ToC figure
RNA tetrahedral nanoparticles with two different sizes are successfully assembled by one-pot bottom-up approach with high efficiency and thermal stability. The reported design principles can be extended to construct higher order polyhedral RNA architectures for various applications.
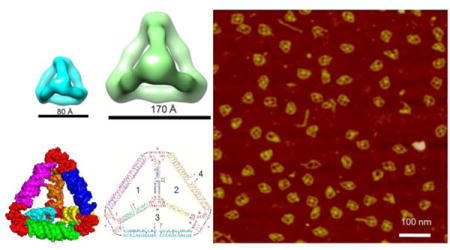
Keywords: RNA nanotechnology, RNA nanoparticle, Phi29 packaging RNA, Three-way junction, Tetrahedron
The Watson-Crick base paring properties of DNA and RNA have led to their development as excellent building blocks for the construction of nanomaterials by bottom up self-assembly [1–6]. For constructing larger architectures by directional or angular extension, it is necessary to extend the building blocks with a defined angle or orientation. This has been challenging for DNA since one helix turn of 360° is 10.5 nucleotides for the regular B-DNA. A non-integer per helix turn will result in the twisting of the extension angle or the restriction in orientation control. In RNA, the number of nucleotides per helix turn is an integer of 11 for A-form RNA, which is a common structure for most RNA sequences. We propose that this unique property of RNA will enable RNA structural growth with precise one directional control, which will facilitate the construction of large size architectures for materials science, computer device, and biomedical applications.
RNA nanotechnology is an emerging field that involves the design, construction and functionalization of nanometer-scale particles composed mainly of RNA for applications in biomedical and material sciences [1]. Previous reports have shown that a variety of RNA nanostructures can be constructed with defined sizes, shapes and stoichiometry, including triangles [7], squares [8–10], bundles [11, 12], 2D arrays [7, 13], hexamers [14–16] and 3D cages [17–19] by bottom-up self-assembly based on intra- and inter-RNA interactions. We have extensively utilized the structural features of bacteriophage phi29 packaging RNA [20] to construct varieties of RNA nanoparticles via loop-loop interactions [11, 14, 21, 22], palindrome sequence mediated foot-to-foot interactions [11, 21], and three-way junction (3WJ) motif [7, 21, 23–25]. More recently, we constructed 2D polygons such as triangle, square and pentagon [25] using the pRNA-3WJ as core scaffold at the vertices. Here we extended our previous 2D work to design and construct 3D RNA nanoparticles with controllable shape and size.
Tetrahedral geometry is attractive because of their intrinsic mechanical rigidity and structural stability. It is a pyramid-like structure with four triangular faces and six edges. Several methods have been used to construct DNA based tetrahedrons, such as an origami approach based on a long single DNA strand [26], complementary hybridization of strands using sticky ends [27, 28], and hierarchical assembly of tiles [29]. RNA is an attractive alternative building block due to its high thermal stability [30, 31] and versatility in structures [1, 32] well beyond the simplistic canonical Watson-Crick base pairing in DNA nanostructures. Herein, we used our well characterized ultrastable 3WJ motif [15, 23] as core scaffold to construct tetrahedral architectures. The RNA tetrahedrons have defined 3D structure as revealed by atomic force microscope (AFM), as well as by single particle cryo-electron microscopy (cryo-EM) that has long been found to be very challenging in imaging pure RNA structures without forming complex with proteins [33]. Moreover, for the functionization of the RNA tetrehedrons, aptamers, ribozyme and siRNA were placed at the edges of the RNA tetrahedrons with high precision without disrupting the overall structure. Importantly, the functional modules were incorporated prior to the assembly of the RNA tetrahedrons to ensure the production of homogeneous nanoparticles with high yield. Biodistribution studies revealed that RNA tetrahedrons functionalized with EGFR targeting RNA aptamer specifically targeted orthotopic breast tumors without detectable accumulation in healthy vital organs. The RNA tetrahedrons are envisioned to have a broad impact in nanotechnology arena, such as for organizing nanoscale materials with high precision, encapsulation of functional materials within its hollow cavity, targeted therapy to diseased cells and as image-guided delivery vectors.
In this study, the pRNA-3WJ motif [15, 23] (Fig. 1a) was used as a core module and placed at each of the four vertices to build RNA tetrahedrons (Fig. 1b). A total of four RNA strands were designed consisting of four pRNA 3WJ core sequences and six linking RNA sequences. A 3D model of the RNA tetrahedron (Fig. 1c) was then generated using computational modeling software UCSF Chimera [34], Swiss PDB Viewer (http://www.expasy.org/spdbv/) and PyMOL Molecular Graphics System (https://www.pymol.org/). The resulting computational model exhibited authentic tetrahedral conformation. For assembling the RNA tetrahedrons, the four RNA strands were synthesized by in vitro transcription and then mixed in stoichiometric ratio and annealed in 1×Tris buffer in a one-pot manner. Step-wise assembly of the complex was observed by native polyacrylamide gel electrophoresis (PAGE) (Fig. 1d).
Figure 1. Design and assembly of 8 nm RNA tetrahedrons.
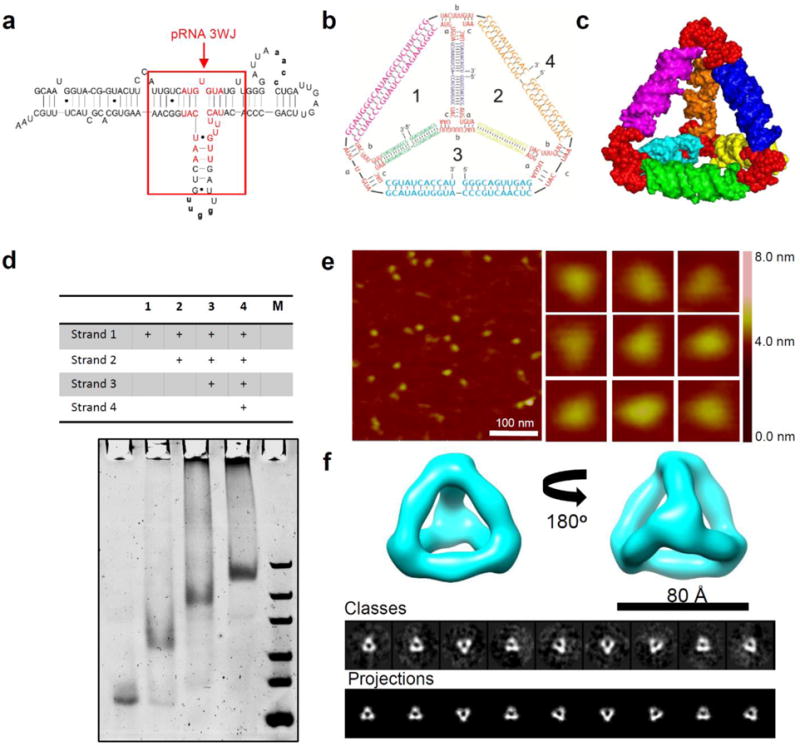
(a) 2D sequence of pRNA monomer showing the central pRNA-3WJ motif. The 22-nucleotide core sequence (with red color) of pRNA-3WJ are used to construct RNA tetrahedrons. (b) 2D sequences and (c) 3D computational model of RNA tetrahedrons. (d) 7% native PAGE gel showing step-wise assembly of RNA tetrahedrons. ‘+’ indicates the presence of the strands. M: ultra low range DNA Ladder. (e) AFM images and (f) Single particle cryo-EM 3D reconstruction of 8 nm RNA tetrahedrons.
For 3D RNA tetrahedrons, the most convincing structural characterization in their native state comes from single particle cryo-EM studies. We firstly analyzed the 3D structure of the 8 nm RNA tetrahedrons with 2-helix turn per edge (Fig. 1f). Single particle 3D reconstruction was applied by analyzing a total of 1254 particles collected from cryo-EM images and achieved a final resolution of 19 Å (gold standard criterion, 0.143 Fourier shell correlation, Suppl. Fig. 1). The 3D reconstruction data revealed that the RNA tetrahedron has a clear overall shape consistent with the computational 3D model (Fig. 1c) which agrees with the predicted size of 8 nm. The 2D computed projections of the reconstructed RNA tetrahedron 3D model showed clear match to the 2D class averages of the raw particles, suggesting the reconstructed 3D model truly represented the native structure and conformation of the designed RNA tetrahedron. The images from cryo-EM accord with the global images obtained by AFM imaging (Fig. 1e). However, the central cavities of each tetrahedron were too small to be resolved by AFM and gave an apparent size of 12.5±0.6 nm. The observed size discrepancy is due to limitations in the diameter of the AFM tip. To further characterize the size of the RNA tetrahedrons, dynamic light scattering (DLS) was performed. DLS assumes that the particles have an average globular geometry in solution. The apparent hydrodynamic size of the RNA tetrahedron was determined to be 8.5±2.4 nm (Fig. 2a).
Figure 2. Physiochemical characterization of RNA tetrahedrons.
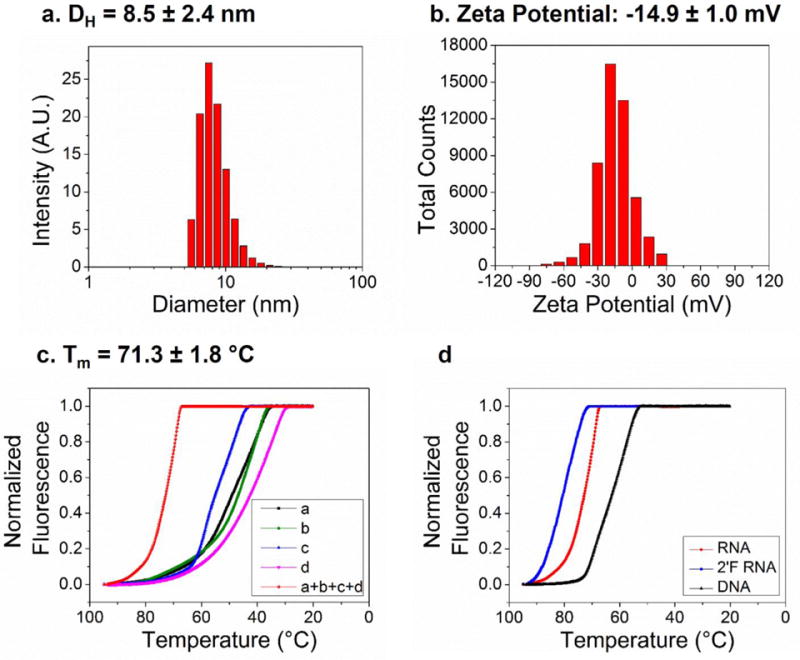
Dynamic Light Scattering (DLS) assay showing (a) the hydrodynamic size and (b) the zeta potential of RNA tetrahedrons. (c) Melting curve of RNA tetrahedron complex and each of the four component strands. (d) Comparison of melting curves for RNA, 2′-F and DNA tetrahedrons.
The surface charge of RNA tetrahedrons, measured as zeta potential, was also evaluated by DLS. As expected, due to the phosphate backbone of nucleotides, RNA tetrahedrons have a negative surface charge with a single peak at ‒14.9 ± 1.0 mV. This negative surface charge is advantageous for the overall colloid stability of RNA nanoparticles and prevents forming aggregation in solution. Moreover, negative surface charge could also reduce the non-specific interaction of the nanoparticles with the reticuloendothelial system (RES) and minimize non-specific cell entry, which is attractive for in vivo targeted drug delivery and theranostic applications [35].
To assess the thermodynamic stability of RNA tetrahedrons, we investigated their melting temperatures (Tm) by measuring their fluorescence intensities in the presence of SYBR Green II dye with the change of temperature on a real-time PCR machine. Melting experiments revealed that the assembled RNA tetrahedron had a very smooth, high-slope temperature dependent melting curve with a Tm of 71.3 ± 1.8 °C. The high slope indicates cooperative assembly of the tetrahedron from its four component strands. Moreover, we compared the melting curve of RNA, 2′-F RNA and DNA tetrahedrons. The results showed that 2′-F RNA tetrahedrons was the most stable with a Tm of 77.7 ± 2.4 °C, followed by RNA tetrahedron with a Tm of 71.3±1.8 °C, and finally DNA tetrahedron with a much lower Tm of 58.3±0.5 °C. These results were in agreement with previously reported thermodynamic stability of nucleic acids with the order of stability: 2′-F RNA > RNA > DNA [30, 31]
To investigate their enzymatic stability, we incubated unmodified and 2′-F modified (U and C nucleotides) RNA tetrahedrons in cell culture medium with 10% FBS (Fetal Bovine Serum) (Suppl. Fig. 2). At specific time points, aliquots were extracted and evaluated by native PAGE. The unmodified RNA tetrahedrons were degraded within 15 minutes, while 2′-F counterparts were stable over an extended period of time, well beyond 24 hrs. The resistance to serum-mediated degradation combined with the high thermodynamic stability is particularly attractive for the in vivo application of these RNA nanoparticles.
To demonstrate the precise tunable sizes of the RNA tetrahedrons, we designed a larger 17 nm RNA tetrahedron in which every edge of the tetrahedron was extended to 55 bp equal to 5 helix turns (Suppl. Fig. 3). This larger nanoparticle has the same overall tetrahedral geometry in our design (Fig. 3a). Upon annealing the four component strands in one pot self-assembly, the larger RNA tetrahedron assembled with high efficiency, as revealed by native PAGE analysis (Fig. 3b). DLS experiments revealed that the hydrodynamic diameter of the larger RNA tetrahedron was 16.9 ± 1.6 nm, which is in agreement with the designed dimensions (Fig. 3c). Moreover, AFM imaging was clearly able to resolve the tetrahedral morphology along with the inner cavities (Fig. 3d). Since the RNA tetrahedrons were dried on the APS-modified mica surface before imaging in air, flattened tetrahedral shapes were observed. The RNA nanoparticles were also highly homogenous in shape and structure, demonstrating the robustness of the self-assembly of RNA tetrahedrons. Cryo-EM image further showed the very clear RNA tetrahedron nanoparticles (indicated by red circles) with the expected sizes and geometries (Fig. 3e). Single particle 3D reconstruction of a total of 1582 particles collected from 131 cryo-EM images achieved a resolution of 23 Å (gold standard criterion, 0.143 Fourier shell correlation, Suppl. Fig. 4).
Figure 3. Design, assembly and characterization of 17 nm RNA tetrahedrons.
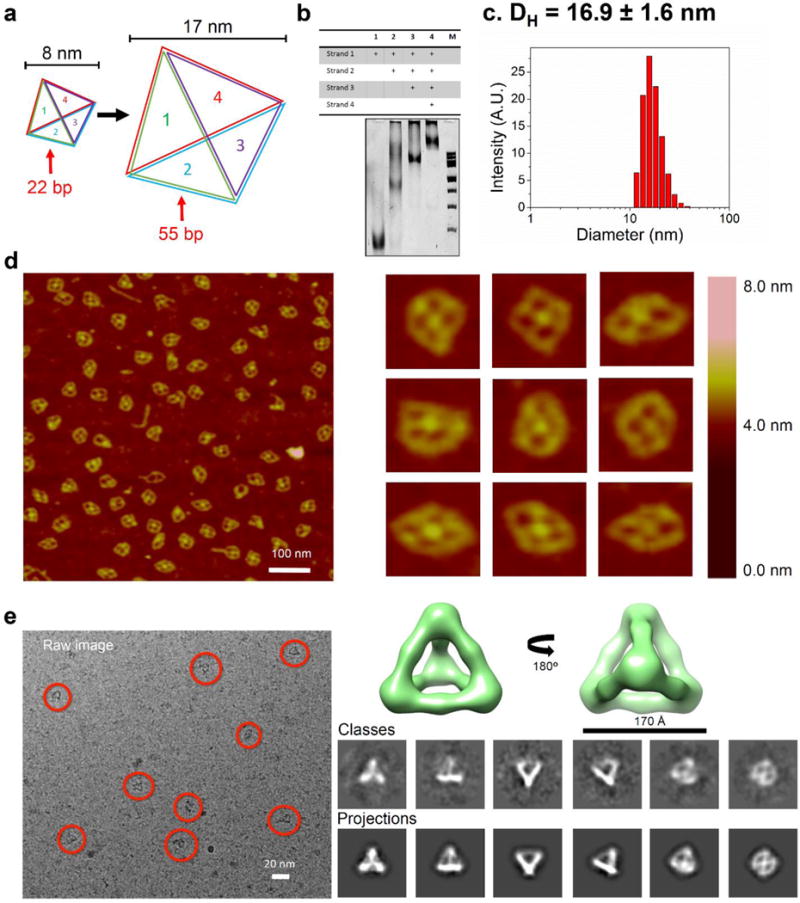
(a) Schematic showing tunable size conversion (from 22 bp per edge to 55 bp per edge) of RNA tetrahedrons. (b) 6% native PAGE gel showing step-wise assembly of larger RNA tetrahedrons. ‘+’ indicates the presence of the strands. M: 100bp DNA ladder. (c) DLS assay showing the hydrodynamic size of larger RNA tetrahedrons. (d) AFM images and (e) Cryo-EM images and 3D reconstruction of RNA tetrahedrons.
To evaluate the application of tetrahedrons, four different functional modules including a hepatitis B virus (HBV) ribozyme, fluorogenic aptamers for Malachite Green (MG) or Spinach, and a streptavidin-binding aptamer (Fig. 4a) were incorporated into the tetrahedron structure. The sequences of the functional modules were simply fused with the sequences of the 3WJ core, and then synthesized by in vitro transcription. After annealing the strands, the step-wise self-assembly was evaluated by native PAGE analysis to confirm the successful assembly of the RNA tetrahedrons (Fig. 4b). Functional assays were then conducted to determine whether the modules retained their authentic folding and functionalities upon incorporation into the RNA tetrahedrons.
Figure 4. Functional characterization of multifunctional RNA tetrahedrons.
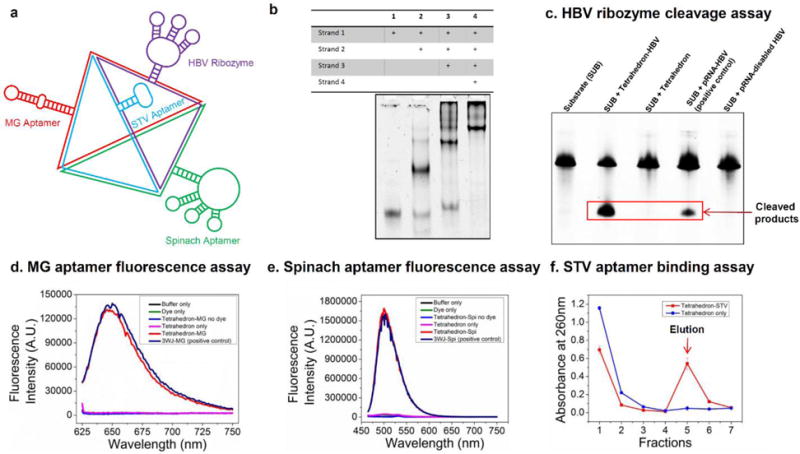
(a) Schematic showing multifunctional RNA tetrahedrons harboring HBV ribozyme, MG aptamer, Spinach aptamer and STV aptamer. (b) 7% native PAGE gel showing step-wise assembly of multifunctional RNA tetrahedrons. ‘+’ indicates the presence of the strands. (c) Ribozyme activity assay showing cleavage of 135 nt substrate. Fluorogemic assay demonstrating fluorescence emission of (d) MG aptamer and (e) Spinach aptamer. (f) Streptavidin (STV) aptamer binding assay using STV affinity column.
The HBV ribozyme is a hammerhead ribozyme that can target and cleavage the 135-nt HBV genomic RNA substrate [36]. The HBV ribozyme was fused to the RNA tetrahedron by extending one of the strands of the nanoparticle. After incubation with the RNA tetrahedron harboring the HBV ribozyme, the HBV substrate was cleaved into fragments with smaller molecular weights, as revealed by PAGE analysis (Fig. 4c). The yield of the cleavage reaction was comparable with the positive control (pRNA harboring HBV ribozyme). In contrast, RNA tetrahedron by itself or harboring disabled HBV ribozyme (G→A mutation in catalytic site) had no catalytic effects. The result confirmed that the designed multifunctional RNA tetrahedron successfully escorted the HBV ribozyme, and the catalytic activity was retained after ribozyme sequence was fused to the tetrahedron.
Both MG [37, 38] and Spinach [39] aptamers are well-characterized for their fluorogenic properties, which emit fluorescence upon binding of their respective dye targets, triphenylmethane and 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI). To verify that the MG and spinach aptamers incorporated into the multifunctional RNA tetrahedron are still functional and folded correctly, fluorescence studies were performed using a fluorospectrophotometer. The fluorescence emission spectrums showed that both the aptamers retained their ability to bind their respective dyes and emitted strong fluorescence at a similar level to the positive control, indicating the retention of the correct folding and functionalities of these fluorogenic aptamers (Fig. 4d–e). The fluorogenic modules fused with tetrahedron nanoparticles can have potential applications for imaging these nanoparticles in cells.
Upon incubation with STV agarose resins, the multifunctional RNA tetrahedron harboring STV aptamer [40] successfully bound to the resin with high affinity and were eluted by biotin (Fig. 4f). In contrast, the negative control tetrahedron did not bind to the resin and also did not show up in the elution fractions. The results indicated that the fusion with RNA tetrahedron did not interfere with the native structure and function of the STV aptamer. Moreover, in PAGE analysis of the eluted multifunctional nanoparticles harboring MG and Spinach aptamer, they still retained their fluorogenic properties, indicating that the RNA tetrahedron structure promoted the correct folding of the two aptamers after fusion into the RNA nanoparticle (Suppl. Fig. 5). The results demonstrated that the fused STV aptamer could be potentially employed as a handle to specifically purify the assembled multifunctional RNA tetrahedrons by using STV agarose resins.
Dual-luciferase reporter assay was utilized to study the gene silencing effects of the RNA tetrahedron harboring four siRNAs targeting the same region of the Firefly luciferase gene (Suppl. Fig. 6). The Renilla luciferase, which was not the target of the luciferase siRNA, served as an internal control. The relative ratio of the expression level of the firefly luciferase to the Renilla luciferase was used to evaluate the targeted gene silencing effect in MCF-7 cells upon transfection. The results revealed that the tetrahedron harboring four luciferase siRNAs with 1 nM concentration achieved ~90% gene silencing which is similar to the positive control of luciferase siRNA only at 4 nM concentration. In contrast, the tetrahedron only and scrambled control did not show any noticeable gene silencing effects (Fig. 5b). These results demonstrated that luciferase siRNA still retained their gene silencing ability after incorporation into the RNA tetrahedrons, suggesting that RNA tetrehedrons could serve as an efficient vehicle for intracellular siRNA delivery.
Figure 5. In vitro and in vivo evaluation of RNA tetrahedrons harboring siRNA and cancer-targeting aptamers.
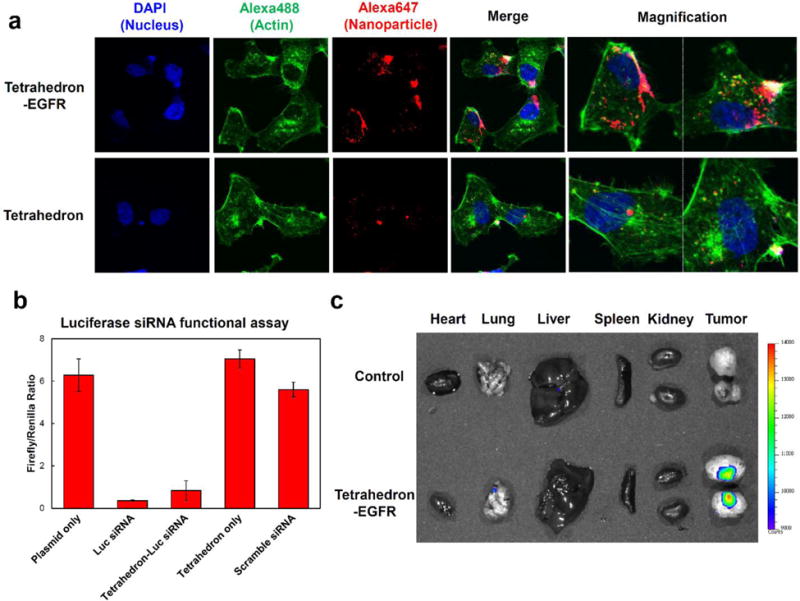
(a) Confocal images showing RNA tetrahedron (with and without EGFR aptamers) binding to MDA-MB-231 cells. (b) Luciferase siRNA silencing effects assayed by dual luciferase assay. Error bars indicate mean ± SD. (c) Biodistribution assay in orthotopic MDA-MB-231 tumor-bearing mice after systemic tail vein injection of RNA tetrahedrons harboring EGFR aptamers.
For effective cancer therapy, it is critical to guide therapeutics to specific cancer cells. The Epidermal Growth Factor Receptor (EGFR) family of Receptor Tyrosine Kinases (RTK) are highly prevalent in both primary tumors and metastatic breast cancer cells, making them an ideal candidate for targeted therapies [41]. RNA aptamers have been developed to bind to EGFR receptors [42–45] with high selectivity and sensitivity. Here we incorporated EGFR targeting RNA aptamers into RNA tetrahedrons (Suppl. Fig. 7) and evaluated their cellular binding by confocal microscopy (Fig. 5a). Alexa647 (red) labeled RNA strand was incorporated into tetrahedrons for fluorescence imaging. Tetrahedrons without EGFR aptamer were used as negative control. Confocal imaging showed that tetrahedron nanoparticles with EGFR aptamers strongly bound to EGFR(+) MDA-MB-231 cells (Fig. 5a), as revealed by the co- localization of the Alexa647-labeled RNA (red) and cellular actin (green) (Fig. 5a). In contrast, negative control “naked” tetrahedrons without the EFGR aptamer showed negligible cellular binding. The results suggest that the EFGR aptamer could facilitate the binding of RNA tetrahedrons to EGFR-expressing cancer cells.
For studying the in vivo cancer-targeting properties of the RNA tetrhedrons, we generated orthotopic breast cancer mouse models by injecting MDA-MB-231 cells directly into the mammary fat pad of athymic nude mice [46] to generate xenografts. Nuclease-resistant 2′-F RNA tetrahedrons harboring EGFR aptamers and Alexa647 were systemically injected into mice and biodistribution monitored by whole body imaging. The mice were sacrificed 8 hours post injection and their organs were collected for ex vivo imaging (Fig. 5c). RNA tetrahedrons were not detected in any other organs except the breast tumor, indicating that the nanoparticles were cleared from normal organs quickly and did not accumulate in the liver, lung, spleen or kidney after systemic injection. In another confirmatory animal trial by using EGFR-expressing KB cells xenograft mouse model, the similar cancer-targeting ability of the RNA tetrahedrons was also observed (Suppl. Fig. 8). The selective cancer-targeting ability of the RNA tetrahedrons would make this delivery system an attractive candidate for future targeted cancer imaging studies and/or cancer therapy.
In conclusion, here we report the design and self-assembly of multifunctional 3D RNA tetrahedrons based on the ultrastable pRNA-3WJ motif. The constructed RNA tetrahedrons have defined 3D structures as revealed by both AFM and single particle cryo-EM. The size of the RNA tetrahedrons can be easily tuned by changing the number of RNA base pairs per edge. Melting experiments revealed its high thermodynamic stability. Aptamers, ribozyme and siRNA were successfully incorporated into the RNA tetrahedron with their correct folding and optimal functionality. Cellular binding and biodistribution study showed that the RNA tetrahedron functionized with EGFR-aptamer targeted orthotopic breast tumors without detectable accumulation in other healthy organs. These results suggest that 3D RNA tetrahedron nanoparticles have the potential to escort imaging modules and therapeutics for in vivo cancer diagnosis and therapy.
Supplementary Material
Acknowledgments
The research was supported by NIH grants U01CA151648 (P.G.), R01EB019036 (P.G.) and DOD grant W81XWH-15-1-0052 (D.S.). Service of Shared Resource Facilities was provided by University of Kentucky Markey Cancer Center grant P30 CA177558. The cryo-EM work was supported by NIH grants P41GM103832 (W.C.) and P50 GM103297 (W.C.). The AFM work was supported by NIH grant P01GM091743 to Yuri Lyubchencko. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. P.G. is a co-founder of Biomotor and RNA Nanotechnology Development Corp. Ltd. ((Supporting Information is available online from Wiley InterScience or from the author)).
Data repository: Cryo-EM maps of the RNA tetrahedrons have been deposited to EMDataBank (8 nm RNA Tetrahedron: EMD-8233; 17 nm RNA Tetrahedron: EMD-8234).
Contributor Information
Hui Li, College of Pharmacy/Division of Pharmaceutics and Pharmaceutical Chemistry; College of Medicine/Department of Physiology & Cell Biology/Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH 43210, USA.
Dr. Kaiming Zhang, Verna and Marrs McLean Department of Biochemistry and Molecular Biology, National Center for Macromolecular Imaging, Baylor College of Medicine, Houston, Texas 77030, USA.
Fengmei Pi, College of Pharmacy/Division of Pharmaceutics and Pharmaceutical Chemistry; College of Medicine/Department of Physiology & Cell Biology/Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH 43210, USA.
Sijin Guo, College of Pharmacy/Division of Pharmaceutics and Pharmaceutical Chemistry; College of Medicine/Department of Physiology & Cell Biology/Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH 43210, USA.
Dr. Luda Shlyakhtenko, Department of Pharmaceutical Sciences, University of Nebraska Medical Center, Omaha, NE 68198, USA
Prof. Wah Chiu, Verna and Marrs McLean Department of Biochemistry and Molecular Biology, National Center for Macromolecular Imaging, Baylor College of Medicine, Houston, Texas 77030, USA
Prof. Dan Shu, College of Pharmacy/Division of Pharmaceutics and Pharmaceutical Chemistry; College of Medicine/Department of Physiology & Cell Biology/Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH 43210, USA
Prof. Peixuan Guo, College of Pharmacy/Division of Pharmaceutics and Pharmaceutical Chemistry; College of Medicine/Department of Physiology & Cell Biology/Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH 43210, USA.
References
- 1.Guo P. Nature Nanotechnology. 2010;5:833. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Lee T, Dziubla T, Pi F, Guo S, Xu J, Li C, Haque F, Liang X, Guo P. Nano Today. 2015;10:631. doi: 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P. Adv Drug Deliv Rev. 2014;66C:74. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeman NC. Annu Rev Biochem. 2010;79:65. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Nangreave J, Liu Y, Yan H. J Am Chem Soc. 2014;136:11198. doi: 10.1021/ja505101a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones MR, Seeman NC, Mirkin CA. Science. 2015;347:1260901. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 7.Khisamutdinov EF, Jasinski DL, Guo P. ACS Nano. 2014;8:4771. doi: 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasinski D, Khisamutdinov EF, Lyubchenko YL, Guo P. ACS Nano. 2014;8:7620. doi: 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Severcan I, Geary C, V E, C A, Jaeger L. Nano Lett. 2009;9:1270. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dibrov SM, McLean J, Parsons J, Hermann T. Proc Natl Acad Sci U S A. 2011;108:6405. doi: 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu D, Moll WD, Deng Z, Mao C, Guo P. Nano Lett. 2004;4:1717. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Nucleic Acids Res. 2006;34:1381. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 14.Guo P, Zhang C, Chen C, Trottier M, Garver K. Mol Cell. 1998;2:149. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. RNA. 2013;19:1226. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Nano Lett. 2011;11:878. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao C, Li X, Tian C, Jiang W, Wang G, Mao C. Nat Commun. 2014;5:3890. doi: 10.1038/ncomms4890. [DOI] [PubMed] [Google Scholar]
- 18.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. Nat Chem. 2010;2:772. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, Heldman E, Reiser J, Chiu W, Freed EO, Shapiro BA. Nano Lett. 2014;14:5662. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo P, Erickson S, Anderson D. Science. 1987;236:690. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 21.Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. RNA. 2013;19:766. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu Y, Shu D, Haque F, Guo P. Nat Protoc. 2013;8:1635. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Nature Nanotechnology. 2011;6:658. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P. Nano Today. 2012;7:245. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khisamutdinov EF, Li H, Jasinski D, Chen J, Fu J, Guo P. Nucleic Acids Res. 2014;42:9996. doi: 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Wei B, Nangreave J, Lin CX, Liu Y, Mi YL, Yan H. J Am Chem Soc. 2009;131:13093. doi: 10.1021/ja903768f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadowski JP, Calvert CR, Zhang DY, Pierce NA, Yin P. ACS Nano. 2014;8:3251. doi: 10.1021/nn4038223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Nat Nanotechnol. 2012;7:389. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iinuma R, Ke YG, Jungmann R, Schlichthaerle T, Woehrstein JB, Yin P. Science. 2014;344:65. doi: 10.1126/science.1250944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Binzel DW, Khisamutdinov EF, Guo P. Biochemistry. 2014;53:2221. doi: 10.1021/bi4017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Biochemistry. 1995;34:11211. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 32.Leontis NB, Westhof E. Curr Opin Struct Biol. 2003;13:300. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 33.Garmann RF, Gopal A, Athavale SS, Knobler CM, Gelbart WM, Harvey SC. RNA. 2015;21:877. doi: 10.1261/rna.047506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. Journal of Computational Chemistry. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Grodzinski P, Torchilin V, editors. Advanced Drug Delivery Reviews: Cancer Nanotechnology. Elsevier; 2014. [Google Scholar]
- 36.Hoeprich S, ZHou Q, Guo S, Qi G, Wang Y, Guo P. Gene Ther. 2003;10:1258. doi: 10.1038/sj.gt.3302002. [DOI] [PubMed] [Google Scholar]
- 37.Baugh C, Grate D, Wilson C. J Mol Biol. 2000;301:117. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 38.Flinders J, Defina SC, Brackett DM, Baugh C, Wilson C, Dieckmann T. Chembiochem. 2004;5:62. doi: 10.1002/cbic.200300701. [DOI] [PubMed] [Google Scholar]
- 39.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srisawat C, Engelke DR. RNA. 2001;7:632. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hynes NE, Lane HA. Nat Rev Cancer. 2005;5:341. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 42.Esposito CL, Passaro D, Longobardo I, Condorelli G, Marotta P, Affuso A, de F V, Cerchia L. PLoS One. 2011;6:e24071. doi: 10.1371/journal.pone.0024071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, Giangrande PH. Nucleic Acids Res. 2012;40:6319. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim MY, Jeong S. Nucleic Acid Ther. 2011;21:173. doi: 10.1089/nat.2011.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CH, Chernis GA, Hoang VQ, Landgraf R. Proc Natl Acad Sci U S A. 100:9226. doi: 10.1073/pnas.1332660100. 3 A.D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, Xu R, Guo P. ACS Nano. 2015;9:9731. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


