Abstract
Purpose
Head and neck squamous cell carcinoma (HNSCC) is commonly treated with radiotherapy, and local failure after treatment remains the major cause of disease-related mortality. To date human papillomavirus (HPV) is the only known clinically validated, targetable biomarkers of response to radiation in HNSCC.
Experimental Design
We performed proteomic and transcriptomic analysis of targetable biomarkers of radioresistance in HPV-negative HNSCC cell lines in vitro, and tested whether pharmacologic blockade of candidate biomarkers sensitized cells to radiotherapy. Candidate biomarkers were then investigated in several independent cohorts of patients with HNSCC.
Results
Increased expression of several targets was associated with radioresistance, including FGFR, ERK1, EGFR, and focal adhesion kinase (FAK), also known as PTK2. Chemical inhibition of PTK2/FAK, but not FGFR, led to significant radiosensitization with increased G2/M arrest and potentiated DNA damage. PTK2/FAK overexpression was associated with gene amplification in HPV-negative HNSCC cell lines and clinical tumors. In two independent cohorts of patients with locally advanced HPV-negative HNSCC, PTK2/FAK amplification was highly associated with poorer disease-free survival (DFS) (P=0.012 and P=0.034). PTK2/FAK mRNA expression was also associated with worse DFS (P=0.03). Moreover, both PTK2/FAK mRNA (P=0.021) and copy number (P=0.063) were associated with DFS in the Head and Neck Cancer subgroup of The Cancer Genome Atlas.
Conclusion
Proteomic analysis identified PTK2/FAK overexpression is a biomarker of radioresistance in locally advanced HNSCC, and PTK2/FAK inhibition radiosensitized HNSCC cells. Combinations of PTK2/FAK inhibition with radiotherapy merit further evaluation as a therapeutic strategy for improving local control in HPV-negative HNSCC.
Keywords: PTK2/FAK, head and neck cancer, radiation, proteomics
Introduction
Head and neck squamous cell carcinoma (HNSCC) kills approximately 200,000 people a year (1), most from locoregional recurrence. One of the primary modes of therapy for this disease is radiation. Although the localized manifestation of HNSCC suggests that proper local therapy can be curative, the disease frequently exhibits intrinsic radioresistance. Indeed disease recurs locally in up to 50% of patients after radiotherapy for locally advanced HNSCC, ultimately leading to their demise (2–4).
Given the importance of radiation in the management of HNSCC, a variety of studies have been performed to identify biomarkers of radioresistance in this disease. A notable success story in this effort is the recognition that human papillomavirus (HPV) -driven HNSCC is associated with much higher cure rates than HPV-negative disease (5). In patients with HPV-negative HNSCC, mutations in TP53 (6) and altered function of EGFR and PI3 kinase/AKT signaling have been implicated in HNSCC radioresistance (6–9). Preclinical studies of EGFR activation in HNSCC ultimately culminated in a clinical trial investigating the use of cetuximab (an EGFR-targeted antibody) in combination with radiotherapy (4). However, in this trial radiotherapy failed in about 50% of patients even with EGFR inhibition. Also, the most recent randomized trial investigating cetuximab showed no improvement in outcome when used in combination with chemoradiation (2). Thus, additional “targetable” biomarkers of radioresistance in HPV-negative HNSCC are needed.
To this end, we conducted a comprehensive analysis of both protein and gene expression in a large panel of HPV-negative HNSCC cell lines. Specifically, we used reverse phase protein array (RPPA) to analyze 177 proteins and phosphoproteins in 49 head and neck cancer cell lines to identify proteins most strongly associated with radioresistance, with similar analyses of mRNA expression. We identified and validated the importance of focal adhesion kinase (FAK/PTK2) in radioresistance, showing that chemical inhibition of its kinase function resulted in consistent radiosensitization. Notably, we further found that PTK2/FAK was associated with treatment failure after radiotherapy in several separate cohorts of patients with HPV-negative HNSCC.
Materials and Methods
Cell lines
The 49 HNSCC cell lines used for this study, generously supplied by Dr. Jeffrey Myers through The University of Texas MD Anderson Cancer Center Head and Neck cell line repository or acquired from the American Type Culture Collection (ATCC, Manassas, VA), are shown in Supplementary Table S1. All cell lines were cultured in Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS), penicillin/streptomycin, glutamine, sodium pyruvate, nonessential amino acids, and vitamins. Each cell line was subjected to short tandem repeat genomic profiling for authentication. HN31 HNSCC cell lines stably expressing either scrambled shRNA or shRNA specific for PTK2/FAK were generated as described previously(6).
Copy number and mRNA expression
Single nucleotide polymorphisms (SNPs) were analyzed with SNP 6.0 or CytoScan HD arrays (Affymetrix), and the data were analyzed with Partek (v6.6, Partek Inc.), ASCAT (v2.1), and R software as described previously (10). Expression of selected mRNAs was analyzed with llumina HumanWG-6 V3 BeadChip human whole-genome expression arrays as described in the Supplementary Methods.
Reverse phase protein array
Protein lysate was collected from each cell line under full-serum (10% FBS) or serum-starvation (0% FBS) conditions for 24 hours before collection, or under serum-stimulated conditions (24 hr serum starvation followed by 30 min of 10% FBS before collection) and subjected to RPPA analysis as described previously (11) (Supplementary Methods). Cell lines were separated into resistant and sensitive groups by median surviving fraction at 2 Gy (SF2). Proteins and phosphoproteins that were expressed at a ≥1.5 fold difference between the two groups, with a false discovery rate of 1%, were examined (12).
In vitro radioresponse
Clonogenic cell survival was assessed in all HNSCC cell lines as described previously (6). Cell cycle was analyzed by flow cytometry of propidium iodide–stained DNA. All experiments were done up to three times for each cell line. Additional details are found in the Supplementary Methods.
Clinical data
Patient data were collected either from patients treated at MD Anderson Cancer Center or from The Cancer Genome Atlas (TCGA) public HNSCC database. All MD Anderson patients received surgical resection followed by postoperative radiotherapy, generally to 60 Gy, from 1990 through 2003. Two separate cohorts of patients were acquired and analyzed by two independent groups at MD Anderson as part of ongoing projects. Tumor sections were stained for p16 where available, and p16-positive tumors were excluded from the study (n=5). Records in the HNSCC cohort of TCGA database are incomplete with regard to therapy, but most patients were treated with surgery, with many receiving postoperative radiotherapy. Patients with known HPV-positive tumors and patients lacking follow-up information were excluded from the analysis. Tumor characteristics are shown in Supplementary Table S2, and other information on clinical data analysis is included in the Supplementary Methods. Univariate analysis was done with Cox regression examining tumor stage, nodal stage, site and PTK2 expression. Variables with a P value <0.1 were included in the multivariate model and forward stepwise Cox regression was performed. For PTK2/FAK mRNA expression, time-dependent receiver operating characteristic (ROC) curves analysis was used to assess the performance of survival prediction by comparing the area under the curves (AUC) between different time points from two different cohorts (MD Anderson and TCGA) (13). Because most events took place within 12 months of treatment in the MD Anderson cohort, the ability of models to predict outcome at and around 12 months was assessed with permutation tests; the P values of the AUC were calculated from 1000 permutations of the survival data. Survival curves were generated by using the method of Kaplan-Meier, with log rank statistics used to determine significance. R software, SPSS statistical software (v.20) and GraphPad Prism were used for clinical data analysis. Additional information on mRNA and copy number expression in the clinical samples is given in the Supplementary Methods.
Results
Cell line analysis identifies proteins associated with radioresistance in HNSCC
To identify novel targets for radiosensitization, we surveyed 49 HNSCC cell lines for their response to radiation and imposed a classification of either sensitive or resistant according to their respective SF2. We used RPPA to survey the expression of 177 proteins and phosphoproteins (Figure 1A). Analyzed proteins include those involved in DNA-damage repair, apoptosis, and the epithelial-to-mesenchymal transition (EMT) as well as targetable kinases. To focus our investigation, we limited our analysis to protein level changes of 1.5-fold or more, with a false discovery rate of <1%. We also performed immunoblot confirmation of the increased levels of PTK2/FAK in radioresistant HNSCC cell lines seen on RPPA (Supplementary Figure S1).
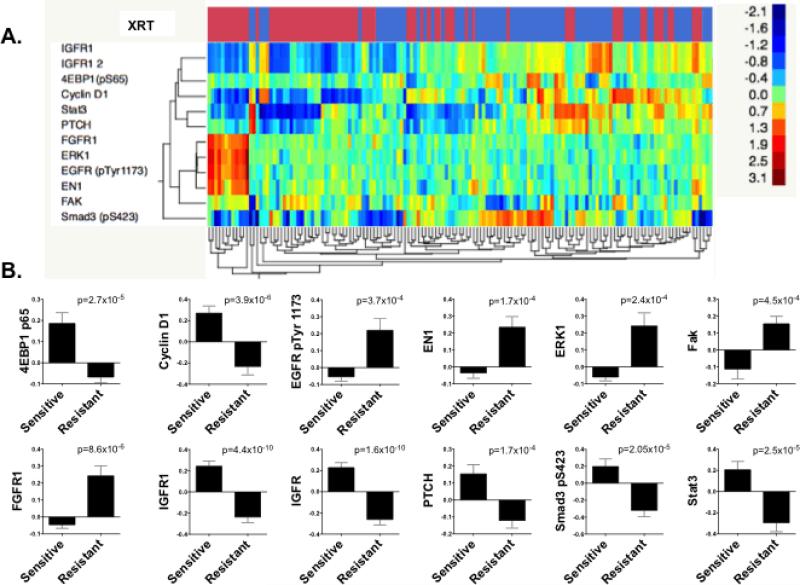
Figure 1. Proteomic profiling identifies markers of radioresistance in HPV-negative HNSCC cell lines.
(A) Reverse phase protein array analysis of 49 HNSCC cell lines, illustrated as unsupervised hierarchical clustering. Shown are proteins and phospho-proteins differentially expressed between radioresistant (red bars) and radiosensitive (blue bars) cell lines (false discovery rate 1%, mean fold difference ≥ 1.5). (B) Graphical representation of proteins and phospho-proteins differentially expressed between groups.
RPPA identified a number of proteins that were differentially expressed between radioresistant and sensitive HNSCC cell lines. Specifically, 4EBP1 p65, cyclin D1, IGFR, PTCH, phosphorylated Smad3, and Stat 3 were all significantly downregulated in radioresistant cell lines (Figure 1B). However, because the primary focus of the current work was to identify radioresistance biomarkers that are directly targetable, we turned our attention to phosphorylated EGFR, EN1, ERK1, FAK, and FGFR1, all of which were upregulated in the radioresistant cell lines. Of these targets, EGFR activation is well known to be associated with radioresistance in HNSCC (14), providing at least partial validation for this RPPA-based approach. EN1 is a homeobox transcription factor primarily regulating neural development (15). Although further investigation of its role in cellular response to radiation may be interesting, at present no drugs directly targeting this protein are available. Thus, ERK1, FAK and FGFR1 seem to be the most promising targetable biomarkers for radioresistance in HNSCC based upon RPPA.
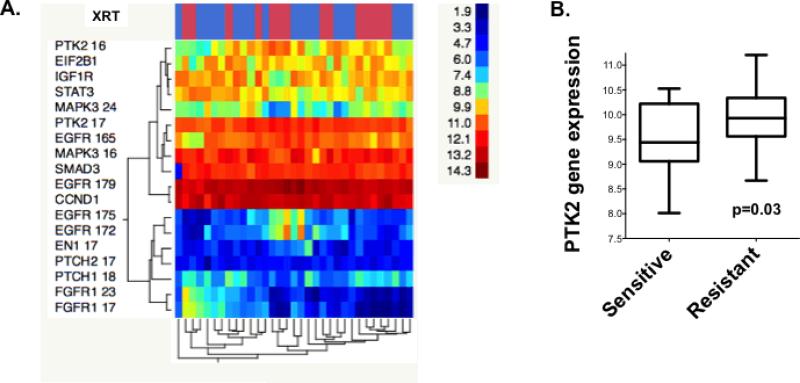
We next compared gene expression by using mRNA array of the proteins differentially expressed between the two groups in our RPPA screen. Similar to our RPPA results, we found increased gene expression of PTK2/FAK in radioresistant compared with sensitive cell lines (P=0.03; Figure 2A and B). However, neither ERK1 nor FGFR1 mRNA were expressed at significantly different levels between resistant and sensitive HNSCC. From this combined analysis, PTK2/FAK emerged as a highly significant marker of radioresistance at both the mRNA and protein expression levels. PTK2/FAK is also interesting as a therapeutic target, as several inhibitors are currently in or have completed phase I/II trials (16).
Figure 2. PTK2/FAK mRNA expression is significantly higher in radioresistant HPV-negative HNSCC.
(A) Genes encoding proteins expressed differently in the RPPA analysis (Fig. 1) were examined similarly, comparing radioresistant (red bars) and radiosensitive (blue bar) groups in hierarchical clustering. (B) PTK2/FAK gene expression was significantly higher in radioresistant HNSCC cell lines.
Inhibition of PTK2/FAK leads to significant radiosensitization and potentiation of DNA damage
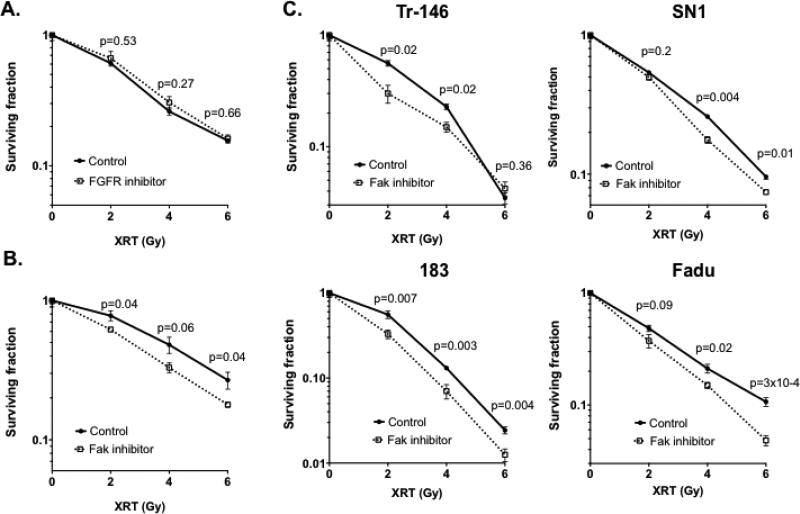
Next, to examine if pharmacologic inhibition of PTK2/FAK leads to radiosensitization in HNSCC, we treated HN5 cells with radiation in combination with PF00562271, which blocks the ATP-binding domain of FAK, rendering it reversibly catalytically inactive. For comparison purposes, we also examined nintedanib, which inhibits FGFR as well as VEGF and PDGF. With colony formation used as our readout of response, we found that nintedanib did not result in increased sensitivity to up to 6 Gy of radiation in HN5 cells (Figure 3A). Conversely, inhibition of PTK2/FAK reduced the surviving fraction in HN5 cells at all three doses of radiation tested (Figure 3B), at statistically significant levels or trending toward significance (p=0.06). Moreover, shRNA-targeting PTK2/FAK showed radiosensitization as well (Supplementary Figure S2A & B). To examine if this phenomenon is independent of cell line, we conducted the identical assay with four additional HPV-negative HNSCC cell lines and found that inhibition of FAK resulted in pronounced radiosensitization (Figure 3C). In addition, Fadu cells exposed to PF00562271 and radiation produced increased levels of phospho-H2AX, indicating unrepaired double-strand DNA damage, as well as G2/M arrest with a subsequent sub-G1 peak (Supplementary Figure S2C & D).
Figure 3. Inhibition of PTK2/FAK, but not FGFR, leads to radiosensitization in HPV negative HNSCC cells.
(A) The FGFR inhibitor nintedanib does not radiosensitize HN5 HNSCC cells. B & C) (B) The FAK inhibitor PF562271 radiosensitizes HN5 HNSCC cells and other HNSCC cell lines (C). P values for each radiation dose comparison are shown.
PTK2/FAK is highly expressed in HNSCC
On the basis of these findings, we investigated the utility of PTK2/FAK as a clinically targetable biomarker of radioresistance. Because a candidate biomarker of radioresistance should be expressed at levels high enough to be routinely detectable in a malignancy of interest in addition to being potentially targetable, we assessed PTK2/FAK gene expression in a large, publically available database of HNSCC cell lines (www.oncomine.com) (17) and HNSCC tumors (TCGA Research Network) (18) (Supplementary Figure S3A & B). Analysis of both cell lines and tumors confirmed high levels of PTK2/FAK expression in compared with other solid tumors, indicating its possible importance in HNSCC.
PTK2/FAK copy number is highly associated with mRNA and protein expression
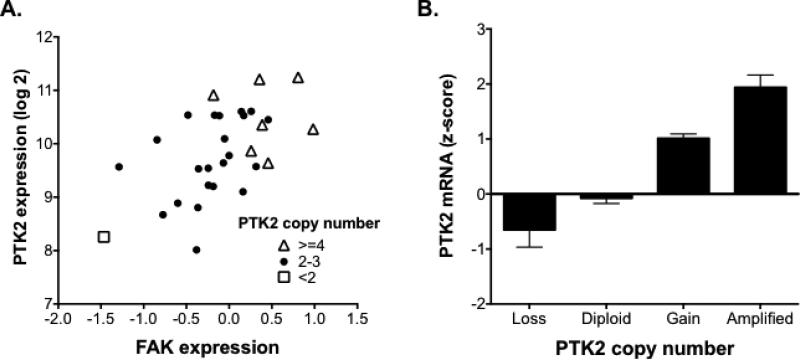
To further investigate the nature of FAK/PTK2 overexpression in HNSCC, we measured PTK2/FAK copy numbers in our panel of HNSCC cell lines. Both PTK2/FAK gene expression and protein levels were found to be highly correlated with FAK/PTK2 copy number in those cell lines (Figure 4A). An additional analysis of HNSCC tumors from the publically available data in TCGA (Figure 4B) also showed that PTK2 copy number was highly associated with gene expression and protein level, with amplifications in PTK2/FAK leading to increased PTK2/FAK expression. These data suggest that PTK2/FAK copy number amplification is a mediator of its overexpression in HNSCC.
Figure 4. PTK2/FAK copy number is highly associated with mRNA expression and protein level in HPV-negative HNSCC.
(A) Both PTK2/FAK gene expression (Pearson r=0.55, P=0.001) and copy number (Pearson r=0.46, P=0.008) are significantly correlated with protein level in the tested HNSCC cell lines. (B) PTK2/FAK copy number is significantly correlated with gene expression in samples from The Cancer Genome Atlas patients with known HPV-negative head and neck cancer (n=243; Pearson r=0.51, P<0.0001).
PTK2/FAK is associated treatment failure in several independent cohorts of patients with HPV-negative HNSCC
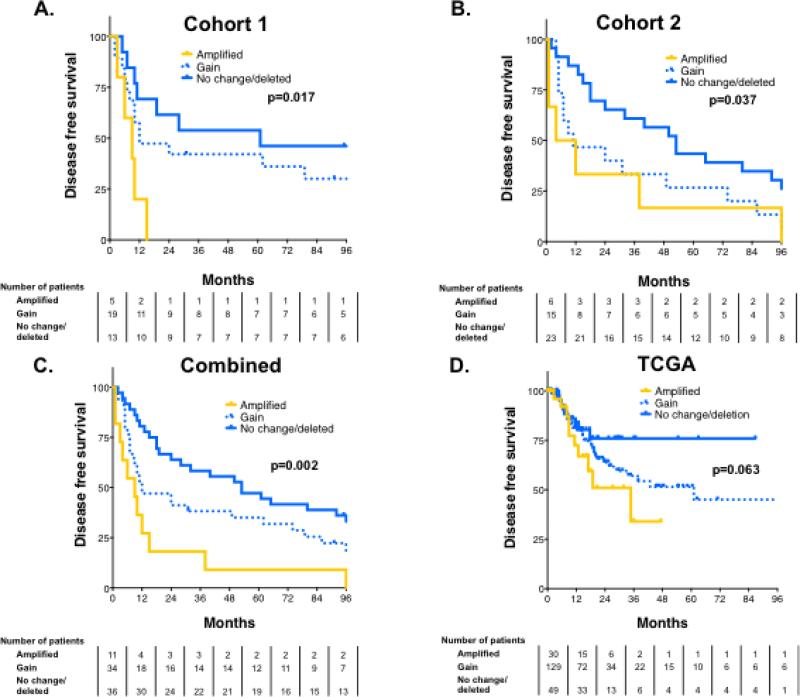
Finally, to determine if FAK/PTK2 was associated with locoregional failure following radiation treatment, we analyzed PTK2/FAK copy number in archival tumors from two separate cohorts of patients with locally advanced HNSCC treated with surgery and postoperative radiotherapy at MD Anderson and determined its relationship to disease-free survival (DFS) (tumor characteristics in Supplementary Table S2). Copy number was determined separately for each cohort by independent investigators blinded to outcome. In the first cohort, PTK2/FAK copy number was significantly associated with DFS on univariate analysis (P=0.015) and independently predicted DFS on multivariate analysis (HR 2.07, P=0.012) (Supplementary Table S3). Specifically, patients with FAK/PTK2 amplification had significantly worse outcomes and experienced disease recurrence significantly sooner than remaining patients. All of the patients with amplified PTK2 had experienced treatment failure by 24 months, whereas 70% of patients with either no alteration in FAK/PTK2 copy number or a deletion were disease-free at that time (Figure 5A). This finding was confirmed in a second cohort of patients, that amplification of PTK2/FAK was a significant negative predictor of DFS (Figure 5B). Again, this finding was significant on both univariate (P=0.039) and multivariate analysis (HR 1.67, P=0.031) (Supplementary Table S3). Combining these two cohorts to increase the statistical power of our analysis further validated that PTK2 was associated with radioresistant head and neck cancer (Figure 5C). As a further, separate validation of this finding, we examined PTK2/FAK copy number in publically available data available from TCGA Research Network (tumor characteristics in Supplementary Table S4) (18). As shown in Figure 5D, PTK2/FAK trended toward association with DFS (p=0.063) in all patients, moreover DFS was significantly different between patients whose tumors expressed amplified PTK2 FAK (median DFS 18.9 mos) compared to those with either a deletion or no change in PTK2/FAK (median DFS 53.1 mos, P=0.05). Again, this finding was significant on multivariate analysis (HR 1.6, P=0.034, Supplementary Table 3).
Figure 5. PTK2/FAK copy number is associated with disease recurrence in HPV-negative HNSCC.
PTK2/FAK copy number is associated with poorer disease-free survival (DFS) in an inpatients with HNSCC treated with radiotherapy at MD Anderson, considered both separately (A and B) and combined (C). PTK2/FAK copy number trends to association with DFS in the Head and Neck Cancer TCGA cohort as well (P=0.063).
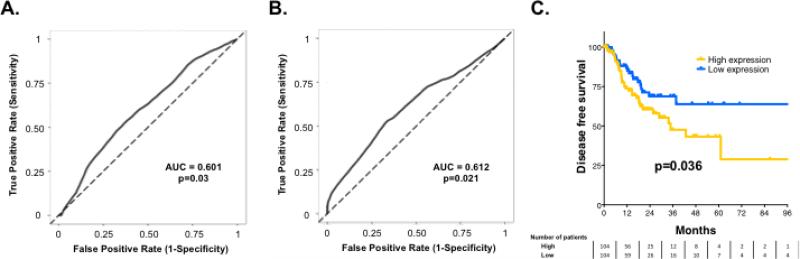
To further extend these findings, we also examined FAK/PTK2 mRNA expression in a group of 102 patients with locally advanced HNSCC treated with surgery and radiotherapy (Figure 6A). Similar to PTK2/FAK copy number, higher levels of FAK/PTK2 mRNA expression were associated with worse DFS (P=0.03). To validate this finding, we examined the Head and Neck Cancer TCGA cohort and again found that PTK2/FAK mRNA expression was associated with DFS (P=0.021) (Figure 6B). Specifically, median DFS was not reached in patients with tumors expressing low levels of PTK2/FAK mRNA, compared with a median DFS of 35 months in patients with tumors expressing high levels of PTK2/FAK mRNA (P=0.036, Figure 6C).
Figure 6. PTK2/FAK mRNA expression is associated with disease recurrence in patients with HPV-negative HNSCC.
PTK2/FAK mRNA expression is associated with DFS in an institutional cohort of 102 patients with HPV-negative HNSCC (A) and in patients from the Head and Neck Cancer TCGA cohort (B). (C) Kaplan Meier curve showing DFS in patients from the TCGA cohort split at median PTK2/FAK expression.
Discussion
We undertook a systematic, high-throughput search to identify biomarkers of radioresistance that can be targeted with agents currently available in clinical trials. By using comprehensive proteomic and genomic analysis, we identified several druggable kinases upregulated in HNSCC cell lines that are resistant to radiation, a finding suggesting that these proteins may be important in recurrence of HNSCC after radiation treatment as well as being novel targets for biologically driven radiosensitization. Our methods were at least partially confirmed by our identification of upregulation of EGFR, a marker of radioresistance whose inhibitor, cetuximab, is the only FDA-approved radiosensitizer in HNSCC. Although other signaling pathways had significant differences in key enzymes between sensitive and resistant cell lines, many of those identified were significantly downregulated in the resistant cells, which does not allow direct targeting for radiosensitization.
Two kinases identified as potential biomarkers for radioresistance in HNSCC were FGFR1 and FAK. Amplification of the FGFR1 gene has been associated with worse outcomes in patients with oral SCC (19). Several tyrosine kinase inhibitors targeting FGFRs are currently under evaluation and one such inhibitor, nintedanib, has been shown to prolong survival when used in combination with docetaxel in lung adenocarcinoma (20). However, in our study pharmacologic inhibition of FGFR1 did not radiosensitize HN5 HNSCC cells, nor was FGFR1 copy number associated with DFS (data not shown). Thus, although we cannot rule out FGFR inhibition as a possible therapy, we focused our attention primarily on PTK2/FAK.
In this study, we identified the importance of PTK2/FAK to in vitro radioresistance in HNSCC at both the proteomic and gene expression level. Inhibition of PTK2/FAK function led to radiosensitization in several cell lines, primarily due to G2/M arrest and unrepaired DNA damage. Most importantly, PTK2/FAK was validated as a marker of failure after radiotherapy in several cohorts of patients with HNSCC. This provides strong evidence of the clinical importance of PTK2/FAK and the significance of targeting this kinase for radiosensitization in this disease.
Although PTK2/FAK is highly expressed in HPV-negative HNSCC, the cause of PTK2/FAK overexpression in is not clear. This gene is not routinely mutated in HNSCC, with mutations being seen in only 2% of HNSCC patients (TCGA Research Network) (18). Although PTK2/FAK copy number gain or amplification is observed in approximately 70% of cases, some work indicates that PTK2/FAK protein expression is not driven by copy number amplification (18,21). However, studies in breast and lung cancer have directly linked PTK2/FAK copy number with both mRNA and protein expression (22,23). In the current study, we provide additional evidence of a direct link between PTK2/FAK amplification and expression in HPV-negative HNSCC.
Notably, the canonical role of PTK2/FAK in cancer progression has concerned metastatic spread. That is indeed logical, as its primary function is in regulation of focal adhesions and cell-to-cell interactions through integrin binding (24). However, HNSCC is far less likely to metastasize than many solid tumors, and death is primarily due to locoregional recurrence. Yet here we identify PTK2/FAK as both an in vitro and clinical marker of radioresistance that seems to be targetable to achieve radiosensitization. Indirect targeting of PTK2/FAK via β1 integrin inhibition and direct targeting via siRNA can achieve radiosensitization, possibly via alteration of JNK kinase signaling (25,26). Selective targeting of PTK2/FAK in endothelial cells also can radiosensitize tumors in preclinical lung carcinoma and melanoma models (27). At least one study, however, has reported that knockout of PTK2/FAK in SCC cells and in a preclinical xenograft model leads to radioresistance (28); the reason for these discrepant findings is not known. Interestingly, the preclinical model for the latter study used SCC expressing wild-type p53, whereas previous studies have examined cells expressing primarily mutant p53, which represent the vast majority of both HNSCC cell lines and tumors (29,30). PTK2/FAK and p53 are known to interact on several levels, with wild-type p53 functioning as a transcriptional repressor of PTK2/FAK (31) as well as directly binding PTK2/FAK protein (32,33). Small-molecule disruption of this binding seems to have some antitumor effect via re-activation of the apoptotic function of p53 (34). Also, in at least one study TP53 mutation was correlated strongly with PTK2/FAK overexpression in breast cancer (35). Thus, the conflicting results of PTK2/FAK inhibition on radiosensitization may be partially p53-driven, as a group of TP53 mutants seem to maintain at least some functionality in the realm of radioresponse (6). The interactions between mutant p53 and PTK2/FAK, and the potential role these interactions have in radioresponse, merit further investigation.
In conclusion, our high-throughput proteomic-genomic method successfully identified novel, targetable biomarkers for radioresistance in HNSCC and validated these biomarkers clinically. Among candidate markers associated with radioresistance, we found that PTK2/FAK blockade sensitized HNSCC cells to radiotherapy and that PTK2/FAK overexpression and amplification were associated with shorter DFS. Taken together, these findings support further evaluation of PTK2/FAK as a marker for identifying patients who are more likely to experience relapse after radiotherapy, and for clinical testing of FAK inhibition in combination with radiation therapy for patients with HPV-negative HNSCC.
Supplementary Material
Translational relevance.
Radiation therapy remains a mainstay of treatment for head and neck cancer (HNSCC), with most patients dying from local failure. To date, human papillomavirus (HPV) is the only known clinical biomarker of radioresistance in this disease. In the current study, we undertook a systemic proteomic analysis of HPV-negative HNSCC to identify markers of radioresistance, with particular focus on proteins that could be targeted with agents currently in clinical trials. Several proteins were found to be associated with radioresistance, including FGFR, ERK1, EGFR, and focal adhesion kinase (FAK), also known as PTK2. Inhibition of PTK2/FAK, but not FGFR, led to significant radiosensitization in several HNSCC cell lines via increased G2/M arrest and the potentiation of DNA damage. We found that PTK2/FAK was highly expressed in both HNSCC cell lines and tumors, and that this expression was highly associated with PTK2/FAK copy number. Notably, PTK2/FAK was highly associated with outcome in several independent cohorts of patients with locally advanced HNSCC treated with radiation. Thus, using a broad proteomic analysis, we have identified both a novel, validated biomarker of radioresponse in HNSCC as well as a potential target for radiosensitization.
Acknowledgments
Funding: This work was supported by following grants from the National Cancer Institute (National Institutes of Health): R01 CA 168484-022 (JH), HNSCC SPORE 5 P50CA070907-16 (JH and JM), CCSG 5 P30 CA01667239 (JH), & R01 CA 168485 (RM). This work was also supported by the Cancer Prevention Institute of Texas (RP150293) (HS).
Footnotes
The authors report no conflicts of interest related to the current manuscript.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ang KK, Zhang Q, Rosenthal DI, Nguyen-Tan PF, Sherman EJ, Weber RS, et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol. 2014;32:2940–50. doi: 10.1200/JCO.2013.53.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang KK, Zhang QE, Rosenthal DI, Nguyen-Tan PF, Sherman EJ, Weber RS, et al. A randomized phase III trial (RTOG 0522) of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III-IV head and neck squamous cell carcinomas (HNC). [2013 Jul 25];J Clin Oncol [Internet] 2011 :29. doi: 10.1200/JCO.2013.53.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 5.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skinner HD, Sandulache VC, Ow TJ, Meyn RE, Yordy JS, Beadle BM, et al. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res. 2012;18:290–300. doi: 10.1158/1078-0432.CCR-11-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ang KK, Berkey BA, Tu X, Zhang H-Z, Katz R, Hammond EH, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–6. [PubMed] [Google Scholar]
- 8.Jedlinski A, Ansell A, Johansson A-C, Roberg K. EGFR status and EGFR ligand expression influence the treatment response of head and neck cancer cell lines. J Oral Pathol Med. 2013;42:26–36. doi: 10.1111/j.1600-0714.2012.01177.x. [DOI] [PubMed] [Google Scholar]
- 9.Bussink J, van der Kogel AJ, Kaanders JH. Activation of the PI3-K/AKT pathway and implications for radioresistance mechanisms in head and neck cancer. Lancet Oncol. 2008;9:288–96. doi: 10.1016/S1470-2045(08)70073-1. [DOI] [PubMed] [Google Scholar]
- 10.Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, et al. Mutational Landscape of Aggressive Cutaneous Squamous Cell Carcinoma. Clin Cancer Res. 2014;20:6582–92. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 13.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–44. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 14.Perri F, Pacelli R, Scarpati GDV, Cella L, Giuliano M, Caponigro F, et al. Radioresistance in head and neck squamous cell carcinoma: Biological bases and therapeutic implications. Head Neck. 2014 doi: 10.1002/hed.23837. [DOI] [PubMed] [Google Scholar]
- 15.Sgaier SK, Lao Z, Villanueva MP, Berenshteyn F, Stephen D, Turnbull RK, et al. Genetic subdivision of the tectum and cerebellum into functionally related regions based on differential sensitivity to engrailed proteins. Development. 2007;134:2325–35. doi: 10.1242/dev.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanthi E, Krishna MH, Arunesh GM, Venkateswara Reddy K, Sooriya Kumar J, Viswanadhan VN. Focal adhesion kinase inhibitors in the treatment of metastatic cancer: a patent review. Expert Opin Ther Pat. 2014;24:1077–100. doi: 10.1517/13543776.2014.948845. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freier K, Schwaenen C, Sticht C, Flechtenmacher C, Mühling J, Hofele C, et al. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC). Oral Oncol. 2007;43:60–6. doi: 10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Reck M, Kaiser R, Mellemgaard A, Douillard J-Y, Orlov S, Krzakowski M, et al. Docetaxel plus nintedanib versus docetaxel plus placebo in patients with previously treated non-small-cell lung cancer (LUME-Lung 1): a phase 3, double-blind, randomised controlled trial. Lancet Oncol. 2014;15:143–55. doi: 10.1016/S1470-2045(13)70586-2. [DOI] [PubMed] [Google Scholar]
- 21.Canel M, Secades P, Rodrigo J-P, Cabanillas R, Herrero A, Suarez C, et al. Overexpression of focal adhesion kinase in head and neck squamous cell carcinoma is independent of fak gene copy number. Clin Cancer Res. 2006;12:3272–9. doi: 10.1158/1078-0432.CCR-05-1583. [DOI] [PubMed] [Google Scholar]
- 22.Yom CK, Noh D-Y, Kim WH, Kim HS. Clinical significance of high focal adhesion kinase gene copy number and overexpression in invasive breast cancer. Breast Cancer Res Treat. 2011;128:647–55. doi: 10.1007/s10549-010-1150-2. [DOI] [PubMed] [Google Scholar]
- 23.Ocak S, Yamashita H, Udyavar AR, Miller AN, Gonzalez AL, Zou Y, et al. DNA copy number aberrations in small-cell lung cancer reveal activation of the focal adhesion pathway. Oncogene. 2010;29:6331–42. doi: 10.1038/onc.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eke I, Deuse Y, Hehlgans S, Gurtner K, Krause M, Baumann M, et al. β₁Integrin/FAK/cortactin signaling is essential for human head and neck cancer resistance to radiotherapy. J Clin Invest. 2012;122:1529–40. doi: 10.1172/JCI61350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hehlgans S, Eke I, Cordes N. Targeting FAK radiosensitizes 3-dimensional grown human HNSCC cells through reduced Akt1 and MEK1/2 signaling. Int J Radiat Oncol Biol Phys. 2012;83:e669–76. doi: 10.1016/j.ijrobp.2012.01.065. [DOI] [PubMed] [Google Scholar]
- 27.Tavora B, Reynolds LE, Batista S, Demircioglu F, Fernandez I, Lechertier T, et al. Endothelial-cell FAK targeting sensitizes tumours to DNA-damaging therapy. Nature. 2014;514:112–6. doi: 10.1038/nature13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham K, Moran-Jones K, Sansom OJ, Brunton VG, Frame MC. FAK deletion promotes p53-mediated induction of p21, DNA-damage responses and radio-resistance in advanced squamous cancer cells. PLoS ONE. 2011;6:e27806. doi: 10.1371/journal.pone.0027806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golubovskaya VM, Finch R, Kweh F, Massoll NA, Campbell-Thompson M, Wallace MR, et al. p53 regulates FAK expression in human tumor cells. Mol Carcinog. 2008;47:373–82. doi: 10.1002/mc.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim S-T, Chen XL, Lim Y, Hanson DA, Vo T-T, Howerton K, et al. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golubovskaya VM, Finch R, Cance WG. Direct interaction of the N-terminal domain of focal adhesion kinase with the N-terminal transactivation domain of p53. J Biol Chem. 2005;280:25008–21. doi: 10.1074/jbc.M414172200. [DOI] [PubMed] [Google Scholar]
- 34.Golubovskaya VM, Ho B, Zheng M, Magis A, Ostrov D, Morrison C, et al. Disruption of focal adhesion kinase and p53 interaction with small molecule compound R2 reactivated p53 and blocked tumor growth. BMC Cancer. 2013;13:342. doi: 10.1186/1471-2407-13-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golubovskaya VM, Conway-Dorsey K, Edmiston SN, Tse C-K, Lark AA, Livasy CA, et al. FAK overexpression and p53 mutations are highly correlated in human breast cancer. Int J Cancer. 2009;125:1735–8. doi: 10.1002/ijc.24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.