The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/DPa complex by physical competition and SUMOylation in the G1/S cell cycle transition.
Abstract
Development requires the proper execution and regulation of the cell cycle via precise, conserved mechanisms. Critically, the E2F/DP complex controls the expression of essential genes during cell cycle transitions. Here, we discovered the molecular function of the Arabidopsis thaliana SUMO E3 ligase METHYL METHANESULFONATE SENSITIVITY GENE21 (AtMMS21) in regulating the cell cycle via the E2Fa/DPa pathway. DPa was identified as an AtMMS21-interacting protein and AtMMS21 competes with E2Fa for interaction with DPa. Moreover, DPa is a substrate for SUMOylation mediated by AtMMS21, and this SUMOylation enhances the dissociation of the E2Fa/DPa complex. AtMMS21 also affects the subcellular localization of E2Fa/DPa. The E2Fa/DPa target genes are upregulated in the root of mms21-1 and mms21-1 mutants showed increased endoreplication. Overexpression of DPa affected the root development of mms21-1, and overexpression of AtMMS21 completely recovered the abnormal phenotypes of 35S:E2Fa-DPa plants. Our results suggest that AtMMS21 dissociates the E2Fa/DPa complex via competition and SUMOylation in the regulation of plant cell cycle.
INTRODUCTION
Cell division and differentiation are precisely controlled by the cell cycle machinery, which monitors critical checkpoints, including the G1/S and G2/M transitions (Hartwell and Kastan, 1994; Dewitte and Murray, 2003). Models of the cell cycle suggest that processes such as mitosis and endoreduplication predominantly rely on checkpoint regulation by cyclin-dependent kinases (CDKs) (Gutierrez, 2009; Satyanarayana and Kaldis, 2009). During the G1/S transition, CDKs phosphorylate retinoblastoma protein (Rb) to suppress its inhibition of the E2F/DP transcription factor complex, pushing the cell into S phase (Weinberg, 1995; Boniotti and Gutierrez, 2001; Nakagami et al., 2002). The E2F family plays a major role in cell cycle regulation by controlling gene expression during the checkpoint (Müller and Helin, 2000). E2F binds with its partner DP, which provides a second DNA binding site, stabilizing the association of E2F with DNA (Bandara et al., 1993; Helin et al., 1993; Krek et al., 1993). The E2F/DP gene family is conserved in higher eukaryotes. In Arabidopsis thaliana, the homologous proteins E2Fa, E2Fb, or E2Fc form heterodimers with DPa or DPb via their dimerization domains (Magyar et al., 2000; Ramirez-Parra and Gutierrez, 2000). The transactivation function of E2F is controlled by its nuclear localization, which is mediated by the interaction between specific E2F and DP partners (Kosugi and Ohashi, 2002). E2Fa controls the G1/S transition by inducing the transcription of genes required for cell cycle progression and DNA replication (Vandepoele et al., 2005). Overexpression of E2Fa and DPa in Arabidopsis induces plant cells to undergo either ectopic cell division or enhanced DNA endoreduplication (De Veylder et al., 2002). Moreover, overexpression of dominant-negative CDKB1;1 enhances the endoreplication phenotypes but represses the extra cell division in E2Fa-DPa-overexpressing plants (Boudolf et al., 2004), suggesting a potential linkage between the G1/S and G2/M checkpoints.
The cell cycle is controlled by a network of posttranslational protein modifications. For instance, CDKs establish a complex phosphorylation network (Malumbres, 2014), whereas cyclins are generally regulated by ubiquitination-mediated degradation (Glotzer et al., 1991). SUMOylation, which transfers the small ubiquitin-like modifier (SUMO) onto protein substrates, is a critical modification of cell cycle regulation (Dasso, 2008). Depletion of the SUMO-conjugating enzyme UBC9 results in a block of the G2/M transition in yeast and failure of embryonic development in mice (al-Khodairy et al., 1995; Nacerddine et al., 2005). Conversely, the SUMO proteases Ulp1 and Ulp2, which remove SUMO from substrates, are also essential for cell cycle progression (Li and Hochstrasser, 2000; Taylor et al., 2002). Multiple key regulators of the cell cycle are subject to SUMOylation. For instance, in human cells, SUMO attaches to Rb to repress its inhibition of the activity of the E2F/DP complex (Ledl et al., 2005; Li et al., 2006), whereas the SUMO E3 ligase PIASy increases pRB-dependent repression via recruitment of E2F-responsive promoters (Man et al., 2006). CDK/Cyclin complexes are also potential targets of SUMOylation. The stability of CDK6, which is critically involved in the G1/S transition, is mediated by SUMO modification in glioblastoma cells (Bellail et al., 2014). Conjugation of SUMO with Cyclin D1 is critical for the nuclear localization of Cyclin D1 in the oncogene-induced senescence signaling pathway (Wang et al., 2011). Recently, a large-scale quantitative proteomics study provided evidence that SUMOylation affects all aspects of cell cycle progression (Schimmel et al., 2014).
Similar to ubiquitination, SUMOylation occurs through a cascade that includes a heterodimeric activating enzyme (E1), a conjugating enzyme (E2), and (usually) a SUMO ligase (E3) (Wilkinson and Henley, 2010). SUMO E3 enzymes stimulate substrate SUMOylation under physiological conditions. Multiple SUMO ligases, including a group of Siz/PIAS family proteins with an SP-RING domain, have been implicated in various cellular pathways in different species. For instance, the five vertebrate PIAS proteins are involved in many processes, including gene expression and signal transduction (Palvimo, 2007), and another member, MMS21, is critical for genome maintenance (Stephan et al., 2011; Jacome et al., 2015). Recent studies have shown that SUMOylation in plants regulates stress tolerance and development via targeting protein components in related signaling pathways (Saracco et al., 2007; Elrouby and Coupland, 2010; Miller et al., 2010). For instance, SUMOylation targets DELLAs in the gibberellin pathway (Conti et al., 2014) and CESTA in the brassinosteroid pathway (Khan et al., 2014). The Arabidopsis SUMO ligase SIZ1 participates in responses to stresses such as drought and freezing (Catala et al., 2007; Miura et al., 2007). The Sugimoto group and our group previously reported that depletion of AtMMS21 (also named HPY2), encoding a SUMO E3 enzyme with an SP-RING domain, impairs stem cell niche maintenance during root development in Arabidopsis (Huang et al., 2009; Ishida et al., 2009). Further dissection of the function of AtMMS21 by our group revealed that AtMMS21 is a conserved component of the SMC5/6 complex and is involved in drought tolerance, DNA damage response, and meiosis (Zhang et al., 2010; Xu et al., 2013; Liu et al., 2014; Yuan et al., 2014). In the absence of AtMMS21, the levels of several G2/M phase transition regulators, such as CYCB1, CDKB1, and CDKB2, are reduced, and the endoreplication level is dramatically increased (Ishida et al., 2009). However, the molecular mechanism underlying how AtMMS21 participates in cell cycle control remains unclear. In this study, we found that AtMMS21 dissociates the E2Fa/DPa complex in plants, identifying a regulatory mechanism for cell cycle progression.
RESULTS
Identification of Cell Cycle Components Interacting with AtMMS21
We previously demonstrated that the mms21-1 mutant displays severe defects in root and leaf development (Huang et al., 2009) (Figure 1A). To uncover the role of AtMMS21 in cell cycle regulation, we examined the ploidy level of the mms21-1 mutant by flow cytometry analysis. In the cotyledons and first pairs of true leaves of 13-d-old plants, mms21-1 had fewer nuclei with 2C DNA contents and more nuclei with higher DNA contents, compared with the wild type (Figure 1B), consistent with the results from hpy2-1, another AtMMS21 mutant allele (Ishida et al., 2009). This phenotype suggests that endoreplication is enhanced in the absence of AtMMS21, implying the important role of AtMMS21 in plant cell cycle regulation.
Figure 1.
The Phenotype of mms21-1.
(A) The phenotypes of 1-week-old (left panel) and 3-week-old (right panel) wild-type and mms21-1 plants. Bar = 1 cm.
(B) The endoreplication levels in wild-type and mms21-1 plants were quantitatively analyzed by flow cytometry. The cotyledons or the first pair of true leaves (leaf 1-2) of 13-d-old plants were collected for analysis. The data are means ± sd from at least three biological replicates.
To elucidate the molecular function of AtMMS21 in cell cycle progression, we next used yeast two-hybrid screens to identify cell cycle-related components that interact with AtMMS21. We tested eight critical regulators, CDKA, CYCB1;1, RBR, E2Fa, E2Fb, E2Fc, DPa, and DPb, in the two-hybrid assay to examine their interactions with AtMMS21. Most of these regulators did not interact with AtMMS21, except for DPa, which forms heterodimers with E2F members in the regulation of checkpoint transition.
DPa Interacts with AtMMS21
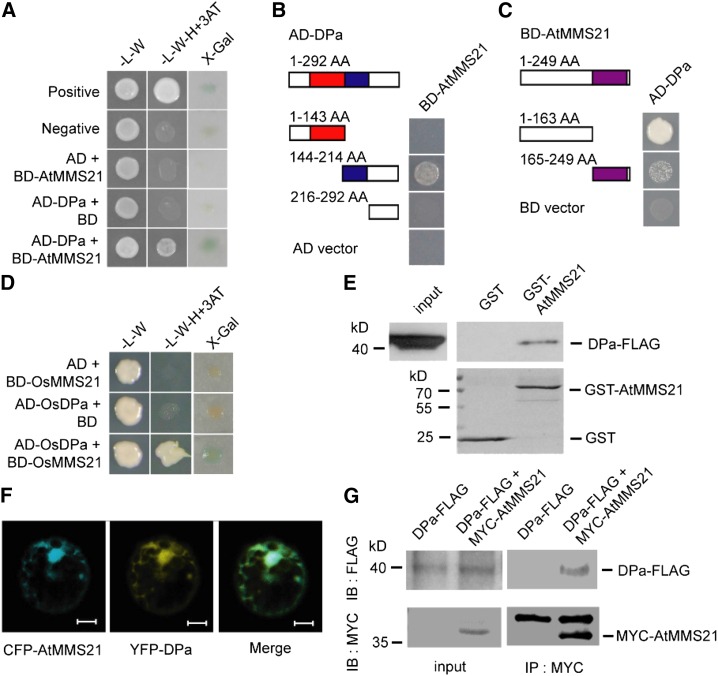
We used an independent yeast two-hybrid experiment to confirm that DPa interacts with AtMMS21 (Figure 2A) and identify the functional domains required for their interaction in yeast. DPa contains a DNA binding domain at the N terminus and a dimerization domain at the C terminus (Magyar et al., 2000). Deletion of the C-terminal region of DPa abolished the interaction with AtMMS21, whereas deletion of its N-terminal domain did not affect this interaction. The C-terminal fragment without the dimerization domain did not interact with AtMMS21, implying that binding to AtMMS21 requires the dimerization domain of DPa (Figure 2B). To determine the functional region of AtMMS21 that interacts with DPa, we investigated two truncated forms of AtMMS21, finding that the N terminus of AtMMS21 specifically interacted with DPa in yeast (Figure 2C). We also observed that the rice (Oryza sativa) homologs OsDP and OsMMS21 interacted in yeast (Figure 2D), suggesting that the DPa-MMS21 interaction is conserved in different plant species. The physical interaction between DPa and AtMMS21 was further confirmed by an in vitro pull-down assay. Compared with the control sample, FLAG-tagged DPa protein was pulled down by GST-AtMMS21, indicating that DPa directly associates with AtMMS21 (Figure 2E).
Figure 2.
DPa Interacts with AtMMS21.
(A) The interaction between DPa and AtMMS21 was confirmed in a yeast two-hybrid assay.
(B) Identification of the domain on DPa required for its interaction with AtMMS21 by yeast two-hybrid analysis. The DNA binding domain (red) and dimerization domain (blue) of DPa and the SP-RING domain (purple) of AtMMS21 are indicated.
(C) Identification of the domain on AtMMS21 required for its interaction with DPa by yeast two-hybrid analysis.
(D) The interaction between rice DP and MMS21 in a yeast two-hybrid assay.
(E) The interaction between DPa and AtMMS21 in an in vitro pull-down assay. The DPa-FLAG proteins were incubated with immobilized GST or GST-AtMMS21, and proteins immunoprecipitated with glutathione sepharose were detected using anti-FLAG antibody. The amounts of GST and GST-AtMMS21 are shown in the bottom panel.
(F) The subcellular colocalization of DPa and AtMMS21. CFP-AtMMS21 and YFP-DPa were coexpressed in protoplasts. After 48 h, the CFP and YFP signals were detected and merged. Bar = 10 µm.
(G) The interaction between DPa and AtMMS21 in an in vivo coimmunoprecipitation assay. Total protein extracts from transgenic plants carrying 35S:DPa-FLAG alone or both 35S:DPa-FLAG and 35S:MYC-AtMMS21 were immunoprecipitated with the immobilized anti-MYC antibody. The proteins from crude lysates (left) and immunoprecipitated proteins (right) were detected using anti-FLAG or anti-MYC antibody.
To further explore the association between DPa and AtMMS21 in plant cells, CFP-fused AtMMS21 and YFP-fused DPa were coexpressed in Arabidopsis protoplasts, and confocal microscopy revealed that the CFP and YFP signals predominantly overlapped (Figure 2F). In addition, the bimolecular fluorescence complementation data indicated that DPa and AtMMS21 interacted with each other in cytoplasm and nucleus (Supplemental Figure 1). Direct evidence for this interaction in vivo was provided by a coimmunoprecipitation assay using transgenic plants expressing FLAG-tagged DPa and MYC-tagged AtMMS21, revealing that DPa-FLAG was specifically immunoprecipitated with MYC-AtMMS21 (Figure 2G). Taken together, these results supported the conclusion that DPa interacts with AtMMS21.
AtMMS21 Interferes with the Interaction between E2Fa and DPa
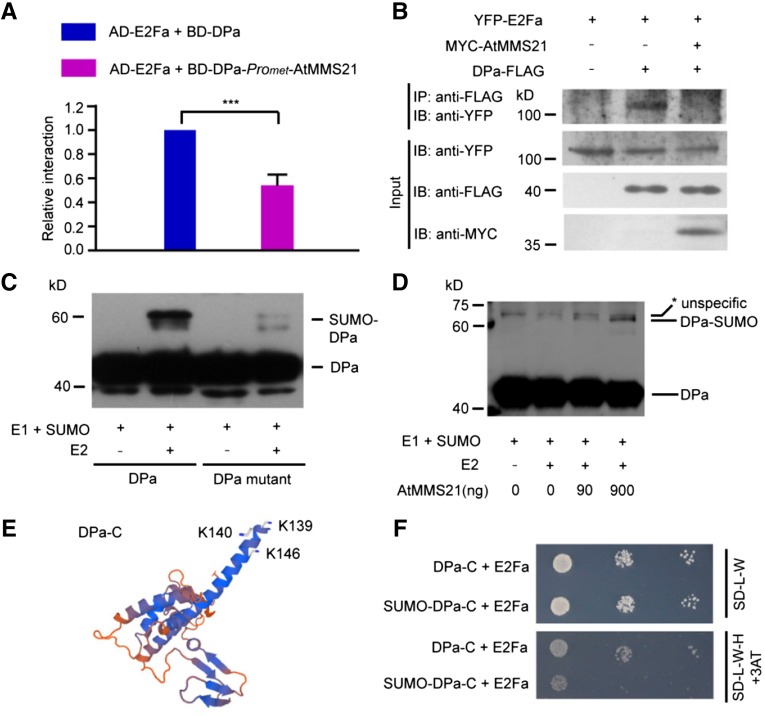
Heterodimer formation between DPa and E2Fa requires the DPa dimerization domain (Magyar et al., 2000), and this dimerization domain interacted with AtMMS21 in our yeast two-hybrid assay. Therefore, we next tested whether the AtMMS21-DPa interaction interferes with the DPa-E2Fa interaction in a yeast three-hybrid assay. A prey construct expressing E2Fa was transformed into yeast with a bait construct expressing DPa with or without AtMMS21. The relative activity was reduced in the presence of AtMMS21 (Figure 3A), indicating that AtMMS21 attenuates the E2Fa-DPa interaction in yeast.
Figure 3.
AtMMS21 Enhances the SUMOylation of DPa and Interferes with the Interaction between E2Fa and DPa.
(A) The effects of AtMMS21 on the interaction between DPa and E2Fa in a yeast three-hybrid assay. Yeast cells were transformed with pBridge-BD-DPa-ProMET-AtMMS21 or pBridge-BD-DPa with pGADT7-E2Fa and cultured on SD-L-W-M medium. The relative α-galactosidase activities are shown. The activity from the sample with BD-DPa and AD-E2Fa was set to 1. The data are means ± sd from three independent experiments. ***P < 0.001, Student’s t test.
(B) The effects of AtMMS21 on the DPa–E2Fa interaction in plant cells. YFP-E2Fa was expressed in protoplasts from wild-type or transgenic plants overexpressing DPa-FLAG alone or both DPa-FLAG and MYC-AtMMS21. The interaction was detected by immunoprecipitation on anti-FLAG resin. The total lysates (input) and the immunoprecipitated proteins (IP) were detected with an antibody recognizing YFP. The expression levels of DPa-FLAG and MYC-AtMMS21 were also detected using the indicated antibodies.
(C) The SUMO conjugation of DPa was detected in a reconstituted SUMOylation system in E. coli. In the presence of E1 and SUMO1, the unconjugated and SUMO-conjugated wild type or the mutant with K139/140/146R substitutions of FLAG-tagged DPa with or without E2 were detected with anti-FLAG antibody.
(D) AtMMS21 mediates the in vitro SUMOylation of DPa. Affinity-purified DPa-FLAG was used as a substrate in an in vitro SUMOylation assay. The SUMOylation levels of DPa in different samples were detected in an immunoblot using anti-FLAG antibody. Asterisk indicates an unspecific signal.
(E) The positions of SUMO attachment sites on the DPa protein. The three-dimensional structure of the C terminus of Arabidopsis DPa was predicted using Swiss Model. The potential SUMOylation sites are indicated.
(F) The effect of SUMO on the interaction between DPa and E2Fa. SUMO1 was fused to DPa-C (144 to 292 amino acids) in BD vector and used in a yeast two-hybrid assay with AD-E2Fa to detect their interaction. BD-DPa-C without SUMO was used as a control.
To confirm the effect of AtMMS21 on the E2Fa/DPa complex in plant cells, YFP-E2Fa was expressed in protoplasts from wild-type or transgenic plants producing DPa-FLAG or both DPa-FLAG and MYC-AtMMS21. YFP-E2Fa was specifically immunoprecipitated with DPa-FLAG, but this interaction was abolished by AtMMS21 in planta (Figure 3B). These data collectively support the conclusion that AtMMS21 interferes with the E2Fa-DPa interaction.
SUMOylation of DPa Mediated by AtMMS21 Dissociates the E2Fa/DPa Complex
Because AtMMS21 is a SUMO E3 ligase, we were interested in determining whether DPa is a substrate for SUMOylation and if so, whether AtMMS21 affects its SUMOylation. First, FLAG-tagged DPa was expressed in a reconstituted Escherichia coli system with Arabidopsis SUMOylation machinery proteins (Okada et al., 2009). In the presence of E1, E2, and AtSUMO1, a DPa-FLAG signal with higher molecular weight was detected in the immunoblot, suggesting that DPa is a potential substrate for SUMOylation. The GPS-SUMO1.0 software (Zhao et al., 2014) predicted Lys-146 of DPa to be a target residue. However, mutating Lys-146 did not affect SUMO conjugation of DPa (data not shown). When we mutated two other lysines (Lys-139 and Lys-140) adjacent to the predicted site, along with Lys-146, the SUMOylation of DPa was almost abolished (Figure 3C), suggesting that the three lysines are major target sites for DPa SUMOylation. We purified the SUMOylation components and used them to determine the effect of AtMMS21 on SUMO modification of DPa. The SUMOylation of DPa was enhanced with increasing AtMMS21 level, indicating that AtMMS21 facilitates the attachment of SUMO to DPa (Figure 3D).
Our structure prediction revealed that the SUMOylation sites on DPa (Lys-139, -140, and -146) reside at the edge of its dimerization domain, which interacts with E2Fa (Figure 3E), suggesting that SUMOylation may affect the DPa-E2Fa interaction. Based on previous studies in mammalian cells that examined the function of SUMOylation by fusing SUMO with its target protein (Ross et al., 2002), we fused AtSUMO1(ΔGG) to the N-terminal truncated version of DPa (DPa-C) at the position adjacent to its SUMOylation sites to mimic the SUMO-conjugated form. The results of yeast two-hybrid analysis indicated that the interaction between E2Fa and SUMO-fused DPa-C was much weaker than that between E2Fa and the sample without SUMO fusion (Figure 3F), suggesting that SUMOylation of DPa interferes with its interaction with E2Fa.
AtMMS21 Affects the Translocation of E2Fa/DPa
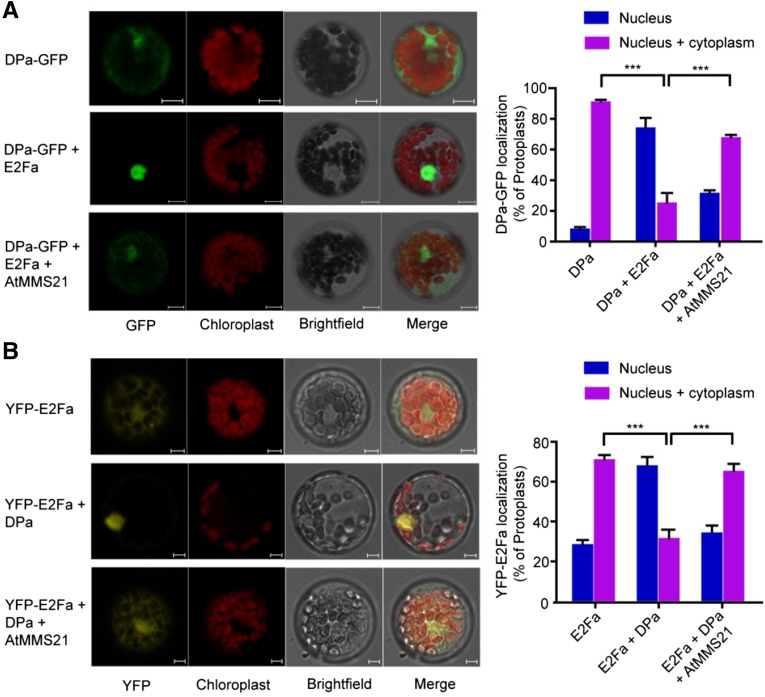
Since we determined that AtMMS21 interferes with the interaction between DPa and E2Fa, we next examined the consequences of this interference in the cell. Previous studies have revealed that when E2Fa or DPa is expressed separately, the protein localizes in the cytoplasm and nucleus; however, c-expression of both proteins drives complete nuclear localization (Kosugi and Ohashi, 2002). We therefore examined the effect of AtMMS21 on the translocation of the E2Fa/DPa complex in protoplasts. Consistent with the previous data, we observed DPa-GFP in the nucleus and cytoplasm in most of the cells. By contrast, we detected DPa-GFP only in the nucleus in ∼80% of the cells coexpressing CFP-E2Fa. Interestingly, when AtMMS21 was also overexpressed in the cells, the percentage of protoplasts with complete nuclear localization of DPa-GFP dramatically decreased, even in the presence of CFP-E2Fa (Figure 4A).
Figure 4.
AtMMS21 Affects the Translocation of E2Fa/DPa.
(A) Subcellular distribution of GFP-fused DPa proteins in protoplasts. Plasmids carrying 35S:DPa-GFP alone, or both 35S:DPa-GFP and 35S:AtMMS21, were transfected into wild-type protoplasts with or without CFP-E2Fa. When coexpressed with CFP-E2Fa, only the protoplasts with both CFP and GFP signals were used for analysis. Representative GFP signals from the majority of protoplasts from DPa-GFP alone, a combination of DPa-GFP and CFP-E2Fa, and a combination of DPa-GFP, AtMMS21, and CFP-E2Fa are shown. The autofluorescence from chloroplasts and bright-field (BF) signals were also detected and merged. Statistical data from means ± sd from three independent biological replicates (n > 100) are shown in the right panel. Bar = 10 µm.
(B) Subcellular distribution of YFP-fused E2Fa in protoplasts. YFP-E2Fa was transiently expressed in protoplasts from wild-type and transgenic plants carrying 35S:DPa alone or both 35S:DPa and 35S:AtMMS21. Representative YFP signals from the majority of the indicated protoplasts are shown. Statistical data from means ± sd from three independent biological replicates (n > 100) are shown in the right panel. ***P < 0.001, Student’s t test. Bar = 10 µm.
We also examined the subcellular localization of E2Fa in protoplasts from transgenic plants. In wild-type protoplasts, the YFP-E2Fa signal was detected throughout the cell. By contrast, YFP-E2Fa localized to the nucleus in ∼80% of cells from plants constitutively overexpressing DPa. However, the strict nucleus location of YFP-E2Fa was only observed in ∼30% of cells from plants co-overexpressing DPa and AtMMS21 (Figure 4B), suggesting that AtMMS21 dramatically disrupted the translocation of E2Fa/DPa, possibly by interfering with the DPa-E2Fa interaction. Because E2Fb also forms heterodimers with DPa and regulates cell cycle progression (Kosugi and Ohashi, 2002), we also performed a similar experiment to detect whether AtMMS21 affects the subcellular localization of E2Fb/DPa. Interestingly, the effect of AtMMS21 on the translocation of E2Fb was not significant (Supplemental Figure 2), implying the specificity of E2Fa in this regulation process. Taken together, these results suggest that AtMMS21 impairs the translocation of E2Fa/DPa from the cytoplasm to the nucleus.
Overexpression of DPa Has Effects on the Root Development of mms21-1
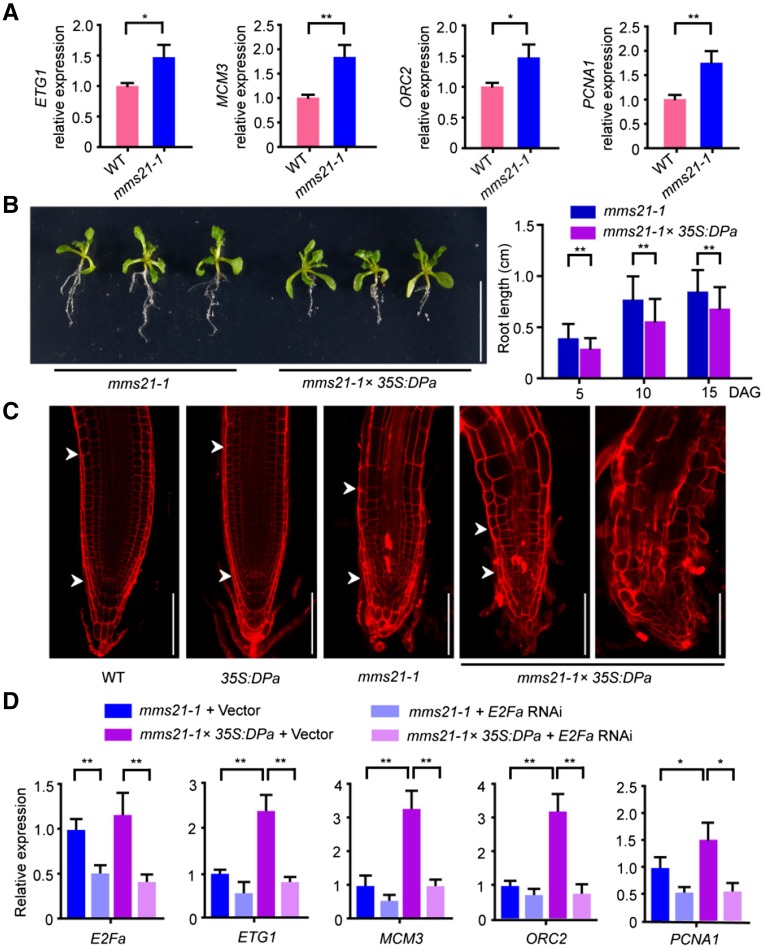
The data from our biochemistry and cell biology experiments suggest that AtMMS21 affects the E2Fa/DPa complex. To test the hypothesis, we compared the expression levels of ETG1, ORC2, MCM3, and PCNA1 (De Veylder et al., 2002; Egelkrout et al., 2002; Stevens et al., 2002; Diaz-Trivino et al., 2005; Takahashi et al., 2008), which are target genes of E2Fa/DPa, in the roots of wild-type and mms21-1 seedlings. The result indicated that the E2Fa target genes were upregulated in the mms21-1 roots (Figure 5A), providing evidence for the functional interaction between AtMMS21 and E2Fa/DPa.
Figure 5.
Overexpression of DPa Affects Root Development in mms21-1.
(A) The relative expression levels of E2Fa target genes in the roots of 4-d-old seedlings were analyzed by real-time PCR. The expression level in the wild type was set to 1. The data are means ± sd from triplicate experiments. *P < 0.05, **P < 0.01, Student’s t test.
(B) The phenotypes of mms21-1 and mms21-1×35S:DPa plants. The photograph was taken 15 d after germination. Statistical data of root length shown in the right panel are means ± sd from at least 30 seedlings in three biological independent experiments. **P < 0.01, Student’s t test. Bar = 1 cm.
(C) Representative root apical meristems from wild-type, 35S:DPa, mms21-1, and mms21-1×35S:DPa plants. Roots from 5-d-old seedlings were stained with PI. The meristem region is indicated by arrowheads; quiescent center is indicated by the lower arrowhead. Bar = 100 µm.
(D) The relative expression levels of E2Fa target genes in the protoplasts of mms21-1 and mms21-1×35S:DPa with or without RNA interference for E2Fa. The protoplasts were collected 60 h after transformation and used for RNA extraction for real time PCR. The expression level in mms21-1 with empty vector was set to 1. The data are means ± sd from three experiments. *P < 0.05, ** P < 0.01, Student’s t test.
Next, we used a genetic approach to explore the functional interaction among these proteins in plants. The physiological functions of the E2Fa/DPa complex can be studied by overexpression in Arabidopsis (De Veylder et al., 2002; Kosugi and Ohashi, 2003; Boudolf et al., 2004). To dissect the effect of AtMMS21 on the in vivo function of DPa, the DPa overexpression construct was introduced into the mms21-1 mutant by a genetic cross. The morphology of control 35S:DPa plants was identical to that of wild-type plants (Supplemental Figure 3), consistent with the results from the Inze group (De Veylder et al., 2002), suggesting that overexpressing DPa alone does not affect the development of wild-type plants. Surprisingly, overexpressing DPa in mms21-1 severely enhanced the mutant defect in root development at various growth stages (Figure 5B).
Analysis of the root meristem region provided an additional clue about the effect of DPa on root development. The meristem cell number was lower in mms21-1 plants than in wild-type or 35S:DPa plants. However, overexpressing DPa in mms21-1 obviously reduced the meristem cell number of this mutant and even completely destroyed the meristem region in some plants (Figure 5C), suggesting that overexpressing DPa in the AtMMS21 mutant enhances cell differentiation.
To uncover the reason for the effects of 35S:DPa in mms21-1, the expression of the E2Fa-target genes was measured in the protoplasts generated from mms21-1 or 35S:DPa×mms21-1 with or without RNA interference for E2Fa. In mms21-1, the RNA levels of target genes were significantly increased in the presence of 35S:DPa. However, this increase was dramatically suppressed when E2Fa was knockdown (Figure 5D), providing evidence that the abnormal cell differentiation is dependent on the E2Fa/DPa complex.
AtMMS21 Interferes with the Function of the E2Fa/DPa Complex in Plants
Previous studies have demonstrated that coexpression of DPa and E2Fa results in early arrested development and extra endoreduplication in Arabidopsis (De Veylder et al., 2002), providing a perfect system for us to detect the effect of AtMMS21 on the in vivo function of this complex. We crossed plants homozygous for 35S:E2Fa with plants homozygous for 35S:DPa and heterozygous for 35S:AtMMS21 (Supplemental Figure 4). Half of the offspring had short roots and curled cotyledons and half of the plants developed normally, like wild-type seedlings (Figure 6A; Supplemental Figure 4). PCR analysis of individual plants confirmed that plants with the arrested development phenotype contained both the 35S:E2Fa and 35S:DPa constructs, whereas the plants with normal phenotypes contained three constructs, 35S:E2Fa, 35S:DPa, and 35S:AtMMS21 (data not shown). The transcript levels of E2Fa and DPa did not change in the presence of AtMMS21 (Supplemental Figure 4), and overexpressing AtMMS21 had no obvious effect on plant development (Supplemental Figure 3), strongly suggesting that the phenotype difference resulted from the interaction among these proteins. The analysis of root length indicated that overexpressing AtMMS21 completely rescued defective root development in seedlings coexpressing E2Fa and DPa (Figures 6A and 6B). The abnormal leaf development and the extensive endoreduplication in cotyledons in the 35S:E2Fa-DPa plants were similar with the previous results from the Inze group (De Veylder et al., 2002). However, these abnormal phenotypes were returned to normal by overexpression of AtMMS21 (Figures 6C and 6D). Additionally, the results from hypocotyl and root cap indicated that abnormal cell division occurred in the E2Fa-DPa transgenic plants (De Veylder et al., 2002; Wildwater et al., 2005) was suppressed by the overexpression of AtMMS21 (Supplemental Figure 5). We found that the expression of E2Fa target genes increased in plants overexpressing E2Fa and DPa but returned to nearly normal levels in the presence of AtMMS21 overexpression (Figure 6E). These results strongly suggest that AtMMS21 has important effects on the function of the E2Fa/DPa complex in vivo.
Figure 6.
AtMMS21 Interferes with the Function of the E2Fa/DPa Complex in Plants.
(A) The phenotypes of the wild type, 35S: E2Fa-DPa, and three independent lines (#1, #2, and #3) from verified 35S:DPa-E2Fa-AtMMS21 plants. The photograph was taken 7 d after germination. Bar = 1 cm.
(B) Statistical data for root lengths of the indicated plants. The results are means ± sd from at least 35 seedlings per line at 7 DAG in three independent experiments. ***P < 0.001, Student’s t test.
(C) The phenotypes of 3-week-old seedlings.
(D) The endoreplication levels in the indicated plants were quantitatively analyzed by flow cytometry. The cotyledons of 8-d-old seedlings were used for analysis. The results are means ± sd from of at least three biological replicates.
(E) The relative expression levels of E2Fa target genes in the indicated plants were analyzed by real-time PCR. The expression level in the wild type was set to 1. The data are means ± sd from triplicate experiments. **P < 0.01, ***P < 0.001, Student’s t test.
(F) The effect of mutated AtMMS21 on the function of E2Fa/DPa. The root length of two independent lines (#1 and #2) of 35S:E2Fa-DPa-AtMMS21mut was compared with that of the wild type, 35S:E2Fa-DPa, or 35S:E2Fa-DPa-AtMMS21. The photograph was taken at 7 d after germination, and the statistical data are means ± sd from at least 20 seedlings. ***P < 0.001, Student’s t test. Bar = 1 cm.
We also evaluated the contribution of physical competition or SUMOylation to the function of E2Fa-DPa in planta. Given the SP-RING domain is required for the SUMO ligase activity of AtMMS21, the Cys-to-Ser (C178S) and His-to-Ala (H180A) mutations in this domain destroy its SUMO E3 activity (Ishida et al., 2009). When we introduced the mutated version of AtMMS21 with C178S and H180A into 35S:E2Fa-DPa plants, the defective root development of these plants was only partially rescued (Figure 6F), suggesting that SUMOylation and physical competition both contribute to the effect of AtMMS21 on the E2Fa/DPa complex.
DISCUSSION
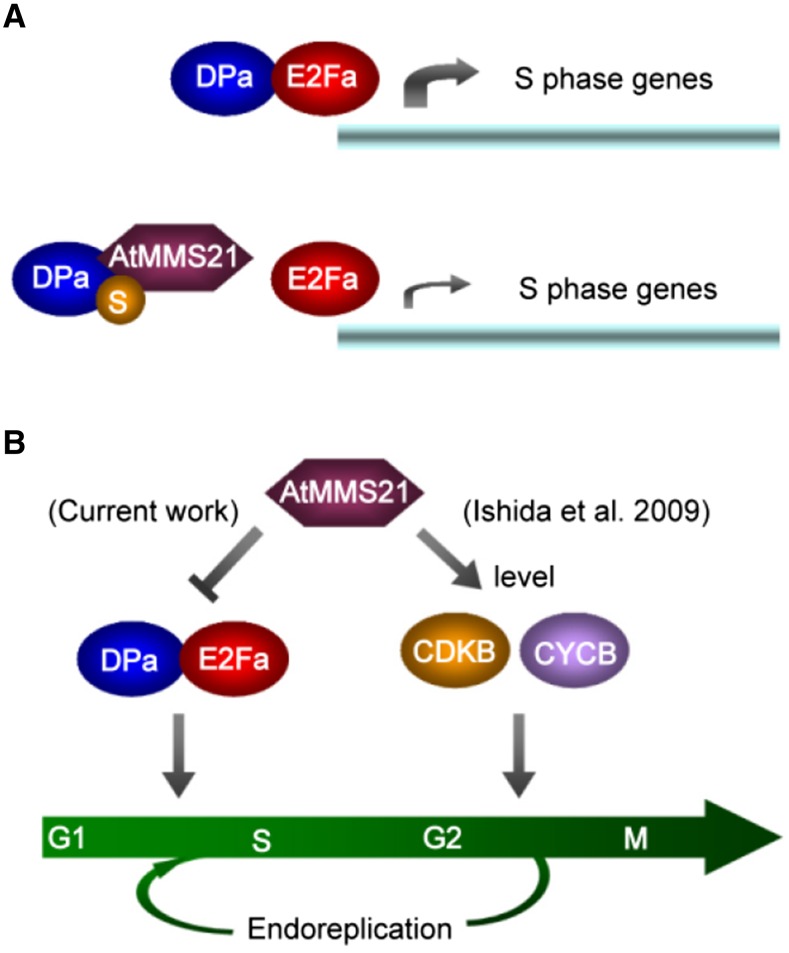
Plant development relies largely on the balance between cell proliferation and differentiation. The G1/S transition is a crucial crossroad at the interface between cell proliferation and differentiation (De Veylder et al., 2003). At the G1/S transition, CDKA/CYCD complexes phosphorylate RBR to dissociate E2F/DP transcription factors from RBR repression and to activate S-phase gene expression (Gutierrez, 2009). DPa forms heterodimers with E2Fa or E2Fb to enhance E2F/DPa translocation to the nucleus (Kosugi and Ohashi, 2002). Our data indicated that AtMMS21 interacts with DPa and significantly suppresses the translocation of E2Fa/DPa but not the E2Fb/DPa complex. This specificity may be a result of the difference in the structures for protein interaction. A previous study indicated that the expression of CDKB1:1 is upregulated in E2Fa-DPa-overproducing plants (Boudolf et al., 2004), suggesting that the E2Fa/DPa complex is also involved in the G2/M transition. The levels of CDKBs at the G2/M transition are reduced (Ishida et al., 2009), while the expression levels of ETG1, ORC2, MCM3, and PCNA1 are increased in the absence of AtMMS21, suggesting that the regulation of E2Fa/DPa by AtMMS21a is critical for the G1/S transition. These results also imply that AtMMS21 may have other potential targets to control the level of CDKB during the G2/M transition. The enhancement of endoreplication in the AtMMS21 mutant may result from stimulating the G1/S transition and blocking the G2/M transition. In a future study, it would be interesting to test whether overexpression of CDKB or CYCB in mms21-1 is able to overcome the defect in the cell cycle progression. Additionally, because E2Fa stimulates cell proliferation or endoreplication in different types of tissues (Magyar et al., 2012), the interaction between DPa and AtMMS21 may affect different types of cell cycle switches in specific tissues.
Our data show that the production of excess AtMMS21 reduces the interaction affinity between DPa and E2Fa in plant cells. Because AtMMS21 functions as a SUMO ligase, this interference may result from physical competition or SUMOylation. On the one hand, the dimerization domain of DPa interacts with E2Fa (Ramirez-Parra and Gutierrez, 2000), and our results indicate that this domain is also required for the interaction with AtMMS21; therefore, these proteins may physically compete, supported by our yeast three-hybrid assays. On the other hand, DPa is a substrate for SUMOylation, and AtMMS21 enhances this reaction. Interestingly, the SUMOylation sites on DPa are adjacent to its dimerization domain. Our SUMO fusion experiment revealed that attachment of SUMO, a 10-kD polypeptide, may interfere with the protein interaction around the modified sites and also has important effects on the E2Fa/DPa complex. Indeed, human Rb, a crucial factor that inhibits the E2F/DP complex, undergoes SUMOylation, which stimulates the release of the E2F/DP complex (Ledl et al., 2005; Li et al., 2006), suggesting that SUMOylation is critical to this pathway at the G1/S transition in various species. The finding that mutated AtMMS21 without SUMO ligase activity only partially rescued the abnormal development of plants overproducing DPa and E2Fa provides evidence that both physical competition and SUMOylation contribute to the dissociation of the E2Fa/DPa complex (Figure 7A). The interaction between Arabidopsis E2F and DP regulates their nuclear translocation and transactivation (Kosugi and Ohashi, 2002), suggesting that AtMMS21 may affect the E2Fa/DPa translocation and the expression of target genes by biochemical interference. A recent study showing that human MMS21 regulates SUMOylation and nuclear-to-cytoplasmic translocation of skNAC-Smyd1 in myogenesis revealed that SUMOylation has a conserved function in the translocation of protein complexes (Berkholz et al., 2014). Given that transcription occurs in the nucleus, the impairment of translocation would reduce the level of E2Fa/DPa in the nucleus, thereby reducing target gene expression.
Figure 7.
Predicted Model of the Role of AtMMS21 in Plant Cell Cycle Regulation.
(A) The effect of physical competition and SUMOylation on the E2Fa/DPa complex.
(B) The role of AtMMS21 in cell cycle checkpoint regulation.
The previous study showed that overexpression of a dominant-negative form of CDKB1;1 enhances the endoreplication in the plants coexpressing E2Fa/DPa, possibly by stimulating G1/S and blocking G2/M transition (Boudolf et al., 2004). Similarly, the E2Fa target genes are upregulated in the roots of mms21-1, suggesting that DPa and E2Fa may form a more stable complex with higher transcriptional activity and stimulate the G1/S transition in the absence of AtMMS21, but at the same time, the G2/M transition is blocked because the levels of CDKBs and CYCBs are reduced in this mutant (Ishida et al., 2009), resulting in enhanced differentiation (Figure 7B). When DPa is overexpressed in the wild-type plants, the excess DPa may have no significant effect on endogenous E2Fa/DPa activity. However, when DPa is overexpressed in mms21-1 plants, the excess DPa may escape from the interaction with AtMMS21 and enhance the interaction with endogenous E2Fa, resulting in a more severe differentiation phenotype than that of the mms21-1 mutant. The real-time PCR result in Figure 5D showed that 35S:DPa dramatically increases the expression of E2Fa/DPa target genes in mms21-1, and these effects are suppressed by interfering with E2Fa expression, supporting the idea that the phenotype is dependent on the higher activity of the E2Fa/DPa complex. As a model, in mms21-1, the G1/S transition is stimulated but the G2/M transition is suppressed. When DPa is overexpressed in mms21-1, the G1/S transition is more active because the excess DPa is not inhibited by AtMMS21 but the G2/M transition is still blocked due to downregulation of CDKB/CYCB in the absence of AtMMS21 (Ishida et al., 2009), resulting in enhanced differentiation.
Furthermore, in the 35S:E2Fa-DPa plants, there may not be sufficient levels of endogenous AtMMS21 to completely dissociate the excess E2Fa/DPa complex, resulting in abnormal cell division or endoreplication. When AtMMS21 is also overexpressed in the plants, the E2Fa/DPa complex is dissociated completely, recovering the abnormal phenotype to normal. Previous studies showed that E2Fa/DPa controls both division and endoreplication in different tissues (De Veylder et al., 2002; Boudolf et al., 2004). Differentiation is enhanced in cotyledons and some root regions in 35S:DPa-E2Fa, these phenotypes are completely similar with that in mms21-1. It is interesting that the root stem cell region in mms21-1 is overdifferentiated (Huang et al., 2009; Ishida et al., 2009), while the number of root stem cells in 35S:E2Fa-DPa is increased (Wildwater et al., 2005). It is possible that, in the stem cell region of 35S:DPa-E2Fa, the G1/S transition is stimulated but the G2/M transition is not inhibited in the presence of AtMMS21, resulting in enhanced cell division. These data support our model that the regulation of E2Fa/DPa by AtMMS21 is important for the G1/S transition.
The data from this work show that DPa is an important target for AtMMS21 in cell cycle control. However, numerous proteins are targets of SUMOylation, but cells have only a few SUMO E3 ligases; therefore, many factors in different pathways may share one SUMO ligase. For instance, AtSIZ1, another SUMO ligase in Arabidopsis, has been reported to regulate more than ten substrates in different signaling pathways (Wilkinson and Henley, 2010). That is why the developmental phenotypes of mms21-1 and 35S:E2Fa-DPa differ slightly. However, the cell cycle-related phenotypes, such as the endoreplication levels in cotyledons, the root developmental defect, and the expression levels of E2Fa-target genes, are similar in these two types of plants, providing evidence for their associated functions in cell cycle regulation. It will be interesting to identify other targets for AtMMS21 in different cellular processes in the future. Given that MMS21, E2F, and DP are conserved among various species, it is possible that the mechanism we discovered in Arabidopsis is also applicable to other organisms. For example, our data provide evidence for an interaction between rice MMS21 and DP proteins, implying that this interaction is conserved, at least in plants. A previous study in human cells indicated that hMMS21 is also involved in the G1/S transition (Ni et al., 2012). Therefore, in the future study, it will be interesting to determine whether the regulatory mechanism mediated by the MMS21-DP interaction is also conserved in mammalian cells.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana Columbia-0 (wild type) and mms21-1 (CS848340) were described previously (Huang et al., 2009). The seeds of the transgenic 35S:E2Fa and 35S:DPa lines (De Veylder et al., 2002) were kindly provided by the Inze Laboratory. Seeds were surface sterilized for 2 min in 75% ethanol followed by 5 min in 1% NaClO solution, rinsed five times with sterile water, plated on Murashige and Skoog medium with 1.5% Suc and 0.8% agar, and then stratified at 4°C in the dark for 2 d. Plants were grown under long-day conditions (16 h of light/8 h of dark, 80 μE s−1 m−2 light intensity) at 22°C.
Generation of Transgenic Plants
For the 35S:MYC-AtMMS21 construct, the coding sequence of AtMMS21 was obtained by PCR amplification and cloned into the PBA002-MYC vector at the BamHI/SpeI sites. To generate the 35S:MYC-AtMMS21mut construct, site-directed mutagenesis was performed using the MutanBest kit (Takara). For the 35S:DPa-FLAG construct, DPa was amplified and fused with a FLAG tag at its C terminus in the vector pPZPY122, and then the fragment containing 35S:DPa-FLAG was subcloned into pCAMBIA1302 at the HindIII/SpeI sites. For the E2Fa-RNAi construct, the fragment containing the inverted repeat sequences of E2Fa spliced by an intron was cloned into the pCanG-myc vector by BamHI and SpeI.
The constructs were transformed into Agrobacterium tumefaciens EHA105, which was then used to transform Arabidopsis (Columbia) by the floral-dip method (Clough and Bent, 1998). Homozygous lines of transgenic plants were used in this study. The 35S:MYC-AtMMS21 construct was introduced into homozygous 35S:DPa plants (De Veylder et al. 2002) for phenotype analysis or 35S:DPa-FLAG plants for biochemistry analysis.
Yeast Two-Hybrid and Three-Hybrid Assays
Yeast two-hybrid assays were performed according to the manufacturer’s instructions for the Matchmaker GAL4-based two-hybrid system 3 (Clontech). AtMMS21 was cloned into the pGBKT7 vector. The coding sequences of CDKA1, CYCB1;1, E2Fa, E2Fb, E2Fc, RBR, DPa, and DPb were cloned into the pGADT7 vector. For the interaction domain characterization, the truncated products were generated as described in the figure legends. For the SUMO1-DPa-C fusion construct, SUMO-1(ΔGG, amino acids 1 to 91) was fused to N terminus of DPa-C (amino acids 144 to 292) by overlap PCR and cloned into pGBKT7. The bait and prey constructs were transformed into the yeast strain AH109 (Clontech). Protein-protein interactions were tested by stringent (SD/–Leu/–Trp/–His) selection supplied with 3-amino-1,2,4-triazole and β-galactosidase activity measurement according to the manufacturer’s protocol (Clontech).
Yeast three-hybrid system (Clontech) was applied to analyze the effect of AtMMS21 on the E2FA-DPa interaction. E2Fa was cloned into pGADT7 and DPa was cloned into the three-hybrid vector pBridge (Clontech) fused with GAL4. AtMMS21 was cloned under the control of a MET25 promoter in the same pBridge vector. The bait and prey pair was cotransformed into the yeast strain Y2HGold. A quantitative analysis of α-galactosidase activity was performed using p-nitrophenylα-d-Galactopyranoside (PNP-α-Gal; Sigma-Aldrich) according to the Yeast Protocols Handbook (Clontech).
In Vitro Pull-Down Assay
GST or GST-AtMMS21 recombinant proteins were incubated with glutathione sepharose (GE Healthcare) in a binding buffer (50 mM Tris, pH 7.4, 120 mM NaCl, 5% glycerol, 0.5% Nonidet P-40, 1 mM PMSF, and 1 mM β-mercaptoethanol), for 2 h at 4°C, then were collected and mixed with supernatant containing His6-DPA-FLAG and incubated at room temperature for 60 min. After being rinsed five times with washing buffer (50 mM Tris, pH 7.4, 120 mM NaCl, 5% glycerol, and 0.5% Nonidet P-40), the bound proteins were boiled in SDS sample buffer and subjected to SDS-PAGE and immunoblotting.
Coimmunoprecipitation
The regular coimmunoprecipitation assay was performed using transgenic plants harboring both 35S:DPa-FLAG and 35S:DPa-FLAG-MYC-AtMMS21. Proteins were extracted from the leaves of 3-week-old transgenic plants in extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 20% glycerol, and 0.1% Nonidet P-40) containing protease inhibitor cocktail (Roche). After centrifugation at 13,000g for 12 min, the supernatant was incubated with anti-MYC antibody (Clontech) at 4°C overnight. Then, 50 μL of protein A agarose beads (Sigma-Aldrich) was added. After 3 h of incubation at 4°C, the beads were centrifuged and washed four times with washing buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 10% glycerol, and 0.1% Nonidet P-40). Proteins were eluted with SDS sample buffer and analyzed by immunoblotting.
For the interference assay, the YFP-E2Fa construct was transformed into Arabidopsis protoplasts (Yoo et al., 2007) from the indicated transgenic plants. After 60 h, protoplasts were harvested for immunoprecipitation following the coimmunoprecipitation protocol described above using FLAG M2 Affinity Gel (Sigma-Aldrich).
SUMOylation Assay
The SUMOylation assay in Escherichia coli was performed as previously described (Okada et al., 2009). DPa fused with a FLAG tag was cloned into pCDFDuet-1 (Novagen) and expressed in bacteria carrying pET28-SAE1a-His-AtSAE2 (E1) (Budhiraja et al., 2009) and pACYCDuet-1-SCE1-SUMO1 (GG) (E2 and SUMO). The transformed cells were cultured in LB medium to OD600 of 0.5 and induced by 0.5 mM IPTG. After incubation for 12 h at 25°C, cells were harvested and used for immunoblotting.
To detect the effect of AtMMS21 on the SUMOylation of DPa, the Arabidopsis SUMO E1 (SAE1a-His6-AtSAE2), E2 (His6-SCE1), His6-SUMO1 (GG), His6-DPa-FLAG, and GST-AtMMS21 were expressed in E. coli BL21(DE3) and purified by Ni-NTA Agarose (Qiagen) or Glutathione Sepharose 4B (GE Healthcare). For the in vitro SUMOylation assay, His6-SAE2/SAE1a (500 ng), His6-SCE1 (0 or 100 ng), His6-SUMO1(GG) (4 μg), substrate protein His6-DPa-FLAG (1 μg), GST-AtMMS21 (0, 90 ng, or 900 ng), and SUMO reaction buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 5% glycerol, 5 mM ATP, and 5 mM MgCl2) were mixed together in a total reaction volume of 25 mL and incubated at 30°C for 3 h. After boiling in SDS sample buffer, the samples were then analyzed by immunoblotting.
Subcellular Localization Detection
For the DPA localization analysis, E2Fa was cloned and fused with CFP under a 35S promoter in a modified pBluescript SK vector. DPa was amplified and fused with GFP in the pCAMBIA1302 vector to generate DPa-GFP. Next, AtMMS21 was cloned into the XhoI site of pCAMBIA1302-DPa by replacing the DNA fragment encoding hygromycin resistance, resulting in a plasmid expressing both DPa-GFP and AtMMS21. The constructs were cotransformed into Arabidopsis protoplasts as described previously (Yoo et al., 2007), and only the protoplasts expressing with both GFP and CFP fluorescence were used for analysis. For E2Fa or E2Fb localization analysis, E2Fa or E2Fb was cloned into the modified pBluescript SK vector containing 35S:YFP. The plasmid YFP-E2Fa or YFP-E2Fb was transformed into the protoplasts from transgenic plants overexpressing DPa-FLAG or DPa-FLAG/MYC-AtMMS21. The fluorescent GFP or YFP signals in transformed protoplasts were examined by confocal microscopy (Leica LSM710).
Root Length Analysis and Microscopy
The primary roots of plants incubated vertically on Murashige and Skoog medium were photographed using a Canon camera, and the lengths of the primary roots at indicated day were determined using Digimizer 3.2 software. For confocal laser imaging of roots, roots were counterstained with 10 μg/mL propidium iodide (PI; Sigma-Aldrich) for 1 min and mounted in water for confocal microscopy. For hypocotyl observation, the tissues were placed overnight in ethanol and subsequently washed in water, then mounted in chloroacetaldehyde:water:glycerol (8:3:1) solution for 10 to 20 min before microscopy.
Confocal images were taken using a Zeiss LSM 710 laser scanning microscope with the following excitation/emission wavelengths: 561 nm/591 to 635 nm for PI, 488 nm/505 to 530 nm for GFP, 514 nm/530 to 600 nm for YFP, and 458 nm/475 to 525 nm for CFP.
Flow Cytometric Analysis
Plants were chopped with a razor blade in 400 μL nuclei extraction buffer (Partec Cystain UV Precise P), the supernatant was filtered over a 30-µm mesh, and 1.6 mL staining buffer (Partec Cystain UV Precise P) was added. The nuclei were analyzed by CyFlow Cube 8 with CyView software (Partec). At least 8000 nuclei were scored in triplicates for each sample.
Gene Expression Analysis
Total RNA was extracted using the Plant RNAprep Pure Kit (Tiangen) with DNase I treatment following the manufacturer’s instructions. Reverse transcription was performed using a PrimeScript RT reagent kit (Takara). After the RT reaction, the cDNA template was subjected to PCR using SYBR Premix Ex Taq (Takara) in a Bio-Rad CFX 96 system (C1000 Thermal Cycler) and detected by Bio-Rad CFX Manager software. RT-qPCR was performed with three replicates and UBQ10 as a reference gene. For the measurement of gene expression in protoplasts, the transformation was performed as described (Yoo et al., 2007). The protoplasts were collected for RNA extraction after 60 h.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AtMMS21 (At3g15150), SAE1 (At3g57870), SAE2 (At2g21470), SCE1 (AT3G57870), SUMO1 (At4G26840), CDKA1;1 (At3g48750), CYCB1;1 (At4g37490), E2Fa (At2g36010), E2Fb (At5g22220), E2Fc (At1g47870), DPa (At5g02470), DPb (At5g03415), RBR (At3g12280), ETG1 (At2g40550), ORC2 (At2g37560), MCM3 (At5g46280), PCNA1 (At1g07370), UBQ10 (At4g05320), OsDP (LOC_Os01g48700), and OsMMS21 (LOC_Os05g48880).
Supplemental Data
Supplemental Figure 1. Detection of DPa-AtMMS21 interaction by bimolecular fluorescence complementation.
Supplemental Figure 2. Subcellular distribution of YFP-fused E2Fb in protoplasts.
Supplemental Figure 3. The phenotypes of wild-type, 35S:DPa, 35S:E2Fa, and 35S:AtMMS21 seedlings.
Supplemental Figure 4. The generation and identification of plants carrying 35S:E2Fa-DPa-AtMMS21.
Supplemental Figure 5. The phenotypes of hypocotyl and root cap of the 35S:E2Fa-DPa-AtMMS21 plants.
Supplemental Data Set 1. Primers used in this study.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31170269 and U1201212), the Major Project from the Education Department of Guangdong Province (2014), the Natural Science Foundation of Guangdong (S2012020011032), and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2010). We thank Lieven De Veylder (Ghent University) and the Arabidopsis Biological Resource Center for the seeds and A. Bachmair for the SUMO E1 plasmid used in this study.
AUTHOR CONTRIBUTIONS
Y.L., J.L., and C.Y. conceived the project and designed the research. Y.L., J.L., M.Y., F.W., J.Z., J.J., H.H., Q.W., G.L., and P.X. performed the research. Y.L., J.L., M.Y., and C.Y. analyzed the data. J.L. and C.Y. wrote the article.
Glossary
- CDK
cyclin-dependent kinase
- PI
propidium iodide
References
- al-Khodairy F., Enoch T., Hagan I.M., Carr A.M. (1995). The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J. Cell Sci. 108: 475–486. [DOI] [PubMed] [Google Scholar]
- Bandara L.R., Buck V.M., Zamanian M., Johnston L.H., La Thangue N.B. (1993). Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 12: 4317–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellail A.C., Olson J.J., Hao C. (2014). SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat. Commun. 5: 4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkholz J., Michalick L., Munz B. (2014). The E3 SUMO ligase Nse2 regulates sumoylation and nuclear-to-cytoplasmic translocation of skNAC-Smyd1 in myogenesis. J. Cell Sci. 127: 3794–3804. [DOI] [PubMed] [Google Scholar]
- Boniotti M.B., Gutierrez C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28: 341–350. [DOI] [PubMed] [Google Scholar]
- Boudolf V., Vlieghe K., Beemster G.T., Magyar Z., Torres Acosta J.A., Maes S., Van Der Schueren E., Inzé D., De Veylder L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhiraja R., Hermkes R., Müller S., Schmidt J., Colby T., Panigrahi K., Coupland G., Bachmair A. (2009). Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. 149: 1529–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Ouyang J., Abreu I.A., Hu Y., Seo H., Zhang X., Chua N.-H. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Conti L., Nelis S., Zhang C., Woodcock A., Swarup R., Galbiati M., Tonelli C., Napier R., Hedden P., Bennett M., Sadanandom A. (2014). Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 28: 102–110. [DOI] [PubMed] [Google Scholar]
- Dasso M. (2008). Emerging roles of the SUMO pathway in mitosis. Cell Div. 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Joubès J., Inzé D. (2003). Plant cell cycle transitions. Curr. Opin. Plant Biol. 6: 536–543. [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T., de Almeida Engler J., Ormenese S., Maes S., Naudts M., Van Der Schueren E., Jacqmard A., Engler G., Inzé D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21: 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Murray J.A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54: 235–264. [DOI] [PubMed] [Google Scholar]
- Diaz-Trivino S., del Mar Castellano M., de la Paz Sanchez M., Ramirez-Parra E., Desvoyes B., Gutierrez C. (2005). The genes encoding Arabidopsis ORC subunits are E2F targets and the two ORC1 genes are differently expressed in proliferating and endoreplicating cells. Nucleic Acids Res. 33: 5404–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout E.M., Mariconti L., Settlage S.B., Cella R., Robertson D., Hanley-Bowdoin L. (2002). Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14: 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N., Coupland G. (2010). Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA 107: 17415–17420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., Kirschner M.W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138. [DOI] [PubMed] [Google Scholar]
- Gutierrez C. (2009). The Arabidopsis cell division cycle. Arabidopsis Book 7: e0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Kastan M.B. (1994). Cell cycle control and cancer. Science 266: 1821–1828. [DOI] [PubMed] [Google Scholar]
- Helin K., Wu C.-L., Fattaey A.R., Lees J.A., Dynlacht B.D., Ngwu C., Harlow E. (1993). Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7: 1850–1861. [DOI] [PubMed] [Google Scholar]
- Huang L., Yang S., Zhang S., Liu M., Lai J., Qi Y., Shi S., Wang J., Wang Y., Xie Q., Yang C. (2009). The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 60: 666–678. [DOI] [PubMed] [Google Scholar]
- Ishida T., Fujiwara S., Miura K., Stacey N., Yoshimura M., Schneider K., Adachi S., Minamisawa K., Umeda M., Sugimoto K. (2009). SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome A., Gutierrez-Martinez P., Schiavoni F., Tenaglia E., Martinez P., Rodríguez-Acebes S., Lecona E., Murga M., Méndez J., Blasco M.A., Fernandez-Capetillo O. (2015). NSMCE2 suppresses cancer and aging in mice independently of its SUMO ligase activity. EMBO J. 34: 2604–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Rozhon W., Unterholzner S.J., Chen T., Eremina M., Wurzinger B., Bachmair A., Teige M., Sieberer T., Isono E., Poppenberger B. (2014). Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat. Commun. 5: 4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 128: 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2003). Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol. 132: 2012–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W., Livingston D.M., Shirodkar S. (1993). Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science 262: 1557–1560. [DOI] [PubMed] [Google Scholar]
- Ledl A., Schmidt D., Müller S. (2005). Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 24: 3810–3818. [DOI] [PubMed] [Google Scholar]
- Li S.-J., Hochstrasser M. (2000). The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20: 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Santockyte R., Shen R.-F., Tekle E., Wang G., Yang D.C., Chock P.B. (2006). Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J. Biol. Chem. 281: 36221–36227. [DOI] [PubMed] [Google Scholar]
- Liu M., Shi S., Zhang S., Xu P., Lai J., Liu Y., Yuan D., Wang Y., Du J., Yang C. (2014). SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis. BMC Plant Biol. 14: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H., Helin K. (2000). The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta 1470: M1–M12. [DOI] [PubMed] [Google Scholar]
- Magyar Z., Atanassova A., De Veylder L., Rombauts S., Inzé D. (2000). Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett. 486: 79–87. [DOI] [PubMed] [Google Scholar]
- Magyar Z., Horváth B., Khan S., Mohammed B., Henriques R., De Veylder L., Bakó L., Scheres B., Bögre L. (2012). Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 31: 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M. (2014). Cyclin-dependent kinases. Genome Biol. 15: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man J.-H., et al. (2006). PIAS3 induction of PRB sumoylation represses PRB transactivation by destabilizing its retention in the nucleus. Nucleic Acids Res. 34: 5552–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.J., Barrett-Wilt G.A., Hua Z., Vierstra R.D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.-J., Hasegawa P.M. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P.P., Dejean A. (2005). The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9: 769–779. [DOI] [PubMed] [Google Scholar]
- Nakagami H., Kawamura K., Sugisaka K., Sekine M., Shinmyo A. (2002). Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni H.-J., Chang Y.-N., Kao P.-H., Chai S.-P., Hsieh Y.-H., Wang D.-H., Fong J.C. (2012). Depletion of SUMO ligase hMMS21 impairs G1 to S transition in MCF-7 breast cancer cells. Biochim. Biophys. 1820: 1893–1900. [DOI] [PubMed] [Google Scholar]
- Okada S., Nagabuchi M., Takamura Y., Nakagawa T., Shinmyozu K., Nakayama J., Tanaka K. (2009). Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 50: 1049–1061. [DOI] [PubMed] [Google Scholar]
- Palvimo J.J. (2007). PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 35: 1405–1408. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E., Gutierrez C. (2000). Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett. 486: 73–78. [DOI] [PubMed] [Google Scholar]
- Ross S., Best J.L., Zon L.I., Gill G. (2002). SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10: 831–842. [DOI] [PubMed] [Google Scholar]
- Saracco S.A., Miller M.J., Kurepa J., Vierstra R.D. (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana A., Kaldis P. (2009). Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28: 2925–2939. [DOI] [PubMed] [Google Scholar]
- Schimmel J., Eifler K., Sigurðsson J.O., Cuijpers S.A., Hendriks I.A., Verlaan-de Vries M., Kelstrup C.D., Francavilla C., Medema R.H., Olsen J.V., Vertegaal A.C. (2014). Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell 53: 1053–1066. [DOI] [PubMed] [Google Scholar]
- Stephan A.K., Kliszczak M., Morrison C.G. (2011). The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett. 585: 2907–2913. [DOI] [PubMed] [Google Scholar]
- Stevens R., Mariconti L., Rossignol P., Perennes C., Cella R., Bergounioux C. (2002). Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J. Biol. Chem. 277: 32978–32984. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Lammens T., Boudolf V., Maes S., Yoshizumi T., De Jaeger G., Witters E., Inzé D., De Veylder L. (2008). The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 27: 1840–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.L., Ho J.C., Oliver A., Watts F.Z. (2002). Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease. J. Cell Sci. 115: 1113–1122. [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Vlieghe K., Florquin K., Hennig L., Beemster G.T., Gruissem W., Van de Peer Y., Inzé D., De Veylder L. (2005). Genome-wide identification of potential plant E2F target genes. Plant Physiol. 139: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.D., Lapi E., Sullivan A., Ratnayaka I., Goldin R., Hay R., Lu X. (2011). SUMO-modified nuclear cyclin D1 bypasses Ras-induced senescence. Cell Death Differ. 18: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R.A. (1995). The retinoblastoma protein and cell cycle control. Cell 81: 323–330. [DOI] [PubMed] [Google Scholar]
- Wildwater M., Campilho A., Perez-Perez J.M., Heidstra R., Blilou I., Korthout H., Chatterjee J., Mariconti L., Gruissem W., Scheres B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.A., Henley J.M. (2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428: 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Yuan D., Liu M., Li C., Liu Y., Zhang S., Yao N., Yang C. (2013). AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol. 161: 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yuan D., Lai J., Xu P., Zhang S., Zhang J., Li C., Wang Y., Du J., Liu Y., Yang C. (2014). AtMMS21 regulates DNA damage response and homologous recombination repair in Arabidopsis. DNA Repair (Amst.) 21: 140–147. [DOI] [PubMed] [Google Scholar]
- Zhang S., Qi Y., Yang C. (2010). Arabidopsis SUMO E3 ligase AtMMS21 regulates root meristem development. Plant Signal. Behav. 5: 53–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., Ren J. (2014). GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42: W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.