Abstract
The Phytophthora genus comprises of some of the most destructive plant pathogens and attack a wide range of hosts including economically valuable tree species, both angiosperm and gymnosperm. Many known species of Phytophthora are invasive and have been introduced through nursery and agricultural trade. As part of a larger project aimed at utilizing genomic data for forest disease diagnostics, pathogen detection and monitoring (The TAIGA project: Tree Aggressors Identification using Genomic Approaches; http://taigaforesthealth.com/), we sequenced the genomes of six important Phytophthora species that are important invasive pathogens of trees and a serious threat to the international trade of forest products. This genomic data was used to develop highly sensitive and specific detection assays and for genome comparisons and to make evolutionary inferences and will be useful to the broader plant and tree health community. These WGS data have been deposited in the International Nucleotide Sequence Database Collaboration (DDBJ/ENA/GenBank) under the accession numbers AUPN01000000, AUVH01000000, AUWJ02000000, AUUF02000000, AWVV02000000 and AWVW02000000.
Keywords: Invasive species, Oomycetes, Forest health
| Specifications | |
|---|---|
| Organism/cell line/tissue | Six Phytophthora species, see Table 1. |
| Sex | Not applicable |
| Sequencer type or array | Illumina Hi-Seq |
| Data format | Analyzed; i.e. raw data filtered and assembled |
| Experimental factors | Genomic sequence of pure microbial cultures |
| Experimental features | Genomic sequence of pure microbial cultures |
| Consent | Not applicable. Data are available without restriction |
| Sample source location | Various; see Table 1 |
1. Direct link to deposited data
http://www.ncbi.nlm.nih.gov/bioproject/PRJNA190823
http://www.ncbi.nlm.nih.gov/bioproject/PRJNA190824
http://www.ncbi.nlm.nih.gov/bioproject/PRJNA190825
http://www.ncbi.nlm.nih.gov/bioproject/PRJNA190826
2. Experimental design, materials and methods
Woodlands and trees are under serious threat from an increasing number of Phytophthora species [1]. Several species of these fungus-like microorganisms may attack over 100 different host species and possess the ability to infect woody tissues, making them potentially destructive in plantations and native forest ecosystems worldwide [2], [3]. Early detection, monitoring and surveillance are important aspects in preventing such outbreaks but are hindered by a lack of genomic resources. Genome sequencing and comparisons should help to develop biosurveillance tools and to predict pathogenic outcome of the interaction of these microorganisms and their host.
Here, we present the draft genome sequence of six Phytophthora species threatening trees, which were selected on the basis of their potential to cause significant economic losses and large-scale damage to forest ecosystems. Phytophthora lateralis is an invasive pathogen that infects Port Orford Cedar (Chamaecyparis lawsoniana) and has spread throughout the natural range of the tree [2], [4]. P. lateralis is a sister species of P. ramorum, a species that has been responsible for the deaths of millions of trees in North America and Europe as a result of Sudden Oak Death and Sudden Larch Death [1], [5], [6]. The P. lateralis epidemic has affected both the wood export market as Port Orford Cedar is a valued species in foreign markets as well as the nursery trade as it is also a valued horticultural species [2]. Phytophthora alni sp. alni, the cause of Alder decline has been a serious threat to riparian ecosystems in Europe over the last 20 years [7]. The emergence of this disease is linked to an interspecific hybridization event, as P. alni subsp. alni. P. alni subsp. uniformis and P. alni subsp. multiformis, initially identified as genetic variants of P. alni sp. alni [8], were shown to be the parental species of the more aggressive hybrid P. alni sp. alni [9]. Phytophthora kernoviae first found in 2003 in the UK, primarily causes bleeding stem lesions on Fagus sylvatica and foliar and stem necrosis on Rhododendron ponticum, but has also been found on other hosts [10]. Phytophthora cambivora originally associated with Castanea species is a widespread root and canker pathogen of many woody hosts, but is most problematic on hardwoods in Europe, in particular on European beech and European chestnut [11], [12], [13]. Phytophthora pinifolia was first described in Chile and caused widespread disease on the needles and shoots of Pinus radiata [14]. Phytophthora cryptogea is a widespread pathogen of numerous ornamental hosts infecting roots, stems and leaves, and is an important pathogen in the nursery industry often isolated during surveys of infected plant material [15].
Genome assemblies were obtained by generating paired-end Illumina reads using the Hiseq 2000 platform at Canada's Michael Smith Genome Sciences Centre or GSC (Vancouver, Canada). For each species, DNA was extracted from pure culture using the DNA extraction procedure of Moller et al. [16]. Two genomic DNA libraries with fragment size of approximately 250 bp and 800 bp were constructed according to British Columbia Cancer Agency Genome Sciences Centre's tube-based paired-end library protocols. One μg of high molecular weight genomic DNA was sonicated (Covaris E210) in 60 μL volume to 200-300 bp. The DNA fragments were end-repaired, phosphorylated and bead purified in preparation for A-tailing. Illumina sequencing adapters were ligated overnight at 16 °C. Adapter ligated products were bead purified and enriched with 10 cycles of PCR using primers containing a hexamer index that enables library pooling. Paired-end 100 base reads were sequenced per pool in a single lane of an Illumina HiSeq2000 instrument.
Illumina chastity failed reads were removed, and the remaining reads were filtered by looking for exact read matches against Penicillium chrysogenum (GCA_000710275.1, GCA_000523475.1, GCA_000816005.1 and GCA_000801355.1) and P. marneffei (GCA_000001985.1, GCA_000227055.2 and GCA_000750115.1) genome sequences to eliminate commensal fungi contaminants. Each library was assembled into contigs using ABySS and a range of k-values from 32 to 96. ABySS was also used to scaffold the contigs, taking care to minimize the duplication of highly repeated sequences that can proliferate on the ends of scaffolds. The best assembly was then selected based on genome size and contiguity (best N50). Completeness of the genome assemblies was assessed using BUSCO (Benchmarking Universal Single-Copy Orthologs).
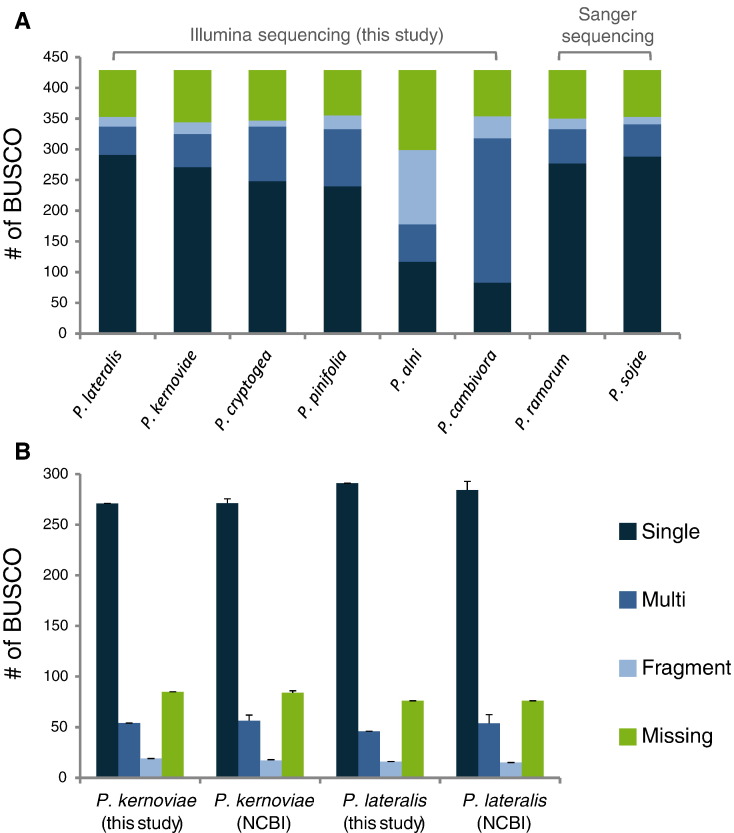
Raw sequence data and the sets of gene and protein models are available using the GenBank and Sequence Read Archive (SRA) accession numbers listed in Table 2. Genome assembly statistics obtained for P. kernoviae and P. lateralis were in the range of those obtained for Illumina de novo assemblies of two same species (P. kernoviae [n = 5], assembly size: 40.3 Mbp ± 3.0; N50: 61,035 ± 2195; length of longest scaffold: 796,176 bp ± 284,587 bp. P. lateralis [n = 4], assembly size: 50.6 Mbp ± 4.8; N50: 22,373 bp ± 6314; length of longest scaffold: 300,607 bp ± 247,251) (Table 2) [17]. Sequencing completeness was estimated using BUSCO based on a set of 429 single-copy ortholog genes common to Eukaryotes [18]. For the P. alni sp. alni assembly, quality control values were largely under those obtained for the other species with an assembly size twice the expected value of 114 Mbp [19] and a N50 under 3Kb (Table 2). Only 299 (69.0%) out of the 429 eukaryotic BUSCOs were found in this genome; a majority of these were fragmented and duplicated BUSCOs (121 fragmented and 61 duplicated; 60.9%), illustrating the difficulty to obtain an accurate de novo assembly for this homoploid hybrid species [19] (Table 2; Fig. 1A). With 344 (80.2%) to 355 (82.8%) BUSCO genes found all the other genomes (Table 2; Fig. 1), assemblies looked complete relative to the published Phytophthora and Oomycete de novo assemblies (Table 2; Fig. 1) [17], [20], [21], [22].
Table 2.
Assembly statistics and gene content for the genome sequences reported in this study.
| Species | Isolate | Genome assembly accession # | Total size (Mbp) | Genome coverage | # of scaffolds | N50 (bp) | Length of longest scaffold (bp) | BUSCO coverage |
|---|---|---|---|---|---|---|---|---|
| P. alni sp. alni | CBS_117376 | GCA_000439335.1 | 236.0 | 113.0 × | 118,474 | 2791 | 47,541 | 299 (69.0%) |
| P. cambivora | CBS_114087 | GCA_000443045.1 | 230.6 | 163.0 × | 72,332 | 4693 | 76,007 | 354 (82.5%) |
| P. cryptogea | CBS_418.71 | GCA_000468175.2 | 63.8 | 345.0 × | 19,533 | 12,607 | 234,588 | 347 (80.9%) |
| P. kernoviae | CBS_122049 | GCA_000448265.2 | 39.4 | 474.0 × | 5026 | 64,601 | 435,012 | 344 (80.2%) |
| P. lateralis | CBS_168.42 | GCA_000500205.2 | 52.4 | 470.0 × | 9039 | 23,425 | 178,165 | 353 (82.3%) |
| P. pinifolia | CBS_122922 | GCA_000500225.2 | 94.6 | 470.0 × | 36,928 | 8087 | 93,857 | 355 (82.8%) |
Fig. 1.
Genome completeness (BUSCO results) for six Phytophthora genomes. Searches for single-copy Eukaryote orthologs (n = 429) were conducted following Augustus gene predictions. A) Comparison of the six Illumina genomes with published genome assemblies of P. ramorum (GCA_000149735.1) and P. sojae (GCA_000149755.2). B) Comparison of the P. kernoviae and P. lateralis assemblies obtained in this study with five P. kernoviae (GCA_000333075.2, GCA_000333095.2, GCA_000333115.2, GCA_000785725.2 and GCA_000785735.2) and four P. lateralis (GCA_000318465.2, GCA_000333055.2, GCA_000338795.2 and GCA_000338815.2) Illumina assemblies downloaded from NCBI.
These genomic data were used to develop highly sensitive and specific detection assays that will have applications in biosurveillance of potentially invasive threats [23]. These genomes complete an initial collection of forest-related Phytophthora species [17], [21], [24] and will be used in comparative studies in conjunction with transcriptomic data, to identify factors related to epidemic traits such as the capacity to attack woody tissues and multiple host species.
Table 1.
Phytophthora species and isolates sequenced.
| Species | Isolate | Host | Location |
|---|---|---|---|
| Phytophthora alni sp. alni | CBS_117376 | Alnus sp., roots | Hungary |
| P. cambivora | CBS_114087 | Abies procera | Oregon, USA |
| P. cryptogea | CBS_418.71 | Gerbera sp. | The Netherlands |
| P. kernoviae | CBS_122049 | Rhododendron sp. | United Kingdom |
| P. lateralis | CBS_168.42 | Chamaecyparis lawsoniana | Oregon, USA |
| P. pinifolia | CBS_122922 | Pinus radiata, needles | Arauco, Chile |
Acknowledgments
We would like to acknowledge members of the TAIGA team at UBC Vancouver Hesther Yueh, Stéphanie Beauseigle, Padmini Herath and from CFIA team Debbie Shearlaw, Miranda Newton for their help with this project. This work was funded by Genome Canada, Genome British Columbia, the Canadian Forest Service (Genomics Research and Development Initiative, GRDI), FP Innovations and the Canadian Food Inspection Agency, through a Large Scale Applied Research Program (LSARP 2112; Genome Canada grant).
Contributor Information
Nicolas Feau, Email: feaunico@mail.ubc.ca.
Richard C. Hamelin, Email: rhamelin@gmail.com.
References
- 1.Grünwald N.J., Garbelotto M., Goss E.M., Heungens K., Prospero S. Emergence of the sudden oak death pathogen Phytophthora ramorum. Trends Microbiol. 2012;20:131–138. doi: 10.1016/j.tim.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Hansen E.M. Alien forest pathogens: Phytophthora species are changing world forests. Boreal Environ. Res. 2008;13:33–41. [Google Scholar]
- 3.Hansen E.M. Phytophthora species emerging as pathogens of forest trees. Curr. For. Reports. 2015;1:16–24. [Google Scholar]
- 4.Hansen E.M., Goheen D.J., Jules E.S., Ullian B. Managing Port-Orford-cedar and the introduced pathogen Phytophthora lateralis. Plant Dis. 2000;84:4–14. doi: 10.1094/PDIS.2000.84.1.4. [DOI] [PubMed] [Google Scholar]
- 5.Brasier C., Webber J. Sudden larch death. Science. 2010;329:824–825. doi: 10.1038/466824a. [DOI] [PubMed] [Google Scholar]
- 6.Robin C., Piou D., Feau N., Douzon G., Schenck N., Hansen E.M. Root and aerial infections of Chamaecyparis lawsoniana by Phytophthora lateralis: a new threat for European countries. For. Pathol. 2011;41:417–424. [Google Scholar]
- 7.Aguayo J., Adams G.C., Halkett F., Catal M., Husson C., Nagy Z.Á. Strong genetic differentiation between North American and European populations of Phytophthora alni subsp. uniformis. Phytopathology. 2012;103:190–199. doi: 10.1094/PHYTO-05-12-0116-R. [DOI] [PubMed] [Google Scholar]
- 8.Brasier C.M., Kirk S.a., Delcan J., Cooke D.E.L., Jung T., Man in't Veld W.A. Phytophthora alni sp. nov. and its variants: designation of emerging heteroploid hybrid pathogens spreading on Alnus trees. Mycol. Res. 2004;108:1172–1184. doi: 10.1017/s0953756204001005. [DOI] [PubMed] [Google Scholar]
- 9.Ioos R., Andrieux A., Marçais B., Frey P. Genetic characterization of the natural hybrid species Phytophthora alni as inferred from nuclear and mitochondrial DNA analyses. Fungal Genet. Biol. 2006;43:511–529. doi: 10.1016/j.fgb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Brasier C.M., Beales P.a., Kirk S.a., Denman S., Rose J. Phytophthora kernoviae sp. nov., an invasive pathogen causing bleeding stem lesions on forest trees and foliar necrosis of ornamentals in the UK. Mycol. Res. 2005;109:853–859. doi: 10.1017/s0953756205003357. [DOI] [PubMed] [Google Scholar]
- 11.Grandall B.S. The distribution and significance of the chestnut root rot phytophthoras, P. cinnamomi and P. cambivora. Plant Dis. Rep. 1950;34:194–196. [Google Scholar]
- 12.Brasier C.M., Kirk S.A. Comparative aggressiveness of standard and variant hybrid alder phytophthoras, Phytophthora cambivora and other Phytophthora species on bark of Alnus, Quercus and other woody hosts. Plant Pathol. 2001;50:218–229. [Google Scholar]
- 13.Jung T., Hudler G.W., Jensen-Tracy S.L., Griffiths H.M., Fleischmann F., Osswald W. Involvement of Phytophthora species in the decline of European beech in Europe and the USA. Mycologist. 2005;19:159. [Google Scholar]
- 14.Durán A., Gryzenhout M., Slippers B., Ahumada R., Rotella A., Flores F. Phytophthora pinifolia sp. nov. associated with a serious needle disease of Pinus radiata in Chile. Plant Pathol. 2008;57:715–727. [Google Scholar]
- 15.Leonberger A.J., Speers C., Ruhl G., Creswell T., Beckerman J.L. A survey of Phytophthora spp. in midwest nurseries, greenhouses, and landscapes. Plant Dis. 2013;97:635–640. doi: 10.1094/PDIS-07-12-0689-RE. [DOI] [PubMed] [Google Scholar]
- 16.Möller E.M., Bahnweg G., Sandermann H., Geiger H.H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992;20:6115–6116. doi: 10.1093/nar/20.22.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studholme D.J., McDougal R.L., Sambles C., Hansen E., Hardy G., Grant M. Genome sequences of six Phytophthora species associated with forests in New Zealand. Genomics Data. 2015;7:54–56. doi: 10.1016/j.gdata.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 19.Husson C., Aguayo J., Revellin C., Frey P., Ioos R., Marçais B. Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genet. Biol. 2015;77:12–21. doi: 10.1016/j.fgb.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Berger H., Yacoub A., Gerbore J., Grizard D., Rey P., Sessitsch A. Draft genome sequence of biocontrol agent Pythium oligandrum strain Po37, an Oomycota. Genome Announc. 2016;4:e00215–e00216. doi: 10.1128/genomeA.00215-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn L., O'Neill P.a., Harrison J., Paskiewicz K.H., McCracken A.R., Cooke L.R. Genome-wide sequencing of Phytophthora lateralis reveals genetic variation among isolates from Lawson cypress (Chamaecyparis lawsoniana) in Northern Ireland. FEMS Microbiol. Lett. 2013;2008:179–185. doi: 10.1111/1574-6968.12179. [DOI] [PubMed] [Google Scholar]
- 22.Tyler B.M., Tripathy S., Zhang X., Dehal P., Jiang R.H.Y., Aerts A. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 23.Lamarche J., Potvin A., Pelletier G., Stewart D., Feau N., Alayon D.I.O. Molecular detection of 10 of the most unwanted alien forest pathogens in Canada using real-time PCR. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Mata Saez L., McCracken A.R., Cooke L.R., O'Neill P., Grant M., Studholme D.J. Draft genome sequences of seven isolates of Phytophthora ramorum EU2 from Northern Ireland. Genomics Data. 2015;6:191–192. doi: 10.1016/j.gdata.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]