Abstract
In recent years, it has been reported that the frequency of DNA-methylation regulatory gene mutations – mutations of the genes that regulate gene expression through DNA methylation – is high in acute myeloid leukemia. The objective of the present study was to elucidate the clinical characteristics and prognosis of acute myeloid leukemia with associated DNA-methylation regulatory gene mutation. We studied 308 patients with acute myeloid leukemia. DNA-methylation regulatory gene mutations were observed in 135 of the 308 cases (43.8%). Acute myeloid leukemia associated with a DNA-methylation regulatory gene mutation was more frequent in older patients (P<0.0001) and in patients with intermediate cytogenetic risk (P<0.0001) accompanied by a high white blood cell count (P=0.0032). DNA-methylation regulatory gene mutation was an unfavorable prognostic factor for overall survival in the whole cohort (P=0.0018), in patients aged ≤70 years, in patients with intermediate cytogenetic risk, and in FLT3-ITD-negative patients (P=0.0409). Among the patients with DNA-methylation regulatory gene mutations, 26.7% were found to have two or more such mutations and prognosis worsened with increasing number of mutations. In multivariate analysis DNA-methylation regulatory gene mutation was an independent unfavorable prognostic factor for overall survival (P=0.0424). However, patients with a DNA-methylation regulatory gene mutation who underwent allogeneic stem cell transplantation in first remission had a significantly better prognosis than those who did not undergo such transplantation (P=0.0254). Our study establishes that DNA-methylation regulatory gene mutation is an important unfavorable prognostic factor in acute myeloid leukemia.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease whose onset involves a variety of chromosomal abnormalities and gene mutations.1,2 To improve the outcome of AML treatment, it is very important to establish a prognosis from cytogenetic analysis and provide accordingly differentiated treatment.1,2 Standard chemotherapy is given to patients with a favorable prognosis according to their cytogenetic profile, whereas allogeneic transplantation in first remission is actively promoted for patients with an unfavorable cytogenetic prognosis.3
Meanwhile, for the approximately 60% of cases that fall into the intermediate prognosis group, clinicians are seeking to provide a clear prognosis and differentiated therapy based on cytogenetic analysis, which have not been available for this group of patients. In the latter half of the 2000s, gene mutations in AML were successively discovered, and attempts have been made to use these to provide a differentiated prognosis. The most important of these gene mutations for differentiated prognosis are FMS-like tyrosine kinase 3 internal tandem duplications (FLT3-ITD), nucleophosmin 1 (NPM1) gene mutation, and CCAAT/enhancer binding protein A (CEBPA) gene mutation.4–9 In 2008, Richard et al. reported that AML cases that were FLT3-ITD-negative and NPM1-mutation-positive, or that were accompanied by CEBPA biallelic mutation, were associated with a favorable prognosis and that allogeneic hematopoietic stem cell transplantation in first remission was not indicated.9,10 It was also reported that FLT3-ITD-positive AML has a very unfavorable prognosis, which might be improved by allogeneic hematopoietic stem cell transplantation in first remission.10 These findings have been integrated as prognostic factors in the European Leukemia Net (ELN) and National Comprehensive Cancer Network (NCCN) guidelines,12,13 which are becoming widely applied in clinical practice.
However, these gene mutations are observed in only around 30% of cases with intermediate cytogenetic prognosis, meaning that differentiation of prognosis is still insufficient. In recent years, the use of next-generation sequencers has facilitated energetic exploration of gene mutations, leading to the discovery of other AML-related gene mutations,14–17 and mutations in the genes that regulate DNA methylation, such as DNA methyltransferase 3 alpha (DNMT3A), Tet methylcytosine dioxygenase 2 (TET2), isocitrate dehydrogenase1 (IDH1), and isocitrate dehydrogenase 2 (IDH2).18–23 It has been suggested that these gene mutations may also have prognostic relevance in some cases of AML, but this view has yet to become established. Moreover, a further series of gene mutations has recently been discovered, so that differentiated prognosis based on gene mutations has actually become more confused.14,16,18,24
The focus of the present study was DNA-methylation regulatory gene mutations (DMRGM), which are mutations of the genes that regulate gene expression through DNA methylation (IDH1, IDH2, DNMT3A, TET2). Studies have been carried out of the various DMRGM and other genes reported as indicating an unfavorable prognosis in AML.18–20,22, 23,25–30 Our group has also reported that AML with DMRGM at onset is associated with a high frequency of FLT3-ITD at relapse and has an unfavorable prognosis.31 However, since comprehensive gene mutation analysis using a next-generation sequencer is costly, there have so far been few reports on DMRGM-based prognostic analysis covering a large number of cases. In particular, there are very few reports on large cohorts in which there is a combined analysis of both TET2 mutation, which is mutually exclusive with IDH1 and IDH2 mutations, and DNMT3A mutation.20 Since IDH1/2 mutations and TET2 mutation are mutually complementary, DMRGM need to be subjected to integrated analysis as a group, but there have so far been no reports of such analysis having been performed. We therefore analyzed the clinical characteristics of a group of AML patients with DMRGM and the prognostic impact of these mutations.
Methods
Patients
We studied 308 patients with de novo AML (excluding M3) treated at Nippon Medical School Hospital or its affiliated institutions. A comprehensive genetic mutational analysis, as described below, was conducted among patients with ≥20% blasts in bone marrow or peripheral blood. The study was conducted in accordance with the Declaration of Helsinki; written informed consent was obtained from the participants, and the patients were analyzed and treated with respect for their welfare and free will. The study protocol was approved by our institutional review board.
Screening for cytogenetic mutations
G-band analysis was performed on bone marrow samples obtained from patients at initial presentation. When it was difficult to obtain bone marrow samples, peripheral blood was used instead. For patients suspected of having M2, M3, or M4e AML based on the French-American-British classification, fluorescence in situ hybridization analysis was used to search additionally for RUNX1-RUNX1T1, PML-RARA, and CBFB-MYH11 mutations. The cytogenetic prognosis was then classified in accordance with the system recommended by the ELN.
Screening for molecular genetic abnormalities in all exons of 20 genes and hotspots of eight genes
An oligonucleotide library was generated by emulsion polymerase chain reaction using order-made probes designed against the exons of the following 20 genes: TET2 (HGNC:25941), DNMT3A (HGNC:2978), ASXL1 (HGNC:18318), KMT2A (HGNC:7132: the HGNC nomenclature of MLL has recently changed to KMT2A), RUNX1 (HGNC:10471), KIT (HGNC:6342), TP53 (HGNC:11998), PTPN11 (HGNC:9644), GATA2 (HGNC:4171), WT1 (HGNC:12796), STAG2 (HGNC:11355), RAD21 (HGNC:9811), SMC1A (HGNC:11111), SMC3 (HGNC:2468), DAXX (HGNC:2681), BCOR (HGNC:20893), BCORL1 (HGNC:25657), NF1 (HGNC:7765), DDX41 (HGNC:18674), and PHF6 (HGNC:18145). The library was sequenced with the next-generation sequencer Ion PGM™. With respect to detected mutations, the NCBI and COSMIC databases were used to search for polymorphisms and cancer-related mutations. For newly identified mutations, genetic polymorphisms were checked using Sanger sequencing with remission-stage samples.
Genes located in known hot spots (FLT3-ITD and FLT3-TKD, NPM1, IDH1, IDH2, NRAS, and KRAS), those for which probe design for emulsion sequencing was difficult (CEBPA), and those for which analysis with Ion PGM™ was difficult (KMT2A-PTD), were analyzed using previously reported methods.31
Statistical analysis
The primary endpoint was overall survival of DMRGM-positive AML patients in the study cohort. A hazard ratio of 0.707 and an overall survival rate of 50% were assumed for the sample size calculation. Recruitment of approximately 300 patients allowed for a power of 80% in detecting a difference of that size with a type I error of 5%. During the study period, there were 80 deaths among the 135 DMRGM-positive AML patients. This provided an 80% statistical power to detect a hazard ratio of 0.542 with a significance level (alpha) of 0.05 (two-tailed) regarding overall survival, which was defined as the time interval from the date of diagnosis to the date of death. Relapse-free survival for patients who had achieved complete remission was calculated as the time interval from the date of complete remission to the date of relapse.
The χ2 test was used to test the association between categorical variables and the presence and absence of mutations. The Fisher exact test was used if the expected frequency of an event was less than five in any cell of a 2 × 2 table. The non-parametric Mann– Whitney U test was used to determine the statistical significance of differences in median values. All statistical tests were two-sided. The Kaplan-Meier method and log-rank test were applied to analyze overall survival and relapse-free survival. With respect to prognostic factors, multivariate analysis was conducted with the Cox proportional hazards model. A stepwise backward procedure selection model was used to extract independent events. Events at a significance level of P≤0.20 were analyzed. Statistical analyses were performed using GraphPad Prism (version 6.00 for Windows, GraphPad Software, La Jolla, CA, USA) and IBM SPSS Statistics (version 21.0 for Windows, IBM Corp., Armonk, NY, USA), while the power calculation was performed using GraphPad StatMate (version 2.00 for Windows).
Results
Patients’ background
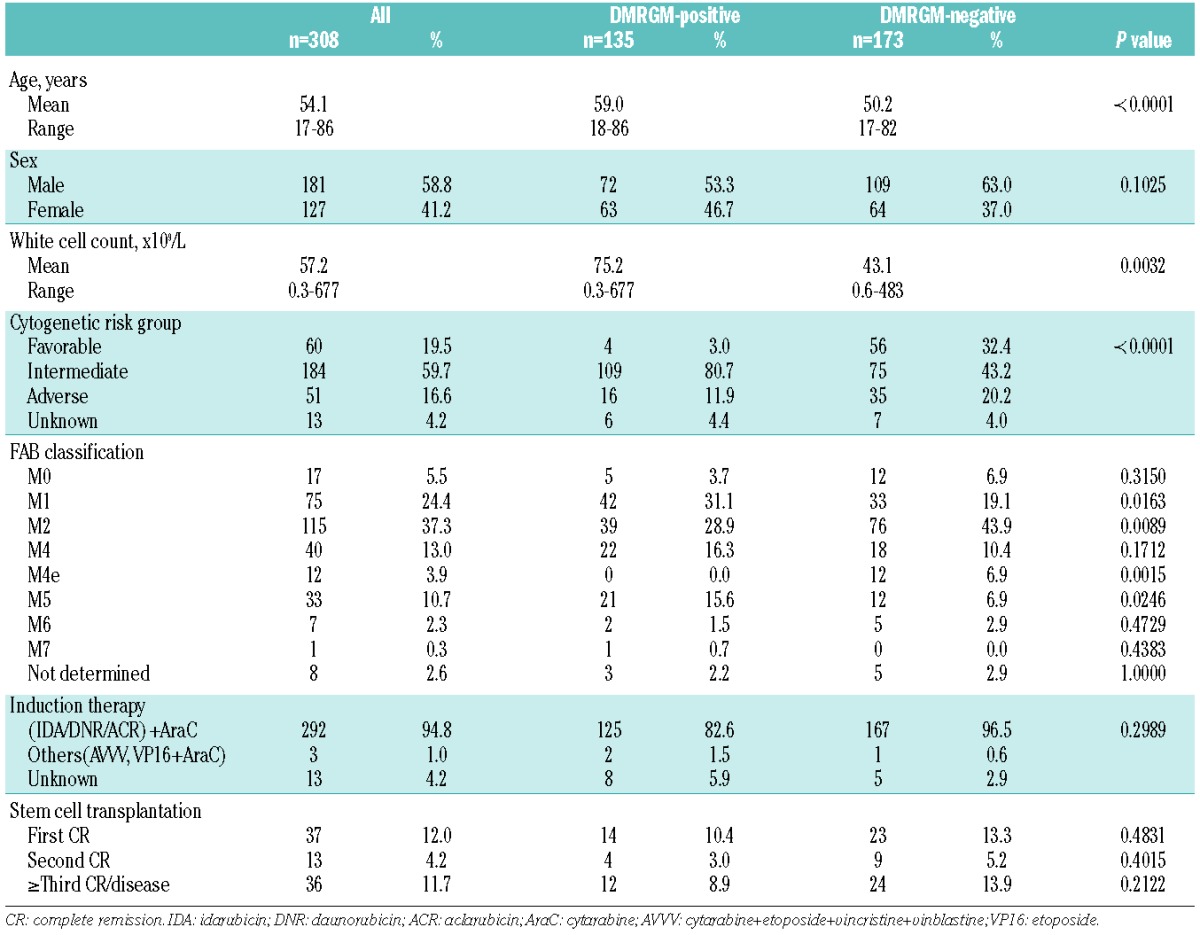
The average age of the 308 patients studied was 54.1 years (range, 17–86 years). There were 181 males (58.8%) and 127 females (41.2%). Based on cytogenetic analysis, 60 cases (19.5%) were assigned to the favorable risk group, 51 (16.6%) to the unfavorable risk group, and 184 (59.7%) to the intermediate risk group. Overall, 148 (48.1%) had a normal karyotype (Table 1, Online Supplementary Table S1). Gene mutations were observed in 265 cases (86.0%), with the most frequent being NPM1 mutation (88 cases, 28.6%), DNMT3A mutation (71, 23.1%), and FLT3-ITD (65, 21.1%) (Online Supplementary Figure S1, Online Supplementary Table S2). Gene mutations with an incidence of ≤3% were excluded from the analysis as they were too infrequent to be studied as prognostic factors (Online Supplementary Figure S1).
Table 1.
Clinical background of the AML patients studied.
Clinical characteristics of patients with DNA-methylation regulatory gene mutations
A DNMT3A mutation was found in 71 cases (23.1%), TET2 mutation in 57 (18.5%), IDH2 mutation in 28 (9.1%), and IDH1 mutation in 17 (5.5%). TET2 mutation was more frequent in older patients (average 63.5 years, P<0.0001) (Online Supplementary Table S3). High white blood cell counts were found in patients with mutations of DNMT3A (average 86.6×109/L, P=0.0028) and TET2 (average 80.7×109/L, P=0.0389). Mutations in DNMT3A (P<0.0001), TET2 (P=0.0087), and IDH2 (P=0.0199) were more frequent in the intermediate cytogenetic risk group (Online Supplementary Table S3).
A DMRGM was found in 135 cases (43.8%), indicating that this group of gene mutations occurs with very high frequency in AML. There were 99 cases (73.3%) with one DMRGM (DMRGM1), 34 (25.2%) with two (DMRGM2), and two (1.5%) with three (DMRGM3). No cases had four or more DMRGM (Figure 1). Compared to AML without DMRGM (non-DMRGM), AML with DMRGM was more frequent in older patients (DMRGM: average 59.0 years; non-DMRGM: average 50.2 years; P<0.0001), in patients with high white blood cell count (DMRGM: average 75.2×109/L; non-DMRGM: average 43.1×109/L; P=0.0032), and in the intermediate cytogenetic risk group (DMRGM: 80.7%; non-DMRGM: 43.4%; P<0.0001) (Table 1).
Figure 1.
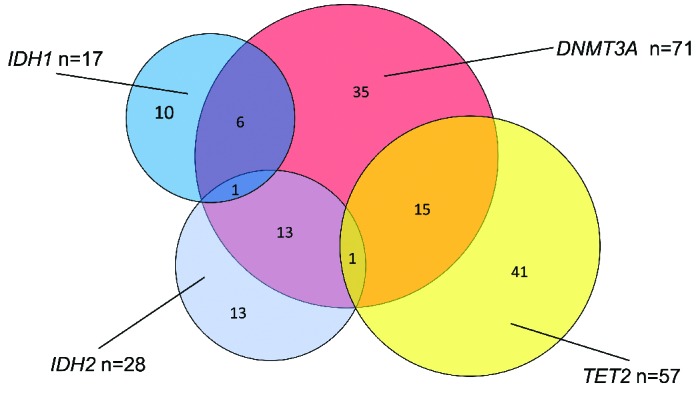
Frequency and overlap of DMRGM. There were 173 cases of DMRGM-negative AML (DMRGM0), 99 cases with one DMRGM (DMRGM1: 10 cases with IDH1, 13 with IDH2, 35 with DNMT3A, and 41 with TET2), 34 cases with two DMRGM (DMRGM2: 6 with IDH1+DNMT3A, 13 with DH2+DNMT3A, and 15 with DNMT3A+TET2), and two cases with three DMRGM (DMRGM3: 1 with IDH1+IDH2+DNMT3A and 1 with IDH2+DNMT3A+TET2).
Overlapping gene mutations
DNMT3A mutation was frequently present together with FLT3-ITD (P=0.0005), FLT3-TKD (P=0.0200), and mutations of PTPN11 (P=0.0254), NPM1 (P<0.0001), and IDH2 (P=0.0002), but was mutually exclusive with NRAS mutation (P=0.0364) and CEBPA double mutation (CEBPA dm, P=0.0265). TET2 mutation was frequently present together with NPM1 mutation (P=0.0031) and CEBPA monoallelic mutation (P=0.0441), but was mutually exclusive with NRAS mutation (P=0.0229), IDH1 mutation (P=0.0497), and IDH2 mutation (P=0.0380) (Online Supplementary Table S4). DMRGM was frequently present together with FLT3-ITD (P=0.0007) and mutations of PTPN11 (P=0.0190) and NPM1 (P<0.0001), but was mutually exclusive with mutations of NRAS (P=0.0035) and WT1 (P=0.0241) (Online Supplementary Table S4).
Prognostic analysis in all subjects
With regards to overall survival, the factors associated with an unfavorable prognosis were age >70 years (P<0.0001), adverse cytogenetic risk (P<0.0001), FLT3-ITD (P<0.0001), KMT2A-PTD (P=0.0152), DNMT3A mutation (P=0.0017), TET2 mutation (P=0.0043), and TP53 mutation (P<0.0001). In recent years, the development of a non-myeloablative regimen for hematopoietic stem cell transplantation has made it possible also for patients aged 65 to 69 years to undergo allogeneic hematopoietic stem cell transplantation. In the present study, analysis of prognosis was therefore stratified between patients aged ≤70 and >70 years. Favorable cytogenetic risk (P<0.0001), allogeneic hematopoietic stem cell transplantation (in first or second remission) (P<0.0001), and CEBPA dm (P=0.0282) were associated with a favorable prognosis (Online Supplementary Table S5).
As regards relapse-free survival, the factors associated with an unfavorable prognosis were adverse cytogenetic risk (P=0.0210), FLT3-ITD (P<0.0001), TET2 mutation (P=0.0219), and TP53 mutation (P=0.0011), whereas allogeneic hematopoietic stem cell transplantation (in first remission) (P<0.0001) and NRAS mutation (P=0.0142) were associated with a favorable prognosis (Online Supplementary Table S5).
Stratified analysis of FLT3-ITD-negative patients aged below 70 years with intermediate cytogenetic prognosis
Age >70 years, adverse cytogenetic risk, and FLT3-ITD were powerful poor prognostic factors. Rates of overall survival and relapse-free survival were therefore analyzed following stratification based on age ≤70 years, intermediate cytogenetic prognosis, and FLT3-ITD negativity. For overall survival, DNMT3A mutation was associated with an unfavorable prognosis (P=0.0429). On the other hand, allogeneic hematopoietic stem cell transplantation (in first or second remission) was associated with a favorable prognosis (P=0.0283) (Online Supplementary Table S5). For relapse-free survival, TP53 mutation was associated with an unfavorable prognosis (P=0.0068), while, allogeneic hematopoietic stem cell transplantation (in first remission) was associated with a favorable prognosis (P=0.0008) (Online Supplementary Table S5).
Prognostic impact of DNA-methylation regulatory gene mutations
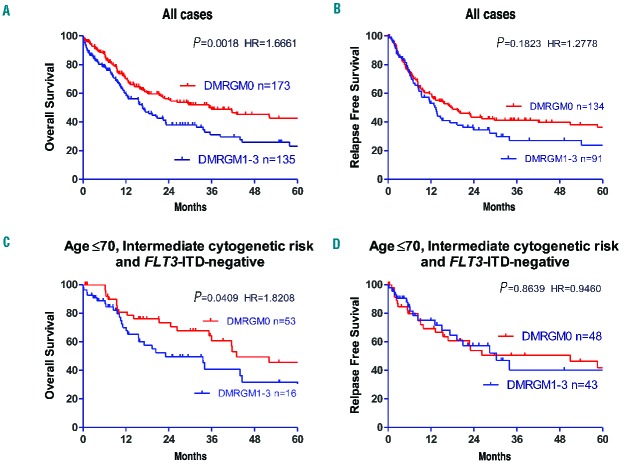
The significance of DMRGM as a prognostic factor was investigated. With regards to overall survival, cases with DMRGM had a significantly poorer prognosis than cases without DMRGM (P=0.0018) (Figure 2A). Furthermore, patients with DMRGM tended to have a higher rate of relapse than patients without DMRGM, but the difference was not statistically significant (Figure 2B). Additionally, overall and relapse-free survival rates were analyzed following stratification based on age ≤70 years, intermediate cytogenetic risk, and FLT3-ITD negativity. With regards to overall survival, cases with DMRGM had a significantly poorer prognosis than cases without DMRGM (P=0.0409) (Figure 2C). However, there was no significant difference in relapse-free survival between patients with and without DMRGM (Figure 2D).
Figure 2.
Overall and relapse-free survival rates in AML cases with and without DMRGM. (A) Overall survival rate for all cases. (B) Relapse-free survival rate for all cases. (C) Overall survival rate in FLT3-ITD-negative cases aged ≤ 70 years with intermediate cytogenetic prognosis. (D) Relapse-free survival rate in FLT3-ITD-negative cases aged ≤ 70 years with intermediate cytogenetic prognosis. HR: Hazard ratio.
Prognostic impact of number of DNA-methylation regulatory gene mutations
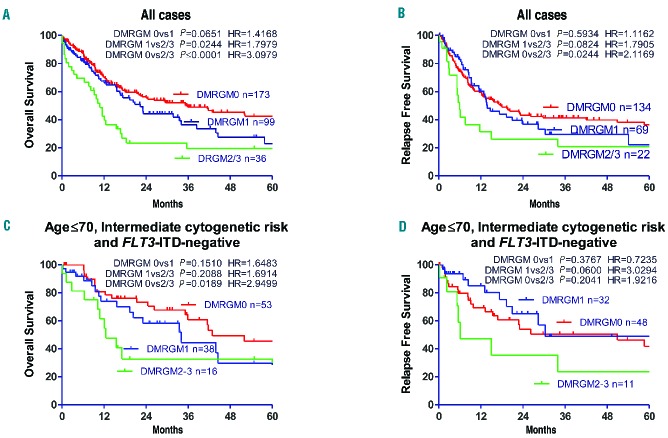
Overall and relapse-free survival rates were studied relative to the number of DMRGM mutations. The greater the number of DMRGM, the poorer the prognosis (DMRGM0 versus DMRGM2+3: P<0.0001; DMRGM1 versus DMRGM2+3: P=0.0244; DMRGM0 versus DMRGM1: P=0.0651) (Figure 3A). As far as concerns relapse-free survival, DMRGM2+3 was associated with a poorer prognosis than DMRGM0 (P=0.0244) and DMRGM1 (P=0.0824) (Figure 3B). Additionally, overall and relapse-free survival rates were analyzed following stratification based on age ≤70 years, intermediate cytogenetic risk, and FLT3-ITD negativity. The overall survival rate of patients with DMRGM2+3 was significantly lower than that of patients with DMRGM0 (P=0.0189) (Figure 3C). The relapse-free survival rate of patients with DMRGM2+3 tended to be worse than that of patients with DMRGM1, but not significantly so (P=0.0600) (Figure 3D).
Figure 3.
Overall and relapse-free survival rates in AML patients with no DMRGM, one DMRGM, and two or more DMRGM (A) Overall survival rate for all cases. (B) Relapse-free survival rate for all cases. (C) Overall survival rate in FLT3-ITD-negative cases aged ≤ 70 years with intermediate cytogenetic prognosis. (D) Relapse-free survival rate in FLT3-ITD-negative cases aged ≤ 70 years with intermediate cytogenetic prognosis. HR: Hazard ratio.
Prognostic significance of DNA-methylation regulatory gene mutation combinations
In the present analysis, prognosis was poor for patients with DMRGM, but was found to be poorer still for those with DMRGM2+3. We, therefore, investigated whether the combinations of DMRGM influence prognosis in patients with DMRGM2+3. The 15 patients with DNMT3A mutation accompanied by TET2 mutation (DNMT3A mutation+/TET2 mutation+) were compared with the 20 patients with DNMT3A mutation accompanied by IDH mutation (DNMT3A mutation+/IDH1 or IDH2 mutation+). No clear significant difference was established in overall survival rate (Online Supplementary Figure S2A), but DNMT3A mutation+/TET2 mutation+ was associated with a poorer relapse-free survival rate (P=0.0349) (Online Supplementary Figure S2B).
Multivariate analysis
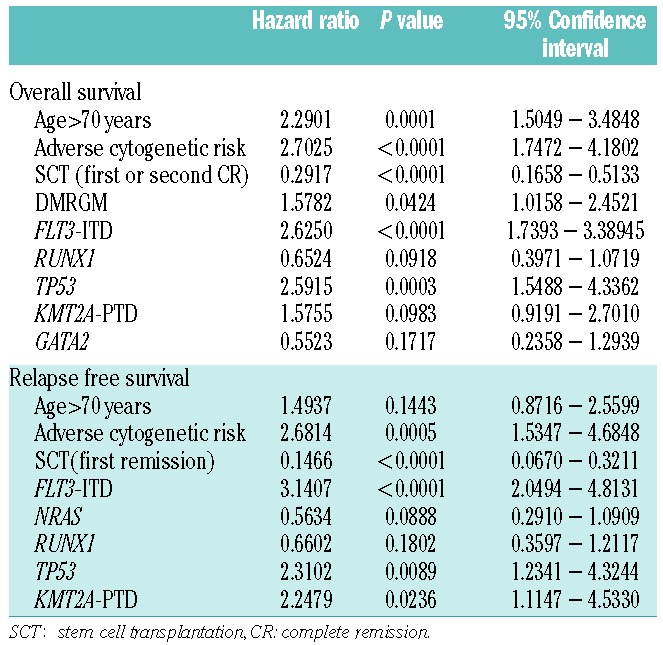
Multivariate analysis (Cox proportional hazard model) via the step-wise method was carried out using the following variables: age >70 years, poor cytogenetic risk, transplantation time (in first or second complete remission), DMRGM, and gene mutation associated with poor prognosis. The following were identified as independent unfavorable prognostic factors for overall survival: age >70 years (P=0.0001); adverse cytogenetic risk (P<0.0001); DMRGM (P=0.0424); FLT3-ITD (P<0.0001); and TP53 mutation (P=0.0003) (Table 2), whereas the independent unfavorable prognostic factors for relapse-free survival were: unfavorable cytogenetic risk (P=0.0005); FLT3-ITD (P<0.0001); TP53 mutation (P=0.0089); and KMT2A-PTD (P=0.0236) (Table 2).
Table 2.
Multivariate analysis of prognostic factors.
Efficacy of allogeneic transplantation in first remission for patients with DNA-methylation regulatory gene mutation-positive acute myeloid leukemia
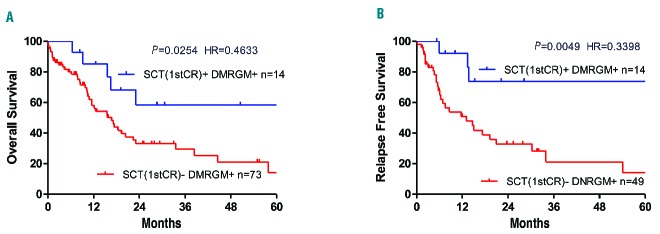
The efficacy of allogeneic transplantation in first remission for patients with DMRGM aged ≤70 years was examined. In terms of overall survival, patients with DMRGM who underwent allogeneic stem cell transplantation in first remission [SCT (1st CR)+/DMRGM+] had a significantly more favorable prognosis than those who did not undergo such transplantation [SCT(1st CR)−/DMRGM+] (P=0.0254) (Figure 4A). Likewise, in terms of relapse-free survival, SCT (1st CR)+/DMRGM+ cases had a significantly more favorable prognosis than SCT (1st CR) −/DMRGM+ cases (P=0.0049) (Figure 4B). A similar analysis was undertaken in cases aged ≤70 years, cases with intermediate cytogenetic risk, and FLT3-ITD-negative cases, but as the number of SCT(1st CR)+/DMRGM+ cases was low, at just seven, the efficacy of allogeneic stem cell transplantation in first remission is not shown here.
Figure 4.
Efficacy of allogeneic stem cell transplantation in first remission for DMRGM-positive AML cases aged 70 years or below. (A) Overall survival rate for all cases. (B) Relapse-free survival rate for all cases. HR: hazard ratio; SCT: stem cell transplantation; 1stCR: first complete remission.
Discussion
It was confirmed that DMRGM are very frequently present in AML, being observed in 135 of the 308 cases (43.8%) studied. A DMRGM was an unfavorable prognostic factor for overall survival in the whole group and in patients aged ≤70 years, in patients with an intermediate cytogenetic risk group, and in FLT3-ITD-negative patients. Allogeneic stem cell transplantation in first remission may improve the prognosis of cases with DMRGM mutation.
Until now, the significance of individual DMRGM as prognostic factors in AML has not been clear. Regarding DNMT3A, the most frequent of the DMRGM, Ley et al. reported that DNMT3A R882 mutation and non-R882 mutation are both associated with unfavorable prognosis, independently of whether FLT3-ITD is present.18 Meanwhile, Patel et al. reported that patients with intermediate cytogenetic risk who have a DNMT3A mutation do not have an unfavorable prognosis even if FLT3-ITD is not present.24
Because of the expense associated with comprehensive gene mutation analysis using a next-generation sequencer, there have so far been few reports on DMRGM-based prognostic analysis involving a large number of patients. Such studies consist of the one by Patel et al., based on 398 cases, and one by Hou et al., based on 500 cases.24,25 Our analysis covered four DMRGM – IDH1, IDH2, DNMT3A, and TET2 – in 308 cases of AML, a large cohort suggesting reliable results. Moreover, ours is the first study in which AML prognosis was explored with division into groups based on DMRGM.
Our study revealed that the greater the number of DMRGM, the poorer the prognosis is. It has been reported that increasing numbers of DMRGM are associated with unfavorable outcome in a number of hematologic malignancies. For example, Papaemmanuil et al. described that leukemia-free survival in patients with myelodysplastic syndromes becomes shorter with increasing number of gene mutations.32 Guglielmelli et al. also reported that, in primary myelofibrosis, the greater the number of mutations of ASXL1, EZH2, SRSF2, and IDH1/2, the poorer the patients’ prognosis is.33 We have also reported recently that three or more gene mutations is an unfavorable prognostic factor in de novo AML.34 According to whole-exon mutation analysis using a next-generation sequencer, there are on average 2.5–5.0 gene mutations per case in AML,14,18,34 and it has been established that the onset of AML requires the combination of a number of gene mutations.16,17 These findings suggest that cases with a large number of gene mutations have a correspondingly high level of genomic instability and point to the strong possibility of mutations in genes not yet examined.34 This is thought to result in a poorer prognosis. Our finding that prognosis in AML worsens with increasing number of DMRGM suggests that to improve prognostic analysis in AML it will be important not only to focus on each individual gene mutation, but also to seek to identify overall genomic instability in individual cases.
In a whole-exon analysis of 12,380 healthy subjects, Genovese et al. found that healthy subjects in whom DMRGM mutations were observed had a high rate of myeloid malignancies in hematopoietic organs a number of years later.35 Shlush et al. demonstrated that, in AML with DNMT3A mutation, the mutation also occurred in hematopoietic stem cells with normal differentiation potential, so that even in cases in which chemotherapy induced remission, the DNMT3A mutation remaining in the stem cells subsequently led the resulting blood cells to proliferate and cause relapse of the leukemia.36 If this finding applies to all DMRGM-positive AML cases, then chemotherapy alone cannot be expected to be effective in DMRGM-positive AML. As indicated by Patel et al., for young patients with DMRGM-positive AML, allogeneic hematopoietic stem cell transplantation in first remission may be the only curative therapy available. The findings above indicate that the prognosis of patients with DMRGM-positive AML is unfavorable, that the outcome worsens with increasing numbers of such mutations and that these patients should be treated with allogeneic hematopoietic stem cell transplantation in first remission.
One of the issues that needs to be considered when going forward with this research is that the subjects in this study were of Japanese ethnicity. So far. there have been no reports on the relation of ethnicity to frequency of gene mutations and prognosis in AML. Whether the present research findings would be replicated in cohorts of patients of other ethnicities is an area that requires investigation in the future. Moreover, the present study did not include patients treated with azacitidine or other demethylating agents. The efficacy of demethylating agents in DMRGM-positive AML is therefore another area requiring investigation.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/101/9/1074
References
- 1.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350(16):1617–1628. [DOI] [PubMed] [Google Scholar]
- 2.Bullinger L, Döhner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16): 1605–1616. [DOI] [PubMed] [Google Scholar]
- 3.Schlenk RF, Döhner K, Mack S, et al. Prospective evaluation of allogeneic hematopoietic stem-cell transplantation from matched related and matched unrelated donors in younger adults with high-risk acute myeloid leukemia: German-Austrian trial AMLHD98A. J Clin Oncol. 2010:28(30):4642–4648. [DOI] [PubMed] [Google Scholar]
- 4.Preudhomme C, Sagot C, Boissel N, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood. 2002;100(8):2717–2723. [DOI] [PubMed] [Google Scholar]
- 5.Pabst T, Eyholzer M, Fos J, Mueller BU. Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer. 2009;100(8):1343–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kottaridis PD, Gale RE, Frew ME, et al. The presence of a FLT3-internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98(6):1752–1759. [DOI] [PubMed] [Google Scholar]
- 7.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106(12):3733–3739. [DOI] [PubMed] [Google Scholar]
- 8.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106(12):3740–3746. [DOI] [PubMed] [Google Scholar]
- 9.Schlenk RF, Döhner K, Krauter J, et al. Mutation and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918. [DOI] [PubMed] [Google Scholar]
- 10.Taskesen E, Bullinger L, Corbacioglu A, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117(8):2469–2475. [DOI] [PubMed] [Google Scholar]
- 11.Brunet S, Labopin M, Esteve J, et al. Impact of FLT3 internal tandem duplication on the outcome of related and unrelated hematopoietic transplantation for adult acute myeloid leukemia in first remission: a retrospective analysis. J Clin Oncol. 2012;30(7):735–741. [DOI] [PubMed] [Google Scholar]
- 12.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. [DOI] [PubMed] [Google Scholar]
- 13.O’Donnell MR, Abboud CN, Altman J, et al. Acute myeloid leukemia. J Natl Compr Canc Netw. 2012;10(8):984–1021. [DOI] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kihara R, Nagata Y, Kiyoi H, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014;28(8):1586–1595. [DOI] [PubMed] [Google Scholar]
- 16.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. [DOI] [PubMed] [Google Scholar]
- 17.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley TJ, Ding L, Walter MJ, et al. , DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thol F, Damm F, Lüdeking A, et al. Incidence and prognostic influence of DNMT3a mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. [DOI] [PubMed] [Google Scholar]
- 20.Chou WC, Chou SC, Liu CY, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118(14):3803–3810. [DOI] [PubMed] [Google Scholar]
- 21.Figueroa ME1, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6): 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcucci G1, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(14):2348–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boissel N, Nibourel O, Renneville A, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28(23):3717–3723. [DOI] [PubMed] [Google Scholar]
- 24.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou HA, Kuo YY, Liu CY, et al. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012;119(2):559–568. [DOI] [PubMed] [Google Scholar]
- 26.Marcucci G, Metzeler KH, Schwind S, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30(7):742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aslanyan MG, Kroeze LI, Langemeijer SM, et al. Clinical and biological impact of TET2 mutations and expression in younger adult AML patients treated within the EORTC/GIMEMA AML-12 clinical trial. Ann Hematol. 2014;93(8):1401–1412. [DOI] [PubMed] [Google Scholar]
- 28.Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119(24):5824–5831. [DOI] [PubMed] [Google Scholar]
- 29.Gaidzik VI, Schlenk RF, Paschka P, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG). Blood. 2013;121(23):4769–4777. [DOI] [PubMed] [Google Scholar]
- 30.Gaidzik VI, Paschka P, Späth D, et al. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol. 2012;30(12):1350–1357. [DOI] [PubMed] [Google Scholar]
- 31.Wakita S, Yamaguchi H, Omori I, et al. Mutations of the epigenetics-modifying gene (DNMT3a, TET2, IDH1/2) at diagnosis may induce FLT3-ITD at relapse in de novo acute myeloid leukemia. Leukemia. 2013;27(5):1044–1052. [DOI] [PubMed] [Google Scholar]
- 32.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guglielmelli P, Lasho TL, Rotunno G, et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: an international study of 797 patients. Leukemia. 2014;28(9):1804–1810. [DOI] [PubMed] [Google Scholar]
- 34.Wakita S, Yamaguchi H, Ueki T, et al. Complex molecular genetic abnormalities involving three or more genetic mutations are important prognostic factors for acute myeloid leukemia. Leukemia. 2016:30(3): 545–554. [DOI] [PubMed] [Google Scholar]
- 35.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]