CurT is required for shaping the thylakoid membrane architecture in Synechocystis and mediates osmotic stress responses.
Abstract
Photosynthesis occurs in thylakoids, a highly specialized membrane system. In the cyanobacterium Synechocystis sp PCC 6803 (hereafter Synechocystis 6803), the thylakoids are arranged parallel to the plasma membrane and occasionally converge toward it to form biogenesis centers. The initial steps in PSII assembly are thought to take place in these regions, which contain a membrane subcompartment harboring the early assembly factor PratA and are referred to as PratA-defined membranes (PDMs). Loss of CurT, the Synechocystis 6803 homolog of Arabidopsis thaliana grana-shaping proteins of the CURVATURE THYLAKOID1 family, results in disrupted thylakoid organization and the absence of biogenesis centers. As a consequence, PSII is less efficiently assembled and accumulates to only 50% of wild-type levels. CurT induces membrane curvature in vitro and is distributed all over the thylakoids, with local concentrations at biogenesis centers. There it forms a sophisticated tubular network at the cell periphery, as revealed by live-cell imaging. CurT is part of several high molecular mass complexes, and Blue Native/SDS-PAGE and isoelectric focusing demonstrated that different isoforms associate with PDMs and thylakoids. Moreover, CurT deficiency enhances sensitivity to osmotic stress, adding a level of complexity to CurT function. We propose that CurT is crucial for the differentiation of membrane architecture, including the formation of PSII-related biogenesis centers, in Synechocystis 6803.
INTRODUCTION
Oxygenic photosynthesis originated in cyanobacteria more than 2.4 billion years ago and went on to transform Earth’s atmosphere and biosphere. The underlying process of light-driven photosynthetic electron transport is mediated by multiprotein/pigment complexes, which are located within a specialized system of membrane sheets termed thylakoids. During the evolutionary transition from cyanobacteria to present-day chloroplasts, this system has undergone substantial diversification (Mullineaux, 2005; Allen et al., 2011). Contemporary forms range from undifferentiated thylakoids in cyanobacteria to elaborate systems that are differentiated into grana and stroma regions in plant chloroplasts (Mullineaux, 2005).
Despite this increase in complexity over the course of evolution, even “simple” cyanobacterial systems exhibit compositional and functional membrane heterogeneity (Nickelsen and Rengstl, 2013). Perhaps the most striking example is the cyanobacterium Gloeobacter violaceus, which lacks internal thylakoids and organizes its photosynthetic complexes in distinct patches within the plasma membrane (Rexroth et al., 2011). Moreover, spatial separation between developing and functional thylakoids has been observed in the model cyanobacterium Synechocystis sp PCC 6803 (hereafter Synechocystis 6803). Immunolocalization of the PSII assembly factor PratA (for processing-associated TPR protein) in fractionated membranes, and examination of ultrathin sections by immunogold electron microscopy have revealed specialized, PratA-defined membrane (PDM) regions forming biogenic centers at peripheral sites in cells where thylakoids converge (Schottkowski et al., 2009b; Stengel et al., 2012).
Some details of the ultrastructure of these centers have begun to emerge (Stengel et al., 2012). Some of the convergence areas are composed of a rod-like structure, previously named the “thylakoid center,” which is in turn surrounded by membranous material within which thylakoid lamellae appear to originate (van de Meene et al., 2006; Stengel et al., 2012; Nickelsen and Zerges, 2013; Rütgers and Schroda, 2013). A current working model for these biogenesis centers postulates that the initial steps in the assembly of photosynthetic complexes, and in particular, PSII, take place at the biogenic PDMs. Subsequently, precomplexes migrate laterally into thylakoid lamellae, where their assembly is completed (Nickelsen and Rengstl, 2013). Recently, evidence based on the subcellular distribution of the D1 degradation-related FtsH protease and the PSII repair factor Slr0151 (Yang et al., 2014; Sacharz et al., 2015; Rast et al., 2016), has been obtained that maintenance, i.e., the repair, of PSII is also localized at or near these areas. However, whether or not plasma and thylakoid membranes fuse at these sites has not yet been resolved (Liberton et al., 2006; van de Meene et al., 2006; van de Meene et al., 2012).
Only limited information is available on the spatial organization of thylakoid membrane biogenesis in land plants. Their chloroplasts harbor a dynamic thylakoid membrane system, which is comprised of nonappressed stromal thylakoids and appressed grana regions. Stromal thylakoids are likely to represent sites where membrane proteins are synthesized and assembled within the membrane (Yamamoto et al., 1981; Danielsson et al., 2006), while the physicochemical forces driving grana formation are still a matter of debate (Nevo et al., 2012; Kirchhoff, 2013; Pribil et al., 2014). However, it has been proposed that stromal moieties of LHCII determine membrane stacking of adjacent thylakoid disks (Fristedt et al., 2009; Daum et al., 2010; Anderson et al., 2012). Moreover, a family of thylakoid-shaping proteins, with four members named CURVATURE THYLAKOID1A-D (CURT1A-D), has been identified in Arabidopsis thaliana (Armbruster et al., 2013). CURT1 proteins localize to grana margins, where they induce membrane bending, thereby determining the architecture of the thylakoid network (Pribil et al., 2014). Intriguingly, cyanobacteria, whose thylakoids do not differentiate into grana regions, also contain a single CURT1 homolog (Armbruster et al., 2013). Here, we report on the characterization of this homolog, CurT, from Synechocystis 6803. Our data reveal that the cyanobacterial protein is essential for the shaping of thylakoid membranes and thereby promotes efficient assembly of PSII at the cell periphery. Our data argue for an ancient membrane-curving activity of CURT1-like proteins, which are necessary to form an efficient thylakoid system in cyanobacteria, as well as having a critical role in the response to osmotic stress.
RESULTS
Inactivation of curT Affects Membrane Architecture
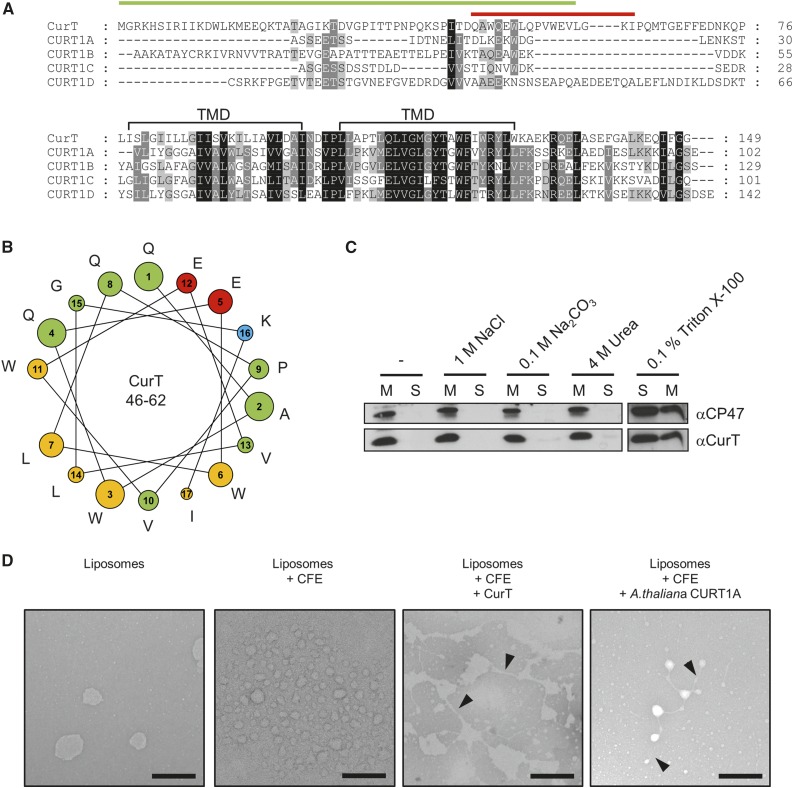
The open reading frame slr0483 encodes the only CURT1-like protein expressed in the cyanobacterium Synechocystis 6803. The corresponding cyanobacterial gene was previously named synCURT1 to emphasize its homology to CURT1 from Arabidopsis (Armbruster et al., 2013; Luque and Ochoa de Alda, 2014). However, in conformity with conventional nomenclature for bacterial genes, we adopt the name curT. The CurT protein is predicted to comprise 149 amino acids and contains within its C-terminal half two putative transmembrane domains, which exhibit a high degree of sequence similarity with the Arabidopsis CURT1A-D proteins. The N-terminal half shows less similarity to its Arabidopsis counterparts, but it harbors a predicted amphipathic α-helix (amino acids 46 to 62) that has been implicated in membrane bending (Figures 1A and 1B; Armbruster et al., 2013). Interestingly, the transmembrane domains of CURT homologs share sequence and structural features with a domain that is found in some thylakoid-associated cyanobacterial aminoacyl-tRNA synthetases and has been hypothesized to be involved in mediating the unusual membrane attachment of these enzymes (Luque and Ochoa de Alda, 2014).
Figure 1.
General Characteristics of CurT.
(A) Sequence alignment of CurT and CURT1A-D from Arabidopsis. Predicted chloroplast transit-peptide sequences of CURT1A-D are omitted (Armbruster et al., 2013). Positions of predicted C-terminal transmembrane domains (TMD) are marked (black bar), as is the position of the predicted amphipathic N-terminal helix in CurT (red bar). The green bar denotes the peptide sequence used for antibody production. Identical and related amino acids that are conserved in 100, 80, and 60% of the sequences are highlighted in black, dark gray, and light gray, respectively.
(B) Helical-wheel representation of the N-terminal amphipathic helix of CurT (red bar in [A]). The color code reflects the physicochemical properties of amino acid side chains: green, polar and uncharged; blue, polar and positively charged; red, polar and negatively charged; yellow, hydrophobic.
(C) Solubility of CurT. Membrane-associated proteins (50 µg) from wild-type cells were extracted with 5 mM HEPES (pH 7.6) containing either 1 M NaCl, 0.1 M Na2CO3, 4 M urea, 0.1% Triton X-100, or no additional solute. After separation of membrane-bound (M) and solubilized (S) proteins by centrifugation, proteins were fractionated by SDS-PAGE and CurT was immunodetected on immunoblots. As a control, the integral membrane protein CP47 from PSII was analyzed in parallel.
(D) Transmission electron micrographs of negatively stained liposomes. Cell-free expression (CFE) of CurT and Arabidopsis CURT1A was performed in the presence of liposomes similar in composition to the lipids of the thylakoid membrane. Liposomes before and after cell-free protein expression in the absence of DNA served as negative controls. Black arrowheads indicate tubular connections between liposomes. Bars = 250 nm.
To verify the predicted membrane association of CurT, total membranes from wild-type cells were exposed to various agents, and the solubility of CurT was assessed using an antibody directed against a fragment comprising its first 58 N-terminal amino acids (Figure 1A). We first confirmed that CurT can be detected in the membrane fractions of cell lysates, and exposure to 0.1 M Na2CO3, 4 M urea, or 1 M NaCl failed to extract it, as would be expected for a membrane-bound protein (Figure 1C). Indeed, treatment of the cell lysate with the nonionic detergent Triton X-100 rendered CurT soluble, as was the case for the PSII inner antenna protein CP47, an integral membrane protein with six transmembrane α-helices (Figure 1C). Thus CurT, like its Arabidopsis counterparts, is likely to be an integral membrane protein.
To test whether the cyanobacterial CurT has similar membrane-tubulating properties to CURT1A of Arabidopsis, we used the same liposome-based assay to probe its ability to form tubules (Armbruster et al., 2013). We expressed CurT in vitro, using a cell-free extract supplemented with liposomes similar in composition to the thylakoid membrane. Subsequently, liposome topology was visualized by transmission electron microscopy (TEM; Figure 1D). Like the grana-forming CURT1A, its Synechocystis 6803 homolog efficiently induced localized “compression” of liposomes into thin tubule-like segments, revealing its strong membrane-curving activity (Figure 1D). Hence, the membrane-shaping function of members of the CURT1 family is conserved from cyanobacteria to plants. Nevertheless, liposome shapes caused by either CurT or CURT1A displayed some differences in the degree of membrane tubulation. These might be due to the variable N termini of both proteins (Figure 1D).
To dissect the function of CurT in vivo, we generated a knockout mutant by inserting a kanamycin resistance cassette into the unique AgeI site in the slr0483 reading frame (Supplemental Figure 1A; see Methods). Complete segregation of the mutation was confirmed by PCR and immunoblot analyses (Supplemental Figures 1B and 1C). Progressively higher levels of antibiotic (up to 400 µg kanamycin/mL) were used for mutant selection, as previous attempts to select a fully segregated curT- mutant had been unsuccessful, probably due to application of insufficient selection pressure (Armbruster et al., 2013). When growth rates of wild-type and mutant cells were compared, 2.0- and 1.5-fold increases in doubling time were observed for curT- grown under photoautotrophic and photoheterotrophic conditions, respectively (Table 1; Supplemental Figure 2).
Table 1. Physiological Characteristics of the curT- Mutant.
| Strain | Doubling Time (h) | Oxygen Evolution (nmol h−1 OD750−1)a | Chlorophyll Content (µg OD750−1)b | Cell Size (µm)c | Cell Number (mL−1 OD750−1)d | |
|---|---|---|---|---|---|---|
| Wild type | 8.17 ± 0.17e | 16.00 ± 0.42f | 1933 ± 187 | 2.51 ± 0.14 | 2.64 ± 0.20 | 3.90 ± 0.06 x 107 |
| curT- | 12.59 ± 0.20e | 32.26 ± 0.56f | 1074 ± 20 | 1.95 ± 0.29 | 2.74 ± 0.44 | 3.67 ± 0.16 x 107 |
Oxygen evolution is expressed in nmol O2 produced per hour per OD750 unit.
Chlorophyll content is expressed in µg OD750−1.
The diameter of nondividing cells is presented in micrometers.
Cell number was determined per ml per OD750. Data are means ± sd of at least three independent experiments.
Doubling times in the presence (e) or absence (f) of 5 mM glucose measured under continuous illumination at 30 µmol photons m−2 s−1 and CO2-limiting conditions.
Strikingly, curT inactivation also led to loss of competence for DNA uptake during both transformation and conjugation-based experiments. Hence, attempts to complement the curT- mutant were not applicable. However, in the course of the work, four independent rounds of transformation of wild-type cells were performed to generate fresh curT- mutants. All mutant strains obtained the same phenotype, strongly suggesting that no secondary sites were involved in its establishment. To rule out the possibility that read-through transcription from the resistance marker gene affects the upstream reading frame slr0482 (of unknown function) in the mutant, RT-PCR with appropriate primers was performed (Supplemental Figure 3). No change in slr0482 mRNA accumulation was observed, confirming that the curT- phenotype is caused by disruption of the curT gene. In agreement with this finding, recent transcriptome analyses of Synechocystis 6803 have clearly shown that the curT mRNA is monocistronic (Kopf et al., 2014).
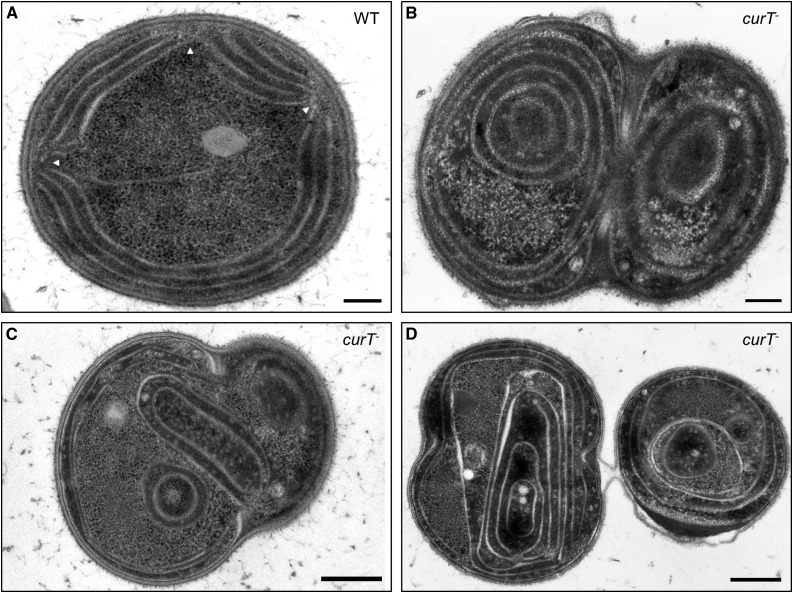
In view of CurT’s in vitro membrane-tubulating activity, the ultrastructure of the curT- mutant was examined by TEM. As shown in Figure 2A, the wild-type strain exhibits the typical thylakoid organization with ordered thylakoid sheets at the cell periphery and convergence zones next to the plasma membrane. In contrast, massive disorganization of the thylakoid membrane system was observed in curT-. In all cells analyzed, the mutant thylakoids appeared as disordered sheets that traversed the cytoplasm and completely lacked the typical convergence zones with the corresponding biogenesis centers at the periphery (Figures 2B to 2D). This indicates a strict requirement of CurT for the establishment of normal thylakoid membrane architecture in Synechocystis 6803.
Figure 2.
Ultrastructural Analysis of curT- Cells.
Transmission electron micrographs of typical wild-type (A) and curT- ([B] to [D]) Synechocystis 6803 cells, taken at 44,000× ([A] and [B]) and 28,000× ([C] and [D]) magnification, respectively. Ultrathin sections (35 to 45 nm) of cryofixed samples were stained with osmium tetroxide and poststained using lead citrate. Arrowheads indicate biogenesis centers. Bars = 200 nm in (A) and (B) and 500 nm in (C) and (D).
Inactivation of curT Affects Photosynthetic Performance
The severely altered morphology of the thylakoid membrane system in curT- cells, and in particular the lack of PSII-related biogenesis centers, prompted us to investigate the photosynthetic performance of the mutant (Stengel et al., 2012). The chlorophyll content was reduced by ∼20%, and absorption spectroscopy also revealed a reduction in phycobilisomes as well as an increase in carotenoid levels (Table 1; Supplemental Figure 4). Despite changes in ultrastructure and pigment levels, size and number of curT- cells at OD750 were not significantly different as judged by t test-based statistical analysis (Table 1).
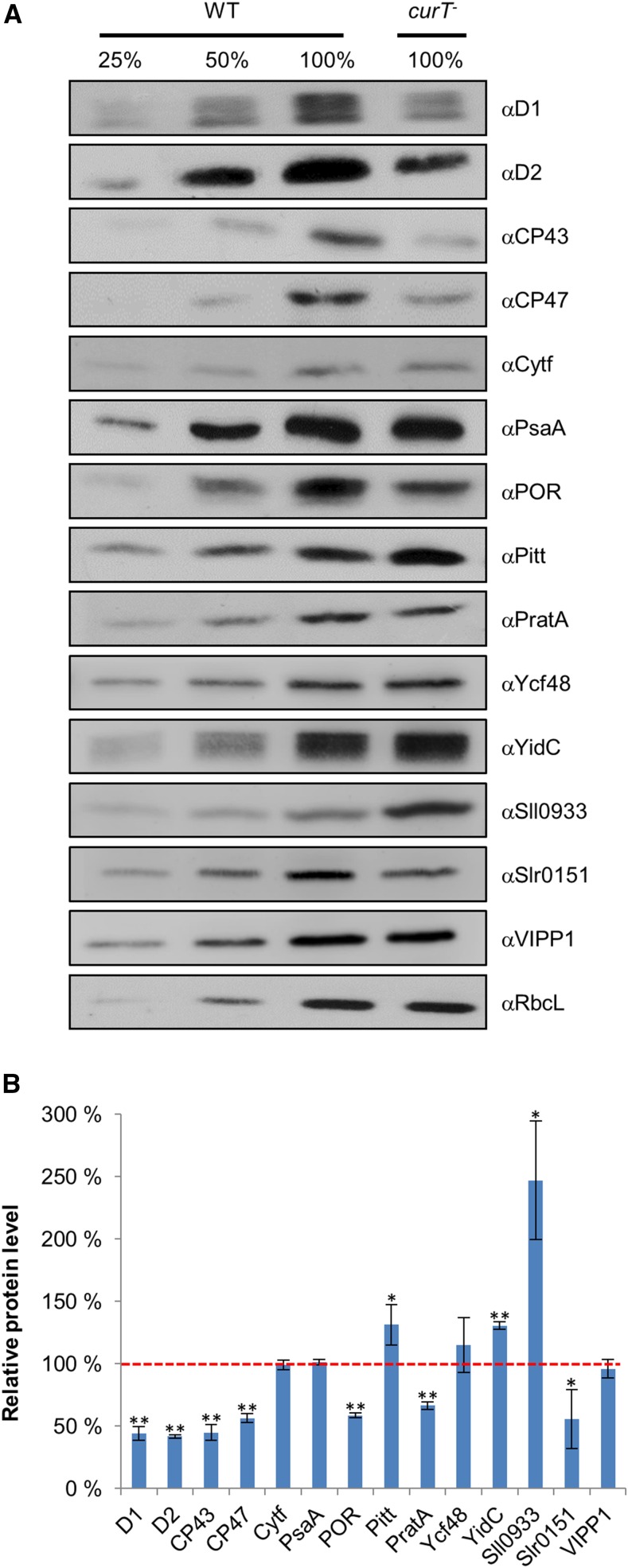
To explore the photosynthetic defects of the mutant in detail, immunoblot analyses were performed using antibodies raised against specific subunits of photosynthetic complexes and some of their biogenesis factors. In curT-, PSII reaction center subunits including D1, D2, CP47, and CP43 were found to be present at 44, 42, 56, and 45% of the wild-type level, respectively (Figure 3). Interestingly, amounts of the PSII assembly/repair factors PratA and Slr0151, as well as the light-dependent protochlorophyllide oxidoreductase (POR) enzyme, were also reduced (Figure 3; Yang et al., 2014; Rast et al., 2016). Levels of the Pitt protein, which interacts with POR (Schottkowski et al., 2009a), and the Oxa homolog YidC (Ossenbühl et al., 2006) were slightly increased in curT- (Figure 3). On the other hand, Sll0933, the homolog of the PSII assembly factor PAM68 in Arabidopsis, accumulated to 250% of the wild-type level (Armbruster et al., 2010). In contrast, amounts of both Cytf and PsaA, which serve as markers for the Cyt(b6f) and PSI complexes, respectively, the RbcL subunit of the Rubisco enzyme, the PSII assembly factor Ycf48 (Komenda et al., 2008), and VIPP1 remained unaltered in curT- (Figure 3). Overall, the alterations in protein accumulation indicate that loss of CurT preferentially affects the accumulation of PSII.
Figure 3.
Accumulation of Components of Photosynthetic Complexes in the curT- Mutant.
(A) Whole cell extracts (30 µg protein) from wild-type and curT- strains were fractionated by SDS-PAGE and analyzed on immunoblots using the indicated antibodies.
(B) The histogram shows levels of the indicated proteins in curT- extracts relative to wild-type samples (dashed red line). Data are means ± sd of three independent experiments. Significant differences from wild-type protein levels according to Student’s t test with error probabilities of 5 and 1% are indicated by one asterisk and two asterisks, respectively.
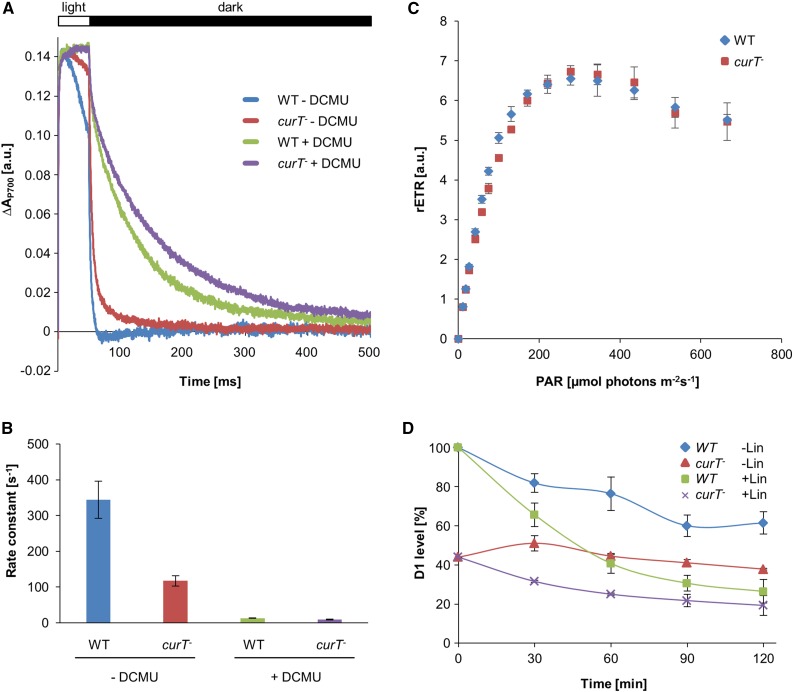
This result was further corroborated by analysis of the photosynthetic performance of curT-. The rate of light-dependent oxygen evolution was reduced by ∼50% in the mutant (Table 1), and analysis of P700+ reduction kinetics revealed impaired electron donation by PSII (Figure 4A). PSII-driven P700+ reduction is slowed down by a factor of three in curT-, whereas the residual cyclic electron transfer activity in the presence of DCMU is less than 5% in both the wild type and curT- (Figure 4B). In contrast, the light intensity-dependent electron transfer capacity, which was measured as relative electron transfer rate, did not differ significantly between the wild type and curT- (Figure 4C). Both wild-type and curT- cells reach a capacity limit at ∼250 µmol photons m−2 s−1 and the decay of relative electron transfer rate beyond the capacity limit is very similar. These results indicate that the point of onset of PSII-related photoinhibition is the same in both strains, which in turn implies that electron flow downstream of PSII is unchanged in the mutant.
Figure 4.
P700+ Reduction Kinetics and High-Light Effects in curT- Cells.
(A) Averaged traces (10 single measurements) of P700+ decay after complete oxidation by a 50-ms multiple turnover pulse (10,000 µmol photons m−2 s−1) in the absence and presence of 10 µM DCMU.
(B) Rate constants for P700+ reduction were obtained by fitting of the data with single exponential functions. Error bars indicate the sd of three independent measurements.
(C) Impact of light intensity on the (relative) electron transport rate (rETR). Cells were exposed to gradually increasing light intensities, which resulted in increasing electron transport until the capacity limit was reached. PAR: light intensity (µmol photons m−2 s−1). Error bars indicate the sd of three independent measurements.
(D) D1 protein level in the wild type and curT- after high-light treatment (800 µmol photons m−2 s−1) in the presence and absence of lincomycin. Samples were taken every 30 min and processed as in Figure 3 to determine the level of D1 present. 100% refers to wild-type D1 level at the beginning of the experiment.
Low PSII levels in curT- could be caused either by reduced synthesis/biogenesis or its enhanced degradation. To measure the rates of D1 repair, the kinetics of D1 accumulation upon induction of photoinhibition by high-light treatment (800 µmol photons m−2 s−1) was assayed by immunological means (Figure 4D). In wild-type cells, the D1 level decreased to 60% (relative to the initial time point) after 90 min of treatment. As previously shown, the drop is much more pronounced (∼35% left) when repair synthesis of D1 is inhibited by the addition of lincomycin (Figure 4D; Komenda et al., 2008). Surprisingly, in curT-, the level of residual D1 is rather stable over the same time period, and lincomycin treatment induces a more modest decrease to 60% relative to the initial time point (Figure 4D). These data clearly indicate that, in absence of CurT, residual D1 is less susceptible to degradation, even though its absolute amount is reduced. In line with this, no effect on growth rates was observed when either wild-type or curT- cells were cultivated for 2 d at 200 µmol photons m−2 s−1 (Supplemental Figure 5).
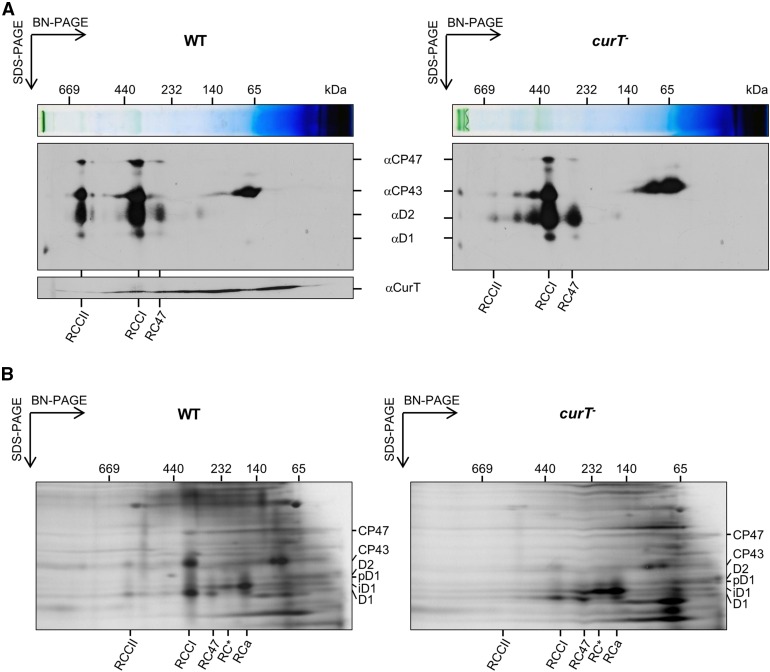
To assess the role of CurT in the assembly of PSII, we compared the two-dimensional profiles of thylakoid membrane proteins from wild-type and mutant cells by Blue Native (BN)/SDS-PAGE. PSII assembly intermediates were subsequently visualized via immunodetection of PSII core subunits. As shown in Figure 5A, dimeric PSII core complexes (RCCII) are drastically underrepresented in curT-, whereas relative levels of earlier assembly intermediates, including the monomeric CP43-less RC47 complex and nonassembled CP43, increase. Parallel detection of CurT itself revealed a “smeared” signal in the size range from 15 up to 500 kD, suggesting that CurT forms part of high molecular mass complexes (Figure 5A). Finally, in vivo 35S protein pulse-labeling experiments confirmed that RC and RC47 complexes in particular are efficiently formed in curT-, but the transition to RCCI complexes is severely delayed (Figure 5B). As a consequence, dimeric RCCII complexes do not incorporate any radioactive label over the experiment’s time course of 25 min (Figure 5B). Taken together, these data show that curT inactivation has a severe impact on PSII biogenesis, but little or no effect on the degradation of the complex.
Figure 5.
PSII Assembly in curT- Cells.
(A) BN/SDS-PAGE of membrane proteins. Membrane fractions from wild-type and curT- cells were solubilized with 1.3% n-dodecyl-β-maltoside, and proteins were fractionated by two-dimensional BN/SDS-PAGE, blotted, and probed with antibodies against the PSII subunits D1, D2, CP47, and CP43. Wild-type blots were also probed with the αCurT antibody. The sizes of marker proteins and the positions of PSII assembly intermediates (RCCII, RCCI, and RC47) are shown above and below each image.
(B) In vivo pulse labeling of membrane proteins with 35S. Pulse-labeled PSII core proteins were separated via BN/SDS-PAGE and visualized by autoradiography.
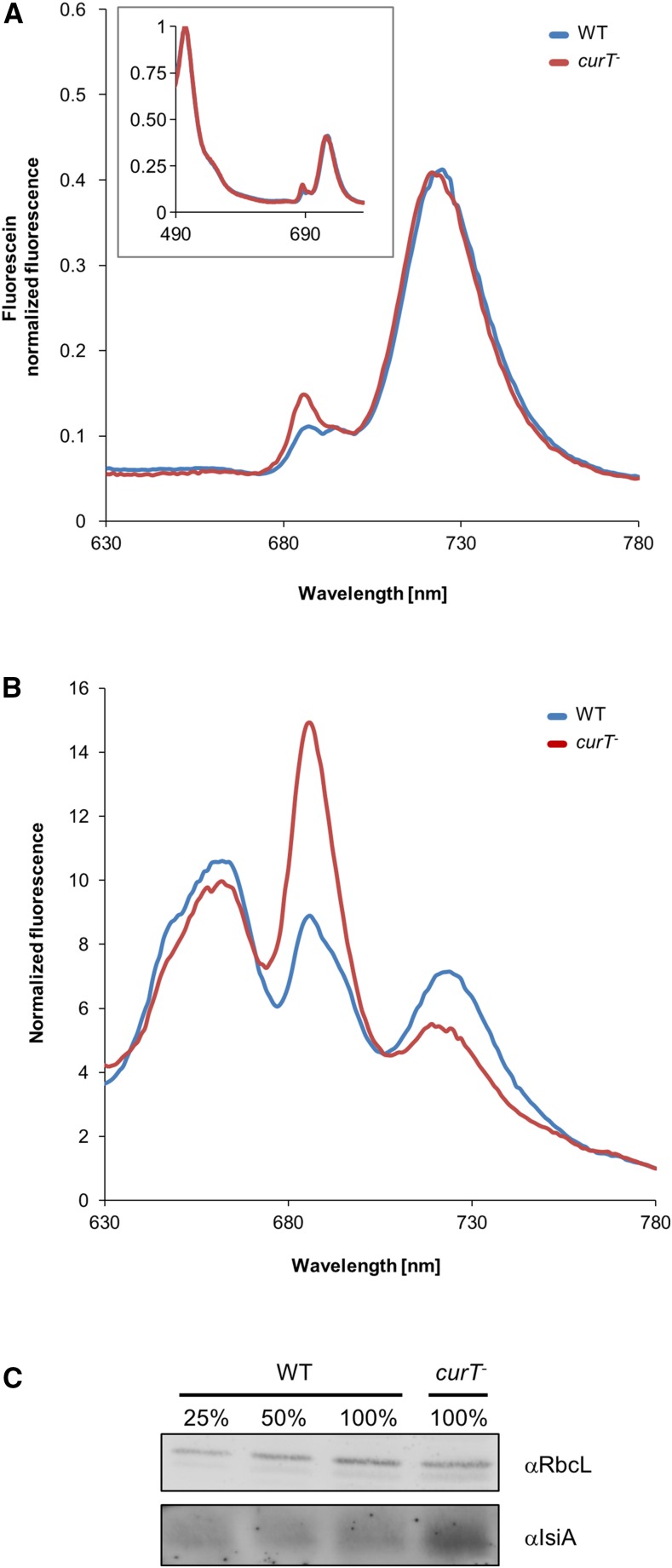
The data presented so far suggest a PSII-related phenotype for curT-, so we explored the effects of curT disruption on photosynthesis further by comparing the low-temperature (77K) fluorescence emission spectra in cell suspensions following excitation of chlorophyll at 440 nm (Figure 6A). The signals were normalized to the emission maximum at 514 nm of the external standard fluorescein (Figure 6A). In curT-, the PSI emission peak amplitude was similar to that of the wild type (Figure 6A). However, the spectra differed around 685 nm. Here, we observed an increase in fluorescence in the mutant strain, an emission signature that is probably related to the inner antenna protein CP43 (Figure 6A; Nilsson et al., 1992). Increased chlorophyll fluorescence at 685 nm has previously been shown to be characteristic for cells that are accumulating the stress-induced IsiA protein, which shares structural similarities with CP43 (Odom et al., 1993; Yeremenko et al., 2004; Wilson et al., 2007). A second indication for accumulation of IsiA is a diagnostic blue shift of the 725-nm fluorescence peak by ∼5 nm, which is indeed observed in the curT- spectrum (Figure 6A; Sandström et al., 2001). We therefore determined the level of IsiA in both the wild type and curT- by immunoblot analysis. As expected, IsiA was strongly induced in curT-, suggesting that the mutant suffers from severe stress (Figure 6C). When fluorescence at 77K was recorded after excitation at 580 nm, which mainly excites the phycobiliproteins, the signal at 685 nm (PSII related) was enhanced in the curT- mutant, most likely reflecting IsiA emission and/or the presence of uncoupled phycobilisomes (Figure 6B; Wilson et al., 2007). In contrast, the 725-nm peak (PSI related) was reduced in curT- relative to the wild type, suggesting less coupling of phycobilisomes to PSI (Figure 6B). The increase in uncoupled phycobilisomes can be directly attributed to the reduction in RCCII levels found in the curT- mutant (Figure 5A), since phycobilisomes are attached to PSII dimers for efficient light harvesting (Watanabe and Ikeuchi, 2013; Chang et al., 2015).
Figure 6.
Loss of CurT Induces Stress.
(A) Low-temperature (77K) fluorescence emission of Synechocystis 6803 wild-type (blue) and curT- (red) cells excited at 440 nm. The spectra were normalized to the added fluorescein standard.
(B) Low-temperature (77K) fluorescence emission of Synechocystis 6803 wild type (blue) and curT- (red) excited at 580 nm. The spectra were normalized to 780 nm.
(C) Level of IsiA in the curT- mutant relative to the wild type. A mean value of 368% ± 96% (sd) was determined from three independent experiments.
Thus, the curT- mutant exhibits a characteristic set of photosynthetic defects. The primary target of the CurT membrane-shaping function appears to be PSII, in particular in its biogenic phase. However, the reduction in PSII content cannot be responsible for the lack of thylakoid convergence zones at the plasma membrane because these structures can still be observed in the psbA- mutant TD41, which is lacking D1 and therefore unable to assemble any PSII complexes (Supplemental Figure 6; Nilsson et al., 1992).
Subcellular Localization of CurT
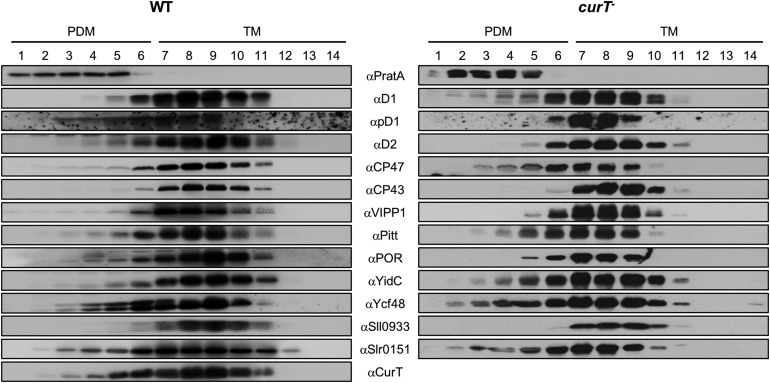
We previously proposed that the initial steps in PSII assembly take place in biogenesis centers located at the interface between plasma and thylakoid membranes (Stengel et al., 2012). These biogenic membranes (PDMs) are marked by the PSII Mn2+ delivery factor PratA and can be separated from thylakoids by a two-step sucrose-gradient centrifugation procedure (Schottkowski et al., 2009b; Rengstl et al., 2011). When the distributions of various proteins within such membrane fractions from the wild type and curT- were compared, two striking changes were detected in the mutant. First, the precursor of the D1 subunit of PSII (pD1) is absent from PDMs in the mutant; second, the inner antenna protein CP47, but not CP43, of PSII shifts toward fractions of lower density (Figure 7). Hence, the organization of PDMs appears to be perturbed in curT-. In wild-type cells, most of the CurT protein was found in thylakoid membranes; only a minor fraction, similar in amount to that of the PSII assembly factors Ycf48, Pitt, and YidC as well as the VIPP1 protein, comigrated with PDMs (Figure 7). According to rough estimates based on densitometric signal analysis, 25% of total cellular CurT is normally found in the PDMs and 75% in the thylakoids (Figure 7).
Figure 7.
Membrane Sublocalization of PSII-Related Factors.
Cell extracts from wild-type and curT- cells were fractionated by two consecutive rounds of sucrose density gradient centrifugation (Schottkowski et al., 2009b). The second linear gradient (20 to 60% sucrose) was apportioned into 14 fractions, which were analyzed by immunoblotting using the indicated antibodies. Fractions 1 to 6 represent PDMs, and fractions 7 to 14 thylakoid membranes (TMs). To facilitate comparison between gradients, sample volumes were normalized to the volume of fraction 7 that contained 40 µg of protein.
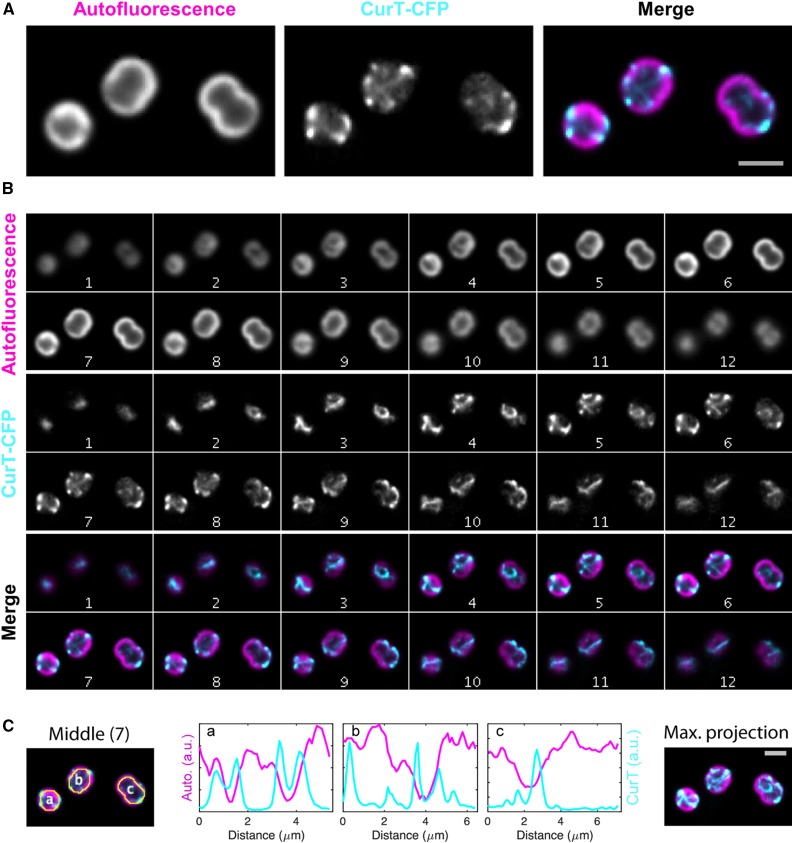
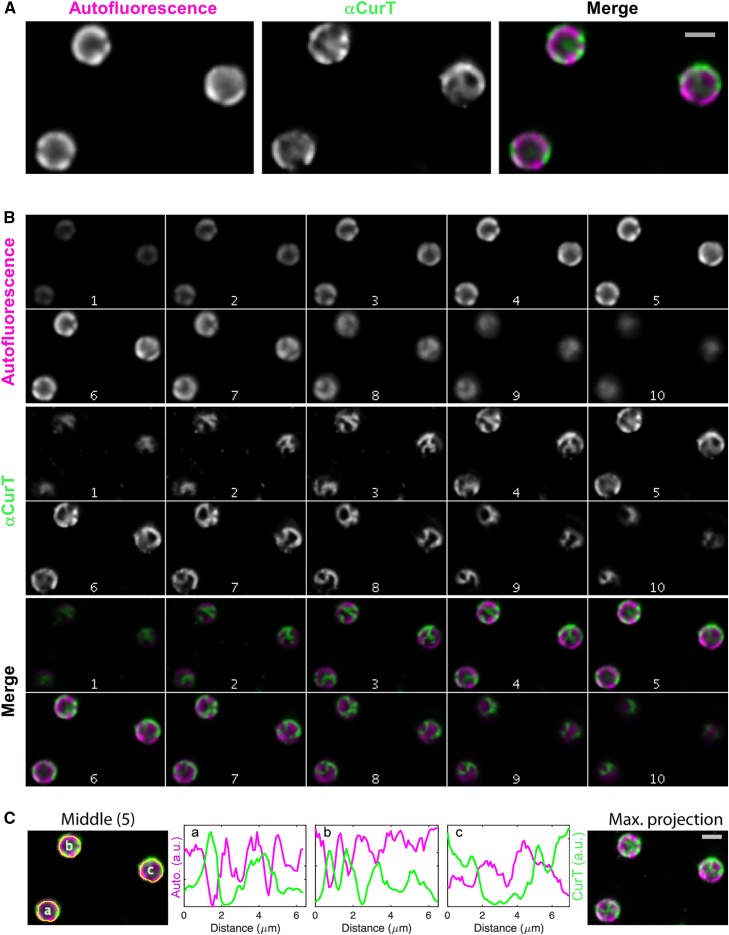
To further analyze the localization of CurT in its cellular context in vivo, we constructed a translational fusion in which the monomeric, enhanced CFP mTurquoise2 (Goedhart et al., 2012) is attached to the C terminus of CurT and is expressed under the control of the native curT promoter. The resulting strain curT-CFP showed a fully restored wild-type growth phenotype, indicating that the CFP tag does not affect CurT’s function (Supplemental Figure 7A). In agreement with this, the fusion protein accumulated to wild-type levels (96% ± 16%; Supplemental Figure 7B) and localized to the same membrane subfractions as does native CurT (Supplemental Figure 7C). The CurT-CFP fluorescence signal was distinctly discernible above the wild-type autofluorescence background (Supplemental Figure 8A) and was distributed in a network-like pattern with concentrated areas at the cell periphery at mid-plane. Analysis of CurT-CFP fluorescence in z axis montages revealed that these signals often appeared as rod-like structures that seemed to follow the spherical inner surface of the cell (Figures 8A and 8B; Supplemental Movies 1 and 2). In some cases, these structures also appear to extend through the cytoplasm (Figures 8A and 8B; Supplemental Movie 3).
Figure 8.
Live-Cell Imaging of the curT-CFP Strain by Fluorescence Microscopy.
In the curT-CFP strain, the native curT gene has been replaced by a CFP-tagged version resulting in the expression of the CurT-CFP fusion protein under the control of the native promoter.
(A) Close-up views of the mid-cell plane in the CFP and far-red autofluorescence channels shown in slice 7 in (B).
(B) Synechocystis 6803 cells expressing CurT C-terminally fused to the mTurquoise2 variant of CFP from its native chromosomal promoter. The two imaging channels are displayed as z axis montages (auto-scaled contrast, step size 250 nm) in grayscale and color composite configuration. The thylakoid autofluorescent channel as imaged in far-red fluorescence is depicted in magenta and CurT signal as imaged in cyan fluorescence is depicted in cyan.
(C) Fluorescence intensity profiles of the two channels along the autofluorescent peripheral cell “ring” are shown for slice 7. The maximum projection of the entire Z montage is also shown. Bars (light gray) = 2 µm.
When chlorophyll autofluorescence indicative of thylakoid membranes was visualized in the same cell, only a partial overlap with the CurT-CFP signal was observed (Figure 8; Supplemental Movies 1 to 3). Strikingly, the peripheral CurT-CFP signals frequently reached their maximal intensity in those areas where chlorophyll fluorescence was low, i.e., where thylakoid convergence zones are expected to form (Sacharz et al., 2015). This becomes even clearer when the intensity of a circumferential profile that follows the thylakoid’s fluorescence signal is quantified separately for each of the two fluorescent channels (Figure 8C).
In contrast, the chlorophyll autofluorescence in the curT- mutant seems to be evenly distributed (Supplemental Figure 9). Following the fluorescence in an intensity profile as in curT-CFP, no regions with a similar decline in fluorescence were found, most likely due to the absence of biogenesis centers. In addition, some circular structures presenting chlorophyll autofluorescence were detected in the interior of the cells. Hence, the disturbed curT- ultrastructure is also reflected in the distribution of chlorophyll pigments (Supplemental Figure 9).
To confirm the network-like distribution of CurT-CFP in the cell, we used immunofluorescence to detect CurT in the wild type in situ. Synechocystis 6803 cells were fixed and treated with the αCurT antibody and an Oregon Green-conjugated secondary antibody. As shown in Figures 9A and 9B, both techniques revealed similar network-like patterns of the CurT signal, which coalesced into rod-like structures at the cell surface and essentially filled up the gaps between the autofluorescent thylakoid regions (negative controls shown in Supplemental Figures 8B and 8C). Again, regions of low chlorophyll fluorescence, typical of biogenesis centers, generally exhibited stronger CurT-related fluorescence (Figure 8C). Occasionally, CurT-CFP signals were also observed near traversing thylakoid lamellae (when these were present in the cell analyzed), but the autofluorescence intensity of such thylakoids is much weaker (Figure 8B, cell b in slice 6; Supplemental Movie 3).
Figure 9.
Immunofluorescence Detection of CurT.
(A) Close-ups of slice 5 in (B).
(B) Wild-type Synechocystis 6803 cells labeled with αCurT antibodies and detected with Oregon Green-conjugated secondary antibodies. The z axis montages are shown as in Figure 8B, except that the step size is 280 nm (1 to 10).
(C) Intensity profiles of the two channels along the autofluorescent peripheral cell “ring” are shown for slice 5. The maximum projection of the entire Z montage is also shown. Bars (light gray) = 2 µm.
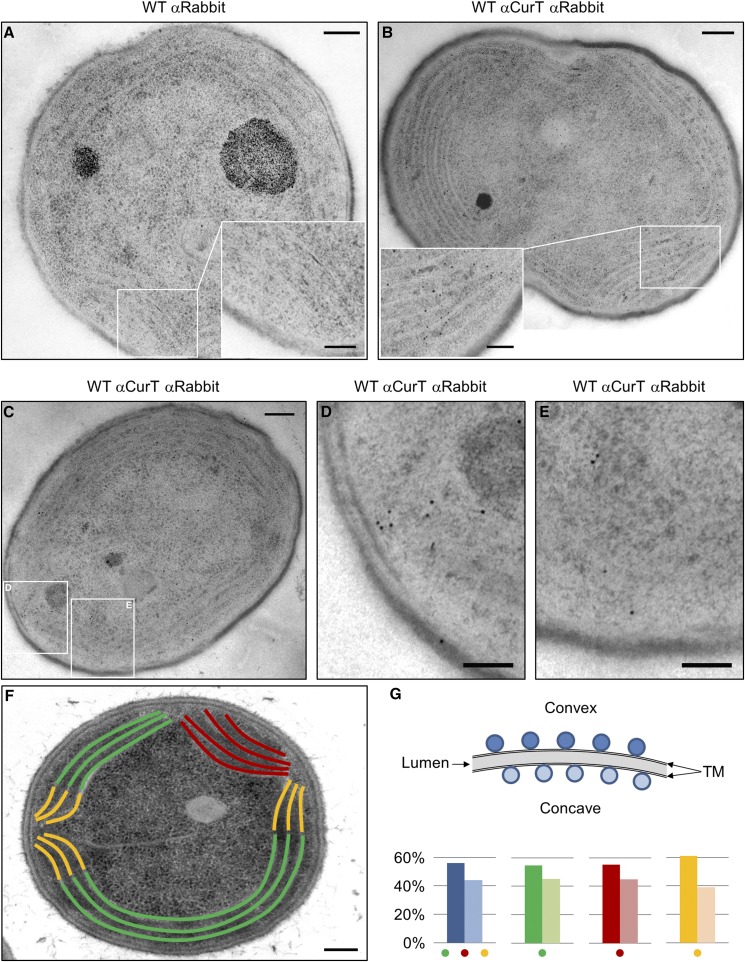
Finally, CurT was localized by immunogold labeling experiments on ultrathin sections of wild-type Synechocystis 6803 cells. Almost no signals were detected when wild-type and curT- sections were processed in the absence of the primary antibody as a negative control (Figure 10A; Supplemental Figure 10A). When mutant curT- cells were processed in the presence of the αCurT antiserum, a low level of randomly distributed background signals was observed (Supplemental Figure 10B and Supplemental Table 1). However, the number of signals located at the thylakoid membrane was clearly reduced relative to the wild type. Therefore, these signals were treated as nonspecific background. Upon incubation of wild-type cell sections with antibodies directed against the N terminus of CurT (Figure 1A), the antigen was detected on the cytoplasmic surface of thylakoid membranes (Figures 10B and 10C). This strongly suggests that its N terminus is oriented toward the cytoplasm. Overall, CurT signals appeared to be distributed along both thylakoids and PDMs. This agrees with the broad distribution of CurT across various fractions in the membrane fractionation experiments (Figure 7). In only a few cases (12%), however, was some clustering of immunogold signals at thylakoid convergence regions found (Figures 10D and 10E). Thus, the high local concentration of CurT at biogenic centers seen in the fluorescence-based approaches (Figures 8 and 9) was not quantitatively reflected in the immunogold labeling data. This is likely due to the fact that immunogold electron microscopy can only detect antigens on the surface of the section. This issue is further complicated by the heterogeneity of the CurT assemblies (see below and Discussion).
Figure 10.
Subcellular Localization of CurT.
Ultrathin sections of wild-type Synechocystis 6803 cells were incubated with rabbit αCurT prior to incubation with gold-conjugated goat anti-rabbit IgG.
(A) Negative control (wild-type processed without addition of primary antibody).
(B) and (C) Immunogold-labeled sections showing CurT localization at the cytoplasmic side of the thylakoid membrane.
(D) Cluster formation of CurT signals at a putative biogenesis center from (C).
(E) Biogenesis center from the same cell (C) without any obvious CurT clustering of gold particles.
(F) Schematic depiction of a wild-type cell (from Figure 2A) with color-coded curved membranes: green = thylakoids that follow the curvature of the cell; red = thylakoids that bend away from the plasma membrane; yellow = thylakoids that bend toward the plasma membrane to converge on sites of biogenesis centers.
(G) Relative distribution of gold signals between convex (dark color) and concave (light color) sides of membranes (n = 1584 signals from a total of 95 cells). The undifferentiated, overall signal distribution is given in blue. Bars = 200 nm (overview) and 100 nm (details).
Nevertheless, closer inspection of the immunogold signals revealed an asymmetric distribution of CurT signals with regard to the two faces of thylakoid sheets. As illustrated in Figure 10G, curved thylakoid sheets possess a longer convex and a shorter concave face. When we analyzed a total of ∼1500 CurT immunogold signals (from a total of 95 cells) that were unambiguously located to one side of the thylakoid membrane (for a representative example, see Supplemental Figure 11), 56.1% were found to be located at the convex face, while the other 43.9% localized to the concave side (Figure 10G; Supplemental Table 2). When differently curved regions (Figure 10F), including thylakoid lamellae that (1) follow the overall shape of the cell (green), (2) bend away from the plasma membrane (red), or (3) bend toward it, i.e., where thylakoids converge to form biogenesis centers (yellow), were examined separately, red and green regions showed a CurT distribution in the range of 55% convex to 45% concave (Figure 10G; Supplemental Table 2). However, at the biogenesis centers (the regions highlighted in yellow in Figure 10F), the uneven distribution of the immunogold signals was more pronounced: 61% of signals were found on the convex side and 39% on the concave face of thylakoid lamellae (Figure 10G; Supplemental Table 2). Statistical analyses confirmed the significance of these differences in signal distributions along the thylakoid membrane types (Supplemental Table 3).
Taken together with the finding that CurT is required for the formation of the convergence sites forming biogenesis centers, these data are consistent with the idea that the thylakoid sheets are shaped by CurT, which induces membrane curvature via asymmetric intercalation on the two sides of thylakoid sheets.
Different Forms of CurT Are Found in PDMs and Thylakoids
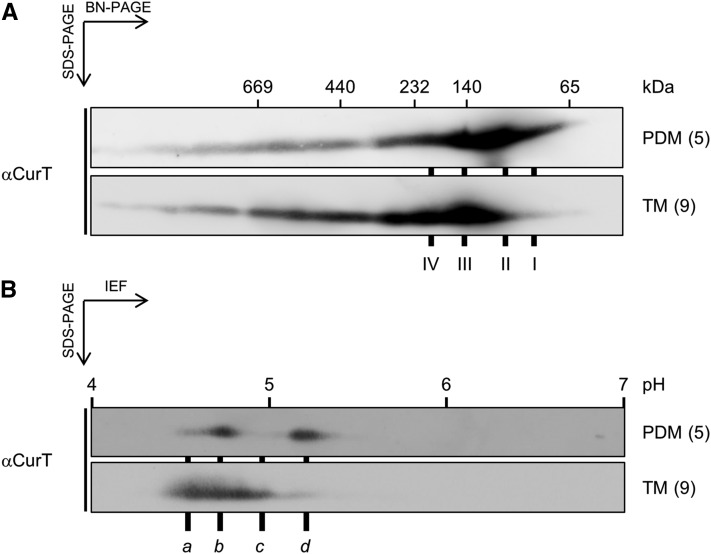
CurT was found to localize to both highly curved PDMs and less bent thylakoids. Its high local concentration at biogenic centers as seen in the fluorescence-based approaches suggests a dosage-dependent effect of CurT on membrane shaping. An alternative possibility is that different CurT variants might exist in the two membrane types, which could potentially serve different functions. To explore this possibility, we asked whether CurT is found in different complexes using 2D BN/SDS-PAGE to characterize membrane material isolated via two-step sucrose gradient centrifugation (Figure 7). In accordance with the data in Figure 5A, we find several low molecular mass complexes containing CurT (Figure 11A). Smaller complexes of ∼80 (complex I) and ∼100 kD (complex II) accumulate in membrane fraction 5 (representing PDMs), but these are far less prevalent in fraction 9 representing thylakoids (Figure 11A). In addition, CurT-containing subcomplexes of ∼140 (complex III) and ∼200 kD (complex IV) are more prominently represented in thylakoids, together with even larger complexes ranging up to 670 kD (Figure 11A).
Figure 11.
Differentially Modified Forms of CurT Are Found in Different Complexes in PDMs and Thylakoid Membranes.
Samples were isolated by fractionation of wild-type membranes via two consecutive sucrose density gradients according to Schottkowski et al. (2009b). After density centrifugation, samples were applied to BN/SDS-PAGE analysis (A) or isoelectric focusing (B). Numbers on the right indicate the respective fraction from the second gradient (see Figure 7). Fraction 5 represents the PDMs and fraction 9 the thylakoid membrane.
Even more strikingly, PDMs and thylakoids also differ with respect to the forms of CurT they contain. Thus, isoelectric focusing (IEF) analysis of both membrane fractions revealed at least four different CurT isoforms (a to d; Figure 11B). PDMs contain mainly forms b and d as well as trace amounts of a, while thylakoids apparently possess very low levels of form d and accumulate forms a, b, and c (Figure 11B). The nature of the underlying CurT modifications still has to be determined. Interestingly, however, CurT has recently been identified as a phosphoprotein in a proteomic study (Spät et al., 2015), and the CurT variants in Figure 11B may differ in their phosphorylation states. At all events, PDMs and thylakoids can be distinguished by their complement of CurT isoforms, which potentially play a role in formation of the CurT subcomplexes that define the two membrane fractions.
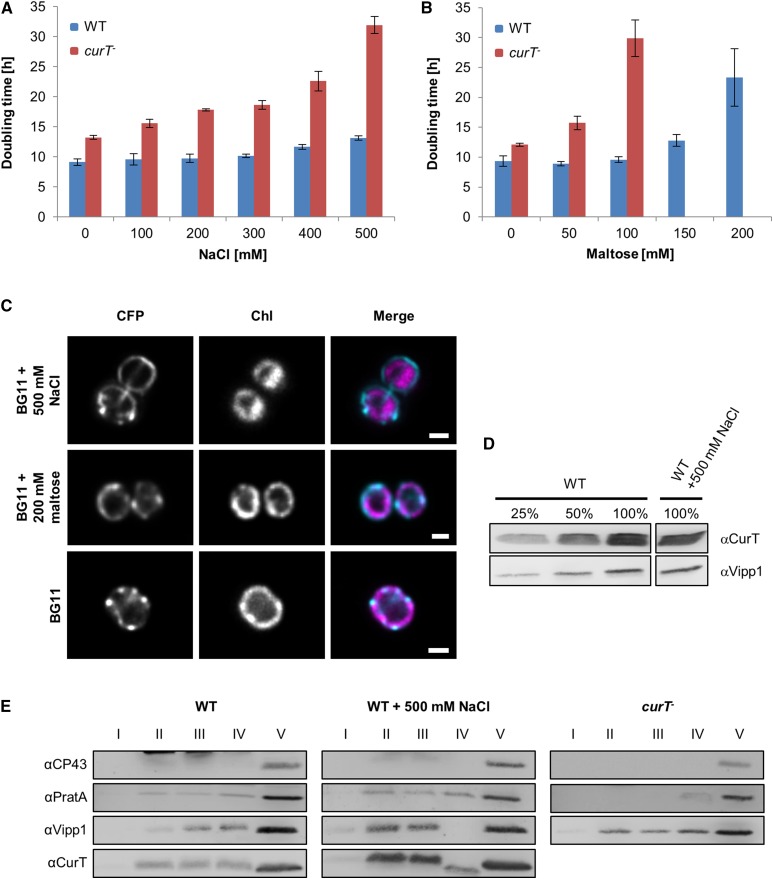
CurT Is Involved in the Osmotic Stress Response
To further explore the stress-related role of CurT (Figure 6) and its link to IsiA induction, the effect of other potential stressors on curT- cells was tested. High light levels had a minor effect on curT- growth and its photosynthetic performance (Figure 4; Supplemental Figure 5; section above), suggesting that other environmental factors induced isiA expression. Our search was also motivated by a previous proteomic survey of plasma membrane proteins of Synechocystis 6803 in high salt conditions in which CurT and VIPP1 were found among the most highly induced targets (Huang et al., 2006). First, we cultured wild-type and curT- cells in different salt concentrations and found that growth of curT- was severely affected in high salt (Figure 12A). Wild-type cells were only slightly affected by the addition of salt to the medium. The deleterious effect of the loss of CurT on growth was even more pronounced when maltose, an osmotically active compound, was present in the medium (Figure 12B). Indeed, curT- cells failed to survive exposure to 150 mM maltose, whereas growth of the wild type was only slightly compromised. These findings assign an additional function to CurT, i.e., a protective role during osmotic stress.
Figure 12.
Loss of CurT Increases Sensitivity to Abiotic Stress.
(A) and (B) Doubling times of wild-type and curT- under salt stress (A) or osmotic stress (B). Doubling times are means ± sd of three independent cultures.
(C) Localization of CurT-CFP under salt and osmotic stress. Samples were examined as described in Figure 8. Bars = 1 µm.
(D) Protein levels of wild-type cells after growth in BG11 medium containing 500 mM NaCl. Samples were analyzed on the same gel, but unrelated samples between the presented signals were omitted.
(E) Membrane preparations obtained from wild-type cells, wild-type cells grown in the presence of 500 mM NaCl, and curT- cells were subjected to the first gradient step in the membrane fractionation scheme. The gradient was divided into five fractions, and 10% of fractions I to IV and 0.2% of fraction V were analyzed by SDS-PAGE. Fraction II includes the plasma membrane and fraction V consists of PratA-defined membrane and thylakoid membrane.
When the curT-CFP strain was analyzed under these stress conditions, enhanced accumulation of CurT at the cell periphery was found, mostly outside the autofluorescent thylakoids and consistent with accumulation at the plasma membrane (Figure 12C). This was further substantiated by membrane fractionation experiments, which also revealed an increased relative abundance of CurT in the plasma membrane fraction in the first sucrose gradient (Figure 12E). Determination of total cellular CurT levels demonstrated that its absolute amount did not increase under stress (Figure 12D). Therefore, its higher abundance in the plasma membrane under osmotic stress is likely to be due to CurT trafficking toward the plasma membrane instead of increased synthesis.
The same appears to hold true for VIPP1, which has been implicated in membrane maintenance in both cyanobacteria and chloroplasts (Zhang and Sakamoto, 2015). At stress conditions, an increase of the VIPP1 signal in the plasma membrane can be monitored in the wild-type (Figure 12E). However, enhanced accumulation of VIPP1 in plasma membranes is unaffected in the curT- mutant, suggesting that thylakoid convergence zones are not strictly required for the localization of VIPP1.
DISCUSSION
CurT Determines Thylakoid Architecture
This study demonstrates that the protein CurT, a homolog of the grana-forming CURT1 proteins in plants, is essential for establishing the proper thylakoid membrane architecture in the cyanobacterium Synechocystis 6803. In particular, CurT’s membrane-bending activity is likely to be required for the formation of thylakoid biogenesis centers at points on the cell periphery where thylakoids converge toward the plasma membrane. Here, PratA-mediated preloading of early PSII assembly intermediates with Mn2+ ions has been proposed to take place, together with PSII repair (Stengel et al., 2012; Sacharz et al., 2015). This idea is supported by the following lines of evidence: (1) The curT- mutant is devoid of any thylakoid sectors that have convergence zones at their distal ends; instead thylakoid membranes are displaced and disposed as disordered or continuous rings. (2) CurT, similarly to CURT1A, possesses a membrane-curving activity in vitro and intercalates asymmetrically into thylakoids. (3) A high local concentration of CurT is observed at regions of high curvature where biogenesis centers are found. (4) Lack of CurT affects the formation and accumulation of PSII complexes, as well as the abundance of some of its assembly factors. The drastic effects of CurT inactivation indicate an important role for this factor in membrane shaping. In contrast to the situation for CURT1A from Arabidopsis, attempts to stably overexpress CurT in Synechocystis 6803 failed. Although short-lived, transient increases in CurT levels were observed when curT was expressed via strong heterologous promoters, wild-type levels of the protein were restored during subsequent rounds of cultivation of transgenic lines. Apparently Synechocystis 6803 cells do not tolerate substantial alterations in CurT dosage and counter-select against these.
This is in agreement with a dosage-dependent membrane-curving activity of CurT, which is suggested by its high local concentration at the edges of thylakoid autofluorescent regions, which mark the sites of the highly curved thylakoid convergence zones (Figures 8, 9, and 13; Supplemental Movies 1 to 3). In addition, some CurT was detected in structures extending through the cytoplasm, which most likely represent thylakoid sheets traversing the cell from one thylakoid convergence zone to another (Figures 2A and 8). CurT might be involved in the formation and stabilization of these structures.
Figure 13.
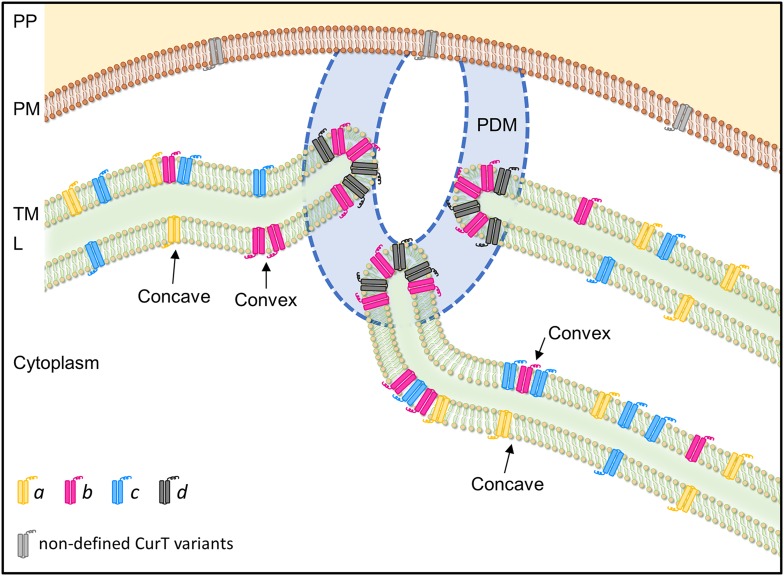
Model of CurT Distribution at Convergence Zones in Synechocystis 6803.
The distribution of CurT in thylakoid (green) and plasma membranes (brown) is shown. The pattern of localization of CurT in the thylakoid membrane is derived from membrane fractionation via sucrose density centrifugation, fluorescence-based detection of the CurT-CFP fusion, and endogenous CurT, and immunogold labeling experiments (Figures 7 to 10). The different CurT variants a, b, c, and d identified in the IEF experiments (Figure 11) are depicted in yellow, pink, blue, and black, respectively. CurT present in the plasma membrane is highlighted in gray, as its precise nature and role remain to be defined (Figure 12). Membranes are represented as lipid bilayers. Convex and concave faces of bent thylakoid sheets are indicated by black arrows. The PDM, which is dedicated to PSII assembly and repair, is depicted in blue. The dotted blue line emphasizes that the ultrastructure in this region has not yet been fully resolved. PP, periplasm; PM, plasma membrane; TM, thylakoid membrane; L, lumen.
Members of the CURT1 family contain an N-terminal amphipathic α-helix that may be involved in the membrane-bending activity of CurT (Armbruster et al., 2013). Several eukaryotic membrane-shaping proteins possess amphipathic helices as part of either an ENTH (epsin N-terminal homology) or an N-BAR (Bin, amphiphysin, Rvs; with N-terminal amphipathic helix) domain (Ford et al., 2002; Gallop and McMahon, 2005; Zimmerberg and Kozlov, 2006). ENTH and N-BAR domains are rather large (∼200 amino acids), and it has been shown that amphipatic helices present in those domains insert into one leaflet of the lipid bilayer and thereby induce curvature of the membrane (reviewed in Gallop and McMahon, 2005; Zimmerberg and Kozlov, 2006). However, CurT itself is a small protein (149 amino acids) that contains two transmembrane domains which insert into both leaflets of the lipid bilayer (Figure 1A), as already suggested by Armbruster et al. (2013). The amphipathic helix faces the cytoplasm in Synechocystis 6803 or the stroma in Arabidopsis and, most likely, it fine-tunes the degree of membrane bending mediated by CurT (Armbruster et al., 2013).
In addition, the increase in curvature correlated with CurT accumulation at convex sides of thylakoid lamella (Figures 10 and 13). Hence, curving might be mediated by asymmetric integration of CurT into the lipid bilayers forming opposite faces of thylakoid membrane sheets (Figure 13). However, the mechanism causing this asymmetry remains elusive.
The presence of different CurT isoforms and complexes in PDMs and thylakoids adds a further level of complexity, and most likely reflects the dynamic regulation of this system (Figure 11). The formation of different complexes might be primed by the phosphorylation of CurT within its N-terminal, cytoplasmic part (Spät et al., 2015). It seems likely that these complexes play distinct roles in mediating the different CurT functions, i.e., membrane bending and maintenance.
However, it remains to be seen how exactly curvature is established, maintained, and fine-tuned. In addition, other determinants of membrane topology and their putative interplay with CurT have to be considered, e.g., lipid and protein (complex) composition of membranes, cellular turgor pressure, and other factors required for membrane biogenesis/maintenance, e.g., the VIPP1 protein, before precise molecular models of CurT’s mode of action can be constructed (Rast et al., 2015; see below). That these other factors can be crucial for membrane architecture is documented by the fact that some marine cyanobacteria, such as Prochlorococcus and Synechococcus, contain curved thylakoids (but no thylakoid convergence zones at the plasma membrane) although they lack a curT gene.
The Role of Biogenesis Centers
The absence of biogenesis centers in the curT- mutant now enables one to answer some questions relating to the role of these thylakoid substructures, which form in some but not all cyanobacteria (Kunkel, 1982; van de Meene et al., 2006). First, they are not essential, since even the curT- mutant still accumulates PSII to ∼50% of wild-type levels. On the other hand, in the mutant assembly of PSII is impaired, as revealed by the increased accumulation of early assembly intermediates. In addition, PSII dimers are almost completely absent in curT-. Whether the reduction in PSII dimers is attributable to an assembly problem linked to the absence of biogenesis centers or to a secondary defect in dimer stabilization caused by changes in overall thylakoid membrane ultrastructure remains to be determined. Thus, biogenesis centers are more likely to represent evolutionary “add-ons” that facilitated efficient thylakoid biogenesis. This is compatible with the fact that some cyanobacteria are devoid of any biogenesis center-related substructures. Second, their absence compromises PSII, but the accumulation of other photosynthetic complexes, i.e., PSI and the Cyt(b6f) complex is not affected by CurT deficiency (Figure 3). This observation agrees with previous findings that neither the PsaA subunit of PSI nor its assembly factor Ycf37 is detectable in isolated PDM fractions (Rengstl et al., 2011).
In addition to reduced de novo PSII assembly, a higher relative stability of D1 in high-light experiments has been detected in curT-, which suggests that D1 degradation during PSII repair is less efficient in the mutant. Photo-damaged D1 is degraded by the FtsH2/FtsH3 complex, which has previously been shown to partly localize to thylakoid convergence zones at the cell periphery, i.e., biogenesis centers (Boehm et al., 2012; Sacharz et al., 2015). It therefore seems possible that the absence of biogenesis centers in curT- leads to mislocalization of the FtsH2/FtsH3 complex and less effective D1 degradation.
Moreover, a PSII-related function for CurT is supported by a meta-analysis of the Synechocystis 6803 transcriptome. When the CyanoEXpress 2.2 database (Hernández-Prieto and Futschik, 2012; Hernández-Prieto et al., 2016) was queried for genes coexpressed with curT (slr0483), genes for five PSII subunits, psbE, psbO, psbF, psbL, and psb30, were found among the first 10 hits (Supplemental Table 4). From this we infer that biogenic centers do not play an essential role in the assembly of all photosynthetic complexes, but serve to enhance PSII assembly/repair, possibly through the efficient delivery of Mn2+. Residual PSII in curT- cells is likely to be supplied with cytoplasmic Mn2+ via a second, independently operating, ABC transporter-based system in the plasma membrane, named the Mnt pathway (Bartsevich and Pakrasi, 1995; Bartsevich and Pakrasi, 1996).
Formally, we cannot exclude that CurT is directly involved in the PSII assembly process. However, considering CurT’s membrane bending capacity and the mutant phenotype (Figures 1D and 2), it appears more likely that CurT forms cellular substructures for efficient PSII biogenesis, i.e., biogenesis centers.
As mentioned above, some ultrastructural features of biogenesis centers have emerged from studies employing electron microscopy-based tomography (van de Meene et al., 2006). To date, however, a high-resolution picture of the precise membrane architecture within these centers remains elusive. In particular, whether or not a direct connection between plasma membrane and thylakoids exists continues to be debated. As recently discussed and in line with this gap in knowledge, different membrane fractionation techniques have revealed different distributions of PSII-related subunits and assembly factors between plasma and thylakoid membranes (Heinz et al., 2016; Liberton et al., 2016; Selão et al., 2016). The overall picture that emerges is that the initial hypothesis that PSII biogenesis is initiated at the plasma membrane is no longer tenable, whereas a specialized thylakoid subfraction like the PDMs that make contact with the plasma membrane could explain many of the available data and thus clarify some aspects of the debate (Zak et al., 2001; Pisareva et al., 2011; Rast et al., 2015). Moreover, it has previously been hypothesized that “ribosome-decorated membrane-like complexes” forming at the innermost thylakoid sheet might represent biogenic compartments (van de Meene et al., 2006; Mullineaux, 2008). Whether these structures are affected in curT- cannot be judged based on our TEM analysis but requires ultrastructural data of higher resolution. However, the accessibility of thylakoids for ribosomes should be increased in the less compressed membrane system of curT- (Figures 2B to 2D).
A scenario similar to that of Synechocystis 6803 holds for the situation in chloroplasts of the green alga Chlamydomonas reinhardtii, in which de novo PSII biogenesis has been shown to be initiated in punctate regions named T-zones, located close to the pyrenoid (Uniacke and Zerges, 2007). Unlike the mRNAs for the PSII components, neither RNAs for PSI subunits nor PSI assembly factors are localized to T-zones, indicating that PSI and PSII assembly processes are spatially separated (Uniacke and Zerges, 2009; Nickelsen and Zerges, 2013; Rast et al., 2015). Interestingly, a recent cryoelectron tomography study demonstrated that, next to T-zones at the chloroplast base-lobe junction, the inner chloroplast envelope exhibits invaginations/connections to thylakoids that structurally resemble the organization of cyanobacterial biogenesis centers (Engel et al., 2015). For future investigations, it will be interesting to see, if a homolog of the CURT1 family is involved in the formation of these structures in Chlamydomonas.
Evolution of CURT1 Function
Homologs of the CURT1 protein family can be found throughout cyanobacteria, green algae, and plants (Armbruster et al., 2013). The Arabidopsis CURT1A-D proteins have recently been shown to induce bending of thylakoid membranes at grana margin regions (Armbruster et al., 2013). Interestingly, and in contrast to the situation in Synechocystis 6803, their inactivation did not result in severe photosynthetic deficiencies, although the thylakoids formed lacked defined grana stacks. Nevertheless, the data available support the idea that the function of thylakoid membrane structures that were regarded as being independent, i.e., cyanobacterial biogenesis centers and grana, are closely related. Intriguingly, both of these thylakoid membrane structures are dedicated to aspects of PSII function but do not involve PSI (Pribil et al., 2014). In both the prokaryotic and eukaryotic systems, CURT1 homologs appear to bend thylakoid membranes, as indicated by (1) the distortion of membrane ultrastructure seen in mutants devoid of curved thylakoids, (2) the localization of CURT1 homologs at grana margins and their local concentration at biogenesis centers, and (3) the in vitro membrane-curving activity of both CURT forms. However, the phenotypic differences between Arabidopsis and Synechocystis 6803 mutants may be attributable to the different functions of the membrane subcompartments affected in the two model organisms. Whereas the role of grana is still under debate, one function of the cyanobacterial biogenesis centers is the above-mentioned high-throughput uptake of Mn2+ ions for efficient assembly of PSII (Stengel et al., 2012; Nickelsen and Rengstl, 2013; Pribil et al., 2014). However, it appears that in both cases the primary function of CURT1 proteins is to generate curved membrane regions. These discoveries suggest that, until now, two independently observed structures, i.e., cyanobacterial thylakoid biogenesis centers and chloroplast grana regions, are closely related. The degree of curvature is much less pronounced in the cyanobacterial system. As previously discussed, it was probably the subsequent evolution of a membrane-localized light-harvesting system in chloroplasts that made the formation of tightly curved grana thylakoids possible (Mullineaux, 2005; Nevo et al., 2012; Pribil et al., 2014).
CurT Is Involved in Osmotic Stress Response
One unexpected phenotype of the curT- mutant is its sensitivity to osmotic stress. These data revealed that, in addition to shaping membranes, CurT also influences their functional integrity. This latter function might be intrinsic to CurT or could be mediated by the VIPP1 protein, which has been shown to be required for membrane maintenance in bacteria and chloroplasts, probably by acting as a supplier of lipids (for an overview, see Zhang and Sakamoto, 2015). Both factors accumulate in the plasma membrane upon osmotic stress, but VIPP1 localization is not severely affected by curT inactivation, which suggests that VIPP1 operates independently of CurT. In agreement with this, repeated coimmunoprecipitations revealed no interaction between both proteins. Therefore, it remains to be established whether a direct functional relationship between them exists.
Previous investigations of osmotic stress in Synechocystis 6803 showed a kidney-shaped form of wild-type cells under very high concentrations of osmotically active compounds (Marin et al., 2006). The cells were severely deformed, indicating changes in the structure of the cellular envelope. Since CurT shifts toward the plasma membrane already under lower osmolyte concentrations, we suggest a stabilizing function of CurT in the plasma membrane. CurT might be necessary to tolerate the forces trying to deform the cells under osmotic stress.
In conclusion, this analysis has identified a crucial determinant for shaping and maintaining the cyanobacterial thylakoid membrane system, i.e., CurT, a homolog of the grana-forming CURT1 protein family from Arabidopsis. In particular, CurT is involved in the formation of specific thylakoid substructures close to the plasma membrane at which PSII is assembled/repaired. Future work will dissect the precise ultrastructure of these centers and enable us to elaborate a molecular model for the spatiotemporal organization of PSII assembly in cyanobacteria.
METHODS
Strains and Growth Conditions
Wild-type and mutant Synechocystis 6803 cells were grown on solid or in liquid BG 11 medium (Rippka et al., 1979) supplemented with 5 mM glucose (unless indicated otherwise) at 30°C under continuous illumination at a photon irradiance of 30 μmol m−2 s−1 of white light. For growth during high-light experiments for investigation of D1 repair, cells were cultivated in a FMT150 photobioreactor (Photon Systems Instruments) at 800 µmol photons m−2 s−1 in the presence or absence of lincomycin (100 µg/mL). Cell density was monitored photometrically at 750 nm. Doubling times were determined after 2 d of growth. To generate the insertion mutant curT-, the curT gene (slr0483) was first amplified from wild-type genomic DNA by PCR using the primer pair 0483/5 (GAAGCCTATTTAGCTAAGGCCGAAAG) and 0483/3 (TAGTACCTGGTCTTCCATGGCGT), and the resulting fragment was cloned into the Bluescript pKS vector (Stratagene). A kanamycin resistance cassette was then inserted into the single AgeI restriction site (159 bp downstream of the start codon), and this disruption construct was used to replace the wild-type gene as described (Wilde et al., 2001).
Bioinformatic and Computational Analysis
CURT1 sequences of Synechocystis 6803 were obtained from CyanoBase (http://genome.kazusa.or.jp/cyanobase/Synechocystis) and CURT1 homologs in other organisms were retrieved from NCBI/BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Numbers and positions of putative transmembrane domains in the predicted proteins were determined with TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The sequence for the N-terminal amphipathic helix was predicted by Jpred 3 (http://www.compbio.dundee.ac.uk/www-jpred/) and plotted using a helical wheel projection script (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi). The programs ClustalW2 (http://www.ebi.ac.uk/tools/clustalw2) and GeneDoc (http://www.nrbsc.org/old/gfx/genedoc/) were used to generate multiple alignments of amino acid sequences. Quantitative analysis of immunoblots was performed with the AIDA Image Analyzer V3.25 (Raytest Isotopenmessgeräte).
Coexpression patterns of curT were analyzed using the CyanoEXpress 2.2 database (http://cyanoexpress.sysbiolab.eu/; Hernández-Prieto and Futschik, 2012; Hernández-Prieto et al., 2016).
Measurement of Chlorophyll Concentration and Oxygen Production
Determination of chlorophyll content was performed according to Wellburn and Lichtenthaler (1984). For oxygen evolution measurements, cells were grown in BG-11 without glucose, harvested in mid-log phase, and diluted to an OD750 = 0.5. Oxygen evolution rates were measured with a Clark-type oxygen electrode (Hansatech Instruments) at 1000 µmol photons m−2 s−1 (cool white light) after 5 min of dark acclimation. Oxygen evolution under saturating light conditions was measured without any additives.
Cell Size and Cell Number Estimation
Synechocystis 6803 strains were grown in liquid BG11 media containing 5 mM glucose. For cell size estimation, the cells were analyzed with a confocal laser scanning microscope TCS SP5 (Leica). The diameter of the cells was measured with the LAS AF Lite software (Leica). Cells that were about to divide were not taken into account. For the determination of cell number, cultures were set to an OD750 = 1 and analyzed using a Neubauer counting chamber (Paul Marienfeld). Statistical significance was determined by Student’s t test.
Measurements of P700+ Reduction Kinetics and Relative Electron Transport Rates
P700+ reduction kinetics and changes in chlorophyll fluorescence yield at different light intensities were measured at a chlorophyll concentration of 5 µg/mL with a Dual-PAM-100 instrument (Heinz Walz). Complete P700 oxidation was achieved by a 50-ms multiple turnover pulse (10,000 µmol photons m−2 s−1) after 2 min of dark incubation. P700+ reduction kinetics were recorded without any additions, as well as in the presence of 10 µM DCMU. Ten technical replicates were averaged and fitted with single exponential functions. Data interpretation was done according to Bernát et al. (2009).
Light saturation measurements were performed according to Xu et al. (2008). The actinic light intensity was increased stepwise from 0 to 665 µmol photons m−2 s−1 with 30-s adaptation periods. Maximal fluorescence yields were obtained by applying saturating light pulses (duration 600 ms, intensity 10,000 µmol photons m−2 s−1).
Low-Temperature Fluorescence Measurements
Fluorescence emission spectra were measured with an AMINCO Bowman Series 2 luminescence spectrometer. Wild-type and curT- cells were grown in the presence of 5 mM glucose to mid log-phase and concentrated to 2.5 µg/mL chlorophyll, adding 2 µM fluorescein as an internal standard. Samples (1 mL) were transferred to thin glass tubes, dark-adapted for 30 min at 30°C, and shock frozen in liquid N2. To quantify chlorophyll/fluorescein and phycobilisome-mediated fluorescence, samples were excited and emission recorded at 440 nm/510 nm (chlorophyll and fluorescein excitation/maximum fluorescein emission) and 580 nm/685 nm, respectively. Emission was scanned at 490 to 800 nm or at 640 to 800 nm, respectively.
Antibody Production and Protein Analysis
For generation of an αCurT antibody, the N-terminal coding region of curT (amino acid positions 1 to 58) was amplified by PCR using primers N0483/5 (AAGGATCCGTGGGCCGTAAACATTCAA) and N0483/3 (AAGTCGACGCACTTCCCACACCGGTTGTA) and cloned into the BamHI and XhoI sites of the pGEX-4T-1 vector (GE Healthcare). Expression of the GST fusion protein in Escherichia coli BL21(DE3) and subsequent affinity purification on Glutathione-Sepharose 4B (GE Healthcare) were performed as described (Schottkowski et al., 2009b). A polyclonal antiserum was raised against this protein fragment in rabbits (Pineda). Other primary antibodies used in this study have been described previously, i.e., αpD1 (Schottkowski et al., 2009b), αD1 (Schottkowski et al., 2009b), αD2 (Klinkert et al., 2006), αPratA (Klinkert et al., 2004), αYcf48 (Rengstl et al., 2011), αPitt (Schottkowski et al., 2009a), αPOR (Schottkowski et al., 2009a), αYidC (Ossenbühl et al., 2006), αVIPP1 (Aseeva et al., 2007), αSll0933 (Armbruster et al., 2010), and αSlr0151 (Rast et al., 2016) or were purchased from Agrisera, i.e., αCP43, αCP47, αPsaA, αCyt f, αRbcL, and αIsiA. The αGFP-HRP antibody was acquired from Miltenyi Biotech.
Protein concentrations were determined according to Bradford (1976) using RotiQuant (Carl Roth). Protein extraction, membrane fractionation on sucrose-density gradients, and quantification of Western signal intensities was performed as described previously (Rengstl et al., 2011). Solubility properties of CurT were determined according to Schottkowski et al. (2009a).
Sucrose-density centrifugation separating PDM and thylakoid membrane fractions by two consecutive gradients was performed as described previously (Schottkowski et al., 2009b, Rengstl et al., 2011).
Two-dimensional fractionations of cyanobacterial membrane proteins via BN/SDS-PAGE and of pulse-labeled proteins by BN/SDS-PAGE were performed as previously reported (Klinkert et al., 2004; Schottkowski et al., 2009b; Rengstl et al., 2013). For the analysis of native complexes in fractions obtained by sucrose-density centrifugation, the volume corresponding to 350 µg of protein in fraction 7 was applied to BN/SDS-PAGE.
For IEF, the volume corresponding to 50 µg of protein from fraction 7 in 200 μL of IEF buffer (8 M urea, 4% CHAPS, and 1% IPG buffer, pH 4 to 7 [GE Healthcare]) was applied to an 11-cm Immobiline DryStrip, pH 4 to 7 (GE Healthcare), in a Protean IEF Cell (Bio-Rad). Strips were rehydrated for 12 h at 20°C and 50 V. Focusing was performed in the following steps: 500 V, rapid, 1500 Vh; 1000 V, linear, 880 Vh; 6000 V, linear, 9680 Vh; 6000 V, rapid, 5390 Vh; 50 µA per gel; focus temperature 20°C. Subsequently, the strips were incubated in equilibration buffer I (6 M urea, 0.375 M Tris, pH 8.8, 20% glycerol, 2% SDS, and 2% DTT) and equilibration buffer II (6 M urea, 0.375 M Tris, pH 8.8, 20% glycerol, 2% SDS, and 2.5% iodoacetamide) for 10 min in each buffer. Then the IEF strips were applied to second-dimension SDS-PAGE.
TEM and Immunogold Labeling
Sample preparation for TEM, including freeze substitution and immunogold labeling of CurT, was performed as described previously (Stengel et al., 2012). Ultrathin sections were cut to thicknesses of 35 to 45 nm and 45 to 65 nm for ultrastructural analyses and immunogold labeling, respectively. For immunogold labeling, sections collected on collodion-coated Ni grids were incubated for 18 h at 4°C with (rabbit) αCurT (diluted 1:100, 1:500, 1:1000, or 1:4000 in blocking buffer [5% fetal calf serum] with 0.05% Tween 20) and further processed as described previously (Stengel et al., 2012). As a negative control, wild-type Synechocystis 6803 cells were incubated without the primary antibody and exposed to the gold-labeled anti-rabbit IgG. To image the ultrastructure and the immunogold-treated samples, a Fei Morgagni 268 transmission electron microscope was used and micrographs were taken at 80 kV.
To avoid errors in the interpretation of the immunogold labeling experiment, only signals that were unambiguously localized to distinctly visible thylakoid membranes were taken into account. For each cell, different regions (Figure 10F) were defined, and in each region the signals on the convex and concave sides of the thylakoid membrane were analyzed separately (see Supplemental Figure 11 for representative counts). Statistical significance was assessed by χ2 test (Zöfel, 1988).
For the analysis of clusters of CurT signals in biogenesis centers, a cluster was defined as a minimum of four signals in close proximity. Overall, 219 biogenesis centers were examined with regard to CurT cluster formation.
Preparation of Proteoliposomes
Liposomes with a lipid mixture of 25% monogalactosyl diacylglycerol, 42% digalactosyl diacylglycerol, 16% sulfoquinovosyl diacylglycerol, and 17% phosphatidylglycerol (Lipid Products) were prepared as described previously (Armbruster et al., 2013). To generate proteoliposomes, CurT and CURT1A proteins were expressed in the presence of liposomes for 3 h at 37°C using a PURExpress In Vitro Protein Synthesis Kit (New England Biolabs). The liposomes were separated from the mix by centrifugation in a discontinuous sucrose gradient (100,000g; 18 h). Subsequently, the fraction containing the liposomes was extracted and dialyzed against 20 mM HEPES (pH 7.6). For transmission electron microscopy, the proteoliposomes were negatively stained with 1% uranyl acetate on a carbon-coated copper grid. Micrographs were taken on a Fei Morgagni 268 transmission electron microscope at 80 kV.
curT-CFP Strain Generation
Gibson assembly was used for single-step cloning (Gibson, 2011) of the slr0483 gene (together with its ∼0.8-kb upstream region), the mTurquoise2 gene fragment, the gentamycin-resistance cassette (aacC1), and an ∼1.7-kb downstream region of slr0483 into pBR322 cleaved with EcoRV and NheI. The slr0483 gene was fused in frame to the codon-optimized mTurquoise2 (DNA2.0) preceded by a linker sequence encoding GSGSG. The resulting plasmid was used to transform Synechocystis 6803 as described previously (Zang et al., 2007). After ∼10 d of incubation on BG11-GelRite (1.5%) medium in the presence of 2 µg/mL gentamycin, several antibiotic resistant colonies were obtained and streaked out on fresh medium. These transformants were then tested for full segregation of the tagged allele by colony PCR with primers flanking the slr0483 gene.
Fluorescence Microscopy
An aliquot of a cell suspension in mid-log phase was placed under a BG11-agarose pad on a glass cover slip (#1.5) and imaged on an inverted microscope (Zeiss AxioObserver.Z1) equipped with an ORCA Flash4.0 V2 camera (Hamamatsu), a Lumenecor SOLA II Light Engine, and a Plan-Apochromat 63×/1.40 Oil DIC M27 (NA 1.4) objective (Zeiss). Image acquisition was done with Zen 2.1 software in cyan and far-red fluorescence channels (Zeiss Filter Sets 47 HE and 50) as Z-series with a step size of 250 nm and effective pixel size of 103 nm. Acquired image files were subsequently deconvolved (constrained iterative method) using a default theoretical Point Spread Function in Zen 2.0 (Zeiss). Extraction of intensities for line profiles was performed by image thresholding the thylakoid autofluorescence channel and eroding the resulting binary mask in Fiji (Schindelin et al., 2012).
For immunofluorescence analysis, cells were grown to mid-log phase as in the case of the cells prepared for live-cell microscopy. A 10-mL culture was fixed for 20 min in 3.7% formaldehyde and, following extensive washing with PBS, cells were first permeabilized in 0.05% Triton X-100 for 20 min and then in lysis buffer (50 mM Tris-HCl, 100 mM NaCl, and 5 mg/mL lysozyme) at 37°C for 2 h with gentle shaking. To block unspecific binding sites, the cell pellet was incubated in 5% BSA in PBS at 30°C for 1 h. The αCurT serum (1:500 dilution) was added to cells in fresh blocking buffer for 1 h at 30°C with gentle shaking. Cells were then washed three times with blocking buffer and incubated with goat anti-rabbit IgG secondary antibodies conjugated to Oregon Green 488 (Invitrogen) (1:1000 dilution) under the conditions as for the primary antibodies. Cells were washed three times with PBS and prepared for microscopy as above. Imaging was done as described for live-cell microscopy, except that a Colibri.2 LED light source was employed for illumination and a CoolSnap HQ camera (Photometrics) was used for image acquisition. Cells were imaged as Z-series (step size 280 nm) in the yellow and far-red fluorescent channels (Zeiss Filter Sets 46 HE and 50) using the 505- and 625-nm LED modules. Effective pixel size in the final image was 102 nm. Deconvolution and image analysis was done as described above. As judged by the staining intensity of the Oregon Green-conjugated antibodies, approximately half of the cells in any given field of view appeared to have been successfully permeabilized, a rate that is common in our experience. No significant signal was detected in the control sample that had not been exposed to the primary antibodies.
Accession Numbers
Sequence data from this article can be found in the CyanoBase and EMBL/GenBank data libraries under accession number slr0483.
Supplemental Data
Supplemental Figure 1. Construction of the curT- mutant.
Supplemental Figure 2. Growth phenotype of curT-.
Supplemental Figure 3. Expression analysis of the open reading frame slr0482 (located upstream of curT) in the curT- strain.
Supplemental Figure 4. Absorbance spectra of curT- and wild-type cells.
Supplemental Figure 5. High-light growth phenotypes of wild-type Synechocystis 6803 and the curT- strain.
Supplemental Figure 6. Ultrastructure of the TD41 mutant, which lacks the psbA transcript and the D1 protein.
Supplemental Figure 7. Expression and localization of CurT-CFP.
Supplemental Figure 8. Localization of CurT and CurT-CFP by fluorescence microscopy.
Supplemental Figure 9. Chlorophyll autofluorescence of curT-.
Supplemental Figure 10. The curT- mutant as a negative control for immunogold labeling.
Supplemental Figure 11. Representative counts of immunogold signals.
Supplemental Table 1. Overview of immunogold signals in curT- cells.
Supplemental Table 2. Overview of immunogold signals in specific regions of Synechocystis 6803 wild-type cells.
Supplemental Table 3. Statistical analysis of immunogold signals for wild-type Synechocystis 6803 cells.
Supplemental Table 4. Coexpression of curT.
Supplemental Movie 1. Scan through the z axis of a curT-CFP cell.
Supplemental Movie 2. Rendered 3D model of a CurT-CFP detection.
Supplemental Movie 3. Scan through the z axis of a curT-CFP cell with a traversing thylakoid lamella.
Supplementary Material
Acknowledgments
We thank Y. Nishiyama for providing αpD1 and J. Soll for αVIPP1 and αYidC antibodies. We also thank Nina Dyczmons-Nowaczyk for support with the fluorescence measurements. This work was supported by grants awarded to J.N. (Ni390/9), M.M.N. (NO 836/3), and D.L. (Le1265/29) by the Deutsche Forschungsgemeinschaft in the context of Research Unit FOR2092 and the Cluster of Excellence RESOLV (EXC 1069). A.G. is a Howard Hughes Medical Institute postdoctoral fellow in the laboratory of Erin O’Shea at Harvard University. L.S. was supported by a PhD fellowship from the Chinese Scholarship Council. We dedicate this article to Fabrice Rappaport, an outstanding colleague and friend who passed away so early.
AUTHOR CONTRIBUTIONS
S.H., A.R., L.S., M.M.N., D.L., and J.N. designed the research. S.H., A.R., L.S., A.G., I.L.G., E.H., M.L., B.R., and S.V. performed the research. All authors contributed to data analysis. S.H., A.R., and J.N. wrote the article.
Glossary
- PDM
PratA-defined membrane
- TEM
transmission electron microscopy
- BN
Blue Native
- IEF
isoelectric focusing
References
- Allen J.F., de Paula W.B.M., Puthiyaveetil S., Nield J. (2011). A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 16: 645–655. [DOI] [PubMed] [Google Scholar]
- Anderson J.M., Horton P., Kim E.-H., Chow W.S. (2012). Towards elucidation of dynamic structural changes of plant thylakoid architecture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3515–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U., Zühlke J., Rengstl B., Kreller R., Makarenko E., Rühle T., Schünemann D., Jahns P., Weisshaar B., Nickelsen J., Leister D. (2010). The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U., et al. (2013). Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25: 2661–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aseeva E., Ossenbühl F., Sippel C., Cho W.K., Stein B., Eichacker L.A., Meurer J., Wanner G., Westhoff P., Soll J., Vothknecht U.C. (2007). Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol. Biochem. 45: 119–128. [DOI] [PubMed] [Google Scholar]
- Bartsevich V.V., Pakrasi H.B. (1995). Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 14: 1845–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsevich V.V., Pakrasi H.B. (1996). Manganese transport in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 271: 26057–26061. [DOI] [PubMed] [Google Scholar]
- Bernát G., Waschewski N., Rögner M. (2009). Towards efficient hydrogen production: the impact of antenna size and external factors on electron transport dynamics in Synechocystis PCC 6803. Photosynth. Res. 99: 205–216. [DOI] [PubMed] [Google Scholar]
- Boehm M., Yu J., Krynicka V., Barker M., Tichy M., Komenda J., Nixon P.J., Nield J. (2012). Subunit organization of a synechocystis hetero-oligomeric thylakoid FtsH complex involved in photosystem II repair. Plant Cell 24: 3669–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Chang L., Liu X., Li Y., Liu C.-C., Yang F., Zhao J., Sui S.-F. (2015). Structural organization of an intact phycobilisome and its association with photosystem II. Cell Res. 25: 726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson R., Suorsa M., Paakkarinen V., Albertsson P.-Å., Styring S., Aro E.-M., Mamedov F. (2006). Dimeric and monomeric organization of photosystem II. Distribution of five distinct complexes in the different domains of the thylakoid membrane. J. Biol. Chem. 281: 14241–14249. [DOI] [PubMed] [Google Scholar]
- Daum B., Nicastro D., Austin J. II, McIntosh J.R., Kühlbrandt W. (2010). Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell 22: 1299–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Schaffer M., Kuhn Cuellar L., Villa E., Plitzko J.M., Baumeister W. (2015). Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4: e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.G.J., Mills I.G., Peter B.J., Vallis Y., Praefcke G.J.K., Evans P.R., McMahon H.T. (2002). Curvature of clathrin-coated pits driven by epsin. Nature 419: 361–366. [DOI] [PubMed] [Google Scholar]
- Fristedt R., Willig A., Granath P., Crèvecoeur M., Rochaix J.-D., Vener A.V. (2009). Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21: 3950–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop J.L., McMahon H.T. (2005). BAR domains and membrane curvature: bringing your curves to the BAR. Biochem. Soc. Symp. 72: 223–231. [DOI] [PubMed] [Google Scholar]
- Gibson D.G. (2011). Enzymatic assembly of overlapping DNA fragments. In Methods in Enzymology, Voigt C., ed (San Diego, CA: Academic Press; ), pp. 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J., von Stetten D., Noirclerc-Savoye M., Lelimousin M., Joosen L., Hink M.A., van Weeren L., Gadella T.W.J. Jr., Royant A. (2012). Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 3: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Liauw P., Nickelsen J., Nowaczyk M. (2016). Analysis of photosystem II biogenesis in cyanobacteria. Biochim. Biophys. Acta 1857: 274–287. [DOI] [PubMed] [Google Scholar]
- Hernández-Prieto M.A., Futschik M.E. (2012). CyanoEXpress: A web database for exploration and visualisation of the integrated transcriptome of cyanobacterium Synechocystis sp. PCC6803. Bioinformation 8: 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Prieto M.A., Semeniuk T.A., Giner-Lamia J., Futschik M.E. (2016). The transcriptional landscape of the photosynthetic model cyanobacterium Synechocystis sp. PCC6803. Sci. Rep. 6: 22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Fulda S., Hagemann M., Norling B. (2006). Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 6: 910–920. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H. (2013). Architectural switches in plant thylakoid membranes. Photosynth. Res. 116: 481–487. [DOI] [PubMed] [Google Scholar]
- Klinkert B., Elles I., Nickelsen J. (2006). Translation of chloroplast psbD mRNA in Chlamydomonas is controlled by a secondary RNA structure blocking the AUG start codon. Nucleic Acids Res. 34: 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert B., Ossenbühl F., Sikorski M., Berry S., Eichacker L., Nickelsen J. (2004). PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 279: 44639–44644. [DOI] [PubMed] [Google Scholar]
- Komenda J., Nickelsen J., Tichý M., Prásil O., Eichacker L.A., Nixon P.J. (2008). The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283: 22390–22399. [DOI] [PubMed] [Google Scholar]
- Kopf M., Klähn S., Scholz I., Matthiessen J.K.F., Hess W.R., Voß B. (2014). Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 21: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel D.D. (1982). Thylakoid centers: Structures associated with the cyanobacterial photosynthetic membrane system. Arch. Microbiol. 133: 97–99. [Google Scholar]
- Liberton M., Howard Berg R., Heuser J., Roth R., Pakrasi H.B. (2006). Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Protoplasma 227: 129–138. [DOI] [PubMed] [Google Scholar]
- Liberton M., Saha R., Jacobs J.M., Nguyen A.Y., Gritsenko M.A., Smith R.D., Koppenaal D.W., Pakrasi H.B. (2016). Global proteomic analysis reveals an exclusive role of thylakoid membranes in bioenergetics of a model cyanobacterium. Mol. Cell. Proteomics 15: 2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque I., Ochoa de Alda J.A.G. (2014). CURT1,CAAD-containing aaRSs, thylakoid curvature and gene translation. Trends Plant Sci. 19: 63–66. [DOI] [PubMed] [Google Scholar]
- Marin K., Stirnberg M., Eisenhut M., Krämer R., Hagemann M. (2006). Osmotic stress in Synechocystis sp. PCC 6803: low tolerance towards nonionic osmotic stress results from lacking activation of glucosylglycerol accumulation. Microbiology 152: 2023–2030. [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W. (2005). Function and evolution of grana. Trends Plant Sci. 10: 521–525. [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W. (2008). Biogenesis and dynamics of thylakoid membranes and the photosynthetic apparatus. In The Cyanobacteria, A. Herrero and E. Flores, eds (Norfolk, UK: Caister Academic Press; ), pp. 289–304. [Google Scholar]
- Nevo R., Charuvi D., Tsabari O., Reich Z. (2012). Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J. 70: 157–176. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Rengstl B. (2013). Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64: 609–635. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Zerges W. (2013). Thylakoid biogenesis has joined the new era of bacterial cell biology. Front. Plant Sci. 4: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson F., Simpson D.J., Jansson C., Andersson B. (1992). Ultrastructural and biochemical characterization of a Synechocystis 6803 mutant with inactivated psbA genes. Arch. Biochem. Biophys. 295: 340–347. [DOI] [PubMed] [Google Scholar]
- Odom W.R., Hodges R., Chitnis P.R., Guikema J.A. (1993). Characterization of Synechocystis sp. PCC 6803 in iron-supplied and iron-deficient media. Plant Mol. Biol. 23: 1255–1264. [DOI] [PubMed] [Google Scholar]
- Ossenbühl F., Inaba-Sulpice M., Meurer J., Soll J., Eichacker L.A. (2006). The synechocystis sp PCC 6803 oxa1 homolog is essential for membrane integration of reaction center precursor protein pD1. Plant Cell 18: 2236–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva T., Kwon J., Oh J., Kim S., Ge C., Wieslander A., Choi J.-S., Norling B. (2011). Model for membrane organization and protein sorting in the cyanobacterium Synechocystis sp. PCC 6803 inferred from proteomics and multivariate sequence analyses. J. Proteome Res. 10: 3617–3631. [DOI] [PubMed] [Google Scholar]
- Pribil M., Labs M., Leister D. (2014). Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 65: 1955–1972. [DOI] [PubMed] [Google Scholar]
- Rast A., Heinz S., Nickelsen J. (2015). Biogenesis of thylakoid membranes. Biochim. Biophys. Acta 1847: 821–830. [DOI] [PubMed] [Google Scholar]
- Rast A., Rengstl B., Heinz S., Klingl A., Nickelsen J. (2016). The role of Slr0151, a tetratricopeptide repeat protein from Synechocystis sp. PCC 6803, during Photosystem II assembly and repair. Front. Plant Sci. 7: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengstl B., Oster U., Stengel A., Nickelsen J. (2011). An intermediate membrane subfraction in cyanobacteria is involved in an assembly network for Photosystem II biogenesis. J. Biol. Chem. 286: 21944–21951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengstl B., Knoppová J., Komenda J., Nickelsen J. (2013). Characterization of a Synechocystis double mutant lacking the photosystem II assembly factors YCF48 and Sll0933. Planta 237: 471–480. [DOI] [PubMed] [Google Scholar]
- Rexroth S., Mullineaux C.W., Ellinger D., Sendtko E., Rögner M., Koenig F. (2011). The plasma membrane of the cyanobacterium Gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell 23: 2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111: 1–61. [Google Scholar]
- Rütgers M., Schroda M. (2013). A role of VIPP1 as a dynamic structure within thylakoid centers as sites of photosystem biogenesis? Plant Signal. Behav. 8: e27037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacharz J., Bryan S.J., Yu J., Burroughs N.J., Spence E.M., Nixon P.J., Mullineaux C.W. (2015). Sub-cellular location of FtsH proteases in the cyanobacterium Synechocystis sp. PCC 6803 suggests localised PSII repair zones in the thylakoid membranes. Mol. Microbiol. 96: 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström S., Park Y.-I., Öquist G., Gustafsson P. (2001). CP43′, the isiA gene product, functions as an excitation energy dissipator in the cyanobacterium Synechococcus sp. PCC 7942. Photochem. Photobiol. 74: 431–437. [DOI] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottkowski M., Ratke J., Oster U., Nowaczyk M., Nickelsen J. (2009a). Pitt, a novel tetratricopeptide repeat protein involved in light-dependent chlorophyll biosynthesis and thylakoid membrane biogenesis in Synechocystis sp. PCC 6803. Mol. Plant 2: 1289–1297. [DOI] [PubMed] [Google Scholar]
- Schottkowski M., Gkalympoudis S., Tzekova N., Stelljes C., Schünemann D., Ankele E., Nickelsen J. (2009b). Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 284: 1813–1819. [DOI] [PubMed] [Google Scholar]
- Selão T.T., Zhang L., Knoppovaá J., Komenda J., Norling B. (2016). Photosystem II assembly steps take place in the thylakoid membrane of the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 57: 878. [DOI] [PubMed] [Google Scholar]
- Spät P., Maček B., Forchhammer K. (2015). Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation. Front. Microbiol. 6: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A., Gügel I.L., Hilger D., Rengstl B., Jung H., Nickelsen J. (2012). Initial steps of photosystem II de novo assembly and preloading with manganese take place in biogenesis centers in Synechocystis. Plant Cell 24: 660–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J., Zerges W. (2007). Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell 19: 3640–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J., Zerges W. (2009). Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc. Natl. Acad. Sci. USA 106: 1439–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Meene A.M., Hohmann-Marriott M.F., Vermaas W.F., Roberson R.W. (2006). The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184: 259–270. [DOI] [PubMed] [Google Scholar]
- van de Meene A.M.L., Sharp W.P., McDaniel J.H., Friedrich H., Vermaas W.F.J., Roberson R.W. (2012). Gross morphological changes in thylakoid membrane structure are associated with photosystem I deletion in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1818: 1427–1434. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Ikeuchi M. (2013). Phycobilisome: architecture of a light-harvesting supercomplex. Photosynth. Res. 116: 265–276. [DOI] [PubMed] [Google Scholar]
- Wellburn A.R., Lichtenthaler H. (1984). Formulae and program to determine total carotenoids and chlorophylls A and B of leaf extracts in different solvents. In Advances in Photosynthesis Research: Proceedings of the VIth International Congress on Photosynthesis, Brussels, Belgium, August 1–6, 1983, C. Sybesma, ed (Dordrecht, The Netherlands: Springer Netherlands), pp. 9–12. [Google Scholar]
- Wilde A., Lünser K., Ossenbühl F., Nickelsen J., Börner T. (2001). Characterization of the cyanobacterial ycf37: mutation decreases the photosystem I content. Biochem. J. 357: 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Boulay C., Wilde A., Kerfeld C.A., Kirilovsky D. (2007). Light-induced energy dissipation in iron-starved cyanobacteria: roles of OCP and IsiA proteins. Plant Cell 19: 656–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Bernát G., Singh A., Mi H., Rögner M., Pakrasi H.B., Ogawa T. (2008). Properties of mutants of Synechocystis sp. strain PCC 6803 lacking inorganic carbon sequestration systems. Plant Cell Physiol. 49: 1672–1677. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Burke J., Autz G., Jagendorf A.T. (1981). Bound ribosomes of Pea chloroplast thylakoid membranes: location and release in vitro by high salt, puromycin, and RNase. Plant Physiol. 67: 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liao L., Bo T., Zhao L., Sun X., Lu X., Norling B., Huang F. (2014). Slr0151 in Synechocystis sp. PCC 6803 is required for efficient repair of photosystem II under high-light condition. J. Integr. Plant Biol. 56: 1136–1150. [DOI] [PubMed] [Google Scholar]
- Yeremenko N., Kouřil R., Ihalainen J.A., D’Haene S., van Oosterwijk N., Andrizhiyevskaya E.G., Keegstra W., Dekker H.L., Hagemann M., Boekema E.J., Matthijs H.C.P., Dekker J.P. (2004). Supramolecular organization and dual function of the IsiA chlorophyll-binding protein in cyanobacteria. Biochemistry 43: 10308–10313. [DOI] [PubMed] [Google Scholar]
- Zak E., Norling B., Maitra R., Huang F., Andersson B., Pakrasi H.B. (2001). The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. USA 98: 13443–13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang X., Liu B., Liu S., Arunakumara K.K.I.U., Zhang X. (2007). Optimum conditions for transformation of Synechocystis sp. PCC 6803. J. Microbiol. 45: 241–245. [PubMed] [Google Scholar]
- Zhang L., Sakamoto W. (2015). Possible function of VIPP1 in maintaining chloroplast membranes. Biochim. Biophys. Acta 1847: 831–837. [DOI] [PubMed] [Google Scholar]
- Zimmerberg J., Kozlov M.M. (2006). How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7: 9–19. [DOI] [PubMed] [Google Scholar]
- Zöfel P. (1988). Statistik in der Praxis. (Stuttgart, Germany: Gustav Fischer Verlag; ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.