Abstract
Hedgehog (Hh) inhibitors have emerged as valid tools in the treatment of a wide range of cancers. Indeed, aberrant activation of the Hh pathway occurring either by ligand-dependent or -independent mechanisms is a key driver in tumorigenesis. The smoothened (Smo) receptor is one of the main upstream transducers of the Hh signaling and is a validated target for the development of anticancer compounds, as underlined by the FDA-approved Smo antagonist Vismodegib (GDC-0449/Erivedge) for the treatment of basal cell carcinoma. However, Smo mutations that confer constitutive activity and drug resistance have emerged during treatment with Vismodegib. For this reason, the development of new effective Hh inhibitors represents a major challenge for cancer therapy. Natural products have always represented a unique source of lead structures in drug discovery, and in recent years have been used to modulate the Hh pathway at multiple levels. Here, starting from an in house library of natural compounds and their derivatives, we discovered novel chemotypes of Hh inhibitors by mean of virtual screening against the crystallographic structure of Smo. Hh functional based assay identified the chalcone derivative 12 as the most effective Hh inhibitor within the test set. The chalcone 12 binds the Smo receptor and promotes the displacement of Bodipy-Cyclopamine in both Smo WT and drug-resistant Smo mutant. Our molecule stands as a promising Smo antagonist able to specifically impair the growth of Hh-dependent tumor cells in vitro and in vivo and medulloblastoma stem-like cells and potentially overcome the associated drug resistance.
Hedgehog (Hh) signaling is a morphogenetic pathway that has a crucial role during embryonic development and tissues homeostasis.1, 2, 3 In vertebrates, Hh pathway activation is mediated by two transmembrane receptors: Patched1 (Ptch1), endowed with inhibitory functions, and Smoothened (Smo), which is the central transducer of Hh pathway and belongs to the class F (Frizzled) G protein-coupled receptor family. In physiological conditions, extracellular Hh ligand (Shh, Ihh, Dhh) binding to Ptch1 protein relieves its repression to Smo allowing signal transduction and activation of the Gli transcription factors, which in turn upregulate target genes involved in the most important cellular processes. Aberrant activation of Hh signaling is deeply involved in tumorigenesis. Indeed, activating germline or somatic mutations of genes encoding Hh pathway components are found in human and murine basal cell carcinoma (BCC) and medulloblastoma (MB).4, 5 Moreover, uncontrolled Hh signaling has been reported to drive tumor progression in several cancers, including lung, breast, stomach, pancreas and hematopoietic malignancies.6 For this reason, the development of Hh inhibitors is eliciting great interest in drug discovery. Vismodegib (GDC-0449/Erivedge) and others Smo antagonists have shown promising results in MB and BCC tumors. However, despite an initial clinical response, a number of drug-resistant Smo mutations were observed in patients also in recent clinical trials.7, 8, 9 Further, some clinical trials have failed so far,10, 11, 12, 13 due to poor pharmacokinetics, low selectivity on cancer stem cells (CSCs), and the presence of bystander co-regulatory mechanisms of the Hh pathway. Indeed, anti-Smo resistance is mediated by hyperactivation of the powerful downstream Gli factors due to Gli2 amplification during Vismodegib or Sonidegib (LDE-225) treatment,4, 14 or upregulation of Gli via a non-canonical Hh signaling activation, such as the induction of phosphoinositide 3-kinase (PI3K) pathway observed during Sonidegib administration.15, 16 Notably, non-canonical Hh mysregulation can also occur through Gli-independent events that include Src kinase activation,17 calcium spike activity at the primary cilium,18 activation of the GTPases Rac1 and RhoA by coupling of Smo to Gi proteins,19 and metabolic reprogramming by cilium-dependent Smo-Ca2+-AMPK axis.20 These findings raise the need for new effective Smo antagonists able to escape drug resistance and to counteract tumor growth.
Natural products are a unique source of remedies and medicines since ancient times, and still have a key role in modern drug discovery.21, 22, 23 The first Hh inhibitor ever discovered has been Cyclopamine, an alkaloid isolated from Veratrum californicum that potently antagonizes Smo and has efficacy against Hh-dependent tumors.24, 25 In recent years, several natural products have been found to impact on Hh transduction by direct or indirect mechanisms.26 Of note, in our previous effort to identify small molecules targeting Gli1/DNA interaction, the isoflavone GlaB has been discovered.27 These evidences clearly indicate that natural products represent a profitable source of chemotypes to modulate the Hh pathway at multiple levels.
To this end, an in house library of natural compounds and their derivatives was screened in silico towards the crystallographic structure of the Smo bound to Cyclopamine.28 Hh functional based assay identified the chalcone 12 as the most effective Hh inhibitor within the test set. 12 binds to Smo, is not sensitive to drug-resistant Smo mutation, and shows anti-oncogenic activity promoting growth arrest of Hh driven tumor cells and primary MB cells from Ptch+/− mice, and inhibiting MB stem-like cells self-renewal.
In summary, in this work we identified the chalcone 12, and other small molecules, which represent novel natural products chemotypes of Hh inhibitors.
Results
Virtual screening
To identify natural products chemotypes of Smo antagonists, an in house library of natural and synthetic compounds was screened in silico against the crystallographic structure of Smo bound to Cyclopamine (PDB: 4O9R).28 Although being of relatively restricted dimensions, the library is endowed with a noticeable chemical diversity and lead-like features, and has been successfully used in some computer-based hits/leads discovery studies.27, 29 The FRED docking program (OpenEye) was used for carrying out docking simulations. Molecules were ranked according to the Chemgauss4 score, and the predicted binding mode of the top 20% molecules was visually inspected. Virtual hits fitting the antagonists binding site of Smo, and interacting with key residues highlighted by crystallographic studies (namely, N219, Y394, K395, R400 and E518)28, 30, 31 were deemed top priority. After a subsequent analysis of chemical diversity, molecules 1–17 (Table 1) were submitted to functional investigation.
Table 1. List of small molecules selected in silico as Smo inhibitors.
Chemistry
The potential Smo antagonists 1–17 identified in silico (see Table 1) show a noticeable range of chemical diversity and differ also for their source, which is either natural or synthetic. Most of them belong to the flavonoids family, which includes flavonol (namely, 1 and 17), flavanone (namely, 2 and 15), isoflavon (namely, 9, 13 and 16) and isoflavanone (namely, 11) derivatives. There are three Diels-Alder type adducts (namely, 5, 6 and 8) and three chalcones (namely, 7, 10 and 12). One steroid-derived alkaloid (namely, 4) and one triterpene (namely, 14) complete the test set, together with a phenylpropanoid glycoside (namely, 3). The molecular weights range from about 312 to 658 Da, the highest values corresponding to the glycosylated (e.g., 2, 3 and 15) and Diels-Alder type adducts (namely 5, 6 and 8). The source of 1–17 are collected in Table 1, together with the reference data.32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44 7, 10 and 12 are of synthetic origin but endowed with a natural scaffold, namely chalcone, and have been synthesized by Claisen–Schmidt reaction, following a slightly modified procedure developed by Professor Bargellini (1879–1963).45, 46, 47
Identification of Smo antagonists: functional screening, predicted binding mode and theoretical affinity
The inhibitory properties of the potential Smo antagonists 1–17 were investigated in a luciferase reporter assay that is widely used for characterizing Hh inhibitors. NIH3T3 Shh-Light II cells, stably incorporating a Gli-responsive firefly luciferase reporter (Gli-RE),25 were treated with the synthetic Smo agonist SAG24 alone or in combination with the selected small molecules to evaluate their dose–response ability to suppress Hh pathway. At the maximum concentration of 30 μM, compounds 1–5 revealed no activity (Supplementary Figure S1a), whereas 6–10 and 11–12 showed mild and high activity, respectively (Supplementary Figure S1b and S1c) with an IC50 range of 4–38 μM (Supplementary Table S1). We excluded the possibility that inhibition activity in this assay was mediated by cytotoxicity because no decrease was shown in the luciferase assay control Renilla as observed instead for 13–17.
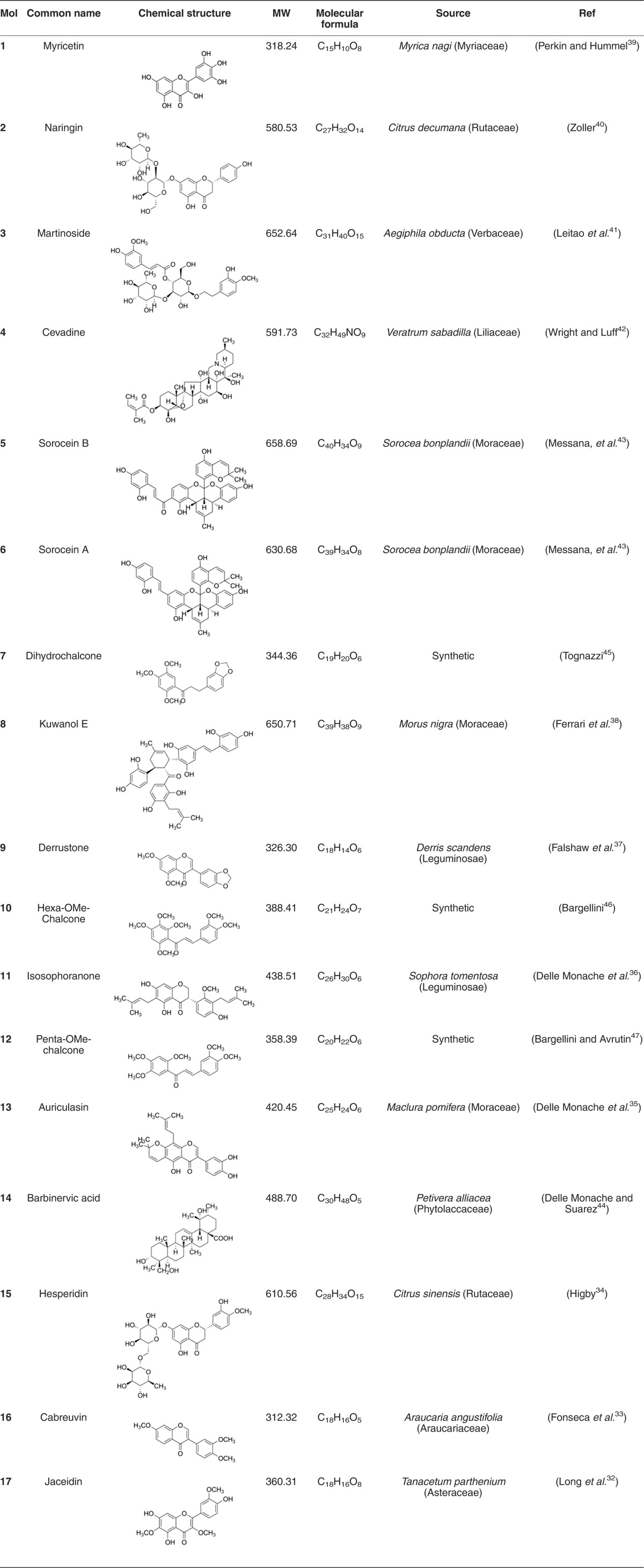
These data were in accordance with the molecular docking simulations showing that 6–12 and, particularly the most active 11 and 12 are able to fit the antagonists' site of Smo, which is located within its heptahelical bundle, and to establish interaction with its key residues. In particular, 11 and 12 bind into the hydrophobic pocket bounded by residues F484, I215, L221, M301, L303, W480, F222 and Y394. This latter residue also establishes π–π stacking interaction and an H-bond with 11 and 12 (Figures 1a and b). Additional H-bonds are established with, N219, Q477, E518 and R400. Notably, these residues have been already identified in X-ray crystallography studies as crucial for small molecules binding to Smo.28, 30, 31 Finally, the binding mode of 11 and 12 is highly comparable and show a noticeable shape and pharmacophoric overlapping each other. The predicted binding mode of molecules 6–10 is reported in Supplementary Figure S2, and is highly consistent with the above description, even if it is characterized by a fewer interactions with Smo residues, as well as with a worse score than 11 and 12 (Supplementary Table S1). For the sake of clarity, molecular docking was performed exclusively against the well-known antagonists site of Smo, occupied by the natural antagonist Cyclopamine in the selected X-ray structure.28 Indeed, our biological data convincingly support that Hh inhibitors 11 and 12 interact with Smo within the same site as Cyclopamine.48
Figure 1.
Inhibition of endogenous Hh signaling in NIH3T3 Shh-Light II cells and predicted binding mode to Smo. (a and b) Docking-based binding conformation of compounds 11 and 12 within the antagonists' site of Smo. The crystallographic structure of Smo encoded by PDB ID: 4O9R was used, and is showed as green cartoon. Residues within 5 Å from the ligands are showed as green lines. Residues involved in binding to well-known crystallographic Smo antagonists are highlighted as green sticks. Small molecules are showed as cyan sticks. H-bond interactions are highlighted by black dashed lines. (c–d) Dose–response curve of compound 12 (c) and compound 11 (d) in SAG-treated in comparison with untreated NIH3T3 Shh-Light II cells. Treatment time was 48 h, and normalization was against Renilla luciferase. Data show the mean±S.D. of three independent experiments. *P<0.05; **P<0.01 versus SAG
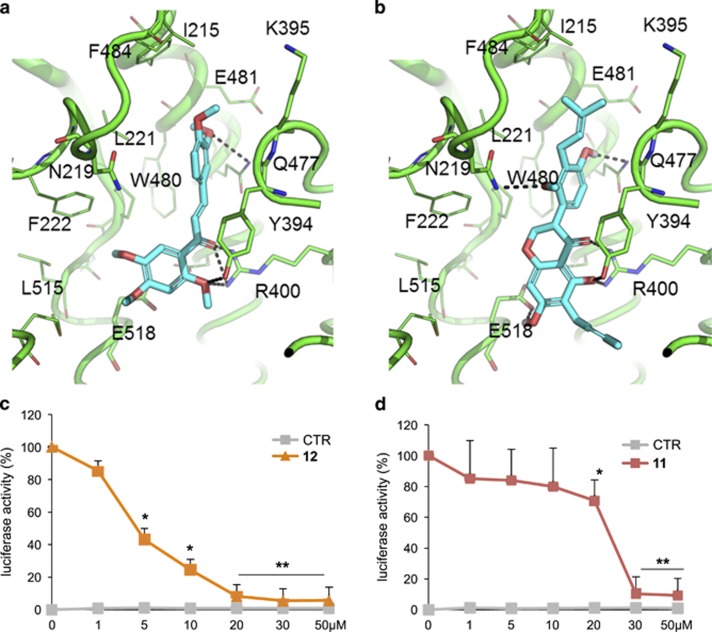
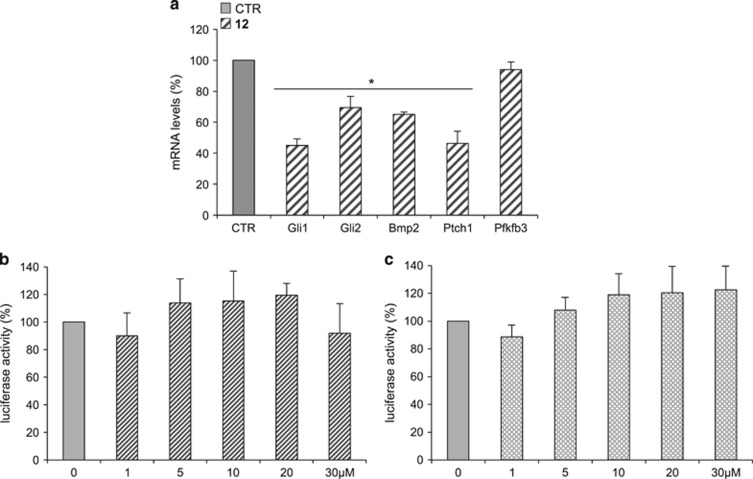
In more detail, a dose–response curve of 11 and 12 in NIH3T3 Shh-light II cells luciferase assay proved that 12 (2',4',5',3,4-pentamethoxychalcone) was the most powerful in inhibiting the Hh pathway, with an IC50 of 4.44 μM (Figure 1c and Supplementary Table S1) while 11, namely isosophoranone, showed a lower effect, with an IC50 value of 22.56 μM (Figure 1d). For this reason, we focused further studies only on 12. To investigate the inhibitory properties of 12 on Hh pathway, we analyzed endogenous Hh target gene activation in genetically defined Ptch1−/− mouse embryonic fibroblasts (Ptch1−/− MEFs) (Figure 2a). In these cells, constitutive activation of the Hh pathway is the consequence of the loss of repressive receptor Ptch1 gene, thus determining high expression levels of Hh target. Compound 12, at a concentration of 5 μM, significantly reduced mRNA levels of several endogenous Hh target genes, including Gli1, the most powerful effector of Hh signaling.5
Figure 2.
Compound 12 inhibits Hh signaling without affecting AP1/Jun and WNT/β-Catenin pathways. (a) The graphs show the Hh target genes expression levels in Ptch1−/− MEFs treated for 48 h with 12 or DMSO as control. mRNA levels were determined by quantitative real-time PCR (qRT-PCR) normalized to endogenous control (β2-microglobulin and HPRT). Pfkfb3 gene was used as negative control. *P<0.05 versus CTR. (b and c) AP1/Jun and WNT/β-Catenin pathways activity were assayed in MEFs WT transfected with MMP1-luciferase reporter and c-Jun (b) or Top Flash-luciferase reporter and β-Catenin (c), respectively, in the presence or absence of increasing concentrations of compound 12. Treatment time was 24 h, and normalization was against Renilla luciferase. Data show the mean±S.D. of three independent experiments
Importantly, 12 revealed specificity of action for Hh signaling without affecting luciferase activity driven by Hh-unrelated (i.e., Jun/AP1) (Figure 2b) and Hh-related (i.e., Wnt/β-catenin) signaling pathways, respectively (Figure 2c).
Docking-based complexes were further relaxed through energy minimization in explicit solvent, and their theoretical affinity to Smo was estimated by means of a rescoring procedure based on multiple functions (Supplementary Table S1). The Chemgauss4 and the Chemscore functions proved to rank correctly the compounds and to discriminate quite well Hh inhibitors from inactives. In contrast, rescoring with XSCORE or the Molecular-Mechanics Generalized Born Surface Area (MM-GBSA) approach provided a worse ranking of tested compounds (Supplementary Table S1).49, 50, 51 This benchmarking study performed on diverse naturally occurring chemotypes may facilitate future structure-based virtual screenings against Smo.
It is worth mentioning that 12 was predicted not to interact with D473 (Supplementary Figure S3), a key residue responsible for drug resistance at the Smo receptor upon mutation to histidine (D473H), as identified in clinical patients treated with Vismodegib.7 Therefore, it is expected that this molecule could be active also against the drug-resistant form of the Smo receptor, thus representing a potential benefit for clinical applications.
Compound 12 binding to cells expressing Smo wild-type or drug-resistant Smo mutant
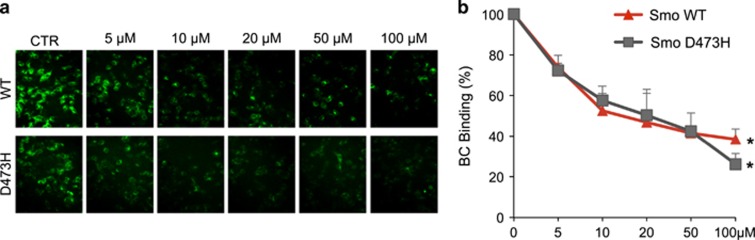
To verify the direct action of compound 12 on Smo receptor, we carried out a displacement assay based on the use of the Bodipy-Cyclopamine (BC), a fluorescent derivative of Cyclopamine that interacts with Smo at the level of its heptahelical bundle.28 Moreover, we used this assay to verify the ability of 12 to bind both Smo WT and Smo D473H mutant, the first described human Smo point mutation, which confers resistance to treatment with the Smo antagonist Vismodegib.7, 52 Indeed, drug-resistance due to Smo mutations has raised the need to develop novel Smo antagonists able to overcome this main limitation for the clinical development of effective anti-Smo molecules.7, 8, 9, 53 To this end, HEK293T cells transfected with a vector expressing Smo WT or Smo D473H mutant, were incubated with BC in the absence or presence of various concentrations of 12. As shown in Figures 3a and b, 12 revealed comparable effects on Smo WT and Smo D473H, showing dose-dependent effects and equal binding affinity corresponding to similar IC50 values (12 on Smo WT IC50=24.50 μM, and on Smo D473H IC50=22.68 μM). Moreover, 12 significantly inhibited BC binding to murine Smo WT and the mouse orthologous Smo D477G mutant (Supplementary Figure S4). Notably, while Vismodegib showed about 1000-fold loss affinity for the resistant Smo mutants in binding assay, 12 conserved the same affinity (Supplementary Table S2). These in vitro findings reveal that 12 acts as Smo antagonist by binding within the Cyclopamine site, and suggest its potential use for the treatment of cancers that are dependent on Hh signaling, including Vismodegib-resistant tumors.
Figure 3.
Compound 12 inhibition of Bodipy-Cyclopamine (BC) binding to whole cell expressing either Smo WT or Smo D473H. (a) Competitive binding of Bodipy-Cyclopamine (BC) in HEK293T cells transfected transiently with human WT or mutant Smo (D473H) was conducted with various concentrations of compound 12. BC binding (green) is visualized using fluorescence microscopy in a representative field. (b) The concentrations–response curves express the percentage of BC incorporation observed after compound 12 treatment in HEK293T cells transfected with human WT or mutant (D473H) Smo, respectively. Data show the mean±S.D. of three independent experiments. *P<0.05 versus CTR. Quantitative data are the average BC intensity from five independent fluorescence microscopy images
Compound 12 inhibits Hh-dependent cell growth of cerebellar granule cell progenitors
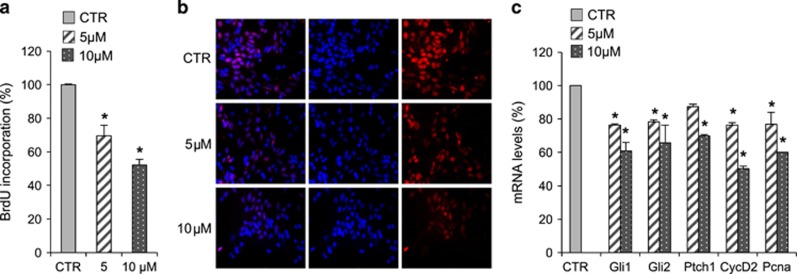
Hh signaling is a critical regulator of cerebellum development controlling the proliferation of granule cell progenitors (GCPs) under Purkinje cell-derived Shh stimuli. Withdrawal of Hh signal, occurring physiologically after the first post-natal week in mice,54 causes cell growth arrest and induces their differentiation into mature granules.55 Importantly, genetic and epigenetic alterations in the Hh pathway are responsible for the lack of GCPs proliferation arrest, leading to the tumorigenic conversion of these progenitors, considered as the cell of origin of MB.56, 57 To investigate the biological effects of 12, we first tested its ability to suppress Hh-dependent growth in 4-day-old mouse cerebellar progenitors. While treatment of GCPs with the Smo agonist SAG-enhanced cell BrdU uptake (Figures 4a and b), the addition of 12 reduced significantly this activity, decreasing the proliferation rate in a dose-dependent way (Figures 4a and b). Accordingly, GCPs treated with 12 displayed reduced mRNA levels of Hh target genes and markers related to cell growth (i.e., Gli1, Gli2, Ptch1, Pcna, cyclin D2) correlating the decrease of GCPs proliferation after 12 treatment with its inhibitory effects on Hh signature (Figure 4c).
Figure 4.
Compound 12 inhibition of Hh-dependent growth of cerebellum granule cell progenitors. (a and b) BrdU assay in cerebellar granule cell progenitors (GCPs). GCPs isolated from 4-day-old mice were treated with SAG alone or in combination with compound 12 at 5 and 10 μM concentrations for 48 h. (a) Inhibition of cell proliferation was measured as percentage of BrdU incorporation in comparison with SAG-treated sample. Data show the mean±S.D. of three independent experiments. *P<0.05 SAG+compound 12 versus SAG (CTR). (b) The immunofluorescence staining of BrdU (red) and nuclear Hoechst staining (blue) show the decrease of BrdU uptake after compound 12 treatment (5 and 10 μM). (c) qRT-PCR analysis show Hh and proliferation targets mRNA expression levels determined in GCPs culture derived from 4-day-old mouse cerebella treated with SAG alone or in combination with compound 12 at 5 and 10 μM concentrations for 48 h. In all qRT-PCR experiments, the results were normalized to endogenous control (β2-microglobulin and HPRT). Shown is the mean of three independent experiments. Error bars indicate S.D. *P<0.05 SAG+compound 12 versus SAG (CTR)
Compound 12 inhibits the growth of Hh-dependent tumor cells in vitro and in vivo
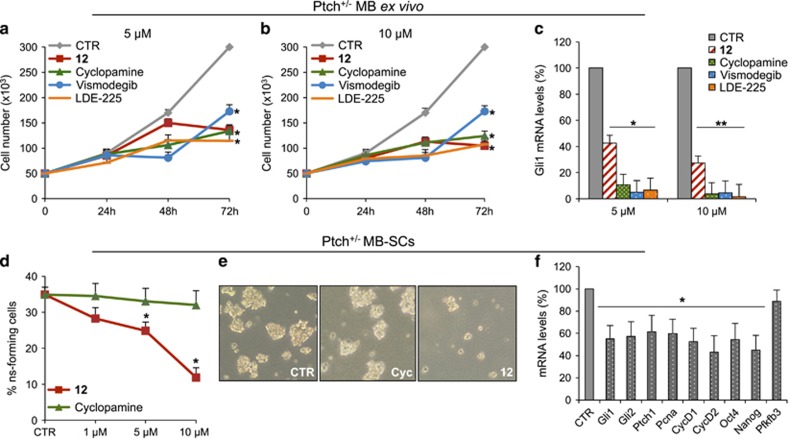
To verify the efficacy of 12 to suppress the proliferation of cancer cells in comparison with other Smo antagonists, we used Hh-dependent tumor cell models, such as MB, BCC and prostate cancer. Primary MB cells freshly isolated from Ptch1+/− mice tumors58, 59, 60 and treated with 12 showed the significant inhibition of the proliferation in comparison with other Smo antagonists (namely, Cyclopamine, Vismodegib and LDE-225), as consequence of the decrease of Gli1 mRNA levels (Figures 5a–c).
Figure 5.
Compound 12 inhibition of Hh-dependent MB tumor cell growth. (a–c) Ex vivo cell cultures from Ptch1+/− mice MBs were treated with compound 12, Cyclopamine, Vismodegib, LDE-225 or DMSO only. (a and b) After the indicated times, a trypan blue count was performed to determine the growth rate of viable cells. (c) Gli1 mRNA expression levels were determined by qRT-PCR normalized to endogenous control (β2-microglobulin and HPRT). (d–f) Compound 12 inhibits MB-SCs self-renewal. (d) Suspension of single MB-SCs isolated from Ptch1+/− mice were cultured in stem cell medium to allow the formation of primary neurospheres. Primary neurospheres were dissociated and treated with increasing concentrations of compound 12, Cyclopamine or DMSO only. After 7 days of treatment, the number of secondary neurospheres derived from a known number of single cells was counted. The self-renewal MB-SCs' capability is expressed as percentage of neurosphere-forming cells. (e) Representative bright-field images of tumor neurospheres after compound 12 or Cyclopamine treatment are also shown. (f) MB-SCs isolated from Ptch1+/− mice MBs were treated for 48 h with compound 12 or DMSO only. qRT-PCR analysis show Hh, proliferation and stemness target mRNA. For qRT-PCR, results were normalized to endogenous control (β2-microglobulin and HPRT). All data show the mean±S.D. of three independent experiments. *P<0.05; **P<0.01 versus DMSO (CTR)
Hh signaling has a central role in stem/progenitor cell maintenance and self-renewal. In several tumors, including MB, the aberrant activation of Hh signaling contributes to CSCs proliferation by the Gli mediated regulation of stemness marker Nanog.61, 62 The presence of CSCs in the tumor mass is a major cause of resistance and favors tumor relapse,63 thus representing an attractive druggable targets for anticancer therapy. Therefore, CSCs appear appealing tools for testing the therapeutic potential of the 12 herein identified. We show that 12, but not Cyclopamine, suppressed the clonogenic self-renewal ability of murine Ptch1+/− MB stem-like cells to form spheres from single-cell suspension (Figure 5d) and they appeared reduced in number and size (Figure 5e). Consistent with these results, 12 reduced Hh pathway activity in MB stem-like cells as evaluated by the decrease of the pathway readouts Gli1, Gli2 and Ptch1 mRNAs, stemness markers (Nanog, Oct4) as well as growth genes expression (cyclins D1 and D2, Pcna) (Figure 5f).
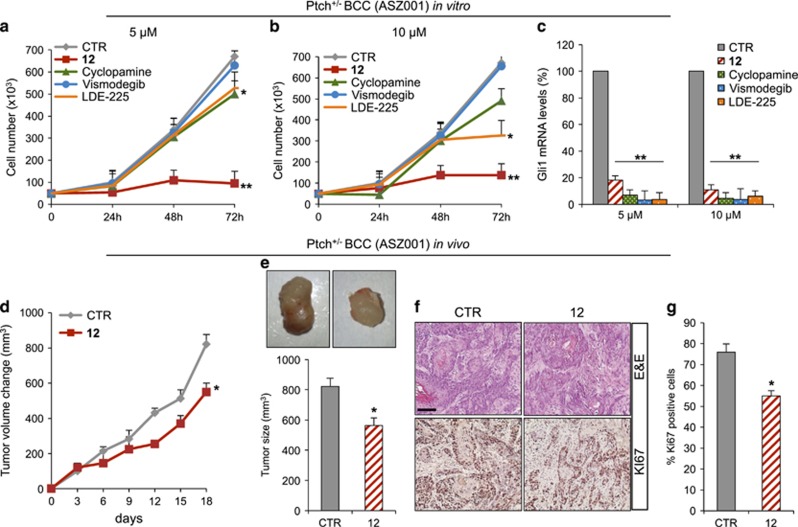
The ability of 12 to inhibit tumor cells proliferation was also tested in mouse ASZ001 BCC cells, previously characterized as a specific Hh-dependent tumor cell line harboring Ptch1 deletion.64 12 showed significant efficacy to impair ASZ001 BCC cell growth in vitro compared with Cyclopamine, Vismodegib and LDE-225 (Figures 6a and b), in agreement with the decrease of Gli1 mRNA levels observed after drug treatment (Figure 6c). The inhibitory effect of 12 on tumor growth was also confirmed in vivo using a BCC allograft model. To this end, NOD/SCID mice were engrafted with ASZ001 BCC cells and treated every second day with s.c. injections of 12 at a concentration of 50 mg/kg or vehicle alone. During the treatment period, we observed reduction of tumor cell growth in 12 treated mice compared with the controls (Figures 6d and e), consistently with decreased percentage of Ki67 labeling (Figures 6f and g).
Figure 6.
Compound 12 inhibition of Hh-dependent BCC cell growth in vitro and in vivo. (a–c) Compound 12 inhibition of Hh-dependent BCC tumor cell growth. ASZ001 BCC cells were treated with compound 12, Cyclopamine, Vismodegib, LDE-225 or DMSO only (a and b). After the indicated times, a trypan blue count was performed to determine the growth rate. Gli1 mRNA expression levels were determined by qRT-PCR after treatment of ASZ001 BCC cells with compound 12, Cyclopamine, Vismodegib, LDE-225 or DMSO only (c). Results were normalized to endogenous control (β2-microglobulin and HPRT). All data show the mean±S.D. of three independent experiments. *P<0.05; **P<0.01 versus DMSO (CTR). (d–g) ASZ001 BCC allografts. Change of tumor volume during compound 12 or vehicle treatment period (d). Representative flank allografts average volumes (e). H&E and immunohistochemical staining of Ki67 of allograft tumor samples (f). Scale bars represent 250 μm for H&E and Ki67. Quantification of Ki67 staining from immunohistochemistry shown in (f). (g) Shown is the mean±S.D. of tumor (n=6) for each treatment. *P<0.05 versus CTR
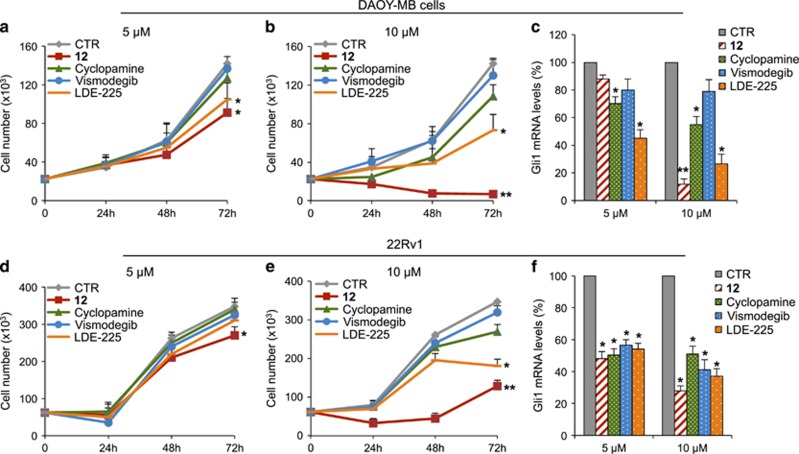
To elucidate the anti-proliferative effects of 12 in human cancer cell lines,65, 66, 67 we investigated its ability to block the growth of MB, BCC and prostate cancer cells, which are convenient model for monitoring the pharmacological inhibition of the Hh pathway.68, 69 As shown in Figure 7, 12 displayed an higher activity than other Smo antagonists in inhibiting DAOY and 22Rv1 cell proliferation (Figures 7a and b, d and e), consistent with the significant decrease of Gli1 mRNA levels after treatment (Figures 7c and f). Similar results were obtained in human BCC TE354T cell line, previously described as Hh-dependent cells,70 in which 12 was compared with Vismodegib (Supplementary Figure S5). Overall, these results confirmed that 12 inhibits the proliferation of Hh-driven tumor cells.
Figure 7.
Compound 12 inhibition of human Hh-dependent tumor cell growth. Human medulloblastoma DAOY (a and b) or human prostate carcinoma epithelial 22Rv1 cells (d and e) were treated with compound 12, Cyclopamine, Vismodegib, LDE-225 or DMSO only. After the indicated times, a trypan blue count was performed to determine the growth rate. Gli1 mRNA expression levels were determined by qRT-PCR after treatment of DAOY (c) or 22Rv1 (f) cells with compound 12, Cyclopamine, Vismodegib, LDE-225 or DMSO only. Results were normalized to endogenous control (β-actin and HPRT). All data show the mean±S.D. of three independent experiments. *P<0.05; **P<0.01 versus DMSO (CTR)
Discussion
In this study, we identified novel naturally occurring chemotypes of Hh inhibitors by mean of an integrated multidisciplinary study mixing molecular modeling with chemistry, molecular and cell biology. Particularly, the 2',4',5',3,4-pentamethoxychalcone (12), was the most potent Hh inhibitor identified herein, and proved to impair the growth of Hh-dependent tumor cells in vitro and in vivo.
In this study, we also pinpointed the relevance of natural products as useful source for drug discovery in cancer therapy. In fact, natural products represent more than one-third of all FDA-approved new molecular entities, and one-quarter of these derived from plants.21 Near the end of the Twentieth century, the use of natural products seriously declined in favor of new emerging technologies to generate drug candidates. However, these strategies did not deliver the expected results and recently there has been a renewed interest in the use of natural products.23, 41, 71, 72 Particularly, chemistry has emerged as the preferred tool to modify natural products up to suitable drug candidates, as underlined by the Cyclopamine derivative IPI–926, which has recently completed phase II clinical trials.73
Cancer is one of the main human diseases for which natural products find therapeutic applications. In this context, the morphogenetic Hh signaling represents a noticeable example of druggable tumorigenic pathway. In the past years, many research efforts have been spent in the identification of Hh antagonists able to suppress the aberrant activation of this signaling occurred in several disparate human tumors. The first Hh inhibitor ever discovered has been Cyclopamine, an alkaloid isolated from Veratrum californicum that antagonizes Smo and has efficacy against Hh-dependent tumors.25 Moreover, the isoflavone genistein, first isolated in Genista tinctoria and naturally occurring in several plants including tobacco and maize, inhibits weakly the Hh pathway.74 Other natural products have been found to impair Hh transduction by acting on the main positive regulators of this signaling, both upstream on Smo69 and downstream on the transcription factor Gli1.12, 27
Recently, the clinical development of Smo antagonists has proved disappointing because their scarce pharmacokinetics, severe side effects, and the emergence of drug resistance due to point mutations that rendered Smo insensitive to the drugs. With the aim to identify natural products as new Smo antagonists able to overcome these limits, we performed a docking-based virtual screening of an in house library of natural compounds and their derivatives, composed of about 1200 small molecules of natural or synthetic origin. This structure-based approach significantly beneficed of the crystallographic structure of Smo bound to the natural antagonist Cyclopamine. Seventeen compounds selected in silico were subsequently screened for their ability to counteract Hh activity by a luciferase functional assay in NIH3T3 Shh-Light II cells. According to this in vitro assay, five compounds (1–5) were inactive, five (6–10) showed mild activity and two (11–12) revealed high activity, whereas 13–17 resulted toxic. The dose–response activity of chalcone 12, the most effective molecule here identified, was supported by the demonstration of its direct binding to Smo receptor, as predicted by molecular modeling and confirmed in a displacement assay with BC. More interestingly, 12 resulted active on the D473H drug-resistant Smo mutant, the main cause of failure of the Vismodegib treatment, suggesting the possible therapeutic applicability of 12 for the treatment of Vismodegib-resistant tumors.75 The strong effect of 12 to inhibit Hh activity compared with BC displacement assay supposed its ability to affect other positive regulators of the Hh signaling. In this regard, 12 suppressed the expression of endogenous Hh target genes in Ptch1−/−, as well as in SuFu−/− MEFs (Supplementary Figure S6),76 two cell models in which the constitutive activation of Hh signaling is consequence of the genetic ablations of the upstream Ptch1 and the downstream SuFu-negative regulators, respectively.
Hh inhibition by 12 was also observed in the physiological cellular context of GCPs, whose proliferation is under Hh pathway controls during the cerebellum development. Indeed, 12 suppressed Hh gene signature in SAG-treated GCPs cells, with the consequent decrease of their proliferation. Noteworthy, 12 demonstrated specificity of action for Hh signaling, resulting inactive on both Hh-related and -unrelated pathways, such as Wnt/β-catenin and Jun/AP1 signaling, respectively. Aberrant activation of Hh pathway leads to tumorigenesis by ligand-independent mutational activation of the Hh signaling or ligand-dependent paracrine signaling.77 In the paracrine model of Hh-dependent oncogenesis, the ligand produced by tumor-derived cells signals to the surrounding stromal cells and indirectly promotes tumor growth. Here, we demonstrate the ability of 12 to selectively inhibit the Gli1 mRNA expression and the proliferation of Hh-dependent tumor cells in vitro and in vivo, including MB and BCC, two tumor models where Hh pathway activity occurs by ligand-independent manner. The effect of our compound in tumor models displaying ligand-dependent Hh pathway activation such as colorectal, pancreatic and bladder cancer, would be interesting to be investigated.
Overall, our findings underline the relevance of natural products as useful source for drug discovery in cancer therapy, and discover a new specific Smo antagonist that inhibits the Hh-dependent tumor cells growth, and stands as a valuable starting point to develop potential therapeutic agents for Vismodegib- or Sonidegib-resistant tumors.
Experimental section
Chemistry
Source of compounds 1–17. All the tested compounds (namely, 1–17) are known structures which belong to our in house library of natural products. Chemical identity of compounds 1–17 was assessed by re-running NMR experiments, which proved to be in agreement with the literature data reported below for each compound. The purity of all compounds, checked by reversed-phase HPLC under the chromatographic conditions reported in the Supporting Information, was always higher than 95%.
Compound 1 (myricetin or 3,5,7-trihydroxy-2-(3,4,5-trihydroxy-phenyl)-4H-chromen-4-one) was purchased from Sigma Aldrich, St. Louis, MO, USA, as used without further modification.
Compound 2 (naringin or (2S)-7-[(2S,3R,4S,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-[(2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxyoxan-2-yl]oxy-5-hydroxy-2-(4-hydroxyphenyl)-2,3-dihydrochromen-4-one was purchased from Sigma-Aldrich, as used without further modification.
Compound 3 (martinoside or [(2R,3R,4R,5R,6R)-5-hydroxy-6-[2-(3-hydroxy-4-methoxyphenyl)ethoxy]-2-(hydroxymethyl)-4-[(2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxyoxan-3-yl] (E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate) showed NMR spectra identical to the literature.41
Compound 4 (cevadine or (3β,4α,9β,16β)-4,12,14,16,17,20-hexahydroxy-4,9-epoxycevan-3-yl (2Z)-2-methyl-2-butenoate) showed NMR spectra identical to the literature.78
Compound 5 (sorocein B or (2E)-1-[(3aS,13bS,13cR)-4,11-dihydroxy-8a-(5-hydroxy-2,2-dimethyl-2H-chromen-8-yl)-2-methyl-1,8a,13b,13c-tetrahydro-3aH-benzo[3,4]isochromeno[1,8-bc]chromen-5-yl]-3-(2,4-dihydroxyphenyl)-2-propen-1-one) showed NMR spectra identical to the literature.43
Compound 6 (sorocein A or (3aS,13bS,13cR)-6-[(E)-2-(2,4-dihydroxyphenyl)vinyl]-8a-(5-hydroxy-2,2-dimethyl-2H-chromen-8-yl)-2-methyl-1,8a,13b,13c-tetrahydro-3aH-benzo[3,4]isochromeno[1,8-bc]chromene-4,11-diol) showed NMR spectra identical to the literature.43
Compound 7 (dihydrochalcone or 3-benzo[1,3]dioxol-5-yl-1-(2,4,5-trimethoxy-phenyl)-propan-1-one) showed NMR spectra identical to the literature.79
Compound 8 (kuwanol E or [2,4-dihydroxy-3-(3-methyl-2-buten-1-yl)phenyl][(1R,2S,6S)-6-(2,4-dihydroxyphenyl)-2-{4-[(E)-2-(2,4-dihydroxyphenyl)vinyl]-2,6-dihydroxyphenyl}-4-methyl-3-cyclohexen-1-yl]methanone) showed NMR spectra identical to the literature.80
Compound 9 (derrustone or 3-(1,3-benzodioxol-5-yl)-5,7-dimethoxy-4H-chromen-4-one) showed NMR spectra identical to the literature.81
Compound 10 (2',3',4',6',3,4-hexamethoxychalcone or 3-(3,4-dimethoxy-phenyl)-1-(2,3,4,6-tetramethoxy-phenyl)-2-propen-1-one) showed NMR spectra identical to the literature.79
Compound 11 (isosophoranone or 5,7-dihydroxy-3-[4-hydroxy-2-methoxy-3-(3-methyl-2-buten-1-yl)phenyl]-6-(3-methyl-2-buten-1-yl)-2,3-dihydro-4H-chromen-4-one) showed NMR spectra identical to the literature.82, 83, 84
Compound 12 (2',4',5',3,4-pentamethoxychalcone or 3-(3,4-dimethoxy-phenyl)-1-(2,4,5-trimethoxy-phenyl)-2-propen-1-one) showed NMR spectra identical to the literature.79
Compound 13 (auriculasin or 7-(3,4-dihydroxyphenyl)-5-hydroxy-2,2-dimethyl-10-(3-methyl-2-buten-1-yl)-2H,6H-pyrano[3,2-g]chromen-6-one) showed NMR spectra identical to the literature.35
Compound 14 (barbinervic acid or (1R,2R,4aS,6aR,6aS,6bR,8aR,9S,10R,12aR,14bS)-1,10-dihydroxy-9-(hydroxymethyl)-1,2,6a,6b,9,12a-hexamethyl-2,3,4,5,6,6a,7,8,8a,10,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid) showed NMR spectra identical to the literature.85
Compound 15 (hesperidin or (2S)-5-hydroxy-2-(3-hydroxy-4-methoxyphenyl)-7-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxymethyl]oxan-2-yl]oxy-2,3-dihydrochromen-4-one) was purchased from Sigma-Aldrich, as used without further modification.
Compound 16 (cabreuvin or 3-(3,4-dimethoxyphenyl)-7-methoxy-4H-chromen-4-one) showed NMR spectra identical to the literature.33
Compound 17 (jaceidin or 5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-3,6-dimethoxy-4H-chromen-4-one) showed NMR spectra identical to the literature.32
Molecular modeling
The main features of the in house library of natural compounds and their derivatives have been already described elsewhere.27, 29 Conformational analysis was performed with OMEGA2 from OpenEye,86, 87 keeping all parameters at their default values and allowing the storage of up to 600 conformers of each molecule in the final database. Hydrogen sampling options were activated. The receptor for molecular docking simulations was prepared with the make_receptor utility of OEDocking. Molecular docking was performed with the FRED docking program (OpenEye),88, 89, 90 using the highest-resolution settings. During virtual screening, only the best pose of each compound was saved and virtual Smo antagonists were ranked according to the Chemgauss4 score. In subsequent accurate docking simulations, up to 10 poses of each molecule were stored. Energy minimization of docking complexed was performed with Amber12.91 In detail, Smo/ligand complexes were solvated in a rectilinear box of TIP3P typed water molecules buffering 4Å from the protein. The solvent was first energy minimized for 100 steps with the Steepest Descent algorithm (S.D.) and for subsequent 200 steps with the Conjugate Gradient algorithm (CG) before to energy minimize the solvated solute for 1000 steps S.D. and subsequent 4000 steps CG.
Rescoring was performed with multiple programs and functions including XSCORE,49 Chemscore,92 and the MM GBSA method implemented in Amber12, using settings previously described.50, 91, 93
Biology
Cell cultures, transfection and treatments
HEK293T, Shh-Light II, Ptch1–/– MEFs, SuFu–/– MEFs, wild-type MEFs and 22Rv1 cells were cultured in DMEM plus 10% FBS. DAOY cells were maintained in Eagle's Minimum Essential Medium (MEM) plus 10% FBS. All media contained l–glutamine and antibiotics. ASZ001 BCC cells were cultured in 154CF medium (Gibco-BRL, Grand Island, NY, USA) plus 2% FBS chelated with Chelex 100 sodium form (Sigma Aldrich), calcium chloride 0.05 mM (Gibco-BRL) and antibiotics. Cerebellar GCPs (from 4-days-old mice) were isolated and cultured as previously described.94 TE354T human BCC cells (ATCC CRL–7762) were cultured in DMEM medium (ATCC 30–2002) plus 10% FBS and antibiotics. Murine MBs were isolated from Ptch1+/– mice (The Jackson Laboratory, Bar Harbor, ME, USA). Tissues were collected as previously described,27 and immediately prepared cell suspensions were used for short-term cultures to keep Hh-sensitivity in vitro.58, 59, 60 Transient transfections were performed using DreamFectTM Gold transfection reagent (Oz Biosciences SAS, Marseille, France). Cells were treated with SAG (200 nM, Alexis Biochemicals Farmingdale, NY, USA), Bodipy-Cyclopamine (5 nM, BioVision Inc., San Francisco, CA, USA), Cyclopamine (Calbiochem, Nottingham, UK), Vismodegib (Selleckchem, Munich, Germany), LDE-225 (Selleckchem, Munich, Germany).
Hh-dependent luciferase reporter assay
The luciferase assay was performed in Shh-Light II cells, stably incorporating a Gli-responsive luciferase reporter and the pRL-TK Renilla (normalization control), treated for 48 h with SAG (200 nM) and the studied compounds. Luciferase and Renilla activity were assayed with a dual-luciferase assay system according to the manufacturer's instruction (Biotium Inc., Hayward, CA, USA). Results are expressed as luciferase/Renilla ratios and represent the mean±S.D. of three experiments, each performed in triplicate.
Bodipy-Cyclopamine (BC) binding assay
Human Myc-DDK-tagged Smo WT or human Myc-DDK-tagged Smo D473H or mouse Flag-tagged Smo WT or Flag-tagged Smo-D477G mutant were transfected in HEK293T cells. Cells were washed in PBS supplemented with 0.5% fetal bovine serum, fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, and incubated for 2 h at 37 °C both in the same medium supplemented with Bodipy-Cyclopamine (5 nM) and the studied compounds. The cells were permeabilized with Triton X100 (Sigma) 0.2%. Dako Fluorescent mounting (Dako, Carpinteria, CA, USA) was used as mounting medium and Hoechst reagent for staining of the cell nuclei. Bodipy (green) and Hoechst (blue) signals were analyzed in three to four representative fields per coverslip (× 20 magnification, 1000 cells/field). Data were expressed as percentage of BC incorporation observed with BC alone.95
mRNA expression analysis
Total RNA was isolated with Trizol (Invitrogen/Life Technologies, Carlsbad, CA, USA) and reverse transcribed with SensiFAST cDNA Synthesis Kit (Bioline Reagents Limited, London, UK). Quantitative real-time PCR (Q-PCR) analysis of Gli1, Gli2, Ptch1, Bmp2, Pfkfb3, CycD1, CycD2, Pcna, Oct4, Nanog, β-2 microglobulin, β-actin and HPRT mRNA expression was performed on each cDNA sample using the VIIA7 Real Time PCR System employing Assay-on-Demand Reagents (Life Technologies). A reaction mixture containing cDNA template, SensiFAST Probe Lo-ROX Kit (Bioline Reagents Limited) and primer probe mixture was amplified using FAST Q-PCR thermal cycler parameters. Each amplification reaction was performed in triplicate and the average of the three threshold cycles was used to calculate the amount of transcript in the sample (using SDS version 2.3 software). mRNA quantification was expressed, in arbitrary units, as the ratio of the sample quantity to the quantity of the calibrator. All values were normalized with two endogenous controls, β-2 microglobulin or β-actin and HPRT, which yielded similar results.
Cell proliferation and MB stem cells neurosphere-forming assay
Cell proliferation was evaluated by BrdU detection (Roche, Welwyn Garden City, UK). Briefly, after the BrdU pulse (24 h for GCPs) cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100, and BrdU detection was performed according to the manufacturer's instructions. Nuclei were counterstained with Hoechst reagent. At least 500 nuclei were counted in triplicate, and the number of BrdU-positive nuclei was recorded. To determine the growth rate of viable cells, a trypan blue count was performed after a treatment period of 24–48–72 h with studied compound. For the neurosphere-forming assay, cells were plated at clonal density (1–2 cells/mm2) into 96-well plates and cultured in selective medium as previously described.62
Allograft experiment
A total of 2 × 106 ASZ001 BCC cells were resuspended in an equal volume of 154CF medium and Matrigel (BD Biosciences, Heidelberg, Germany) and injected s.c. at the posterior flank of female NOD/SCID mice (Charles River Laboratories, Lecco, Italy), as previously described.96 Tumors were grown until a median size of ~200 mm3. Animals were randomly divided into two groups (n=6) and treated with solvent only (DMSO-Miglyol, 1:5) (Miglyol 812N by CREMER OLEO GmbH & Co. KG, Hamburg, Deutschland) or 12 in solvent (50 mg/kg) for 18 days. Tumor growth was monitored by measuring the size by caliper. Tumor volumes change was calculated by the formula length × width × 0.5 × (length+width).97 All animal experiments were approved by local ethics authorities.
Immunohistochemistry
For immunohistochemical staining tissues were formalin fixed and paraffin embedded. Sections were incubated with (1:100) rabbit monoclonal Ki67 antibody (Thermo Fisher Scientific, MA, USA) diluted in PBS. Detection was carried out with the mouse-to-mouse HRP (DAB) staining system (ScyTek Laboratories, Logan, UT, USA) accordingly to the manufacturer's instructions.
Statistical analysis
Statistical analysis was performed using StatView 4.1 software (Abacus Concepts Inc., Piscataway, NJ, USA). Statistical differences were analyzed with the Mann–Whitney U-test for nonparametric values, and a P<0.05 was considered significant. Results are expressed as mean±S.D. from an appropriate number of experiments (at least three biological replicas).
Acknowledgments
We thank F. Bufalieri and M. Ruat for their support during the revision of this paper. This research was supported by grants AIRC IG14723, PRIN 2012-2013 (2012C5YJSK002), Progetti di Ricerca di Università Sapienza di Roma, and Pasteur Institute/Cenci Bolognetti Foundation. The authors wish to thank the OpenEye Free Academic Licensing Program for providing a free academic license for molecular modeling and chemoinformatics software.
Author contributions
PI, RA, CI, FG, MM, DQ, FB and LDM performed research. PI, MM, BB, AG and LDM designed research and analyzed data. IDA, GC, IS and BB analyzed data. PI, MM and LDM wrote the paper.
Glossary
- Hh
Hedgehog
- DMSO
dimethyl sulfoxide
- SAG
Smoothened agonist
- GCPs
Granule Cell Progenitors
- BrdU
5'-Bromo-2'-deoxy-Uridine
- DMEM
Dulbecco's modified Eagle medium
- FBS
fetal bovine serum
- PBS
phosphate buffered saline
- IF
immunofluorescence
- BC
Bodipy-Cyclopamine
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by R Johnstone
Supplementary Material
References
- Robbins DJ, Fei DL, Riobo NA. The Hedgehog signal transduction network. Sci Signal 2012; 5: re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001; 15: 3059–3087. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 2013; 14: 416–429. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med 2009; 361: 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 1997; 277: 1109–1113. [DOI] [PubMed] [Google Scholar]
- Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med 2013; 19: 1410–1422. [DOI] [PubMed] [Google Scholar]
- Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 2009; 326: 572–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Sarin KY, Whitson RJ, Li JR, Kim G, Rezaee M et al. Smoothened variants explain the majority of drug resistance in Basal cell carcinoma. Cancer Cell 2015; 27: 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Seguin N, Modrusan Z, Januario T et al. Genomic analysis of smoothened inhibitor resistance in Basal cell carcinoma. Cancer Cell 2015; 27: 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieran MW. Targeted treatment for sonic hedgehog-dependent medulloblastoma. Neuro Oncol 2014; 16: 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Magno L, Coni S, Di Marcotullio L, Canettieri G. Digging a hole under Hedgehog: downstream inhibition as an emerging anticancer strategy. Biochim Biophys Acta 2015; 1856: 62–72. [DOI] [PubMed] [Google Scholar]
- Infante P, Alfonsi R, Botta B, Mori M, Di Marcotullio L. Targeting GLI factors to inhibit the Hedgehog pathway. Trends Pharmacol Sci 2015; 36: 547–558. [DOI] [PubMed] [Google Scholar]
- Pandolfi S, Stecca B. Cooperative integration between HEDGEHOG-GLI signalling and other oncogenic pathways: implications for cancer therapy. Expert Rev Mol Med 2015; 17: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Hatton BA, Villavicencio EH, Khanna PC, Friedman SD, Ditzler S et al. Hedgehog pathway inhibitor saridegib (IPI-926) increases lifespan in a mouse medulloblastoma model. Proc Natl Acad Sci USA 2012; 109: 7859–7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med 2010; 251ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against smoothened antagonists. Cancer Res 2011; 71: 5057–5061. [DOI] [PubMed] [Google Scholar]
- Yam PT, Langlois SD, Morin S, Charron F. Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron 2009; 62: 349–362. [DOI] [PubMed] [Google Scholar]
- Belgacem YH, Borodinsky LN. Sonic hedgehog signaling is decoded by calcium spike activity in the developing spinal cord. Proc Natl Acad Sci USA 2011; 108: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizio AH, Chinchilla P, Chen X, Kim S, Manning DR, Riobo NA. Heterotrimeric Gi proteins link Hedgehog signaling to activation of Rho small GTPases to promote fibroblast migration. J Biol Chem 2011; 286: 19589–19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R, Amann S, Bayer M, McGee SL, Loipetzberger A, Connor T et al. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell 2012; 151: 414–426. [DOI] [PubMed] [Google Scholar]
- Patridge E, Gareiss P, Kinch MS, Hoyer D. An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov Today 2015; 21: 204–207. [DOI] [PubMed] [Google Scholar]
- Cragg GM, Newman DJ. Natural products: a continuing source of novel drug leads. Biochim Biophys Acta 2013; 1830: 3670–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 2015; 14: 111–129. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev 2002; 16: 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L et al. Effects of oncogenic mutations in smoothened and patched can be reversed by cyclopamine. Nature 2000; 406: 1005–1009. [DOI] [PubMed] [Google Scholar]
- Manetti F, Taddei M, Petricci E. Structure–activity relationships and mechanism of action of small molecule smoothened modulators discovered by high-throughput screening and rational design. Top Med Chem 2015; 16: 43–108. [Google Scholar]
- Infante P, Mori M, Alfonsi R, Ghirga F, Aiello F, Toscano S et al. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. EMBO J 2015; 34: 200–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weierstall U, James D, Wang C, White TA, Wang D, Liu W et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat Commun 2014; 5: 3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarello A, Mori M, Chiaradia-Delatorre LD, Menegatti ACO, Delle Monache F, Ferrari F et al. Discovery of Mycobacterium tuberculosis protein tyrosine phosphatase B (PtpB) inhibitors from natural products. PLoS One 2013; 8: e77081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wu H, Evron T, Vardy E, Han GW, Huang XP et al. Structural basis for smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun 2014; 5: 4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature 2013; 497: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Sauleau P, David B, Lavaud C, Cassabois V, Ausseil F et al. Bioactive flavonoids of Tanacetum parthenium revisited. Phytochemistry 2003; 64: 567–569. [DOI] [PubMed] [Google Scholar]
- Fonseca FN, Ferreira AJS, Sartorelli P, Lopes NP, Floh EIS, Handro W et al. Phenylpropanoid derivatives and biflavones at different stages of differentiation and development of Araucaria angustifolia. Phytochemistry 2000; 55: 575–580. [DOI] [PubMed] [Google Scholar]
- Higby RH. The chemical nature of hesperidin and its experimental medical use as a source of vitamin P - a review. J Am Pharm Assoc 1941; 30: 629–635. [Google Scholar]
- Delle Monache G, Scurria R, Vitali A, Botta B, Monacelli B, Pasqua G et al. Two Isoflavones and a flavone from the fruits of Maclura pomifera. Phytochemistry 1994; 37: 893–898. [Google Scholar]
- Delle Monache G, Delle Monache F, Marini-Bettolo GB, De Albuquerque MMF, De Mello JF, De Lima OG. Flavonoids of Sophora tomentosa (Leguminosae). I. Sophoronol, a New 3-Hydroxyflavanone. Gazz Chim Ital 1976; 106: 935–945. [Google Scholar]
- Falshaw CP, Harmer RA, Ollis WD, Wheeler RE, Lalitha VR, Subba Rao NV. Natural occurrence of 3-aryl-4-hydroxycoumarins. Part II. Phytochemical examination of Derris scandens (roxb.) Benth. J Chem Soc 1969: 374–382.
- Ferrari F, Monacelli B, Messana I. Comparison between in vivo and in vitro metabolite production of Morus nigra. Planta Med 1999; 65: 85–87. [DOI] [PubMed] [Google Scholar]
- Perkin AG, Hummel JJ. The colouring principle contained in the bark of Myrica nagi. J Chem Soc Trans 1896; 69: 1287–1294. [Google Scholar]
- Zoller HF. Some constituents of the american grapefruit (Citrus decumana. Ind Eng Chem 1918; 10: 364–374. [Google Scholar]
- Leitao SG, Kaplan MA, delle Monache F. Phenylpropanoid glucosides from Aegiphila obducta. J Nat Prod 1994; 57: 1703–1707. [DOI] [PubMed] [Google Scholar]
- Wright CR, Luff AP. The alkaloids of Veratrum sabadillaAsagroea officinalis. J Chem Soc Trans 1878; 33: 338–359. [Google Scholar]
- Messana I, Ferrari F, Delle Monache F, Yunes RA, Calixto JB, Bisognin T. Three new diels-alder type adducts from the roots of Sorocea bonplantii Baillon. Heterocycles 1991; 32: 1287–1296. [Google Scholar]
- Delle Monache F, Suarez LEC. 6-C-Formyl and 6-C-hydroxymethyl flavanones from Petiveria alliacea. Phytochemistry 1992; 31: 2481–2482. [Google Scholar]
- Tognazzi V. Calconi e Idrocalconi. Gazz Chim Ital 1924; 54: 697–704. [Google Scholar]
- Bargellini G. Contributo alla Conoscenza della Costituzione della Scutellareina. Gazz Chim Ital 1919; 49: 47–63. [Google Scholar]
- Bargellini G, Avrutin G. Sopra alcuni derivati dell'ossidrochinone. Gazz Chim Ital 1910; 40: 342–347. [Google Scholar]
- Hoch L, Faure H, Roudaut H, Schoenfelder A, Mann A, Girard N et al. MRT-92 inhibits Hedgehog signaling by blocking overlapping binding sites in the transmembrane domain of the smoothened receptor. FASEB J 2015; 29: 1817–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RX, Lai LH, Wang SM. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput-Aided Mol Des 2002; 16: 11–26. [DOI] [PubMed] [Google Scholar]
- Mori M, Manetti F, Botta M. Predicting the binding mode of known NCp7 inhibitors to facilitate the design of novel modulators. J Chem Inf Model 2011; 51: 446–454. [DOI] [PubMed] [Google Scholar]
- Christodoulou MS, Mori M, Pantano R, Alfonsi R, Infante P, Botta M et al. Click reaction as a tool to combine pharmacophores: the case of Vismodegib. Chempluschem 2015; 80: 938–943. [DOI] [PubMed] [Google Scholar]
- Dijkgraaf GJ, Alicke B, Weinmann L, Januario T, West K, Modrusan Z et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res 2011; 71: 435–444. [DOI] [PubMed] [Google Scholar]
- Jimeno AWeiss GJ, Miller WH Jr., Gettinger S, Eigl BJ, Chang AL et al. Phase I study of the Hedgehog pathway inhibitor IPI-926 in adult patients with solid tumors. Clin Cancer Res 2013; 19: 2766–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, De Smaele E, Screpanti I, Gulino A. Suppressors of hedgehog signaling: linking aberrant development of neural progenitors and tumorigenesis. Mol Neurobiol 2006; 34: 193–204. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 1999; 22: 103–114. [DOI] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 2008; 14: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M et al. Medulloblastoma can be initiated by deletion of patched in lineage-restricted progenitors or stem cells. Cancer Cell 2008; 14: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science 2002; 297: 1559–1561. [DOI] [PubMed] [Google Scholar]
- Sasai K, Romer JT, Lee Y, Finkelstein D, Fuller C, McKinnon PJ et al. Shh pathway activity is down-regulated in cultured medulloblastoma cells: implications for preclinical studies. Cancer Res 2006; 66: 4215–4222. [DOI] [PubMed] [Google Scholar]
- Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 2014; 25: 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz I, Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol 2007; 17: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Po A, Ferretti E, Miele E, De Smaele E, Paganelli A, Canettieri G et al. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J 2010; 29: 2646–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoranjan B, Venugopal C, McFarlane N, Doble BW, Dunn SE, Scheinemann K et al. Medulloblastoma stem cells: modeling tumor heterogeneity. Cancer Lett 2013; 338: 23–31. [DOI] [PubMed] [Google Scholar]
- Aszterbaum M, Epstein J, Oro A, Douglas V, LeBoit PE, Scott MP et al. Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat Med 1999; 5: 1285–1291. [DOI] [PubMed] [Google Scholar]
- Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 2003; 425: 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature 2004; 431: 707–712. [DOI] [PubMed] [Google Scholar]
- Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol 2006; 208: 17–25. [DOI] [PubMed] [Google Scholar]
- Bar EE, Chaudhry A, Farah MH, Eberhart CG. Hedgehog signaling promotes medulloblastoma survival via Bc/II. Am J Pathol 2007; 170: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci 2009; 30: 303–312. [DOI] [PubMed] [Google Scholar]
- Salto-Tellez M, Peh BK, Ito K, Tan SH, Chong PY, Han HC et al. RUNX3 protein is overexpressed in human basal cell carcinomas. Oncogene 2006; 25: 7646–7649. [DOI] [PubMed] [Google Scholar]
- Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov 2005; 4: 206–220. [DOI] [PubMed] [Google Scholar]
- Waters AL, Hill RT, Place AR, Hamann MT. The expanding role of marine microbes in pharmaceutical development. Curr Opin Biotechnol 2010; 21: 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay MR, Nesler M, Weatherhead R, Castro AC. Recent patents for Hedgehog pathway inhibitors for the treatment of malignancy. Expert Opin Ther Pat 2009; 19: 1039–1056. [DOI] [PubMed] [Google Scholar]
- Slusarz A, Shenouda NS, Sakla MS, Drenkhahn SK, Narula AS, MacDonald RS et al. Common botanical compounds inhibit the hedgehog signaling pathway in prostate cancer. Cancer Res 2010; 70: 3382–3390. [DOI] [PubMed] [Google Scholar]
- Ruat M, Hoch L, Faure H, Rognan D. Targeting of Smoothened for therapeutic gain. Trends Pharmacol Sci 2014; 35: 237–246. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell 2006; 10: 187–197. [DOI] [PubMed] [Google Scholar]
- Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP et al. A paracrine requirement for hedgehog signalling in cancer. Nature 2008; 455: 406–410. [DOI] [PubMed] [Google Scholar]
- Niitsu A, Harada M, Yamagaki T, Tachibana K. Conformations of 3-carboxylic esters essential for neurotoxicity in veratrum alkaloids are loosely restricted and fluctuate. Bioorg Med Chem 2008; 16: 3025–3031. [DOI] [PubMed] [Google Scholar]
- Avila HP, Smania Ede F, Monache FD, Smania A Jr. Structure-activity relationship of antibacterial chalcones. Bioorg Med Chem 2008; 16: 9790–9794. [DOI] [PubMed] [Google Scholar]
- Hano Y, Nomura T, Ueda S. 2 New diels-alder type adducts, mulberrofuran-T and kuwanol-E, from callus tissues of Morus-Alba L. Heterocycles 1989; 29: 2035–2041. [Google Scholar]
- Subba Rao NV, Khan WA. Structural studies on Scandenin. I. A study of its physical and chemical properties. Indian J Chem 1963; 2: 74–77. [Google Scholar]
- Delle Monache G, Delle Monache F, Marini Bettolo GB, De Albuquerque MMF, De Mello JF, De Lima OG. Flavonoids of Sophora tomentosa (Leguminosae). II. Isosophoranone, a new diprenylated isoflavanone. Gazz Chim Ital 1977; 107: 189–193. [Google Scholar]
- Komatsu M, Yokoe I, Shirataki Y. Studies on the constituents of sophora species. XIII. Constituents of the aerial parts of Sophora tomentosa L. Chem Pharm Bull (Tokyo) 1978; 26: 3863–3870. [DOI] [PubMed] [Google Scholar]
- Russell GB. New isoflavonoids from root bark of kowhai (Sophora microphylla. Aust J Chem 1997; 50: 333–336. [Google Scholar]
- Delle Monache F, Menichini F, Cuca Suarez LE. Petiveria alliacea: II. further flavonoids and triterpenes. Gazz Chim Ital 1996; 126: 275–278. [Google Scholar]
- Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model 2010; 50: 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OpenEye.. OMEGA 2.5.1.4: OpenEye Scientific Software, Santa Fe, NM. Available at:http://www.eyesopen.com.
- McGann M. FRED pose prediction and virtual screening accuracy. J Chem Inf Model 2011; 51: 578–596. [DOI] [PubMed] [Google Scholar]
- McGann M. FRED and HYBRID docking performance on standardized datasets. J Comput Aided Mol Des 2012; 26: 897–906. [DOI] [PubMed] [Google Scholar]
- OpenEye. FRED 3.0.1 OpenEye Scientific Software, Santa Fe, NM. Available at: http://www.eyesopen.com.
- Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J et al. AMBER 12, University of California. San Francisco 2012.
- Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des 1997; 11: 425–445. [DOI] [PubMed] [Google Scholar]
- Miller BR, McGee TD, Swails JM, Homeyer N, Gohlke H, Roitberg AE. MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput 2012; 8: 3314–3321. [DOI] [PubMed] [Google Scholar]
- Argenti B, Gallo R, Di Marcotullio L, Ferretti E, Napolitano M, Canterini S et al. Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J Neurosci 2005; 25: 8338–8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti F, Faure H, Roudaut H, Gorojankina T, Traiffort E, Schoenfelder A et al. Virtual screening-based discovery and mechanistic characterization of the acylthiourea MRT-10 family as smoothened antagonists. Mol Pharmacol 2010; 78: 658–665. [DOI] [PubMed] [Google Scholar]
- Eberl M, Klingler S, Mangelberger D, Loipetzberger A, Damhofer H, Zoidl K et al. Hedgehog-EGFR cooperation response genes determine the oncogenic phenotype of basal cell carcinoma and tumour-initiating pancreatic cancer cells. EMBO Mol Med 2012; 4: 218–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA 2007; 104: 8455–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.