al., 2001; Jasinski et al., 2005; Bolduc et al., 2012). Wing formation bears marked similarities with leaf margin outgrowths, such as serrations and lobes (Barkoulas et al., 2008; Bilsborough et al., 2011), suggesting similar underlying mechanisms may be involved. For example, both serrations and lobes involve PIN1 convergence points in the leaf margin and changes in margin growth rate and orientation. Overexpression of KNOX genes can also influence lobe phenotype (Chuck et al., 1996), supporting a possible role for the stripe of BKn3 expression in modulating wing shape. Perhaps BKn3 near the distal edge of the wing outgrowth is involved in boundary formation or is involved in modulating growth rate, reminiscent of the function of other homeodomain genes such as RCO (REDUCED COMPLEXITY) (Vlad et al., 2014). Thus, through modulating tissue cell polarity, BKn3 is able to influence growth orientations and rates, leading to marginal outgrowths. This may explain the formation of marginal leaf flaps in other class 1 KNOX overexpression mutants, such as maize Kn1 (Ramirez et al., 2009). In the Kn1 mutant, these leaf flaps exhibit a change in regional polarity, as they form in the margin of the blade and have sheath-like identity (Ramirez et al., 2009).
In addition to these effects on tissue cell polarity and growth, BKn3 induces a reversal in regional polarity in the Hd lemma: The first ectopic flower has a distal palea, while the second ectopic flower has a proximal palea. Tissue cell polarity does not match regional polarity, as both of these ectopic floral meristems form in the context of an initially proximodistal SoPIN1 polarity field. Thus, in this case, regional polarity may not be a consequence of tissue cell polarity but may instead be established through a separable mechanism. Several hypotheses might account for the inversion of regional polarity (Figure 9). One is that BKn3 expression (Figure 9A, dark-blue hatching) initiates the formation of an ectopic inflorescence meristem on the adaxial surface of the Hd lemma (Figure 9A, pink) (Williams-Carrier et al., 1997). The ectopic floral meristems would then form on the flanks of the inflorescence meristem, 180° to each other, recapitulating the distichous patterning of both the vegetative and inflorescence meristem (Figure 9A, orange). The palea may be initiated closest to the center of the inflorescence meristem (Figure 9A, green), for example, where BKn3 expression is highest. This hypothesis predicts that BKn3 expression spans a domain that encompasses both floral meristems. This view is also consistent with the region between the ectopic flowers having a more rachis-like rather than lemma identity (Stebbins and Yagil, 1966). Another hypothesis is that identities within the lemma determine regional polarity (Figure 9B). For example, the ectopic floral meristems (Figure 9B, orange) may be initiated on the flanks of a lemma/awn boundary domain (Figure 9B, light blue), with the palea (Figure 9B, green) forming toward this boundary domain. This hypothesis is supported by the observation that the awnless mutation, lks2, is epistatic to Hd, suggesting that the awn identity region is required for the Hd mutant phenotype (Roig et al., 2004). In practice, it is hard to distinguish between this and the previous hypothesis, as the development of the second floral meristem is variable and it is difficult to assess the expression pattern of BKn3 across the lemma in 3D. According to either of these hypotheses, floral (regional) polarity results from an interaction between identities through signaling that does not depend on tissue cell polarity.
Figure 9.
Hypotheses for Inverted Regional Polarity in Hd.
Diagrams of the adaxial surface of the Hd lemma (black) outlining the hypotheses.
(A) Induction of an inflorescence meristem. The expression of BKn3 (dark-blue hatch) results in the formation of an inflorescence meristem (pink), which then initiates floral meristems (orange) on its flanks; the palea (green) are initiated on the side of the floral meristem closest to the center of the inflorescence meristem.
(B) Underlying lemma properties. Ectopic BKn3 expression (dark-blue hatch) occurs in the lemma/awn boundary (light blue) and triggers the formation of ectopic floral meristems (orange). Palea (green) develop on the side of the ectopic floral meristem closest to the lemma/awn boundary.
Interactions between identities may also explain the changes in regional polarity observed in the snapdragon (Antirrhinum majus) Hirzina mutant outgrowth, which exhibits a proximal-distal-proximal pattern of epidermal features (Golz et al., 2002). Similarly, alterations to the proximal-distal axis in Kn1 leaves (Ramirez et al., 2009) may reflect changes in identity, as KN1 protein at the base of the leaf is thought to confer proximal identity (Jackson, 2002).
Previous studies have shown that class 1 KNOX genes are essential for meristem identity (Barton and Poethig, 1993; Kerstetter et al., 1997) and have a diverse range of phenotypic effects when overexpressed. These studies indicate that class 1 KNOX genes regulate differentiation and growth (Stebbins and Yagil, 1966; Smith et al., 1992; Sinha et al., 1993; Lincoln et al., 1994; Janssen et al., 1998; Shani et al., 2009). KNOX genes may also have a role in leaf outgrowth, as indicated by the presence KNOX at the base of developing wild-type maize leaves and in the margin of compound leaves (Bharathan et al., 2002; Shani et al., 2009; Hay and Tsiantis, 2006) and by the loss of leaf formation in knox null mutants (Barton and Poethig, 1993; Long et al., 1996; Kerstetter et al., 1997; Vollbrecht et al., 2000). Our analysis of the Hd barley mutant suggests that KNOX genes may influence organ outgrowth through modulating tissue cell polarity patterns, which influence oriented cell differentiation and growth patterns, as well as alterations in regional polarity. Such modulations may also explain other KNOX overexpression phenotypes, such as the formation of knots and spurs. Thus, class 1 KNOX gene activity likely involves an interplay between identity, regional polarity, and tissue cell polarity.
METHODS
Plants
Barley (Hordeum vulgare cv Bowman) seeds (Hd BW341) were stratified at 4°C for 48 h and germinated at room temperature. Five days after coleoptile emergence, the seedlings were planted in P15 trays in John Innes Cereal Mix and grown in standard summer greenhouse conditions; time-course samples were taken beginning at 17 d after coleoptile emergence.
Photography
Mature wild-type and Hd inflorescences were photographed using an Olympus XZ XZ-1.
OPT
Inflorescences collected in 100% ethanol were prepared for OPT as described (Sharpe et al., 2002; Lee et al., 2006). Specimens larger than 1.5 cm were not embedded in agarose. Samples were imaged 400 times on an x/y rotation, either on a Prototype OPT scanner (Lee et al., 2006) (<1 cm), the Commercial Scanner Bioptonics 3001 (SkyScan) (<1.5 cm), or a Macro OPT scanner (<4 cm) in BABB (1:2 benzyl alcohol:benzyl benzoate; Sigma-Aldrich). On the prototype scanner, we used white or UV light through the GFP1 filter, and UV light through the TXR filter. On the commercial scanner, we used white light through an infrared filter and UV light through a GFP1, GFP+, or Cy3 filter. On the Macro scanner, we used white light through a GFP filter or UV light through a GFP or TXR filter. Images were aligned using NRecon (NRecon, version 1.6.3.3; SkyScan 2010). Volviewer (http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/VolViewer#Description) was used to reconstruct and edit the images.
Time-Course Staging
Two time courses were collected: the first covering 240 h and the second covering 380 h of wild-type and Hd development. Initiation of flowering varied due to fluctuating growth conditions; therefore, to align both time courses, a time zero morphology state (M0) was defined. To define M0, images in the first time point of time course 1 were measured using FIJI (Schindelin et al., 2012), and the average dimensions of the fifth floret from the base of the inflorescence were calculated. The mean ± 2 sds defined a range of values that described the M0 reference. Comparing the average dimensions of floret five in the second time course allowed both time courses to be combined. Each time point was defined in hours since M0 (h) based on the time of harvest. The natural logarithm (ln) of floret 5 width was used to develop a growth graph, as ln(floret 5 width) did not vary significantly (P = 0.76) between the wild type and Hd. The equation of the line of best fit was used to stage all experimental data in hours since M0 (h).
Confocal Microscopy of Hd Lemmas
Hd inflorescences that had been fixed in 100% ethanol were imaged under bright-field light on a Leica M205C stereomicroscope with a DFC495 camera. Floret width was measured using FIJI (Schindelin et al., 2012), and each floret was allocated to a 20 h timeslot (80<t>100 h, 100<t>120 h, etc.) using the growth curve equation. Each lemma was fixed (adaxial surface up) to a slide, rehydrated, and stained in 0.1% calcofluor (fluorescent brightener 28, F3543; Sigma-Aldrich) for 20 min. Lemmas were imaged using a 25× multi-immersion lens on a Zeiss 780 confocal microscope (violet laser diode [405 nm] excitation laser, PMT detectors 400 to 480 nm). Larger images were reconstructed in Adobe Photoshop from multiple tiles. Each figure image is a representative of the typical morphology in the 20 h timeslot, and the time stated is the median of the timeslot.
RNA in Situ Hybridization Probes
Probes targeted to unique sequences in the coding sequence of each gene were obtained from the published barley genome (Mayer et al., 2012): BKn3 (AK376780), HvLAX1 (AK369586), HvNAM (MLOC_65286.1), and HvSoPIN1 (MLOC_293.1). Probe templates were amplified from wild-type Bowman cDNA using the following primers: BKn3 (F:GCCATCAAGGCCAAGATCATCTCC, R:GGTAGAAGACGAGCGTCTTCTTCATGCC, to give 600 bp), HvLAX1 (F:CTACCTCATCAGCGTCCTCTACGTCG, R:CTAGCCGAACCTGTAGAGCCCGCTG, to give 530 bp), HvNAM (F:AGATGGAGCGGTACGGTTCTCTG, R:GCAGGTAGATGTACTTGAACTTGGC, to give 399 bp), and HvSoPIN1 (F:TTCCACTTCATCTCCTCCA, R:CCTCCGCCTGCAGGTCCG, to give 413 bp). These products were cloned into pCR 4-TOPO (Invitrogen Life Technologies TOPO TA Cloning kit and Maximum Efficiency One Shot OmniMAX 2 T1 Phage-Resistant Chemically competent E. coli) according to the manufacturer’s guidelines. Plasmids were sequenced using a BigDye Terminator v3.1 cycle sequencing kit and Eurofins Genomics. RNA probes were made using the protocol from Coen et al. (1990) with the following modifications: The probe template with the T7 or T3 transcription start site was amplified using PCR and purified using a QIAquick PCR purification kit (Qiagen) followed by phenol-chloroform extraction. One microgram of purified PCR was used to make the digoxigenin-UTP-labeled RNA using T7 or T3 RNA polymerase. All probes were hydrolyzed in 200 mM carbonate buffer, pH 10.2, at 60°C for 1 h.
RNA in Situ Hybridization Tissue Fixation
Barley inflorescences were collected in ice-cold 4% paraformaldehyde (PBS, pH 7, 16% paraformaldehyde solution [Electron Microscopy Sciences], 4% DMSO, and 0.1% Triton X-100), placed under vacuum 3 × 10 min, and fixed overnight at 4°C. Fixed samples were washed in 0.85% saline for 30 min at 4°C, 50% ethanol/0.85% saline for 3 h at 4°C, and 70% ethanol/0.85% saline for 3 h at 4°C and stored at 4°C in 70% ethanol/0.85% saline. Samples were transferred to Tissue-Tek mesh biopsy cassettes and loaded into a Tissue-Tek TEC VIP vacuum wax infiltrator (Sakura) with the following program: 4 h in 70% ethanol at 35°C, 4 h in 80% ethanol at 35°C, 4 h in 90% ethanol at 35°C, three cycles of 4 h in 100% Xylene at 35°C, and four cycles of 4 h in 100% paraffin wax at 60°C; all steps were performed with agitation and under vacuum. Samples were transferred to hot paraffin in a Tissue-Tek TEC (Sakura) embedding machine and embedded, followed by storage at 4°C. Blocks were sliced in 8-µm-thick ribbons using a Reichet-Jung 2030 microtome at room temperature. The tissue slices were mounted on Polysine microscope slides (VWR 631-0107) with water, dried on a 37°C hotplate for 48 h, and stored at 4°C.
RNA in Situ Hybridization Protocol
The RNA in situ hybridization protocol was as described (Coen et al., 1990) with the following modifications: 80 μL hybridization buffer and 2 to 4 μL of RNA probe were used per slide and covered with HybriSlip Hybridization Covers (Grace Bio-Labs). Slides were washed with 0.2% SSC (saline sodium citrate buffer) at 50°C before washing in NTE (0.5 M NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA) at 37°C and RNase treatment. The samples were washed in NTE buffer, followed by Buffer 1 (100 mM of Tris-HCl and 150 mM of NaCl) at room temperature before incubating with blocking reagent (Roche) for 1 h. Antidigoxygenin-AP (Roche) was used at 1:3000 dilution in 1% BSA, 0.3% Triton X-100, and Buffer 1 and incubated for 1.5 h. Subsequent washes were first performed with Buffer 1 with 0.3% Triton X-100, then once with Buffer 1 only. Localization of the antidigoxygenin was visualized by incubating with 0.15 mg mL−1 NBT (nitro blue tetrazolium; Promega) and 0.075 mg mL−1 BCIP (5-bromo-4-chloro-3-indolyl-phosphate; Promega) in 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2 at room temperature overnight.
RNA in Situ Hybridization Imaging
Slides were imaged in water on a Leica DM600 microscope with a DFC420 digital camera under DIC light. Imaged slides were dried and mounted with VectaMount AQ (aqueous mounting medium, H-5501; Vector Laboratories).
Antibodies
The SoPIN1 primary antibody was provided by Devin O’Connor (The Sainsbury Laboratory, Cambridge University; O’Connor et al., 2014) and used at a 1:200 dilution. Standard anti-guinea pig Alexa 488 secondary antibodies from Life Technologies (A11073, lot 1235789) were used to detect the SoPIN1 primary antibody at a 1:200 dilution.
Immunolocalization Tissue Fixation
Barley inflorescences were fixed in FAA (50% ethanol, 5% acetic acid, and 3.7% formaldehyde; Sigma-Aldrich) with 1% DMSO and 0.5% Triton X-100, placed under vacuum for 3 × 10 min, then fixed overnight at 4°C. Fixed samples were washed with 50% ethanol for 3 h at 4°C, followed by 70% ethanol for 3 h at 4°C and stored at 4°C in 70% ethanol.
Whole-Mount Immunolocalization
The whole-mount immunolocalization protocol was adapted from Conti and Bradley (2007) with the following modifications: Digestion in 2% Driselase (from Basidiomycetes sp, D9515; Sigma-Aldrich) and 1% Pectolyase Y-23 (Yakult Pharmaceutical) for 30 min at 37°C and washing in PBS before the citrate boiling step. Permeabilization was performed with a 2-h incubation in 1% Triton X-100 and 5% DMSO in PBS before blocking for 1 h in 1% BSA with 0.3% Triton X-100. For all antibodies, 1:200 dilutions were used. Samples were stained in 0.1% calcofluor for 40 min.
Whole-Mount Immunolocalization Imaging
Lemmas were imaged using a 25× dip lens on an SP5 (II) Leica confocal microscope: calcofluor, violet laser diode (405 nm) excitation laser and PMT detectors, 400 to 480 nm; Alexa-488, argon ion (488 nm) excitation laser and PMT detectors, 500 to 575 nm.
Scanning Electron Microscopy Imaging
Cryo-scanning electron microscopy images were taken using a Zeiss Supra 55 VP FEG with a Gatan Alto2500 cryo system and were generated by JIC Bio-Imaging Service (Elaine Barclay). Images were of the adaxial surface of mature Hd lemmas below the ectopic flower (which was removed).
Image Processing
Figures were assembled in Adobe Photoshop 2015; the brightness and contrast were altered for better visualization where appropriate. For large images, tiles were aligned in Adobe Photoshop and merged. The scale bars in every image were standardized to 100 µm, 200 µm, 250 µm, or 1 cm where appropriate. Arrows and highlighted cells were added using Adobe Photoshop 2015.
GPT Framework Modeling
Modeling was performed using the GPT framework (Growth toolbox rev. 4813) (Kennaway et al., 2011) run on MatLab R2013b. The canvas at the start of the wing simulations presented in this article represents the morphology of the lemma at 170 h (0.66 mm high and 0.5 mm wide with 2786 finite elements; more elements are added to the wing region at 340 h to make 3352) with a fixed base, mimicking attachment to the base of the floret (m.fixedDFmap(id_base_p==1,2)=true, where 2 defines the y axis). The model takes 20 min to run from −50 to 380 h on an INTEL Core i7-2600 CPU processor. The model stops at 350 h when the canvas is 1.7 mm × 0.9 mm. Each step in the simulation represents 2 h.
There are two model phases: setup (−50 to 170 h) and wing (170 to 380 h). The setup phase generates the starting shape of the canvas for the wing simulations (the wing simulation phase is focused on in this article). In the setup phase, the canvas deforms from a semicircular canvas (0.15 × 0.1 mm) with a layer of dividing virtual cells to a shape similar to that of a lemma at 170 h of development. If a random selection of virtual cells is labeled early in the model, the resulting clonal sectors are proximodistally elongated by 170 h. In the wing phase, hypotheses for how wing outgrowths could form in the margin of the canvas based on changes in polarity and growth rates are tested. Virtual clonal sectors in this phase of the model are activated at 170 h, and the resulting sector shapes are observed at 350 h.
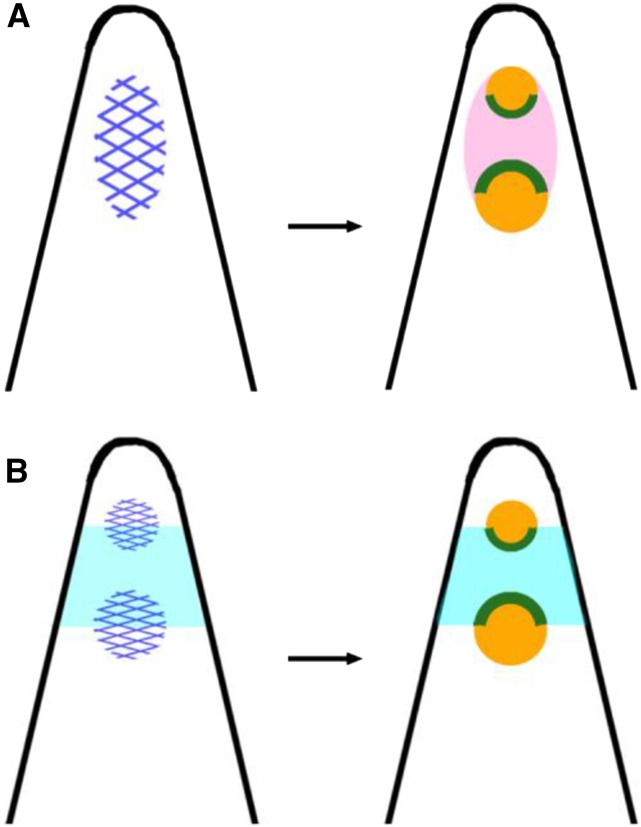
The setup phase defines factors (Table 1) that remain the same throughout the model. There are signal factors, denoted by “s”, which have production (P), diffusion (D), and decay (De) parameters (Table 2), and identity factors denoted by “i,” which have a value of 1, and if defined by a signal factor, they have the same gradient as the signal factor at that point in time. Signal factors diffuse according to the following equation:
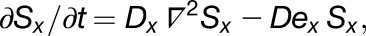 |
where x denotes the specific signal factor (S). The gradient of diffusible factors is frozen using the following function:
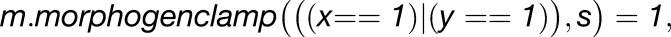 |
where x and y define regions of the canvas and s is the signal factor. The diffusion and decay parameters of signal factors are set to 0 after a period of time to maintain the gradient pattern.
Table 1. Identity and Signaling Factors Used in the Model.
| Factor | Phase | Position | Role |
|---|---|---|---|
| iBASE | Setup | Proximal base | Base of the canvas |
| iPLUSORG | Setup | Proximal base | POL source |
| iMINUSORG | Setup | Gradient from tip | POL sink |
| iEDGE | Setup | Margin of canvas | Defines iWINGTIP |
| iBOUNDARY | Setup | Below sBKN3 | New iPLUSORG region |
| iWINGTIP | Setup | Lateral points | Produces sWING |
| iCENTRE | Setup | Distally shifted, middle | Produces sBKN3 |
| sBKN3 | Setup | Produced by iCENTRE | POL sink |
| sPGRAD | Setup | Produced by iBASE | Promotes growth rates |
| sWING | Wing | Produced by iWINGTIP | Promotes growth rates in wings |
| POL | Setup | Produced by iPLUSORG | Defines growth axis |
Outlines of the name and category of each factor, when it is activated, the canvas position, and its role in the model are provided. i, identity factor; s, signaling factor.
Table 2. Parameters for Signaling Factors.
| Factor | Parameter | Value |
|---|---|---|
| POL | DPOL | 0.001 |
| DePOL | 0.100 | |
| PPOL | 1.000 | |
| sWING | DsWING | 0.001 |
| DesWING | 0.100 | |
| PsWING | 1.000 | |
| sBKN3 | DsBKN3 | 0.001 |
| DesBKN3 | 0.100 | |
| PsBKN3 | 1.000 | |
| sPGRAD | DsPGRAD | 0.001 |
| DesPGRAD | 0.100 | |
| PsPGRAD | 1.000 |
All of the parameters relevant for the signal factors in the model, including diffusion rate (D), decay rate (De), and production (P; maximum value).
In this framework, a polarity field is used to define the orientations of growth, K (growth can be defined parallel, Kpar, perpendicular, Kper, or normal, Knor [thickness], to the polarity field). This polarity field is based upon the local concentration gradient of a signal factor, POL. POL is produced at sources, defined by iPLUSORG, and degraded at sinks, defined by iMINUSORG. This forms the basis of the PRN. During the setup phase, iPLUSORG is at the base of the canvas and iMINUSORG is at the distal tip of the canvas. During the wing phase, the POL gradient is reset when sBKN3 is added as a sink and when iBOUNDARY is introduced as an additional iPLUSORG region to the PRN.
Growth rates are influenced by the factors in model. Promotion (pro) and inhibition (inh) of growth are defined using the following functions:
 |
where x is the factor influencing growth and n is the constant that defines how much the factor promotes or inhibits growth rates.
During the setup phase growth is defined with the following equations:
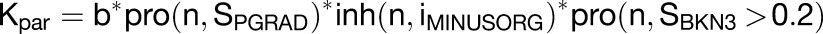 |
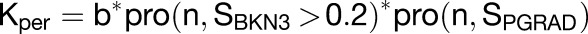 |
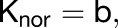 |
where b is the basic growth rate, n is the magnitude, S is the signal factor, and i is the identity factor.
During the wing phase of the most complex model (outlined in Figures 8N to 8R), growth is defined with the following equations:
 |
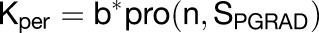 |
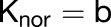 |
The values of all of the parameters at different times in the model are outlined in Table 3. The full code for the models is available for download at http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/OpenSourceDownload_PlantCell_Richardson_2016/GPT_HoodedBarleyWingModels.zip.
Table 3. Parameters for the Growth Regulatory Network at Different Times in the Model.
| Parameter | Description | Parameter Value at Different Stages |
|||
|---|---|---|---|---|---|
| 0–125 h | 125–170 h | 170-240 h | 240–380 h | ||
| Kpar b | Basic growth rate | 0.011 | 0.012 | 0.011 | 0.0019 |
| Kpar PsPGRAD | Promotion of Kpar by sPGRAD | 0.600 | 0.010 | 1.300 | 1.0000 |
| Kpar IiMINUSORG | Inhibition of Kpar by iMINUSORG | 1.200 | 1.000 | 1.000 | 1.0000 |
| Kpar PsBKN3 | Promotion of Kpar by sBKN3 | 0.000 | 0.010 | 0.000 | 0.0000 |
| Kpar PsWING | Promotion of Kpar by sWING | 0.000 | 0.000 | 0.000 | 8.0000 |
| Kpar IsWING | Inhibition of Kpar by sWING >0.65 | 0.000 | 0.000 | 0.000 | 10.0000 |
| Kper b | Basic growth rate | 0.005 | 0.006 | 0.002 | 0.0060 |
| Kper PsBKN3 | Promotion of Kper by sBKN3 | 0.000 | 1.000 | 0.000 | 0.0000 |
| Kper PsPGRAD | Promotion of Kper by sPGRAD | 0.000 | 1.500 | 5.000 | 5.0000 |
| Knor | Basic growth rate | 0.010 | 0.010 | 0.001 | 0.0010 |
Accession Numbers
Sequences used to design the RNA in situ hybridization probes can be obtained from the published barley genome sequence (Mayer et al., 2012): BKn3 (AK376780), HvLAX1 (AK369586), HvNAM (MLOC_65286.1), and HvSoPIN1 (MLOC_293.1).The seeds used in this work were Bowman barley wild-type seeds and Bowman Hooded mutant (Hd BW341) obtained from Arnis Druka (James Hutton Institute, UK).
Supplemental Data
Supplemental Figure 1. Setup phase of the model and the effect of modulating different features.
Supplementary Material
Acknowledgments
We thank Catherine Taylor for plant care, Karen Lee for help with OPT imaging and processing, Grant Calder for help with confocal microscopy, Elaine Barclay for cryo-scanning electron microscopy imaging, Richard Kennaway for help with computational modeling, Desmond Bradley for extensive help with methods and useful discussion, and Chris Whitewoods and Catherine Mansfield for useful discussions on the manuscript. We also thank Sarah Hake and Devin O’Connor for the maize SoPIN1 antibody and for useful discussions. We also thank Arnis Druka at the James Hutton Institute for the Bowman barley seeds. This work was supported by the John Innes Centre Rotation PhD program funded by BBSRC and the John Innes Society (A.R.). This work was also supported by BBSRC Grants BB/M023117/1 and BB/F005997/1.
AUTHOR CONTRIBUTIONS
E.C., A.B.R., and A.R. designed the research. A.R. generated and analyzed the data and built the computational models. A.B.R. and A.R. worked on method development and cloning. E.C. and A.R. wrote the article. A.B.R. edited the article. E.C. provided funding.
Glossary
- OPT
optical projection tomography
- GPT
growing polarized tissue
- PRN
polarity regulatory network
Footnotes
Articles can be viewed without a subscription.
References
- Abley K., De Reuille P.B., Strutt D., Bangham A., Prusinkiewicz P., Marée A.F.M., Grieneisen V.A., Coen E. (2013). An intracellular partitioning-based framework for tissue cell polarity in plants and animals. Development 140: 2061–2074. [DOI] [PubMed] [Google Scholar]
- Abley K., Sauret-Güeto S., Marée A.F., Coen E. (2016). Formation of polarity convergences underlying shoot outgrowths. eLife 5: e18165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler P.N. (2002). Planar signaling and morphogenesis in Drosophila. Dev. Cell 2: 525–535. [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40: 1136–1141. [DOI] [PubMed] [Google Scholar]
- Barton M.K., Poethig R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831. [Google Scholar]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860. [DOI] [PubMed] [Google Scholar]
- Bilsborough G.D., Runions A., Barkoulas M., Jenkins H.W., Hasson A., Galinha C., Laufs P., Hay A., Prusinkiewicz P., Tsiantis M. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 108: 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M.K., Morohashi K., O’Connor D., Grotewold E., Hake S. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26: 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnett O.T. (1938). Hood and supernumerary spike development in barley. J. Agric. Res. 57: 371–377. [Google Scholar]
- Calderón Villalobos L.I., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Lincoln C., Hake S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E.S., Romero J.M., Doyle S., Elliott R., Murphy G., Carpenter R. (1990). floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell 63: 1311–1322. [DOI] [PubMed] [Google Scholar]
- Conti L., Bradley D. (2007). TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19: 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorani F., Beemster G.T.S. (2006). Quantitative analyses of cell division in plants. Plant Mol. Biol. 60: 963–979. [DOI] [PubMed] [Google Scholar]
- Golz J.F., Keck E.J., Hudson A. (2002). Spontaneous mutations in KNOX genes give rise to a novel floral structure in Antirrhinum. Curr. Biol. 12: 515–522. [DOI] [PubMed] [Google Scholar]
- Hake S., Smith H.M., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20: 125–151. [DOI] [PubMed] [Google Scholar]
- Harlan H.V. (1931). The origin of Hooded barley. J. Hered. 22: 265–272. [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165. [DOI] [PubMed] [Google Scholar]
- Jackson D. (2002). Double labeling of KNOTTED1 mRNA and protein reveals multiple potential sites of protein trafficking in the shoot apex. Plant Physiol. 129: 1423–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B.-J., Lund L., Sinha N. (1998). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117: 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565. [DOI] [PubMed] [Google Scholar]
- Jönsson H., Heisler M.G., Shapiro B.E., Meyerowitz E.M., Mjolsness E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennaway R., Coen E., Green A., Bangham A. (2011). Generation of diverse biological forms through combinatorial interactions between tissue polarity and growth. PLOS Comput. Biol. 7: e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R.A., Laudencia-Chingcuanco D., Smith L.G., Hake S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054. [DOI] [PubMed] [Google Scholar]
- Lawrence P.A., Struhl G., Casal J. (2007). Planar cell polarity: one or two pathways? Nat. Rev. Genet. 8: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Avondo J., Morrison H., Blot L., Stark M., Sharpe J., Bangham A., Coen E. (2006). Visualizing plant development and gene expression in three dimensions using optical projection tomography. Plant Cell 18: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Long J., Yamaguchi J., Serikawa K., Hake S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Mayer K.F., Waugh R., Brown J.W., Schulman A., Langridge P., Platzer M., Fincher G.B., Muehlbauer G.J., Sato K., Close T.J., Wise R.P., Stein N.; International Barley Genome Sequencing Consortium (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716. [DOI] [PubMed] [Google Scholar]
- Mitchison G.J. (1980). A model for vein formation in higher plants. Proc. R. Soc. Lond. B Biol. Sci. 207: 79–109. [Google Scholar]
- Mitchison G.J., Hanke D.E., Sheldrake A.R. (1981). The polar transport of auxin and vein patterns in plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 295: 461–471. [Google Scholar]
- Müller K.J., Romano N., Gerstner O., Garcia-Maroto F., Pozzi C., Salamini F., Rohde W. (1995). The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374: 727–730. [DOI] [PubMed] [Google Scholar]
- Nelissen H., Rymen B., Jikumaru Y., Demuynck K., Van Lijsebettens M., Kamiya Y., Inzé D., Beemster G.T. (2012). A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr. Biol. 22: 1183–1187. [DOI] [PubMed] [Google Scholar]
- O’Connor D.L., Runions A., Sluis A., Bragg J., Vogel J.P., Prusinkiewicz P., Hake S. (2014). A division in PIN-mediated auxin patterning during organ initiation in grasses. PLOS Comput. Biol. 10: e1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza P., et al. (2010). Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr. Biol. 20: 2223–2228. [DOI] [PubMed] [Google Scholar]
- Péret B., et al. (2012). AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (1984). Cellular parameters of leaf morphogenesis in maize and tobacco. In Contemporary Problems in Plant Anatomy, R.A. White and W.C. Dickison, eds (Orlando, FL: Academic Press), pp. 235–259. [Google Scholar]
- Ramirez J., Bolduc N., Lisch D., Hake S. (2009). Distal expression of knotted1 in maize leaves leads to reestablishment of proximal/distal patterning and leaf dissection. Plant Physiol. 151: 1878–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig C., Pozzi C., Santi L., Müller J., Wang Y., Stile M.R., Rossini L., Stanca M., Salamini F. (2004). Genetics of barley hooded suppression. Genetics 167: 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. (1969). Polarity and the induction of organized vascular tissues. Ann. Bot. (Lond.) 33: 263–275. [Google Scholar]
- Sachs T. (1981). The control of the patterned differentiation of vascular tissues. Adv. Bot. Res. 9: 151–262. [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger R.G., Becraft P.W., Hake S., Freeling M. (1995). Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev. 9: 2292–2304. [DOI] [PubMed] [Google Scholar]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., Sharon E., Ori N. (2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J., Ahlgren U., Perry P., Hill B., Ross A., Hecksher-Sørensen J., Baldock R., Davidson D. (2002). Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296: 541–545. [DOI] [PubMed] [Google Scholar]
- Sinha N.R., Williams R.E., Hake S. (1993). Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7: 787–795. [DOI] [PubMed] [Google Scholar]
- Smith L.G., Greene B., Veit B., Hake S. (1992). A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116: 21–30. [DOI] [PubMed] [Google Scholar]
- Smith R.S., Guyomarc’h S., Mandel T., Reinhardt D., Kuhlemeier C., Prusinkiewicz P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E., van Houwelingen A., Kloos D., Mol J., Koes R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85: 159–170. [DOI] [PubMed] [Google Scholar]
- Stebbins G.L., Yagil E. (1966). The morphogenetic effects of the hooded gene in barley. I. The course of development in hooded and awned genotypes. Genetics 54: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoma S., Lucas M., Chopard J., Schaedel M., Traas J., Godin C. (2008). Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLOS Comput. Biol. 4: e1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester A.W., Cande W.Z., Freeling M. (1990). Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000. [DOI] [PubMed] [Google Scholar]
- Teale W.D., Paponov I.A., Palme K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7: 847–859. [DOI] [PubMed] [Google Scholar]
- Vlad D., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Veit B., Sinha N., Hake S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243. [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R.E., Lie Y.S., Hake S., Lemaux P.G. (1997). Ectopic expression of the maize kn1 gene phenocopies the Hooded mutant of barley. Development 124: 3737–3745. [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.