Abstract
Background
The mobilization of transposable elements (TEs) is suppressed by host genome defense mechanisms. Recent studies showed that the cis-regulatory region of Arabidopsis thaliana COPIA78/ONSEN retrotransposons contains heat-responsive elements (HREs), which cause their activation during heat stress. However, it remains unknown whether this is a common and potentially conserved trait and how it has evolved.
Results
We show that ONSEN, COPIA37, TERESTRA, and ROMANIAT5 are the major families of heat-responsive TEs in A. lyrata and A. thaliana. Heat-responsiveness of COPIA families is correlated with the presence of putative high affinity heat shock factor binding HREs within their long terminal repeats in seven Brassicaceae species. The strong HRE of ONSEN is conserved over millions of years and has evolved by duplication of a proto-HRE sequence, which was already present early in the evolution of the Brassicaceae. However, HREs of most families are species-specific, and in Boechera stricta, the ONSEN HRE accumulated mutations and lost heat-responsiveness.
Conclusions
Gain of HREs does not always provide an ultimate selective advantage for TEs, but may increase the probability of their long-term survival during the co-evolution of hosts and genomic parasites.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-016-1072-3) contains supplementary material, which is available to authorized users.
Keywords: Brassicaceae, COPIA, Evolution, Heat stress, ONSEN
Background
Transposable elements (TEs) are ubiquitous components of eukaryotic genomes. Their functions and roles range from DNA parasites, through regulators of gene transcription to facilitators of genome evolution (reviewed in [1, 2]). Together with other types of repeats, TEs comprise 10–80 % of plant genome content and specific families of long terminal repeat (LTR) retrotransposons can reach thousands of copies per genome [1, 3]. Plants evolved several layers of sophisticated epigenetic silencing mechanisms in order to suppress TE activity. Their transcripts are degraded by the post-transcriptional gene silencing (PTGS) pathway, which greatly reduces possible transposition events [4]. In parallel, transcriptional gene silencing (TGS) stably silences TEs by deposition of DNA methylation via RNA directed DNA methylation (RdDM) mechanism (reviewed in [5, 6]). The repressed state is further stabilized by accumulation of specific histone modifications and faithfully transmitted in a DNA replication-dependent manner to the next generations. External or internal factors [7, 8] can lead to transient loss of silencing, but the epigenetic control will be re-established through tissue-specific RdDM activity [9]. In addition to the nimble epigenetic silencing system, entire TEs can be physically removed from the host genome by deletion-biased homologous recombination processes [10].
In spite of the multi-layer amplification barriers, many TE families show signs of recent transpositions [11–13], suggesting that TEs occasionally escape epigenetic surveillance. There is increasing evidence that stress treatments affect chromatin structure and may lead to transposon activation (reviewed in [1, 14]). A possible mechanism was proposed based on the analysis of stress-induced TEs. LTRs of medicago cold-inducible repetitive element (MCIRE) retrotransposon contain a putative cold-responsive element (CRE) in alfalfa (Medicago sativa) [15]. The CRE is specified by a conserved 5-bp core sequence (CCGAC) typical for C-repeat (CRT)/dehydration-responsive elements (DRE) that are recognized by cold-specific transcription factors (TFs) [16]. LTRs of heat-responsive COPIA78/ONSEN (used as synonyms in this study) retrotransposon in Arabidopsis thaliana [7, 8, 17], contain a cluster of four nGAAn motifs forming a heat-responsive element (HRE) [18]. During heat stress (HS), the ONSEN HRE is bound by heat shock factor A 2 (HSFA2), which triggers its transcriptional activity. This regulation is very specific and greatly independent of TGS control as the loss of decreased DNA methylation 1 (DDM1) in mutant plants did not trigger ONSEN transcriptional activation [7], in contrast to other typical LTR retrotransposons [19].
Presence of HRE and CRT/DRE motifs in ONSEN and MCIRE, respectively, suggested that the TEs’ response to stresses may be mediated by specific TF binding motifs. HREs were previously classified into four types based on their structure and, most likely, also activity [20]. The strongest 4P HRE contains at least four adjacent nGAAn motifs and is bound by two HSFA2 trimers. The 3P HRE is bound by a single HSFA2 trimer and represents a moderately responsive HRE. In contrast, gap and step HREs with irregularly and more distantly spaced nGAAn motifs have on average lower HRE activity. Therefore, the HRE composition needs to be considered in order to define the strength of transcriptional response.
Here we identified multiple heat-responsive COPIA families in Arabidopsis lyrata and A. thaliana, two closely related species, using RNA sequencing (RNA-seq). Subsequently, we extended our analysis to five other Brassicaceae species and reconstructed putative HREs, their evolutionary history, and validated our predictions by transcriptional analysis after HS treatment.
Results
Identification of heat-responsive TE families in A. thaliana and A. lyrata
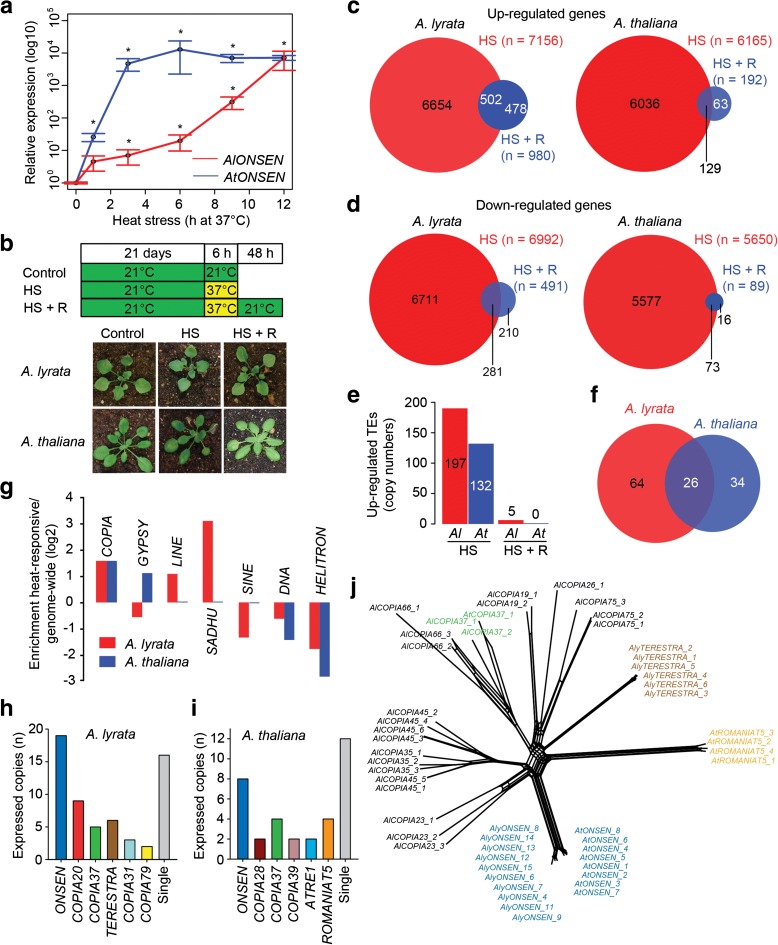
First, we determined HS conditions that would be effective and comparable for A. lyrata MN47 and A. thaliana Col-0 plants. As the A. lyrata genome contains sequences with high homology to the A. thaliana ONSEN retrotransposon, we quantified ONSEN transcripts in both species by reverse transcription quantitative polymerase chain reaction (RT-qPCR) during a HS (37 °C) time series using soil-grown plants. Transcripts accumulated faster in A. thaliana, but to comparable amounts in both species after 12 h of HS (Fig. 1a). We selected 6 h at 37 °C, leading to a significant and reproducible ONSEN transcript accumulation in both species (T-test, P <0.05), as the standard HS treatment. Subsequently, samples of control, heat-stressed (6 h HS), and recovered (6 h HS + 48 h 21 °C) plants were RNA-sequenced (Fig. 1b).
Fig. 1.
Transcriptome analysis of heat-stressed A. lyrata and A. thaliana plants. a Effects of HS on ONSEN heat-responsiveness in A. thaliana and A. lyrata. Both species were stressed at 37 °C for the indicated number of hours (h) and subsequently analyzed for the amount of ONSEN transcript (log10) by RT-qPCR relative to GAPD-H transcript amounts. * significant (t-test, P <0.05) transcript enrichment relative to 0 h control. Error bars indicate standard deviation of three biological replicates. b Design of plant HS treatment for RNA-seq and representative phenotypes of control, 6 h heat-stressed at 37 °C and recovered plants. c, d Number of significantly (c) upregulated or (d) downregulated protein-coding genes after 6 h at 37 °C and 48 h recovery at non-stress conditions in both species. e, f Number of significantly upregulated (e) TEs and (f) TE families after 6 h HS and 48 h recovery. g Identification of TE groups enriched for heat-responsive copies. Retrotransposons were divided into SINE, SADHU, LINE, COPIA, and GYPSY family members. The relative enrichment of heat-activated TEs was calculated as ratio between % of all heat-activated to % of all TEs genome-wide and expressed on a log2 scale. The major heat-responsive COPIA families in (h) A. lyrata and (i) A. thaliana. The families containing a single HRE are displayed as “single copies.” j RT amino acid sequences (Additional file 12)-based phylogenetic network of selected heat-responsive (colored) and non-responsive (black) A. lyrata and A. thaliana COPIA families. The data are also provided as un-rooted three in Additional file 5: Figure S2
To assess the extent of plant responses to HS, we monitored transcript levels from 32,793 A. lyrata and 32,678 A. thaliana protein-coding genes. This revealed significant upregulation (adjusted P <0.05; DESeq) of 21.8 % A. lyrata genes (n = 7156) and 18.9 % A. thaliana genes (n = 6165) after 6 h HS (Fig. 1c; Additional files 1 and 2). After recovery, we found only 2.9 % (n = 980) of genes still upregulated in A. lyrata and 0.6 % (n = 192) in A. thaliana. A. lyrata showed 21.3 % (n = 6992) downregulated genes after HS and 1.5 % (n = 491) after recovery (Fig. 1d). There were 17.3 % (n = 5650) significantly downregulated genes after HS and only 0.3 % (n = 89) after recovery in A. thaliana. Hence, HS treatment induced a similar degree of transient transcriptional changes in both species.
Because there is no publicly available A. lyrata TE annotation, we prepared custom-made catalogues of 53,089 A. lyrata and 17,009 A. thaliana repetitive elements (Additional files 3 and 4, respectively). Although the two species differed threefold in their TEs numbers, their spectra of TE families were similar (Additional file 5: Figure S1). The multi-copy nature of many TEs hinders RNA-seq analysis using standard protocols. Therefore, we developed the COMparative EXpression of TEs (COMEX) method, which allows quantification of transcripts derived from individual TE copies and effective removal of the RNA-seq reads mapping across TE families (see “Methods;” Additional files 6 and 7). We found 197 and 132 significantly (adjusted P <0.05; DESeq) upregulated TEs, representing 90 and 60 families (26 in common), after 6 h HS in A. lyrata and A. thaliana, respectively (Fig. 1e, f; Additional files 8 and 9). Comparing the major upregulated TE groups versus those in the whole genome revealed general under-representation of DNA transposons and HELITRONs and A. lyrata-specific under-representation of SINEs. In contrast, we found an over-representation of heat-responsive SADHU and LINE retrotransposons in A. lyrata, GYPSY elements in A. thaliana, and COPIA TEs in both species (Fig. 1g). Heat-responsive AlCOPIAs (n = 60; 100 %) comprised six families with at least two heat-inducible elements (Fig. 1h): AlCOPIA31 (n = 3; 3 %), AlCOPIA79 (n = 2; 3 %), AlCOPIA37 (n = 5; 11 %), AlCOPIA20 (n = 9; 14 %), AlONSEN (n = 19; 37 %), and a so far unknown family which we named TEMPERATURE RESPONSIVE TRANSPOSON (TERESTRA, n = 6; 10 %), as well as a bulk of single copies from different families (n = 16; 22 %). A. thaliana heat-responsive COPIAs (n = 34) were represented by six families with more than one heat-responsive TE. However, only AtONSEN (n = 8; 29 %) and AtCOPIA37 (n = 4; 12 %) were common between both species (Fig. 1i). A prominent A. thaliana-specific family was ROMANIAT5, comprising 12 % (n = 4) of all heat-responsive AtCOPIAs. After recovery, all TEs were re-silenced in A. thaliana and only five (AlCOPIA37, AlRE1, SADHU6-1, AlATN9_1, and AlLINE1_3A) showed increased transcript amounts in A. lyrata (Fig. 1e; Additional file 8). Surprisingly, ONSEN was fully silenced after two days of recovery, most likely owing to a shorter HS applied here compared to the previous study [7]. The families representing at least 10 % of heat-responsive COPIA elements in each species were considered for further analysis (Fig. 1h).
Next, we tested whether heat-responsive COPIA families represent a particular COPIA clade. We reconstructed phylogeny of HS-responsive COPIA37, ONSEN, TERESTRA, ROMANIAT5, and seven HS-non-responsive COPIA families (19, 23, 26, 35, 45, 66, 75) based on their RT sequences (Fig. 1j; Additional file 5: Figure S2). The coding sequence was preferred over LTRs for the similarity analysis because this is strongly influenced by length of the input sequences, which may vary drastically in case of LTRs from different families. All heat-responsive families formed distinct and early separated branches, suggesting multiple independent origins of COPIA heat-responsiveness.
The structure and evolution of ONSEN heat-responsiveness
There are 24 COPIA78 elements in A. thaliana Col-0 (TAIR10) including eight full-length copies and 16 fragments (Table 1, Additional file 5: Table S1). However, only the eight full-length ONSEN copies were found to be heat-responsive (Additional file 9). We performed in silico reconstruction of the putative HREs using a proposed classification [20], which suggested two HREs in all heat-responsive AtONSENs: a low efficiency gap HRE and the highest efficiency 4P HRE (Fig. 2a; Additional file 5: Figure S3). While the gap HRE is present in all eight A. thaliana full-length ONSENs, the 4P was changed into a 3P HRE with moderate efficiency in AtONSEN4, due to loss of the fourth motif (Additional file 5: Figure S3). In contrast, none of the 16 fragments or solo LTRs contains functional HREs nor shows heat-responsiveness according to RNA-seq (Additional file 9).
Table 1.
Copy numbers of elements within analyzed COPIA families in Brassicaceae species
| Species | ONSEN | COPIA37 | HATE | ROMANIAT5 | ||||
|---|---|---|---|---|---|---|---|---|
| Total | Full | Total | Full | Total | Full | Total | Fulla | |
| Arabidopsis lyrata | 55 | 10 | 57 | 5 | 6 | 6 | 131 | 0 |
| Arabidopsis thaliana | 24 | 8 | 32 | 1 | 0 | 0 | 49 | 0 |
| Ballantinia antipoda | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Boechera stricta | 2 | 2 | 0 | 0 | 14 | 7 | 53 | 0 |
| Brassica rapa | 6 | 2 | 2 | 0 | 2 | 0 | 7 | 0 |
| Capsella rubella | 0 | 0 | 2 | 1 | 0 | 0 | 0b | 0 |
| Eutrema salsugineum | 2 | 1 | 2 | 0 | 6 | 1 | 65 | 0 |
aAll ROMANIAT5 elements lacked integrase domain
bOnly three solo LTRs were found in C. rubella
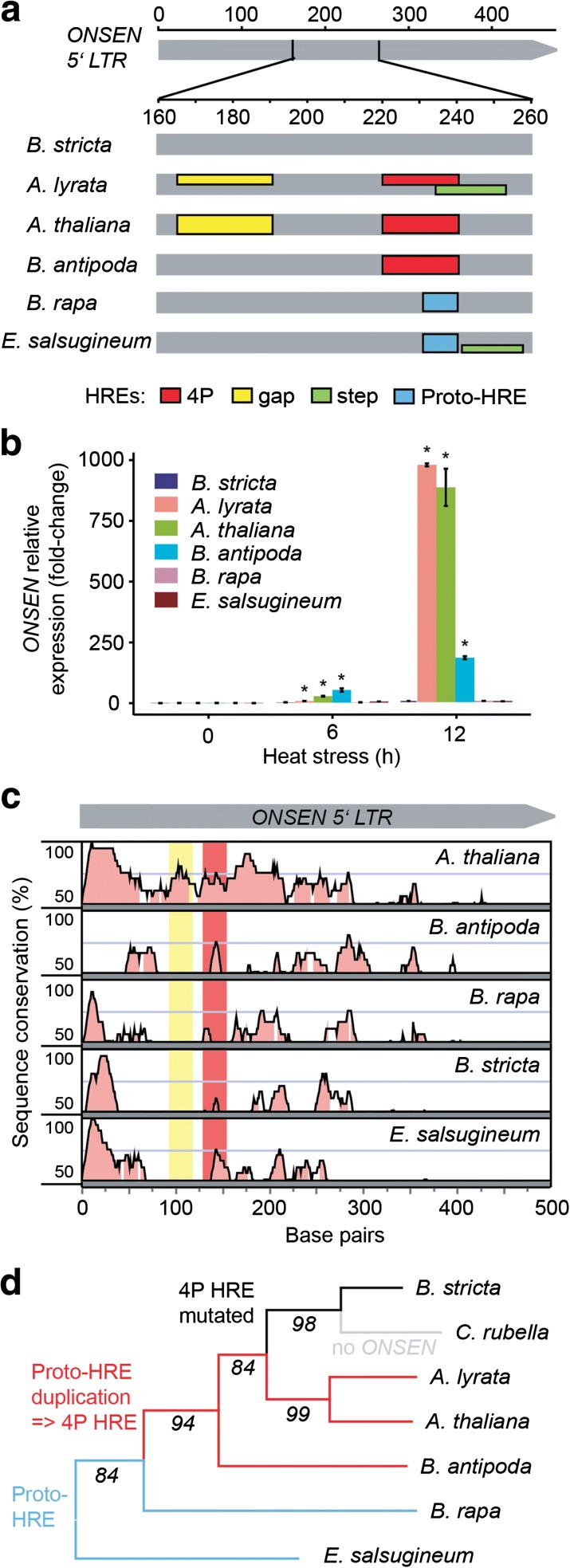
Fig. 2.
Evolution of ONSEN heat-responsiveness. a Schematic representation of in silico reconstruction of putative HREs in ONSEN 5’ LTR in different Brassicaceae species. HRE reconstruction follows criteria proposed by [20]. Colored boxes spanning the entire height of the gray field indicate HREs found in ≥50 % of the heat-responsive copies in A. thaliana and A. lyrata or all copies in other species. The lower boxes represent less frequent (<50 %) variants. Detailed information including sequences underlying individual HREs can be found in Additional file 5: Figure S2. b Transcript levels of ONSEN elements in Brassicaceae after 6 h and 12 h at 37 °C. Quantitative PCR values were obtained using species-specific primer pairs and normalized to UBC28. Error bars indicate standard deviation of three biological replicates and * P <0.05 in Student’s t-test. c Sequence conservation over the ONSEN 5’ LTR. Species-specific 5’ LTR consensus sequences were compared to A. lyrata query using 20 bp sliding window and 7 bp minimum consensus length. The y-axis for each species shows 50–100 % sequence conservation. Regions with ≥70 % similarity (pink-filled) were considered as conserved. Red and yellow background colors indicate the A. lyrata 4P and gap HRE regions. d Reconstruction of ONSEN HRE evolution. The phylogenetic tree was developed using the CHALCONE SYNTHASE gene of each individual species. The numbers at branches indicate bootstrap values. Blue lines show species with proto-HREs, red shows those carrying 4P HREs, black shows loss of HREs in B. stricta, and gray shows the COPIA78 family in C. rubella
We found 55 COPIA78 TEs in A. lyrata. Ten are full-length elements and 45 are fragments, either solo LTRs or incomplete according to the gaps in the genome assembly (Table 1; Additional file 5: Figure S3 and Table S2). In total, 15 copies contain at least one putative HRE with three or more adjacent (≤5 bp) nGAAn motifs. Remarkably, a high number of AlONSENs carry HREs identical to A. thaliana copies (Fig. 2a; Additional file 5: Figure S3). AlONSEN 2 and 8 have A. thaliana-like gap HREs; the 4P type is present in AlONSEN 10 and both co-occur in AlONSEN 6, 7, 9, 11, 14, 15, 17. In addition, we observed putative low efficiency gap/step HREs substituting the 4P HRE in AlONSEN 1, 4, 5, 12, and 13. All AlONSENs with predicted 4P HREs were upregulated after 6 h HS (Additional file 8). AlONSEN 3 and 16 were also found upregulated although they did not contain putative HREs. This was most likely caused by ambiguity in RNA-seq analysis, as 100 % of the reads mapping to these elements were multiply mapping to other ONSENs. Hence, there is a high correlation between the predicted HREs and RNA-seq results.
Conservation of the most frequent HRE haplotype between the Arabidopsis species raised the question about the evolutionary history of ONSEN heat-responsiveness in the Brassicaceae. Therefore, we searched for COPIA78 elements in whole-genome assemblies of Boechera stricta v1.2, Brassica rapa FPsc v1.3 (both JGI; Phytozome), Capsella rubella [21], Eutrema salsugineum [22], and low coverage assembly of Ballantinia antipoda (Vu, Finke, and Pecinka; unpublished data) using genome-wide BLAST searches. We confirmed the absence of COPIA78 in Capsella [23], but found at least one ONSEN copy in all other species (Table 1). RT nucleotide sequence identity was >80 % (Additional file 5: Figure S4), fitting previously proposed criteria for a single TE family [24]. The LTR identity was lower (typically <70 %) due to the presence of insertions and deletions and decreased with phylogenetic distance (Additional file 5: Figure S5). Nevertheless, it allowed us reconstructing putative HREs. None of the other species contained the A. thaliana-like gap HRE (Fig. 2a; Additional file 5: Figure S3). However, there was a perfectly conserved 4P HREs in two out of three ONSENs in B. antipoda. ONSENs of other species either did not contain any HREs (B. stricta) or they represented only lower efficiency types and were non-homologous to the Arabidopsis HREs (Fig. 2a). To challenge the predicted HREs, we grew all species in vitro and quantified ONSEN transcript levels after 6 h and 12 h of HS (Fig. 2b). In agreement with RNA-seq results, we found massive 884–976-fold upregulation in A. lyrata and A. thaliana. There was also high (185-fold) upregulation in B. antipoda containing the putative 4P HRE, but lacking an additional gap HRE (Fig. 2a, b). In contrast, B. stricta, B. rapa, and E. salsugineum predicted to have no or only low efficiency HREs did not show strongly increased ONSEN transcript amounts (Fig. 2b).
To test whether HREs represent a major cis-regulatory element in ONSEN LTRs, we performed phylogenetic shadowing of the LTR consensus sequences (Fig. 2c). Although the 4P HRE region was partially conserved, there are several other similarly conserved regions. The longest stretch of conserved LTR sequence comprises approximately the first 25–30 bp (Fig. 2c), which may be required for TE RT.
By anchoring the structural information on the Brassicaceae chalcone synthase-based phylogeny, we reconstructed the evolutionary trajectory of ONSEN HREs (Fig. 2d). The nTTCnnGAAn motif, which can be considered as the non-functional sequence preceding the 4P HRE (proto-HRE), is present in B. rapa and E. salsugineum (Fig. 2a; Additional file 5: Figure S3), suggesting that it existed already at the onset of Brassicaceae evolution. Later, proto-HRE became duplicated and instantly created the high affinity 4P HRE. Molecular dating of the split of the B. antipoda lineage [25] suggests that this motif was maintained over 6–9 million years of evolution. However, the 4P HRE was occasionally lost due to accumulation of the point mutations (B. stricta) or deletion of whole elements (Capsella).
Species-specific gain of HREs in COPIA37 and the novel family TERESTRA
The other TE family found to be heat-responsive in both Arabidopsis species was COPIA37 (Fig. 1h, i). However, this phenotype was restricted to fewer copies as only 8.8 % (five out of 57) and 12.5 % (four out of 32) of A. lyrata and A. thaliana COPIA37s, respectively, showed upregulation upon HS (Additional files 8 and 9). The 5’ LTRs of all heat-responsive copies contained putative low affinity binding gap and step HREs (Fig. 3a; Additional file 5: Figure S6). In addition, we found putative 3P HREs in three AtCOPIA37s and two AlCOPIA37s. These HREs originated from a common nTTCn rich LTR region, but were not identical. Search in other species revealed the presence of COPIA37 in B. rapa (n = 2), C. rubella (n = 2), and E. salsugineum (n = 2; Table 1), but here we found only low affinity binding gap and/or step HREs in the latter two species (Fig. 3a; Additional file 5: Figure S6). To test whether the predicted HREs correlate with heat-responsiveness, we exposed all species to 6 and 12 h HS and quantified the transcript amounts by RT-qPCR (Fig. 3b). We observed up to 25-fold activation for A. lyrata COPIA37 and a weaker (fivefold) activation for A. thaliana, both carrying putative 3P HREs. The amount of COPIA37 transcript reached its peak at 6 h and decreased in spite of continued HS. Other species, carrying only lower efficiency gap and or step HREs, did either not accumulate the transcript or only at a single experimental point. Hence, the most effective 3P HREs evolved independently in A. lyrata and A. thaliana and also COPIA37 elements of other species carry diverse set of HREs.
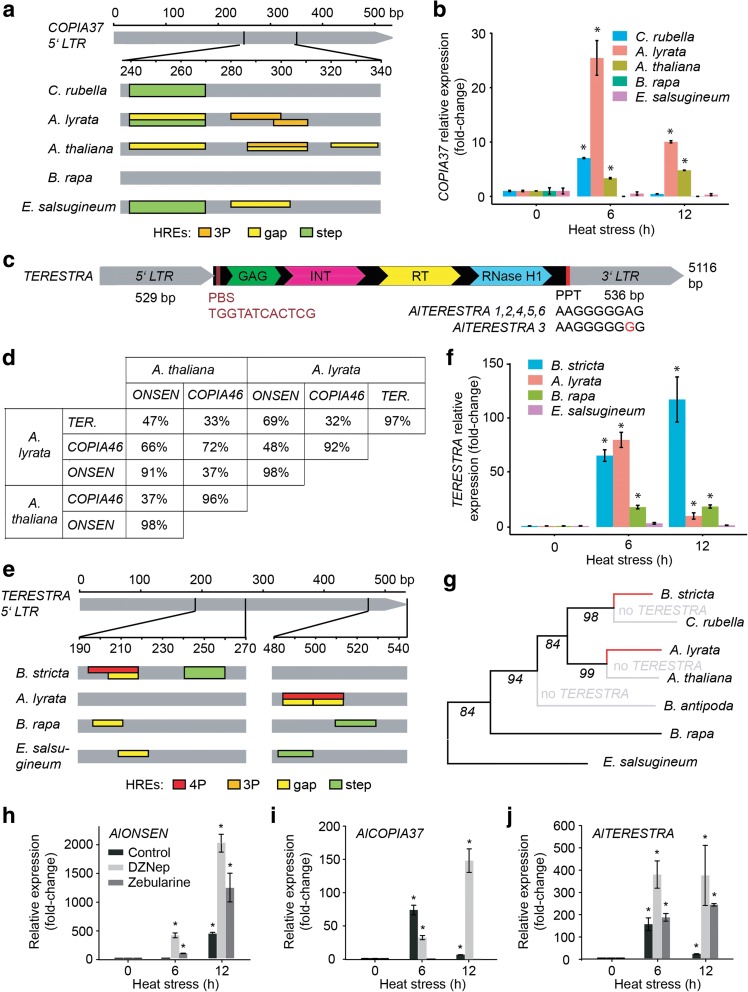
Fig. 3.
COPIA37 and TERESTRA are novel heat-responsive COPIA families. a In silico reconstruction of putative HREs in the 5’ LTR of COPIA37 in different species. HRE classification follows criteria proposed by [20]. Colored boxes spanning the entire height of the gray field indicate HREs found in ≥50 % of the heat-responsive copies in A. thaliana and A. lyrata or all copies in other species. The lower boxes represent less frequent (<50 %) HREs. Detailed information including sequences underlying individual HREs can be found in Additional file 5: Figure S5. b Transcript levels of COPIA37 in Brassicaceae after 6 and 12 h 37 °C HS. The values were normalized to transcript levels of UBC28. Error bars indicate standard deviation between three biological replicates and * P <0.05 in Student’s t-test. c Schematic representation of A. lyrata TERESTRA (TERESTRA). LTRs are indicated in gray. Capsid protein (GAG), integrase (INT), RT, and RNAse H1 domains are shown within the light-blue-labeled TERESTRA protein-coding part. Primer binding sequence (PBS) and polypurine tract (PPT) are indicated by red boxes. d Sequence similarities within pair-wise LTR alignments between A. lyrata and A. thaliana TERESTRA, ONSEN, and COPIA46 families. More than 70 % similarity was expected for members of the same family. TERESTRA is absent in A. thaliana. e In silico reconstruction of putative HREs in the 5’ LTR of TERESTRA. The criteria were as described for Fig. 3a. Detailed information including sequences underlying individual HREs can be found in Additional file 5: Figure S9. f Transcript levels of TERESTRA in response to 6 and 12 h 37 °C HS in different Brassicaceae. The experiment was performed as described in (b). g Reconstruction of TERESTRA HRE evolution. The phylogenetic tree was developed using a chalcone synthase gene of each individual species. The numbers at the base of the branches indicate bootstrap values. Black lines show species with low efficiency HREs and red lines highlight independently evolved high efficiency HREs in A. lyrata and B. stricta. Gray lines denote species where TERESTRA could not be found. TE transcript accumulation of (h) ONSEN, (i) COPIA37, and (j) TERESTRA after 0, 6, and 12 h 37 °C HS preceded by 48 h control (no inhibitor), 10 μM 3-deazaneplanocin A (DZNep), or 40 μM zebularine treatment. Transcript amounts were normalized to UBC28 mRNA and signals from drug and heat-treated samples were recalculated as fold-changes relative to 0 h. Error bars indicate variation between two biological replicates and * P <0.05 in Student’s t-test
We also identified TERESTRA as a new retrotransposon heat-responsive family. The A. lyrata genome contains six TERESTRA copies sharing 97 % similarity (Fig. 3d). BLAST searches using TERESTRA sequences revealed only local similarities to ONSEN LTRs and COPIA46 GAG and POL domains and no other significant hits. Therefore, we performed de novo TERESTRA analysis. Based on the order of GAG and POL, TERESTRA was unambiguously identified as Ty1/COPIA LTR-retrotransposon (Fig. 3c). Based on only 70 % similarity in an alignment of TERESTRA to COPIA46 and ONSEN elements (Fig. 3d), we defined TERESTRA as a novel COPIA family. The consensus length of the complete AlTERESTRA element was 5116 bp and the 5’ and the 3’ LTR were 529 and 536 bp long, respectively (Fig. 3c; Additional file 5: Figure S7). Sequence analyses of AlTERESTRAs revealed that all copies are full length, contain a tRNA primer binding site and a polypurine tract, suggesting their autonomy (Fig. 3c). TERESTRA LTRs are relatively A-T-rich (69 %) and the consensus sequence contained only a small number of cytosines in symmetrical contexts (CG = 5, CHG = 0; H = A, T or C), which resembles LTR nucleotide composition of ONSEN [18]. TERESTRA was missing in the A. thaliana Col-0 genome. Therefore, we extended our search to 50 A. thaliana accessions by genotyping them with TERESTRA-specific primers (Additional file 5: Table S3). This screen also gave negative results and suggested the absence of TERESTRA in A. thaliana. However, we found TERESTRA TEs in Arabidopsis cebennensis (95 % identity; Additional file 5: Figure S8) and Arabidopsis halleri (91 % identity; Additional file 5: Figure S9) using the NCBI sequence database. Furthermore, there were TERESTRAs in B. stricta (n = 14), B. rapa (n = 2), and E. salsugineum (n = 6; Table 1), but not outside of the Brassicaceae.
All six AlTERESTRAs were heat-responsive (Fig. 1h; Additional files 8). Screening of AlTERESTRA LTRs for possible HREs revealed a cluster of six nGAAn motifs, which can assemble either two partially overlapping gap HREs or a 4P HRE (Fig. 3e; Additional file 5: Figure S10). Based on the high AlTERESTRA transcriptional heat response (Fig. 3f), we favor the latter possibility. Another species with high TERESTRA transcriptional activation after 6 h and 12 h HS was B. stricta (Fig. 3f). By BLAST we found 14 TERESTRA copies in the B. stricta genome (Table 1). Eleven copies among them contain a complex cluster of up to five adjacent nGAAn motifs in their 5’ LTRs (Fig. 3d; Additional file 5: Figure S10). According to a conservative approach, three nGAAn motifs within this cluster can putatively form a low affinity gap HRE, but high TERESTRA activation in B. stricta suggests that all five motifs can establish 4P HREs as compatible with a more relaxed prediction (Fig. 3e, f). Importantly, all predicted B. stricta HREs are at positions different from those in A. lyrata HREs (Fig. 3e), highlighting their species-specific evolution (Fig. 3g). In B. rapa, one TERESTRA copy carries a putative gap HRE and another one a step HRE (Fig. 3e). Out of six TERESTRAs in E. salsugineum, three had putative step and one also an additional gap HRE. However, none of the HREs found in B. rapa and E. salsugineum was homologous to Arabidopsis or Boechera HREs and their predicted low HSF binding efficiency was congruent with the absence of heat-responsiveness in the RT-qPCR experiments (Fig. 3f).
Decreased AlCOPIA37 and AlTERESTRA transcript amounts after 12 h versus 6 h HS (Fig. 3b, f) contrasted with continuous transcript accumulation for AlONSEN (Fig. 2b). We hypothesized that the failure to maintain high transcript level could be caused by the TGS. Due to a lack of A. lyrata TGS mutants, we used a pharmacological approach to interfere with TE silencing [26]. We treated 14-day-old A. lyrata plants with 10 μM 3-deazaneplanocin A (DZNep) and 40 μM zebularine, including control plants without treatment. DZNep is an S-adenosylhomocysteine synthesis inhibitor, which blocks the production of SAM, the methyl group donor required for DNA and histone methylation. Zebularine is a cytidine analog leading to DNA de-methylation and loss of silencing from specific transposons [27–29]. After two days of drug treatment, plants were heat-stressed for 0, 6, and 12 h and the amount of transcript analyzed by RT-qPCR (Fig. 3h–j). Control DZNep and zebularine treatment increased COPIA37 and ONSEN transcript tenfold and fivefold, respectively (Additional file 5: Figure S11), suggesting that both TEs can be weakly activated by TGS interference also without HS treatment. TERESTRA was not activated by the drug treatment. A combination of HS with drug treatments had strong additive effects in all cases, except for zebularine and HS-treated COPIA37 (Fig. 3h–j). Both ONSEN and TERESTRA transcripts accumulated at much higher levels that were not decreasing at 12 h HS (Fig. 3h, i). The effect was generally stronger for DZNep and weaker for zebularine. This suggests that the heat-induced TE transcript accumulation is rapidly suppressed by epigenetic means, in particular for TEs carrying lower affinity binding HREs.
AtROMANIAT5 contributes to transcriptional regulation of APUM9 under HS
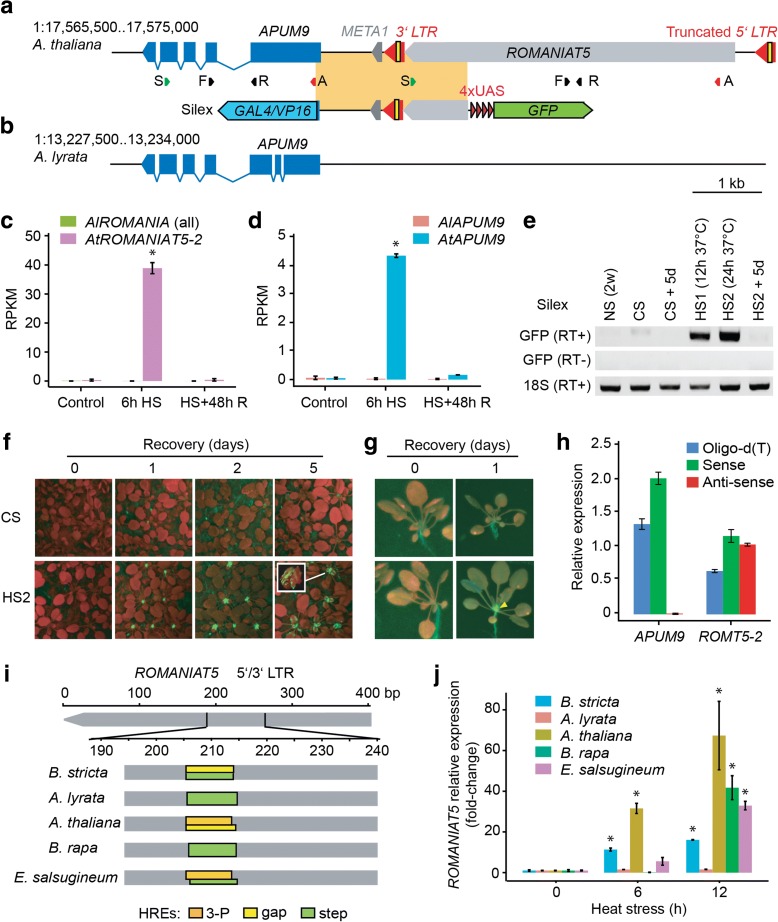
There are four heat-responsive ROMANIAT5 TEs in A. thaliana but none in A. lyrata (Fig. 1h, i; Additional files 8 and 9). All AtROMANIAT5 elements lack an integrase domain, suggesting that these elements are incomplete and non-autonomous (Table 1). A previous study revealed that one of the heat-responsive copies AtROMANIAT5-2 (At1g35735) is under complex epigenetic control by Morpheus molecule 1 (MOM1) and RdDM pathways, and loss of this control causes upregulation of the Arabidopsis PUMILIO9 (APUM9; At1g35730) gene located directly downstream of the TE [30]. To better understand the potential role of ROMANIAT5 in regulating APUM9 during HS, we reconstructed their loci in A. thaliana and A. lyrata (Fig. 4a, b) and also retrieved the number of reads mapping to both loci under different experimental conditions (Fig. 4c, d). Interestingly, we observed significant (t-test, P <0.05) upregulation of APUM9 upon HS in A. thaliana but not in A. lyrata (Fig. 4c, d), where the nearby ROMANIAT5 is missing. This suggested that ROMANIAT5-2 controls APUM9 transcription under HS. To validate this observation, we used a reporter line (called Silex) which contains the APUM9 upstream region and the ROMANIAT5-2 3’ LTR upstream of a GFP reporter (Fig. 4a) [31]. The Silex reporter construct is silenced during entire A. thaliana development, except for developed siliques, but the reporter activity can be restored in the background of MOM1 RdDM double mutants and histone deacetylase 6 mutants [31]. We grew Silex reporter plants under controlled conditions with and without HS. GFP transcripts were missing in the control plants but present after 12 and 24 h at 37 °C (Fig. 4e). GFP accumulated in the apical meristem after 24 h of HS recovery and remained detectable for at least five days (Fig. 4f, g), although GFP transcript was not present anymore (Fig. 4e). Heat-responsiveness of Silex transgene in the absence of ROMANIAT5-2 5’ LTR suggested that the locus may be at least partially controlled by a bi-directional heat-responsive promoter activity of the 3’ LTR. Indeed, we found putative 3P/gap HREs within the 3’ LTRs (and also the 5’ LTRs) of all heat-responsive AtROMANIAT5 TEs. However, transcription from the 3’ LTR could result in ROMANIAT5-2 antisense transcript. To test this, we isolated A. thaliana RNA after HS and performed complementary DNA (cDNA) synthesis using strand-specific RT primers (Fig. 4a). Control cDNA from RT with oligo-d(T) primers gave signals for both genetic elements (Fig. 4h). Strand-specific RT-qPCR revealed HS-induced sense transcript, but no antisense transcript, for APUM9. In contrast, both types of primers resulted in amplification of ROMANIAT5-2 transcripts, suggesting that it is transcribed in both directions under HS. The distribution of RNA-seq reads did not indicate large amounts of a read through transcription from ROMANIAT5-2 to APUM9 (Additional file 5: Figure S12). Altogether, this confirms the 3’ LTR as bi-directional HS-responsive promoter.
Fig. 4.
ROMANIAT5-2 controls heat-responsiveness of APUM9 in A. thaliana. a Schematic representation of the ROMANIAT5-2 – APUM9 region in A. thaliana. The yellow block within the 3’ LTR represents a 3P/Gap heat responsive element (HRE). S position of primers for RT of the sense transcripts, A position of primers for RT of the anti-sense transcripts, F and R forward and reverse quantitative PCR primers. META1 is a transposon fragment flanking ROMANIAT5-2 3’ LTR. Silex: the orange block corresponds to the genomic fragment cloned upstream of the 4× “upstream activating sequence” (UAS, violet) and green fluorescent protein (GFP; green). b Schematic representation of the A. lyrata APUM9 locus. Reads per kilobase per million reads (RPKM) for (c) ROMANIAT5 and (d) APUM9 under control, 6 h at 37 °C HS and HS with 48 h recovery at control conditions (HS + R). * P <0.05 in t-test. e RT-PCR analysis of Silex reporter construct response to HS. NS non-stressed control plants, CS and HS control- or heat-stressed plants, respectively, +0 and +5d days of recovery at non-stress conditions, RT+ and RT– samples with and without RT, respectively. 18S rRNA transcript serves as positive control. f GFP signal in control and 24 h heat-stressed (HS2) Silex, detected after 0, 1, 2, or 5 days of recovery. Red – chlorophyll, green – GFP. g Close-up view of plants treated as described in (f). h Strand-specific RT-qPCR of APUM9 and ROMANIAT5-2 in A. thaliana after 6 h HS. i Putative HREs in ROMANIAT5 LTRs in Brassicaceae. j RT-qPCR for ROMANIAT5 in Brassicaceae after 6 and 12 h at 37 °C HS. The values were normalized to UBC28. Error bars indicate standard deviation between three biological replicates and * P <0.05 in Student’s t-test
We found ROMANIAT5 elements in genomes of all species except for B. antipoda (Table 1). Putative HREs were present in at least some copies of ROMANIAT5 in all species except for C. rubella that contained only solo LTRs. There were step and gap HREs in A. lyrata, B. rapa, and B. stricta, 3P/gap HREs in E. salsugineum, and 3P HREs in A. thaliana (Fig. 4i; Additional file 5: Figure S13). The predicted HSF binding affinity of individual HREs correlated well with the amount of ROMANIAT5 transcripts found after 6 and 12 h of HS (Fig. 4j). The only exception was B. rapa, which showed 42-fold upregulation after 12 h HS but the analyzed copies carried at most only low affinity step HRE. This could be due to the presence of heat-responsive ROMANIAT5 copies in the part of the B. rapa genome that is not yet assembled.
Discussion
Transpositions and insertions of TEs may lead to loss of gene functionality [32, 33]. Therefore, TEs activity and mobility are tightly controlled by epigenetic means throughout the entire plant development [5, 6]. On the other hand, new insertions contribute to genome evolution and regulation of gene transcription [2]. Therefore, it was already suggested in the early days of transposon research that, under conditions when diversity of regulatory patterns in a population may provide a better basis for selection, limited transposon activation could be beneficial [34]. However, how occasional TE expression is provoked and how control is regained later is still a matter of debate. There is a rapidly increasing number of reports showing transient TE activation under various stress conditions reviewed in [1]. Therefore, it was hypothesized that stresses may open a window for transpositions. Here, we introduced the ONSEN (COPIA78) family as a model for understanding TE control and behavior under HS. ONSEN shows massive transcriptional upregulation upon HS in A. thaliana and new insertions in progenies of heat-stressed Pol IV mutant [7, 8, 17]. The molecular basis of ONSEN heat-responsiveness was puzzling until recently, when a typical HSFA2 TF binding HRE was identified in its cis-regulatory region [18]. Presence of canonical TF binding motifs in TE promoters was described for D. melanogaster and M. truncatula [15, 35]. However, the frequency of such activation strategy among TEs was unknown.
We analyzed LTRs of A. thaliana and A. lyrata heat-responsive COPIA TEs ONSEN, COPIA37, TERESTRA, and ROMANIAT5 for putative HREs. A minimum of three adjacent (<5 bp) nGAAn motifs can form a basal HRE, whose activity will depend on their distance and the total number [20]. Heat-responsive COPIAs featured the whole spectrum of HREs ranging from the 4P types in ONSEN and TERESTRA, through 3P types in COPIA37 and ROMANIAT5 to a dozen of variable gap and step HREs in all these families. By comparing predicted HREs with transcriptional data, we conclude that gap and step HREs are mostly not sufficient to trigger HS-induced TE upregulation. This is congruent with their proposed low HSF binding efficiency [36]. Predicted 3P HREs correlated with up to a hundred-fold (COPIA37, ROMANIAT5) and 4P HREs with up to a thousand-fold (ONSEN, TERESTRA) transcript accumulation upon HS. This suggests a strong correlation between putative HREs and the transcriptional response of the TEs.
Previously it was shown that the TGS machinery antagonizes the TE activation [7, 17]. We found that the speed of re-silencing during or after HS depends on the HRE type. While ONSEN, with the strong 4P HRE, accumulated transcript during entire HS exposure, TEs carrying lower affinity HREs typically showed a maximum transcript amount at 6 h HS and lower levels at 12 h HS. This silencing can be reduced by treatment with DNA methylation inhibitors. Hence, stressed plants take active measures to prevent TE transpositions already during ongoing HS treatment. However, HS-induced TE activation must not always aim at transposition, but can be part of the plant regulome [2]. In A. thaliana, we found that heat-responsive AtROMANIAT5-2 controlled transcription of the APUM9 gene located downstream of the element. As we did not observe any evidence for high amount of a read-through transcript from ROMANIAT5-2 towards APUM9, we hypothesize that this transcriptional activation may be mediated rather by a specific three-dimensional chromatin organization at this locus. APUM9 gene was previously shown to be under control by HDA6 and synergistically by MOM1 and RdDM pathways, but not DDM1 and MET1 [30, 31]. Therefore, AtROMANIAT5-2 may represent a domesticated transposon with fine-tuned HS-regulated activation, contributing to transcriptional control of APUM9.
To challenge the hypothesis that HREs could be beneficial for TE amplification (but not necessarily for the host genome stability), we reconstructed evolutionary trajectories for HREs of ONSEN, COPIA37, TERESTRA, and ROMANIAT5 in the Brassicaceae. ONSEN was not heat-responsive in the early separated lineages represented by B. rapa and E. salsugineum, because its LTRs contained only one half of the 4P HRE (proto-HRE), which does not constitute a functional HRE. The proto-HRE became duplicated approximately 6–9 millions of years ago [25] and directly formed the present days 4P HRE found in the genus Arabidopsis and in the Australian species B. antipoda. Hence, ONSEN 4P HRE represents an evolutionary conserved cis-regulatory element. However, it should be noted that there are several other similarly or even more conserved regions within the ONSEN LTR. Whether they represent other TF binding sites and/or enhancers remains currently unknown. Furthermore, the ONSEN example shows that even high affinity HREs do not allow a TE to overrule the host genome defense, because their heat-responsiveness was lost in B. stricta, and the whole family became vanished from the C. rubella genome. In TERESTRA, high affinity 4P HREs evolved independently at two different LTR regions in the closely related species A. lyrata and B. stricta, while 3P HREs of COPIA37 emerged multiple times from a common nTTCn-rich LTR region. In contrast to ONSEN, HREs of these families are evolutionary young and species-specific. Whether they will be evolutionary successful, is an open question, but we speculate this to be the case for A. lyrata TERESTRA, where all genomic copies are full length, carry strong HRE, and respond to heat.
At present it is unknown whether higher temperatures in southern latitudes lead to greater amplification of heat-responsive TEs in subtropical relative to temperate zones. Although this is possible, there are also several factors that may act against such correlation. First, southern populations may reduce effects of HS by adaptation and growth at favorable microclimatic and/or temporal conditions [37]. Second, the genomes are subject to purification mechanisms and the higher transposition rate may be opposed by a greater frequency of TE removal [10]. Indeed, HS was shown to increase frequency of DNA sequence removal by a single strand annealing type of homologous recombination in transgenic constructs structurally resembling a LTR retrotransposon [38, 39]. Therefore, the final number of stress responsive TEs per genome may be the result of multiple effects acting in a complex network.
Conclusions
TEs evolve cis-regulatory elements, such as HREs, rapidly and independently in many groups. This may represent a strategy to produce new copies, constantly challenging the host defense system by searching for potential weak points. Successful regulatory elements may become evolutionary conserved and spread by new TE insertions in a self-reinforcing loop. However, these copies will be silenced and frequently removed from the genome. Hence, stress-mediated TE activation is likely not an unequivocal and straightforward winning principle, but rather a necessary strategy to survive under the pressure of the host defense systems. It is also likely that the host genome can benefit to some extent, and in specific cases, from cis-regulatory elements spread by TEs.
Methods
Plant materials and growth conditions
We used: Arabidopis thaliana Col-0 and Silex [31], Arabidopsis lyrata subsp. lyrata MN47, Ballantinia antipoda, Boechera stricta ES9, Brassica rapa FPSc, Capsella rubella, and Eutrema salsugineum. Before standard HS experiments, A. thaliana and A. lyrata seeds were placed on wet soil, stratified for one week at 4 °C, and then grown in a growth chamber (Percival) at 21 °C during the day and 16 °C during the night (16 h light/8 h dark) until plants reached approximately the five-leaves stage. Subsequently, a part of the plants was transferred to 37 °C for 6 h. RNA samples for sequencing were collected from some of the stressed plants and the controls directly after stress. The remaining stressed plants were allowed to recover at control conditions and collected after 48 h. Later, HS and drug-treatment experiments were performed with in vitro grown plants. First, the seeds were surface-sterilized with 8 % sodium hypochlorite for 6 to 12 min, washed with copious amounts of sterile water, dried under sterile conditions, and spread on sterile ½ Murashige-Skoog medium. After one week of stratification at 4 °C, the Petri dishes with seeds were transferred to growth chamber with a long day regime (16 h light/8 h dark) and constant temperature of 21 °C. Plates with rosettes at the pre-bolting stage were then placed in another chamber with 37 °C for 6 h. For combined drug and heat treatments, A. lyrata plants were grown as described above, then transferred to plates with no inhibitor, 10 μM DZNep, or 40 μM zebularine (both Sigma-Aldrich) for 48 h and then exposed to 0, 6, or 12 h at 37 °C HS. Aerial plant tissues were harvested immediately after the stress, flash frozen in liquid nitrogen, and stored at –80 °C.
Seeds of the Silex reporter line were sown directly on potting soil and stratified at 4 °C for 48 h. The pots were then placed in a Percival CU-22 L chamber at 21 °C with 12 h light (140 mmol m−2s−1) and 12 h dark. When the plants turned 14 days old, the pots were placed at 6 °C under the same light conditions for 24 h. At this time, control plants were moved back to the 21 °C chamber while HS plants underwent 24 h HS at 37 °C with light conditions as before. Immediately after the HS treatment, all pots were placed again at 21 °C. Fluorescence pictures of control and HS plants were taken at 0 and after 1, 2, and 5 days of recovery. Fluorescence imaging was performed using an Aequoria dark box with a mounted ORCAII CCD camera (Hamamatsu, Japan).
Nucleic acids extraction, cDNA synthesis, and RT-qPCR
Genomic DNA was isolated using the Phytopure gDNA Kit (GE Healthcare). Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) with an on column DNaseI (Roche) digestion or by the standard Trizol method with additional DNaseI (Thermo Scientific) digestion. cDNA was synthesized from 1 μg total RNA per sample using the Revert Aid H-Minus First Strand cDNA synthesis kit with the oligo-d(T) primer (all Thermo Scientific). For strand-specific RT, total RNA of 6 h HS A. thaliana plants was divided into five aliquots which were converted into cDNA using (1) oligo-d(T) primer, APUM9 (2) sense and (3) antisense transcript primer, and ROMANIAT5-2 (4) sense and (5) antisense transcript primers. RT-qPCR analysis was performed on three biological replicates with at least two technical replicates in a CFX384 instrument (BIO-RAD) using the SensiMix Plus SyBr Kit (PEQLAB). Expression values were calculated relative to control-treated samples using the standard curve method [40] and normalized using the glyceraldehyde-3-phosphate-dehydrogenase C2 (GAPC-2) or the UBC28 gene with a stable expression under mock, HS, and recovery conditions. Primers used in this study are listed in Additional file 5: Table S4.
RNA sequencing
One μg total RNA per sample with RIN >8.0 (Agilent Bioanalyzer 2100) was used to construct strand non-specific sequencing libraries with the Illumina TruSeq RNA Library Kit v2 according to the manufacturer’s instructions. Library quality was tested on a Bioanalyzer and high-quality libraries were subsequently sequenced in the 100 bp single-end read mode using a HiSeq 2500 sequencer (Illumina). Adaptor sequences and low quality bases were trimmed and low quality reads were filtered out with the FAST-X toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) using custom-made scripts. Subsequently, reads were mapped to the corresponding reference genome (TAIR 10 genome assembly or A. lyrata genome assembly v1.0) using tophat2 [41] with default settings. On average, >15 million sequencing reads per library passed trimming and quality filtering. The numbers of reads mapping to specific genomic positions were retrieved using Qualimap and the latest A. thaliana genome annotation TAIR10 and A. lyrata genome annotation v2 [42] for genes and custom-made repeat annotations for TEs. The TE data were further processed with COMEX (see below) and data for genes were analyzed directly using the DESeq package in R software [43, 44].
COMparative EXpression of transposable elements (COMEX)
Accurate quantification of TEs expression using short read sequences is hampered by high similarity of potentially many genomic copies. We developed a simple protocol called COMEX (https://github.com/bpietzenuk/COMEX) that partially overcomes this problem and allows analysis of TE transcription from RNA-seq data. Out of >10 million reads per average sequencing library, 0.12 % and 0.73 % high-quality mappable reads corresponded to TEs within our custom made A. thaliana and A. lyrata, respectively, TE annotations. This suggests that TE expression analysis using RNA-seq can be made more sensitive by high sequencing depth. The reads were processed via a shell-script that merges the pipeline as follows. First, the binary mapping.bam file is converted into a readable .sam file. Subsequently, ends are printed (ToPrint_end1.py) to the .sam file and mapping errors are removed (Selectnonrepeated1.py). In the following step (Selectmultiplymapped1.py), the output files for the uniquely mapping and the multiply mapping reads are created. The high-quality uniquely mapping (UM) TE reads were directly accepted for expression analysis. High-quality multiply mapping TE reads were analyzed to identify those providing usable information. We classified multi-mapping reads into two categories: (1) informative reads mapping to multiple members of the same TE family (Specifically Multiply Mapped – SMM); and (2) non-informative reads mapping across TE families (Non-specifically Multiply-Mapped – NMM) using the TE annotation gff-file. Reads of the second category were discarded (new_cases1.py). Afterwards, UM and SMM are merged into a single .sam-file and converted into a binary .bam-file. Subsequently, the output file of the COMEX2.0-pipeline (filename.output.final.bam) containing the number of SMM and UM reads from the same TE family was retrieved using a strand non-specific protocol in Qualimap. To avoid a bias by repeated counting of SMM reads, we used the proportional read count method that divides the power of a read by the number of mapped positions. This provided the number of reads per individual TE families and TEs, which were subjected to statistical analysis using the DESeq package in R software [43]. To avoid considering potentially large number TEs with minimal transcriptional changes, which would be later difficult to validate experimentally, we considered only those which had at least 0.55 RPKM in one of the experimental time points.
In silico sequence analysis
Sequences of interest were extracted from corresponding TE annotation files using bedtools [45]. LTR reconstruction was carried out in LTR-Finder [46] or manually by pairwise and multiple alignments of the 3'end to the 5'ends of TE annotated regions using MUSCLE or multalin with the DNA 5–0 comparison table option. Structural analysis and annotation of TERESTRA was performed using LTR Finder and blastx using NCBI non-redundant protein sequences database. LTR_Finder was used in both analyses with the threshold option set to 2.0 using the tRNA database of A. thaliana to predict PBS. The LTR length range was set from 100–3500 and the minimum LTR distance was set to 1000. Other parameters were left at default settings. Search for ONSEN sequences within genomes of various Brassicaceae was done using BLASTN within Phytozome 10 [47, 48]. Hits with a sequence identity of >70 % were extracted and manually investigated. Positive hits with a query coverage <70 % were analyzed manually for sequence similarity with Multialign using the DNA 5–0 comparison table option. The input ONSEN RT and LTR sequences are provided in Additional files 10 and 11, respectively.
Phylogenetic analysis
To analyze the evolutionary distance of the Ty1/COPIA LTR-retroelements, multiple sequence alignments of the RT domains were performed using the genomic nucleotide sequences in MUSCLE [49]. RT protein sequences used for construction of the network and the tree (Fig. 1j and Additional file 5: Figure S2, respectively) are provided in Additional file 12. The evolutionary history was inferred using the Neighbor-Joining method (Kimura-2-Parameter method) with 1000 bootstrap replicates. Positions containing missing data and gaps were removed (pairwise deletion option) leading to a total of 862 position in the final dataset. The tree was visualized as an unrooted tree. Phylogenetic network of genomic RT domain sequences from Ty1/COPIA LTR-retroelements was constructed using Neighbor-Net [50] within the splitstree 4.0 package [51, 52]. The phylogenetic distances were calculated by LogDet-pairwise genetic distances using LDDIST [53] with imputed missing matrix entries. Multiple sequence alignments of CHS genomic sequences were performed using MUSCLE [49]. The CHS phylogeny was inferred using the Maximum Likelihood tree based on the Kimura-2-parameter model with 1000 bootstrap replicates. CHS sequences were retrieved from [25]. All positions containing gaps and missing data were eliminated. There were a total of 1267 positions in the final dataset. All phylogenetic trees were constructed within MEGA 7 [54]. Phylogenetic shadowing and analysis of motif conservation was performed with mVISTA [55, 56] using LTR consensus sequences of different species prepared in BioEdit [57], allowing fasta ambiguity codes for low conserved positions. Sequences were aligned using AVID [58]. The cutoff was defined as ≥70 % conservation over a 20 bp sliding window with the minimal consensus of 7 bp relative to A. lyrata 5’ LTR sequence.
Accession numbers
Short sequence reads were deposited in the NCBI GEO archive under accession number GSE69077.
Acknowledgements
We thank J. de Meaux, E. Schranz, S. Woody, and K.S. Schumaker for seed material. We are grateful to B. Eilts, R. Gentges, P. Pecinkova, and D. Hamacher for plant care and technical assistance, A. Abdelsamad for basic identification of TEs using RepeatMasker, J. Ali for preparation of the shell script, O. Mittelsten Scheid for critical reading of the manuscript, and M. Koornneef, V. Cavrak, K. Schneeberger, and J. Jimenez-Gomez for numerous discussions.
Funding
This work was supported by the German Research Foundation (DFG) grant AP1859-2 to A.P within the Science Priority Program 1529 Adaptomics and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) fellowship BEX 10896/14-7 to C.M.
Availability of data and materials
RNA-sequencing data have been uploaded to the Gene Expression Omnibus and are available under accession numbers [GEO: GSE69077]. Lists of A. lyrata and A. thaliana TEs are provided in Additional files 3 and 4. Primers for PCR analyses are available in Additional file 5: Table S4. The COMEX method is described in detail and is available to download from https://github.com/bpietzenuk/COMEX; this software is licensed under GNU General Public License version 3. The seeds of plants used in this work can be requested from the corresponding author.
Authors’ contributions
Experiments in this study were conceived by AP, EB, and AMJ. Experiments were performed by BP, CM, HG, NB, and AA. RNA sequencing was analyzed by BP and NB. Phylogenetic analyses were performed by BP. Transcript quantifications were done by CM, BP, and HG. Silex experiments were performed by HG. AP wrote the manuscript with contribution of all authors. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Additional files
Table listing transcriptional changes of genes after 6 h HS and recovery in A. lyrata. (XLSX 6774 kb)
Table listing transcriptional changes of genes after 6 h HS and recovery in A. thaliana. (XLSX 6407 kb)
List of A. lyrata TEs in general feature format. (GFF 4554 kb)
List of A. thaliana TEs in general feature format. (GFF 1310 kb)
Number of TE families in A. thaliana (n = 364) and A. lyrata (n = 376) as identified by RepeatMasker. Figure S2. Unrooted phylogenetic tree of heat-responsive and -non-responsive COPIA TEs. Table S1. List of ONSEN elements in A. thaliana Col-0 genome. Figure S3. HREs found in 5’ LTRs of ONSEN elements. Table S2. List of COPIA78/ONSEN elements in A. lyrata MN47 genome. Figure S4. Percentage identity matrix of the RT nucleotide sequences of ONSEN elements from different species. Figure S5. Percentage identity matrix of LTR nucleotide sequences of ONSEN elements from different species. Figure S6. HREs found in the 5’ LTRs of COPIA37 elements. Figure S7. Consensus DNA sequence of A. lyrata TERESTRAs. Table S3. List of A. thaliana accessions negatively tested for presence of TERESTRA elements. Figure S8. The fragment of TERESTRA from A. cebennensis clone 44. Figure S9. A. halleri TERESTRA reconstructed based on NCBI BLASTs using A. lyrata TERESTRA consensus sequence. Figure S10. HREs found in 5’ LTRs of TERESTRA elements. Figure S11. Transcriptional response of ONSEN, COPIA37, and TERESTRA to DNA methylation inhibitor treatments in A. lyrata. Figure S12. Density of RNA-seq reads mapping over APUM9 – ROMANIAT5-2 region. Figure S13. Putative HREs in 5’/3’ LTRs of ROMANIAT5 elements. (PDF 2864 kb)
Table listing transcriptional changes of all TEs after 6 h HS and recovery in A. lyrata. (XLSX 6086 kb)
Table with transcriptional changes of all TEs after 6 h HS and recovery in A. thaliana. (XLSX 1850 kb)
Table showing significantly upregulated and downregulated TEs after 6 h HS and recovery in A. lyrata. (XLSX 63 kb)
Table listing significantly upregulated and downregulated TEs after 6 h HS and recovery in A. thaliana. (XLSX 825 kb)
Contains RT sequences of ONSENs from different species and was used to generate Additional file 5: Figure S4. (FASTA 15 kb)
Includes LTR sequences of ONSENs from different species and was used to generate Additional file 5: Figure S5. (FASTA 14 kb)
Contains RT amino acid sequences of COPIA TEs used for to generate Fig. 1j and Additional file 5: Figure S2. (FASTA 73 kb)
References
- 1.Grandbastien M-A. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2015;1849:403–16. doi: 10.1016/j.bbagrm.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Lisch D. How important are transposons for plant evolution? Nat Rev Genet. 2013;14:49–61. doi: 10.1038/nrg3374. [DOI] [PubMed] [Google Scholar]
- 3.Vu GTH, Schmutzer T, Bull F, Cao HX, Fuchs J, Tran TD, et al. Comparative genome analysis reveals divergent genome size evolution in a carnivorous plant genus. Plant Genome. 2015;8:0021. doi:10.3835/plantgenome2015.04.0021. [DOI] [PubMed]
- 4.Mari-Ordonez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet. 2013;45:1029–39. doi: 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- 5.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matzke MA, Mosher RA. Rna-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 7.Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Scheid OM. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22:3118–29. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I. Stress-induced activation of heterochromatic transcription. PLoS Genet. 2010;6:e1001175. doi: 10.1371/journal.pgen.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baubec T, Finke A, Mittelsten Scheid O, Pecinka A. Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 2014;15:446–52. doi: 10.1002/embr.201337915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devos KM, Brown JKM, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–9. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark RM, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–81. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 2014;10:e1004785. doi: 10.1371/journal.pgen.1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willing E-M, Rawat V, Mandáková T, Maumus F, James GV, Nordström KJV, et al. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nature Plants. 2015;1:14023. doi: 10.1038/nplants.2014.23. [DOI] [PubMed] [Google Scholar]
- 14.Gutzat R, Mittelsten Scheid O. Epigenetic responses to stress: Triple defense? Current Opin Plant Biol. 2012;15:568–73. doi: 10.1016/j.pbi.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivashuta S, Naumkina M, Gau M, Uchiyama K, Isobe S, Mizukami Y, et al. Genotype-dependent transcriptional activation of novel repetitive elements during cold acclimation of alfalfa (Medicago sativa) Plant J. 2002;31:615–27. doi: 10.1046/j.1365-313X.2002.01383.x. [DOI] [PubMed] [Google Scholar]
- 16.Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472:115–9. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 18.Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten Scheid O. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 2014;10:e1004115. doi: 10.1371/journal.pgen.1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed-Williams L, Thao K, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakurai H, Enoki Y. Novel aspects of heat shock factors: DNA recognition, chromatin modulation and gene expression. FEBS J. 2010;277:4140–9. doi: 10.1111/j.1742-4658.2010.07829.x. [DOI] [PubMed] [Google Scholar]
- 21.Slotte T, Hazzouri KM, Agren JA, Koenig D, Maumus F, Guo Y-L, et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 2013;45:831–5. doi: 10.1038/ng.2669. [DOI] [PubMed] [Google Scholar]
- 22.Yang R, Jarvis DJ, Chen H, Beilstein M, Grimwood J, Jenkins J, et al. The reference genome of the halophytic plant Eutrema salsugineum. Frontiers Plant Sci. 2013;4:46. doi: 10.3389/fpls.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito H, Yoshida T, Tsukahara S, Kawabe A. Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene. 2013;518:256–61. doi: 10.1016/j.gene.2013.01.034. [DOI] [PubMed] [Google Scholar]
- 24.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 25.Mandakova T, Joly S, Krzywinski M, Mummenhoff K, Lysak MA. Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell. 2010;22:2277–90. doi: 10.1105/tpc.110.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pecinka A, Liu C-H. Drugs for plant chromosome and chromatin research. Cytogenet Genome Res. 2014;143:51–9. doi: 10.1159/000360774. [DOI] [PubMed] [Google Scholar]
- 27.Baubec T, Pecinka A, Rozhon W, Mittelsten SO. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 2009;57:542–54. doi: 10.1111/j.1365-313X.2008.03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foerster AM, Dinh HQ, Sedman L, Wohlrab B, Mittelsten SO. Genetic rearrangements can modify chromatin features at epialleles. PLoS Genet. 2011;7:e1002331. doi: 10.1371/journal.pgen.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C-H, Finke A, Diaz M, Rozhon W, Poppenberger B, Baubec T, et al. Repair of DNA damage induced by the cytidine analog zebularine requires ATR and ATM in Arabidopsis. Plant Cell. 2015;27:1788–800. doi: 10.1105/tpc.114.135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokthongwattana C, Bucher E, Čaikovski M, Vaillant I, Nicolet J, Scheid OM, et al. MOM1 and POL-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 2010;29:340–51. doi: 10.1038/emboj.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hristova E, Fal K, Klemme L, Windels D, Bucher E. HISTONE DEACETYLASE 6 controls gene expression patterning and DNA methylation-independent euchromatic silencing. Plant Physiol. 2015;168:1298–308. doi: 10.1104/pp.15.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, et al. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461:427–30. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- 33.Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–4. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- 34.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 35.Guerreiro MPG. What makes transposable elements move in the Drosophila genome? Heredity. 2012;108:461–8. doi: 10.1038/hdy.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enoki Y, Sakurai H. Diversity in DNA recognition by heat shock transcription factors (HSFS) from model organisms. FEBS Lett. 2011;585:1293–8. doi: 10.1016/j.febslet.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Wilczek AM, Cooper MD, Korves TM, Schmitt J. Lagging adaptation to warming climate in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2014;111:7906–13. doi: 10.1073/pnas.1406314111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyko A, Filkowski J, Kovalchuk I. Homologous recombination in plants is temperature and day-length dependent. Mut Res. 2005;572:73–83. doi: 10.1016/j.mrfmmm.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 39.Pecinka A, Rosa M, Schikora A, Berlinger M, Hirt H, Luschnig C, et al. Transgenerational stress memory is not a general response in Arabidopsis. PLoS One. 2009;4:e5202. doi: 10.1371/journal.pone.0005202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larionov A, Krause A, Miller W. A standard curve based method for relative real time pcr data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rawat V, Abdelsamad A, Pietzenuk B, Seymour DK, Koenig D, Weigel D, et al. Improving the annotation of Arabidopsis lyrata using RNA-seq data. PLoS One. 2015;10:e0137391. doi: 10.1371/journal.pone.0137391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Core Team . R: A language and environment for statistical computing. Vienna: Foundation for Statistical Computing; 2013. [Google Scholar]
- 45.Quinlan AR, Hall IM. Bedtools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Z, Wang H. Ltr_finder: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–8. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–86. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bryant D, Moulton V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–65. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 51.Huson DH. Splitstree: Analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 52.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 53.Thollesson M. Lddist: A perl module for calculating logdet pair-wise distances for protein and nucleotide sequences. Bioinformatics. 2004;20:416–8. doi: 10.1093/bioinformatics/btg422. [DOI] [PubMed] [Google Scholar]
- 54.Kumar S, Stecher G, Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, et al. Vista: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–7. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 56.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. Vista: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall TA. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 58.Bray N, Dubchak I, Pachter L. Avid: A global alignment program. Genome Res. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-sequencing data have been uploaded to the Gene Expression Omnibus and are available under accession numbers [GEO: GSE69077]. Lists of A. lyrata and A. thaliana TEs are provided in Additional files 3 and 4. Primers for PCR analyses are available in Additional file 5: Table S4. The COMEX method is described in detail and is available to download from https://github.com/bpietzenuk/COMEX; this software is licensed under GNU General Public License version 3. The seeds of plants used in this work can be requested from the corresponding author.