Abstract
BACKGROUND
After 3 years of individual-donation nucleic acid test (ID-NAT) screening by the South African National Blood Service (SANBS), a repository of 73 human immunodeficiency virus antibody (anti-HIV)-negative window period (WP)-yield samples and 28 anti-HIV–positive, HIV-RNA–negative elite controllers (ECs) became available for comparison of a p24 antigen (p24 Ag) assay (Innogenetics), two viral load assays (Siemens branch DNA [bDNA] 3.0 and Abbott real-time polymerase chain reaction [RT-PCR]), and three triplex NAT assays (Novartis Diagnostics Ultrio and Ultrio-Plus and Roche TaqScreen) by replicate testing of dilutions.
STUDY DESIGN AND METHODS
Viral loads were assessed by bDNA and RT-PCR assays and if below 100 copies (cps)/mL, by Ultrio limiting dilution probit analysis. The probability of virus transmission by WP and EC donations was estimated for different levels of the 50% minimum infectious dose (ID50) using Poisson distribution statistics.
RESULTS
The equal distribution of WP donations plotted by log HIV-RNA levels indicated a random appearance of donors in the ramp-up phase. The HIV p24 Ag assay detected 45% of WP samples and the cutoff crossing point was estimated at 8140 (bDNA)/ 22,710 (RT-PCR) cps/mL. On replicate retesting of 40 HIV p24 Ag–negative ID-NAT WP-yield samples Ultrio minipool (MP)8, Ultrio-Plus MP8, and TaqScreen MP6 detected 79, 81, and 78%, respectively. Modeling with an estimated ID50 of 31.6 virions/RBC indicated that 15% of p24 Ag–negative ID-NAT WP-yield donations would have transmitted HIV if MP6–8 NAT had been used. Only 2% of RBC transfusions from ECs are estimated to be infectious with a worst-case ID50 estimate of 316 virions.
CONCLUSION
Our analysis of viremia and infectivity of WP and EC donations enables comparison of the efficacy of NAT options in preventing HIV transmission risk.
Simultaneous screening of 2,147,446 South African blood donations over a 3-year period for human immunodeficiency virus (HIV)-RNA by individual-donation nucleic acid testing (ID-NAT) and anti-HIV by chemiluminescence immunoassay has detected a total of 3067 HIV-positive donations, which can be distinguished into three categories of infections:1 1) 2965 concordantly HIV-RNA and antibody-reactive donors (96.7%); 2) 81 antibody-negative, HIV-RNA–positive window period (WP) donations (2.6%); and 3) 21 anti-HIV–positive donors with undetectable HIV-RNA, so-called elite controllers (ECs;2 0.7%). The latter two categories of infection require further confirmatory testing, that is, ideally ID-NAT and anti-HIV on a follow-up sample along with viral load and p24 antigen (Ag) testing as well as replicate triplex NAT or discriminatory (d)HIV assays for the NAT-yield samples and Western blot assay and replicate ID-NAT assays for the EC samples on the index plasma bag. To enable this supplemental testing on potential WP and EC samples, aliquots are taken from the fresh-frozen plasma (FFP) units when available and are frozen at −80°C in a biorepository.
The South African National Blood Service (SANBS) collects and tests between 700,000 and 800,000 donations per annum of which the majority, 80%, are from regular repeat donors. The HIV prevalence in repeat donors (0.019%) is 50-fold lower than that in first-time donors (0.98%). The majority of HIV infections in South Africa are of the Clade C subtype.3 Upon testing of 76 WP donations with sufficient viral load for sequence analysis, 72 (95%) were of Clade C, two of Clade B, one of Clade A, and one of CRF02_AG (unpublished data). Of the 81 WP donations confirmed by anti-HIV seroconversion and/or p24 antigen (p24 Ag) and viral load testing, 73 had sufficient quantity of plasma available for further characterization by 1) a p24 Ag assay (Innogenetics, Ghent, Belgium); 2) two viral load assays (Siemens Versant branch DNA [bDNA] 3.0 assay; Abbott real-time polymerase chain reaction [RT-PCR] assay); and 3) three NAT blood screening assays, that is, the Ultrio and Ultrio Plus assays (Novartis Diagnostics, Emeryville/Gen-Probe, San Diego, CA) and the TaqScreen assay (Roche Molecular Systems, Pleasanton, CA). Twenty-eight WP samples with a viral load below 500 copies/mL (cps/mL) in the bDNA assay were used for preparing twofold dilution panels, which were tested in multiple replicates using the Ultrio and Ultrio Plus assays and the TaqScreen assay (testing performed by Creative Testing Solution, Tampa, FL). The parallel testing of the WP sample dilution panels against HIV-1 Subtype B and C standard dilution panels calibrated in cps/mL allowed us to 1) estimate the HIV-RNA concentration in each of 23 WP samples with viral load below the detection limit of the bDNA assay, 2) compare the relative sensitivity of the three NAT blood screening assays, and 3) compare the sensitivity of p24 antigen testing with that of ID and minipool (MP)-NAT options for the three assays, that is, MP4, MP8, and MP16 for the Ultrio and Ultrio Plus assays and MP6 for the TaqScreen assay.
The mathematical risk analysis methods currently used4,5 assume that donors present randomly during the early WP at the blood bank. However, so far there are no studies confirming that HIV-infected blood donors indeed appear randomly over time in the seronegative window phase. The accurate estimation of the viral loads in the individual WP samples, even below the detection limit of the quantitative HIV-RNA assays, enabled us to examine whether or not the viral loads in the ramp-up phase of viremia are equally distributed relative to the previously documented doubling time.
During 4 years of ID-NAT screening SANBS also identified 28 EC donations for which plasma samples were available in enough volume for testing in multiple replicate Ultrio and Ultrio Plus assays. The comparison of the proportion of reactive results, relative to detection rates on HIV-1 Subtype B and C standard dilution panels, enabled us to estimate the very low concentrations of HIV-RNA in the plasma of these ECs.
In this study, we also estimate the probability of infectivity of the individual WP and EC donations using formulas published by Weusten and coworkers,5 taking the transfused plasma volume, different assumptions for 50% minimum infectious dose (ID50), and the observed viral load distributions of WP and EC donations into account. This made it possible to estimate the transmission risk of the actual WP donations for each of six screening options that were evaluated in the study, that is, p24 Ag, ID-NAT, MP4, MP6, MP8, and MP16 NAT and compare this with the probability of EC donations to be infectious in a theoretical testing scenario based on ID-NAT alone without screening of anti-HIV and p24 Ag.
MATERIALS AND METHODS
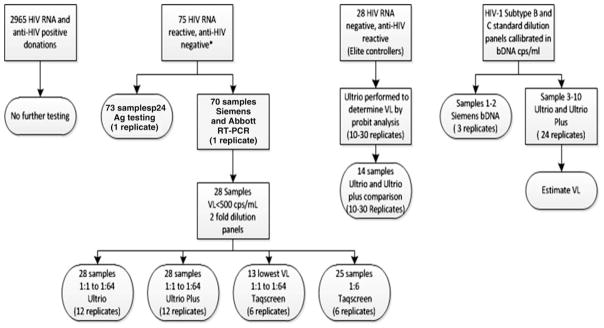
The following paragraphs describe the testing algorithms employed by SANBS from October 2005 to September 2008, to confirm presence of HIV in the samples that were selected for the preparation of test panels used for the comparison of HIV assays. Thereafter, the assays and statistical analyses are described. The flow diagram in Figure 1 summarizes the different HIV assays that were performed on the undiluted and diluted NAT- and serology-yield samples. For this study approval was obtained from the SANBS Human Research Ethics Committee
Fig. 1.
Overview of HIV assays performed on HIV WP and EC samples. VL = viral load.
Testing algorithm and selection of samples
SANBS tests all donations in parallel for HIV-1 using the Abbott Prism HIV-1/2 antibody assay (Abbott Diagnostics, Delkenheim, Germany) and the Procleix Ultrio assay (Novartis Diagnostics) on the Tigris instrument. Any reactive results are repeated in duplicate on Prism and Tigris-Ultrio assays as well as in singlate on the dHIV assay developed to resolve and confirm Ultrio-reactive donations. Concordantly reactive donations in Ultrio, dHIV, and anti-HIV Prism assay are considered confirmed infections and were not part of this study.
HIV WP NAT yields
A potential HIV WP NAT yield is a donation that is Ultrio repeat-reactive and/or dHIV reactive, but anti-HIV nonreactive on Prism. The potential yield case becomes a confirmed HIV-NAT–yield case when the recalled donor seroconverts to anti-HIV and/or p24 Ag in follow-up samples, and/or the index donation plasma is reactive in the p24 Ag assay and/or a viral load assay. Of 81 confirmed WP NAT yields in 3 years of ID-NAT screening 75 samples were tested for both p24 Ag (Innotest, Innogenetics) and viral load in either Siemens Versant bDNA 3.0 or Roche TaqMan PCR assays, as well as in replicate Ultrio and dHIV assays. Forty-two of the 75 WP samples (56%) were negative for p24 Ag and 19 (25%) were also negative in the viral load assay routinely used for confirmation. For these 19 bDNA- or qPCR-negative samples, the presence of HIV-RNA could be confirmed by five replicate Ultrio and dHIV assays on samples taken from the frozen plasma unit. Of the 75 samples two HIV p24 antigen–negative WP samples could not be used for the study because of lack of sample in one case and because of double infection with HBV in another case. Hence, of the 75 p24 antigen and viral load tested WP samples, 73 samples were available for comparing different NAT blood screening options and the p24 Ag assay. Of these samples 70 samples were available for comparison of two viral load assays (Fig. 1).
ECs
A potential “serology-yield” case is a donation that is repeat reactive for HIV antibody on Prism and nonreactive or non–repeat reactive on NAT. Such a donor was classified as an EC when he or she was established to not be on antiviral therapy and when the Western blot was positive but qPCR is negative or yielded a very low viral load. Although the definition of an EC is an HIV-infected person who is not on antiretroviral therapy and has a viral load of fewer than 50 cps/mL for a period of 2 years while anti-HIV is confirmed positive in Western blot,2 we could not follow these donors for 2 years to confirm persistent low-level viremia but nonetheless believe classification as ECs is reasonable. The samples from the plasma units (when available) from ECs were tested in qPCR and in 10 to 30 replicate dHIV and/or Ultrio assays to confirm whether HIV-RNA was detectable. After 4 years of testing 28 confirmed EC cases had available FFP units from which plasma samples were prepared for estimating viral load on the basis of the proportion of dHIV reactive assays. Of these 14 samples were also available in enough quantity for comparison of the reactivity rates in the Ultrio and Ultrio Plus assays.
Preparation of HIV-positive donation samples and dilution panels
SANBS maintains a national plasma repository, which aliquots and stores each virus-positive FFP unit detected during routine blood screening. The frozen plasma bags of the NAT and serology yields are thawed and aliquoted into 3.5-mL aliquots and stored at −80°C. Seventy-three WP samples were available for the comparison studies of HIV viral load and triplex NAT blood screening assays. Each WP NAT–yield plasma sample had already been tested on the bDNA quantitative assay or Roche TaqMan assay by Toga Laboratories (Edenvale, Gauteng, South Africa). Twenty-eight samples that had a viral load less than 500 cps/mL were used to make dilution panels. A doubling dilution was made from 1:1 to 1:64 for each dilution panel. In addition a 1:6 dilution was made of each of the 28 WP donations with a viral load below 500 cps/mL. The dilutions were made with negative human plasma that was filtered through a 113-mm strengthened 24-cm filter (Whatman, Catalog No. 1113-240, GE Healthcare, Waukesha, WI). Each twofold dilution panel comprised of seven members and the 1:6 diluted samples were divided into multiple vials each containing 4.5 mL of plasma. The panels were frozen at −80°C until replicate testing in the NAT assays.
Standard dilution panels
Two commercially available panels with dilution series with known viral concentrations of HIV-RNA Subtype B and Subtype C were supplied by Biologicals Quality Control (BioQControl, DDL Laboratories, Rijswijk, the Netherlands). The HIV-1 RNA concentrations in these panels have been calibrated using multiple replicate tests in the Versant bDNA 3.0 assay (Siemens, Tarrytown, NY).
HIV assays performed on study panels
Viral load assays
The two highest concentrations of each DDL panel were tested in three replicates on the bDNA assay to confirm the quantification of the BioQControl panels in cps/mL. The WP NAT yields that had sufficient plasma stored (n = 70) were tested in one replicate on the bDNA assay. One aliquot of these WP samples was shipped to Abbott Laboratories (Chicago, IL) for testing in the Abbott HIV RT-PCR assay (Abbott Diagnostics).
Triplex NAT assays
The sample Members 3 to 10 of the BioQControl Genotype B and C panel were tested in 24 replicates on the Ultrio and Ultrio Plus assays (Novartis Diagnostics). Each of 28 WP NAT–yield samples that had a viral load below 500 cps/mL was made into a dilution panel and each panel member was tested in 12 replicates on the Ultrio and Ultrio Plus assay. Twenty-eight EC samples were tested undiluted in 10 to 30 replicates in the Ultrio assay. Fourteen of these samples in enough supply were also tested undiluted in 10 to 30 replicate assays in the Ultrio Plus assay. All Ultrio and Ultrio Plus testing was performed at the SANBS virology laboratory.
Thirteen of the twofold dilution panels of the WP NAT yields with the lowest viral loads also had each panel member tested in six replicates with the TaqScreen assay on the s201 system (Roche Molecular Systems), along with the 1:6 dilutions (recommended MP size for the TaqScreen assay) of 25 HIV WP samples with viral loads below 100 cps/mL. To enable sixfold replicate TaqScreen assays two 4.5-mL aliquots of the samples were shipped on dry ice to CTS Laboratories (Tampa, FL). Each aliquot was tested in three separate TaqScreen test runs performed within 48 hours after thawing of the samples. To allow for replicate tests on the MPX s201 TaqScreen system each sample needed to be relabeled before testing with donation bar code and test series numbers.
HIV p24 Ag assay
Each of the WP NAT–yield donations samples were tested on the Innogenetics Innotest mAB p24 Ag assay (Innogenetics) as part of the routine confirmatory assays during operational supplemental testing. This assay is known to be one of the most sensitive p24 Ag assays.6
Statistical analyses and risk modeling
Using the data of all HIV assays entered into spreadsheets we performed a series of statistical analyses described in the following paragraphs.
Sensitivity of NAT options
For each of the 28 WP NAT–yield samples tested by Ultrio and Ultrio Plus in twofold dilutions (undiluted, 1:2, 1:4, 1:8, 1:16, 1:32, and 1:64) we counted the number of reactive results out of 12 replicate assays per sample for ID, MP4, MP8, and MP16 test options. Samples containing viral loads above 500 cps/mL were not tested in dilutions, since it was assumed they would be 12 of 12 (100%) reactive at all MP sizes including MP16 (this assumption was confirmed in dilution panels of WP samples with viral loads between 100 and 500 cps/mL in the bDNA assay). For each of the ID-and MP-NAT options the sum of the total number of reactive results in the 73 anti-HIV–negative WP samples was calculated and divided by the total number of replicates (73 × 12) tested. The same was also done for the ID and MP6 NAT options for the TaqScreen assay, but in this method each undiluted and 1:6 dilution was tested in six replicates. The proportion of reactive results in each of the ten evaluated NAT options was also calculated for the 40 HIV p24 Ag–negative samples to mimic a situation where the anti-HIV screening assay would be replaced by a p24 Ag/anti-HIV combo assay. The proportions of reactive results between the test options were compared by chi-square test. Differences were considered significant when the p value was below 0.05.
Probit analysis
We used probit analysis in a statistical package (SPSS, SPSS, Inc., Chicago, IL) to determine the 95 and 50% detection limits of the NAT methods on the HIV Subtype B and C standard dilution panels that were tested in 24 replicate assays at each concentration of the standards (see Appendix, Table A1). We compared the relative sensitivities of Ultrio, Ultrio Plus, and TaqScreen assays by parallel line probit analysis. For each of the WP samples the sensitivity of Ultrio and Ultrio Plus relative to TaqScreen was determined. If the 95% confidence limits overlap the value 1.0 the difference between the two assays was not significant for that particular sample. For WP donation samples that had RNA levels below the limit of quantitation by the viral load assays the HIV-RNA concentrations were estimated from the replicate qualitative NAT results based on the shift in probit curves in parallel line probit analysis. The very low viral loads in EC samples were estimated by comparing the percentage of Ultrio reactives on the undiluted samples with the probit curve of either the HIV Subtype B or Subtype C standard dilution panels. Finally, parallel line probit analysis was used as a way to estimate the viral load (in cps/mL) at the 50 and 95% HIV p24 Ag seroconversion points based on the results from 75 WP samples. This was done to obtain limit of detection (LOD) estimates for the p24 Ag assay, similar to those used for NAT systems, to enable using the risk analysis method according to Weusten and colleagues5 for comparison of residual risk for the NAT options and p24 Ag testing in the South African donor population.
Transmission risk analysis
We calculated the probability of infection by red blood cells (RBCs) and FFP units with inferred 50% minimum infectious dose (N50) of 3.16, 31.6, 316, and 3160 virions. The infectivity is determined by Poisson statistics and calculated by the formula:
with N the expected number of viral particles in the blood product and pv the individual probability of a viral particle to cause infection. From this equation a simple relation can be derived between the viral load corresponding to aN50 and pv:
From this equation it can be estimated that pv is 0.22, 0.022, 0.0022, and 0.00022 when the ID50 (or N50) is 3.16, 31.6, 316, and 3160 virions, respectively. For RBCs and FFP units we assumed that the transfused plasma volume was 20 and 200 mL, respectively. So, if the viral load in a donation was estimated at 0.5 cps/mL by probit analysis for an RBC unit N was (0.5 × 20)/2 or 5 virions. The probability that an RBC transfusion is infectious can then be calculated using the equation above. For example, for an ID50 of 3.16 virions and 5 transfused virions
for RBC units. However, for a FFP unit from the same donation with N = 50 virions in 200 mL plasma,
For each of the 73 WP donations the probability of an RBC unit to be infectious was estimated using the formulas described above. However, to determine the probability that the lower viral load WP donations would indeed transmit HIV to a recipient the probability of infectivity (calculated as described above) was multiplied with the probability of nondetection in the NAT option evaluated. For this latter variable we did not use the observed proportion of nonreactives in the NAT option, but used the normalized probability of detection deduced from the HIV Subtype C standard probit curve at the viral load that was found by parallel line probit analysis. Hence, for each of the estimated viral loads the probability of nondetection was determined from the probit formula that was entered in a spreadsheet. The product of the two probabilities of infectivity and of non-detection was then calculated for RBC units (20 mL of plasma) for each of the NAT options and for ID50 values of 3.16, 31.6, and 316 virions. We then calculated the sum of the probabilities of each WP donation to be infectious and not detected by the NAT option to estimate the number of undetected transmission cases for these ID50 values. One has to bear in mind that this is the transmission risk on retest of the ID-NAT WP donations by the different NAT options and does not include the residual transmission risk of the WP donations with viral loads below the Ultrio detection limit that were not detected during the 3 years of ID-NAT screening.
For the 28 ECs undetected by routine ID-NAT we used the equation above using ID50 levels of 31.6, 316, or 3160 virions for each individual case for which the HIV-RNA concentration was determined by probit analysis. The overall risk of all EC donations to be infectious is the sum of the probabilities, so it becomes possible to estimate an average probability of a random EC transfusion to be infectious for different ID50 values.
Comparison of quantitative HIV-RNA and p24 Ag assays
The log values of the HIV-RNA concentration reported by the bDNA and Abbott RT assay were plotted against each other, and the slope, correlation coefficient, and intercept were determined by linear regression analysis. The conversion factor between the copy numbers reported by the two assays was deduced from the difference between the intercepts. Similarly, the logit-transformed7 p24 Ag signal-to-cutoff ratio (S/CO) values were plotted against the log of bDNA cps/mL. The sensitivity of the p24 Ag assay, or the S/CO = 1 cutoff crossing point, in cps/mL of WP ramp-up samples was calculated from the point where the linear regression line intersected with the log HIV-RNA scale.
RESULTS
Comparison of quantitative HIV-RNA assays in WP samples
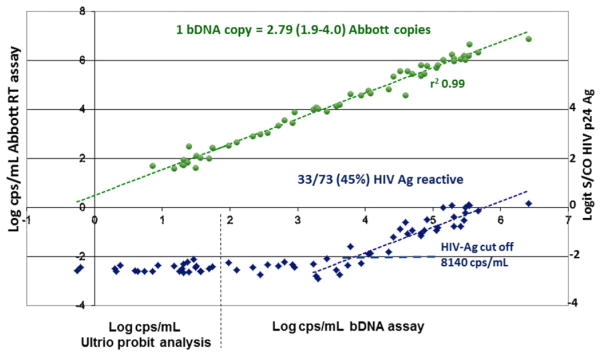
Of 70 SANBS NAT-yield (WP) samples 51 (72.8%) were bDNA reactive and 65 (92.9%) were reactive in the Abbott RT assay, of which seven had viral loads below the 40 cps/mL quantification limit. For 23 WP samples below 100 cps/mL in the bDNA assay, the viral loads were quantified by replicate Ultrio assays on twofold dilutions. Parallel line probit analysis against the DDL HIV Subtype C standards showed that the viral loads in these samples varied between 0.78 and 97.1 cps/mL. The Abbott RT assay reported 2.8 (1.9–4.0)-fold higher concentrations on the quantifiable HIV WP samples than the bDNA assay, according to linear regression analysis (Fig. 2). Apart from a probable systematic underquantification in the bDNA assay for Subtype C according to limiting dilution analysis (see standardization details in the Appendix), there was good agreement between the two viral load assays (r2 = 0.99), which indicates that the WP samples were efficiently detected by both the Siemens bDNA and the Abbott RT assays.
Fig. 2.
Log-linear correlation between HIV-RNA concentration in 70 ID-NAT WP–yield samples measured in cps/mL by Siemens bDNA 3.0 assay (>100 cps/mL) or by Ultrio limiting dilution probit analysis on DDL HIV Subtype C standards calibrated in bDNA cps/mL (<100 cps/mL) and viral load in cps/mL found by Abbott RT-PCR assay (green circles) and Logit S/CO values in Innogenetics HIV p24 Ag assay (blue diamonds).
Comparison of quantitative HIV-RNA and p24 Ag assays on WP samples
Of 73 WP samples, 33 (45%) were reactive in the Innogenetics p24 Ag assay. We performed a linear regression analysis plotting the logit-transformed S/CO values of the p24 Ag assay against the log HIV-RNA concentration and found a cutoff crossing point at 8140 cps/mL (Fig. 2). This crossing point was influenced by one positive p24 Ag result (S/CO, 1.71) on a sample with 5988 cps/mL in the bDNA assay that might have been false reactive on the p24 Ag assay. The two samples with the closest viral loads to this sample had 7394 and 8689 cps/mL and were p24 Ag nonreactive with S/CO values of 0.49 and 0.59. The three samples with the nearest higher viral loads of 14,333, 16,123, and 22,365 cps/mL were borderline p24 Ag reactive, with S/CO values of 1.06, 1.00, and 1.17, respectively. We also approached estimation of the p24 Ag detection limit in the WP ramp-up samples differently by performing a parallel line probit analysis against the HIV Subtype C standard dilution series. The 95% LOD was 64,000 cps/mL and the 50% LOD was 10,000 cps/mL, which is in good agreement with the cutoff crossing point (S/CO, 1.0) of 8140 cps/mL found by regression analysis. The p24 Ag S/CO of 1 cutoff crossing point in the HIV Subtype C WP ramp-up samples would be 22,710 cps/mL when converted to Abbott RT cps/mL (Fig. 2).
Random appearance of donors in the WP
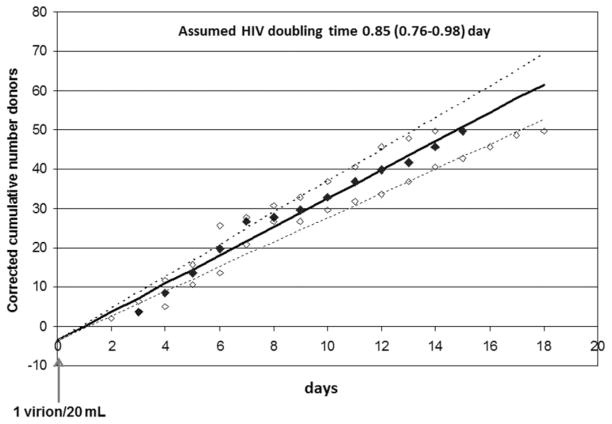
For the analysis of the viral load distribution and the related time that donors donate blood during the WP ramp-up phase we decided to exclude the majority (29/33) of HIV p24 Ag–positive WP samples and restrict the analysis to 47 donations with a bDNA test result below 27,000 cps/mL, which is the end of log linear ramp-up stage according to seroconversion panel studies published by Fiebig and colleagues.8 Up to this level it is assumed that the HIV-RNA growth curve is exponential and not attenuated by the antibody response or lack of target cells to sustain maximal viral replication. The viral loads of the 47 ID-NAT yields in the early ramp-up phase varied from 0.8 to 84 cps/mL for the 25 donors with undetectable viral load in the bDNA assay and from 96 to 26,157 cps/mL in 22 donors with measurable viral load in the bDNA assay. The 15 donors with the lowest viral loads increasing from 0.8 to 17.0 cps/mL were detected with 49.3% to 99.6% probability by replicate ID-NAT testing, according to interpolation based on the HIV Subtype C standard probit curve. To correct for the nondetected cases each donor case with low viral load was divided by the probability of detection. The HIV-RNA concentration was converted into days by using an HIV doubling time of 0.85 days (95% confidence interval [CI], 0.76–0.98) according to Fiebig and colleagues.8 Since each virion particle has two HIV-RNA copies, a concentration of 2 cps/20 mL in an RBC unit or 0.1 cps/mL was arbitrarily set as the start of the infectious WP or t = 0. By translating the viral load into the time of appearance based on the log linear increase of viremia, the number of corrected donor appearances per day (whereby Day 1 was defined as 0.5–1.5, Day 2 as 1.5–2.5, etc.) could be established. We then plotted the corrected cumulative appearance of the donors as the number of cases over the days for doubling times (95% CI) of 0.85 (0.76–0.98) days and found a correlation of 0.988 with linear regression analysis (Fig. 3). The linear increase of the number of donors over time in the WP supports the conclusion that the donors who are in an early stage of HIV infection present to give blood randomly over the first 15 days of the WP.
Fig. 3.
Cumulative number of HIV-infected donors appearing in the early WP ramp-up phase (<27,000 cps/mL) during 3 years of ID-NAT screening corrected for the reduced probability of detection by Ultrio during the initial infection phase (<20 cps/mL). Cumulative appearance of donors at viral doubling time of 0.85 days (◆) and at doubling times of 0.76 and 0.98 days (◇) representing 95% CIs.8
Length of HIV antigen–negative and –positive WPs
Given that we excluded high-viral-load donations from the above analysis because the log-linear relationship between viral load and time no longer exists as antibody seroconversion approaches, the question remained as to whether donors also appear randomly to give blood in the later HIV p24 Ag–positive high-viral-load phase of infection. If we assume donors present at the blood bank at the same rate during the p24 Ag–positive and –negative phases of the WP, the length of these phases can be estimated from the ratio of the number of donors identified in these two stages, which were 45.4 and 35.6, respectively, when corrected for six of 81 incompletely characterized WP infections. The length of the early HIV-RNA+/p24 Ag–WP is 11.5 days based on a doubling time of 0.85 days and using seroconversion points of 0.8 and 10,000 cps/mL (according to calibration in bDNA copies) or 2.3 and 27,900 cps/mL (according to calibration in Abbott RT copies) for the Ultrio and p24 Ag assays, respectively. From the ratio of p24 Ag–positive to p24 Ag–negative WP donors, the p24Ag+ WP is then estimated at 9.0 days. Hence, based on random donor appearance in the acute phase, an HIV-RNA+/anti-HIV (Prism)–negative WP of 20.5 days was estimated assuming that the mean viral doubling time of Genotype C is the same as for Genotype B (0.85 days).8 Note that the pre–ID-NAT WP is not included in this estimate. If the ID50 is one virion/20 mL, the pre–ID-NAT WP is 2.9 or 4.1 days based on the LODs of the Ultrio assay on the Subtype C standard expressed in bDNA and Abbott RT cps/mL, respectively (see Appendix, Table A1).
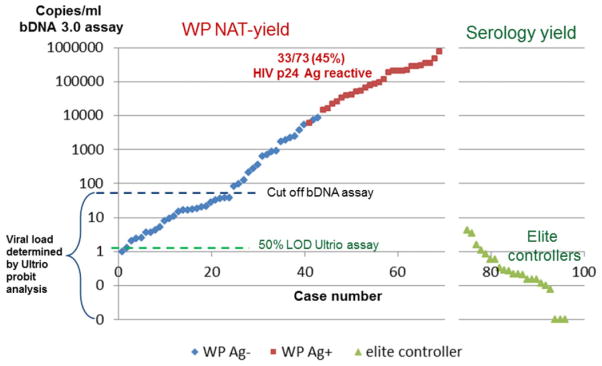
Comparison of viral load distribution in WP and EC samples
Figure 4 compares the viral load distribution as found by probit analysis against the HIV Subtype B standard probit curve in WP and EC samples. Viral load in WP samples varied between 1.2 and more than 500 cps/mL. Nine of 73 WP samples had viral loads below 10 cps/mL (range, 1.2–8.3 cps/mL), which was detectable with 49% to 95% probability in replicate Ultrio assays. Four of 28 samples from ECs (all with viral load <40 cps/mL in Roche TaqMan assay) had similar low viral loads varying from 1.2 to 5.5 cps/mL, detected 48% to 90% of the time by Ultrio in 20 to 25 replicate assays. Twelve of the 28 ECs had even lower viral loads varying between 0.3 and 1.0 cps/mL and were detected 10% to 40% of the time in 10 to 30 replicate Ultrio assays. Two cases were reactive in only one of 19 and one of 29 dHIV replicates and had estimated viral loads of 0.1 to 0.2 cps/mL. Finally, samples from seven donors classified as ECs that had strongly reactive chemiluminescence results and unequivocal positive Western blot patterns were never reactive in 10 to 33 replicate ID-NAT assays. These seven donors were tested in the dual target (LTR/gag) Roche TaqMan assay as well as the single target (LTR) TaqScreen assay to exclude test failure of the dual target (LTR/pol) Ultrio assays, and all had undetectable viral load.
Fig. 4.
Equal distribution of HIV-RNA load (bDNA cps/mL) in samples from WP donations and ECs observed during 3 years of ID-NAT screening.
Relative sensitivity of blood screening NAT assays on WP samples
The relative sensitivities of the Ultrio and Ultrio Plus assays were compared by performing replicates of 12 assays on twofold dilutions of the 23 bDNA assay–negative WP samples. These assays were found to be equally sensitive for detection of low-level HIV-RNA in parallel line probit analysis, except for one sample containing 28 cps/mL that was detected with 2.7 (1.0–7.4)-fold higher sensitivity by the Ultrio Plus assay. The 13 samples with the lowest viral loads were also tested on the same twofold dilution panels in six replicate TaqScreen assays. Four samples were detected at 2.3- to 3.0-fold lower concentrations by Ultrio Plus than by TaqScreen, whereas in nine samples the difference in sensitivity was not significant. When all 13 samples were analyzed together in a parallel line probit analysis, the difference in analytical sensitivity between the Ultrio and TaqScreen assays in ID-NAT configuration was not significant, whereas Ultrio Plus was marginally but significantly (p <0.05) more sensitive (Table 1).
TABLE 1.
Sensitivity of Ultrio and Ultrio Plus relative to TaqScreen on twofold dilution panels of the 13 HIV WP samples with the lowest viral load as determined by parallel line probit analysis from the proportions of reactive results on 12 Ultrio (Plus) and six TaqScreen replicate assays
| WP NAT yield sample | cps/mL to Subype B standard | cps/mL to Subtype C standard | Relative sensitivity (95% CI)
|
|
|---|---|---|---|---|
| Ultrio relative to TaqScreen | Ultrio Plus relative to TaqScreen | |||
| 1 | 1.2 | 0.8 | 1.04 (0.3–3.7) | 1.15 (0.4–4.4) |
| 2 | 1.6 | 1.0 | 2.88 (0.3–346) | 1.73 (0.1–88.3) |
| 3 | 3.2 | 2.0 | 0.61 (0.2–1.5) | 1.23 (0.5–3.2) |
| 4 | 3.8 | 2.4 | 0.88 (0.5–1.7) | 1.42 (0.8–2.3) |
| 5 | 5.7 | 3.6 | 1.74 (0.5–9.0) | 1.16 (0.3–5.0) |
| 6 | 5.8 | 3.7 | 0.85 (0.4–1.7) | 1.38 (0.7–2.8) |
| 7 | 8.3 | 5.3 | 2.43 (1.3–5.7) | 1.17 (0.6–2.2) |
| 8 | 11.4 | 7.3 | 1.42 (0.7–3.1) | 2.98 (1.4–7.9) |
| 9 | 12.5 | 8.0 | 2.95 (1.4–7.5) | 2.29 (1.1–5.4) |
| 10 | 17.1 | 10.9 | 0.39 (0.2–0.8) | 0.85 (0.4–1.7) |
| 11 | 23.0 | 14.7 | 1.24 (0.7–2.3) | 1.28 (0.7–2.4) |
| 12 | 25.9 | 16.5 | 1.64 (0.8–3.6) | 2.46 (1.2–6.2) |
| 13 | 26.2 | 16.7 | 2.93 (1.4–7.9) | 2.36 (1.1–6.0) |
| Overall | all panels | 1.08 (0.86–1.37)* | 1.34 (1.06–1.69)† | |
Difference not significant.
Difference significant (p < 0.05).
Sensitivity of blood screening NAT assays on EC samples
Table 2 shows the proportion of reactive results on multiple replicate Ultrio and Ultrio Plus assays on 14 EC samples. Ultrio and Ultrio Plus detected the samples with equal sensitivity. Overall Ultrio Plus detected 32% and Ultrio 28% of replicates as reactive (p = 0.31).
TABLE 2.
Proportion of reactive results in 14 samples from ECs tested in 10 to 30 replicate Ultrio and Ultrio Plus assays
| Sample | cps/mL | Ultrio
|
Ultrio Plus
|
||||
|---|---|---|---|---|---|---|---|
| Reactive | Number | Percent | Reactive | Number | Percent | ||
| 1 | 0.1 | 1 | 30 | 3 | 2 | 30 | 7 |
| 2 | 0.1 | 1 | 19 | 5 | 2 | 20 | 10 |
| 3 | 0.1 | 0 | 20 | 0 | 2 | 20 | 10 |
| 4 | 0.2 | 2 | 20 | 10 | 1 | 20 | 5 |
| 5 | 0.2 | 2 | 20 | 10 | 3 | 20 | 15 |
| 6 | 0.2 | 2 | 20 | 10 | 4 | 20 | 20 |
| 7 | 0.2 | 3 | 20 | 15 | 5 | 20 | 25 |
| 8 | 0.2 | 4 | 25 | 16 | 3 | 20 | 15 |
| 9 | 0.3 | 5 | 25 | 20 | 4 | 20 | 20 |
| 10 | 0.3 | 5 | 20 | 25 | 8 | 20 | 40 |
| 11 | 0.9 | 13 | 25 | 52 | 13 | 20 | 65 |
| 12 | 1.6 | 7 | 10 | 70 | 10 | 10 | 100 |
| 13 | 3.5 | 20 | 23 | 87 | 15 | 20 | 75 |
| 14 | 4.3 | 18 | 20 | 90 | 17 | 20 | 85 |
| All | 83 | 297 | 28 | 89 | 280 | 32 | |
Sensitivity of ID- and MP-NAT options on WP samples
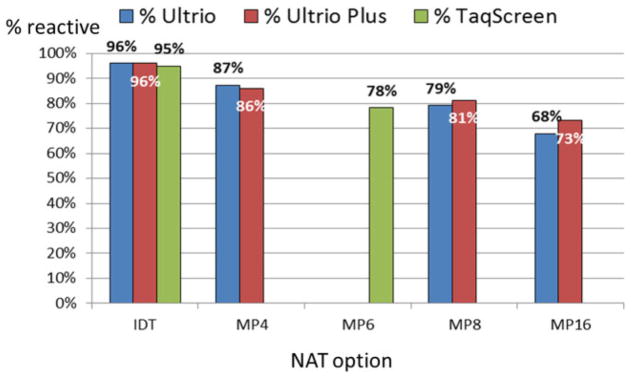
Table 3 shows the proportion of 73 anti-HIV–negative WP samples that tested reactive on 12 or six replicate tests per sample in each of 10 NAT options evaluated. On retest of the eight lowest viral load samples (1.2–12.5 cps/mL) the ID-NAT options detected them 17% to 92% of the time. This explains why the three NAT assays in ID-NAT configuration detected all 73 ID-NAT–yield samples with 97.2% to 97.8% sensitivity on 12 replicate Ultrio and Ultrio Plus assays and six replicate TaqScreen assays per WP sample. However, on diluted samples that mimicked MP4, MP8, and MP16 NAT on Ultrio and Ultrio Plus and MP6 NAT on TaqScreen, the proportions of reactive results on WP samples declined significantly. TaqScreen MP6 detected 88.1% of replicates and had comparable sensitivity to Ultrio MP8 and Ultrio Plus MP8 that detected 88.6 and 89.7% of replicates, respectively. Ultrio MP16 and Ultrio Plus MP16 were significantly less sensitive (82.4 and 85.3%), but were significantly more sensitive than the Innotest p24 Ag assay, which detected 45.2% of samples as reactive. To evaluate a situation in which SANBS would introduce a p24 Ag/anti-HIV assay for serologic testing reducing the incremental yield of NAT, we also compared the relative proportions of reactive assays on replicate testing of 40 samples that were nonreactive on the p24 Ag assay (Fig. 5). Not surprisingly the differences in the proportions of reactive replicate assays between the pool sizes became larger, ranging from 87% for Ultrio MP4, to 79% for Ultrio MP8, and to 68% for Ultrio MP16.
TABLE 3.
Proportion of ID-NAT WP samples detectable by HIV p24 Ag assay and on retest of several ID-NAT and MP-NAT options*
| Pool size | Number of 73 WP samples reactive† (%) in replicate assays
|
|||
|---|---|---|---|---|
| HIV p24 Ag (single) | Ultrio (12 replicates) | Ultrio Plus (12 replicates) | TaqScreen (six replicates) | |
| IDT | 33.0 (45.2) | 71.4 (97.8) | 71.4 (97.8) | 70.8 (97.2) |
| MP4 | 67.9 (93.0) | 67.4 (92.3) | ||
| MP6 | 64.1 (88.1) | |||
| MP8 | 64.7 (88.6) | 65.5 (89.7) | ||
| MP16 | 60.2 (82.4) | 62.2 (85.3) | ||
Data are reported as number (%).
Number of replicates reactive divided by 12 for Ultrio and Ultrio Plus and divided by six for TaqScreen.
Fig. 5.
Proportion of ID- or MP-NAT reactives on retest in 12 or six replicate assays per sample in Ultrio (Plus) and TaqScreen, respectively, of 40 HIV p24 Ag–negative WP donations found during 3 years of ID-NAT screening.
Modeling transmission risk of NAT options in WP
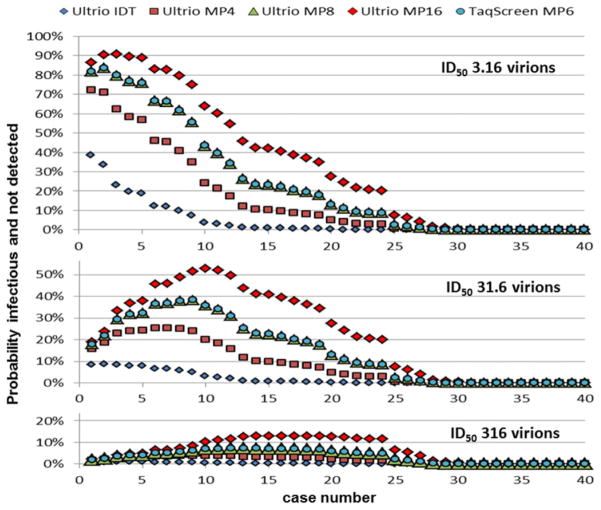
We estimated the probability of HIV transmission from the probability of nondetection and the probability of infectivity given the amount of virus infused by 20 mL of plasma in RBC transfusions using Poisson distribution formulas (see example described under Materials and Methods). The graphs in Figure 6 show the probability of virus transmission on retest of the 40 p24 Ag–negative ID-NAT–yield cases with increasing viral loads ranging from 1.2 to 8689 cps/mL by the different ID- and MP-NAT options for different assumed minimum infectious doses. Table 4 gives the sum of the total number and percentage of HIV transmissions by RBC transfusions that are expected on retest of the 40 p24 Ag–negative WP cases for these ID50 values. For example with a not unlikely scenario of an ID50 of 31.6 virions (see Discussion), MP16 NAT would allow 9.1 transmissions derived from the 40 ID-NAT–positive but p24 Ag–negative donations (23%) to occur. With Ultrio MP8 or TaqScreen MP6 this number would reduce to 5.4 of 40 cases (15%). This implies that 5.4 of 8.9 (66%) and 5.4 of 8.3 (71%) of ID-NAT–yield cases that were not detected by Taqscreen MP6 and Ultrio MP8, respectively, are expected to be infectious with an ID50 of 31.6 virions. With this infectivity level the risk of p24 Ag–negative WP samples transmitting HIV on retest by Ultrio ID-NAT would be 2.0%. Note that the predicted numbers of HIV transmissions in the modeling of Table 4 start from 40 p24 Ag–negative WP samples with viral loads ranging from 1.2 to 8700 cps/mL and do not include the risk of lower viral load WP donations missed by ID-NAT screening. In 2,190,836 donations screened during the 3-year ID-NAT screening period, the risk modeling4 predicts 3.3 undetected HIV transmission cases with an ID50 of 31.6 virions (1.5 ID-NAT–negative infectious RBC units/million).
Fig. 6.
Probability of RBC transfusions from 40 HIV p24 Ag–negative ID-NAT WP–yield donations with increasing viral load from 1.2 to 8700 cps/mL to be infectious and not detected on retest by ID and MP-NAT options calculated for three levels of the minimum infectious dose.
TABLE 4.
Modeling of the number (and percentage) of HIV transmission cases by RBCs from 40 p24 Ag–negative ID-NAT WP–yield donations and 28 ECs as a function of the viral load, the screening option, and the minimum infectious dose*
| RBC from | Range HIV-RNA cps/mL | Number | Screening option | Estimated number (%) of transmission cases according to ID50 in virions
|
|||
|---|---|---|---|---|---|---|---|
| 3.16 | 31.6 | 316 | 3160 | ||||
| p24 Ag–negative ID-NAT–positive WP | 1–10,000 | 40 | Anti-HIV + Ultrio IDT | 1.9 (4.8) | 0.8 (2.0) | 0.1 (0.2) | |
| Anti-HIV + Ultrio MP4 | 6.4 (16.0) | 3.5 (8.8) | 0.7 (1.7) | ||||
| Anti-HIV + Ultrio MP8 | 9.8 (24.5) | 5.9 (14.8) | 1.4 (3.5) | ||||
| Anti-HIV + TaqScreen MP6 | 9.8 (24.5) | 5.9 (14.8) | 1.4 (3.5) | ||||
| Anti-HIV + Ultrio MP16 | 13.6 (34.0) | 9.1 (22.8) | 2.5 (6.2) | ||||
| Ultrio NR EC | 0.1–10 | 28 | Ultrio IDT alone | 4.3 (15.5) | 0.6 (2.2) | 0.06 (0.23) | |
Data are reported as number (%).
Modeling infectivity of EC blood
As described under Materials and Methods the viral load of each of the ECs can be translated to a probability of infectivity by transfusion of 20 mL of plasma by RBC units or 200 mL of plasma by FFP units using Poisson distribution formulas. When the sum of the probabilities is divided by the 28 cases for different assumptions of the minimum infectious dose one can calculate the mean probability of an EC RBC unit being infectious. Table 4 shows the probability of EC transfusions transmitting HIV modeled for different ID50 values. If the ID50 is 316 virions the probability of HIV transmission would be 2.2% for RBC transfusions and 15.5% for FFP transfusions. If, however, the ID50 is 3160 virions, the infectivity would only be 0.2 and 2.2% for RBC and FFP transfusions, respectively. Even if the ID50 of EC blood was similar to the estimated infectivity of HIV by WP transfusions, that is, 31.6 virions (see Discussion), the probability of virus transmission by RBC transfusions of ECs would be only 15.5%.
DISCUSSION
The first objective of our study was to compare the sensitivity of the HIV p24 Ag assay with the currently used triplex ID- and MP-NAT systems for detection of anti-HIV–negative WP donations in South Africa. It was reassuring to establish that there were no important differences in analytical sensitivity between the currently used commercial NAT methods (Ultrio, Ultrio Plus, and TaqScreen) when applied in ID-NAT configuration (Table 1). Only 33 of 73 (45%) ID-NAT WP–yield donations were detected by the Innogenetics p24 Ag assay and 82% to 90% were detectable by MP6- to MP16-NAT (Table 3). This latter proportion reduced to 68% to 81% when only p24 Ag–negative ID-NAT yields were assessed (Fig. 5).
The testing of relatively large numbers of HIV WP and EC donor samples using either undiluted or diluted samples in the different NAT methods allowed us also to determine the viral load distribution in these two categories of HIV-infected donors with discrepant results in NAT and serology assays (Fig. 4). The viral loads in p24 Ag–negative WP samples ranged from 1.2 to 8700 cps/mL, whereas it ranged from 6000 to more than 500,000 cps/mL in p24 Ag–reactive WP samples. By contrast, the viral load in the EC samples was extremely low and ranged from less than 0.1 to 5.5 cps/mL. There was a good correlation between the Siemens bDNA and Abbott RT assays in detecting the Clade C WP samples, but the latter assay reported 2.8-fold higher copy numbers (Fig. 2 and Appendix). However, when IU/mL values are compared the level of consensus between the two viral load assays improves (see Standardization details, Appendix).
The examination of the viral load distribution and comparison of different methods for HIV-RNA quantification provided insight into the fundamentals of the currently used mathematical models for estimation of residual transmission risk,4,5 that is, 1) strong evidence that recently infected donors present randomly over time during the infectious WP (Fig. 3) and 2) accurate quantification in HIV-RNA copies,9 which have a direct relationship with potentially infectious virions (see Appendix). In this context Kleinman and coworkers10 recently reviewed infectivity studies that are relevant for transmission risk analyses.5 The ID50 of HIV-1 in the early WP was estimated to be 3.1 (1–10) virions or 6.2 (2–20) RNA copies if the results obtained by experimental inoculation of simian immunodeficiency virus (SIV) in macaques11 are applicable to human infection. In the anti-SIV–positive phase (viral set point) the infectivity was approximately 100-fold less, due to the neutralizing effect of SIV antibody.11 These authors proposed that for worst-case risk analyses modeling should use an ID50 of 3.1 (1–10) virions in plasma for seronegative WP donations and of 316 (100–1000) virions in plasma for EC donations. However, there are data from human HIV transmission cases by transfusion of stored blood components not detected by MP-NAT that would indicate that the minimum infectious dose of HIV in stored components prepared from WP donations could very well be 10- or even 100-fold higher than the ID50 established in the SIV model, that is, 31.6 (10–100) or even 316 (100–1000) plasma virions per transfused component.12
The p24 Ag concentration in plasma is known to correlate with the logit-transformed S/CO ratios6 and linear regression analysis of results on the SANBS Clade C WP samples showed a parallel increase with the viral load in the ramp-up phase starting at 10,000 cps/mL according to quantification in the bDNA assay9 and 28,000 cps/mL in the Abbott RT assay (Fig. 2). When applying the mathematical formulas for the infectivity of individual WP donations of known viral load in cps/mL as determined in this study, we were able to predict that 15 and 23% of p24 Ag–negative WP donations or 66% to 71% of WP donations missed by MP6-16NAT options are likely to transmit the virus (Fig. 5, Table 4), even if the minimum infectious dose in stored RBC transfusions is 10-fold higher than 3.16 (1–10) virions established in a macaque-SIV infectivity model.11 This analysis confirms the utility of ID-NAT screening for HIV in a high-incidence setting like South Africa, which has interdicted low-level viremic donations at an increasing rate as the donor pool has expanded to include larger proportions of first-time and black donors who are at increased risk of HIV infection.
Our comparative analysis indicates that if MP16-NAT instead of ID-NAT had been used by SANBS 12.8 of 40 (32%) of the p24 Ag–negative WP donations identified during the initial 3 years of ID-NAT screening would have gone unrecognized, and 9.1 of 40 (23%) would likely have transmitted HIV if the ID50 is 31.6 virions. If the infectivity of HIV in stored RBCs from WP donations is 10-fold lower (ID50 of 316 virions), as roughly estimated by Kleinman and colleagues,10 on only a few MP-NAT breakthrough transmission cases, the modeling predicts that only 2.5 of 40 (6%) of p24 Ag–negative ID-NAT–yield donations missed by MP16-NAT would have been infectious.
Not surprisingly, the correct estimation of the minimum infectious dose is crucial for HIV transmission risk analysis and therefore we also looked at data from the United States to shed some light on this issue: Over the past decade MP16-NAT has been used to screen 66 million donations by the American Red Cross, and 32 HIV WP donations have been interdicted.13 Recently the viral load distribution of 29 of these donations was studied in the Abbott RT assay14 and 17 had viral loads below 10,000 cps/mL, which is the level of viremia above which p24 Ag is detectable. If, like in SANBS, 32% of the projected p24 Ag–negative ID-NAT–yield cases would be undetectable by MP16-NAT, an additional eight WP donations would have been detectable had ID-NAT been employed. During the past decade only three donations have been implicated as MP16-NAT breakthrough infections after lookback investigations of seroconverting donors documented by the American Red Cross.15–17 Hence, 3 of 25 (11%) (2/17 [11%] according to ARC data of Dr Susan Stramer, personal communication) projected to be p24 Ag–negative ID-NAT yields in the United States were documented to be infectious. So, if the observed MP16-NAT breakthrough cases in the United States would represent all cases (assuming that no additional unrecognized transmission events have occurred) one could predict that the ID50 of HIV is somewhere between 31.6 virions and 316 virions as can be deduced from our modeling data (compare with Table 4). If, however, half of the MP16-NAT breakthrough HIV transmissions were not recognized, the ID50 is expected to be closer to 31.6 (10–100) virions.
We, therefore, believe that the ID50 of 3.16 (with a range of 1–10) virions, as deduced from the SIV-macaque model11 could be used for worst-case WP risk analysis, but that an ID50 of 31.6 (range of 10–100) virions is probably a better estimate for HIV infectivity in stored blood components from WP donations. With this infectivity level, the residual pre–ID-NAT WP transmission risk estimated according to the method of Weusten and colleagues5 based on 5 years of SANBS data18 is 5.5 and 1.5 transmissions per million donations with ID50 estimates of 3.16 or 31.6 virions, respectively (and Ultrio LODs on Subtype B standard; see Appendix). If p24 Ag testing would have been still in use instead of ID-NAT the risk modeling predicts that 31 HIV transmissions would have occurred per million RBC transfusions with an ID50 of 31.6 virions. To date, after 7 years of screening of 5.3 million donations no HIV transmission events have been established in South Africa following lookback investigations and recipient hemovigilance. Before ID-NAT was introduced the lookback program identified on average two documented transfusion transmissions per year. This observed transmission rate is approximately 15-fold lower than the current risk estimate in case NAT would not have been implemented. The relatively low number of established transmission events before implementing NAT may be due to several factors: 1) the fact that transfusion of blood from black donors known to be at higher risk was avoided at that time, 2) lack of efficiency of lookback programs in general, and 3) a lower infectivity level than the ID50 of 31.6 (10–10) virions used in the risk modeling above. Obviously the introduction of ID-NAT has reduced HIV transmission risk more than 10-fold, documenting the value of this intervention for blood safety on the African continent.
The simultaneous detection of HIV infection by serologic and ID-NAT screening showed that 0.7% of mainly first-time donations in South Africa that tested confirmed HIV antibody positive were negative by ID-NAT. These seropositive donors had viral loads below the 95% LOD of the Ultrio assay and hence were confirmed as ECs. These ECs have been identified among HIV-infected blood donors in several countries and the proportion varies between 1 and 4% depending on the sensitivity of the NAT screening systems employed.19 None of these EC donations in South Africa had quantifiable viral load results in the Roche TaqMan dual target (LTR/gag) viral load assay, confirming that the Ultrio dual target (LTR/pol) assay did not fail to detect any strains of HIV-1.20,21 In a considerable proportion (25%) of EC donors HIV-RNA could not be detected despite 10 to 33 replicate Ultrio assays. Theoretically, these replicate Ultrio-nonreactive donors could have aborted the infection, and further studies are required to examine whether proviral DNA is detectable in their peripheral blood mononuclear cells. Quantification of HIV-RNA by probit analysis showed that the viral load of these ECs varied between 0 and 80 virions/20 mL plasma in RBC units. Although the blood of these donors is not transfused because they are anti-HIV reactive, one wonders whether blood from ECs would be infectious. This question is relevant for a theoretical testing scenario in which p24/anti-HIV combo assays would be replaced by ID-NAT alone (a scenario not likely to be approved of by regulators). Indeed, blood of anti-HIV–positive individuals is not 100% infectious, as shown in studies looking back into recipients of seropositive components before introduction of antibody testing in the United States.22 Eleven percent of antibody-positive transfusions were not infectious, and low viral loads (<1000 cps/mL) and longer storage times (>2 weeks) of RBCs at 4°C were significantly associated with nontransmission. Since the viral load in ECs is extremely low (<10 cps/mL) one could predict that the risk of infection would also be low because the infectivity of the virus in plasma is expected to be 100-fold reduced by neutralizing antibody.11 However, lymphocytes from ECs have been demonstrated to harbor integrated, intact proviral genomes and virus has been isolated from EC lymphocytes using sensitive coculture methods;23 indeed SANBS has been able to detect proviral DNA in blood lymphocytes in a few EC cases (unpublished observations).
To support efficacy and cost-effectiveness analysis of alternative screening scenarios, we thought that it was useful to estimate the potential infectivity of RBC and FFP units of ECs based on the low plasma viral loads found in these infected donors. The mean probability of HIV transmission of the individual EC donations can then be used in further transmission risk modeling and efficacy analysis of different screening scenarios, such as 1) p24 Ag/anti-HIV combo assay, 2) anti-HIV/MP-NAT, 3) anti-HIV/ID-NAT, and 4) ID-NAT alone. Our infectivity modeling study assuming a 100-fold reduced ID50 of 316 virions, as established for transfusions of seropositive blood in the SIV-macaque model,11 estimated an overall probability of 2.2% that HIV in RBCs from ECs is infectious. We propose that this probability be used in modeling the efficacy of alternative screening scenarios comparing p24 Ag/anti-HIV combo assay with and without MP-NAT versus the option of using ID-NAT alone. Since the vast majority of ECs are first-time donors, the latter option of using a dual-target ID-NAT–only screening strategy in repeat and lapsed donors deserves consideration in cost effectiveness models.
Acknowledgments
We thank Novartis Diagnostics for funding this study.
ABBREVIATIONS
- bDNA
branch DNA
- cps
copies
- dHIV
discriminatory human immunodeficiency virus (assay)
- EC(s)
elite controller(s)
- ID
individual donation
- ID50
50% minimum infectious dose
- LOD(s)
limit(s) of detection
- MP
minipool
- N50
50% minimum infectious dose
- p24 Ag
p24 antigen
- SANBS
South African National Blood Service
- S/CO
signal-to-cutoff ratio
- SIV
simian immunodeficiency virus
- WP(s)
window period(s)
APPENDIX
Standardization details
HIV-RNA quantification
The viral loads of the ECs and WP samples of fewer than 100 cps/mL used for the transmission risk analysis in this study were determined by parallel line probit analysis against the BioQControl HIV-1 Subtype B and C reference panels. The manufacturer of these reference panels has tested the HIV subtype stock solutions from which these dilution panels were prepared in multiple bDNA assays in parallel test runs for the calibration in cps/mL (Grabarczyk et al., manuscript submitted for publication). When testing the two highest concentrations of the HIV Subtype B and C standard dilution panels in triplicate assays together with the WP samples in two DNA assay runs we were able to confirm the concentrations given in the package insert of the BioQControl dilution panels (Table A1). The mean bDNA assay value in cps/mL of the total of six replicate assays was 11% lower on the two HIV Subtype B panel members and 6% higher on the Subtype C panel members and thus in line with the values provided by the manufacturer (Table A1). We were therefore confident to combine the HIV-1 concentrations of WP samples of more than 100 cps/mL measured by the bDNA assay with those of fewer than 100 cps/mL determined by probit analysis in the different statistical analyses performed in this study. The 95 and 50% LODs of the Ultrio assay on the HIV Subtype B standard were somewhat higher when expressed in bDNA cps/mL than when expressed in Abbott RT cps/mL in contrast to the HIV Subtype C standard dilutions on which the LODs were higher in the Abbott RT assay (Table A1). According to Poisson distribution the 63% LOD on the Ultrio assay should correspond with one detectable HIV-RNA molecule per assay, which translates to a 63% LOD of 2 cps/mL if the Ultrio would have 100% detection efficiency, since the input volume in the Ultrio assay is 0.5 mL. We found, however, 63% LOD (95% CI) values of 1.8 (1.3–2.6) and 1.2 (0.8–1.7) cps/mL on the HIV Subtype B and C standards, respectively, which indicates that the bDNA assay slightly but significantly underestimates the concentration of HIV-RNA Subtype C by at least a factor of 1.72 (1.18–2.41), assuming that the Ultrio and Ultrio Plus assays operate at 100% detection efficiency on both the HIV Subtype B and the HIV Subtype C standards. We therefore decided to rely on the quantification against the Subtype B standard for the transmission risk analyses in this study. However, to be consistent with the bDNA assay results on the Clade C WP samples we used quantification against the Genotype C standard for the comparison of the viral load assays and the random appearance of donors in the WP (Figs. 2 and 3 of the main article).
TABLE A1.
Analytical sensitivity of Ultrio and Ultrio Plus on HIV-1 Subtype B and C reference panels calibrated in cps/mL by multiple bDNA3.0 or Abbott RT assays*
| cps/mL DDL HIV Subtype B standard | % Ultrio reactive | Geomean cps/mL bDNA (n = 3) | cps/mL DDL HIV subtype C Standard | % Ultrio reactive | % Ultrio Plus reactive | Geomean cps/mL bDNA (n = 3) |
|---|---|---|---|---|---|---|
| 2583 | 2182 | 2882 | 2845 | |||
| 861 | 815 | 961 | 1096 | |||
| 258.3 | 24/24 (100%) | 288.22 | 24/24 (100%) | 24/24 (100%) | ||
| 86.1 | 24/24 (100%) | 96.11 | 24/24 (100%) | 24/24 (100%) | ||
| 25.8 | 24/24 (100%) | 28.82 | 24/24 (100%) | 24/24 (100%) | ||
| 8.6 | 24/24 (100%) | 9.61 | 24/24 (100%) | 23/24 (96%) | ||
| 2.6 | 16/24 (67%) | 2.88 | 22/24 (92%) | 23/24 (96%) | ||
| 0.86 | 8/24 (33%) | 0.96 | 9/24 (38%) | 13/24 (54%) | ||
| 0.26 | 3/24 (13%) | 0.29 | 5/24 (21%) | 7/24 (29%) | ||
| 0.09 | 0/24 (0%) | 0.10 | 2/24 (8%) | 3/24 (13%) | ||
| 95% LOD | 8.3 (5.3–15.3) | In bDNA cps/mL* | 95% LOD | 6.9 (4.2–13.3) | 5.0 (3.1–9.6) | In bDNA cps/mL* |
| 50% LOD | 1.3 (0.89–1.8) | 50% LOD | 0.8 (0.5–1.2) | 0.6 (0.4–0.9) | ||
| 95% LOD | 6.2 (4.0–11.4) | In Abbott RT cps/mL* | 95% LOD | 13.1 (8.0–25.4) | 9.6 (5.9–18.2) | In Abbott RT cps/mL* |
| 50% LOD | 0.9 (0.7–1.3) | 50% LOD | 1.5 (1.0–2.2) | 1.1 (0.8–1.6) |
Abbott RT to bDNA assay conversion factors are 0.75 and 1.91 on HIV Subtype B and C, respectively, as measured on replicate assays on BioQControl-DDL standards.
The regression analysis showed that the Abbott RT assay reported 2.8 (1.9–4.0)-fold higher values than the bDNA assay. This result was not significantly different from a 1.9-fold difference in quantitative values when stock solutions of the BioQControl Subtype C standard were compared in multiple bDNA and RT-PCR assays (data not shown). By contrast when the BioQControl HIV Subtype B standard was compared by replicate testing in these viral load assays the Abbott RT assay reported on average 0.75-fold lower values than the bDNA assay. Similar results were found when the Abbott RT assay was compared with the bDNA assay on a large number of clinical samples of different subtypes in an international multicenter validation study.24
For our comparison study we decided to quantify the HIV-RNA concentration in cps/mL and not in international units (IUs). The BioQControl HIV-1 subtype standard has been calibrated against the WHO standard in multiple bDNA 3.0 assays (Grabarczyk et al., manuscript submitted for publication) and it was found that with replacement of the WHO HIV-1 97/656 standard by the HIV-1 97/650 standard the conversion factor (95% CI) shifted from 0.39 (0.34–0.44) to 0.58 (0.51–0.66) cps/IU. The manufacturers of the Siemens bDNA and Abbott RT-PCR assays have independently determined these conversion factors on only the first WHO HIV-1 97/656 standard and report 0.306 and 0.58 cps/IU, respectively, in the package inserts. Hence if we would have compared the correlation between the two viral load assays on the SANBS Clade C WP samples in IU/mL the 2.8 (1.9–4.0)-fold difference would decrease 1.9-fold to a 1.5 (1.0–2.1)-fold difference. Likewise, the 1.9-fold difference in the concentration in cps/mL in the BioQControl HIV Subtype C standard would disappear when IU/mL values are compared. However, when quantifying the BioQControl Genotype B standard by the two viral load assays in IU/mL the level of consensus would diminish from an Abbot RT/bDNA factor of 0.75 with cps/mL to 0.40 when IU/mL are compared. Further studies are required to confirm that there is a difference in the level of consensus between the two viral load assays for Subtype B and C, a phenomenon that was also observed in another performance evaluation study.24 Nevertheless, in our study on a large number of Clade C WP samples, quantification in IU/mL would have improved the level of consensus between the two viral load assays, showing the value of using a common international standard for quantification.
Importance of accurate quantification of HIV-RNA in cps/mL for transmission risk analysis
Although the introduction of the WHO standard for HIV-1 RNA has improved the comparability of quantitative results between laboratories using different NAT methods, many clinical and blood screening laboratories still express viral load in cps/mL. For blood safety testing there is an important reason why quantification of HIV-RNA in cps/mL is preferred above quantification in arbitrary IUs. There is a direct relationship between HIV-RNA copies and potentially infectious virions. The risk of virus transmission in the early WP ramp-up phase starts when one infectious virus is present in the plasma of a transfused blood component and is determined by the Poisson distribution of infectious viruses in the blood of the early infected donor and the probability that units with that level of viremia would not be detected by the screening assays employed. Weusten and colleagues4 used Poisson distribution statistics to develop a mathematical model for calculating the transmission risk of a WP donation to cause infection in a recipient, which is determined by 1) the amount of plasma in the transfused component, 2) the 95 and 50% LOD of the ID-NAT assay and the MP size, 3) the doubling time of the virus in the WP ramp-up phase, and 4) the estimated infectivity of the virus expressed as the 50% minimum infectious dose (ID50). For this risk model it is essential that both the 95 and 50% lower LODs of the NAT system and the minimum infectious dose (ID50) are expressed in the same RNA copy numbers. The authors are well aware of the fact that the viral load assays are not 100% precise and accurate in the measurement of the number of HIV-RNA molecules, but the in vitro RNA standards used for calibration of the bDNA 3.0 assay have been thoroughly quantified by three different physico-chemical methods.9 Since HIV is a diploid virus we assumed in the risk calculations that one virion carries two genome copies as measured by the bDNA assay. To be able to quantify HIV-RNA in WP and EC samples with viral loads below the detection limit of the bDNA assay we used HIV Genotype B and C standard dilution panels (BioQ-Control, DDL Laboratories, Rijswijk, the Netherlands) that were calibrated in cps/mL using multiple bDNA 3.0 assays and were also quantified by the Abbott RT-PCR assay. Our data indicate that the Siemens bDNA assay underestimates the viral load for HIV Subtype C RNA copies more than 1.7-fold and that the Abbott RT assay is likely to slightly overestimate true genome copy numbers 1.6-fold based on limiting dilution analysis of the Ultrio assay. However, there was no indication of a significant under-detection of the true viral load in cps/mL by the bDNA assay for HIV Subtype B (Table A1). Probably the quantification of viral load in cps/mL by probit analysis against the BioQControl standard dilution panels in our article deviates less than a factor of 2 from true values. Bearing the limitations of standardization of HIV-RNA in true genome copies in mind, we believe that the estimates of the 95 and 50% LODs and ID50 in this article are accurate enough for the purpose of the transmission risk analyses performed in this article.5
Footnotes
CONFLICTS OF INTEREST
NL is contracted by Novartis Diagnostics. The other authors have no conflict of interest.
References
- 1.Vermeulen M, Lelie PN, Sykes W, Crookes R, Swanevelder J, Gaggia L, le Roux M, Kuun E, Gulube S, Reddy R. Impact of individual donation nucleic acid testing on the risk of human immunodeficiency virus, hepatitis B virus and hepatitis C virus transmission in South African donors. Transfusion. 2009;49:1115–25. doi: 10.1111/j.1537-2995.2009.02110.x. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable viral control in the absence of antiretroviral therapy. Immunity. 2007;27:406–16. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Bredell H, Martin DP, Van Harmelen J, Varsani A, Sheppard HW, Donovan R, Gray CM, Williamson C HIVNET028 Study Team. HIV type 1 subtype C gag and nef diversity in Southern Africa. AIDS Res Hum Retroviruses. 2007;23:477–81. doi: 10.1089/aid.2006.0232. [DOI] [PubMed] [Google Scholar]
- 4.Busch MP, Glynn SA, Stramer SL, Strong DM, Caglioti S, Wright DJ, Pappalardo B, Kleinman SH NHLBI-REDS NAT Study Group. A new strategy for estimating risks of transfusion-transmitted viral infections based on rates of detection of recently infected donors. Transfusion. 2005;45:254–64. doi: 10.1111/j.1537-2995.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 5.Weusten J, Vermeulen M, Van Drimmelen H, Lelie N. Refinement of a viral transmission risk model for blood donations screened by NAT in different pool sizes and repeat test algorithms. Transfusion. 2011;51:203–15. doi: 10.1111/j.1537-2995.2010.02804.x. [DOI] [PubMed] [Google Scholar]
- 6.Ly TD, Ebel A, Faucher V, Fihman V, Laperche S. Could the new HIV combined p24 antigen and antibody assays replace p24 antigen specific assays? J Virol Methods. 2007;143:86–94. doi: 10.1016/j.jviromet.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Plikaytis BD, Turner SH, Gheesling LL, Carlone GM. Comparison of standard curve-fitting methods to quantitate Neisseria meningitides group A polysaccharide antibody levels by enzyme-linked immunosorbent assay. J Clin Microbiol. 1991;29:1439–46. doi: 10.1128/jcm.29.7.1439-1446.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–9. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 9.Collins ML, Zayati C, Detmer JJ, Daly B, Kolberg JA, Cha TA, Irvine BD, Tucker J, Urdea MS. Preparation and characterization of RNA standards for use in quantitative branched DNA hybridization assays. Anal Biochem. 1995;226:120–9. doi: 10.1006/abio.1995.1199. [DOI] [PubMed] [Google Scholar]
- 10.Kleinman SH, Lelie PN, Busch MP. Infectivity of human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus and risk of transmission by transfusion. Transfusion. 2009;49:2454–89. doi: 10.1111/j.1537-2995.2009.02322.x. [DOI] [PubMed] [Google Scholar]
- 11.Ma ZM, Stone M, Piatak M, Jr, Schweighardt B, Haigwood NL, Montefiori D, Lifson JD, Busch MP, Miller CJ. High specific infectivity of plasma virus from the pre-ramp up and ramp up stages of acute simian immunodeficiency virus infection. J Virol. 2009;83:3288–97. doi: 10.1128/JVI.02423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinman SH, van Drimmelen H, Lelie N, Busch MP. Minipool NAT HIV-breakthrough transmission case and probability of interdiction by current small pool or individual NAT screening systems [abstract] Vox Sang. 2009;96(Suppl 1):53. 3D-S23-03. [Google Scholar]
- 13.Zou S, Dorsey KA, Notari EP, Foster GA, Krysztof DE, Musavi F, Dodd RY, Stramer SL. Prevalence, incidence, and residual risk of human immunodeficiency virus and hepatitis C virus infections among United States blood donors since the introduction of nucleic acid testing. Transfusion. 2010;50:1495–504. doi: 10.1111/j.1537-2995.2010.02622.x. [DOI] [PubMed] [Google Scholar]
- 14.Tobler LH, Vermeulen M, Stramer SL, Wu S, Busch MP. Distribution of viral loads in US (Clade B) and South Africa (Clade C) NAT yield donors [abstract] Transfusion. 2010;50(Suppl):3A. S1-010A. [Google Scholar]
- 15.Stramer SL, Eder AF, Foster GA, Dy BA, Krysztol D, Trouen-Trend J, Dorsey K, Zou S, Benjamin RJ, Dodd RY. Investigation of reported cases of transfusion transmission of HIV [abstract] Transfusion. 2009;49(Suppl):4A. S2-010A. [Google Scholar]
- 16.Laffoon B, Levi M, Bower WA, Kuehnert M, Brooks JT, Selik RM, Switzer WM, Heneine W, Shankar A, Iuliano AD. HIV transmission through transfusion. Missouri and Colorado, 2008. MMWR Morb Mortal Wkly Rep. 2010;59:1335–6. [PubMed] [Google Scholar]
- 17.Stramer S, Wend U, Candotti D, Foster GA, Hollinger FB, Dodd RY, Allain JP, Gerlich W. Nucleic acid testing to detect HBV infection in blood donors. N Engl J Med. 2011;364:236–47. doi: 10.1056/NEJMoa1007644. [DOI] [PubMed] [Google Scholar]
- 18.Vermeulen M, Coleman C, Gaggia L, Sykes W, Crookes R, Lelie N, Gulube S, Busch M, Reddy R. The effect of change in testing strategy and increasing the black donor base on the HIV window period transmission risk in South Africa: a five year analysis [abstract] Vox Sang. 2011;101(Suppl 1):41. 3D-S12-01. [Google Scholar]
- 19.Laperche S, Morel P, Deschaseaux M, Bouchardeau F, Alimardani G, Guillaume N, Rouger P, Lefrère JJ. HIV antibody screening remains indispensable for ensuring viral safety of blood components despite NAT implementation. Transfusion. 2003;43:1428–32. doi: 10.1046/j.1537-2995.2003.00541.x. [DOI] [PubMed] [Google Scholar]
- 20.Assal A, Barlet V, Deschaseaux M, Dupont I, Gallian P, Guitton C, Morel P, David B, De Micco P. Comparison of the analytical and operational performance of two viral nucleic acid test blood screening systems: Procleix Tigris and cobas s 201. Transfusion. 2009;49:289–300. doi: 10.1111/j.1537-2995.2008.01965.x. [DOI] [PubMed] [Google Scholar]
- 21.Chudy M, Weber-Schehl M, Pichl L, Jork C, Kress J, Heiden M, Funk MB, Nübling CM. Blood screening nucleic acid amplification tests for human immunodeficiency virus type 1 may require two different amplification targets. Transfusion. 2012;52:431–9. doi: 10.1111/j.1537-2995.2011.03281.x. [DOI] [PubMed] [Google Scholar]
- 22.Busch MP, Operskalski EA, Mosley JW, Lee TH, Henrard D, Herman S, Sachs DH, Harris M, Huang W, Stram DO. Factors influencing human immunodeficiency virus type 1 transmission by blood transfusion. Transfusion Safety Study Group. J Infect Dis. 1996;174:26–33. doi: 10.1093/infdis/174.1.26. [DOI] [PubMed] [Google Scholar]
- 23.Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, Lai J, Gandhi SK, Siliciano JD, Williams TM, Siliciano RF. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol. 2007;81:2508–18. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schutten M, Peters D, Back NKT, Beld M, Beuselinck K, Foulongne V, Geretti A-M, Pandiani L, Tiemann C, Niesters HGM. Multicenter evaluation of the new Abbott RealTime assays for quantitative detection of human immunodeficiency virus Type 1 and hepatitis C virus RNA. J Clin Microbiol. 2007;45:1712–7. doi: 10.1128/JCM.02385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]