Abstract
Posttranslational modification (PTM) of proteins and peptides is important for diverse biological processes in plants and animals. The paucity of heterologous expression systems for PTMs and the technical challenges associated with chemical synthesis of these modified proteins has limited detailed molecular characterization and therapeutic applications. Here we describe an optimized system for expression of tyrosine-sulfated proteins in Escherichia coli and its application in a bio-based crop protection strategy in rice.
Tyrosine sulfation is an important posttranslational modification involved in diverse biological processes including immunity and development.1 For example, in humans, tyrosine sulfation of cellular co-receptors mediates their interaction with the gp120 (glycoprotein 120) coat protein of HIV (Human Immunodeficiency Virus).2 The therapeutic potential of tyrosine-sulfated proteins is reflected in a recent report of an engineered sulfated immunoglobulin that neutralized 100% of a diverse panel of neutralization-resistant HIV isolates.3 Historically, tyrosine sulfation was thought to be restricted to eukaryotic biology. However, we recently demonstrated that the plant bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo) harbors a functional tyrosine sulfotransferase, called RaxST (required for activation of XA21-mediated immunity sulfotranserase), that sulfates the 60 amino acid protein RaxX (required for activation of XA21-mediated immunity X), on its central tyrosine (Y41).4,5 Tyrosine sulfated RaxX, but not unsulfated RaxX, triggers an immune response in rice plants carrying the receptor XA21.5 This growing body of research solidifies the role of tyrosine sulfation as a mediator of protein-protein interactions and immune recognition1,6 and demonstrates the relevance of this PTM to human and plant health.
The therapeutic application of posttranslationally modified peptides or proteins requires efficient synthesis. Peptide synthesis of tyrosine-sulfated proteins is technically challenging especially for longer peptides and proteins.1 An alternative approach is to express recombinant proteins together with the corresponding sulfotransferase in E. coli or perform in vitro sulfation assays.1,4 However these strategies often result in heterogeneous sulfation of the target, limiting applications.
In 2006, we described a first-generation system to express sulfated proteins in E. coli that overcomes these drawbacks.7,8 This system relies on an expanded genetic code that enables E. coli cells to direct the incorporation of sulfotyrosine (sY) at UAG (amber) codons during translation. Highly specific incorporation of sY is achieved through a specially-engineered tRNA/aminoacyl-tRNA synthetase (tRNA/aaRS) pair that recognizes sY and the amber codon without cross reacting with endogenous aaRSs and tRNAs.7 This expanded genetic code system has enabled a number of applications including the recombinant production of the therapeutic anticoagulant sulfo-hirudin7,9 and phage display evolution studies on sulfated anti-gp120 antibodies10,11. Several advances in expanded genetic code technology have taken place since these studies were carried out.12,13 Here, we incorporate two main advances into an improved second-generation system for the recombinant expression of tyrosine-sulfated proteins. We demonstrate the high-quantity expression and characterization of highly purified, sulfated RaxX proteins and show that these proteins can induce immunity in rice plants carrying the XA21 immune receptor. This bio-based strategy provides a new avenue for protecting crops from disease.
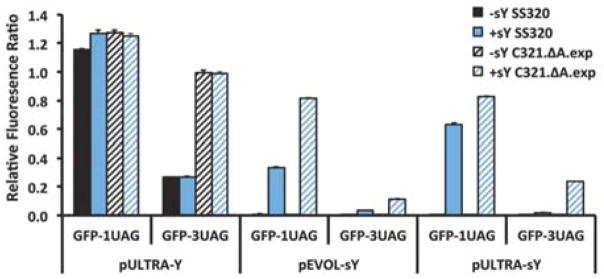
The Schultz group recently developed the pULTRA system for efficient unnatural amino acid incorporation through expanded genetic codes. pULTRA encodes an optimized amber suppressor tRNACUA/aaRS pair that increases the level of tRNA aminoacylation, minimizes tRNA toxicity, and optimizes aaRS expression levels.14 To host tRNACUA/aaRS pairs, the Church and Isaacs groups created a recoded E. coli strain, C321.ΔA.exp, that has all genomic amber stop codons replaced with the alternate stop codon UAA. This strain carries a deletion of release factor 1 (RF1), eliminating active termination at amber stop codons.13 To improve sulfated protein expression, we first cloned our sY-specific aaRS into the pULTRA system. The resulting plasmid, pULTRA-sY (Figure S1), was tested for its ability to insert one or three sYs into GFP (Green Fluorescence Protein) specified by either one or three UAG codons (Figure S1). Comparison of pULTRA-sY with a previous plasmid system for sY incorporation, pEVOL-sY15, revealed that pULTRA-sY achieved substantially higher incorporation of a single sY into GFP in standard SS320 E. coli cells. This advantage was lost when we attempted to incorporate three sYs into GFP, presumably because RF1 competition for UAG codons increases when multiple sY incorporation events are required. However, when we used pULTRA-sY in E. coli strain C321.ΔA.exp, high levels of sY incorporation at single or multiple UAG codons were achieved (Figure 1).
Figure 1. Highly efficient sY protein production using a second-generation sY protein expression system.
Relative fluorescence ratio, with respect to wild type GFP expression, of GFP with one (GFP-1UAG) or three (GFP-3UAG) amber codons. GFPs were expressed in the presence of a control plasmid or plasmids encoding sY incorporation systems (pULTRA-Y and pEVOL-sY or pULTRA-sY) in two E. coli cell lines (SS320 or C321.ΔA.exp). -sY and +sY indicate the absence and presence of 20 mM sY in the growth media. Bars indicate the mean ± SD (n=3).
We next used our improved second-generation expression system for tyrosine-sulfated proteins to produce full-length sulfated RaxX, a protein relevant to crop protection.5 We designed a C-terminal his-tagged RaxX fused to maltose binding protein (MBP) at the N-terminus followed by a 3C protease cleavage site (MBP-3C-RaxX60-His). Because RaxX is normally sulfated at position 41 in Xoo, we specified tyrosine sulfate using an UAG codon at the corresponding position. We expressed MBP-3C-RaxX60-His under the control of the araBAD promoter and induced expression by the addition of arabinose during growth in minimal media. We obtained similar expression levels for RaxX60-Y-His (unsulfated) and RaxX60-sY-His (containing a sulfated tyrosine at position 41) in the presence of 5 mM sY. We obtained up to 4 mg of RaxX60-Y/sY per liter of culture after a three-step purification process including the removal of the N-terminal fusion tag by treatment with 3C protease. The resulting peptides were of a purity > 90%, as estimated by standard LC-MS/MS analysis of trypsin digested protein preparations (Figure S2, Table S1).
Next we assessed the sulfation status of RaxX60-sY by selected reaction monitoring mass spectrometry (SRM-MS). Because of the different physiochemical properties of the sulfated and unsulfated peptide variants, trypsin peptides covering the central tyrosine (Y41) eluted with markedly different retention times (ΔRT ~ 1.1 min). This difference in retention time enabled us to quantify the relative sulfation status in the positive ion-mode despite the loss of sulfate groups from tyrosines during ionization.16 Using our improved sulfated protein expression system, more than 99.5% of the RaxX60-sY tryptic peptides carried a sulfate on Y41, which is well above the sulfation status of three shorter commercially synthesized RaxX-sY peptide variants (Table S2). We were unable to detect any sY-containing peptides derived from RaxX60-Y samples. These results indicate that our optimized second-generation recombinant expression system produces tyrosine-sulfated proteins that are superior to standard commercially synthesized sY peptides.
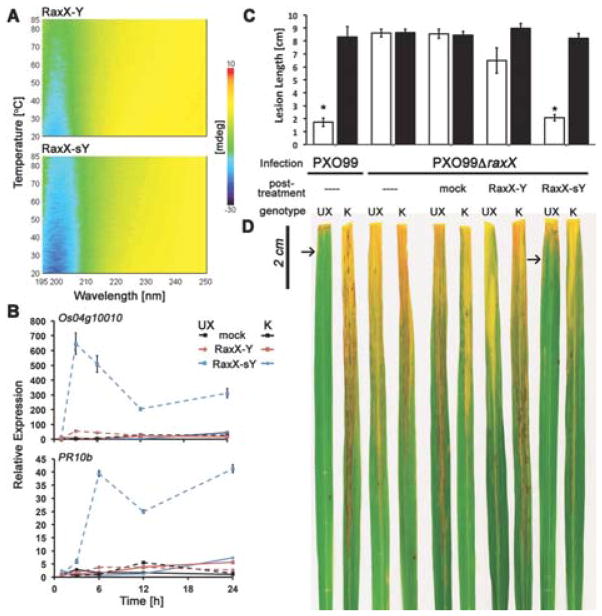
The impact of tyrosine sulfation on protein structure is unpredictable. In some cases it has been shown that sY modifications stabilize intermolecular interactions.9,17 We therefore investigated the structural effect of RaxX60 tyrosine sulfation. Using our highly purified RaxX60-Y and RaxX60-sY, we performed circular dichroism (CD) spectroscopy of RaxX60.18 Standard CD analysis at 20°C indicated that tyrosine sulfation had little impact on the overall fold of RaxX60 with both protein versions being largely disordered with a 20–30% β-strand content, most likely corresponding to a single β-sheet (Figure S3, Table S3).19 Next we tested if sulfation has an impact on protein stability and recorded CD spectroscopy-melting curves. Indeed sulfation had a significant impact on RaxX60 stability with RaxX60-sY keeping its overall fold at much higher temperatures when compared to RaxX60-Y (Figure 2A). This finding suggests that tyrosine sulfation of RaxX60 positively impacts protein stability.
Figure 2. Recombinantly produced tyrosine-sulfated RaxX60-sY induces rice immunity.
(A) Tyrosine 41 sulfation leads to increased thermal stability of the native fold of RaxX60 as measured by CD spectroscopy. (B) Application of RaxX60-sY (500 nM) leads to temporal immune gene activation (Os04g10010 and PR10b) in detached rice leaves in an immune receptor kinase dependent manner (XA21). All data points depict means ± SE (n=3). (C and D) Post-treatment of RaxX60-sY after initial bacterial infection (XooΔraxX) halts disease progression. (C) Bars indicate the mean lesion length ± SE measured 13 days after infection (n ≥ 5). The “*” indicates statistically significant difference from no treatment control (---) using Dunnett’s test (α = 0.01). (D) Rice leaves displaying water-soaked lesions 13 days after infection. Only rice leaves carrying XA21 (UX) treated with RaxX60-sY and the non-infectious control display significantly reduced disease symptoms as highlighted by arrows. UX and K indicates the plant genotypes Ubi::MYC::XA21 and wild type Kitaake control, respectively.
We previously demonstrated that sulfated RaxX variants of different lengths, including a 21 amino acid fragment called RaxX21-sY are immunogenic on rice plants carrying the immune receptor XA21.5 We tested if recombinantly produced sulfated RaxX60-sY is able to induce a similar set of responses. We found that at a concentration of 500 nM, RaxX60-sY activates the rice immune response in an XA21-dependent manner as measured by immune gene expression over time (Figure 2B). RaxX60-sY induced the expression of two well-established rice marker genes (Os04g10010 and PR10b) as early as 3 hours post treatment with a prolonged transcriptional activation observed over 24 hours.
We previously demonstrated that sulfated RaxX variants of different lengths, including a 21 amino acid fragment called RaxX21-sY are immunogenic on rice plants carrying the immune receptor XA21.5
We found that at a concentration of 500 nM, RaxX60-sY activates the rice immune response in an XA21-dependent manner as measured by immune gene expression over time (Figure 2B). RaxX60-sY induced the expression of two well-established rice marker genes (Os04g10010 and PR10b) as early as 3 hours post treatment with a prolonged transcriptional activation observed over 24 hours.
In rice production, resistance to Xoo is agronomically important. Given our observed induction of immunity by RaxX60-sY, we aimed to test whether exogenous application of RaxX60-sY on XA21 rice plants could confer immunity to infectious strains of Xoo. To explore this possibility, we used a variation of a recently established assay20. In this experiment we inoculated rice plants with a virulent strain of Xoo (PXO99AΔraxX) that is not recognized by XA21.5 Two days post-infection, we treated rice leaves with a 1 μM solution of RaxX60-Y or RaxX60-sY. We found that post-treatment with RaxX60-sY dramatically slowed disease progression and symptom development (Figure 2C and D).
Conclusions
We developed an improved second-generation high efficiency expression system for tyrosine sulfated proteins in E.coli and demonstrated its application in immune receptor-mediated crop protection using the model activator-receptor pair RaxX-sY-XA21 in rice. The application of recombinantly produced RaxX60-sY in real field conditions could in principal lead to a reduction of disease progression even after initial infection.
Footnotes
Electronic Supplementary Information (ESI) available: Material and Methods, and Supplementary Figures. See DOI: 10.1039/x0xx00000x
References
- 1.Seibert C, Sakmar TP. Biopolymers. 2008;90:459–477. doi: 10.1002/bip.20821. [DOI] [PubMed] [Google Scholar]
- 2.Stone MJ, Chuang S, Hou X, Shoham M, Zhu JZ. New Biotechnology. 2009;25:299–317. doi: 10.1016/j.nbt.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Gardner MR, Kattenhorn LM, Kondur HR, von Schaewen M, Dorfman T, Chiang JJ, Haworth KG, Decker JM, Alpert MD, Bailey CC, Neale ES, Fellinger CH, Joshi VR, Fuchs SP, Martinez-Navio JM, Quinlan BD, Yao AY, Mouquet H, Gorman J, Zhang B, Poignard P, Nussenzweig MC, Burton DR, Kwong PD, Piatak M, Lifson JD, Gao G, Desrosiers RC, Evans DT, Hahn BH, Ploss A, Cannon PM, Seaman MS, Farzan M. Nature. 2015;519:87–91. doi: 10.1038/nature14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han SW, Lee SW, Bahar O, Schwessinger B, Robinson MR, Shaw JB, Madsen JA, Brodbelt JS, Ronald PC. Nature communications. 2012;3:1153. doi: 10.1038/ncomms2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJG, Luu DD, Chen H, Bahar O, Daudi A, Vleesschauwer DD, Caddell D, Zhang W, Zhao X, Li X, Heazlewood JL, Ruan D, Majumder D, Chern M, Kalbacher H, Midha S, Patil PB, Sonti RV, Petzold CJ, Liu CC, Brodbelt JS, Felix G, Ronald PC. Science Advances. 2015;1:e1500245. doi: 10.1126/sciadv.1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludeman JP, Stone MJ. Br J Pharmacol. 2014;171:1167–1179. doi: 10.1111/bph.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu CC, Schultz PG. Nat Biotech. 2006;24:1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 8.Liu CC, Cellitti SE, Geierstanger BH, Schultz PG. Nat Protocols. 2009;4:1784–1789. doi: 10.1038/nprot.2009.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CC, Brustad E, Liu W, Schultz PG. J Am Chem Soc. 2007;129:10648–10649. doi: 10.1021/ja0735002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu CC, Mack AV, Tsao ML, Mills JH, Lee HS, Choe H, Farzan M, Schultz PG, Smider VV. PNAS. 2008;105:17688–17693. doi: 10.1073/pnas.0809543105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu CC, Choe H, Farzan M, Smider VV, Schultz PG. Biochemistry. 2009;48:8891–8898. doi: 10.1021/bi9011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu CC, Schultz PG. Annual Review of Biochemistry. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 13.Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. Biochemistry. 2013;52:1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young TS, Ahmad I, Yin JA, Schultz PG. Journal of Molecular Biology. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 16.Monigatti F, Hekking B, Steen H. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2006;1764:1904–1913. doi: 10.1016/j.bbapap.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Huang C, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenfield NJ. Nat Protocols. 2007;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gellman SH. Current Opinion in Chemical Biology. 1998;2:717–725. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 20.Wei T, Chen M, Thomas N, Ronald PC. in preparation. [Google Scholar]