Abstract
BACKGROUND
Several comparison studies showed that the Ultrio assay (Novartis Diagnostics) used in individual-donation nucleic acid amplification testing (ID-NAT) format was as sensitive as the TaqScreen assay (Roche) on minipools of six donations (MP6), but the sensitivity of HBV DNA detection has been improved in the new Ultrio Plus version of the assay. A head-to-head comparison study was designed to compare the clinical sensitivity of the Ultrio and Ultrio Plus assay in ID, MP4, and MP8 formats using TaqScreen MP6 as a reference assay.
STUDY DESIGN AND METHODS
Plasma samples of 107 hepatitis B surface antigen (HBsAg)-negative, HBV ID-NAT (Ultrio) positive-yield samples and 29 HBV DNA–negative, HBsAg-positive samples were used for comparison of NAT options in replicate testing of dilutions. Viral loads and relative sensitivities were determined by probit analysis against the Eurohep standard.
RESULTS
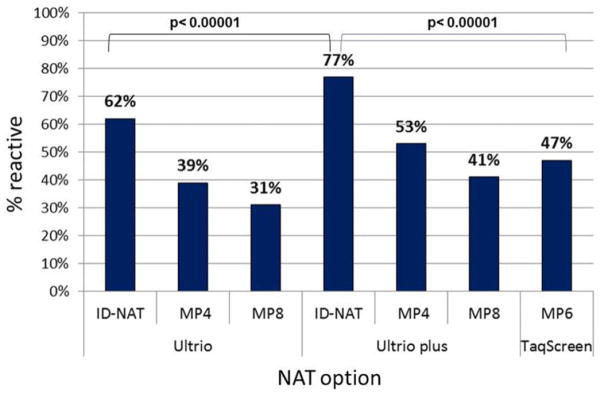
Ultrio Plus detected a significantly (p < 0.00001) higher proportion of replicate assays on HBV NAT yields (77%) than Ultrio ID (62%) and TaqScreen MP6 (47%), whereas Ultrio Plus MP4 and MP8 detected 53 and 41%, respectively. On HBsAg-yield samples missed by Ultrio screening, the reactivity rate increased significantly (p < 0.0001) from 23% in Ultrio to 65% in Ultrio Plus and further to 72% (p = 0.10) in the TaqScreen assay. The overall improvement factor of the analytical sensitivity offered by the target enhancer reagent in the Ultrio Plus assay was 2.5 (2.0–3.1)-fold on the Ultrio yield samples, but 43 (11–350)-fold on the HBsAg yields. In ID-NAT format the analytical sensitivity of TaqScreen relative to Ultrio Plus was 2.0 (1.0–4.2), 0.9 (0.7–1.3), and 1.6 (0.9–3.0) on the Eurohep standard, HBV NAT–, and HBsAg-yield samples respectively.
CONCLUSION
The clinical sensitivity of the currently available commercial NAT methods is mainly driven by the pool size.
There have been at least six studies1–6 that have compared the sensitivity of two commercial triplex nucleic acid amplification test (NAT) systems for detection of hepatitis B virus (HBV), that is, the Ultrio assay on the Tigris instrument, usually applied in individual donation (ID) format and the TaqScreen assay (Roche, Indianapolis, IN) on the s201 MPX system (Roche), commonly used in minipools of six donations (MP6). The first head-to-head comparison studies in France1,2 showed comparable sensitivity of Ultrio individual-donation nucleic acid amplification testing (ID-NAT) to TaqScreen MP6-NAT on HBV genotype dilution panels1 and HBV Genotype A seroconversion panels2 although with a higher degree of variability of results in the Ultrio assay. A comparison study performed in Australia3 found an equal number of occult HBV infections (OBIs) when testing donations from Hong Kong in these two NAT systems. A later comparison study in Thailand4,5 compared the number of NAT yields detected by the two NAT methods in different screening and confirmation test algorithms as well as in different donor populations. The Thai authors suggested that TaqScreen MP6 was more sensitive than Ultrio in ID format,4 but a recent study in the United States using the same Thai NAT-yield samples in a head-to-head comparison study did not show a significant difference in sensitivity between these two NAT configurations.6
Recently the improved Ultrio Plus assay has become available, and the manufacturer of this assay claims a higher sensitivity of HBV detection thanks to lithium hydroxide, a target enhancer reagent (TER) that accelerates the disruption of the viral particles and enhances access to single-stranded DNA for the specific HBV capture probes on the magnetic beads. Before the decision was made to introduce the Ultrio Plus assay the South African National Blood Service (SANBS) performed a head-to-head study for comparison of the Ultrio and Ultrio Plus assays. In this study archive samples taken from stored fresh-frozen plasma (FFP) units of HBV NAT–yield donations have been tested in dilutions for direct comparison of the two assay versions in ID, MP4, and MP8 formats against the TaqScreen MP6 s201 system as a reference assay.
In South Africa the predominant subgenotype is A17 and a relatively high number of NAT-yield cases (HBV DNA positive, hepatitis B surface antigen [HBsAg] negative) have been found, both in the acute phase and in the later stages of OBI.8 One-third of these HBV NAT–yield samples had very low viral load with discrepant results in the duplicate repeat Ultrio and the discriminatory HBV assay, which are routinely performed on the primary test tube of initial reactive donations. Plasma archive samples are routinely taken from the FFP unit for confirmation testing in quantitative polymerase chain reaction (PCR), antibody to hepatitis B core antigen (anti-HBc), antibody to hepatitis B surface antigen (anti-HBs) assays, and replicate Ultrio testing, and the majority of samples had inconsistent Ultrio reactivity, making them suitable for this comparison study. Since these HBV NAT–yield samples were selected by reactivity in the Ultrio assay we also included a number of HBsAg–confirmed-positive samples that were missed by Ultrio screening. The results of this study supported the decision of SANBS to introduce the Ultrio Plus assay.
MATERIALS AND METHODS
Test samples
For this study the HBV-yield samples taken from the FFP unit and stored at −80°C were used for preparing undiluted and 1:4, 1:6, and 1:8 diluted test samples. The dilutions were made with negative human plasma that was filtered through a 113-mm strengthened 24-cm filter (Catalog No. 1113–240, Whatman, GE, Buckinghamshire, UK). Diluted samples were divided in multiple vials each containing 4.5 mL of plasma. The panels were frozen at −80°C until testing in the NAT assays. A number of human immunodeficiency virus (HIV) and HBV double infections in OBI donors were first identified and excluded from the analysis. After 3 years of ID-NAT screening 107 HBV NAT–yield samples and 29 HBsAg-positive, but ID-NAT–nonreactive samples were available for the study. Of the 107 HBV NAT–yield samples only 39 (36%) of samples were consistently reactive on multiple replicate Ultrio assays (Novartis Diagnostics, Emeryville, CA). Therefore, this set of 107 low viral load–yield samples was very instrumental in evaluating the sensitivity of the ID- and MP-NAT options compared in this study. Confirmatory testing in the index and follow-up samples of the donors allowed for further classification of the 107 HBV NAT–yield samples in the following categories:8 1) Twenty-two pre-HBsAg window period (WP) samples; 2) 10 probable vaccine breakthrough samples; 3) 15 immunoglobulin M anti-HBc–positive late-acute-phase WP samples; 4) 29 anti-HBc–positive, anti-HBs–negative OBI samples; 5) 27 anti-HBc– and anti-HBs–positive OBI samples; and 6) three unclassified HBV NAT–yield samples. For analytical sensitivity and viral load determination a dilution panel of the Eurohep Genotype A2 standard calibrated in copies/ mL9 was prepared by BioQControl, DDL Diagnostic Laboratories (Rijswijk, the Netherlands). The 29 HBsAg-positive, HBV DNA–negative samples were confirmed by performing HBsAg neutralization, anti-HBc, anti-HBs, and replicate testing in the Ultrio assay.
Replicate NAT assays
After thawing the undiluted, 1:4, and 1:8 frozen aliquots of the HBV NAT–yield samples they were tested in six replicate assays in both the Ultrio and the Ultrio Plus assays on two separate TIGRIS instruments (Novartis Diagnostics/ Gen-Probe, San Diego, CA) in the SANBS screening laboratory, whereas the 1:6 dilutions were tested by the TaqScreen assay at CTS Laboratories (Tampa, FL). The HBsAg-positive, but Ultrio initially nonreactive samples were tested undiluted in 12 replicate assays on the two Ultrio assay versions and in six replicates on the TaqScreen assay and if 100% reactive in the respective assays also in the same number of replicates on a 1:8 dilution. To enable sixfold replicate TaqScreen assays of HBV NAT– and HBsAg-yield samples two aliquots of the undiluted and diluted samples were shipped on dry ice for testing at CTS Laboratories. Each 4.5-mL aliquot of the TaqScreen assay was tested in three separate test runs performed within 48 hours after thawing of the samples. To allow for replicate tests on the MPX s201 TaqScreen system each sample needed to be relabeled before testing with donation barcode number and test series number. The Eurohep HBV Genotype A2 standard dilution panel was tested in 24 replicates in the Ultrio and Ultrio Plus assay and in 12 replicates in the TaqScreen assay.
Clinical sensitivity
The proportions of replicate test results in the Ultrio HBV NAT–yield and HBsAg-yield samples that were reactive with the different ID- and MP-NAT options were compared by McNemar’s paired chi-square test. In our study the proportion of reactive results in the replicate assays for each NAT option was regarded as a measure of clinical sensitivity in the different categories of HBV NAT samples (e.g., in WP or OBI).
Viral load determination
To compare the analytical sensitivity of the assays on the South African HBV NAT– and HBsAg-yield donations we needed to estimate the viral load in each clinical sample. Since 83% of the 107 confirmed HBV NAT–yield samples was quantitative PCR negative we used the replicate test results in the more sensitive Ultrio Plus assay on the undiluted, 1:4, and 1:8 diluted samples to estimate the viral load. For this purpose the proportion of positive results in the undiluted and diluted samples were compared with those on dilutions of the Eurohep HBV DNA reference standard. Since TaqScreen was only tested on 1:6 diluted HBV NAT–yield samples we could not use these results for viral load determination. The method used to estimate the viral load in copies/mL was parallel line probit analysis using a statistical package (SPSS, SPSS, Inc., Chicago, IL). Once the criteria for parallel slopes of the probit curves have been accepted the relative potency (or concentration) of the test samples against the Eurohep reference standard was deduced from the shift between parallel lines. In the case where Ultrio Plus was zero of six reactive in the undiluted sample the concentration could not be determined and by assuming an Ultrio Plus reactive rate of one of 12 a value of 0.62 copies/mL was imputed. The low viral loads in the HBsAg-positive, Ultrio-nonreactive undiluted or 1:8 diluted samples were also determined by probit analysis by comparing the proportion of positive results in both the Ultrio Plus and the TaqScreen assays against the respective standard probit curves on the Eurohep standards. The geometric mean concentration was calculated from the values obtained in the analyses on the Ultrio Plus and TaqScreen data, respectively, and this was used for comparing the analytical sensitivity of the ID NAT systems on the HBsAg-yield samples.
Analytical sensitivity
The 95 and 50% limits of detection (LODs) and the relative sensitivity of the Ultrio, Ultrio Plus, and TaqScreen assays were determined by parallel line probit analysis using the data on the Eurohep HBV DNA Genotype A standard dilution panel. We also compared the analytical sensitivity of these assays from the proportion of positive results on (the dilutions of) the 107 HBV NAT–yield and 29 HBsAg-yield samples. For the probit analysis on the latter two clinical sample panels the log viral load of the yield sample dilutions (see above) was plotted on the X-axis and the proportion of reactive results on the Y-axis in a similar manner as for the Eurohep standard. For the probit analysis on HBsAg-positive and Ultrio-nonreactive samples the proportions of reactive results on the 1:8 diluted sample were used instead of those on the undiluted sample if these were 12 of 12 or six of six reactive in the Ultrio Plus or TaqScreen assays, respectively.
RESULTS
Clinical sensitivity of NAT options
The clinical sensitivity of the Ultrio and Ultrio Plus assays in ID, MP4, and MP8 format has been compared to TaqScreen MP6 as a reference system on 107 HBV NAT–yield samples initially detected by the Ultrio assay (HBsAg negative in PRISM) and 29 HBsAg-yield samples (HBV DNA negative in Ultrio).
HBV NAT–yield samples
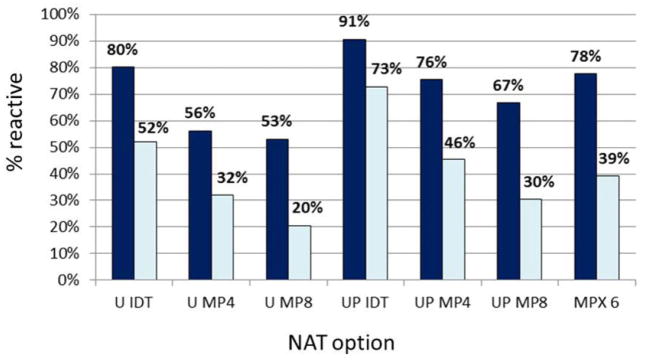
Table 1 compares the percentage of reactive results of the NAT options in replicate testing of different categories of HBV NAT–yield samples, the majority of which had viral loads below the 95% LOD of Ultrio. The bar diagram in Fig. 1 visualizes the difference in sensitivity in all 107 HBV NAT–yield samples taken together. On replicate retesting of the yield samples by the Ultrio assay in ID format only 62% was reactive on the archive samples taken from the FFP units. The proportion of reactive results increased significantly (p < 0.00001) to 77% in replicate Ultrio Plus testing. The proportion of both the Ultrio and the Ultrio Plus replicate test results was significantly (p < 0.00001) higher than the 47% reactivity in the TaqScreen MP6 system. However, TaqScreen MP6 was slightly but significantly (p < 0.03) more sensitive than the 41% reactivity in Ultrio Plus MP8, whereas the 53% reactivity in Ultrio Plus MP4 was more sensitive (p < 0.03) than TaqScreen MP6. Not surprisingly, TaqScreen MP6 was significantly (p < 0.00001) more sensitive than Ultrio in MP4 and MP8 format (Fig. 1). The viral load in WP samples is generally higher than in OBI samples, and this affects the relative clinical sensitivity of the NAT options (Fig. 2). In WP samples Ultrio ID and TaqScreen MP6 have comparable sensitivity (80 and 78%, respectively), but Ultrio Plus detected a significantly (p = 0.0005) higher proportion (91%) of replicates. In the WP samples TaqScreen MP6 and Ultrio Plus MP4 have comparable sensitivity (78 and 76%, respectively), whereas Ultrio Plus MP8 was significantly (p = 0.017) less sensitive (78% vs. 67%). However, in the lower-viral-load OBI samples the relative sensitivities of the different NAT options mimic those in all HBV samples (Fig. 1), although with lower proportions of reactive replicate test results.
TABLE 1.
Comparison of percentage positive results in replicate tests (n = 6 per sample) of different categories of HBV NAT–yield samples in different ID- and MP-NAT options
| Clinical status | Donations | Number of tests | Ultrio
|
Ultrio plus
|
TaqScreen
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID-NAT
|
MP4
|
MP8
|
ID-NAT
|
MP4
|
MP8
|
MP6
|
||||||||||
| Positive | % | Positive | % | Positive | % | Positive | % | Positive | % | Positive | % | Positive | % | |||
| First WP | 22 | 132 | 110 | 83 | 78 | 59 | 75 | 57 | 123 | 93 | 105 | 80 | 93 | 70 | 111 | 84 |
| Vaccine breakthrough* | 10 | 60 | 44 | 73 | 30 | 50 | 27 | 45 | 51 | 85 | 40 | 67 | 35 | 58 | 38 | 63 |
| Second WP | 15 | 90 | 55 | 61 | 27 | 30 | 20 | 22 | 57 | 63 | 32 | 36 | 22 | 24 | 13 | 14 |
| OBI† | 29 | 174 | 87 | 50 | 51 | 29 | 30 | 17 | 129 | 74 | 78 | 45 | 48 | 28 | 68 | 39 |
| OBI a-HBs‡ | 28 | 168 | 91 | 54 | 58 | 35 | 40 | 24 | 120 | 71 | 78 | 46 | 56 | 33 | 66 | 39 |
| Unknown | 3 | 18 | 8 | 44 | 5 | 28 | 4 | 22 | 12 | 67 | 6 | 33 | 7 | 39 | 4 | 22 |
| All | 107 | 642 | 395 | 62 | 249 | 39 | 196 | 31 | 492 | 77 | 339 | 53 | 261 | 41 | 300 | 47 |
Probable vaccine breakthrough infections.
Anti-HBs–negative OBI.
Anti-HBs–positive OBI.
Fig. 1.
Comparison of percentage positive results in replicate tests (n = 6 per sample) on 107 HBV NAT–yield samples in different ID- and MP-NAT options.
Fig. 2.
Comparison of percentage positive results in replicate tests (n = 6 per sample) on 32 acute-phase WP donations (including 10 probable vaccine breakthrough samples) and 57 OBI donations. (
 ) WP (n = 32); (
) WP (n = 32); (
 ) OBI (n = 57).
) OBI (n = 57).
HBsAg-yield samples
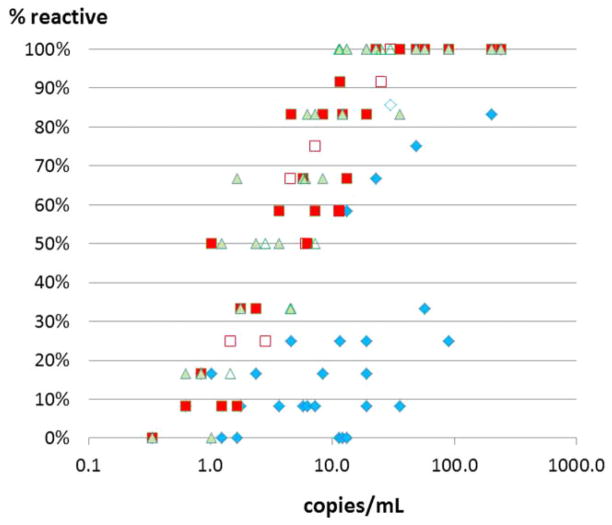
One can imagine that in the HBsAg-yield samples that were selected because of absence of reactivity in the Ultrio assay the relative sensitivities of the NAT options will be different from those in the HBV NAT–yield samples that were selected because of presence of Ultrio reactivity. Indeed the overall proportion of reactive test results in replicate (n = 12) testing of 29 HBsAg-yield samples increased from 80 of 348 (23%) in Ultrio to 225 of 348 (65%) in the Ultrio Plus assay (p < 0.00001), whereas the TaqScreen assay detected a slightly but not significantly (p = 0.0997) higher proportion of 125 of 174 (72%). When comparing reactivity rates between Ultrio Plus and TaqScreen on the 29 individual samples, there were two samples detected significantly more sensitive by either assay (Table 2). Eight samples that were 100% reactive in Ultrio Plus and TaqScreen were also tested in 1:8 dilution and these were detected in 59 of 96 (61%) and 31 of 48 (65%) of replicate assays, respectively. Figure 3 shows the proportion of reactive results in the individual HBsAg-yield samples and the 1:8 diluted samples plotted against the viral load estimated by probit analysis (see Materials and Methods). The graph shows a comparable dose–response relation for Ultrio Plus and TaqScreen but variable lower response rates in the Ultrio assay. These data were used for determining the relative analytical sensitivity of the three NAT methods in this subset of HBsAg-yield samples (see below). One HBsAg- and anti-HBc–confirmed-positive sample was not detected in any of the replicate Ultrio, Ultrio Plus, and TaqScreen assays and thus had a viral load of less than 0.6 copies/mL.
TABLE 2.
Proportion of replicate assays that were reactive on 29 Ultrio initially nonreactive HBsAg carrier samples and individual results on four samples with significantly different reactivity rates between the Ultrio Plus and TaqScreen assays
| Sample | copies/mL | Ultrio† | Ultrio Plus† | TaqScreen† | p value* |
|---|---|---|---|---|---|
| 1 | 1.6 | 0/12 (0) | 1/12 (8) | 4/6 (67) | 0.009 |
| 2 | 4.6 | 3/12 (25) | 10/12 (83) | 2/6 (33) | 0.034 |
| 3 | 1.2 | 0/12 (0) | 1/12 (8) | 3/6 (50) | 0.045 |
| 4 | 1.0 | 2/12 (17) | 6/12 (50) | 0/6 (0) | 0.003 |
| 5–29 | 0.3–240 | Not shown | Not shown | Not shown | NS |
| All (n = 29) | 0.3–240 | 80/348 (23)‡ | 225/348 (65)‡ | 125/174 (72) | 0.0997 |
p value for comparison Ultrio Plus versus TaqScreen.
Data are reported as number (%).
p < 0.0001.
NS = not significant.
[Correction added after online publication 29-Apr-2013: Footnotes have been updated.]
Fig. 3.
Comparison of percentage positive results in replicate tests (n = 12 per sample) of the Ultrio, Ultrio Plus, and TaqScreen assays on 29 initially Ultrio ID-NAT–nonreactive HBsAg-positive donor samples. (
 ) Ultrio neat; (
) Ultrio neat; (
 ) Ultrio 1:8; (
) Ultrio 1:8; (
 ) Ultrio Plus neat; (
) Ultrio Plus neat; (
 ) Ultrio Plus 1:8; (
) Ultrio Plus 1:8; (
 ) TaqScreen neat; (
) TaqScreen neat; (
 ) TaqScreen 1:8.
) TaqScreen 1:8.
Analytical sensitivity of NAT options
Tables 3 and 4 compare the analytical sensitivity of Ultrio, Ultrio Plus, and TaqScreen on three panels: 1) the Eurohep HBV DNA standard, 2) 107 Ultrio NAT-yield sample dilutions, and 3) 29 HBsAg-positive and Ultrio-negative samples. By plotting the HBV DNA concentrations of (diluted) samples on the X-axis and the proportion of reactive results on the Y-axis, the 95 and 50% LODs and the relative sensitivities were determined by parallel line probit analysis. The HBV DNA concentrations in the individual clinical yield samples had first been determined by probit analysis of Ultrio Plus and TaqScreen response rates against the Eurohep Genotype A2 standard (see Materials and Methods). Comparison of replicate tests by Ultrio and Ultrio Plus on the Eurohep standard and on the clinical Ultrio NAT-yield sample dilution series showed a 2.1 (1.2–4.0)- and 2.5 (2.0–3.1)-fold enhancement of the analytical sensitivity by the TER reagent, respectively. However, a much larger 43 (11–350)-fold improvement of the sensitivity of Ultrio Plus relative to Ultrio was observed when testing the HBsAg-positive samples that were missed by Ultrio ID-NAT screening. In this analysis the Ultrio Plus response rates on 1:8 dilutions were taken instead of the rates on the undiluted samples when 100% Ultrio Plus reactivity rates were found on the undiluted samples. This was the case for seven samples with HBV DNA concentrations varying between 11.6 to 201 copies/mL that were 8% to 83% of the time reactive in the Ultrio assay and another sample of 240 copies/mL that was 100% Ultrio reactive. The TaqScreen assay was marginally (2.0 [1.0–4.2]-fold, p = 0.05) more sensitive than the Ultrio Plus assay on the Eurohep standard dilutions, but the difference in analytical sensitivity was not significant on the HBV NAT– and HBsAg-yield samples (Table 4).
TABLE 3.
Analytical sensitivity (50% and 95% detection limits) of NAT systems on (dilutions of) the Eurohep HBV DNA genotype A2 standard, Ultrio HBV NAT–yield samples, and HBsAg-positive and Ultrio initially nonreactive samples as determined by parallel line probit analysis
| LOD (copies/mL)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of
|
Ultrio
|
Ultrio Plus
|
TaqScreen
|
||||||
| Samples | Dilutions | Replicates | 50% (CI) | 95% (CI) | 50% (CI) | 95% (CI) | 50% (CI) | 95% (CI) | |
| Eurohep standard | 1 | 10 | 24 | 9.3 (6.4–13.7) | 90.2 (55.9–165) | 4.5 (3.0–6.6) | 43.1 (26.8–77.9) | 2.2 (1.3–3.9) | 21.4 (11.7–43.8) |
| HBV NAT yields | 107 | 3 | 6 | 10.6 (9.2–12.3) | 120 (94.8–157) | 4.3 (3.7–5.0) | 48.7 (39.3–62.2) | 4.6 (3.6–5.9) | 51.7 (38.3–71.7) |
| HBsAg yields | 32 | 1* | 12 | 375 (156–1267) | 1248 (564–3974) | 4.0 (2.8–5.6) | 34.8 (26.8–139) | 2.5 (1.4–4.1) | 37.9 (20.6–89.2) |
Eight samples that were 100% reactive in Ultrio Plus and TaqScreen were tested in 1:8 dilution to compare percentage response in these assays with that on the undiluted samples in the Ultrio assay.
TABLE 4.
Relative analytical sensitivity of NAT systems on (dilutions of) the Eurohep HBV DNA genotype A2 standard, Ultrio HBV NAT–yield samples, and HBsAg-positive and Ultrio initially nonreactive samples as determined by parallel line probit analysis
| Panel | Sensitivity (CI)
|
||
|---|---|---|---|
| TaqScreen relative to Ultrio | Ultrio Plus relative to Ultrio | TaqScreen relative to Ultrio Plus | |
| Eurohep standard | 4.21 (2.01–9.98) | 2.09 (1.21–3.87) | 2.01 (1.02–4.23) |
| HBV NAT yields | 2.32 (1.72–3.18) | 2.46 (1.97–3.13) | 0.94 (0.71–1.26) |
| HBsAg yields | 93.9 (17.8–1135) | 43.1 (10.7–350) | 1.60 (0.89–3.10) |
DISCUSSION
The introduction of ID-NAT screening with the Ultrio assay in South Africa has been successful not only in eliminating HIV transmission risk10 but also in interdicting a large number of HBV WP and OBI donations.8 Over 4 years of ID-NAT screening the acute-phase and chronic HBV NAT–yield rates were 1:19,600 and 1:16,500, respectively, and in first-time donors even 1:6800 and 1:3500.8 Unlike in most other countries where ID-NAT is introduced, the yield rate in our donor population was almost as high in the acute phase as in the chronic phase and the pre–ID-NAT WP risk was estimated at 1:36,800.8 In this environment, one can benefit from more sensitive HBV DNA detection.
We performed this head-to-head comparison study to understand the impact of replacing the Ultrio assay with the Ultrio Plus assay. For this purpose the relative sensitivity of the Ultrio and Ultrio Plus assay was evaluated on dilutions of multiple Ultrio ID-NAT–yield samples (n = 107), mimicking ID, MP4, and MP8 configurations of the assays and was compared to the TaqScreen MP6 method as a reference assay. The first observation was that the old Ultrio assay performed quite well on replicate repeat testing of the South African HBV NAT–yield samples. On sixfold replicate assays per sample Ultrio detected 62% whereas TaqScreen MP6 detected 47% (p < 0.0001; p < 0.03 if tested in singlet). This result was in contrast with the findings of a similar comparison study of Stramer and colleagues,6 who recently reported a slightly (but in singlet assays not significantly) higher sensitivity of TaqScreen MP6 than of Ultrio ID-NAT in detecting HBV DNA in 129 Thai MP6-NAT (TaqScreen)-yield samples (57% vs. 52%, p = 0.53). This tells us that the relative sensitivity of the NAT systems on the Thai HBV genotypes (predominantly Genotypes B and C), or the South African (predominantly Subgenotype A1) strains cannot be simply translated to the performance of the NAT systems on other HBV genetic variants prevalent around the world. One also must bear in mind that there may be a bias in our comparison study because of the selection of the yield samples by screening with the Ultrio assay and likewise by the TaqScreen assay in the study with the Thai samples.4–6 In this context it was interesting to see that the proportion of replicate test results increased threefold from 23 to 65% in Ultrio Plus when 29 HBsAg-yield samples were tested that were initially missed by the Ultrio screening assay (and thereby introducing a negative selection bias). The clinical sensitivity of the Ultrio Plus assay in the HBsAg-yield samples was comparable to the TaqScreen assay in ID format that detected 72% of the replicates.
When comparing the analytical sensitivity of the two Ultrio assay versions on the Eurohep Genotype A2 standard, we found only a modest 2.1-fold improvement in 50% LODs from 9.4 to 4.5 copies/mL, which would reduce the infectious WP according to the mathematical formulas described by Weusten and colleagues11 from 15.3 to 12.6 days.8 A similar 2.5-fold improvement of the analytical sensitivity was found when the response rates of the two assay versions were compared on the dilutions of the clinical Ultrio ID-NAT–yield samples, the vast majority of which is of HBV Subgenotype A1. However, when probit analysis was performed on the proportions of reactive replicate test results on 29 HBsAg-positive samples missed by routine screening with Ultrio the overall improvement factor by the TER in the Ultrio Plus assay was estimated to be 43 (11–350)-fold, whereas the TaqScreen assay was even 93 (18–1135)-fold more sensitive than Ultrio (Table 4). The dramatic increase in the proportion of reactive results in this subset of HBsAg-yield samples indicates that there may be a subpopulation of HBV strains in South African donors that is poorly detected by the Ultrio assay and will be picked up with more than 10-fold higher sensitivity by the addition of the TER in the Ultrio Plus assay. A similar hypothesis was formulated by Tsoi and coworkekrs12 who used mathematical modeling to explain a more than twofold higher WP NAT yield in Hong Kong blood donors after the Ultrio assay was replaced by the Ultrio Plus assay. Since similar results were found by us in South Africa13 our earlier reported residual risk estimates based on 95 and 50% LODs of the Ultrio assay on the Eurohep HBV Genotype A2 standard8 need to be reassessed. A closer look at the proportion of reactive results of replicate Ultrio, Ultrio Plus, and TaqScreen assays on individual HBsAg-yield samples (Table 2) and on NAT-yield samples (data not shown) confirms the variation in relative sensitivity between the three NAT methods. However, the largest variability in relative sensitivity for individual samples was found with the Ultrio assay, as was also found by other investigators. For example, Grabarczyk and colleagues14 comparing the analytical sensitivity of Ultrio Plus relative to Ultrio on dilutions of different HBV samples of Genotypes A through G found improvement factors varying between 1.3- and 8.7-fold depending on the genotype (or strain within a genotype).
One of the explanations of the differences in relative sensitivity enhancement factors caused by the alkaline shock during the target capture reaction step in the Ultrio Plus assay is the variable length of the double-stranded (ds) DNA region of the circular HBV genome.15 One hypothesis is that in a relatively high proportion of Genotype A1 viruses in the samples selected for this study the target region of the capture probes was single-stranded so that the improvement factor provided by the TER (lithium hydroxide) in Ultrio Plus assay is mainly the effect of a more efficient disruption of viral capsids and release of HBV DNA. By contrast in other strains the dsDNA portion may be longer and here TER could also contribute to the reaction efficiency by the unwinding of dsDNA, so increasing access of the target capture probes to single-stranded HBV DNA. It is therefore possible that the modest enhancement of the sensitivity in detecting HBV Genotype A1 NAT–yield samples in our study from 62% by Ultrio to 77% by Ultrio Plus would be higher when unselected samples of different genotypes would have been used for comparison. Probably the quickest way to shed light on this issue is by comparing the analytical sensitivity of the three NAT methods on a larger panel of Ultrio Plus–yield samples13 and HBsAg-positive samples of different subgenotypes, such as the WHO HBV genotype reference panel, which is composed of multiple samples and sub-genotypes per genotype.16
When using the dilutions of the clinical Genotype A1 NAT–yield and HBsAg-yield samples in our study for comparison of the analytical sensitivity we found that Ultrio Plus and TaqScreen in ID-NAT configuration had equal HBV detection capacity, although the TaqScreen assay was marginally (twofold) more sensitive on the Eurohep Genotype A2 reference panel, possibly reflecting differing relative sensitivities between the A1 and A2 subgenotypes. As a consequence the difference in clinical sensitivity of the Ultrio Plus and TaqScreen NAT options on the South African yield samples is mainly determined by the pool size. Although it is hoped that this also holds for other genotypes than A1, this cannot be claimed unless a comparison study proves equal analytical sensitivity of the Ultrio Plus and the TaqScreen assay on a series of samples of multiple subgenotypes,16 not selected by one of the commercial assays.
In conclusion, this head-to-head comparison study of the sensitivity of NAT options showed the superiority of Ultrio Plus and TaqScreen ID-NAT screening for HBV DNA. Since the Ultrio Plus and TaqScreen assays were found to be equally sensitive in the South African yield samples, it is expected that the efficacy of the currently available commercial NAT systems in preventing HBV transmission is mainly a function of the pool size. [Correction added after online publication 29-April-2013: Last paragraph has been updated.]
Acknowledgments
Funding: Novartis Diagnostics funded this study.
ABBREVIATIONS
- dsDNA
double-stranded DNA
- ID
individual donation
- LOD(s)
limit(s) of detection
- MP4 (-6, -8)
minipools of four (six or eight) donations
- OBI
occult hepatitis B virus infection
- SANBS
South African National Blood Service
- TER
target enhancer reagent
- WP
window period
Footnotes
CONFLICTS OF INTEREST
Dr Nico Lelie is currently working as a consultant for Novartis Diagnostics and all the other authors have no conflicts of interest.
References
- 1.Assal A, Barlet V, Deschaseaux M, Dupont I, Gallian P, Guitton C, Morel P, David B, De Micco P. Comparison of the analytical and operational performance of two viral nucleic acid test blood screening systems: Procleix Tigris and cobas s 201. Transfusion. 2009;49:289–300. doi: 10.1111/j.1537-2995.2008.01965.x. [DOI] [PubMed] [Google Scholar]
- 2.Assal A, Barlet V, Deschaseaux M, Dupont I, Gallian P, Guitton C, Morel P, Van Drimmelen H, David B, Lelie N, De Micco P. Sensitivity of two hepatitis B virus, hepatitis C viris (HCV) and human immunodeficiency virus (HIV) nucleic acid test systems relative to hepatitis B surface antigen, anti-HCV, anti-HIV and p24/anti-HIV combination assays in seroconversion panels. Transfusion. 2009;49:301–10. doi: 10.1111/j.1537-2995.2008.01966.x. [DOI] [PubMed] [Google Scholar]
- 3.Margaritis AR, Brown SM, Seed CR, Kiely P, D’Agostino B, Keller AJ. Comparison of two automated nucleic acid testing systems for simultaneous detection of human immunodeficiency virus and hepatitis C RNA and hepatitis B DNA. Transfusion. 2007;47:1783–93. doi: 10.1111/j.1537-2995.2007.01343.x. [DOI] [PubMed] [Google Scholar]
- 4.Phikulsod S, Oota S, Tirawatnapong T, Sakuldamrongpanich T, Chalermchan W, Louisirirotchanakul S, Tanprasert S, Chongkolwatana V, Kitpoka P, Phanuphak P, Wasi C, Nuchprayoon C. One-year experience of nucleic acid technology testing for human immunodeficiency virus Type 1, hepatitis C virus, and hepatitis B virus in Thai blood donations. Transfusion. 2009;49:1126–35. doi: 10.1111/j.1537-2995.2009.02176.x. [DOI] [PubMed] [Google Scholar]
- 5.Louisirirotchanakul S, Oota S, Khuponsarb K, Chalerm-chan W, Phikulsod S, Chongkolwatana V, Sakuldamrong-panish T, Kitpoka P, Chielsilp P, Tanprasert S, Tirawatnapong T, Wasi C Working Group for NAT Study in Thai Blood Donations. Occult hepatitis B virus infection in Thai blood donors. Transfusion. 2011;51:1532–40. doi: 10.1111/j.1537-2995.2010.03023.x. [DOI] [PubMed] [Google Scholar]
- 6.Stramer SL, Krysztof DE, Brodsky JP, Fickett TA, Reynolds B, Phikulsod S, Oota S, Lin M, Saldanha J, Kleinman SH. Sensitivity comparison of two Food and Drug Administration–licensed, triplex nucleic acid test automated assays for hepatitis B virus DNA detection and associated projections of United States yield. Transfusion. 2011;51:2012–22. doi: 10.1111/j.1537-2995.2011.03140.x. [DOI] [PubMed] [Google Scholar]
- 7.Allain JP, Belkhiri D, Vermeulen M, Crookes R, Cable R, Amiri A, Reddy R, Bird A, Candotti D. Characterization of occult hepatitis B virus strains in South African blood donors. Hepatology. 2009;49:1868–76. doi: 10.1002/hep.22879. [DOI] [PubMed] [Google Scholar]
- 8.Vermeulen M, Dickens C, Lelie N, Walker E, Coleman C, Keyler M, Reddy R, Crookes R, Kramvis A. Hepatitis B virus transmission by blood transfusion during four years of individual donation nucleic acid testing in South Africa: estimated and observed window period risk. Transfusion. 2011;52:880–92. doi: 10.1111/j.1537-2995.2011.03355.x. [DOI] [PubMed] [Google Scholar]
- 9.Heermann KK, Gerlich WH, Chudy M, Schaefer S, Thomssen R. Quantitative detection of hepatitis B virus DNA in two international reference preparations. J Clin Microbiol. 1999;37:68–73. doi: 10.1128/jcm.37.1.68-73.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeulen M, Coleman C, Mitchel J, Reddy R, Van Drimmelen H, Ficket T, Busch M, Lelie N. Comparison of HIV assays in window phase and elite controller samples: viral load distribution and implications for transmission risk. Transfusion. 2013;53:2384–98. doi: 10.1111/trf.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weusten J, Vemeulen M, Van Drimmelen H, Lelie N. Refinement of a viral transmission risk model for blood donations in serconversion window phase screened by nucleic acid testing in different pool sizes and repeat test algorithms. Transfusion. 2011;51:203–15. doi: 10.1111/j.1537-2995.2010.02804.x. [DOI] [PubMed] [Google Scholar]
- 12.Tsoi WC, Lelie N, Lin CK. Enhanced detection of hepatitis B virus in Hong Kong blood donors after introduction of a more sensitive transcription mediated amplification assay. Transfusion. 2013;53:2477–88. doi: 10.1111/trf.12165. [DOI] [PubMed] [Google Scholar]
- 13.Vermeulen M, Coleman C, Lelie N, Van Drimmelen H, Reddy R. Enhancement of HBV NAT yield after introduction of the Ultrio Plus assay. Vox Sang. 2012;103(Suppl):161. (Abstract P-295) [Google Scholar]
- 14.Grabarczyk P, Van Drimmelen H, Kopacz A, Gdowska J, Liszewski G, Piotrowski D, Górska J, Kuśmierczyk J, Candotti D, Lętowska M, Lelie N, Brojer E. Head to head comparison of two transcription mediated amplification assay versions for detection of hepatitis B virus, hepatitis C virus and human immunodeficiency virus type 1 in blood donors. Transfusion. 2013;53:2512–24. doi: 10.1111/trf.12190. [DOI] [PubMed] [Google Scholar]
- 15.Landers TA, Greenberg AB, Robinson WS Structure of Hepatitis B. Dane particle DNA and nature of the endogenous DNA polymerase reaction. J Virol. 1977;23:368–76. doi: 10.1128/jvi.23.2.368-376.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chudy M, Hanschmann KM, Kress J, Nick S, Campos R, Wend U, Gerlich W, Nübling CM. First WHO International Reference Panel containing hepatitis B virus genotypes A-G for assays of the viral DNA. J Clin Virol. 2012;55:303–9. doi: 10.1016/j.jcv.2012.08.013. [DOI] [PubMed] [Google Scholar]