DDB2 influences de novo DNA methylation by interacting with ARGONAUTE4 and controlling the local abundance of 24-nucleotide short interfering RNAs.
Abstract
In eukaryotes, DNA repair pathways help to maintain genome integrity and epigenomic patterns. However, the factors at the nexus of DNA repair and chromatin modification/remodeling remain poorly characterized. Here, we uncover a previously unrecognized interplay between the DNA repair factor DNA DAMAGE BINDING PROTEIN2 (DDB2) and the DNA methylation machinery in Arabidopsis thaliana. Loss-of-function mutation in DDB2 leads to genome-wide DNA methylation alterations. Genetic and biochemical evidence indicate that at many repeat loci, DDB2 influences de novo DNA methylation by interacting with ARGONAUTE4 and by controlling the local abundance of 24-nucleotide short interfering RNAs (siRNAs). We also show that DDB2 regulates active DNA demethylation mediated by REPRESSOR OF SILENCING1 and DEMETER LIKE3. Together, these findings reveal a role for the DNA repair factor DDB2 in shaping the Arabidopsis DNA methylation landscape in the absence of applied genotoxic stress.
INTRODUCTION
Plants produce gametes from somatic lineages late during development. Therefore, genomic and epigenomic alterations occurring in these somatic cells can potentially be transmitted to the next generation. As photosynthetic organisms, plants need to prevent light and more specifically UV irradiation from inducing a large set of irreversible DNA damage. In plants, UV-induced DNA lesions are predominantly repaired by direct repair, catalyzed by photolyases (Kimura and Sakaguchi, 2006). In addition to direct repair, nucleotide excision repair (NER) also contributes to the repair of UV-induced DNA lesions (Kimura and Sakaguchi, 2006). NER comprises two subpathways, global genome repair (GGR) and transcription-coupled repair, both of which process bulky DNA lesions throughout the genome or along the transcribed strand of active genes, respectively (Schärer, 2013). The DNA DAMAGE BINDING2 (DDB2) protein is a key factor for the recognition of UV-induced DNA lesions during GGR. DDB2 binds photolesions as well as “compound” lesions leading to DNA helix distortions (Wittschieben et al., 2005). Mutations of the human and Arabidopsis thaliana DDB2 lead to UV hypersensitivity (Nichols et al., 2000; Molinier et al., 2008). DDB2’s turnover is crucial for efficient removal of bulky DNA lesions and depends on the ubiquitin E3-ligase complex CULLIN4-DNA DAMAGE BINDING protein 1 (CUL4-DDB1; Nag et al., 2001; Molinier et al., 2008).
Epigenomic alterations can also occur during plant development and can potentially be inherited. One such heritable component of plant epigenomes is cytosine methylation, which is required for the stable silencing of transposable elements (TEs) and the regulation of some genes (reviewed in Law and Jacobsen, 2010). In Arabidopsis, TEs and other repeats are methylated throughout their entire length at CG, CHG, and CHH sites (where H = A, T, or C), whereas ∼30% of genes are methylated at CG sites over part of their transcribed region (reviewed in Law and Jacobsen, 2010). Four main DNA methyltransferases are responsible for maintaining these methylation patterns during the plant life cycle and across generations. These are METHYLTRANSFERASE1 (MET1), a homolog of mammalian DNMT1; CHROMOMETHYLASE2 (CMT2) and CMT3, which are unique to plants; and DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2), a homolog of mammalian DNMT3. MET1 and CMT3 are responsible for maintaining methylation in CG and CHG sequence contexts, respectively (reviewed in Law and Jacobsen, 2010). CMT2 is to some extent redundant with CMT3 in methylating CHG sites but has a main function in CHH methylation together with DRM2 (Stroud et al., 2014). However, CMT2 and DRM2 act on different targets and only DRM2 is involved in de novo DNA methylation of repeat sequences, which is mediated by the RNA-directed DNA methylation (RdDM) pathway (Stroud et al., 2014). RdDM involves RNA POL IV and RNA POL V, two plant-specific DNA-dependent RNA polymerases evolutionarily related to RNA POL II (Matzke and Mosher, 2014). RNA POL IV together with RNA-DEPENDENT RNA POLYMERASE2 (RDR2) and the DICER-LIKE3 RNase allow the biogenesis of short interfering RNAs (siRNAs; reviewed in Law and Jacobsen, 2010; Matzke and Mosher, 2014), in a one-precursor/one-siRNA mode (Blevins et al., 2015; Zhai et al., 2015). Processed 24-nucleotide siRNAs loaded on ARGONAUTE4 (AGO4) protein are thought to interact with scaffold transcripts produced by RNA POL V, thereby targeting DRM2 to cognate DNA sequences (Zhong et al., 2014). RdDM is important for the transcriptional repression of TEs and for heterochromatin formation (reviewed in Law and Jacobsen, 2010). Alternative RdDM mechanisms have also been identified that rely on RNA POL II and RDR6 as well as on 21-nucleotide-long siRNAs (Pontier et al., 2012; Nuthikattu et al., 2013).
DNA methylation can be lost either through active DNA demethylation or passively across cell divisions, if de novo and maintenance DNA methylation activities are no longer present (Franchini et al., 2012). In Arabidopsis, enzymatic demethylation is mainly achieved during vegetative growth by the DNA glycosylases REPRESSOR OF SILENCING1 (ROS1), DEMETER LIKE2 (DML2), and DML3. These DNA glycosylases are thought to prevent spreading of DNA methylation away from loci targeted by RdDM (Zhu, 2009). Their activities are biochemically related to the base excision repair pathway, a process with major implications in the removal of non-helix-distorting base lesions from the genome (Zhu, 2009). In both plants and animals, factors at the nexus of DNA repair and DNA methylation dynamics remain poorly characterized. Although RNA POL IV and V are involved in double-strand break (DSB) repair through regulating DSB-induced small RNA biogenesis and functioning (Wei et al., 2012), the link with DNA methylation remains to be further investigated. Indeed, DSB-induced small RNA-mediated DSB repair does not involve the RdDM pathway (Wei et al., 2012).
Active DNA demethylation leads to transient changes in DNA topology, which are caused by the creation of either abasic sites or G/T mismatches upon 5-mC processing (Zhu, 2009; Franchini et al., 2012). Hence, DNA demethylation could theoretically recruit factors (e.g., DDB2, which binds to abasic sites) with affinity for bulky DNA lesions, such as those resulting from G/T mismatches (Wittschieben et al., 2005). Consequently, the ability of DDB2 to recognize changes in DNA structure may potentiate interplays between active DNA demethylation and GGR. Consistent with this scenario is the observation that NER contributes to active DNA demethylation in mammals (Le May et al., 2012; Naegeli and Sugasawa, 2011). GROWTH ARREST AND DNA DAMAGE INDUCIBLE ALPHA PROTEIN influences DNA demethylation activities and physically associates with XPG (XERODERMA PIGMENTOSUM; complementation group G), an essential factor during NER (Barreto et al., 2007). Conversely, XPG and XPF (XERODERMA PIGMENTOSUM; complementation groups G and F) contribute both to DNA demethylation and to the formation of transcriptionally permissive chromatin (Le May et al., 2012). In Arabidopsis, depletion of the plastid-localized Mismatch Repair factor MutS HOMOLOG1 induces heritable alterations of DNA methylation patterns, presumably through a chloroplast-to-nucleus retrograde signaling pathway (Virdi et al., 2015). However, a direct link between DNA repair effectors and DNA methylation/demethylation machineries remains to be established.
Here, we report that Arabidopsis mutants impaired in DDB2 exhibit genome-wide alterations of DNA methylation patterns in the absence of applied genotoxic stress. Furthermore, we provide evidence that DDB2 likely acts as a chaperoning factor to control AGO4-siRNA chromatin accessibility and, thus, de novo DNA methylation at target loci. Moreover, DDB2 influences the efficiency of DNA methylation maintenance. Finally, we show that DDB2 impacts active DNA demethylation by regulating ROS1 and DML3 expression. Together, these findings identify a previously unrecognized interaction between the DNA repair factor, DDB2, and the DNA methylation machinery.
RESULTS
DDB2 Shapes the DNA Methylation Landscape
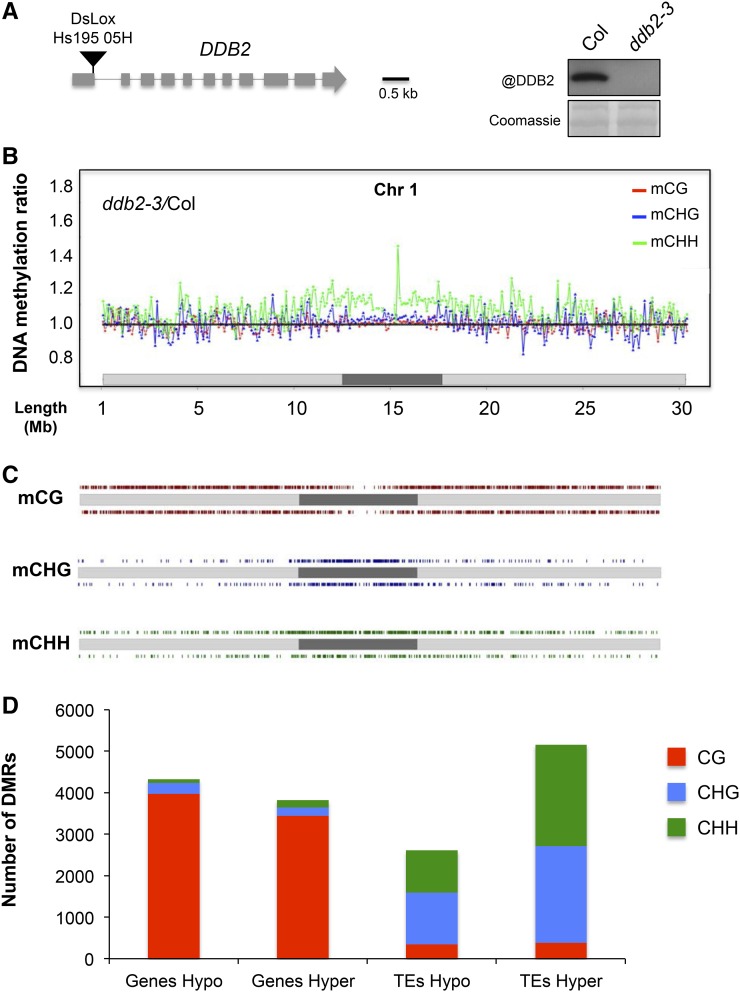
To assess the putative role of DDB2 in DNA methylation, we compared the methylome of Arabidopsis wild-type plants with loss-of-function mutants for DDB2. Mutant plants are sensitive to various DNA damaging agents but do not exhibit obvious developmental phenotypes under standard laboratory growth conditions. The DNA methylome was determined by MeDIP-chip (methyl DNA-immunoprecipitation-chromatin immunoprecipitation) for the ddb2-2 allele in a Nossen (No) accession (Molinier et al., 2008) and by whole-genome bisulfite sequencing (WGBS) for a newly identified ddb2-3 allele in the Columbia-0 background (Col; Figure 1A). Comparison of the DNA methylation profiles of the mutants with those of the relevant wild type (No or Col) revealed numerous differentially methylated regions (DMRs; Figures 1B and 1C; Supplemental Figures 1A, 1B, and 2A). Remarkably, DDB2 loss-of-function plants exhibited a large number of hypermethylated DMRs (hyper-DMRs; Figure 1D; Supplemental Figure 2B). These DMRs are concentrated within centromeric and pericentromeric regions that are highly enriched in TEs and other repeated elements (Figures 1B and C; Supplemental Figures 1A, 1B, 2A, and 2B). Conversely, hypomethylated-DMRs (hypo-DMRs) were more evenly distributed throughout the genome (Figures 1B and 1C; Supplemental Figures 1A, 1B, 2A, and 2B). Single-nucleotide resolution analyses of WGBS data further identified that methylation changes in the CG context were mostly located over genes, while CHG and CHH methylation changes were predominantly over TEs (Figure 1D).
Figure 1.
Context-Dependent DNA Methylation Differences Induced by the ddb2-3 Mutation.
(A) Schematic representation of the DDB2 locus. The black triangle represents the DsLox insertion site, which is located at the 3′ end of the first exon. Exons and introns are shown as gray boxes and lines, respectively. Immunodetection of DDB2 in wild-type (Col) and ddb2-3 plants. Coomassie blue staining was used as loading control.
(B) Differential DNA methylation levels between ddb2-3 and wild-type plants in the CG (red), CHG (blue), and CHH (green) sequence contexts in consecutive 100-kb windows along chromosome 1 (light gray, chromosome arms; dark gray, pericentromeric regions), as determined by WGBS.
(C) Distribution of DMRs between ddb2-3 and wild-type along chromosome 1 for the three sequence contexts (light gray, chromosome arms; dark gray, pericentromeric region; CG, red; CHG, blue; CHH, green). Hyper- and hypo-DMRs (relative to the wild type) are shown above and below each chromosome, respectively.
(D) Total number and identity of CG-, CHG-, and CHH-DMRs (hyper and hypo) between ddb2-3 and wild-type plants.
Representative hyper-/hypo-DMRs overlapping genes and TEs in ddb2-2 and ddb2-3 were confirmed independently by McrBC digestion coupled to quantitative PCR (McrBC-qPCR) on individual loci (Supplemental Figures 1C and 2C). However, while the MeDIP-chip and WGBS data revealed similar trends, there was little overlap between the DMRs identified in No and Col (Supplemental Data Set 1), suggesting accession-specific effects (Schmitz and Ecker, 2012). Moreover, it cannot be excluded that different ecotypes differ in their global 5-mdC contents as already reported (Rozhon et al., 2008). Therefore, to assess putative ecotype-specific epigenomic features, we measured the 5-mdC levels of wild-type No, Col plants as well as ddb2-2 and ddb2-3 mutant plants using ultraperformance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS). Surprisingly, we found that the No ecotype contains significantly reduced 5-mdC levels compared with the Col ecotype (Supplemental Figure 3A). Furthermore, ddb2-2 plants exhibit a significantly higher 5-mdC level than their corresponding wild-type control (No ecotype), while ddb2-3 does not (Supplemental Figure 3A). This analysis reveals that variability in global 5-mdC levels between ecotypes exists and may contribute to the limited number of DMRs overlapping between the two ddb2 mutant lines.
To assess the potential functional significance of the observed DNA methylation changes, we measured the RNA levels of TEs and genes harboring the most significant hypo- and hyper-DMRs by RT-qPCR (Supplemental Data Set 1). Loss and gain of DNA methylation were associated with higher and lower transcript levels, respectively, for the TEs tested (Supplemental Figure 2D), in agreement with the role of DNA methylation in TE silencing in Arabidopsis (Teixeira and Colot, 2010; Law and Jacobsen, 2010). By contrast, no consistent change in RNA levels was observed for hypo- or hyper-DMRs located within genes (Supplemental Figure 2E), as could be expected given the more complex relationship between DNA methylation and gene expression in Arabidopsis (Coleman-Derr and Zilberman, 2012).
Collectively, our data revealed that DDB2 loss of function leads to genome-wide alterations of DNA methylation patterns with a pronounced effect on regions enriched in TEs.
DDB2 Loss of Function Leads to Reduced Efficiency of DNA Methylation Maintenance Processes
DNA methylation changes observed in ddb2 mutant plants may result from local effects in cis as well as from indirect effects such as those caused by expression changes of genes involved in DNA methylation. To explore the latter possibility, the transcript levels of the major components of Arabidopsis DNA methylation pathways were tested by RT-qPCR. No significant change in mRNA levels were detected in ddb2-2 nor ddb2-3 plants compared with wild-type isogenic lines for any of the genes tested, ruling out a general misregulation of DNA methylation pathways in the mutant plants (Supplemental Figure 3B). To further assess the potential link between DDB2 and specific DNA methylation pathways, we compared the methylation levels of met1-, cmt2-, cmt3-, and nrpd1a-derived hypo-DMRs (Stroud et al., 2013) with their corresponding genomic positions in ddb2-3. We found that ddb2-3 plants did not exhibit the DNA hypomethylation levels resembling those found in met1, cmt2, cmt3, or nrpd1a plants (Supplemental Figure 3C), suggesting that DDB2 does not play a direct role in DNA methylation maintenance processes. However, we cannot exclude the possibility that DDB2 indirectly affects DNA methylation maintenance efficiency. We therefore compared the DNA methylation levels of ddb2-induced hypo-DMRs with their corresponding genomic positions in met1-, cmt2-, cmt3-, and nrpd1a-derived hypo-DMRs (Stroud et al., 2013). We found that in met1, cmt3, and nrpd1a plants, DNA methylation levels are strongly reduced compared with wild-type plants (Supplemental Figure 3D). Conversely, in cmt2 plants, CHH methylation did not show a significant reduction compared with wild-type plants (Supplemental Figure 3D). Therefore, loss of DDB2 causes weak, but significant, alterations of DNA methylation mediated by MET1, CMT3 and RdDM. These results suggest a potential relationship between DDB2 and the DNA methylation maintenance processes.
DDB2 Loss of Function Partially Impacts Gene Methylation through ROS1 and DML3 Misregulations
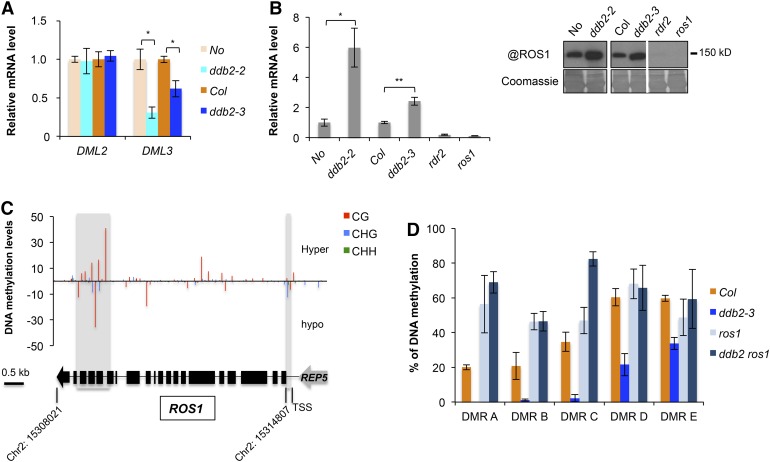
DNA methylation changes identified in ddb2 mutant plants may also result from expression changes of genes involved in active DNA demethylation. While DML2 expression remained unchanged in ddb2 plants, DML3 was down-regulated (Figure 2A). Interestingly, elevated ROS1 mRNA and protein levels were detected in ddb2 plants (Figure 2B).
Figure 2.
DDB2 and the DNA Demethylation Pathway.
(A) RT-qPCR analysis of transcript levels (±sd) of genes encoding the DNA glycosylases DML2 and DML3, in wild-type (No), ddb2-2, wild-type (Col), and ddb2-3 plants. t test, *P = 1.46 10−3 ;**P = 6.95 10−4; ***P = 2.26 10−3. Data are representative of three biological replicates.
(B) Left panel: RT-qPCR analysis of transcript levels (±sd) of genes encoding the DNA glycosylase ROS1 in wild-type (No), ddb2-2, wild-type (Col), ddb2-3, rdr2, and ros1 plants. t test, *P = 1.31 10−3; **P < 0.01. Data are representative of three biological replicates. Right panel: Immunodetection of the ROS1 protein in wild-type (No), ddb2-2, wild-type (Col), ddb2-3, rdr2, and ros1 plants. Coomassie blue staining of the blot is shown.
(C) Top panel: DNA methylation level for the ROS1 locus in ddb2-3 plants. Regions exhibiting significant changes in DNA methylation levels are shaded. Bottom panel: Schematic representation of the ROS1 locus (Chr2: 15,308,021..15314807). Exons and introns are shown as black boxes and lines, respectively. The REP5 transposon is shown in gray. TSS, transcription start.
(D) Genetic interaction between ddb2 and ros1. Percentage of DNA methylation in Col, ddb2, ros1, and in ddb2 ros1 plants for five hypo-DMRs. Data are presented as percentage of methylation (±sd) determined by McrBC-qPCR and are representative of three biological replicates.
Examination of the ddb2-3 methylation profiles identified one CHG hypomethylated region in ddb2-3 mutant plants, over the transcription start site of ROS1 (Figure 2C). This observation is in agreement with data indicating that DNA demethylation of a monitoring sequence (MEMS) in the ROS1 promoter is part of a cis-autoregulatory control (Lei et al., 2015; Williams et al., 2015). In addition, a distinct CG methylation profile was detected at the 3′ end of the gene (Figure 2C). These observations suggest that ROS1 expression is under a complex DNA methylation-based regulatory mechanism that involves, directly or indirectly, DDB2 in addition to RdDM.
Considering that upregulation of ROS1 in ddb2 mutant plants might generate hypo-DMRs, we compared the ddb2-induced hypo-DMRs with those generated by ros1 (Qian et al., 2012). We found that only 6.5 and 7.1% of the ddb2-induced hypo-DMRs overlapped with ros1-derived hyper-DMRs in all cytosine contexts, on protein-coding genes and TEs, respectively (Supplemental Figure 4A). This suggests that only a small proportion of the ddb2-induced hypo-DMRs might directly result from increased ROS1 activity. To test this hypothesis, ddb2-derived hypomethylated regions overlapping with ros1-hyper-DMRs on protein-coding genes were analyzed by McrBC-qPCR in ros1 ddb2 double mutant plants. In agreement with this hypothesis, DNA hypomethylation at these loci was released in the double mutant plants, thus demonstrating that the hypomethylation induced by ddb2 at these genic regions results from ROS1 misregulation (Figure 2D).
The ddb2 mutant plants also exhibit downregulation of the related DNA glycosylase gene DML3 (Figure 2A). This finding prompted us to examine whether gain of DNA methylation in ddb2 could result from this downregulation. To this end, we compared the ddb2-hyper-DMRs with the hyper-DMRs of the triple mutant for ROS1, DML2, and DML3 genes (rdd mutant; Stroud et al., 2013). Overall, more than 50% and ∼23% of ddb2-induced hyper-DMRs overlapped with rdd-derived hyper-DMRs in all cytosine contexts, on protein-coding genes and TEs, respectively (Supplemental Figure 4B).
Collectively, these analyses suggest that alterations of DNA methylation in ddb2 plants is in part caused by complex misregulation of ROS1 and/or DML3 genes.
Control of Proper DNA Methylation Patterns Relies on DDB2 DNA Binding Capacity
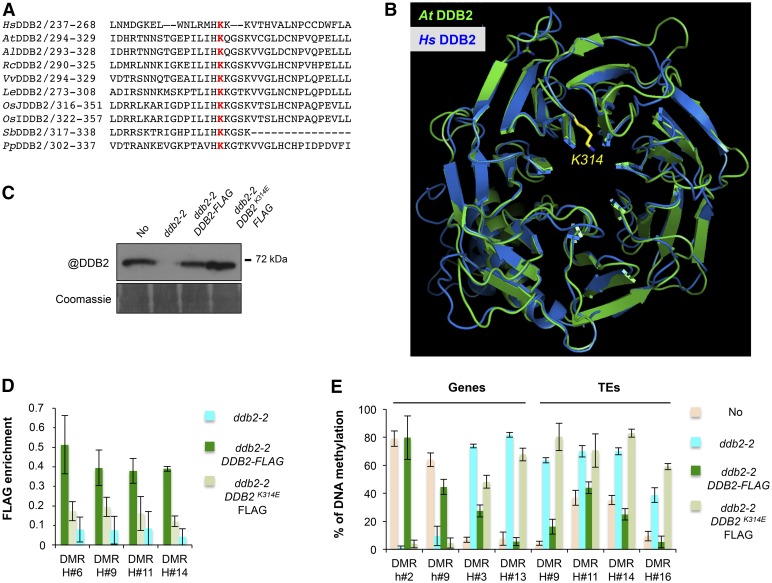
The ability of human DDB2 to bind damaged DNA relies on an internally localized lysine residue that protrudes in the DDB2-WD40 β propeller (K244; Cleaver et al., 1999; Scrima et al., 2008). The position of this residue appears to be evolutionarily conserved from plants to human, as estimated using amino acid sequence alignments and modeling of Arabidopsis DDB2 folding (Scrima et al., 2008; Figures 3A and 3B). To test for a possible functional relationship between DDB2 DNA binding capacity and its effect on DNA methylation, we generated ddb2-2 mutant plants stably expressing a wild-type or a K314E mutated DDB2 form (Figures 3B and 3C). Chromatin Immunoprecipitation (ChIP) of C-terminal FLAG-tagged DDB2 proteins revealed a significant enrichment of wild-type DDB2-FLAG over ddb2-2-derived hyper-DMRs, which was much reduced for DDB2K314E-FLAG, indicating an involvement of K314 in Arabidopsis DDB2 DNA binding at these loci (Figure 3D). We then analyzed by McrBC-qPCR the DNA methylation levels of the same ddb2-2-induced DMRs in the complemented plants. While introduction of nonmutated DDB2-FLAG largely restored proper DNA methylation levels at these loci, this was not the case for DDB2K314E-FLAG (Figure 3E). We concluded from these analyses that functional DNA binding capacity is required for DDB2 to influence local DNA methylation at its target sites independently of exogenous induction of DNA damage.
Figure 3.
Functional Link between DDB2 DNA Binding Capacity and DNA Methylation.
(A) Amino acid alignment of the human DDB2 protein domain surrounding K244 with various plant homologs. Hs, Homo sapiens; At, Arabidopsis thaliana; Al, Arabidopsis lyrata; Rc, Ricinus communis; Vv, Vitis vinifera; Le, Lycopersicon esculentum; OsJ, Oryza sativa group Japonica; OsI, Oryza sativa group Indica; Sb, Sorghum bicolor; Pp, Populus populi. K314, involved in DNA binding and highlighted in red, is highly conserved among all analyzed sequences.
(B) In silico prediction of the structural conservation of Arabidopsis DDB2 protein (green) upon modeling with the human (blue) DDB2 protein structures. K314 is shown in yellow.
(C) Immunodetection of the DDB2-FLAG proteins in ddb2-2/DDB2-FLAG and in ddb2-2/DDB2K314E-FLAG-expressing plants using an anti-DDB2 antibody. Wild-type (No) and ddb2-2 plants were used control. Coomassie blue staining of the blot is shown.
(D) ChIP-qPCR analysis of DDB2 enrichment at hyper-DMR over four TEs in ddb2-2/DDB2-FLAG and ddb2-2/DDB2K314E-FLAG-expressing plants using an anti-FLAG antibody. ChIP performed with ddb2-2 plants with anti-FLAG antibody was used as the negative control. Data are presented as enrichment of FLAG signal (±sd) and are representative of three biological replicates.
(E) Complementation assay for DMRs overlapping two genes and four TEs in ddb2-2/DDB2-FLAG and ddb2-2/DDB2K314E-FLAG expressing plants. Wild-type (No) and ddb2-2 mutant plants were used as controls. Data are presented as percentage of methylation (±sd) and are representative of three biological replicates measured by McrBC-qPCR. H, hyper-DMR; h, hypo-DMR.
DDB2 Impacts the Methylation of TEs through a siRNA-Based Process
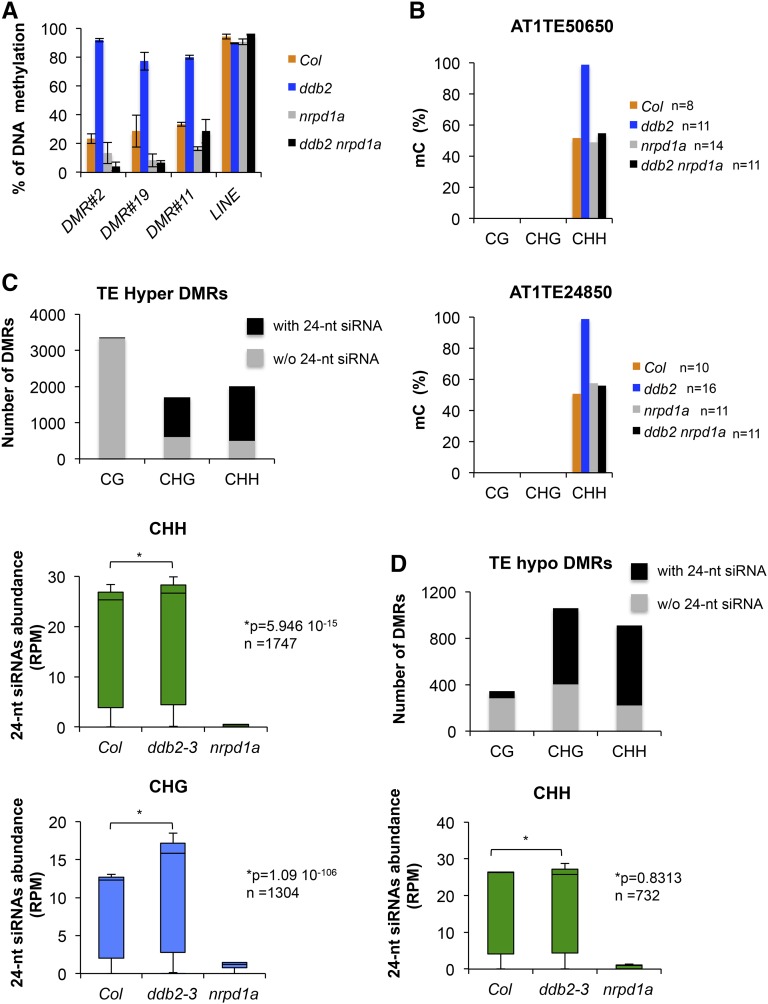
A remarkable feature of the ddb2-induced alterations is the predominant gain of DNA methylation on TEs (Figure 1D; Supplemental Figure 2B). Given the local influence of DDB2 on DNA methylation, we further assessed whether DNA hypermethylation in ddb2 plants involves RdDM. In a first approach, we tested whether representative ddb2-derived hyper-DMRs were maintained upon crossing with RNA POL IV (NRPD1A) mutant plants. McrBC-qPCR analyses showed that hypermethylation was suppressed in ddb2 nrpd1a double mutant plants for the tested DMRs (Figure 4A). In a second approach, we confirmed by Sanger bisulfite sequencing that such gain of DNA methylation (CHH) was suppressed in ddb2 nrpd1a double mutant plants on representative TEs (Figure 4B). This indicates that DNA methylation is acquired and maintained in a RNA POL IV-dependent manner.
Figure 4.
Functional Link between DDB2 and the RdDM Pathway.
(A) Percentage of DNA methylation (±sd) of three ddb2-derived hypermethylated TEs in wild-type (Col), ddb2, nrpd1a, and ddb2 nrpd1a plants determined by McrBC-qPCR. Data are representative of three biological replicates. LINE TE was used as control.
(B) Percentage of DNA methylation of two ddb2-derived hypermethylated TEs for each cytosine context in wild-type (Col), ddb2, nrpd1a, and ddb2 nrpd1a plants determined by Sanger bisulfite sequencing. n = number of clones analyzed.
(C) Top panel: Histograms representing the number of identified ddb2-induced hyper-DMRs overlapping with and without 24-nucleotide siRNA at TEs. Bottom panels: Box plots representing the abundance of 24-nucleotide siRNAs mapping to hypermethylated TEs in wild-type (Col), ddb2-3, and nrpd1a plants for CHG and CHH contexts. The abundance of 24-nucleotide siRNAs is normalized against global small RNA content and expressed as reads per million (RPM). P values were calculated according to Wilcoxon matched-pairs signed ranks test. n = number of hyper-DMRs overlapping with 24-nucleotide siRNA for all cytosine contexts.
(D) Top panel: Histograms representing the number of identified ddb2-induced hypo-DMRs overlapping with and without 24-nucleotide siRNA at TE. Bottom panel: Box plots representing the abundance of 24-nucleotide siRNAs mapping to hypomethylated TEs in wild-type (Col), ddb2-3, and nrpd1a plants in the CHH context. The abundance of 24-nucleotide siRNAs is normalized against global small RNA content and expressed as reads per million. P value is calculated according to Wilcoxon matched-pairs signed ranks test. n = number of hypo-DMRs overlapping with 24-nucleotide siRNA.
To examine the involvement of the RdDM pathway on a larger scale, we then compared the distribution of 24-nucleotide siRNAs over ddb2-derived hyper-DMRs overlapping with TEs in ddb2-3 and nrpd1a seedlings. This revealed that a large proportion (over 50%) of the TEs hypermethylated in CHG or CHH but not in CG (<1%) contexts were associated with 24-nucleotide siRNAs in ddb2-3 mutant plants, in agreement with the targeting of these TEs by RdDM (Figure 4C). For non-CG methylated domains, the abundances of 24-nucleotide siRNAs were significantly increased in ddb2-3 plants whereas, as expected, abundance dramatically decreased in nrpd1a mutant plants (Figure 4C). Accordingly, we further observed that ddb2-induced hyper-DMRs overlap with preexisting DNA methylation in wild-type plants for all sequence contexts, a known prerequisite for siRNA-mediated methylation (Law and Jacobsen, 2010).
Importantly, among the CHH and CHG hyper-DMRs overlapping with 24-nucleotide siRNAs, only a small proportion (15%) exhibited a significant accumulation of 24-nucleotide siRNAs to a higher level than they were before, again in agreement with the local influence of siRNA abundance on DNA methylation levels. This further suggests that other factors or differential local availability of RdDM effectors may have led to enhanced DNA methylation levels in ddb2 mutant plants.
Given that 24-nucleotide siRNAs also guide reestablishment of DNA methylation by DRM2 (Stroud et al., 2014), we tested whether their abundances were affected in ddb2-3 over hypo-DMRs overlapping with TEs. In agreement with this hypothesis, a large majority of the CHH hypomethylated TEs corresponded to domains targeted by 24-nucleotide siRNAs in wild-type plants (around 75%; Figure 4D). Interestingly, their abundances over these hypo-DMRs remained unchanged in the ddb2-3 mutant background (Figure 4D). Consistent with this, more than 94% of the hypo-DMRs did not exhibit decreases in siRNA abundance. Consequently, other factors or local changes in RdDM effector availability may have led to inefficient reestablishment of DNA methylation patterns.
Collectively, these results suggest that DDB2 and RdDM factors cooperate in a complex process that regulates de novo DNA methylation and its reestablishment through siRNA biogenesis, change in abundance, and/or modulation of the activity of effectors.
DDB2 and AGO4 Assemble with siRNAs within High Molecular Weight Complexes
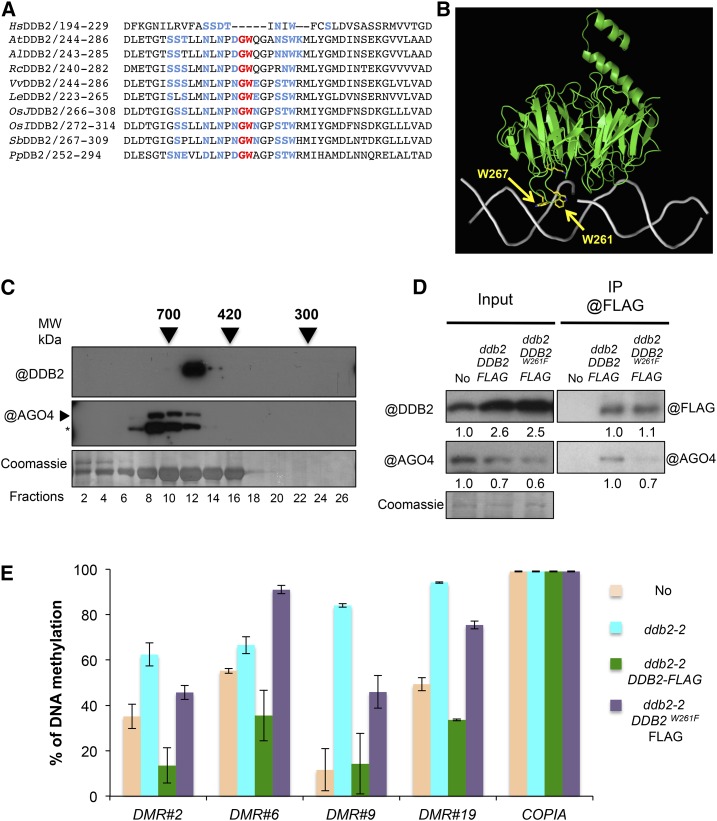
Considering its DNA binding capacity and its link with siRNAs, we wondered whether DDB2 could physically associate with factors triggering de novo DNA methylation. Examination of plant DDB2 amino acid sequences identified a conserved GW motif (Figure 5A) that could potentially be part of an AGO binding domain (El-Shami et al., 2007; Haag and Pikaard, 2011). Additional surrounding amino acids that are key to defining AGO hooks (rich in G, W, S, T, D, E, N, and K and poor in M, C, H, F, and Y; Karlowski et al., 2010) were also found in all plant DDB2 protein sequences examined (Figure 5A). Modeling of DDB2 protein folding positioned the W261 and W267 residues outside the surface of the protein in the vicinity of the bound DNA, close to K314, which is involved in DNA binding (Figure 5B). In view of these observations, we first tested whether DDB2 and AGO4 proteins eluted in fractions of similar sizes in gel filtration analyses. As shown in Figure 5C, DDB2 and AGO4 coeluted in fractions of ∼600 kD. Moreover, the DDB2-FLAG protein coimmunoprecipitated with endogenous AGO4 protein from plant whole-cell extracts (Figure 5D). Finally, transgenic ddb2-2 plants expressing a DDB2W261F-FLAG version were generated and used to test the dependency on the candidate AGO binding domain for association with AGO4. The W261 point mutation slightly reduced AGO4 coimmunoprecipitation efficiency (Figure 5D). Taken together, these results provide compelling evidence that DDB2 can assemble with AGO4 in one or more high molecular weight complexes, presumably through its GW261 motif. We analyzed by McrBC-qPCR the DNA methylation levels of some ddb2-2-induced DMRs in the complemented plants. While introduction of nonmutated DDB2-FLAG largely restored proper DNA methylation levels at these loci, this was not the case for DDB2W261F-FLAG (Figure 5E).
Figure 5.
Characterization of the DDB2-AGO4 Protein Complex.
(A) Amino acid alignment of the Arabidopsis DDB2 protein domain surrounding the GW motif with various plant and human homologs. The GW motif (red) absent in human but conserved among plant DDB2 proteins. Surrounding key amino acids that define the AGO hook are shown in blue. Species abbreviations are given as in Figure 3.
(B) In silico modeling of the Arabidopsis DDB2 protein structure (green) bound to a DNA helix (gray). Tryptophan residues (W) surrounding the GW motif and K314 are exposed to the DNA helix (gray) and shown in yellow.
(C) Size-exclusion chromatography analysis of the DDB2 complex in soluble protein extracts from wild-type (Col) plants. The indicated fractions were analyzed by immunoblot using anti-DDB2 and anti-AGO4 antibodies. Arrows indicate elution peaks of molecular weight standards in the same conditions. Coomassie blue staining of the blot is shown. Asterisk indicates cross-reacting signal.
(D) In vivo pull-down of AGO4 with DDB2 protein. Wild-type (No), ddb2-2/DDB2-FLAG, and ddb2-2/DDB2W261F-FLAG expressing plants were used for immunoprecipitation assays using anti-FLAG antibody. Coomassie blue staining of the blot is shown. Signal intensity relative to control is given below each lane.
(E) Complementation assay for four DMR overlapping TEs in ddb2-2/DDB2-FLAG and ddb2-2/DDB2W261F-FLAG expressing plants. Wild-type (No) and ddb2-2 mutant plants were used as controls. Data are presented as percentage of methylation (±sd) and are representative of three biological replicates measured by McrBC-qPCR. The COPIA TE was used as control.
In a complementary approach, we used immunofluorescence to determine the nuclear DDB2-AGO4 colocalization pattern. As shown in Supplemental Figure 5, DDB2 and AGO4 exhibited a discrete colocalization signal in DDB2-FLAG expressing plants, consistent with the DDB2-AGO4 coimmunoprecipitation. Conversely, DDB2W261F-FLAG and DDB2K314E-FLAG expressing plants did not exhibit obvious DDB2-AGO4 colocalization signal (Supplemental Figure 5). This suggests that both point mutations likely disturbed the DDB2-AGO4 association/mode of action, in agreement with the lack of complementation of DNA methylation profiles reported in the previous sections of the study. Together, these observations emphasize that DDB2 is likely not a general cofactor of AGO4, but rather may be a chaperone.
Moreover, we found that both DDB2K314E-FLAG and DDB2W261F-FLAG expressing plants failed to complement UV hypersensitivity of ddb2-2 plants (Supplemental Figure 6A). Collectively, these set of data suggest that the DDB2 DNA binding ability and the DDB2 GW261 motif, presumably involved in its the association with AGO4, not only contribute to the control of a correct DNA methylation profile but also contribute to maintain genome integrity.
Given that RdDM relies on both biogenesis and loading of siRNAs by AGO4, we tested whether these two steps of the RdDM pathway could affect DDB2-AGO4 complex formation or stability. To this end, we stably expressed DDB2-FLAG in dcl3 plants, which cannot process double-stranded siRNA precursors into 24-nucleotide siRNAs (Law and Jacobsen, 2010). AGO4 coimmunoprecipitated with DDB2-FLAG less efficiently from dcl3 mutant extracts than from wild-type extracts (Supplemental Figure 6B). This suggested that proper 24-nucleotide siRNA biogenesis is required for efficient DDB2-AGO4 complex formation/stability. We also tested the effect of two point mutations in the AGO4 PAZ domain (AGO4Y370AF371A, also referred to as AGO4YAFA) that affect siRNA loading (Ye et al., 2012). GFP-AGO4 and GFP-AGO4YAFA transgenes were stably introduced into the DDB2-FLAG plant line by crossing. Again, coimmunoprecipitation of DDB2-FLAG was less efficient with GFP-AGO4YAFA than with GFP-AGO4 (Supplemental Figure 6C). Due to the prominent localization of the GFP-AGO4YAFA protein fusion in the cytoplasm (Ye et al., 2012), it cannot be excluded that lower coimmunoprecipitation efficiency mainly reflected DDB2-AGO4YAFA differential localization rather than complex formation/stability. Nevertheless, proper siRNA biogenesis and/or, to a lesser extent, loading by AGO4 appear to be important for the DDB2-AGO4 interaction. In turn, this implies that DDB2 could be part of multiple AGO4-siRNA complexes in planta.
Together these data confirm the existence of functional DDB2-AGO4-siRNA complexes potentially influencing de novo DNA methylation at multiple loci.
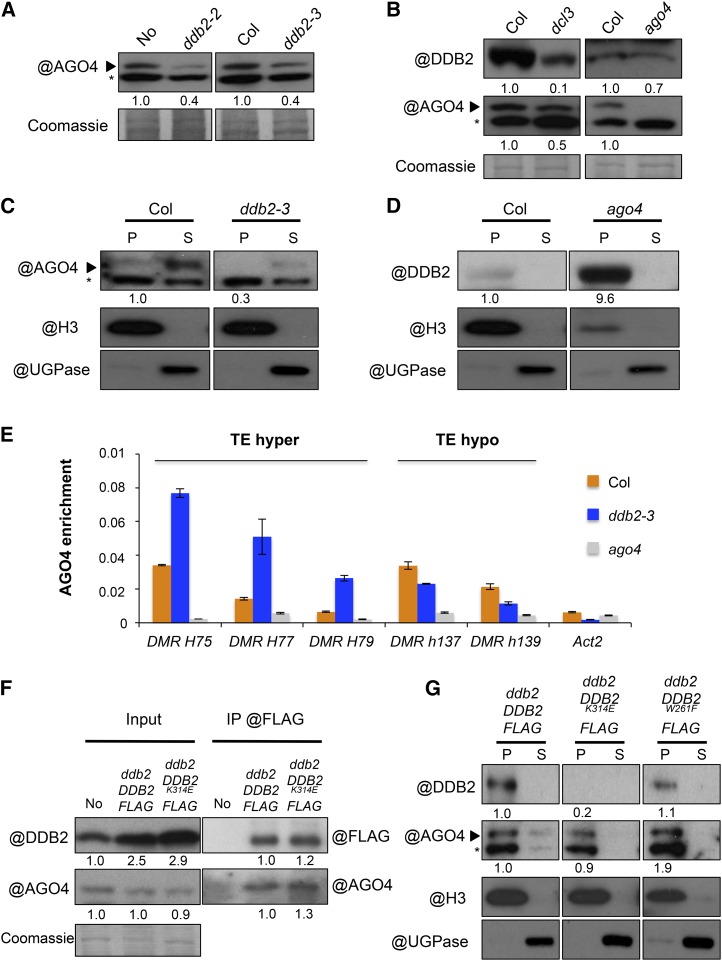
DDB2 Influences AGO4 Protein Content and Association with Chromatin
Low levels of AGO4 are reproducibly observed in ddb2 mutant plant extracts (Figure 6A), which, together with our other analyses, suggested that DDB2 could influence AGO4 stability. In a reciprocal manner, AGO4 or siRNA biogenesis deficiency in ago4 or dcl3 plants also appeared to affect DDB2 protein content (Figure 6B). Considering that DDB2 and AGO4 may act together on chromatin, we tested whether the absence of one partner influences the chromatin association of the other one. Immunoblot analyses of cellular extracts showed that AGO4 enrichment in chromatin fractions was decreased in ddb2-3 (Figure 6C). Reciprocally, DDB2 chromatin content was strongly increased in ago4 plants as compared with wild-type plants (Figure 6D). This suggested that DDB2 association with AGO4 may influence its own stability and that DDB2 and AGO4 differentially regulate their association with chromatin.
Figure 6.
AGO4 Protein Contents.
(A) Immunodetection of AGO4 in cellular extracts of wild-type (No and Col), ddb2-2, and ddb2-3 mutant plants (*, cross-reacting signal). Coomassie blue staining of the blot is shown.
(B) Immunodetection of AGO4 and DDB2 in wild-type Col, dcl3, and ago4 mutant plants. Coomassie blue staining was used as loading control (*, cross-reacting signal).
(C) Immunoblot analysis of AGO4 content in chromatin extracts from wild-type (Col) and ddb2-3 mutant plants. Anti-histone H3 and anti-UGPase antibodies were used as controls for insoluble (chromatin) and soluble fractions, respectively. P, pellet (insoluble fraction); S, supernatant (soluble fraction). Signal intensity relative to H3 is indicated below each lane (*, cross-reacting signal).
(D) Immunoblot analysis of DDB2 protein content in chromatin extracts from wild-type (Col) and ago4 mutant plants. Anti-histone H3 antibody was used as control for chromatin (insoluble fraction) and anti-UGPase antibody was used as control for soluble fraction. P, pellet (insoluble fraction); S, supernatant (soluble fraction). Signal intensity relative to H3 is mentioned below each lane.
(E) ChIP of AGO4 at three hyper-DMRs (TE) and two hypo-DMRs (TE) in wild-type (Col) and ddb2-3 plants using anti-AGO4 antibody. As negative controls, ago4 plants were used as well as Actin2. Data are presented as enrichment of AGO4 signal (±sd) and are representative of three biological replicates.
(F) In vivo pull-down of AGO4 with DDB2 protein. No, ddb2-2 DDB2-FLAG, and ddb2-2 DDB2K314E-FLAG-expressing plants were used for immunoprecipitation assays using anti-FLAG. Signal intensity relative to control is mentioned below each lane.
(G) Amount of DDB2 and AGO4 in chromatin, as analyzed by immunodetection in ddb2-2 DDB2-FLAG, ddb2-2 DDB2K314E-FLAG, and ddb2-2 DDB2W261F-FLAG-expressing plants. Anti-histone H3 antibody was used as control for chromatin (insoluble fraction) and anti-UGPase antibody was used as control for soluble fraction. P, pellet (insoluble fraction); S, supernatant (soluble fraction). Signal intensity relative to H3 is mentioned below each lane (*, cross-reacting signal).
We further explored a possible link between AGO4-DDB2 association and the hypermethylated patterns induced by ddb2 mutations. ChIP-qPCR analyses revealed an enrichment of AGO4 at various hyper-DMRs in ddb2-3 plants, while it was reduced at hypo-DMRs (Figure 6E). This observation is consistent with a possible direct relationship between differential local AGO4 content and changes in DNA methylation levels induced by ddb2. It further indicated a complex effect of DDB2 loss of function on AGO4 homeostasis. Indeed, AGO4 might also locally associate with chromatin independently of DDB2. We therefore investigated the relationship between DDB2 and AGO4 in chromatin using DDB2K314E and DDB2W261F mutations, which impair DDB2 DNA binding and DDB2-AGO4 association, respectively. First, efficient coimmunoprecipitation of AGO4 with DDB2K314E-FLAG showed that DDB2 DNA binding activity is not required for the DDB2-AGO4 interaction (Figure 6F). Interestingly, this mutation impaired DDB2 enrichment in chromatin, as shown previously by ChIP-qPCR at specific loci (Figure 3D), but here this was observed on a more global scale (Figure 6G). More surprisingly, while disturbing the DDB2-AGO4 interaction (Figure 6D), the DDB2 GW motif mutation did not significantly decrease the global enrichment of AGO4 in chromatin fractions (Figure 6G). Taken together, these results indicated that AGO4 association with chromatin either occurs independently of its interaction with DDB2 or is triggered by other factors. Finally, to assess whether the association of AGO4 and DDB2 with chromatin could rely on siRNAs, similar analyses were performed in dcl3 mutant plants. In dcl3 plants, global DDB2 and AGO4 protein contents were slightly reduced (Figure 6B). Additionally, AGO4 but not DDB2 enrichment in chromatin fractions appeared to be affected (Supplemental Figure 6D).
Taken together, this set of experiments sheds light on three aspects: (1) the role of DDB2 on AGO4 homeostasis, (2) a differential enrichment of AGO4 at ddb2-derived hyper- and hypo-DMRs, and (3) the requirement on siRNA biogenesis for AGO4 chromatin loading. These findings suggest that DDB2 acts as a local AGO4 chaperone, possibly regulating its availability on chromatin for DNA methylation effectors.
DISCUSSION
This study revealed that the DNA repair factor DDB2 is involved in shaping the Arabidopsis DNA methylation landscape, a previously uncharacterized feature that we determined in the absence of exogenously applied genotoxic stress. We observed that de novo DNA methylation is influenced by DDB2 acting in a complex with AGO4 to control the abundance of 24-nucleotide siRNAs at TEs and other repeated elements. We also identified that DDB2 can influence the DNA demethylation pathway mediated by ROS1 and/or DML3. Finally, we found that loss of DNA methylation in ddb2 mutant plants appeared to result mainly from defects in the maintenance or in the re-establishment processes.
Deficiency in DDB2 Induces Altered DNA Methylation Patterns through Complex Mechanisms
The most prominent effect on the DNA methylation landscape observed upon DDB2 loss of function was a frequent hypermethylation of heterochromatic repeats and TEs. While the mechanisms triggering some DNA hypomethylation on genes and TEs are still unclear, those responsible for limiting DNA methylation over TEs and other repeats are likely to rely on at least two pathways, possibly based on different affinities of DDB2 for particular chromatin and/or DNA topology changes generated during the methylation or the active demethylation processes.
The majority of the hypermethylated regions observed in DDB2-deficient plants occurred over TEs in all sequence contexts. These DMRs were found to be initially methylated in wild-type plants, to be usually dependent on RNA POL IV/V activities, and to globally associate with increased 24-nucleotide siRNA abundance in ddb2 plants. Hence, these loci may have been subjected to either a reinforcement of preexisting POL IV/V-dependent DNA methylation or to have acquired it de novo in ddb2 mutant plants (reviewed in Fultz et al., 2015). Given that DNA methylation can locally spread over short distances (Ahmed et al., 2011), we cannot exclude that gain of DNA methylation on these TEs is also mediated by RdDM from flanking regions. Epigenomic studies have provided evidence that DNA methylation of some repeated sequences in Arabidopsis is also regulated by a RNA POL II-mediated pathway, independently of RNA POL IV/V (Stroud et al., 2013). This process is mediated by RDR6 and involves 21/22-nucleotide siRNAs, which further activate canonical RdDM and exacerbate DNA methylation (Marí-Ordóñez et al., 2013; Nuthikattu et al., 2013). Our sRNA-seq analysis showed that hypermethylation of TEs in ddb2 mutant plants is not associated with an increased abundance of cognate 21/22-nucleotide siRNAs, suggesting that this secondary mechanism is not at play here. Thus, TEs could be hypermethylated through several coordinated mechanisms that may differ from an exclusive siRNA-dependent process.
A surprising feature of DDB2-deficient plants is the gain of CG methylation over many protein-coding genes. Of particular note, these loci were associated with a slight but significant increased abundance of 21-nucleotide siRNAs. Considering that DNA methylation of RNA POL II transcribed genes relies on 21-nucleotide siRNAs (Bond and Baulcombe, 2015), increased genic DNA methylation could result from the stimulation of transcription-coupled CG methylation. The emerging picture of the functional links between DDB2 and DNA methylation pathways is complex, and at this stage we cannot rule out that other processes may influence CG methylation.
Loss of DNA methylation in ddb2 plants predominantly occurred in a CG context at genic regions and in a non-CG context at TEs. This appeared to partially result from enhanced DNA demethylation by ROS1, presumably acting on all three sequence contexts (Zhu, 2009). Importantly, ddb2-induced hypo-DMRs were much less pronounced than the ones resulting from defective DNA methyltransferase activities, suggesting that DDB2 may indirectly contribute to the processes related to maintenance/reestablishment of DNA methylation in all cytosine contexts. Importantly, the ddb2-induced loss of CHH methylation mainly relies on RdDM, consistent with the existence of a DDB2-AGO4 complex. Therefore, DDB2 may act on siRNA-mediated CHH methylation via putative local regulation of RdDM effector availability.
DDB2, a Putative AGO4 Protein Chaperone
Our analyses showed that Arabidopsis DDB2 W261 residue is required for AGO4 association and stabilization, while K314 is essential for DNA binding. Prediction of the protein structure highlighted that W261 and W267 are positioned in close proximity to the bound DNA together with K314, and that these features are well conserved in other plant species. Moreover, we observed that DDB2 influences AGO4 protein homeostasis. Taken together, these findings suggest that DDB2 docks to AGO4 and regulates its function at particular loci in plants. The absence of a clear GW consensus in human DDB2, but also in other metazoan DDB2 proteins, suggests that DDB2-AGO4 interaction may only be evolutionarily conserved in plants. A potential cooperativity between DDB2 and AGO4-mediated DNA methylation in other organisms may therefore involve different mechanisms.
In Arabidopsis, we found that formation/stabilization of DDB2-AGO4 complexes relies on siRNA biogenesis and partially on AGO4 capacity to load them. The drop of AGO4 protein content observed in DDB2-deficient plants might therefore result from impaired formation of DDB2-AGO4 complexes, by which DDB2 may directly or indirectly stabilize AGO4. While it is well established that AGO4 loads siRNAs in the cytoplasm in the presence of the HSP90 chaperone (Ye et al., 2012), our study reveals a DDB2-dependent AGO4-siRNA regulatory pathway in the nucleus. However, DDB2 is likely not a general AGO4 cofactor. Specifically, it is tempting to speculate that DDB2 and its dynamics influence AGO4-siRNA availability at particular loci to regulate both de novo and DNA methylation maintenance. More generally, it is likely that factors regulating DDB2 homeostasis and dynamics by protein degradation, such as the E3 ligase CUL4-DDB1A (Molinier et al., 2008), also impact DNA methylation, as observed in Neurospora crassa and Schizosaccharomyces pombe in the context of heterochromatin formation (Horn et al., 2005; Jia et al., 2005; Lewis et al., 2010).
DDB2-Dependent Genome/Epigenome Surveillance
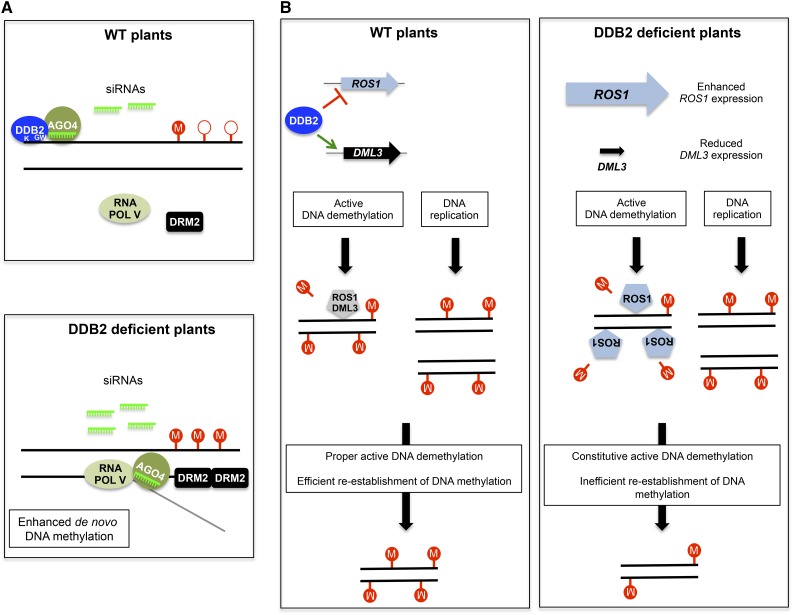
In nature, obligate photosynthetic organisms such as plants are strongly exposed to the damaging effects of excess light and of UV wavebands, which can impact genome and epigenome dynamics by inducing nucleotidic and chromatin variations (reviewed in Rigal and Mathieu, 2011). For example, 5-mC adjacent to cytosines or thymines can efficiently trigger the formation of pyrimidine dimers upon UV exposure (Pfeifer, 2006). Plants may have benefited evolutionarily from combining genome and epigenome surveillance processes, to efficiently deal with the deleterious effects of UV radiation and to optimally control light-dependent developmental programs. Factors influencing both DNA repair and DNA methylation theoretically have the potential to strongly impact plant (epi)genome integrity or, reciprocally, its plasticity. Accordingly, several chromatin factors, such as DDM1 or BRU1, impact the sensitivity of Arabidopsis plants to UV and genotoxic agents (reviewed in Donà and Mittelsten Scheid, 2015). Finally, formation of DNA DSBs induces siRNA biogenesis through a RNA POL IV/V-dependent process, suggesting that different DNA alterations could influence the pool of siRNAs mediating genome repair and/or epigenome maintenance (Wei et al., 2012). Our study unveils DDB2 as another such element at the nexus of DNA repair and epigenetic processes acting to control genome and epigenome integrity. In light of this study, we propose that DDB2 impacts DNA methylation by interconnecting several pathways, such as (1) the RdDM pathway (Figure 7A) and (2) active DNA demethylation and/or reestablishment of DNA methylation (Figure 7B).
Figure 7.
Model of DDB2-Based Control of DNA Methylation Dynamics.
(A) Control of de novo DNA methylation. Top panel: In the vicinity of poorly methylated DNA regions, DDB2 and AGO4 protein complexes would constantly be loaded onto chromatin. Stability of DDB2-AGO4 complexes relies on the DDB2 GW motif, on siRNA biogenesis, and on their loading by AGO4. DDB2-dependent chaperoning of AGO4 would prevent its inappropriate association with RNA POL V C-terminal domain (CTD), thus preventing the generation of undesirable DNA methylation patterns. Bottom panel: In the absence of DDB2, enhanced AGO4 association with the siRNA-RNA POL V transcript-DRM2 complex would trigger DNA hypermethylation. K, K314 involved in DNA binding; GW, AGO-hook motif; circled M and open circles represent methylated and unmethylated cytosines, respectively.
(B) Active DNA demethylation and DNA methylation maintenance. Left panel: DDB2 directly/indirectly acts as regulator of ROS1 and DML3 expression. Therefore, ROS1 and DML3 actively demethylates particular DNA regions followed by efficient reestablishment of initial DNA methylation patterns. In parallel, after DNA replication, efficient maintenance of DNA methylation processes is also necessary to preserve proper DNA methylation profiles. Right panel: In the absence of DDB2, ROS1 expression is upregulated and DML3 expression is downregulated, leading to misregulation of active DNA demethylation. In addition, inefficient reestablishment of DNA methylation could also occur either after ROS1 constitutive active DNA demethylation or after DNA replication.
METHODS
Plant Material
Arabidopsis thaliana mutant plants used in this study are in a No ecotype for ddb2-2 (Molinier et al., 2008) and in the Columbia-0 ecotype (Col) for ddb2-3 (DsLoxHs195_05H), nrpd1a (SALK _583051), rdr2 (SAIL_1277H08), dcl3 (SALK_005512), ago4-1 (Zilberman et al., 2003), met1-3 (Saze et al., 2003), and ros1 (SALK _045303).
Generation of Transgenic Plants
The cDNA of Arabidopsis DDB2 was amplified by PCR using primers described in Supplemental Table 1. For generating the DDB2K314E-FLAG and DDB2W261F-FLAG constructs, overlap PCR extension was performed using primers described in Supplemental Table 1. Both cDNAs were sequenced and cloned into the pOEX2 vector, between the NcoI and AvrII sites (Molinier et al., 2004). The resulting plasmids, pOEX2 DDB2-FLAG, pOEX2 DDB2K314E-FLAG, and pOEX2 DDB2W261F-FLAG, were mobilized into Agrobacterium tumefaciens and used to transform No and ddb2-2 Arabidopsis plants. pAGO4 GFP-AGO4 and pAGO4 GFP-AGO4YAFA transgenic plants have been described by Ye et al. (2012).
Root Growth Assay
To evaluate the UV-C (λ = 254 nm) sensitivity, 6-d-old in vitro-germinated wild-type, homozygous mutant, and transgenic plants were transferred to square plates containing GM medium (MS salts [Duchefa], 1% sucrose, and 0.8% agar-agar Ultrapur [Merck], pH 5.8) and grown vertically for 24 additional hours in the light or in darkness. Root length was measured 24 h after UV-C exposure (900 J/m2) using the Stratalinker 2400 (Stratagene). The relative root growth was calculated as follows: (root length treated/root length untreated) ×100 (±sd). Eight plants per replicate were used. Experiments were performed in triplicates.
WGBS and DMR Calling
Genomic DNA was prepared from 10-d-old seedlings grown in vitro as described by Roudier et al. (2011). Purified genomic DNA was bisulfite-treated and sequenced by the BGI Company with a Hi-Seq apparatus (Illumina).
DMR calling was defined as follows: For each sample type, methylation level was expressed as percentage of methylated reads over total reads per window. DMRs were identified using the difference between identical windows (same start-same stop) and upon filtering (P < 0.01 according Fisher’s exact test) using 100-bp consecutive windows. Consecutive windows exhibiting the same change (hyper/hypo, 1 window gap) were joined and called DMRs. DMRs were called for a DNA methylation difference of higher than 0.4 between mutant and wild type for CG methylation and of 0.1 for non-CG contexts.
MeDIP-Chip and DMR Calling
MeDIP-chip was performed as previously described (Colomé-Tatché et al., 2012). Two micrograms of genomic DNA extracted using the Qiagen MaxiPrep kit was sonicated using a Diagenode Bioruptor (18 times 30 s) and denaturated at 95°C for 10 min in 600 μL of Buffer 1 (10 mM Tris HCl, pH 7.5, 500 mM NaCl, and 1 mM EDTA). Immunoprecipitation was performed by adding 5 µg of anti-5mC mouse monoclonal antibody (Diagenode) to the DNA solution and by incubating the resulting mix overnight at 4°C with gentle agitation. Forty microliters of washed M280 Dynabeads (Invitrogen) was added and the suspension was incubated at 4°C for 4 h with gentle agitation. The IP pellet was repeatedly washed four times using 600 μL of Buffer 1, with 10-min incubations at room temperature with gentle agitation between washes, before elution with 300 μL of Buffer 2 (30 mM Tris-HCl, pH: 8.0) and 7 μL of Proteinase K (NEB; 20 µg/µL) for 1 h at 42°C. DNA from the IP and input fractions was purified by phenol-chloroform extraction and ethanol precipitation and resuspended in 100 µL. IP and input (150 ng) DNA were amplified using the GenomePlex Complete Whole Genome Amplification kit (Sigma-Aldrich) following the manufacturer’s instructions. Cy3 and Cy5 labeling was performed using the Nimblegen Dual color DNA labeling kit (Roche NimbleGen) and cohybridizations were performed using a custom NimbleGen 3x720K array designed according to Col-0 genome sequence, as described (Roudier et al., 2011). An additional MeDIP-chip analysis was performed with wild-type Col-0 plants grown in parallel, and a comparative genome hybridization analysis was performed to eliminate tiles that gave non-equal hybridization signals between wild-type No and Col-0. Segments with log2 (input/input) ≤0.4 were removed for subsequent analyses. Out of 570 defined comparative genome hybridization segments, 317 segments comprising polymorphic 39,012 probes on the array were filtered out. Two independent biological replicates were analyzed (two dye-swaps) for each genotype (wild-type No, wild-type Col-0, and ddb2-2 in a No background). DMRs were defined as segments of at least three consecutive tiles with significantly different methylation levels between the wild type and ddb2-2.
Comparison of DMRs from MeDIP-Chip and WGBS
For each DMR (hyper and hypo) identified in MeDIP-chip, WGBS data from both Col and ddb2-3 were extracted. Cytosines with no coverage or coverage >100 were skipped. The methylation level for Col and ddb2-3 was computed as follows: methylation level = Cm/(Cm + Cu); Cm, number of methylated reads in DMR; Cu, number of unmethylated reads in DMR; and Cm + Cu, total number of reads in DMR. Statistically significant differences in methylation values between the wild type and ddb2-3 were identified using Fisher’s exact test (P value < 0.05). DMRs identified in MeDIP-chip (hyper and hypo) were determined based on their methylation levels found in the WGBS. MeDIP-chip DMRs that were validated with WGBS data were called as true positives.
Identification of 5-mdC by Chromatography Coupled to Mass Spectrometry
Characterization of 5-mdC from plant extracts was performed by comparing retention times, MS transitions, and MS/MS analysis using UPLC-MS/MS. Genomic DNA was prepared from 10-d-old seedlings grown in vitro as described by Roudier et al. (2011). DNA was fragmented as described by Rozhon et al. (2008). All analyses were performed using a Waters Quattro Premier XE equipped with an electrospray ionization source and coupled to an Acquity UPLC system (Waters). Chromatographic separation was achieved using an Acquity UPLC BEH HSST3 column (100 × 2.1 mm, 1.8 µm; Waters), coupled to an Acquity UPLC BEH HSST3 precolumn (2.1 × 5 mm, 1.8 µm; Waters). The mobile phase consisted of (A) water with 0.1% formic acid and (B) methanol with 0.1% formic acid. The following gradient was used: 100% of (A) was maintained for 2 min; a linear gradient of 100% of (A) to 30% of (B) over 8 min; followed by a linear gradient of 30% of (B) to 100% of (B) over 1 min. The return to initial conditions (i.e., to 100% of [A]) was made over 0.5 min and maintained for 3.5 min. The total run time was 15 min. The column was operated at 35°C with a flow rate of 0.42 mL/min. The samples were reconstituted in 120 μL of water and centrifuged for 2 min at 13,000 rpm at 5°C just before injection (sample injection volume 3 µL). Nitrogen generated from pressurized air in a N2G nitrogen generator (Mistral; Schmidlin-dbs-AG) was used as the drying and nebulizing gas. The nebulizing gas flow was set to ∼50 L/h, and the desolvation gas flow to 900 l/h. The interface temperature was set at 400°C and the source temperature at 135°C. 5-mdC standard molecule was used to establish chromatography and mass spectrometry conditions. The selected ion recording MS mode was used to determine parent mass transition of 5-mdC (m/z: 242.42). Fragmentation was performed by collision-induced dissociation with argon at 1.0 × 10−4 mbar. The collision energy was optimized using daughter scan monitoring, and multiple reaction monitoring was then performed for 5-mdC identification. Mass spectrometry conditions for 5-mdC metabolites were set after optimization as follows: polarity ES+, capillary 3 kV, and cone 15 V. Low mass and high mass resolution were 13 for both mass analyzers, ion energies 1 and 2 were 0.5 V, entrance and exit potential were 2 and 1 V, and detector (multiplier) gain was 650 V. Collision-induced dissociation of protonated parent ions was accomplished with a collision energy of 10 V. Data acquisition and analysis were performed with the MassLynx software (version 4.1) running under Windows XP professional on a Pentium PC.
Small RNA Sequencing
Small RNAs were prepared from 10-d-old in vitro-grown seedlings using Tri-Reagent (Sigma-Aldrich) and used for library preparation and Illumina Hi-Seq sequencing (Fasteris).
Gel Filtration
Ten-day-old in vitro-grown seedlings (∼1 g) were ground in liquid nitrogen and resuspended in 2 mL of extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% Nonidet P‐40, 10% glycerol, and EDTA-free Protease Inhibitor Cocktail [Roche; one tablet/50 mL]). The solution was filtered through Miracloth membrane to remove large cell debris and the lysate was centrifuged for 1 h at 13,000g. One milliliter of cleared extract was then injected in a precalibrated Superdex 200 (Amersham) gel filtration column with the same extraction buffer at 0.4 mL/min using an AKTA FPLC system. Forty fractions of 0.5 mL were collected, and 50 μL of the even numbered fractions was analyzed by immunoblotting using anti-DDB2 or anti-AGO4 antibodies.
Modeling of Arabidopsis DDB2
The model of At_DDB2 (At5g58760) was built using MODELER (Sali and Blundell, 1993) with the crystallographic structure of Homo sapiens as template (PDB entry 4E54 chain B), which was selected after a NCBI BLAST of the At_DDB2 sequence against PDB sequences with a score value of 145, a coverage of 58 and 30% of sequence identity.
Chromatin Preparation for Immunoblotting
Fractions of soluble/insoluble proteins were extracted from 1 g of 10-d-old seedlings using Nonidet P-40 lysis buffer (25 mM Tris-HCl, pH 8.0, 0.3 M NaCl, 1 mM EDTA, 10% [v/v] glycerol, Nonidet P-40 1% [v/v], 0.2 mM phenylmethylsulfonyl fluoride, and EDTA-free Protease Inhibitor Cocktail [1 tablet/50 mL]). After grinding, powder was resuspended in 6 mL of Nonidet P-40 lysis buffer and incubated for 30 min on a rotating wheel at 4°C (8 rpm), and the solution was Miracloth-filtered. Removal of extra cell debris was performed by centrifugation (2000g, 5 min, 4°C). Free chromatin-unbound proteins were recovered in the soluble fraction after centrifugation (13,000g, 10 min, 4°C). The pellet containing insoluble and chromatin-bound proteins was resuspended in 75 μL of Nonidet P-40-containing resuspension buffer (Molinier et al., 2008). Variable amounts (25 to 50%) of the insoluble fraction and 2% of the soluble fraction were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies.
Immunoprecipitation Assays
Total soluble proteins were extracted from 0.5 g of 10-d-old seedlings using 3 mL of IP buffer (Pazhouhandeh et al., 2011). Immunoprecipitation was performed using anti-FLAG gel affinity (Sigma-Aldrich). The precipitate was washed four times in IP buffer and resuspended in 50 μL of SDS sample buffer and heated for 3 min at 100°C prior to immunoblotting. The DDB2-FLAG, DDB2K314E-FLAG, and DDB2W261F-FLAG fusion proteins were detected using the anti-FLAG HRP (A8592; Sigma-Aldrich) at a 1:5000 (v: v) dilution in PBST (1× PBS, nonfat dry milk [5%, w/v], and Tween 20 [0.1%, v/v; Sigma-Aldrich]).
Protein Extraction and Immunoblotting
Whole protein extracts were prepared using a denaturing buffer (Molinier et al., 2008). Twenty micrograms of total protein was separated on an 8% SDS gel (15% for detecting histone H3) and blotted onto an Immobilon-P membrane (Millipore). Anti-peptidic anti-AtDDB2 antibody (Molinier et al., 2008) was used at a 1:2000 dilution (v/v); anti-AGO4 (Garcia et al., 2012) at a 1:4000 dilution (v/v); anti-ROS1Ct (R3288-2; Abiocode) at a 1:2000 dilution (v/v); polyclonal anti-H3 (06-755; Millipore) at a 1:10,000 dilution (v/v); and anti-UGPase antibody (AS05 086; Agrisera) at a 1:10,000 dilution (v/v) in PBST.
Immunofluorescence
Leaves of 3-week-old in vitro-germinated plants were fixed under vacuum during 20 min in 4% formaldehyde and 1× PEM (50 mM PIPES, pH 7.3, 5 mM MgSO4, and 5 mM EGTA, pH 7.1). Samples were washed twice in ice-cold 1× PEM (10 min) and chopped with razor blade in ice-cold Galbraith buffer (45 mM MgCl2, 20 mM MOPS, 30 mM sodium citrate, and 0.3% Triton X-100, pH 7). The suspension was Miracloth-filtered and centrifuged for 5 min at 500g (4°C). The nuclear samples were spread onto poly-lysine-treated slides. Slides were postfixed at −20°C during a 10-min incubation in methanol-acetone (1:1, v/v) and blocked with PEMSB (1× PEM, 0.05%Saponin, and 2% BSA) for 2 h at room temperature in a humid chamber. Anti-FLAG (1/100) and anti-AGO4 (1/100), diluted in PEMSB, were added and slides were incubated for 16 h at 4°C. After three 20-min washes with PEMSB, slides were incubated for 2 h at room temperature with secondary antibodies (anti-mouse Alexa 568, 1:300 dilution [A-11031; Thermo Fisher] and anti-rabbit Alexa 488, 1:200 dilution [A-11034; Thermo Fisher]). Slides were washed 3 × 10 min with PEMSB and 1 × 10 min with PEMS. ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (Molecular Probes) was added and slides were observed under a confocal microscope (Zeiss LSM 780).
McrBC Treatment
Genomic DNA (1.5 µg) was digested for 8 h at 37°C with the McRBC enzyme (New England Biolabs), and 1.5 µg of undigested genomic DNA was used as control. DNA methylation levels at specific loci were determined by real-time PCR. For each experiment, three technical replicates were used to calculate the percentage of DNA methylation as follows: 100-log (Tm undigested-Tm digested) × 100. Experiments were, at least, duplicated using independent biological samples. Three technical replicates were performed for each independent biological sample. Primers are listed in Supplemental Table 1.
Sanger Bisulfite Sequencing
Analysis of DNA methylation by Sanger bisulfite sequencing was performed according to procedures described by Foerster and Mittelsten Scheid (2010). Primers were designed using an online tool (http://katahdin.mssm.edu/kismeth/primer_design.pl). Data were analyzed using CyMATE.
Reverse Transcription
Reverse transcription was performed on total RNA extracted using an RNeasy Plant Minikit (Qiagen) from 10-d-old seedlings. The reverse transcription reaction was performed on 5 µg of total RNA using a mixture of random hexamer-oligo d(T) primers and a cDNA reverse transcription kit (Applied Biosystems). One microliter of the reverse transcription reaction was used for qPCR.
ChIP
ChIP experiments were performed as described by Pazhouhandeh et al. (2011) using 10-d-old in vitro-grown seedlings. Preparation of chromatin, sonication, and immunoprecipitation using anti-FLAG or anti-AGO4 antibodies were performed as described by Pazhouhandeh et al. (2011). The immunoprecipitated DNA was analyzed by quantitative PCR (Supplemental Table 1). Data analysis was done as described by Mutskov and Felsenfeld (2004). Experiments were duplicated using independent biological samples. Three technical replicates were performed for each independent biological sample.
qPCR
qPCR was performed using a LightCycler 480 and LightCycler 480 SYBR green I Master mix (Roche) following the manufacturer’s instructions. Data were normalized relative to Actin2, Ubiquino-cytochrome C Reductase, and Hexokinase 1 genes (Supplemental Table 1).
Accession Numbers
The Arabidopsis Genome Initiative accession number for DDB2 is At5g58760. The Gene Expression Omnibus accession number for the bisulfite sequencing and the small RNA deep sequencing data reported in this article is GSE76651.
Supplemental Data
Supplemental Figure 1. Context-dependent DNA methylation differences induced by the ddb2-3 mutation in the Col accession (extended data supporting Figure 1).
Supplemental Figure 2. MeDIP-chip-based DNA methylation profiles of wild-type and ddb2-2 mutant plants of the No accession.
Supplemental Figure 3. Global 5-mdC contents, expression levels of DNA methylation-related genes, and DNA methylation maintenance.
Supplemental Figure 4. ddb2, ros1, and rdd DMRs.
Supplemental Figure 5. Immunolocalization of DDB2 and AGO4.
Supplemental Figure 6. DDB2-AGO4 complex (extended data supporting Figures 5 and 6).
Supplemental Table 1. List of primers used in this study.
Supplemental Data Set 1. ddb2-2 DMRs (TAIR 8) and ddb2-3 DMRs (TAIR10) for each cytosine context.
Supplementary Material
Acknowledgments
We thank Barbara Hohn for critical comments on the manuscript and Olivier Voinnet for valuable support during the course of this work. This work was supported by grants from the French Agence Nationale pour la Recherche (ANR): ANR BLAN07-3_188961 to C.B., V. Colot, and P.G. and ANR-11-JSV2-003-01 to F.B. Additional support was provided through the European Union Seventh Framework Programme Network of Excellence EpiGeneSys (Award 257082 to V. Colot), the Investissements d’Avenir ANR-10-LABX-54 MEMO LIFE, as well as ANR-11-IDEX-0001-02 PSL* Research University (to V. Colot). We acknowledge Y.J. Qi for providing pAGO::GFP-AGO4 and pAGO4::GFP-AGO4YAFA transgenic plants.
AUTHOR CONTRIBUTIONS
J.M., V. Colot, F.B., P.G., and C.B. designed experiments. C.S. performed McrBC-qPCR, RT-qPCR, and ChIP analyses. J.M. performed genetic analyses, immunoblots, and immunolocalization. S.D. and F.B. performed MeDIP-chip. A.K., M.K., I.A., V. Cognat, and S.G. performed bioinformatic analyses. M.B. performed DDB2 modeling. N.B. performed gel filtration. D.H. performed UPLC-MS/MS. J.M., F.B., and V. Colot wrote the article.
Glossary
- GGR
global genome repair
- TE
transposable element
- RdDM
RNA-directed DNA methylation
- siRNA
short interfering RNA
- MeDIP-chip
methyl DNA-immunoprecipitation-chromatin immunoprecipitation
- WGBS
whole-genome bisulfite sequencing
- DMR
differentially methylated region
- McrBC-qPCR
McrBC digestion coupled to quantitative PCR
- DSB
double-strand break
- UPLC-MS/MS
ultraperformance liquid chromatography coupled to tandem mass spectrometry
References
- Ahmed I., Sarazin A., Bowler C., Colot V., Quesneville H. (2011). Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 39: 6919–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G., Schäfer A., Marhold J., Stach D., Swaminathan S.K., Handa V., Döderlein G., Maltry N., Wu W., Lyko F., Niehrs C. (2007). Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445: 671–675. [DOI] [PubMed] [Google Scholar]
- Blevins T., Podicheti R., Mishra V., Marasco M., Wang H., Rusch D., Tang H., Pikaard C.S. (2015). Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4: e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D.M., Baulcombe D.C. (2015). Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J.E., Thompson L.H., Richardson A.S., States J.C. (1999). A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum. Mutat. 14: 9–22. [DOI] [PubMed] [Google Scholar]
- Coleman-Derr D., Zilberman D. (2012). DNA methylation, H2A.Z, and the regulation of constitutive expression. Cold Spring Harb. Symp. Quant. Biol. 77: 147–154. [DOI] [PubMed] [Google Scholar]
- Colomé-Tatché M., et al. (2012). Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc. Natl. Acad. Sci. USA 109: 16240–16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donà M., Mittelsten Scheid O. (2015). DNA damage repair in the context of plant chromatin. Plant Physiol. 168: 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. (2007). Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 21: 2539–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster A.M., Mittelsten Scheid O. (2010). Analysis of DNA methylation in plants by bisulfite sequencing. Methods Mol. Biol. 631: 1–11. [DOI] [PubMed] [Google Scholar]
- Franchini D.M., Schmitz K.M., Petersen-Mahrt S.K. (2012). 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu. Rev. Genet. 46: 419–441. [DOI] [PubMed] [Google Scholar]
- Fultz D., Choudury S.G., Slotkin R.K. (2015). Silencing of active transposable elements in plants. Curr. Opin. Plant Biol. 27: 67–76. [DOI] [PubMed] [Google Scholar]
- Garcia D., Garcia S., Pontier D., Marchais A., Renou J.P., Lagrange T., Voinnet O. (2012). Ago hook and RNA helicase motifs underpin dual roles for SDE3 in antiviral defense and silencing of nonconserved intergenic regions. Mol. Cell 48: 109–120. [DOI] [PubMed] [Google Scholar]
- Haag J.R., Pikaard C.S. (2011). Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12: 483–492. [DOI] [PubMed] [Google Scholar]
- Horn P.J., Bastie J.N., Peterson C.L. (2005). A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 19: 1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S., Kobayashi R., Grewal S.I. (2005). Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 7: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Karlowski W.M., Zielezinski A., Carrère J., Pontier D., Lagrange T., Cooke R. (2010). Genome-wide computational identification of WG/GW Argonaute-binding proteins in Arabidopsis. Nucleic Acids Res. 38: 4231–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Sakaguchi K. (2006). DNA repair in plants. Chem. Rev. 106: 753–766. [DOI] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N., Fradin D., Iltis I., Bougnères P., Egly J.M. (2012). XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Mol. Cell 47: 622–632. [DOI] [PubMed] [Google Scholar]
- Lei M., Zhang H., Julian R., Tang K., Xie S., Zhu J.K. (2015). Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: 3553–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis Z.A., Adhvaryu K.K., Honda S., Shiver A.L., Knip M., Sack R., Selker E.U. (2010). DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 6: e1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V., Voinnet O. (2013). Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Mosher R.A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Molinier J., Lechner E., Dumbliauskas E., Genschik P. (2008). Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet. 4: e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ramos C., Fritsch O., Hohn B. (2004). CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell 16: 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov V., Felsenfeld G. (2004). Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 23: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli H., Sugasawa K. (2011). The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair (Amst.) 10: 673–683. [DOI] [PubMed] [Google Scholar]
- Nag A., Bondar T., Shiv S., Raychaudhuri P. (2001). The Xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21: 6738–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols A.F., Itoh T., Graham J.A., Liu W., Yamaizumi M., Linn S. (2000). Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem. 275: 21422–21428. [DOI] [PubMed] [Google Scholar]
- Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M., Molinier J., Berr A., Genschik P. (2011). MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 3430–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G.P. (2006). Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 301: 259–281. [DOI] [PubMed] [Google Scholar]
- Pontier D., et al. (2012). NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol. Cell 48: 121–132. [DOI] [PubMed] [Google Scholar]
- Qian W., et al. (2012). A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336: 1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal M., Mathieu O. (2011). A “mille-feuille” of silencing: epigenetic control of transposable elements. Biochim. Biophys. Acta 1809: 452–458. [DOI] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhon W., Baubec T., Mayerhofer J., Mittelsten Scheid O., Jonak C. (2008). Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal. Biochem. 375: 354–360. [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815. [DOI] [PubMed] [Google Scholar]
- Saze H., Mittelsten Scheid O., Paszkowski J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34: 65–69. [DOI] [PubMed] [Google Scholar]
- Schärer O.D. (2013). Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 5: a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Ecker J.R. (2012). Epigenetic and epigenomic variation in Arabidopsis thaliana. Trends Plant Sci. 17: 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A., Konícková R., Czyzewski B.K., Kawasaki Y., Jeffrey P.D., Groisman R., Nakatani Y., Iwai S., Pavletich N.P., Thomä N.H. (2008). Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135: 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F.K., Colot V. (2010). Repeat elements and the Arabidopsis DNA methylation landscape. Heredity (Edinb) 105: 14–23. [DOI] [PubMed] [Google Scholar]
- Virdi K.S., Laurie J.D., Xu Y.Z., Yu J., Shao M.R., Sanchez R., Kundariya H., Wang D., Riethoven J.J., Wamboldt Y., Arrieta-Montiel M.P., Shedge V., Mackenzie S.A. (2015). Arabidopsis MSH1 mutation alters the epigenome and produces heritable changes in plant growth. Nat. Commun. 6: 6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.G., Qi Y. (2012). A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112. [DOI] [PubMed] [Google Scholar]
- Williams B.P., Pignatta D., Henikoff S., Gehring M. (2015). Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 11: e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben B.Ø., Iwai S., Wood R.D. (2005). DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J. Biol. Chem. 280: 39982–39989. [DOI] [PubMed] [Google Scholar]
- Ye R., Wang W., Iki T., Liu C., Wu Y., Ishikawa M., Zhou X., Qi Y. (2012). Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol. Cell 46: 859–870. [DOI] [PubMed] [Google Scholar]
- Zhai J., et al. (2015). A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Du J., Hale C.J., Gallego-Bartolome J., Feng S., Vashisht A.A., Chory J., Wohlschlegel J.A., Patel D.J., Jacobsen S.E. (2014). Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2009). Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43: 143–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.