Two RING E3 ligases mediate the degradation of the phosphatase PP2CA, a repressor of abscisic acid signaling, thereby modulating the activation of the ABA pathway in Arabidopsis.
Abstract
Abscisic acid (ABA) is an essential hormone for plant development and stress responses. ABA signaling is suppressed by clade A PP2C phosphatases, which function as key repressors of this pathway through inhibiting ABA-activated SnRK2s (SNF1-related protein kinases). Upon ABA perception, the PYR/PYL/RCAR ABA receptors bind to PP2Cs with high affinity and biochemically inhibit their activity. While this mechanism has been extensively studied, how PP2Cs are regulated at the protein level is only starting to be explored. Arabidopsis thaliana RING DOMAIN LIGASE5 (RGLG5) belongs to a five-member E3 ubiquitin ligase family whose target proteins remain unknown. We report that RGLG5, together with RGLG1, releases the PP2C blockade of ABA signaling by mediating PP2CA protein degradation. ABA promotes the interaction of PP2CA with both E3 ligases, which mediate ubiquitination of PP2CA and are required for ABA-dependent PP2CA turnover. Downregulation of RGLG1 and RGLG5 stabilizes endogenous PP2CA and diminishes ABA-mediated responses. Moreover, the reduced response to ABA in germination assays is suppressed in the rglg1 amiR (artificial microRNA)-rglg5 pp2ca-1 triple mutant, supporting a functional link among these loci. Overall, our data indicate that RGLG1 and RGLG5 are important modulators of ABA signaling, and they unveil a mechanism for activation of the ABA pathway by controlling PP2C half-life.
INTRODUCTION
The plant hormone abscisic acid (ABA) regulates many key processes in plants, including seed germination and development and various biotic and abiotic stress responses (Cutler et al., 2010; Finkelstein, 2013). The ABA signaling pathway is initiated by ABA perception through the PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) family of proteins (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Nishimura et al., 2010). This is followed by interaction with and inactivation of clade A protein phosphatase type 2Cs (PP2Cs), such as ABA INSENSITIVE1 (ABI1) and ABI2, HYPERSENSITIVE TO ABA1 (HAB1) and HAB2, and PROTEIN PHOSPHATASE 2CA/ABA-HYPERSENSITIVE GERMINATION3 (PP2CA/AHG3), thereby releasing their inhibition on three ABA-activated SNF1-related protein kinases (SnRK2s), i.e., SnRK2.2/D, 2.3/I and 2.6/E/OST1 (Umezawa et al., 2009; Vlad et al., 2009). These SnRK2s then activate downstream signaling by phosphorylating numerous players, including ABA-responsive transcription factors (Fujii et al., 2009; Fujii and Zhu, 2009; Nakashima et al., 2009), ion channels (Geiger et al., 2009; Lee et al., 2009), and other mediators/effectors involved in ABA signaling and action (Umezawa et al., 2013; Wang et al., 2013).
To optimize the allocation of resources between growth/development and stress responses, plants need to control the timing and extent of ABA pathway activation. Previous work has indicated that posttranscriptional modifications such as phosphorylation (Kobayashi et al., 2005; Hubbard et al., 2010; Cai et al., 2014) and ubiquitination (Zhang et al., 2005; Liu and Stone, 2010, 2011; Kelley and Estelle, 2012) are important mechanisms to modulate ABA signaling. The ABA-PYR/PYL/RCARs-PP2Cs-SnRK2s core ABA pathway involves multiple members with redundant and nonredundant functions (Park et al., 2009; Fujii and Zhu, 2009; Nakashima et al., 2009; Rubio et al., 2009; Antoni et al., 2013; Zhao et al., 2014), which might function in different combinations depending on various environmental stimuli, developmental stages, or cell types (Santiago et al., 2009; Szostkiewicz et al., 2010; Gonzalez-Guzman et al., 2012; Antoni et al., 2012). Recently, studies that address protein dynamics of core ABA signaling components have been published (Bueso et al., 2014; Irigoyen et al., 2014; Kong et al., 2015; reviewed in Yu et al., 2016). However, a comprehensive understanding of the mechanisms and components that regulate receptor and clade A PP2C protein levels is still lacking, as well as their contribution to the modulation of ABA signaling at different times and developmental stages. For instance, the transcription of some PYR/PYL/RCARs is repressed, whereas that of PP2Cs is stimulated, in response to ABA (Santiago et al., 2009; Szostkiewicz et al., 2010), indicating the existence of a negative feedback transcriptional mechanism to modulate ABA signaling by controlling transcript levels of core elements. In the case of ABI1, it has been demonstrated that ABA induces degradation of this PP2C through PUB12/13 E3 ligases but subsequently upregulates ABI1 expression and protein levels (Kong et al., 2015). PUB13-mediated ABI1 ubiquitination in the presence of PYR1 is strictly dependent on ABA, whereas in the presence of monomeric receptors, ABA only increases ABI1 ubiquitination levels (Kong et al., 2015). Recent work also indicates that ABA receptor proteins, e.g., PYR1, PYL4, and PYL8, can be degraded via an ubiquitination-dependent mechanism through single subunit and CUL4-based E3 ligases, although PYL8 can be protected from degradation by ABA (Bueso et al., 2014; Irigoyen et al., 2014). Ubiquitination of ABA receptors in the nucleus leads to proteasome-dependent degradation, whereas ubiquitination at the plasma membrane leads to vacuolar degradation (Irigoyen et al., 2014; Belda-Palazon et al., 2016).
In Arabidopsis thaliana, the RING-type E3 ubiquitin ligase family, named RGLG (RING DOMAIN LIGASE), is composed of five members, i.e., RGLG1 to 5 (Yin et al., 2007). RGLG1 and RGLG2 have been reported to affect hormone signaling, since the rglg1 rglg2 double mutant shows altered auxin and cytokinin levels (Yin et al., 2007). RGLG2 catalyzes the formation of ubiquitin Lys-63-linked chains when combined with the heterodimeric E2 enzyme formed by MMZ2 and UBC35 (Yin et al., 2007). Recently, it was reported that RGLG3 and RGLG4 are essential regulators of the jasmonate pathway (Zhang et al., 2012, 2015). However, in these studies, the molecular targets of RGLG1-4 were not identified.
In this study, we found that PP2CA, a key negative regulator of ABA signaling (Sheen, 1998; Kuhn et al., 2006; Yoshida et al., 2006; Rubio et al., 2009; Lee et al., 2009; Brandt et al., 2015), is a target of RGLG1 and RGLG5. ABA enhances the interaction of RGLG1 and RGLG5 with PP2CA, ABI2, and HAB2 and mediates in vitro ubiquitination of these PP2Cs. Both loss-of-function and gain-of-function phenotypes of RGLG1 and RGLG5 reveal their role as positive regulators of ABA signaling in different ABA responses. In particular, we show that RGLG1 and RGLG5 mediate the ubiquitination of PP2CA in vitro and in vivo, which leads to ABA-enhanced turnover of PP2CA. Using PP2CA antibodies, we demonstrate that in seedlings treated with the protein synthesis inhibitor cycloheximide (CHX), ABA induces the degradation of endogenous PP2CA. Thus, our work further uncovers a mechanism to control the activation of the ABA pathway by degradation of repressors of ABA signaling.
RESULTS
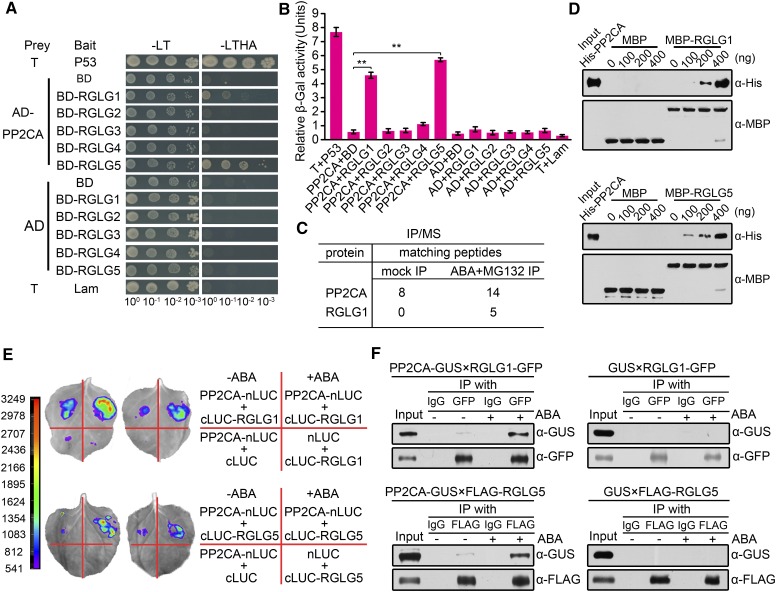
Identification of PP2CA as Interacting Partner of Ubiquitin Ligases RGLG1 and RGLG5
RGLG5 belongs to the RGLG E3 ligase family (Yin et al., 2007). To date, the biological function and targets of RGLG5 remain unclear. We aimed to identify its interacting partners to elucidate RGLG5 function and accordingly, we employed the yeast two-hybrid (Y2H) approach to screen an Arabidopsis cDNA library using RGLG5 as bait. As a result, PP2CA, a key repressor of the ABA pathway (Kuhn et al., 2006; Yoshida et al., 2006; Rubio et al., 2009), was identified as an interacting partner of RGLG5. To ascertain whether other RGLG family members interact with PP2CA, the five RGLGs were each coexpressed with PP2CA in yeast for Y2H screening. Both growth assays in selective medium (Figure 1A) and β-galactosidase quantitative assays (Figure 1B) revealed that PP2CA interacted with both RGLG1 and RGLG5. Expression in yeast cells of PP2CA fused to the GAL4 activation domain (AD) and RGLG1-5 proteins fused to the GAL4 DNA binding domain (BD) was verified by immunoblot analysis (Supplemental Figure 1A). Interestingly, other PP2Cs, namely, ABI2 and HAB2, also showed interactions with RGLG1 and RGLG5, but not ABI1 and HAB1, indicating a certain selectivity of these E3 ligases to recognize clade A PP2Cs (Figure 2A; Supplemental Figure 1B). To better address the functional importance of these interactions, we focused on PP2CA for in-depth study. To investigate whether endogenous RGLGs form an in vivo complex with PP2CA, we utilized a coimmunoprecipitation (coIP)/mass spectrometry approach to identify proteins that coimmunoprecipitated with FLAG-tagged PP2CA expressed in Arabidopsis (35Spro:FLAG-PP2CA). In protein extracts obtained from 2-week-old seedlings after 6 h ABA and 30 h of treatment with the proteasome inhibitor MG132, but not in mock-treated samples, native RGLG1 coimmunoprecipitated with FLAG-PP2CA, suggesting ABA promotes an in vivo association of PP2CA and RGLG1 (Figure 1C; Supplemental Table 1). RGLG1 expression is induced by ABA treatment and is markedly higher than RGLG5 in seedlings (Supplemental Figure 2), which might explain the failure to recover RGLG5 peptides. However, both RGLG1 and RGLG5 expression was induced to similar levels by cold, osmotic, or salt stress in roots (Supplemental Figure 2), suggesting that both genes play a relevant role in this tissue.
Figure 1.
In Vitro and in Vivo Interaction of PP2CA with Ubiquitin Ligases RGLG1 and RGLG5.
(A) Interaction of PP2CA with RGLGs in yeast two-hybrid assays. Photographs were taken after 5 d of yeast cell growth on Leu–Trp–/Yeast Nitrogen Base (YNB) or Leu–Trp–His–Ade–/YNB medium. T+Lam and T+P53 were used as negative and positive controls for the yeast two-hybrid system, respectively.
(B) Quantitative analysis of interactions in (A) by measuring β-galactosidase (β-gal) activities. o-Nitrophenyl-β-d-galactopyranoside was used as substrate. Bars indicate the mean ± sd from three replicates. Asterisks indicate a significant difference from the negative control pair AD-PP2CA+BD (Student’s t test: **P < 0.01).
(C) Number of PP2CA and RGLG peptides identified by coIP/mass Spectrometry. Total proteins were extracted from 35Spro:3×FLAG-PP2CA seedlings treated without (mock) or with ABA and MG132. Immunoprecipitation (IP) was performed using anti-FLAG and the immunoprecipitation products were analyzed by mass spectrometry.
(D) In vitro interaction of PP2CA with RGLG1 and RGLG5. Serial concentrations of His-tagged PP2CA were incubated with immobilized MBP-RGLG1, MBP-RGLG5, or MBP proteins. Bound proteins were detected by immunoblot using anti-His and anti-MBP antibodies.
(E) Interaction between PP2CA and RGLG1 or RGLG5 in firefly luciferase complementation imaging assay. Construct pairs PP2CA-nLUC/RGLGs-cLUC, PP2CA-nLUC/cLUC, and nLUC/RGLGs-cLUC were coexpressed in N. benthamiana leaves. ABA (50 µM) was injected 3 h before LUC observation. Two representative results are shown, and color scale reflects LUC activity.
(F) Coimmunoprecipitation of PP2CA with RGLG1 and RGLG5. GUS-tagged PP2CA and GFP-tagged RGLG1 or FLAG-tagged RGLG5 were coexpressed in N. benthamiana. Protein extracts were incubated with (+) or without (−) 10 µM ABA for 3 h before immunoprecipitation using GFP or FLAG antibodies bounded to Protein G-Sepharose beads. IgG was added in parallel as negative controls.
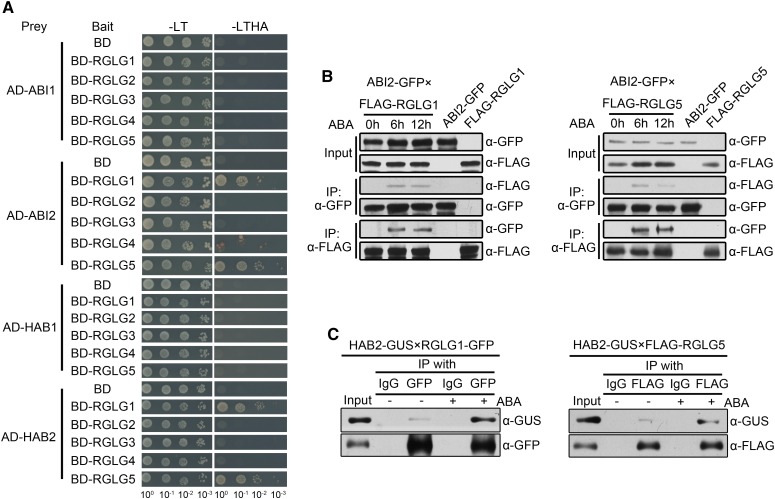
Figure 2.
Interaction Assays of Clade A PP2Cs with RGLGs.
ABI2 and HAB2 interact with RGLG1 and RGLG5 in an ABA-dependent manner.
(A) Yeast-two-hybrid analysis of interactions between PP2Cs and RGLGs. Full-length PP2Cs were each cloned into pGADT7Rec plasmid and used as preys; full-length RGLGs were inserted separately into pGBKT7 plasmid and used as baits. Each prey and bait pair was then cotransformed into yeast. Photographs were taken after 5 d of yeast cell growth on Leu–Trp–/Yeast Nitrogen Base (YNB) and Leu–Trp–His–Ade–/YNB medium.
(B) ABA-enhanced interaction of ABI2 with RGLG1 and RGLG5 in Arabidopsis. Two-week-old transgenic Arabidopsis seedlings coexpressing ABI2-GFP and FLAG-RGLG1 or FLAG-RGLG5 were transferred to liquid MS medium supplemented with 50 µM ABA and sampled at the indicated time points. Immunoprecipitation was performed using anti-FLAG (IP: α-FLAG) or anti-GFP (IP: α-GFP) antibodies bounded to Protein G-Sepharose beads, separately. Anti-GFP and anti-FLAG antibodies were used to detect the proteins in total extracts (Input) and immunoprecipitates (IP).
(C) ABA-enhanced interaction of HAB2 with RGLG1 and RGLG5. HAB2-GUS and RGLG1-GFP or FLAG-RGLG5 fusions were coexpressed in N. benthamiana leaves by infiltration. Protein extracts were incubated with (+) or without (−) 10 µM ABA for 3 h before immunoprecipitation with anti-GFP or anti-FLAG antibodies bound to Protein G-Sepharose beads. IgG was added in parallel as negative controls. Total extracts (Input) and immunoprecipitates (IP) were subjected to immunoblot analysis using anti-GUS, anti-GFP, or anti-FLAG antibodies.
We employed additional strategies to confirm the PP2CA-RGLG1 interaction detected by Y2H and coIP/mass spectrometry analysis and to further study the interaction of RGLG5 with PP2CA. First, PP2CA interaction with RGLG1 and RGLG5 was confirmed by pull-down assays using recombinant purified proteins (Figure 1D). These in vitro assays were performed in the absence of ABA and revealed a basal affinity of PP2CA for RGLG1 and RGLG5. Then, to test whether they associate in plant cells, PP2CA and RGLG1 or RGLG5 were fused to the N-terminal fragment of luciferase (LUC) (nLUC-PP2CA) and the C-terminal fragment of LUC (RGLG1-cLUC or RGLG5-cLUC), respectively, and coexpressed in wild tobacco (Nicotiana benthamiana). Compared with the negative control that expressed only nLUC-PP2CA+cLUC or nLUC+RGLG1/5-cLUC, RGLG1 and RGLG5 showed an interaction with PP2CA in the absence of exogenous ABA addition, but after ABA supplementation, this interaction was markedly enhanced, suggesting ABA favored PP2CA-RGLG1 and PP2CA-RGLG5 interactions in planta (Figure 1E).
Although the in vitro interaction of RGLG1/RGLG5 and PP2CA does not require exogenous ABA addition, results obtained in planta suggest that additional plant components might be involved in the ABA-enhanced interaction observed in LUC assays. To further substantiate this concept, when GUS-tagged PP2CA was coexpressed with either GFP-tagged RGLG1 or FLAG-tagged RGLG5 in N. benthamiana, coIP assays indicated that in the absence of ABA, the interaction was weak, but it was dramatically enhanced by incubating the protein extracts with 10 µM ABA during coIP (Figure 1F). Similar results were obtained after testing their interaction with ABI2 and HAB2 (Figures 2B and 2C), which suggests that RGLG1 and RGLG5 might target several clade A PP2Cs in an ABA-dependent manner. Altogether, our data suggest that the in planta interaction of the E3 ligases RGLG1 and RGLG5 with some clade A PP2Cs is strongly enhanced by ABA.
Loss of Function of RGLG1 and RGLG5 Represses ABA-Mediated Responses
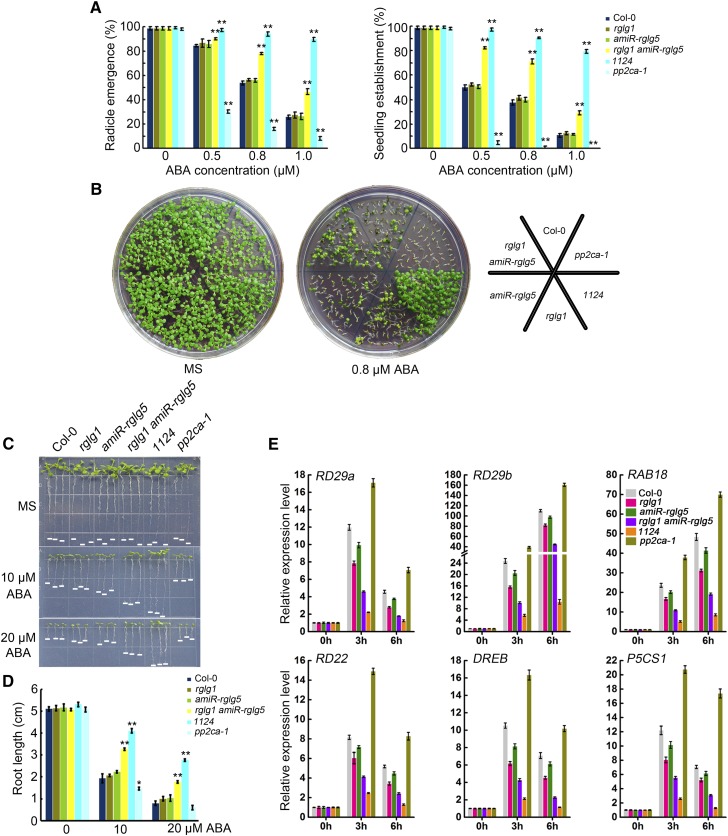
The above-mentioned interactions suggest that RGLG1 and RGLG5 might play roles in the ABA pathway as regulators of some clade A PP2Cs. Since these PP2Cs are key negative regulator of ABA signaling and a potential target of these E3 ligases, we predicted that impaired or enhanced expression of RGLG1/5 might affect ABA signaling. Thus, to test whether RGLG1 and RGLG5 modulate ABA-mediated responses, loss-of-function mutants and overexpression lines were used. rglg1 was kindly provided by A. Bachmair (Yin et al., 2007), but for RGLG5, we could not confirm T-DNA insertion in several lines requested from public stock centers (Salk_021379 and CS812692 from ABRC, and 751D11 from MPIZ), and gene expression analyses indicated that RGLG5 transcript levels were not reduced in these lines. Therefore, we generated knockdown mutant lines of RGLG5 by designing a 35S artificial microRNA (amiR) construct (Supplemental Figure 3A). We examined the transcript levels of RGLG5 in several lines by RT-qPCR and selected two lines (#2 and #4) whose RGLG5 expression was below 20% of the wild type, whereas the expression of RGLG1-4 was not significantly affected (Supplemental Figures 3B and 3C). We generated an rglg1 amiR-rglg5 double mutant and analyzed several ABA-mediated responses, including inhibition of seed germination, seedling establishment, and root growth in rglg1 amiR-rglg5 compared with other genetic backgrounds (Figures 3A to 3E). The well-studied pp2ca-1 and pyr1 pyl1 pyl2 pyl4 (1124) mutants served as internal controls of ABA-hypersensitive and ABA-insensitive phenotypes, respectively, and demonstrated the effectiveness of the ABA treatment (Kuhn et al., 2006; Yoshida et al., 2006; Park et al., 2009). None of the single rglg1 or amiR-rglg5 mutants had obvious differences in the above-mentioned ABA responses compared with the wild type (Figures 3A to 3E). However, the rglg1 amiR-rglg5 double mutant showed diminished ABA sensitivity compared with the wild type, suggesting partial functional redundancy of RGLG1 and RGLG5 in ABA-mediated responses.
Figure 3.
Diminished ABA Sensitivity of the rglg1 amiR-rglg5 Double Mutant.
(A) Seed germination of Col-0, rglg1, rglg5, amiR-rglg5, rglg1 amiR-rglg5, pp2ca-1, and 1124. Radicle emergence was scored after 72-h growth on MS plates containing the indicated concentrations of ABA. Seedling establishment was scored as the percentage of seeds that developed fully green expanded cotyledons after 7-d growth on MS plates containing the indicated concentrations of ABA. Bars are mean ± sd of three replications. Asterisks indicate a significant difference from wild type (Student’s t test: **P < 0.01).
(B) Photographs of representative seedlings as in (A) taken 9 d after sowing on MS plates with or without 0.8 µM ABA.
(C) ABA-mediated inhibition of root growth in Col-0, rglg1, rglg5, amiR-rglg5, rglg1 amiR-rglg5, pp2ca-1, and 1124. Photos were taken 10 d after transferring 3-d-old seedlings to MS plates supplemented without or with the indicated concentrations of ABA.
(D) Quantification of primary root lengths in the indicated genetic backgrounds after ABA treatment indicated in (C). Bars indicate mean ± sd of 30 seedlings. Asterisks indicate a significant difference from wild type (Student’s t test: *P < 0.05; **P < 0.01).
(E) Expression profiles of ABA-responsive genes in Col-0, rglg1, rglg5, amiR-rglg5, rglg1 amiR-rglg5, pp2ca-1, and 1124. Seven-day-old Arabidopsis seedlings were transferred to liquid MS medium containing 50 µM ABA and sampled at the indicated time points. Gene expression was examined using quantitative RT-PCR. UBQ10 was used as the internal control, and expression levels were normalized to that measured at time 0. Data represent the mean ± sd of four technical replicates.
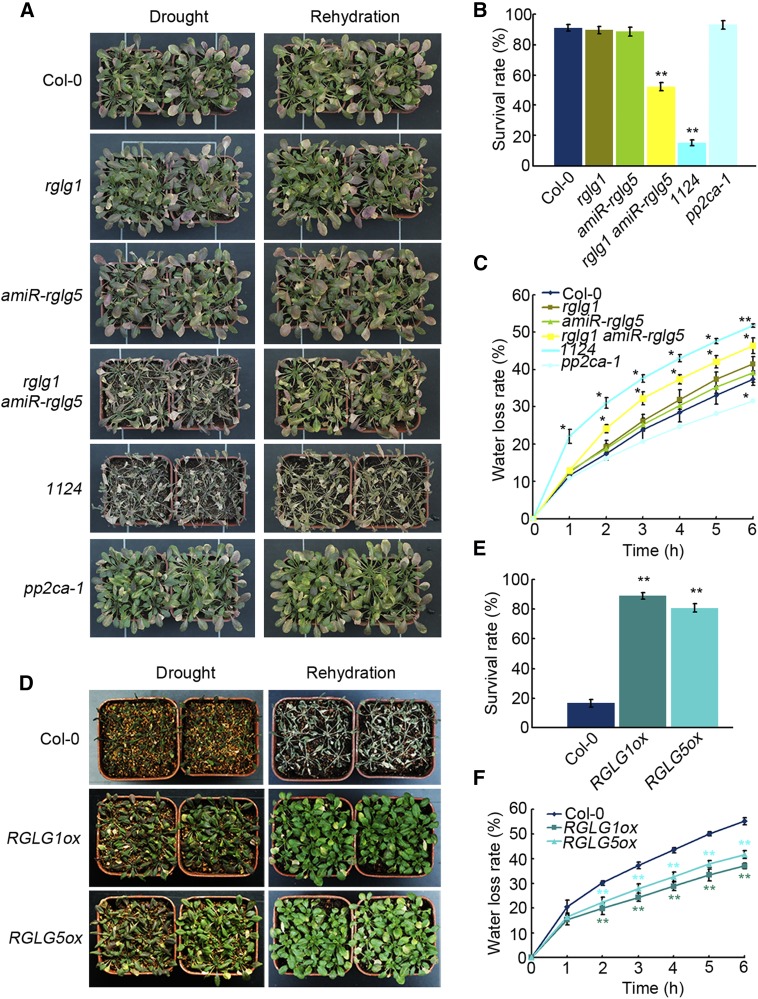
Adult plants of the wild type, single rglg1, or amiR-rglg5 and rglg1 amiR-rglg5 double mutant were grown under well-watered conditions, and no obvious growth differences were observed among them (Figure 4; Supplemental Figure 3D). However, detached leaves of rglg1 amiR-rglg5 showed higher water loss compared with the wild type or single mutants, and under drought stress conditions, the rglg1 amiR-rglg5 double mutant showed reduced survival (Figures 4A to 4C), which is in agreement with its reduced ABA sensitivity. Conversely, the overexpression of either RGLG1 or RGLG5 reduced water loss and markedly enhanced plant survival under drought stress compared with the wild type (Figures 4D to 4F). Overexpression of either RGLG1 or RGLG5 led to enhanced ABA sensitivity in seed germination, seedling establishment, and root growth assays, as well as increased ABA responses under NaCl stress compared with the wild type (Supplemental Figures 4A to 4F). Finally, the induction of several ABA-responsive genes, including RD29b (Nordin et al., 1993), RD22 (Abe et al., 2003), RAB18 (Lång and Palva, 1992) and P5CS1 (Strizhov et al., 1997), was diminished by downregulation of both RGLG1 and RGLG5 (Figure 3E), while it was enhanced by RGLG1 and RGLG5 overexpression (Supplemental Figure 4G). Altogether, these data demonstrate that both RGLG1 and RGLG5 are positive regulators of ABA signaling in different ABA-mediated responses.
Figure 4.
Water Loss and Drought Tolerance of rglg1 amiR-rglg5 and RGLG1/5 Overexpressing Plants.
Enhanced water loss and drought sensitivity were observed in rglg1 amiR-rglg5 compared with the wild type or single mutants, whereas diminished water loss and enhanced drought resistance were observed in plants overexpressing RGLG1 or RGLG5 compared with Col-0.
(A) Drought sensitivity of Col-0, rglg1, amiR-rglg5, rglg1 amiR-rglg5, pp2ca-1, and 1124 plants. Three-week-old plants were subjected to drought stress by withholding watering (Drought) for 12 d (when the lethal effect was observed in the double mutant plants), followed by rewatering (Rehydration).
(B) Survival rates of plants in (A). Survival rates were recorded 3 d after rehydration. Bars indicate sd calculated from three replicated experiments. Asterisks indicate significant difference from the wild type (Student’s t test: **P < 0.01).
(C) Water loss rates of detached leaves of 4-week-old plants as indicated in (A). Fresh weights were monitored at the indicated time. Bars indicate sd calculated from three replications. Asterisks indicate a significant difference from wild type (Student’s t test: *P < 0.05; **P < 0.01).
(D) Enhanced drought tolerance of Arabidopsis plants overexpressing RGLG1 or RGLG5 compared with Col-0. Three-week-old plants were subjected to drought stress by withholding water (Drought) for 19 d (when the lethal effect was observed in the wild-type plants), followed by rewatering (Rehydration).
(E) Percentage of plants that survived the treatment mentioned in (D). Survival rate was recorded 3 d after rehydration. Bars indicate sd calculated from three independent experiments. Asterisks indicate significant difference from the wild type (Student’s t test: **P < 0.01).
(F) Water loss rate of detached leaves of 4-week-old RGLG1 or RGLG5 overexpressing plants compared with the wild type (Col-0). Fresh weight was monitored at the indicated time. Bars indicate sd calculated from three replications. Asterisks indicate a significant difference from wild type (Student’s t test: **P < 0.01).
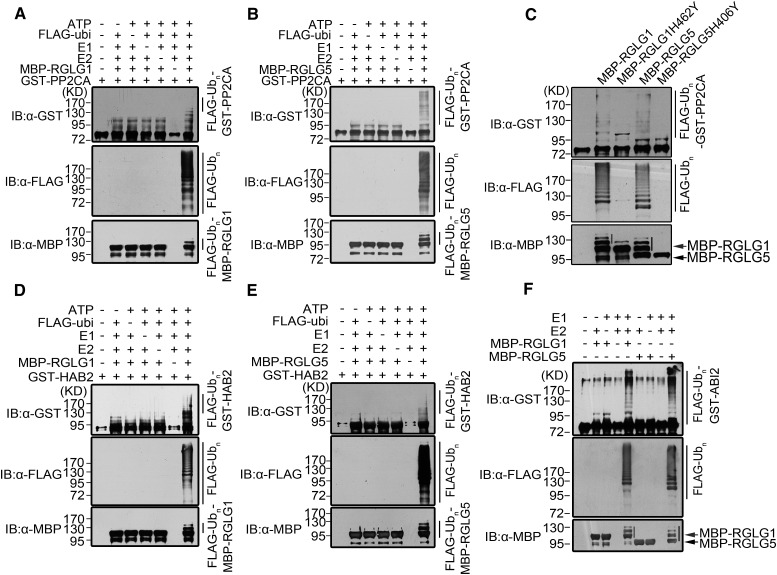
RGLG1 and RGLG5 Mediate Ubiquitination of the Repressor PP2CA in Vitro
Since not all RING-domain proteins possess E3 ligase activity (Deshaies and Joazeiro, 2009), it was important to determine whether RGLG5 indeed has ubiquitin ligase activity. Using purified GST-RGLG5, we showed that RGLG5 could target itself for ubiquitination and that the intensity of the ubiquitination band could be enhanced by increasing the reaction time (Supplemental Figure 5A). Moreover, RGLG5 self-ubiquitination was abolished either by the lack of any component of the ubiquitination reaction or by mutating (H406Y) or deleting the RING domain (Supplemental Figure 5B). Self-ubiquitination of RGLG1 was also confirmed in subsequent experiments (for instance, Supplemental Figure 5C, panel anti-MBP). We then asked whether RGLG1 and RGLG5 could directly target PP2CA for ubiquitination. Purified MBP-RGLG1 and MBP-RGLG5 were incubated with GST-PP2CA in the presence of all other ubiquitination components. As a result, ubiquitination of PP2CA was promoted by MBP-RGLG1 and MBP-RGLG5 in a dose-dependent manner (Figures 5A and 5B; Supplemental Figure 5C), whereas the lack of any component of the reaction abolished PP2CA ubiquitination (Figures 5A and 5B; Supplemental Figure 5C). Therefore, both RGLG1 and RGLG5 promoted ubiquitination of PP2CA in vitro, whereas RING domain-mutated forms of RGLG1 (H462Y) and RGLG5 (H406Y) lost the ability to promote PP2CA ubiquitination (Figure 5C). Similar results were obtained after incubating ABI2 or HAB2 with these two RGLGs (Figures 5D to 5F). Taken together, these data indicate that PP2CA as well as ABI2 and HAB2 can be ubiquitinated in vitro by the E3 ligases RGLG1 and RGLG5.
Figure 5.
RGLG1- and RGLG5-Mediated in Vitro Ubiquitination of PP2CA.
(A) and (B) In vitro ubiquitination of PP2CA by RGLG1 and RGLG5, respectively. Purified GST-PP2CA was incubated with ATP, FLAG-Ubiquitin, E1, E2, and MBP-RGLG1 or MBP-RGLG5 in an in vitro ubiquitination reaction. Each component was omitted from the assays as a control. The ubiquitination products were examined by immunoblot (IB) using anti-GST, anti-FLAG, and anti-MBP antibodies.
(C) In vitro ubiquitination of PP2CA by wild-type RGLG1 and RGLG5 compared with their Ring domain-mutated forms. Ubiquitinated bands were detected as in (A). The first lane contains uniquely purified GST-PP2CA. Arrows (in [C] and [F]) indicate the position of MBP-RGLG1 (red) and MBP-RGLG5 (black). IB using anti-MBP detects self-ubiquitination of MBP-RGLG1 (red line) and MBP-RGLG5 (black line).
(D) and (E) In vitro ubiquitination of HAB2 by RGLG1and RGLG5, respectively.
(F) In vitro ubiquitination of ABI2 by RGLG1 and RGLG5.
Since the in planta interaction of the E3 ligases RGLG1 and RGLG5 with PP2CA is strongly enhanced by ABA (Figure 1E), we investigated whether ABA or the ABA receptor PYL4 could modify in vitro ubiquitination of PP2CA (Supplemental Figure 6). In vitro ubiquitination of GST-PP2CA was not affected by the addition of ABA, PYL4, or both (Supplemental Figure 6). This result confirms the basal affinity of RGLG1 and RGLG5 for PP2CA, and it suggests that additional plant components, changes in their subcellular location, or changes in protein levels induced by ABA (for instance ABA-induced upregulation of PP2CA transcripts) might influence the interaction of PP2CA and RGLG1/RGLG5 in vivo.
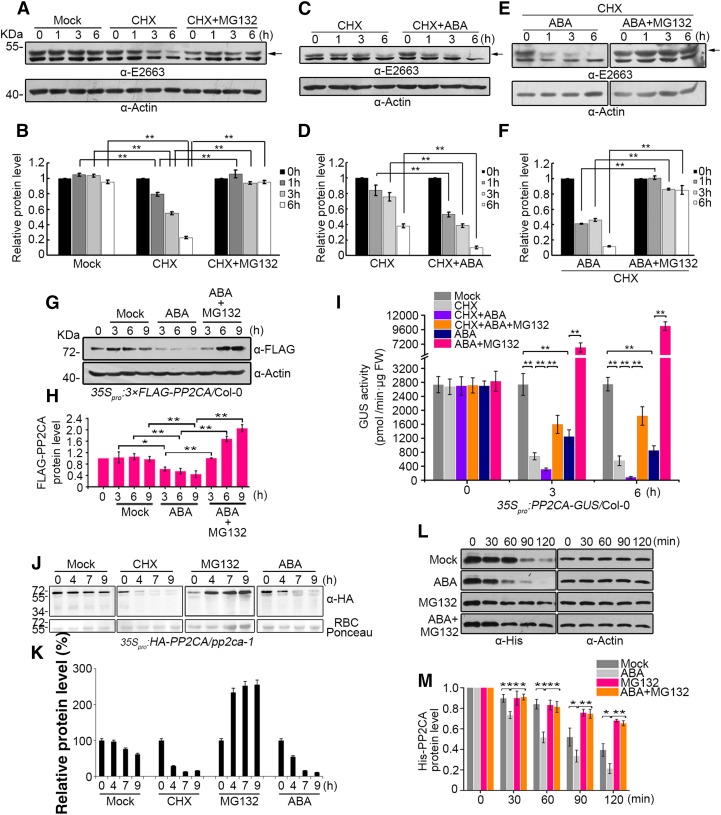
PP2CA Is Subjected to 26S Proteasome Degradation and ABA Enhances PP2CA Degradation
The ubiquitination of PP2CA promoted by RGLG1 and RGLG5 might induce its degradation via the 26S proteasome. Despite its key role in ABA signaling, the protein levels and stability of PP2CA have not been investigated. It is well known that ABA induces transcriptional upregulation of clade A PP2Cs (Santiago et al., 2009; Fujita et al., 2009; Szostkiewicz et al., 2010; Supplemental Figure 7), and recent results from Kong et al. (2015) confirmed that ABI1 protein levels are strongly increased by a 6-h ABA treatment. Since clade A PP2Cs are key negative regulators of ABA signaling, their ABA-mediated upregulation acts as a negative feedback mechanism to reset and desensitize ABA signaling (Santiago et al., 2009). Both GUS histochemical staining and immunoblot analysis revealed low expression of the PP2CA promoter in the absence of ABA, which was markedly upregulated in both leaf and root tissues upon ABA treatment (Supplemental Figures 7A and 7B). This might make it difficult to detect endogenous PP2CA under normal growth conditions and, additionally, the expected molecular weight of PP2CA is close to that of the Rubisco large subunit. We therefore generated a PP2CA polyclonal antibody using truncated (1 to 200 amino acid residues) PP2CA as antigen to immunize rabbit, and by immunoblot analysis, we could detect PP2CA in etiolated seedlings (see Supplemental Figure 8 for PP2CA antibody specificity). We found that ABA treatment upregulated PP2CA protein levels, which is in agreement with the upregulation of PP2CA transcripts and the activation of PP2CA promoter (Supplemental Figures 7 and 8). To investigate PP2CA protein dynamics, we analyzed endogenous PP2CA levels in mock-, CHX-, and CHX+MG132-treated seedlings (Figures 6A and 6B). These experiments revealed a turnover of PP2CA mediated by the 26S proteasome pathway. Next, we compared PP2CA protein stability in CHX-treated seedlings in the absence or presence of exogenous ABA. ABA treatment, in the absence of protein synthesis, enhanced PP2CA degradation (Figures 6C and 6D). Addition of the proteasome inhibitor MG132 blocked PP2CA degradation, which indicated that ABA-enhanced PP2CA degradation also occurs in the 26S proteasome (Figures 6E and 6F).
Figure 6.
ABA Enhances PP2CA Degradation Mediated by the 26S Proteasome Pathway.
(A) PP2CA is degraded by the 26S proteasome. Two-week-old etiolated seedlings were transferred to liquid MS medium and were mock, 100 µM CHX, or 100 µM CHX+50 µM MG132 treated for the indicated time period. Total protein extracts were prepared for immunoblot analysis. The α-E2663 polyclonal antibody of PP2CA was used to determine the endogenous PP2CA protein (arrow) level in (A), (C), and (E). Actin was used as a loading control in (A), (C), and (E) and to normalize PP2CA protein levels.
(B) Quantification of anti-PP2CA and anti-actin signal intensity in (A) using Image J software. The relative protein levels of PP2CA at 0 h were set as 1. Bars show mean PP2CA levels normalized to actin protein. Values are averages ± sd of three independent experiments. Asterisks indicate a significant difference between the indicated comparisons (Student’s t test: **P < 0.01).
(C) ABA enhances PP2CA degradation. Two-week-old etiolated seedlings were treated with 100 µM CHX or 100 µM CHX+50 µM ABA for the indicated time period.
(D) Quantification of anti-PP2CA and anti-actin signal intensity in (C) using Image J software.
(E) ABA enhances PP2CA degradation through the 26S proteasome. Two-week-old etiolated Col-0 seedlings were treated with 100 µM CHX+50 µM ABA or 100 µM CHX+50 µM ABA+50 µM MG132 for the indicated time period.
(F) Quantification of anti-PP2CA and anti-actin signal intensity in (E) using Image J software.
(G) PP2CA protein dynamics after different treatments determined by immunoblot analysis. Two-week-old seedlings constitutively overexpressing 3×FLAG-PP2CA were transferred to liquid MS medium and were mock, 50 µM ABA, or 50 µM ABA+50 µM MG132 treated and sampled at the indicated time points. Immunoblot was performed using anti-FLAG antibody. Actin was used to normalize PP2CA protein levels.
(H) Three-independent experiments as in (G) were performed and the signal intensity was determined by ImageJ software. The protein levels of FLAG-PP2CA without treatment (0 h) were set as 1. Bars show mean FLAG-PP2CA levels normalized to actin protein. Values are averages ± sd of three independent experiments. Student’s t tests were used to determine significant differences in the indicated comparisons. *P < 0.05; **P < 0.01.
(I) Quantified GUS activities in protein extracts prepared from plants expressing PP2CA-GUS submitted to different treatments. Seven-day-old seedlings were mock treated or treated with 50 µM ABA, 50 µM MG132, 100 µM CHX, or the indicated combinations. Samples were harvested at the indicated time points for measuring GUS activity. Bars indicate the mean ± sd from three independent experiments. Student’s t tests were used to determine significant differences in the indicated comparisons. *P < 0.05; **P < 0.01.
(J) Effect of CHX, MG132, and ABA treatment on HA-PP2CA protein levels in the pp2ca-1 background. Ten-day-old seedlings expressing HA-tagged PP2CA were either mock, 100 µM CHX, 50 µM MG132, or 50 µM ABA treated for the indicated time period. Immunoblot analysis using anti-HA was performed to quantify HA-PP2CA levels.
(K) Quantification of anti-HA and Ponceau staining of Rubisco in (J) using Image J software. Bars show mean HA-PP2CA levels normalized to Rubisco protein. Values are averages ± se of three independent experiments.
(L) In vitro degradation of His-PP2CA. Whole cell extracts prepared from 7-d-old Col-0 seedlings were incubated with purified His-PP2CA at 15°C supplemented with or without different combinations of ABA and MG132 and sampled at the indicated time points. Anti-His antibody was used to detect protein dynamics, and anti-actin antibody was used to normalize His-PP2CA protein levels.
(M) Quantification of anti-HisPP2CA and anti-actin signal intensity in (L) using Image J software. The protein levels of His-PP2CA in Col-0 extracts with the indicated treatments at 0 min were set as 1. Bars show mean HisPP2CA levels normalized to actin protein. Values are averages ± sd of three independent experiments. Student’s t tests were used to determine significant differences in the indicated comparisons. *P < 0.05; **P < 0.01.
In order to avoid the use of CHX prior to ABA-treatment, which was required above to prevent ABA-mediated upregulation of the PP2CA promoter, we took advantage of 35Spro:FLAG-PP2CA transgenic lines. Direct measurements of PP2CA protein levels by immunoblot analysis showed that ABA stimulated PP2CA degradation via the 26S proteasome pathway, since coincubation with MG132 prevented this effect (Figures 6G and 6H). Similar results were obtained in a GUS activity assay using Arabidopsis protein extracts prepared from 35Spro:PP2CA-GUS transgenic lines submitted to different treatments (Figure 6I; Supplemental Figure 9). Moreover, we also generated 35Spro:HA-PP2CA lines in the pp2ca-1 background, and protein stability studies also revealed ABA-enhanced degradation of PP2CA, while continuous synthesis of PP2CA in the absence of degradation led to accumulation of the protein (Figures 6J and 6K). Taken together, these results reveal the turnover of PP2CA through the 26S proteasome, which is enhanced by ABA. Finally, we set up an in vitro cell-free degradation assay using purified His-PP2CA. We found that degradation of His-PP2CA was enhanced by exogenous addition of ABA compared with mock-treated plants, which was attenuated by adding MG132 (Figures 6L and 6M).
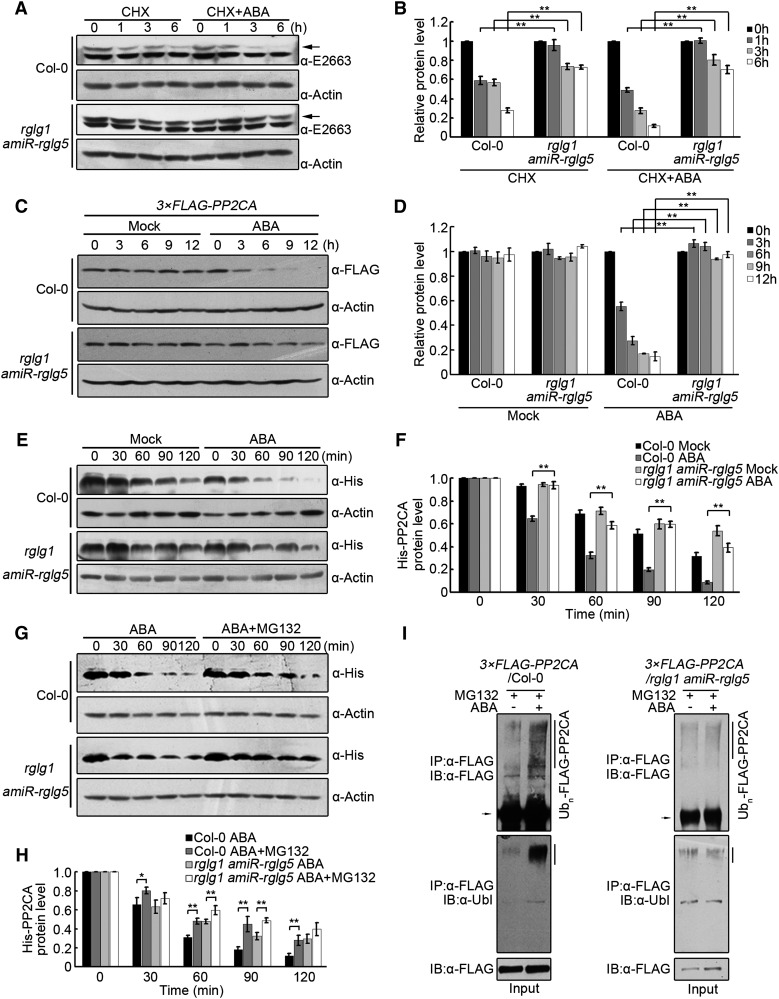
RGLG1 and RGLG5 Mediate ABA-Dependent PP2CA Degradation
To determine whether RGLG1 and RGLG5 mediate protein turnover of PP2CA, endogenous PP2CA protein levels were compared in wild-type and rglg1 amiR-rglg5 seedlings that were treated with CHX in the absence or presence of exogenous ABA. The turnover of PP2CA was markedly diminished in rglg1 amiR-rglg5 compared with the wild type, either in the absence or presence of exogenous ABA; therefore, RGLG1 and RGLG5 mediate PP2CA degradation (Figures 7A and 7B). To avoid the use of CHX together with ABA treatment, protein dynamics of FLAG-PP2CA expressed in rglg1 amiR-rglg5 were compared with that in the wild type (Figure 7C). ABA-promoted PP2CA degradation was abolished by downregulation of the two RGLGs (Figures 7C and 7D). Conversely, ABA-enhanced degradation of FLAG-PP2CA was increased in RGLG1ox or RGLG5ox plants (Supplemental Figures 10A and 10B), suggesting that degradation of PP2CA in response to ABA is dependent on RGLG1 and RGLG5 function. We also conducted in vitro cell-free degradation assays by monitoring the level of His-PP2CA after incubation with protein extracts prepared from rglg1 amiR-rglg5, RGLG1ox, RGLG5ox, or the wild type. Protein extracts prepared from rglg1 amiR-rglg5 mitigated ABA’s effect on the stability of purified His-PP2CA (Figures 7E and 7F), whereas overexpression of RGLG1 or RGLG5 in these extracts enhanced the degradative effect of ABA on PP2CA (Supplemental Figures 10C to 10F). Addition of MG132 mitigated PP2CA degradation; however, MG132-resistant vacuolar proteases are released in cell-free degradation assays, which might explain the partial effect of this compound in preserving PP2CA protein levels (Figures 7G and 7H; Supplemental Figures 10C to 10F).
Figure 7.
RGLG1- and RGLG5-Mediated Protein Turnover of PP2CA.
(A) PP2CA protein levels in Col-0 versus rglg1 amiR-rglg5 in response to ABA. Two-week-old etiolated Col-0 and rglg1 amiR-rglg5 seedlings were treated with 100 µM CHX or 100 µM CHX+50 µM ABA for 0, 1, 3, and 6 h. Samples were harvested for immunoblot analysis using α-E2663 to detect endogenous PP2CA protein levels (arrow) in Col-0 and rglg1 amiR-rglg5. Actin was analyzed as a loading control.
(B) Quantification of anti-PP2CA and anti-actin signal intensity in (A) using Image J software. The relative protein levels of FLAG-PP2CA at 0 h were set as 1. Bars show mean PP2CA levels normalized to actin protein. Values are averages ± sd of three independent experiments. Asterisks indicate a significant difference between the indicated comparisons (Student’s t test: **P < 0.01).
(C) ABA enhances PP2CA protein degradation. Two-week-old transgenic Arabidopsis seedlings constitutively expressing 3×FLAG-PP2CA in Col-0 or rglg1 amiR-rglg5 backgrounds were transferred to liquid MS medium without (Mock) or with 50 µM ABA for the indicated time period. Anti-FLAG was used to detect FLAG-PP2CA. Actin was analyzed as a loading control.
(D) Signal intensity in (C), as determined by Image J software. The relative protein levels of FLAG-PP2CA at 0 h were set as 1. Bars show mean PP2CA levels normalized to actin protein. Values are averages ± sd of three independent experiments. Asterisks indicate a significant difference between the indicated comparisons (Student’s t test: **P < 0.01).
(E) and (G) In vitro degradation of His-PP2CA by protein extracts prepared from 7-d-old Col-0 or rglg1 amiR-rglg5 seedlings. Purified His-PP2CA was incubated for the indicated time period in the absence (Mock) or presence of ABA (E) or in the presence of ABA or ABA+MG132 (G). Protein dynamics of PP2CA was analyzed by immunoblot using anti-His antibody. Actin was analyzed as a loading control.
(F) and (H) The signal intensity was determined by Image J software from three independent experiments as in (E) and (G), respectively. The relative protein levels of His-PP2CA in Col-0 and rglg1 amiR-rglg5 at time point 0 min were set as 1. Bars indicate the mean ± sd from three independent experiments. Student’s t tests were used to determine significance of the indicated comparisons. **P < 0.01; *P < 0.05.
(I) In vivo ubiquitination of PP2CA in different genetic backgrounds. FLAG-PP2CA protein was immunoprecipitated from 2-week-old 35Spro:3×FLAG-PP2CA/Col-0 and 35Spro:3×FLAG-PP2CA/rglg1 amiR-rglg5 seedlings treated with MG132 or ABA+MG132. Immunoprecipitated proteins were detected by immunoblotting using anti-FLAG and anti-ubiquitin antibodies, respectively.
Finally, we investigated whether RGLG1 and RGLG5 mediate PP2CA ubiquitination in planta. FLAG-tagged PP2CA from either the wild type or rglg1 amiR-rglg5 was separately immunoprecipitated after mock or ABA treatment in the presence of MG132. No obvious difference in FLAG-PP2CA ubiquitination level was observed in rglg1 amiR-rglg5 after treatment with ABA, while the addition of exogenous ABA increased the level of ubiquitinated PP2CA in Col-0 (Figure 7I). This result suggests that RGLG1 and RGLG5 are required for ABA-promoted ubiquitination of PP2CA in planta. Altogether, the above data support the notion that RGLG1 and RGLG5 contribute to in vivo degradation of PP2CA.
Functions of RGLG1 and RGLG5 in ABA Signaling Are Dependent on PP2CA
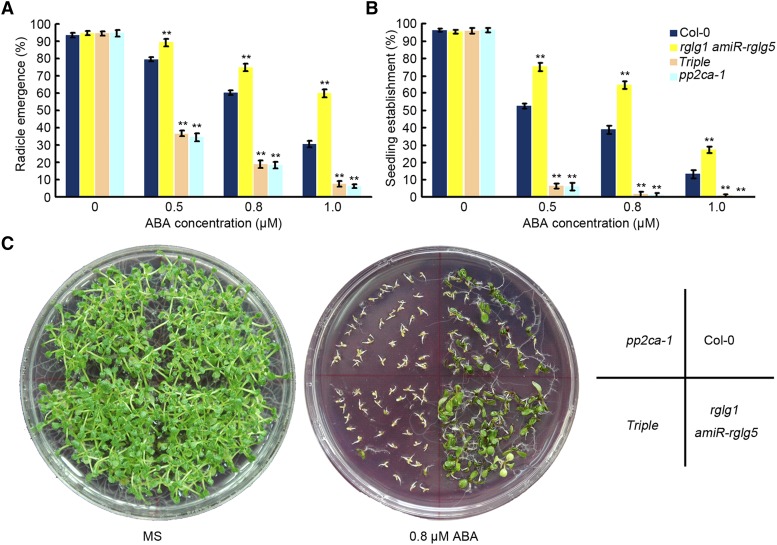
To determine the genetic relationship between RGLG1/5 and PP2CA, we crossed the pp2ca-1 loss-of-function mutant to the rglg1 amiR-rglg5 mutant. Since our results support the notion that RGLG1/5 positively regulate ABA signaling by targeting PP2CA for degradation, RGLG1/5 should act genetically upstream of PP2CA, and the pp2ca loss-of-function mutant should attenuate or abolish some ABA-insensitive phenotypes of the rglg1 amiR-rglg5 mutant. PP2CA/AHG3 encodes a phosphatase that strongly blocks ABA signaling during germination, and pp2ca-1 shows the strongest ABA hypersensitivity in seed germination assays among loss-of-function mutants of clade A PP2Cs (Yoshida et al., 2006; Rubio et al., 2009). Therefore, we analyzed ABA-mediated inhibition of seed germination and seedling establishment in the pp2ca-1 rglg1 amiR-rglg5 triple mutant, since PP2CA plays a predominant role in this ABA response, whereas other putative PP2C targets, HAB2 or ABI2, play less relevant roles in this assay. As shown in Figure 8, ABA-mediated inhibition of radicle emergence, seedling establishment, and early seedling growth in the triple mutant became similar to that of pp2ca-1, reflecting full recovery of ABA sensitivity in rglg1 amiR-RGLG5 by pp2ca-1. This genetic analysis confirms a functional link between RGLG1/5 and PP2CA and indicates that RGLG1/5 act upstream of PP2CA in the ABA signaling pathway.
Figure 8.
ABA Insensitivity of rglg1 amiR-rglg5 Is Dependent on PP2CA Activity.
(A) and (B) Seed germination (A) and seedling establishment (B) of Col-0, rglg1 amiR-rglg5 double, rglg1 amiR-rglg5 pp2ca-1 triple, and pp2ca-1 mutants. Radicle emergence was scored after 72-h growth on MS plates containing the indicated concentrations of ABA (A). Seedling establishment was scored as the percentage of seeds that developed fully green expanded cotyledons after 7-d growth on MS plates containing the indicated concentrations of ABA (B). Bars are mean ± sd of three independent experiments. Asterisks indicate a significant difference from the wild type (Student’s t test: **P < 0.01).
(C) Photographs of representative plates were taken 12 d after sowing the indicated genetic backgrounds on MS plates lacking or containing 0.8 µM ABA.
DISCUSSION
The ABA-PYR/PYL/RCARs-PP2Cs-SnRK2s core ABA signaling pathway has been widely studied in the past several years, and ABA-dependent PYR/PYL-mediated inhibition of PP2Cs has been extensively documented (Cutler et al., 2010; Finkelstein, 2013). Our work, together with recent results from Kong et al. (2015), unveils a second level of regulation during ABA signaling, i.e., the effect of ABA on PP2C protein levels. In this study, we provide evidence that RGLG1/5 E3 ubiquitin ligases positively regulate various ABA responses by targeting PP2CA for degradation in the presence of ABA. In addition to PP2CA, we also found that HAB2 and ABI2 are candidate targets of RGLG1/5. Thus, ABA promoted the interaction of RGLG1/5 with HAB2 and ABI2 in vivo (Figures 2B and 2C), RGLG1/5 mediated ubiquitination of HAB2 and ABI2 in vitro (Figures 5D to 5F), and HAB2 was subjected to ABA-mediated degradation via the 26S proteasome pathway (Supplemental Figure 9). A recent report from Kong et al. (2015) describes the 26S proteasome degradation of the PP2C ABI1 mediated by PUB12/13 U-box E3 ligases, which belong to a different E3 family. In this latter case, only ABI1 was targeted for degradation by PUB12/13 ligases, whereas other PP2Cs closely related to ABI1 were not recognized by these E3 ligases. Therefore, it seems RGLG1/5 might play a more general function in the ABA pathway by regulating more than one PP2C. Another key difference between PUB12/13 and RGLG1/5 is that ubiquitination of ABI1 was absolutely dependent on the presence of ABA receptors, whereas PP2CA, HAB2, and ABI2 were ubiquitinated in the absence of ABA receptors (Figure 5). This suggests that the conformation of PP2CA, HAB2, and ABI2 allows their ubiquitination in vitro by RGLG1/5; however, we found that ABA somehow enhances PP2C-RGLG1/5 interaction in vivo, increasing the efficiency of ubiquitination and subsequent degradation. Altogether, these studies reveal that both the inhibition and degradation of clade A PP2Cs are important for the activation of ABA signaling. Moreover, since degradation of ABI1 (Kong et al., 2015) and PP2CA (this work) was ABA-dependent, the role of PYR/PYL ABA receptors might extend beyond the reversible inhibition of the PP2Cs. Finally, the ABA-insensitive phenotype of rglg1 amiR-rglg5 is not as strong as that of pyr/pyl combined mutants (Gonzalez-Guzman et al., 2012), which suggests that ABA signaling can occur through inhibition of PP2C activity even in the absence of PP2CA ubiquitination. However, the rglg1 amiR-rglg5 mutant shows reduced ABA sensitivity even in the presence of a full set of PYR/PYLs, suggesting that PP2C degradation is required for full activation of ABA signaling.
The role of the 26S proteasome in the turnover of PP2Cs (Kong et al., 2015; this work) and ABA receptors (Irigoyen et al., 2014; Bueso et al., 2014) is further extended by the observation that maize (Zea mays) OST1 seems to be targeted for proteasome degradation by casein kinase 2-mediated phosphorylation at the ABA box (Vilela et al., 2015). Whereas inhibition of PP2C activity is a reversible mechanism that regulates ABA signaling according to fluctuating ABA levels in response to environmental cues, degradation of PP2Cs is an irreversible decision that might lead to sustained ABA signaling when a stress situation persists. However, the degradation of PP2Cs or ABA receptors relies on their ubiquitination, which is a reversible process by the action of deubiquitinating enzymes that can further regulate the stability from core ABA signaling components (Zhao et al., 2016). In nature, both transient and sustained forms of stress occur; therefore, a double mechanism to abolish PP2C function seems to be better prepared to cope with different environmental stresses. In this way, monitoring the abundance of repressor protein provides an additional mechanism for regulating ABA signaling, which complements the biochemical inhibition of phosphatase activity through PYR/PYLs. Thus, ABA, like other hormones such as auxin, jasmonate, and gibberellin, follows a relief of repression mechanism that degrades negative regulators via the 26S proteasome (Santner and Estelle, 2009; Kelley and Estelle, 2012; Kong et al., 2015).
A model integrating RGLG1/5 into the ABA signaling pathway is shown in Figure 9, where RGLG-mediated PP2CA degradation could be auxiliary to PYR/PYL-mediated inhibition of PP2CA activity. This model illustrates the idea that ubiquitin-mediated degradation of PP2CA might be enhanced by ABA perception or facilitated by the upregulation of PP2CA transcripts induced by ABA. The latter hypothesis suggests that RGLG1/5-PP2CA interaction might be simply facilitated by the higher PP2CA protein level induced by upregulation of PP2CA transcripts. Paradoxically, whereas ABA inhibits PP2C activity through ABA receptors and promotes ABI1 and PP2CA degradation, it also upregulates ABI1 and PP2CA transcripts as a feedback mechanism. However, ABA signaling might facilitate the RGLG1/5-PP2CA interaction by other mechanisms. Both RGLG1 and RGLG5 have a predicted N-terminal myristoylation site, and RGLG1-GFP decorates the plasma membrane under nonstress conditions (Yin et al., 2007). ABA treatment might modify the subcellular localization of RGLG1/5 by promoting their translocation to other cell compartments where PP2Cs are more abundant (Antoni et al., 2012). For instance, ABA-activated kinases might modify the subcellular location of RGLG1/5 and facilitate their interaction with PP2Cs. Additionally, although the formation of an ABA-Receptor-PP2CA ternary complex was not required in vitro for ubiquitination of PP2CA, we cannot rule out the possibility that in vivo, such a ternary complex enhances PP2CA ubiquitination, for instance by generating additional contact points between the E3 ligase and the ABA receptor or by limiting PP2C diffusion. Protein concentration is not usually a limiting factor for biochemical assays in vitro, but under in vivo conditions, the actual protein concentration in specific subcellular locations might notably affect the ubiquitination reaction. Indeed, Figure 7I shows that ABA treatment enhances in vivo ubiquitination of PP2CA. Future experiments will be aimed at further elucidating how ABA enhances ubiquitination of PP2CA and facilitates their interaction with RGLG1/5 in vivo.
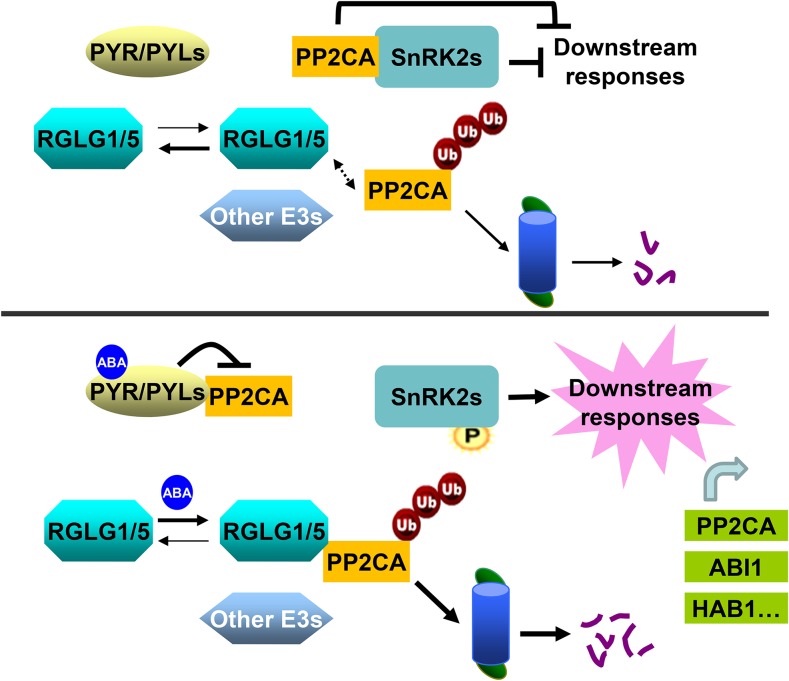
Figure 9.
Proposed Model for the Enhanced Degradation of PP2CA via the 26S Proteasome Mediated by ABA and RGLG1/5.
Under nonstress conditions (top, low ABA levels), RGLG1 and RGLG5 show less interaction with PP2CA (dashed arrow) or there is less PP2CA available for interaction because PP2CA transcripts are upregulated by ABA. PP2CA interacts with SnRK2s, which prevents their activation and leads to the inhibition of downstream ABA responses. PP2Cs themselves also inhibit downstream targets such as TFs and SLAC1. PP2CA levels under nonstress conditions are regulated by RGLG1/5 or additional unidentified E3s. When plants are challenged (bottom, high ABA levels), ABA promotes the interaction of PYR/PYLs with PP2CA, inhibiting its phosphatase activity. ABA facilitates RGLG1/5 interaction with PP2CA, which leads to enhanced ubiquitination and degradation via the 26S proteasome pathway, resulting in full activation of ABA signaling. As a negative feedback mechanism, ABA also increases PP2CA and clade A PP2C transcripts. See main text for additional details.
METHODS
PP2CA Polyclonal Antibodies and Immunoblot Detection of Endogenous PP2CA
The polyclonal antibody of PP2CA was generated by Shanghai ImmunoGen Biological Technology (ABclonal). To avoid cross-reaction with other PP2Cs, a fragment of truncated PP2CA (1 to 200 amino acids) was fused with GST tag and the recombinant protein expressed in Escherichia coli was used as an antigen to raise polyclonal antibody in rabbit. The resulting antiserum was purified by IgG-affinity chromatography. To ensure specific binding to PP2CA, the antibody was further isolated by membrane strip affinity purification. Briefly, 200 μg purified His-PP2CA was loaded onto a SDS-PAGE gel and then transferred to nitrocellulose membrane. The membrane containing His-PP2CA was stained with Ponceau S solution and cut into small strip. The strip was preeluted with 2 mL 0.2 M glycine (pH 2.7) for 2 min and then blocked with 5% nonfat milk in TBST for 1 h. After washing the membrane with TBST three times, it was incubated with antibody solution (200 μL antibody in 3 mL TBS) overnight and then washed again with TBST three times. The antibody was eluted by incubating with 100 to 200 μL 0.2 M glycine (pH 2.7) for 2 min. The resulting antibody solution was neutralized with 2 M Tris-HCl (pH 8.5), generating a final concentration of 150 mM Tris-HCl. The elution and neutralization steps were repeated two more times. The final antibody solution was dialyzed against 1× PBS (pH 7.4), concentrated to a volume of 150 μL with ultrafiltration column, and tested by immunoblot analysis. To prepare protein extracts, seedlings were grown vertically on Murashige and Skoog (MS) solid medium under dark condition for 2 weeks and then transferred into liquid MS medium supplemented with different chemicals under dark condition for the indicated time points. Samples were harvested and total proteins were extracted by homogenizing the seedlings in the lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM DTT, 1 mM PMSF, and plant-specific protease inhibitor cocktail) at a ratio of 1:2 (m/v). The concentration of total protein was determined by Bradford assays, and equal amounts of total proteins were mixed with 4× SDS loading buffer. Boiled samples were separated by SDS-PAGE and analyzed by immunoblot. The polyclonal antibody of PP2CA α-E2663 was used (1:3500 dilution) to detect endogenous PP2CA protein levels. Commercial antibodies were used at a 1:10,000 dilution unless a different dilution was indicated. Actin was detected as loading control.
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as the wild type. The rglg1 (SALK_062384) mutant was obtained from Andreas Bachmair. RGLG5 knockdown mutants were generated based on an artificial microRNA strategy. Seeds of pyr1 pyl1 pyl2 pyl4 (1124) were obtained from Sean R. Cutler. Seeds of pp2ca-1 were kindly provided by Julian I Schroeder; ABI2-GFP transgenic seeds were a gift from Dapeng Zhang. Arabidopsis plants were grown as previously described (Zhang et al., 2012). Nicotiana benthamiana plants were grown under a 16-h-light (80 to 100 μE m−2 s−1)/8-h-dark photoperiod. Philips bulbs were used (TL-D Super 80 36W, white light 840, 4000K light code).
Knockdown of RGLG5 by Artificial MicroRNA-Based Strategy
Overlapping PCR was performed using specific primers designed by Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) and plasmid pBSK as template to replace the miR319a sequence with the 21-nucleotide sequence GTCGTTCACTTATAGGAAACT designed to target RGLG5 transcript. After verification by sequencing, the amiRNA precursor was digested with XhoI/SpeI and cloned into pJim19 digested with the same restriction enzymes. The constructs were introduced into Agrobacterium tumefaciens strain EHA105 by electroporation, and wild-type plants were transformed using the floral dip method as described previously (Clough and Bent, 1998). Seeds of the transformed plants were selected using hygromycin. Homozygous T3 transgenic seeds or plants were used for further studies.
To obtain the rglg1 amiR-rglg5 double mutant, homozygous rglg1 was crossed with amiR-rglg5 (4#). Seeds were sowed on MS medium containing 25 μg/mL hygromycin, and the T-DNA insertion in rglg1 was confirmed by PCR using primers LBa1 and 323A with genomic DNA as template.
Generation of Transgenic Plants
RGLG1, RGLG5, and PP2CA cDNAs were each cloned into XhoI/SpeI-digested pJim19, resulting in a fusion with 3× FLAG tag at the N terminus. To construct RGLG1-GFP and RGLG5-GFP, the GUS sequence of pBI121 was replaced by GFP cDNA via BamHI/SacI sites. The pBI121-RGLG1-GFP and pBI121-RGLG1-GFP plasmids were then created by linking the RGLG1 or RGLG5 coding region without a stop codon to GFP (C terminus) via BamHI and KpnI. To construct PP2CA-GUS and HAB2-GUS fusions, entire coding sequences of PP2CA and HAB2 with no stop codons were amplified by PCR and separately inserted into the pBI121-GUS vector upstream of GUS. The primers used above are listed in Supplemental Data Set 1. All of the genes were expressed are under the control of the CaMV 35S promoter. These constructs were introduced into Agrobacterium strain EHA105 by electroporation, and wild-type or mutant plants were transformed using the floral dip method as reported previously (Clough and Bent, 1998). Seeds of the transformed plants were selected using hygromycin for the pJIM19 constructs or kanamycin for the pBI121 constructs. Homozygous T3 transgenic seeds or plants were used for further studies. The generation of 35Spro:3HA-PP2CA transgenic lines in the pp2ca-1 background was performed as described (Antoni et al., 2012). Transgenic Arabidopsis FLAG-PP2CA×RGLG1-GFP and FLAG-PP2CA×RGLG5-GFP plants were obtained by crossing FLAG-PP2CA with either RGLG1-GFP or RGLG5-GFP transgenic lines. Transgenic Arabidopsis ABI2-GFP×FLAG-RGLG1 and ABI2-GFP×FLAG-RGLG5 plants were produced by crossing ABI2-GFP with transgenic lines FLAG-RGLG1 and FLAG-RGLG5, respectively.
To construct the PP2CApro:GUS gene, a fragment comprising 2 kb 5′ upstream of the ATG start codon from PP2CA was amplified by PCR using the following primers: FproPP2CA (5′-AAGCTTGGTTTTACCCGAACTTAACCCAAATGC-3′, including a HindIII site) and RproPP2CA (5′-GAGCTCCATTTGATCTCTAACAAAACTTCTCCA-3′, including a SacI site). The PCR product was cloned into pCR8/GW/TOPO. Next, it was recombined by Gateway LR reaction into the pMDC163 destination vector (Curtis and Grossniklaus, 2003), in-frame with the GUS gene. The pMDC163-based construct carrying the PP2CApro:GUS gene was transferred to Agrobacterium pGV2260 (Deblaere et al., 1985) by electroporation and used to transform Col-0 wild-type plants by the floral dip method (Clough and Bent, 1998). Seeds of transformed plants were harvested and plated on hygromycin (20 μg/mL) selection medium to identify T1 transgenic plants, and T3 progenies homozygous for the selection marker were used for further studies. Imaging of GUS was performed as previously described (Gonzalez-Guzman et al., 2012).
Y2H Assay
RGLGs were cloned into the pGBKT7 vector as bait, and PP2Cs were inserted into pGADT7-Rec vector as prey, respectively. For interaction assays, each pair of bait and prey constructs was cotransformed into yeast strain AH109 based on the manufacturer’s instructions of the MatchMaker GAL4 Two-Hybrid System (Clontech). Transformants growing well on SD/-Leu/-Trp medium were taken as positive clones harboring both plasmids. Interactions between two proteins were determined by growing transformants on SD/-Leu/-Trp/-His/-Ade medium with serial dilution and quantifying β-galactosidase activity using o-nitrophenyl-β-d-galactopyranoside (Sigma-Aldrich) as substrate. Yeast protein extracts were analyzed by immunoblotting using anti-HA antibody to detect PP2C proteins fused to the GAL4 activation domain (AD) and anti-myc antibody to detect RGLG proteins fused to the GAL4 DNA binding domain (BD) according to the manufacturer’s instructions (Clontech).
Firefly Luciferase Complementation Imaging Assay
The full-length PP2CA was fused upstream of N-Luc in the pCAMBIA1300-NLuc vector, and RGLG1 or RGLG5 was fused downstream of C-Luc in the pCAMBIA1300-CLuc vector. The resulting constructs were transferred into Agrobacterium strain EHA105. To determine the interaction between PP2CA and RGLG1 or RGLG5, Agrobacterium harboring the indicated vectors were resuspended in infiltration buffer containing 10 mM MgCl2, 10 mM MES, 0.5 g/L glucose, and 150 μM acetosyringone to a final concentration of OD600 = 0.5. Equal volumes of different combinations were mixed and coinfiltrated into Nicotiana benthamiana leaves using a needleless syringe. Plants were first incubated in the dark for 24 h, followed by 16 h light/8 h dark for another 24 to 48 h at 22°C. MG132 was infiltrated into the same region 12 h before inspection. To test the effects of ABA treatment, 50 μM ABA was infiltrated into the same area on leaves 3 h before observation. N. benthamiana leaves were infiltrated with 0.1 mg/mL luciferin and placed in the dark for 5 min before CCD imaging. LUC activity was observed with a low-light cooled CCD imaging apparatus (Berthold Technologies). The exposure time of LUC was 1 to 10 min depending on the signal intensity.
Expression and Purification of Recombinant Protein
Full-length RGLG5, PP2CA, HAB2, and ABI2 coding sequences were amplified by PCR and cloned into the prokaryotic expression vector pGEX-4T-1 (Amersham Biosciences) to generate GST fusions. To produce MBP fusion proteins, the full-length coding regions of RGLG1 and RGLG5 were amplified and cloned into the pMAL-c2x vector (New England Biolabs). Primers used are listed in Supplemental Data Set 1. A QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies) was used to produce Ring-domain mutated form of RGLG1 (H462Y) and RGLG5 (H406Y) by PCR amplification with specific primers and the indicated plasmid as template. Recombinant His-PP2CA was produced as described (Antoni et al., 2012). To generate His-PYL4, the full-length coding sequence of PYL4 was amplified, digested by EcoRI and XhoI, and inserted into pET-28a. The recombinant constructs were then transformed into E. coli strain BL21 (DE3). For expression, 1 mM IPTG was added to the cell culture to induce GST-tagged proteins, whereas 0.3 and 0.4 mM IPTG were needed to efficiently induce MBP and His fusion proteins, respectively. The soluble GST-tagged proteins were purified using the MagneGST protein purification system (Promega) according to the manufacturer’s protocols. Purification of MBP fusion proteins was performed following the manufacturer’s instructions of the pMAL Protein Fusion and Purification System (New England Biolabs). The soluble His-fusion protein was extracted and immobilized onto Ni-NTA agarose beads (Qiagen), and the purification procedures were conducted according to the protocols for purification under native conditions of the QIAexpressionist kit.
In Vitro Pull-Down Assay
Equal amounts of purified MBP, MBP-RGLG1, or MBP-RGLG5 proteins were incubated with equal volumes of amylose resin beads (New England Biolabs) in buffer A1 containing 1× TBS, 1 mM PMSF, and protease inhibitor cocktail at 4°C for 2 h with gentle rotation. The beads were then washed three times with buffer A1 and aliquoted equally into four parts. The indicated amount of purified His-PP2CA was added to the mixture, followed by incubation overnight at 4°C. After washing five times with buffer A2 containing 1× TBS, 1 mM PMSF, 0.05% Triton X-100, and protease inhibitor cocktail, the bound proteins were eluted with 1× SDS loading buffer with boiling for 5 min at 100°C and examined by immunoblot using anti-His or anti-MBP antibodies.
Cell-Free Protein Degradation Assay
Cell-free protein degradation assay was conducted as previously described (Wang et al., 2009). Briefly, 7-d-old Arabidopsis seedlings of different genotypes were harvested and ground into a fine powder in liquid nitrogen. Total proteins were extracted in degradation buffer containing 25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 5 mM DTT, and 10 mM ATP. The debris was removed by two centrifugations at 16,000g for 10 min at 4°C, and the supernatants were transferred to new tubes. Protein concentration was determined using the Bradford method (Bio-Rad). Then, total extracts of each genotype were adjusted to equal concentrations with degradation buffer. To monitor the degradation of the recombinant His-PP2CA protein, 100 ng of the purified protein was incubated in 100 μL total extracts (containing 500 μg total proteins) at 15°C for each assay. ABA and MG132 were added to the in vitro degradation assays as indicated. Reactions were stopped by adding 4× SDS loading buffer. Samples were taken at the indicated intervals to determine the abundance of the remaining His-PP2CA by immunoblot using anti-His antibody (Bioeasy, 1:1000 dilution). The bands intensity was quantified using Image J software (http://imagej.nih.gov/ij/).
In Vitro Ubiquitination Assay
The self-ubiquitination assay was performed as described (Zhang et al., 2012). For the in vitro substrate ubiquitination assay, a total reaction volume of 30 μL was produced by combining 500 ng purified MBP-RGLG1 (MBP-RGLG1H462Y) or MBP-RGLG5 (MBP-RGLG55H406Y), 300 ng GST-PP2CA or GST-HAB2 or GST-ABI2, 50 ng E1 (Sigma-Aldrich), 100 ng E2 UbcH5b (Enzo Life Sciences), 3 μg FLAG-tagged ubiquitin (Sigma-Aldrich), 10 mM phosphocreatine, and 0.1 unit of creatine kinase in ubiquitination buffer (50 mM Tris-HCl, pH 7.5, 3 mM ATP, 5 mM MgCl2, and 0.5 mM DTT) and incubated at 37°C for 2 h. The reaction was stopped by adding 4× SDS loading buffer. Samples were separated by 8% SDS-PAGE and analyzed by immunoblot using anti-GST (Abmart), anti-FLAG (Sigma-Aldrich), or anti-MBP (Earthox) antibody.
In Vivo Immunoprecipitation Assays
For coimmunoprecipitation assays using ABI2-GFP×FLAG-RGLG1 or ABI2-GFP×FLAG-RGLG5 transgenic Arabidopsis, 2-week-old seedlings were transferred from selective MS medium to liquid MS containing 50 μM ABA for the indicated time. Plant materials (∼300 mg) were harvested and total proteins were extracted in 3 volumes of lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1% Nonidet P-40, 50 μM MG132, 1 mM DTT, 1 mM PMSF, and plant-specific protease inhibitor cocktail). The concentration of total protein was determined by Bradford assays (Bio-Rad), and each lysate (1 mL) with equal amounts of protein was immunoprecipitated by incubating with 20 μL anti-GFP mAb-Agarose (MBL) beads for 4 h at 4°C with gentle rotation. For immunoprecipitation with anti-FLAG antibody, the lysates were incubated with 0.5 μL anti-FLAG and 25 μL Protein G-Sepharose (Invitrogen) beads for 4 h at 4°C with gentle rotation. After incubation, the beads were washed four times with 1 mL of lysis buffer and eluted by adding 30 μL 1× SDS protein loading buffer and boiling for 5 min at 100°C. Immunoprecipitated proteins were separated by 10% SDS-PAGE and checked using anti-FLAG or anti-GFP (Abmart) antibody.
For coimmunoprecipitation in N. benthamiana, 4- to 5-week-old N. benthamiana leaves were infiltrated with Agrobacterium harboring the indicated constructs. The agroinfiltration was performed as described above (see Firefly luciferase complementation imaging assay). MG132 (50 μM) was also infiltrated into the same region 12 h before sample collection to inhibit protein degradation. Protein extracts were prepared as indicated above except that the lysates were incubated with or without 10 μM ABA at 4°C for 3 h before immunoprecipitation. Polyclonal anti-GFP (1 mg/mL; MBL) or anti-FLAG antibody coupled with 25 μL Protein G-Sepharose was then added to immunoprecipitate the total protein extracts. As negative controls, IgG was added in parallel to eliminate nonspecific binding. After elution as above, samples were subjected to immunoblot analysis, and anti-GUS (Sigma-Aldrich), anti-GFP, or anti-FLAG antibody were used to detect fusion proteins.
To detect the ubiquitination form of PP2CA in vivo, 2-week-old transgenic Arabidopsis 35S:3×FLAG-PP2CA/Col-0 and 35S:3×FLAG-PP2CA/rglg1 amiR-rglg5 were treated with MG132 with or without ABA for 6 h. Samples were harvested and total proteins were extracted as indicated above, except that deubiquitinating enzymes inhibitor PR619 (Sigma-Aldrich) was also included in the lysis buffer. The concentration of total proteins was determined by Bradford assays, and each lysate with equal amount of protein (500 mg) was immunoprecipitated by incubating with 0.5 μL anti-FLAG and 25 μL Protein G-Sepharose beads for 6 h at 4°C with gentle rotation. Immunoprecipitated products were obtained as mentioned above and analyzed by immunoblotting. Anti-FLAG antibody was used to detect total FLAG-PP2CA, and anti-ubiquitin was used to detect the ubiquitinated PP2CA in the plant.
coIP/Mass Spectrometry Analysis
Two-week-old transgenic Arabidopsis seedlings overexpressing FLAG-PP2CA were treated with 50 μM MG132 for 24 h and 50 μM ABA for 6 h. Four to five grams of material was harvested for total protein extraction. Anti-FLAG immunoprecipitates were prepared as described above (see In Vivo Immunoprecipitation Assays). FLAG-tagged proteins were eluted from the beads by adding 400 μg/mL 3×FLAG peptides in 1× PBS buffer, with gentle rotation at 4°C for 1 h three times. The eluted proteins were then collected and lyophilized. A final volume of 30 μL 1× PBS buffer was added to resuspend the powder. The resulting products were resolved on a 4 to 12% NuPAGE Bis-Tris MiNi Gel (Invitrogen) and visualized using a colloidal blue staining kit (Invitrogen). The protein bands from 15 to 100 kD were cut from the gel and digested with trypsin. The extracted peptides were subjected to LC-MS/MS analysis using an Easy nLC 1000 system (Thermo Scientific) connected to a Velos Pro Orbitrap Elite mass spectrometer (Thermo Scientific) equipped with a nano-ESI source. The raw data files were converted to mascot generic format (.mgf) using MSConvert before being submitted for database searches. Protein identification was carried using Mascot server v. 2.3.02 (Matrix Science) against the TAIR protein database.
In Vivo Protein Degradation Assay
Seedlings were grown vertically on MS solid medium for 2 weeks and transferred in liquid MS medium supplemented with 50 μM ABA for the indicated time points. Samples were harvested and total proteins were extracted by homogenizing the seedlings in the lysis buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM DTT, 1 mM PMSF, and plant-specific protease inhibitor cocktail at a ratio of 2:1 (m/v). The concentration of total protein was determined by Bradford assays, and equal amount of total proteins were mixed with 2× SDS loading buffer. Boiled samples were separated by SDS-PAGE and analyzed by immunoblot. Anti-FLAG was used to detect FLAG-PP2CA, and anti-GFP was used to confirm the expression of RGLG fusions. Anti-Actin was detected as a loading control.
GUS Assays
For protein stability analysis, transgenic Arabidopsis seedlings carrying pBI-PP2CA-GUS, pBI-HAB2-GUS or pBI-GUS (negative control) were germinated and grown on MS plate supplemented with 50 μg/mL kanamycin for 7 d followed by transfer to liquid MS medium with or without 50 μM MG132 and/or 100 μM CHX (Sigma-Aldrich), as well as 50 μM ABA for the indicated time. Treated seedlings were collected for GUS staining, which was performed as described previously (Zhang et al., 2012), and stained plants were visualized under a Leica E24HD stereo microscope. GUS activity was measured as reported recently (Zhang et al., 2015). Briefly, total protein was extracted from 7-d-old seedling tissues using GUS extraction buffer (50 mM sodium phosphate buffer, pH 7.5, 10 mM β-mercaptoethanol, 0.1% Triton X-100, and 10 mM EDTA), followed by quantification by Bradford assays. Then, 50 mg total protein was incubated at 37°C for 5 min, followed by the addition of 1 mM 4-methylumbelliferyl-β-d-glucuronide (Sigma-Aldrich). After 60 min of reaction, a 100 μL sample was taken, and 2.4 mL 0.2 M Na2CO3 was added to terminate the reaction. Each sample was quantified for absorbance at excitation 365/emission 455 with a fluorospectrophotometer (Hitachi) to calculate 4-methylumbelliferone production. The final GUS activity was expressed as pmol (4-methylumbelliferone)⋅min−1⋅µg−1 (fresh weight).
Phenotype Analysis
For germination assay, ∼100 sterilized seeds of each genotype were sown on MS plates supplemented without or with different concentrations of ABA. After stratification in the dark at 4°Cfor 3 d, plates were transferred to a greenhouse at 22°C under a 16-h/8-h photoperiod. Germination rate indicates the percentage of seeds that showed obvious emergence of a radicle after 3 d. Seedling establishment was scored after 7 d as the percentage of seedlings that developed fully green expanded cotyledons (Pizzio et al., 2013).
For the root growth assay, seeds were grown vertically on MS medium for 3 to 5 d, and 10 seedlings of each genotype were transferred to MS plates in the presence or absence of different concentrations of ABA for additional periods of time (Pizzio et al., 2013). Photographs were taken and primary root length was monitored and analyzed using ImageJ software.
To study the effect of NaCl on germination and postgermination growth, seeds were sown on MS plates with or without various concentrations of NaCl and the plates were kept in the dark at 4°C for 3 d. Afterward, the plates were moved to a greenhouse under long-day conditions (16-h-light/8-h-dark cycle). Photographs were taken 7 d later to record different sensitivities to NaCl of each genotype.
To measure leaf water loss, rosette leaves of similar developmental stages from 4-week-old plants grown under short-day conditions (8-h-light/16-h-dark cycle) were excised from their roots, placed in open Petri dishes, and kept on the lab bench for the indicated time, and then their fresh weights were monitored. Water loss was expressed as a percentage of weight loss at the indicated time versus initial fresh weight (Zhu et al., 2007). For the drought treatment experiment, 7-d-old plants were transferred from MS medium to water-saturated soil and the plants were grown under short-day conditions until they were 3 weeks old. Watering was withdrawn until severe damage was observed in wild-type plants (18 to 20 d). Survival rate was recorded 3 d after rehydration (Zhu et al., 2007). To minimize experimental variations, the same numbers of plants was grown in the same tray.
RNA Extraction and RT-PCR
For RT-qPCR analysis of ABA-responsive genes, 7-d-old seedlings grown under long-day conditions were transferred to liquid MS medium supplemented with or without 50 μM ABA for the indicated time. Total RNA extraction, cDNA synthesis, and RT-qPCR were performed as described (Zhang et al., 2012). PCR analysis was performed using SYBR Green PCR Master Mix (Toyobo) in the DNA Engine Opticon2 Continuous Fluorescence Detector (MJ Research). Three independent experiments were performed, and four technical replicates of each experiment were performed. UBQ10 was used as an internal control for normalization of transcript levels (Zhang et al., 2012). Data were analyzed using Opticon Monitor 2 software (MJ Research). Primers used for gene expression analyses are listed in Supplemental Data Set 1.
Accession Numbers
The Arabidopsis Genome Initiative numbers for genes mentioned in this article are as follows: RGLG1 (At3G01650), RGLG2 (At5G14420), RGLG3 (At5G63970), RGLG4 (At1G79380), RGLG5 (At1G67800), ABI1 (AT4G26080), ABI2 (AT5G57050), PP2CA (AT3G11410), HAB1 (AT1G72770), HAB2 (AT1G17550), RD29a (AT5G52310), RD29b (AT5G52300), RAB18 (AT5G66400), RD22 (AT5G25610), DREB2A (AT5G05410), P5CS1 (AT2G39800), ERD10 (AT1G20450), ADH1 (AT1G77120), and PYL4 (AT2G38310).
Supplemental Data
Supplemental Figure 1. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay.
Supplemental Figure 2. Relative expression of RGLG1 and RGLG5 in seedlings.
Supplemental Figure 3. RGLG5 transcript levels were reduced in amiRNA transgenic plants compared with the wild type.
Supplemental Figure 4. ABA hypersensitivity in Arabidopsis transgenic lines overexpressing either RGLG1 or RGLG5.
Supplemental Figure 5. E3 ligase activity of RGLG5 and in vitro ubiquitination of PP2CA by RGLG1 and RGLG5.
Supplemental Figure 6. In vitro ubiquitination of PP2CA by RGLG1 and RGLG5 is not affected by ABA and PYL4.
Supplemental Figure 7. Expression of the PP2CA promoter is strongly induced by ABA.
Supplemental Figure 8. α-E2663 is a specific antibody for PP2CA.
Supplemental Figure 9. GUS staining of seedlings expressing PP2CA-GUS or HAB2-GUS reveals ABA-induced degradation of the phosphatases via the 26S proteasome pathway.
Supplemental Figure 10. Overexpression of RGLG1 or RGLG5 enhances ABA-promoted degradation of FLAG-PP2CA.
Supplemental Table 1. List of peptides identified for PP2CA and RGLG1 in anti-FLAG immunoprecipitates obtained from 35Spro:3×FLAG-PP2CA Arabidopsis seedlings mock-treated or treated with ABA and MG132.
Supplemental Data Set 1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Andreas Bachmair for the rglg1 mutant, Sean R. Cutler for the pyr1 pyl1 pyl2 pyl4 seeds, Dapeng Zhang for the transgenic material harboring ABI2, Hongwei Guo and Jianmin Zhou for the pCAMBIA1300-Nluc and pCAMBIA1300-Cluc vectors, and John Olson for assistance in English editing. Work in C.A.’s laboratory was supported by grants from the National Key Basic Science “973” Program (Grant 2012CB114006), the National Natural Science Foundation (Grants 31272023, 31170231, and 90817001) of the Chinese government, and by the State Key Laboratory of Protein and Plant Gene Research, Peking University. Work in P.L.R.’s laboratory was supported by Ministerio de Ciencia e Innovacion, Fondo Europeo de Desarrollo Regional, and Consejo Superior de Investigaciones Cientificas (Grant BIO2014-52537-R).
AUTHOR CONTRIBUTIONS
Q.W., X.Z., M.P.-L., B.B.-P., X.Y., C.A., and P.L.R. designed the experiments. Q.W., X.Z., M.P.-L., B.B.-P., X.W., and S.C. performed the experiments. Q.W., X.Z., C.A., and P.L.R. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Glossary
- ABA
abscisic acid
- CHX
cycloheximide
- Y2H
yeast two-hybrid
- coIP
coimmunoprecipitation
- MS
Murashige and Skoog
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Rodrigues A., Pizzio G.A., Rodriguez P.L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 158: 970–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Peirats-Llobet M., Pizzio G.A., Fernandez M.A., De Winne N., De Jaeger G., Dietrich D., Bennett M.J., Rodriguez P.L. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 161: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazon B., et al. (2016). FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 28: 10.1105/tpc.16.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B., Munemasa S., Wang C., Nguyen D., Yong T., Yang P.G., Poretsky E., Belknap T.F., Waadt R., Aleman F., Schroeder J.I. (2015). Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife 4: e03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E., Rodriguez L., Lorenzo-Orts L., Gonzalez-Guzman M., Sayas E., Muñoz-Bertomeu J., Ibañez C., Serrano R., Rodriguez P.L. (2014). The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 80: 1057–1071. [DOI] [PubMed] [Google Scholar]
- Cai Z., Liu J., Wang H., Yang C., Chen Y., Li Y., Pan S., Dong R., Tang G., Barajas-Lopez Jde.D., Fujii H., Wang X. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 9651–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Deblaere R., Bytebier B., De Greve H., Deboeck F., Schell J., Van Montagu M., Leemans J. (1985). Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 13: 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., Joazeiro C.A. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78: 399–434. [DOI] [PubMed] [Google Scholar]
- Finkelstein R. (2013). Abscisic acid synthesis and response. Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106: 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50: 2123–2132. [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A., Romeis T., Hedrich R. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 106: 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G.A., Antoni R., Vera-Sirera F., Merilo E., Bassel G.W., Fernández M.A., Holdsworth M.J., Perez-Amador M.A., Kollist H., Rodriguez P.L. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K.E., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24: 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen M.L., et al. (2014). Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26: 712–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.R., Estelle M. (2012). Ubiquitin-mediated control of plant hormone signaling. Plant Physiol. 160: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., Yamamoto A., Hattori T. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44: 939–949. [DOI] [PubMed] [Google Scholar]
- Kong L., Cheng J., Zhu Y., Ding Y., Meng J., Chen Z., Xie Q., Guo Y., Li J., Yang S., Gong Z. (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6: 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.M., Boisson-Dernier A., Dizon M.B., Maktabi M.H., Schroeder J.I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140: 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lång V., Palva E.T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20: 951–962. [DOI] [PubMed] [Google Scholar]
- Lee S.C., Lan W., Buchanan B.B., Luan S. (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 106: 21419–21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2011). E3 ubiquitin ligases and abscisic acid signaling. Plant Signal. Behav. 6: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshik A., Nito K., Park S.Y., Wang A., Carvalho P.C., Lee S., Caddell D.F., Cutler S.R., Chory J., Yates J.R., Schroeder J.I. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K., Vahala T., Palva E.T. (1993). Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 21: 641–653. [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzio G.A., Rodriguez L., Antoni R., Gonzalez-Guzman M., Yunta C., Merilo E., Kollist H., Albert A., Rodriguez P.L. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163: 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S., Rodrigues A., Saez A., Dizon M.B., Galle A., Kim T.H., Santiago J., Flexas J., Schroeder J.I., Rodriguez P.L. (2009). Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150: 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S.Y., Márquez J.A., Cutler S.R., Rodriguez P.L. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60: 575–588. [DOI] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078. [DOI] [PubMed] [Google Scholar]
- Sheen J. (1998). Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl. Acad. Sci. USA 95: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizhov N., Abrahám E., Okrész L., Blickling S., Zilberstein A., Schell J., Koncz C., Szabados L. (1997). Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 12: 557–569. [DOI] [PubMed] [Google Scholar]
- Szostkiewicz I., Richter K., Kepka M., Demmel S., Ma Y., Korte A., Assaad F.F., Christmann A., Grill E. (2010). Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 61: 25–35. [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., Shinozaki K. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6: rs8. [DOI] [PubMed] [Google Scholar]
- Vilela B., Nájar E., Lumbreras V., Leung J., Pagès M. (2015). Casein Kinase 2 negatively regulates abscisic acid-activated SnRK2s in the core abscisic acid-signaling module. Mol. Plant 8: 709–721. [DOI] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., Leung J., Rodriguez P.L., Laurière C., Merlot S. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhu D., Huang X., Li S., Gong Y., Yao Q., Fu X., Fan L.M., Deng X.W. (2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y.J., Van Oosten M.J., Zhang H., Tao W.A., Zhu J.K. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 110: 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.J., et al. (2007). Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19: 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Nishimura N., Kitahata N., Kuromori T., Ito T., Asami T., Shinozaki K., Hirayama T. (2006). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 140: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Wu Y., Xie Q. (2016). Ubiquitin-proteasome system in ABA signaling: From perception to action. Mol. Plant 9: 21–33. [DOI] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu Q., Ren J., Qian W., He S., Huang K., Yu X., Gao Y., Huang P., An C. (2012). Two novel RING-type ubiquitin ligases, RGLG3 and RGLG4, are essential for jasmonate-mediated responses in Arabidopsis. Plant Physiol. 160: 808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu Q., Cui S., Ren J., Qian W., Yang Y., He S., Chu J., Sun X., Yan C., Yu X., An C. (2015). Hijacking of the jasmonate pathway by the mycotoxin fumonisin B1 (FB1) to initiate programmed cell death in Arabidopsis is modulated by RGLG3 and RGLG4. J. Exp. Bot. 66: 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Xing L., Wang X., Hou Y.J., Gao J., Wang P., Duan C.G., Zhu X., Zhu J.K. (2014). The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7: ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhou H., Zhang M., Gao Y., Li L., Gao Y., Li M., Yang Y., Guo Y., Li X. (2016). Ubiquitin-specific protease 24 negatively regulates abscisic acid signalling in Arabidopsis thaliana. Plant Cell Environ. 39: 427–440. [DOI] [PubMed] [Google Scholar]
- Zhu S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.