Abstract
Background
How canonical Wnt/β-catenin signals in adult hearts, especially in different diseased states, remains unclear. The proto-oncogene, c-Myc, is a Wnt target and an early response gene during cardiac stress. It is not clear if c-Myc is activated or how it is regulated during heart failure.
Methods and Results
We investigated canonical Wnt/β-catenin signaling and how it regulated c-Myc expression in failing hearts of human ischemic heart disease (IHD), idiopathic dilated cardiomyopathy (IDC), and murine desmin-related cardiomyopathy (DES). Our data demonstrated that canonical Wnt/β-catenin signaling was activated through nuclear accumulation of β-catenin in IDC, IHD and DES when compared to non-failing controls and Transcription Factor 7-like 2 (TCF7L2) was the main β-catenin partner of the T-cell factor (TCF) family in adult hearts. We further revealed that c-Myc mRNA and protein levels were significantly elevated in failing hearts by real-time RT-PCR, Western blotting, and immunohistochemical staining. Immunoprecipitation and confocal microscopy further showed that β-catenin interacted and co-localized with TCF7L2. More importantly, chromatin immunoprecipitation confirmed that β-catenin and TCF7L2 were recruited to the regulatory elements of c-Myc. This recruitment was associated with increased histone H3 acetylation and transcriptional upregulation of c-Myc. With lentiviral infection, TCF7L2 overexpression increased c-Myc expression and cardiomyocyte size while shRNA mediated knockdown of TCF7L2 suppressed c-Myc expression and cardiomyocyte growth in cultured neonatal rat cardiomyocytes.
Conclusions
This study indicates that TCF7L2 mediates canonic Wnt/β-catenin signaling and c-Myc upregulation during abnormal cardiac remodeling in heart failure and suppression of Wnt/β-catenin to c-Myc axis can be explored for preventing and treating heart failure.
Keywords: acetylation, cardiomyopathy, cell signaling/signal transduction, desmin, heart failure
Heart failure is a major health problem that affects over 5 million Americans 1. With the aging population, this problem has become epidemic with over 500,000 new cases diagnosed each year. Despite new therapeutic interventions, heart failure has a poor prognosis with high morbidity and mortality. Identifying key molecular drivers leading to heart failure will be essential to designing effective therapy2-4. The proto-oncogene, c-Myc, is misregulated or genetically altered in a variety of neoplastic processes. Its role in chronic non-neoplastic diseases such as heart failure is not clearly defined. As an immediate early response gene, c-Myc is upregulated within hours of hypertrophic stimulation, but returns to normal levels within 48 hours 5, 6. However, a recent investigation has shown that c-Myc protein elevation can persist for days after mechanical challenge 7. Whether c-Myc is also activated or how it is regulated in diseased hearts remain important unanswered questions.
The canonical Wnt pathway controls cardiac progenitor cell renewal and differentiation 8. In the nucleus, β-catenin forms a complex with T-cell factor/lymphoid enhancer factor (TCF/LEF) family transcriptional factors, activating transcription of target genes such as c-Myc9. It is not clear which TCF/LEF family member is expressed in adult hearts. Furthermore, it is debatable whether canonical Wnt/β-catenin signaling is active in cardiomyocytes as recent studies with the TCF/LEF responsive promoter controlling β-gal expression in transgenic mice revealed little enzyme activity in the myocardium 10. On the other hand, β-catenin is widely expressed in the developing myocardium as well as in adult hearts 11, 12. It is well known that the lack of reporter activity does not mean the absence of direct β-catenin targets since thousands of TCF/LEF consensus sequences have been identified in the genome, but only a small portion of them are selectively occupied by TCF/LEF and β-catenin in a given cell type in a context-dependent manner 13. Epigenetic modifications and sequences surrounding the core consensus site have been shown to drive tissue-specific and developmental stage-dependent engagement of β-catenin with potential Wnt responsive genes.
In this study, we tested the hypothesis that canonical Wnt/β-catenin signaling directly controls c-Myc expression in failing hearts. Our results showed that β-catenin translocated to the nucleus and interacted with Transcription Factor 7-like 2 (TCF7L2) in failing human and mouse hearts. More importantly, TCF7L2 and β-catenin co-occupied the regulatory element of c-Myc gene, resulting in elevated histone H3 acetylation and transcriptional activity. Furthermore, overexpression and knockdown of TCF7L2 in cultured neonatal rat cardiomyocytes positively and negatively control c-Myc expression and cardiomyocyte hypertrophy, respectively. These results indicate that Wnt/β-catenin signaling is activated in heart failure and can directly regulate target gene expression.
Methods
For an extended Methods sections, please refer to the online-only Data Supplement.
Human samples
The study was approved by Institutional Review Boards at University of Pennsylvania and University of Rochester. Written informed consent was obtained from participating individuals or next kins of heart donors. The clinical information of the patients used in this study was summarized in the Table and listed in Supplemental Table 1. All researches were conducted in compliance with Good Clinical Practice standards and all applicable NIH research requirements.
Table.
Patient's information of heart tissues used in this study
| NFH | IHD | IDC | P-value | |
|---|---|---|---|---|
| N=10 | N=10 | N=10 | ||
| Gender (M/F) | 5/5 | 8/2 | 6/4 | |
| Age | 49(4) | 54(3) | 52(2) | 0.571 |
| HW (g) | 381(17) | 473(35) | 429(24) | 0.062 |
| BW (kg) | 71.5(4.3) | 84.9(5.1) | 77.9(5.2) | 0.172 |
| HW/BW (g/kg) | 5.5(0.3) | 5.6(0.4) | 5.6(0.2) | 0.921 |
| EF % | 60.7(3.4) | 17.4(2.3) | 16.6(2.1) | < 0.001* |
M, male; F, female; HW, heart weight; BW, body weight; EF: ejection fraction; NFH, non-failing heart; IHD, ischemic heart disease; IDC, idiopathic dilated cardiomyopathy.
Data are presented as mean (sem).
P < 0.05 indicates statistically significant differences among NFH, IHD and IDC groups.
Animals
The mouse model of desmin-related cardiomyopathy was generated by a 7-amino acid (R172 through E178) deletion in the desmin gene (D7-Des) according to the previous study. Mice were maintained in the FVB/N inbred background and five 14- to 16-week-old D7-Des mice and their non-transgenic littermates of mixed sex were used in this study. One to two day old Sprague Dawley rats were used for the isolation of neonatal rat cardiomyocytes (NRCMs). Animal experiments performed in this study were approved by the Animal Care and Use Committee of the University of South Dakota and University of Rochester.
Viral production and infection of NRCMs
A full length short form of Tcf7l2 obtained from Dr. Elaine Fuchs (The Rockefeller University, New York, NY) 15 was cloned to lentiviral vector (pLVX-IRES-zsGreen, Clonetech, Mountain View, CA). Tcf7l2 shRNA was acquired from Sigma (St. Louis, MO). These viruses were propagated and amplified in 293T cells according to standard protocols. The virus-containing supernatant was collected at 48 hours after transfection for infecting cultured NRCMs that were isolated with the Neonatal Cardiomyocyte Isolation System from Worthington (Lakewood, NJ) according to the manufacturer's instruction based on Simpson's method 16. After 24 hours of culture, NRCMs were incubated with supernatant of lentivirus with full length Tcf7l2, Tcf7l2 shRNA, or GFP (pGIPZ). Forty-eight hours after infection, cells were measured for their size and perimeter by NIH ImageJ and extracted for real time RT-PCR and Western blotting. Each experiment was repeated at least five times independently.
Statistics
All data were presented as mean ± SEM. Differences among means were analyzed with SPSS 18.0 software (IBM Corporation, Armonk, NY). The Levene tests for equal variances were performed before all t-tests or one-way ANOVA. Student t test was used for 2-group analysis with equal variances. If equal variance was not assumed, the Satterthwaite approximate t test was used for 2-group analysis. For differences of more than two means, One-way ANOVA followed by Bonferroni test was used for multiple groups comparison when equal variances were present. If equal variances were not assumed, Tamhane T2 test was used for multiple comparison tests. A P value smaller than 0.05 was considered statistically significant.
Result
Canonical Wnt/β-catenin signaling was activated in FHHs
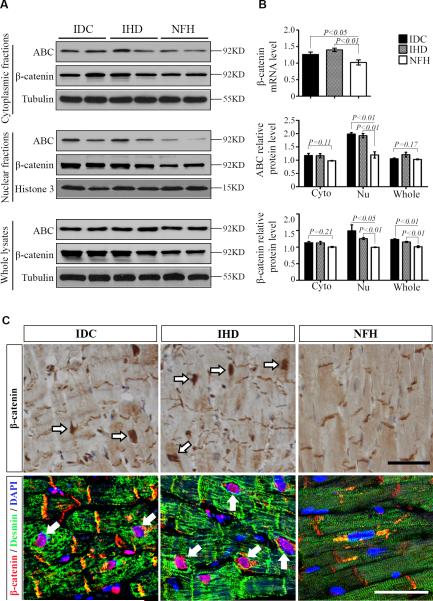
The main difference among ischemic heart disease (IHD), idiopathic dilated cardiomyopathy (IDC), and nonfailing heart (NFH) was ejection fraction by echocardiography (Table). In cardiomyocytes, β-catenin mainly binds to the cadherin cytoplasmic domain at adherens junctions of the intercalated disk and anchors the contractile apparatus to the cell-cell adhesion through α-catenin 11, 12. In the cytoplasm, β-catenin interacts with adenomatous polyposis coli (APC), glycogen synthase kinase-3β (GSK-3β), and Axin in the so called destruction complex which mediates β-catenin phosphorylation and subsequent degradation 17. A hallmark of canonical Wnt/β-catenin signaling is β-catenin nuclear accumulation. Therefore, we performed cellular fractionation to analyze total, cytoplasmic, and nuclear β-catenin levels. Western blots with histone H3 and β-tubulin antibodies confirmed good separation of nuclear and cytoplasmic proteins (Supplemental Figure 1). Both total and active β-catenin (ABC) which is not phosphorylated at serines 33/37 and threonine 41 indeed accumulated significantly (Fig. 1A and 1B, P<0.01) in the nuclear fraction of heart lysates from IDC and IHD compared with NFH. Total β-catenin was also higher in the cytoplasmic fraction and whole lysate from IDC and IHD than from NFH, but only the difference in whole lysate had statistical significance (Fig. 1B, P<0.05). The changes of ABC in cytoplasmic and whole lysates showed no statistical significance (Fig. 1B, P>0.05). The mRNA levels of β-catenin were significantly increased FHHs (Fig. 1B). Immunohistochemical and immunofluorescent staining revealed more nuclear β-catenin in cardiomyocytes from IDC and IHD than from NFH (Fig. 1C). These β-catenin positive nuclei were enlarged and had irregular contours (Fig. 1C).
Figure 1.
Accumulation of β-catenin in the nucleus of CMs from human failing hearts. A: Representative Western blots of total (β-catenin) and non-phosphorylated active β-catenin (ABC) in the cytoplasmic (25 μg), nuclear (25 μg), and whole (12.5 μg) lysates from idiopathic dilated cardiomyopathy (IDC), ischemic heart disease (IHD), and non-failing heart (NFH). B: Fold increases in β-catenin mRNA levels by real-time RT-PCR and in protein levels of total β-catenin and ABC by relative densitometric values of Western blots (N=10 in each group).C: Immunohistochemical staining (top row) revealed that there was no significant difference in β-catenin at intercalated disks (IDs) of CMs from IDC, IHD, and NFH; but increased nuclear β-catenin was observed in CMs of IDC and IHD compared to that of NFH. Representative confocal microscopy of triple stains with desmin, β-catenin, and 4′,6-diamidino-2-phenylindole (DAPI, bottom row). Desmin increased in CMs from IDC and IHD relative to that of NFH. Again, all CMs in IDC, IHD, and NFH showed β-catenin positivity at IDs. Nuclear β-catenin was strong in CMs of IDC and IHD, but not readily detected in CMs of NFH. Representative images from 5 sets of staining were shown. Data were mean ± SEM, P<0.05 indicated statistically significant differences compared with NFH. Scale bar, 50μm.
Desmin related cardiomyopathy also showed canonical Wnt/β-catenin signaling activation
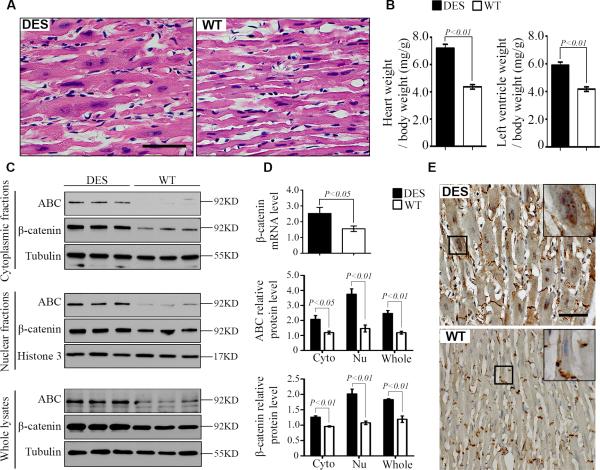
Desmin is increased in human and animal models of heart failure 18, 19. Mouse hearts with the D7-Des mutation (DES) develop cardiomyopathy similar to human hearts with the desmin mutation 14. In this study, we found that cardiomyocytes from 14- to 16-week DES mice demonstrated nuclear enlargement (Fig. 2A) and hypertrophy (Fig. 2B) similar to failing human cardiomyocytes (Fig. 1C). Total β-catenin and ABC demonstrated significant increases in all 3 fractions: whole, cytoplasmic and nuclear from DES relative to that from wild type (WT) littermates (Fig. 2C and 2D). Immunohistochemical staining showed more nuclear β-catenin in cardiomyocytes from DES than from WT (Fig. 2E).
Figure 2.
Accumulation of β-catenin in the nucleus of CMs from 14- to 16-week-old mice with D7-Des mutation (DES). A: Histology demonstrated cardiac hypertrophy with nuclear enlargement in DES hearts similar to human failing hearts compared with wild type (WT) mice. B: Heart weight/body weight ratio and left ventricle weight/body weight ratio were increased in DES hearts. C: Representative Western blots of total (β-catenin) and non-phosphorylated active β-catenin (ABC) in the cytoplasmic (25 μg), nuclear (25 μg), and whole (12.5 μg) lysates from DES and WT hearts. D: Fold increases in β-catenin mRNA levels by real-time RT-PCR and in protein levels of total β-catenin and ABC by relative densitometric values of Western blots from DES over WT hearts. N=5 in each group. E: Representative images of immunohistochemical staining from 5 mice in each group revealed similar β-catenin intensity at intercalated disks (IDs) between DES and WT CMs; but obvious nuclear β-catenin was present in CMs from DES, but not detected that from WT. Data were mean ± SEM, P<0.05 indicated statistically significant differences between DES and WT. Scale bar, 50μm.
TCF7L2 was a main TCF/LEF isoform expressed in the heart and showed upregulation in heart failure
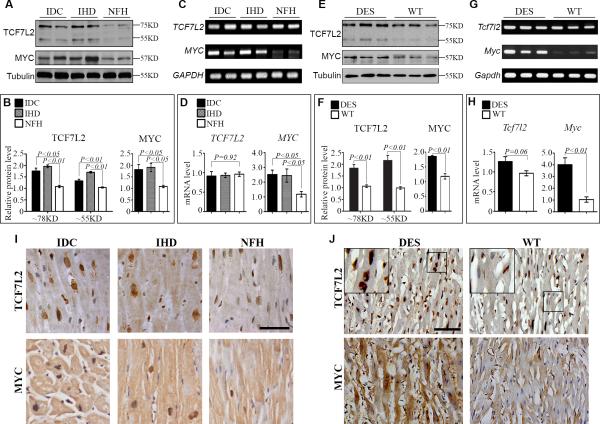
Although β-catenin does not have any DNA binding domain, it is a strong transcriptional activator and controls specific target gene expression through interaction with other DNA binding transcriptional factors, especially TCF/LEFs. Among all four TCF/LEF members, LEF1, TCF7, and TCF7L1 are not detectable in most adult mouse tissues including the heart by Northern blotting or in-situ hybridization, except the presence of TCF7 and LEF1 in lymphoid organs 20, 21. By immunohistochemistry, we did not detect LEF1, TCF7, and TCF7L1 in cardiomyocytes of adult human and mouse hearts (Supplemental Figure 2). Rare lymphocytes in FHH are positive for LEF1 and TCF7. A low level of TCF7L2 mRNA has been reported in the outer compact myocardium by RT-PCR during embryonic development 10. When using enriched Poly(A)+ mRNA, TCF7L2 is found by Northern blotting in adult mouse hearts 22. Genetic tracing further confirms that there are adult mouse cardiomyocytes expressing TCF7L2 or deriving from TCF7L2 active precursors 23. We detected TCF7L2 by RT-PCR and Western blotting in adult human and mouse hearts (Fig.3). Although TCF7L2 mRNA levels were only slightly upregulated, its protein levels were significantly increased in FHHs with IDC and IHD and DES hearts compared with their controls (Fig. 3). Two major bands of TCF7L2 around 78 and 55KD representing long and short isoforms were detected and both were dramatically increased. More importantly, TCF7L2 was mainly present in cardiomyocytes by immunolabeling (Fig. 3I and 3J, Supplemental Figure 2). Immunohistochemical staining also revealed an increase of nuclear TCF7L2 in failing cardiomyocytes compared to controls (Fig. 3I and 3J). These data indicate that TCF7L2 is most likely the main binding partner of β-catenin in the TCF/LEF family during cardiac development and in response to stresses.
Figure 3.
Upregulation of TCF7L2 and c-Myc in human failing and mouse DES hearts. A and B: Representative Western blots (A) of whole lysate and fold increases (B) in relative densitometric values of c-Myc and total long (78 kDa) and short (55 kDa) isoforms of TCF7L2 in idiopathic dilated cardiomyopathy (IDC) and ischemic heart disease (IHD) over non-failing heart (NFH). C and D: Semi-quantitative RT-PCR (C) and fold increases by real-time (D) RT-PCR of TCF7L2 with primers common to both long and short isoforms and c-Myc in IDC, IHD, and NFH. E and F: Representative Western blots (E) and fold increases (F) in relative densitometric values of c-Myc and total long (78 kDa) and short (55 kDa) isoforms of TCF7L2 in mouse DES and WT hearts. G and H: Semi-quantitative RT-PCR (G) and fold increases by real-time (H) RT-PCR of TCF7L2 with primers common to both long and short isoforms and c-Myc in mouse DES and WT hearts. Ten hearts were included in IDC, IHD and NFH while 5 hearts were used in DES and WT groups for every analysis.I and J: Representative images of immunohistochemical staining of TCF7L2 and c-Myc in 5 sets of human and mouse hearts in each group. Data were mean ± SEM, P<0.05 indicated statistically significant differences compared to control groups. Scale bar, 50μm.
Protein and mRNA levels of c-Myc were increased in failing human and mouse hearts
The proto-oncogene, c-Myc, is not detectable by Northern blotting in normal adult hearts and its mRNA is transiently induced during hypertrophic response to stresses 5, 24. In idiopathic hypertrophic cardiomyopathy (IHC), there is also a significant increase in c-Myc expression 25. It is not clear whether c-Myc is expressed in FHH. Western blots clearly showed that there was a significant upregulation of c-Myc protein in FHH (Fig. 3A and B) and DES (Fig. 3E and F) compared with NFH or WT controls. Real time quantitative RT-PCR data also demonstrated that c-Myc mRNA was dramatically elevated in FHH (Fig. 3C and D) and DES (Fig. 3G and H) in contrast to their controls. Furthermore, immunohistochemical staining revealed that c-Myc was mainly present in the nuclei of cardiomyocytes from FHH (Fig. 3I) and DES (Fig. 3J). Cardiomyocyte nuclei with elliptical or ovoid shape and blunt ends were larger than non-cardiomyocyte nuclei with spindle shape and pointed ends. Cardiomyocyte nuclei of FHH and DES often had irregular contours.
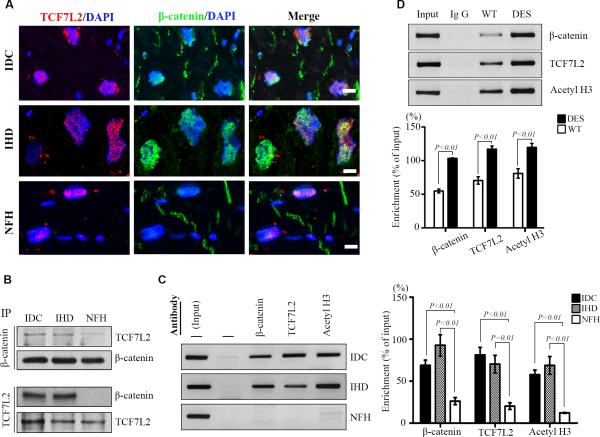
β-Catenin interacted with TCF7L2 and was recruited to the regulatory element of c-Myc
To determine if there was an increase of β-catenin and TCF7L2 interaction in failing hearts, we conducted immunoprecipitation with human hearts. More TCF7L2 was pulled down by β-catenin immunoprecipitation in FHH (Fig. 4B) than in NFH. Similarly, more β-catenin was immunoprecipitated by TCF7L2 antibody in FHH than in NFH. This suggests that more β-catenin is associated with TCF7L2 in failing than non-failing hearts. Double labeling confirmed that both β-catenin and TCF7L2 colocalized in cardiomyocyte nuclei of human hearts (Fig. 4A). To determine whether β-catenin and TCF7L2 directly regulate gene transcription in adult hearts, we performed chromatin immunoprecipitation (ChIP)-PCR with primers (Supplemental Table 2) targeting the TCF/LEF consensus binding site (Supplemental Figure 3) in human and mouse c-Myc promoters 9. As shown in Fig. 4, both TCF7L2 and β-catenin were recruited to the regulatory element of c-Myc in FHH (Fig. 4C) and DES (Fig. 4D), but not in controls. This recruitment was associated with increased histone H3 acetylation indicating transcriptional activation (Fig. 4C and 4D). These data strongly imply that canonical Wnt/β-catenin signaling directly control c-Myc expression in failing hearts (Supplemental Figure 4).
Figure 4.
Colocalization and co-recruitment of β-catenin with TCF7L2. A: Colocalization of TCF7L2 (red) with β-catenin (green) by confocal microscopy in idiopathic dilated cardiomyopathy (IDC, top row), ischemic heart disease (IHD, middle row), and non-failing heart (NFH, bottom row); nuclear counterstaining of DAPI (blue) and bar=10μm. Representative images are from 5 sets of staining in each group. B: Immunoprecipitation and Western blotting of TCF7L2 and β-catenin in IDC, IHD and NFH. Ten human samples were used in each group. C and D: Semi-quantitative PCR of c-Myc promoter after chromatin immunoprecipitation with β-catenin, TCF7L2, and acetylated histone H3 (acetylH3) in human (C) and mouse (D) hearts. Data are representative of PCR and the densitometric analysis from 10 human and 5 mouse hearts in each group. DES indicates D7-Des mutation; and WT, wild-type.
TCF7L2 levels regulated c-Myc expression and controlled hypertrophy in NRCM
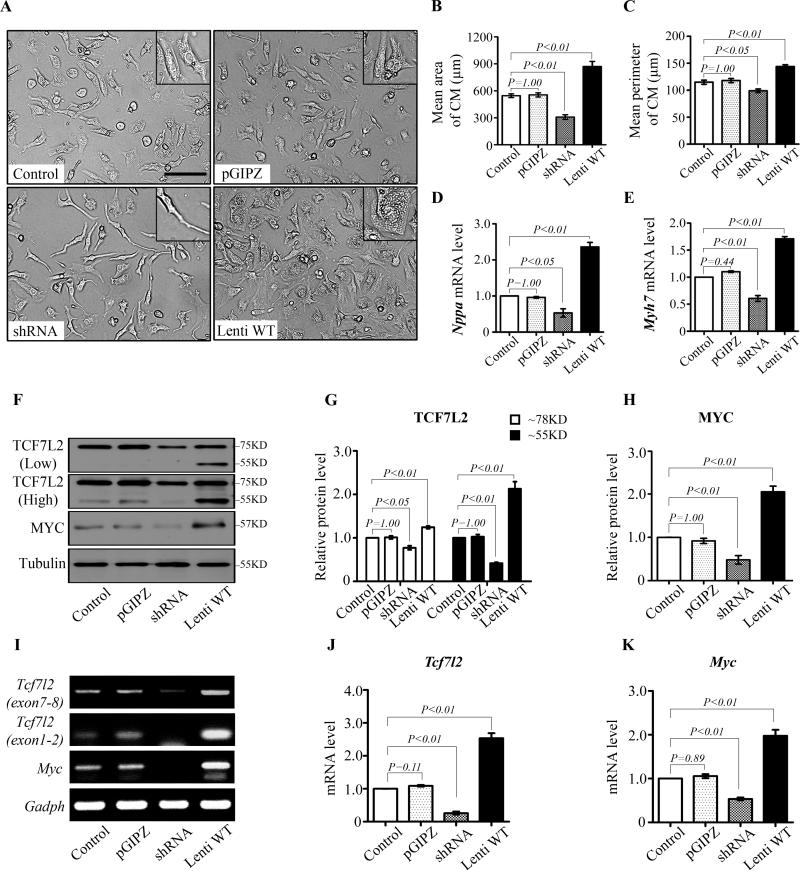
Cultured NRCM is a good in vitro model to study cardiomyocyte hypertrophy 16. We found that TCF7L2 levels modulated cellular morphology of cultured NRCMs. No differences in profile area and perimeter were detected between non-infected and pGPIZ-infected NRCM. Tcf7l2 shRNA infection effectively reduced TCF7L2 levels and led to smaller cell size compared with non-infected or pGPIZ controls (Fig. 5A). Forty-eight hours after shRNA mediated TCF7L2 knockdown, the mean profile area and perimeter of NRCMs decreased by 43.6% and 13.9% (Fig. 5B and 5C). Conversely, the size and perimeter of NRCMs infected with a full-length short form of Tcf7l2 increased significantly to 870.8±55.4 μm2 and 143.7±3.4 μm respectively compared with 545.8±20.6 μm2 and 114.9±3.7 μm in the non-infected controls (Fig. 5B and 5C). By real-time RT-PCR, hypertrophic markers: natriuretic peptide A (Nppa) and β-myosin heavy chain (Myh7), were accordingly downregulated by Tcf7l2 shRNA and upregulated by overexpression of full-length Tcf7l2 (Fig. 5D and 5E). In parallel, TCF7L2 overexpression increased c-Myc protein and mRNA levels. On the other hand, downregulation of TCF7L2 by shRNA decreased c-Myc protein and mRNA levels (Fig. 5F-K). These results further confirm that TCF7L2 can modulate cardiac gene expression and regulate cardiac remodeling.
Figure 5.
TCF7L2 levels control c-Myc expression in cultured neonatal rat cardiomyocytes (NRCMs). A to C: Morphological changes of NRCMs by TCF7L2 transfection. Cells transferred with Tcf7l2 shRNA were narrow and thin spindle-like shape (A, left lower) with smaller size (B) and perimeter (C) compared with non-infected control group. NRCMs enlarged and varied from round-spindle to polygonal by transfection with short form full-length Tcf7l2 (Lenti WT) (A, right lower), while the size and perimeter of cells dramatically increased (B and C). D and E, Nppa and Myh7 mRNA levels were downregulated by Tcf7l2 shRNA, but upregulated by infection with full-length Tcf7l2. F to H: Representative TCF7L2 and c-Myc Western blots (F) as well as fold changes of TCF7L2 (G) and c-Myc (H) in NRCMs transfected with no virus, control virus (pGPIZ), Tcf7l2 shRNA, and full-length short form Tcf7l2. I to K: Semi-quantitative RT-PCR (I) and fold increases by real-time RT-PCR of Tcf7l2 (J) and c-Myc (K) in NRCMs transfected with no virus, control virus (pGPIZ), Tcf7l2 shRNA, and full-length short form Tcf7l2. Data are representative of five independent repeats. P<0.05 indicated statistically significant differences compared to control groups. Scale bar, 100μm.
Discussion
Many signaling pathways have been implicated in the transition from cardiac hypertrophy to failure 2, 3. The canonical Wnt/β-catenin pathway and its target, c-Myc, are activated in animal models of cardiac hypertrophy 5-7. Their roles in human heart diseases, especially end-stage heart failure remain poorly understood. Our results have shown that β-catenin is stabilized and translocated into the nucleus of cardiomyocytes in failing human hearts with IHD and IDC and mouse hearts with DES. More importantly, our data indicate that β-catenin interacts with TCF7L2 and is recruited by TCF7L2 to the promoter region of c-Myc leading to histone H3 acetylation and transcriptional activation. Furthermore, TCF7L2 levels control c-Myc expression and hypertrophy in NRCMs. All these findings strongly support our hypothesis that canonical Wnt/β-catenin signaling is activated and controls cardiac remodeling and Wnt target gene expression in heart failure.
In adults, β-catenin is not required for the mechanic coupling of adjacent cardiac myocytes due to the upregulation of γ-catenin, a close relative of β-catenin 12. Most, but not all studies suggest that canonical Wnt/β-catenin signaling promotes pathologic hypertrophy. Haq et al has shown that β-catenin is stabilized in cardiomyocytes in response to hypertrophic stimuli in vitro and in vivo26. Furthermore, cardiac expression of a stabilized mutant β-catenin was sufficient to promote hypertrophic growth. More importantly, Wnt/β-catenin inhibition can effectively attenuate cardiac hypertrophy 27. Cardiac specific homozygous or even heterozygous deletion of β-catenin significantly attenuates pathological cardiac response to pressure overload by transverse aortic constriction 28, 29. These findings clearly demonstrate the importance of β-catenin signaling during cardiac remodeling in response to hypertrophic stimuli. Myocardial hypertrophy is an independent risk factor for heart failure as well as mortality, Wnt/β-catenin inhibition can be explored as potential therapeutic targets for intervention. Wnt inhibition has also been shown to reduce myocardial infarct size and alleviated cardiac dysfunction 30, 31. However, β-catenin overexpression reduced myocardial infarct size in a rat myocardial infarction model32. An opposite role of β-catenin has been observed in the cardiac response to angiotensin II infusion 33. These contradictory findings uncover the complexity of the Wnt pathway in term of its role in different disease stages as well as cell-autonomous vs non-cell-autonomous actions. The other limitation is that most investigations have only explored the short-term effects of β-catenin activation and inhibition in the heart. Therefore, low level and short term activation of β-catenin in the heart may not be harmful, its prolonged and sustained stimulation is potentially detrimental and can lead to cardiac dysfunction. Long-term follow-up investigations in animal models as well as human clinical trials are required to determine whether Wnt or β-catenin inhibition can prevent adverse cardiac remodeling and pump dysfunction in a variety of heart diseases.
TCF/LEF transcription factors are major nuclear β-catenin partners during Wnt signaling. We have shown that TCF7L2 is expressed in the epicardium and is involved in pericardial fibrosis in heart allografts 34. Here, we further reveal that TCF7L2 is the only TCF/LEF family member detected in adult cardiomyocytes and its protein levels are significantly increased in failing human and mouse hearts. More importantly, TCF7L2 interacts with β-catenin and co-occupies the c-Myc promoter to activate its transcriptional activity. It has been widely accepted that c-Myc plays an important role in initiation as well as the maintenance of cardiac hypertrophy 5, 35-39. As a traditional effector of Wnt pathway, c-Myc has been found possessing TCF/LEF binding elements within its promoter region and can be activated by Wnt/β-catenin signaling through these elements in tumor cells40, 41. Our results indicate that c-Myc is dramatically upregulated in FHH and DES hearts. Moreover, TCF7L2 levels control c-Myc expression in NRCMs. Whether c-Myc activation is beneficial or harmful remains controversial. In mouse models, the outcome of c-Myc activation varies dependent on dosage, duration, and timing. Transgenic c-Myc overexpression driven by the α-myosin heavy chain (MHC) promoter enhances cardiac proliferation, but also accelerates the cell cycle withdrawal of cardiomyocytes during the perinatal period 42. Tamoxifen-activated c-Myc driven by the same MHC promoter results in cell cycle reentry and cardiac hypertrophy and has protective function in mitochondria and energy metabolism after tamoxifen injection 7. On the other hand, c-Myc controlled by a tetracycline-inducible promoter causes heart failure and mitochondrial dysfunction two weeks after induced overexpression by doxycycline 39. Despite this controversy, c-Myc inhibition has been proposed as a therapeutic approach to attenuate cardiac hypertrophy because cardiac specific c-Myc deletion reduces hypertrophic growth without adverse effect in heart function 35.
In summary, our results demonstrate that c-Myc is upregulated by the canonical Wnt/β-catenin pathway through TCF7L2 in FHHs and mouse hearts with desmin-related cardiomyopathy. Both Wnt/β-catenin and c-Myc inhibition have been targeted for inhibition to treat neoplastic diseases. Similar approaches can be harnessed to treat heart diseases. Experimental animal models have shown c-Myc or β-catenin inhibition can prevent cardiac hypertrophy without compromising heart function 29, 30, 35.
Supplementary Material
Clinical Perspective.
The canonical Wnt/β-catenin pathway has been implicated in many disease processes and its activation can cause heart failure in animal models. It is not clear whether this pathway is involved in human heart failure. Moreover, how β-catenin is transmitted to the nucleus in the heart remains undefined. In this study, we demonstrate that β-catenin is stabilized and translocated into the nucleus of cardiomyocytes in human ischemic and dilated cardiomyopathy and mouse desmin cardiomyopathy. More importantly, we show that TCF7L2 is the main TCF/LEF family member expressed in adult human and mouse hearts. TCF7L2 is upregulated in dysfunctional human and murine hearts and interacts with β-catenin to upregulate Wnt target gene, c-myc. Furthermore, TCF7L2 can sufficiently promote hypertrophy, fetal gene expression, and c-myc upregulation in cultured neonatal rat cardiomyocytes. Conversely, TCF7L2 knockdown significantly suppresses these activities. All these findings support our hypothesis that TCF7L2 mediates canonical Wnt/β-catenin signaling and c-Myc upregulation during abnormal cardiac remodeling in heart failure, and that suppression of Wnt/β-catenin to c-Myc axis can be explored for preventing and treating heart failure.
Acknowledgments
Sources of Funding
This research was supported by the National Institute of Health (NIH) grant R01HL111480 and a Grant-in-Aid award 15GRNT22890003 from the American Heart Association Greater River Affiliate to FL, R01HL105993 to KBM, R01 HL122793 to HX and R01 HL072166 to XW.
Footnotes
Disclosures
None.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Greenlund KJ, Hailpern SM, Heit JA, Ho PM, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, McDermott MM, Meigs JB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Rosamond WD, Sorlie PD, Stafford RS, Turan TN, Turner MB, Wong ND, Wylie-Rosett J. American Heart Association Statistics C and Stroke Statistics S. Heart disease and stroke statistics--2011 update: a report from the American Heart Association. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mann DL, Bristow MR. Mechanisms and models in heart failure: the biomechanical model and beyond. Circulation. 2005;111:2837–2849. doi: 10.1161/CIRCULATIONAHA.104.500546. [DOI] [PubMed] [Google Scholar]
- 3.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 4.Chien KR, Knowlton KU, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- 5.Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci U S A. 1988;85:339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komuro I, Kurabayashi M, Takaku F, Yazaki Y. Expression of cellular oncogenes in the myocardium during the developmental stage and pressure-overloaded hypertrophy of the rat heart. Circ Res. 1988;62:1075–1079. doi: 10.1161/01.res.62.6.1075. [DOI] [PubMed] [Google Scholar]
- 7.Ahuja P, Zhao P, Angelis E, Ruan H, Korge P, Olson A, Wang Y, Jin ES, Jeffrey FM, Portman M, Maclellan WR. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J Clin Invest. 2010;120:1494–1505. doi: 10.1172/JCI38331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, Caron L, Wu X, Clarke J, Taketo MM, Laugwitz KL, Moon RT, Gruber P, Evans SM, Ding S, Chien KR. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linask KK, Knudsen KA, Gui YH. N-cadherin-catenin interaction: necessary component of cardiac cell compartmentalization during early vertebrate heart development. Dev Biol. 1997;185:148–164. doi: 10.1006/dbio.1997.8570. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Qu J, Yi XP, Graber K, Huber L, Wang X, Gerdes AM, Li F. Upregulation of gamma-catenin compensates for the loss of beta-catenin in adult cardiomyocytes. Am J Physiol Heart Circ Physiol. 2007;292:H270–H276. doi: 10.1152/ajpheart.00576.2006. [DOI] [PubMed] [Google Scholar]
- 13.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, Witt S, Kimball T, Gulick J, Robbins J. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, Pasolli HA, Fuchs E. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson P, McGrath A, Savion S. Myocyte hypertrophy in neonatal rat heart cultures and its regulation by serum and by catecholamines. Circ Res. 1982;51:787–801. doi: 10.1161/01.res.51.6.787. [DOI] [PubMed] [Google Scholar]
- 17.Haq S, Choukroun G, Kang ZB, Ranu H, Matsui T, Rosenzweig A, Molkentin JD, Alessandrini A, Woodgett J, Hajjar R, Michael A, Force T. Glycogen synthase kinase-3beta is a negative regulator of cardiomyocyte hypertrophy. J Cell Biol. 2000;151:117–130. doi: 10.1083/jcb.151.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heling A, Zimmermann R, Kostin S, Maeno Y, Hein S, Devaux B, Bauer E, Klovekorn WP, Schlepper M, Schaper W, Schaper J. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circ Res. 2000;86:846–853. doi: 10.1161/01.res.86.8.846. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Li F, Campbell SE, Gerdes AM. Chronic pressure overload cardiac hypertrophy and failure in guinea pigs: II. Cytoskeletal remodeling. J Mol Cell Cardiol. 1999;31:319–331. doi: 10.1006/jmcc.1998.0885. [DOI] [PubMed] [Google Scholar]
- 20.Oosterwegel M, van de Wetering M, Dooijes D, Klomp L, Winoto A, Georgopoulos K, Meijlink F, Clevers H. Cloning of murine TCF-1, a T cell-specific transcription factor interacting with functional motifs in the CD3-epsilon and T cell receptor alpha enhancers. J Exp Med. 1991;173:1133–1142. doi: 10.1084/jem.173.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected]. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 22.Lee YJ, Swencki B, Shoichet S, Shivdasani RA. A possible role for the high mobility group box transcription factor Tcf-4 in vertebrate gut epithelial cell differentiation. J Biol Chem. 1999;274:1566–1572. doi: 10.1074/jbc.274.3.1566. [DOI] [PubMed] [Google Scholar]
- 23.Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:4914–4919. doi: 10.1073/pnas.1102300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busk PK, Bartkova J, Strom CC, Wulf-Andersen L, Hinrichsen R, Christoffersen TE, Latella L, Bartek J, Haunso S, Sheikh SP. Involvement of cyclin D activity in left ventricle hypertrophy in vivo and in vitro. Cardiovasc Res. 2002;56:64–75. doi: 10.1016/s0008-6363(02)00510-2. [DOI] [PubMed] [Google Scholar]
- 25.Kai H, Muraishi A, Sugiu Y, Nishi H, Seki Y, Kuwahara F, Kimura A, Kato H, Imaizumi T. Expression of proto-oncogenes and gene mutation of sarcomeric proteins in patients with hypertrophic cardiomyopathy. Circ Res. 1998;83:594–601. doi: 10.1161/01.res.83.6.594. [DOI] [PubMed] [Google Scholar]
- 26.Haq S, Michael A, Andreucci M, Bhattacharya K, Dotto P, Walters B, Woodgett J, Kilter H, Force T. Stabilization of beta-catenin by a Wnt-independent mechanism regulates cardiomyocyte growth. Proc Natl Acad Sci U S A. 2003;100:4610–4615. doi: 10.1073/pnas.0835895100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Schans VA, van den Borne SW, Strzelecka AE, Janssen BJ, van der Velden JL, Langen RC, Wynshaw-Boris A, Smits JF, Blankesteijn WM. Interruption of Wnt signaling attenuates the onset of pressure overload-induced cardiac hypertrophy. Hypertension. 2007;49:473–480. doi: 10.1161/01.HYP.0000255946.55091.24. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Shevtsov SP, Hsich E, Cui L, Haq S, Aronovitz M, Kerkela R, Molkentin JD, Liao R, Salomon RN, Patten R, Force T. The beta-catenin/T-cell factor/lymphocyte enhancer factor signaling pathway is required for normal and stress-induced cardiac hypertrophy. Mol Cell Biol. 2006;26:4462–4473. doi: 10.1128/MCB.02157-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu J, Zhou J, Yi XP, Dong B, Zheng H, Miller LM, Wang X, Schneider MD, Li F. Cardiac-specific haploinsufficiency of beta-catenin attenuates cardiac hypertrophy but enhances fetal gene expression in response to aortic constriction. J Mol Cell Cardiol. 2007;43:319–326. doi: 10.1016/j.yjmcc.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barandon L, Couffinhal T, Ezan J, Dufourcq P, Costet P, Alzieu P, Leroux L, Moreau C, Dare D, Duplaa C. Reduction of infarct size and prevention of cardiac rupture in transgenic mice overexpressing FrzA. Circulation. 2003;108:2282–2289. doi: 10.1161/01.CIR.0000093186.22847.4C. [DOI] [PubMed] [Google Scholar]
- 31.Zelarayan LC, Noack C, Sekkali B, Kmecova J, Gehrke C, Renger A, Zafiriou MP, van der Nagel R, Dietz R, de Windt LJ, Balligand JL, Bergmann MW. Beta-Catenin downregulation attenuates ischemic cardiac remodeling through enhanced resident precursor cell differentiation. Proc Natl Acad Sci U S A. 2008;105:19762–19767. doi: 10.1073/pnas.0808393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahn JY, Cho HJ, Bae JW, Yuk HS, Kim KI, Park KW, Koo BK, Chae IH, Shin CS, Oh BH, Choi YS, Park YB, Kim HS. Beta-catenin overexpression reduces myocardial infarct size through differential effects on cardiomyocytes and cardiac fibroblasts. J Biol Chem. 2006;281:30979–30989. doi: 10.1074/jbc.M603916200. [DOI] [PubMed] [Google Scholar]
- 33.Baurand A, Zelarayan L, Betney R, Gehrke C, Dunger S, Noack C, Busjahn A, Huelsken J, Taketo MM, Birchmeier W, Dietz R, Bergmann MW. Beta-catenin downregulation is required for adaptive cardiac remodeling. Circ Res. 2007;100:1353–1362. doi: 10.1161/01.RES.0000266605.63681.5a. [DOI] [PubMed] [Google Scholar]
- 34.Ye B, Ge Y, Perens G, Hong L, Xu H, Fishbein MC, Li F. Canonical Wnt/beta-catenin signaling in epicardial fibrosis of failed pediatric heart allografts with diastolic dysfunction. Cardiovasc Pathol. 2013;22:54–57. doi: 10.1016/j.carpath.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong W, Mao S, Tobis S, Angelis E, Jordan MC, Roos KP, Fishbein MC, de Alboran IM, MacLellan WR. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J. 2006;25:3869–3879. doi: 10.1038/sj.emboj.7601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dzimiri N, Al-Bahnasi K, Al-Halees Z. Myocardial hypertrophy is not a prerequisite for changes in early gene expression in left ventricular volume overload. Fundam Clin Pharmacol. 2004;18:39–44. doi: 10.1046/j.0767-3981.2003.00212.x. [DOI] [PubMed] [Google Scholar]
- 37.Taketani S, Sawa Y, Ichikawa H, Ohtake S, Nishimura M, Kawaguchi N, Matsuda H. Change of c-Myc expression and cardiac hypertrophy in patients with aortic valve replacement. Ann Thorac Surg. 2001;71:1154–1159. doi: 10.1016/s0003-4975(00)02656-4. [DOI] [PubMed] [Google Scholar]
- 38.Xiao G, Mao S, Baumgarten G, Serrano J, Jordan MC, Roos KP, Fishbein MC, MacLellan WR. Inducible activation of c-Myc in adult myocardium in vivo provokes cardiac myocyte hypertrophy and reactivation of DNA synthesis. Circ Res. 2001;89:1122–1129. doi: 10.1161/hh2401.100742. [DOI] [PubMed] [Google Scholar]
- 39.Lee HG, Chen Q, Wolfram JA, Richardson SL, Liner A, Siedlak SL, Zhu X, Ziats NP, Fujioka H, Felsher DW, Castellani RJ, Valencik ML, McDonald JA, Hoit BD, Lesnefsky EJ, Smith MA. Cell cycle re-entry and mitochondrial defects in myc-mediated hypertrophic cardiomyopathy and heart failure. PLoS One. 2009;4:e7172. doi: 10.1371/journal.pone.0007172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolly C, Zakher A, Strauss C, Suter MM, Muller EJ. Keratinocyte transcriptional regulation of the human c-Myc promoter occurs via a novel Lef/Tcf binding element distinct from neoplastic cells. FEBS Lett. 2007;581:1969–1976. doi: 10.1016/j.febslet.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 41.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 42.Jackson T, Allard MF, Sreenan CM, Doss LK, Bishop SP, Swain JL. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol Cell Biol. 1990;10:3709–3716. doi: 10.1128/mcb.10.7.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.