Abstract
The availability of bromodomain and extra-terminal inhibitors (BETi) has enabled translational epigenetic studies in cancer. BET proteins regulate transcription by selectively recognizing acetylated lysine residues on chromatin. BETi compete with this process leading to both downregulation and upregulation of gene expression. Hypoxia enables progression of triple negative breast cancer (TNBC), the most aggressive form of breast cancer, partly by driving metabolic adaptation, angiogenesis and metastasis through upregulation of hypoxia-regulated genes (for example, carbonic anhydrase 9 (CA9) and vascular endothelial growth factor A (VEGF-A). Responses to hypoxia can be mediated epigenetically, thus we investigated whether BETi JQ1 could impair the TNBC response induced by hypoxia and exert anti-tumour effects. JQ1 significantly modulated 44% of hypoxia-induced genes, of which two-thirds were downregulated including CA9 and VEGF-A. JQ1 prevented HIF binding to the hypoxia response element in CA9 promoter, but did not alter HIF expression or activity, suggesting some HIF targets are BET-dependent. JQ1 reduced TNBC growth in vitro and in vivo and inhibited xenograft vascularization. These findings identify that BETi dually targets angiogenesis and the hypoxic response, an effective combination at reducing tumour growth in preclinical studies.
Introduction
Epigenetic regulators are promising targets in cancer as transcriptional dysregulation and mutations in chromatin modulators and transcription factors (TF) are common in many malignancies. The bromodomain and extra-terminal (BET) proteins are lysine acetylation readers that mediate gene expression, including oncogenes.1 BET inhibitors (BETi) demonstrate many anticancer effects by downregulating gene expression of oncogenic factors.2 Breast cancer is the most common female cancer and triple negative breast cancer (TNBC) is its most aggressive subtype. Low oxygen (hypoxia) can drive TNBC progression,3 promoting adaptation through genes within the major hallmarks of cancer.4 Hypoxia can control gene expression recruiting chromatin remodelling complex5 and histone deacetylases.6 Thus, we investigated whether BETi JQ1 could impair the hypoxia response in TNBC and exert therapeutic effects.
Hypoxia is found in >50% of breast tumours and arises from high metabolic and proliferative rates and aberrant tumour vascularization. Clinically, hypoxia is associated with chemo-radiotherapy resistance, metastasis and poor survival,7 being a key area for targeted therapeutic development.3, 8 Most hypoxic responses are mediated by the hypoxia-inducible factors 1α and 2α (HIF-1α and HIF-2α), which in the absence of O2 stabilize and heterodimerise with HIF-1β.7, 9 This heterodimer binds to the hypoxia response element in gene promoters and induces transcription of genes, which drive molecular adaptation through many pathways, including pH regulation (CA9), angiogenesis (VEGF-A), metabolism (LDHA) and metastasis (LOX).7, 8 Other pathways including the unfolded protein response, XBP1 and ATF4 are also important.8, 10 TNBC shows overexpression of HIF target genes and is the breast cancer subtype most frequently associated with hypoxia.11, 12 Targeting HIF directly is a major challenge, while targeting HIF downstream targets such as VEGF has proven more feasible, although targeting just one protein has had less effect on overall survival than expected.13, 14, 15
The BET proteins (BRD2-4 and BRDT) regulate transcription by ‘reading' acetylated histones and recruiting TFs and epigenetic regulators.1, 2 JQ1 is a BETi16 that showed effects on tumour growth and survival,17 cell cycle arrest, and differentiation.16, 18, 19, 20 Although many attributed JQ1 effect to its MYC downregulation,18, 21 it is unlikely that this is the sole mechanism17 and some studies do not corroborate this,22 as MYC downregulation is not always sufficient to inhibit cell growth23 and JQ1 effects are observable without MYC alteration.24 BET proteins can associate with many TFs21, 25 and other genes are regulated by BETi, such as p21, BCL-xl, BCL2, AKT, FOSL1 and RUNX2.20, 21, 22, 24 Although the oncogenic driver varies, tumour addiction to BET activation seems common to many malignancies. These results led to clinical evaluation and there are 13 BETi clinical trials currently underway (www.clinicaltrials.gov).
Given the clinical investigation of BETi, we assessed whether the BETi JQ1 could alter the hypoxia response, exerting an anti-tumour effect. JQ1 modulated 44% of hypoxia-responsive genes, of which two-thirds were downregulated including CA9 and VEGF-A. JQ1 reduced TNBC growth in monolayer and spheroid culture. Furthermore, JQ1 prevented HIF binding to the CA9 promoter. Finally, JQ1 downregulated CA9 and VEGF-A expression and reduced growth and vascularization (CD31 positivity) in a TNBC xenograft model. These data show that JQ1 impairs tumour response to hypoxia.
Results
JQ1 downregulates the hypoxic transcriptome response
We performed gene array analysis after JQ1 treatment in hypoxia or normoxia in the TNBC cell line MDA-MB-231 (Figure 1) and the ER+ MCF-7, as the hypoxic transcriptome of this cell line is well-documented26 (Supplementary Figure S1). JQ1 alone profoundly affected gene expression (fold change (FC) log2⩾1 and P<0.05, n=3): 2338 genes were differentially expressed (DEG) in MDA-MB-231 and 2993 in MCF-7; while hypoxia induced 119 DEG in MDA-MB-231 and 1286 in MCF-7. Within the DEG in hypoxia, a considerable proportion were modulated by JQ1 in hypoxia, 44% in MDA-MB-231 and 29% in MCF-7 (Figure 1a and Supplementary Figure S1A). Interestingly, JQ1 had a greater impact on hypoxia-sensitive genes than those that were not hypoxia-sensitive (P=0.046; median logFC=1.22 × 1.02) (Supplementary Figure S1C).
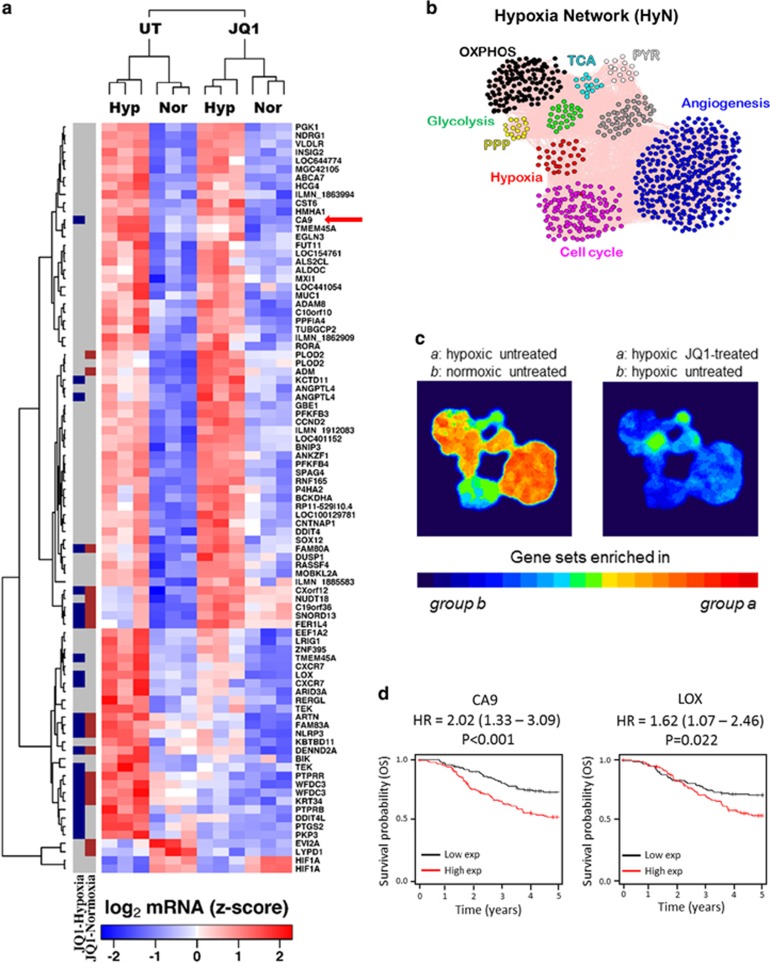
Figure 1.
JQ1 downregulates the expression of several hypoxia-regulated genes, especially CA9. (a) List of DEG under hypoxia in MDA-MB-231 cells obtained from microarray. Columns at the left denote DEG under JQ1 treatment, either in normoxia (red blocks) or hypoxia (blue dots). CA9 is the most prominently downregulated gene in hypoxia (red arrow). (b) Hypoxia Network (HyN) created including pathways regulated by hypoxia. Grey dots are linking genes, included to the network remains stable. (c) Most components of HyN are upregulated by hypoxia and downregulated by JQ1 treatment in MDA-MB-231 cells. (d) Kaplan–Meier curves demonstrating the prognostic value of two genes consistently inhibited by JQ1 in both cell lines tested (MDA-MB-231 and MCF-7) for triple negative breast cancer patients. OXPHOS, oxidative phosphorylation; TCA, tricarboxylic acid cycle; PYR, pyruvate metabolism; PPP, pentose phosphate pathway.
To further investigate the effect of JQ1 on tumour response to hypoxia, we defined which pathways were hypoxia-regulated and evaluated their expression under JQ1 treatment. Then, we developed a Hypoxia Network (HyN) (Figure 1b) containing the Hypoxia Signature27 and a list of genes for each pathway found to be hypoxia-regulated (obtained from KEGG, Supplementary Table S1). Hypoxia upregulated most of the HyN clusters in both cell lines (Figure 1c and Supplementary Figure S1B). Gene Set Enrichment Analysis showed that hypoxia upregulates angiogenesis, glycolysis, oxidative phosphorylation and pentose phosphate pathway in MDA-MB-231 (Figure 1c and Supplementary Table S2). The Hypoxia Signature27 set of genes was upregulated in hypoxia as expected (MDA-MB-231: ES=−0.79, normalized enrichment score=−2.68, P<0.001, false discovery rate q-value<0.001; MCF-7: ES=−0.83, normalized enrichment score=−2.63, P<0.001, false discovery rate q-value<0.001, n=3). JQ1 treatment prevented hypoxic upregulation of the hypoxia signature, angiogenesis, oxidative phosphorylation and pentose phosphate pathway gene data sets, but did not alter glycolysis or MYC expression. In addition, JQ1 treatment downregulated the cell cycle and TCA sets of genes in hypoxia (Figure 1c and Supplementary Table S2).
MCF-7 results confirmed most of these findings (Supplementary Figure S1B and Supplementary Table S3). CA9 was the most significantly downregulated gene in both cell lines (MDA-MB-231: logFC=−1.40, P=2.3 × 10−3; MCF-7: logFC=−4.22, P=1.51 × 10−9, n=3) (Figure 1a). The prognostic value in TNBC of the consistently DEG by JQ1 in hypoxia in both tested cell lines was evaluated (Figure 1d and Supplementary Figure S2) on analysis of TCGA data sets http://cancergenome.nih.gov/. Two of them are associated with poor prognosis in TNBC: CA9 (hazard ratio=2.02; P=0.001) and LOX (hazard ratio=1.62; P=0.022) (Figure 1d).
JQ1 reduces TNBC monolayer and spheroid growth
JQ1 dose-dependently reduced cell growth in monolayer cultures in all the four TNBC cell lines tested, in hypoxia and normoxia (Figure 2a and Supplementary Figure S3A). The non-active enantiomer (−)-JQ116 showed no effect on cell growth (Figure 2a and Supplementary Figure S3A).
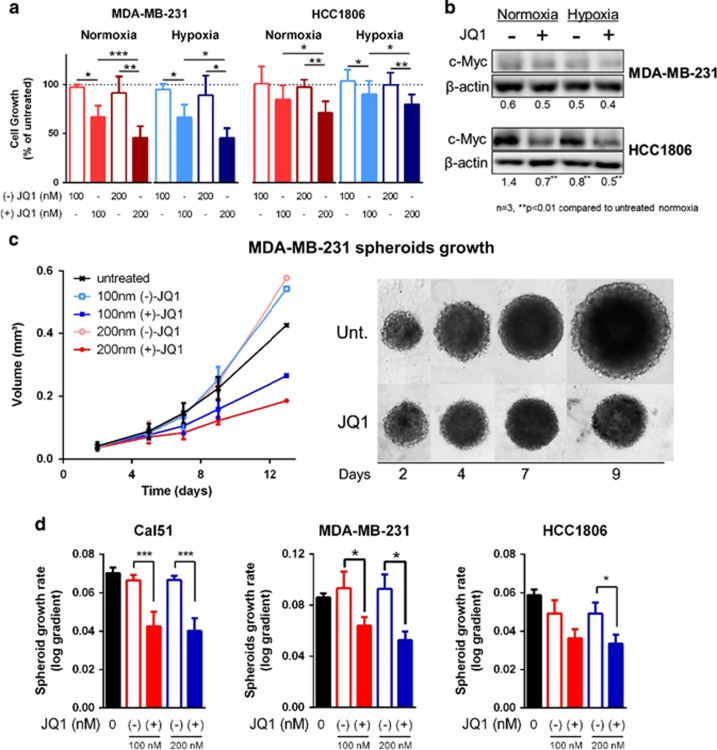
Figure 2.
JQ1 reduces TNBC monolayer and spheroid growth, regardless of MYC. (a) Cell cultures were grown in 96-well plates for 72 h in each condition. (+)-JQ1 dose-dependently reduces monolayer (two-dimensional) cell growth of TNBC cell lines MDA-MB-231 and HCC1806 in normoxia and hypoxia, while (−)-JQ1 does not. (b) JQ1 reduced c-Myc immunocontent only in the MYC-amplified cell line HCC1806, while the non-mutated cell line MDA-MB-231 showed no difference in c-Myc. (c) Representative spheroid growth curve and pictures of MDA-MB-231 spheroids following JQ1 treatment. (d) (+)-JQ1 reduces spheroid growth in MDA-MB-231 and other TNBC cell lines, while (−)-JQ1 does not. One-way analysis of variance, n=3, *P<0.05, **P<0.01, ***P<0.001.
MYC amplification was not a predictor of JQ1 sensitivity (Figure 2b). JQ1 reduced growth and c-Myc expression of HCC1806 (P<0.01, n=3), which has MYC amplification28 and the highest MYC expression among the investigated cell lines. However, JQ1 did not alter c-Myc, yet induced a similar growth inhibition in MDA-MB-231 (Figure 2b and Supplementary Figure S3B), which has no MYC amplification (cBioPortal).28, 29 JQ1 induced a similar growth inhibition in two additional MYC-non-amplified TNBC cell lines: Cal51 and SUM159 (Supplementary Figure S3A). No differential effect of JQ1 on cell growth in two-dimensional culture in normoxia or hypoxia was identified.
JQ1 inhibited growth of tumour spheroids in all the cell lines (Figures 2c and d and Supplementary Figure S3C). This model is more physiologically relevant, as it creates the oxygen, nutrient and pH gradients found in tumours, and improves the translation of drug candidates.30 SUM159 did not grow as spheroids. JQ1 reduced spheroid growth rate in MDA-MB-231 (P<0.05, n=3), HCC1806 (P<0.05, n=3) and Cal51 (P<0.01, n=3). Once again, the non-active enantiomer (−)-JQ1 did not cause any significant effect (Figures 2c and d and Supplementary Figure S3C).
JQ1 reduces CA9 and Ki67 expression in TNBC spheroids
Immunohistochemistry showed that untreated spheroids express CA9 in their hypoxic cores, while JQ1-treated spheroids did not (MDA-MB-231: P<0.001, n=3; HCC1806: P<0.01, n=3) (Figure 3). JQ1-treated spheroids had lower Ki67 staining, indicating an anti-proliferative effect (MDA-MB-231: P<0.001, n=3; HCC1806: P<0.01, n=3) (Figure 3). This is concordant with the downregulation of cell cycle genes by JQ1 in the expression array analyses (Figure 1c, Supplementary Figure S1B and Supplementary Tables S2 and S3).
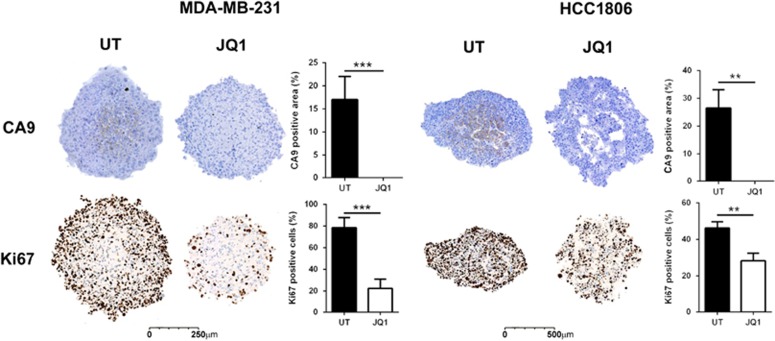
Figure 3.
JQ1 reduces CA9 and Ki67 stain in TNBC spheroids. CA9 expression is visible in untreated (UT) spheroids, but undetectable in JQ1-treated spheroids. The proliferative marker Ki67 was also reduced in spheroids treated with JQ1. For immunohistochemistry, spheroids were fixed with formalin 10%, embedded in agarose, processed, embedded in wax and cut with a microtome. Student t-test, n=3, **P<0.01, ***P<0.001.
JQ1 reduces expression of CA9, VEGF-A and additional hypoxia upregulated genes
Quantitative PCR (qPCR) confirmed the JQ1-induced downregulation of hypoxia-responsive genes. CA9 expression was consistently inhibited by JQ1, without alteration of HIF expression (mRNA and protein) in all cell lines tested (Figure 4 and Supplementary Figure S4A).
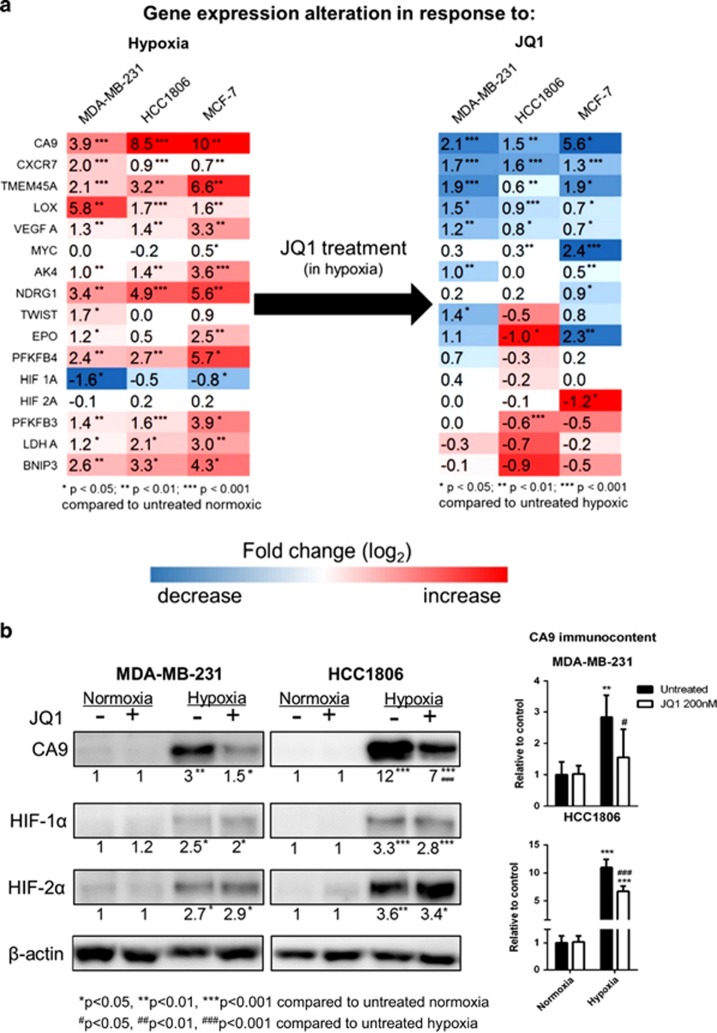
Figure 4.
JQ1 reduces CA9 expression in TNBC cell lines. (a) Hypoxia upregulates several genes and JQ1 downregulates a group of them, CA9 being the most prominent across the three cell lines. HIF expression is not altered by JQ1. (b) CA9 is consistently downregulated by JQ1 in hypoxia, without any effect on HIF. Cells were treated with JQ1 for 24 h prior to RNA or protein extraction, and then gene expression was assessed by RT–qPCR and protein immunocontent was assessed by western blot. Two-way analysis of variance, n=3, *P<0.05, **P<0.01, ***P<0.001.
A panel of 16 genes was investigated including CA9, HIF-1α, HIF-2α, MYC and genes differentially expressed in the array analysis (Figure 1). All the HIF targets included in this panel are upregulated in hypoxia (Figure 4a) and many of them are downregulated by JQ1 treatment (Figure 4b). Within this panel, five genes were consistently downregulated by JQ1 treatment in hypoxia in all cell lines investigated, of which CA9 was the most prominent (MDA-MB-231: logFC=−2.1, P<0.001; HCC1806: logFC=−1.5, P<0.01; MCF-7: logFC=−5.6, P<0.05). The other four genes consistently downregulated are VEGF-A, CXCR7, TMEM45A and LOX. (Figure 4b). Other HIF-regulated genes such as LDHA or BNIP3 were not affected by JQ1 (Figure 4a), indicating a specific effect on a subset of the hypoxia transcriptome. MYC mRNA was downregulated in response to JQ1 treatment in normoxia and hypoxia, but only in MYC-amplified cell lines HCC1806 and MCF-7 (Figure 2b, Supplementary Figure S3B, Figure 4a and Supplementary Figure S4).
Additional BETi were tested (I-BET151 and I-BET762) and confirmed these findings, also downregulating CA9, VEGF, CXCR7, TMEM45A and LOX in hypoxia (Supplementary Figure S5). In contrast, I-BET151 in hypoxia induced VEGF-A in HCC1806 cell line and PFKFB3 in both cell lines tested. No effect was observed on HIF-1α expression, but there was a significant upregulation of HIF-2α at the RNA level.
Immunoblot analysis further confirmed that JQ1 induced a significant reduction of CA9 protein induction in hypoxia in both TNBC cell lines (P<0.01, n=3) (Figure 4b). JQ1 did not significantly alter HIF-1α and HIF-2α protein expression (Figure 4b).
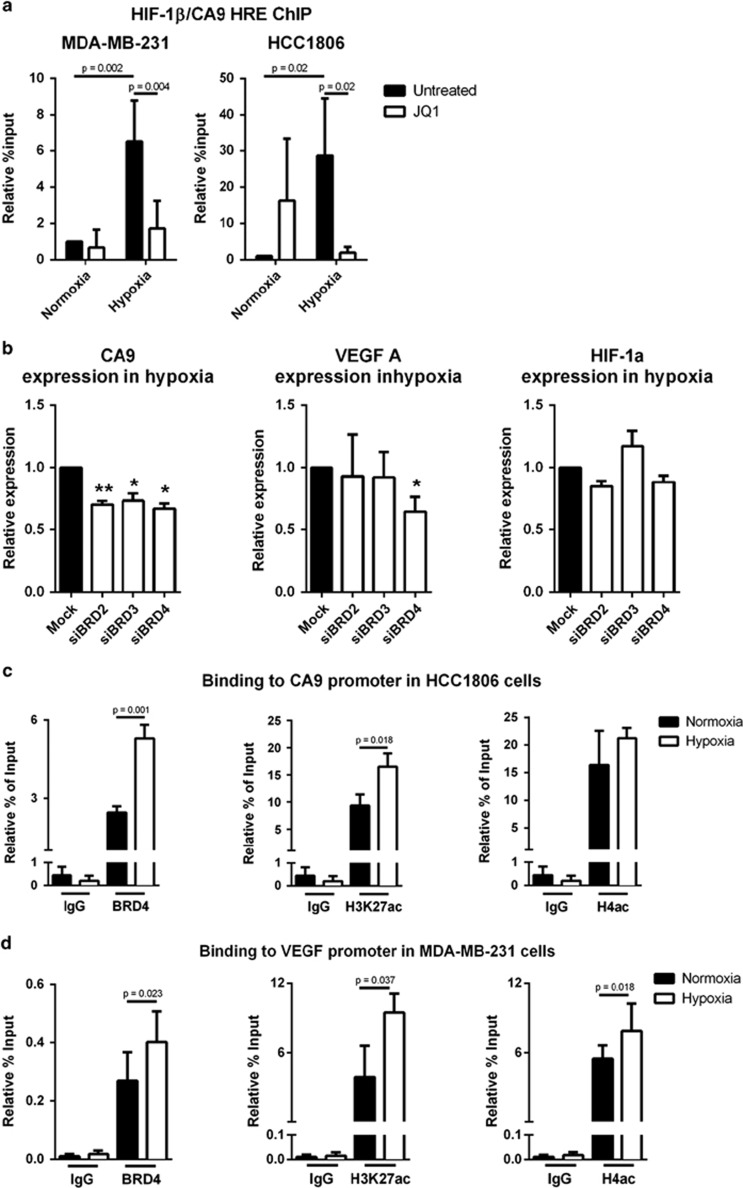
JQ1 reduces HIF binding to the CA9 promoter
To investigate how JQ1 prevents hypoxia-responsive gene expression, we evaluated chromatin immunoprecipitation (ChIP) of HIF-1β to the hypoxia response element of the CA9 promoter, as HIF-1α dimerizes with HIF-1β prior to transcription induction. As expected, hypoxia increased HIF binding in both the MDA-MB-231 (P=0.002, n=3) and HCC1806 (P=0.02, n=3) (Figure 5a). JQ1 treatment in hypoxia reduced HIF binding to the CA9 promoter to normoxic levels in both MDA-MB-231 (P<0.01, n=3) and HCC1806 (P<0.05, n=3) (Figure 5a). This suggests HIF-1 is BET-dependent for binding/recruitment in some of its downstream targets, explaining how BET inhibition reduces the expression of hypoxia-induced genes.
Figure 5.
JQ1 prevented HIF binding to the CA9 promoter and BET protein knockdowns phenocopy JQ1 treatment. (a) ChIP assay for CA9 promoter region with HIF-1β immunoprecipitation in two TNBC cell lines. Two-way analysis of variance, n=3. (b) Expression of CA9, VEGF-A and HIF-1α in hypoxia following siRNA knockdown of BET proteins. One-way analysis of variance, n=3. (c) ChIP assay for CA9 promoter region with BRD4, H3K27 acetylation or H4 acetylation immunoprecipitation in HCC1806. T-test, n=3. (d) ChIP assay for VEGF-A promoter region with BRD4, H3K27 acetylation or H4 acetylation immunoprecipitation in MDA-MB-231. T-test, n=3 (H3K27 acetylation) n=5 (BRD4 and H4 acetylation), *P<0.05; **P<0.01.
In addition to this, expressions of BRD2–4 are increased in hypoxia (Supplementary Figure S6A), suggesting a role for BET proteins in hypoxia. Although JQ1 is a potent inhibitor for all the BET proteins,16 some of its effects are attributed to a specific isoform. We performed siRNA knockdown to investigate which isoform was responsible for these effects (Supplementary Figure S6B). BRDT was excluded as its expression was below the limit of detection. The BRDT siRNA did not change CA9, VEGF or HIF-1α expression. CA9 expression in hypoxia was reduced after BRD2 (P<0.01, n=3), BRD3 (P<0.05, n=3) and BRD4 (P<0.05, n=3) knockdown in MDA-MB-231 cells, without affecting HIF-1α level (Figure 5b). Only BRD4 (P<0.05, n=3) reduced VEGF-A expression in hypoxia (Figure 5b). Therefore, some HIF targets were shown to be BET-dependent (CA9 and VEGF-A), while others, such as LDHA are BET-independent.
To better comprehend how JQ1 can have these effects, BRD4 binding and acetylation of H3K27 and H4 in hypoxia at the promoters of VEGF and CA9 in HCC1806 and MDA-MB-231 were investigated by ChIP qPCR (Figures 5c and d and Supplementary Figures S6C and D). A significant increase of BRD4 binding was identified at the CA9 promoter in hypoxia in HCC1806 cells (P<0.01, n=3) (Figure 5c) and at the VEGF promoter in hypoxia in MDA-MB-231 cells (P<0.05, n=5) (Figure 5d). A significant increase for acetylation of H3K27 was found in response to hypoxia at the CA9 (P<0.05, n=3) and VEGF (P<0.01, n=3) promoters of HCC1806 cells (Figure 5c and Supplementary Figure S6C) and at the VEGF promoter of MDA-MB-231 (P<0.05, n=3) (Figure 5d). There was increased H4 acetylation in response to hypoxia at the VEGF promoter of MDA-MB-231 (P<0.05, n=5) (Figure 5d).
We investigated available BRD4 ChIP-Seq data for MCF-7 in normoxia31 and compared these with those genes that were downregulated by JQ1 in hypoxia and found that: 61% had BRD4 binding by ChIP-Seq in normoxia (Supplementary Tables S5 and S6). We investigated the BRD4 binding to genes that were downregulated by JQ1 in hypoxia and also upregulated in response to hypoxia; 39% of these genes showed direct binding by BRD4 (Supplementary Tables S5 and S6). Finally, we investigated the overlap between BRD4 binding in normoxia in MCF-731 and HIF-1α or HIF-2α binding in hypoxia26 in MCF-7 published data sets. These data confirmed BRD4 and HIF-1 or HIF-2 binding in 14% of the genes that we identified are increased in hypoxia and downregulated by JQ1.
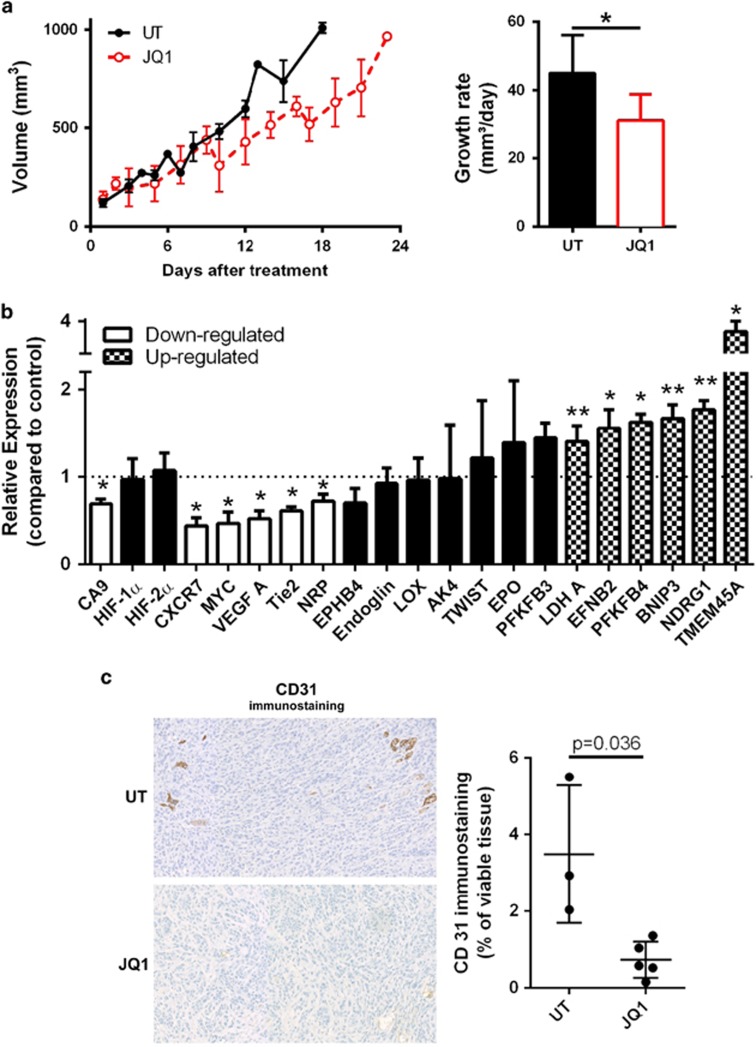
JQ1 reduces tumour growth, CA9 and VEGF-A expression, and vascularization in TNBC xenografts
JQ1 (50 mg/kg) reduced HCC1806 xenograft growth (P<0.05, n=5) (Figure 6a) and expression of CA9 (P<0.05, n=3), VEGF-A (P<0.05, n=3), CXCR7 (P<0.05, n=3) and MYC, but did not affect the levels of HIF-1α or HIF-2α (Figure 6b).
Figure 6.
JQ1 reduces tumour growth, CA9, VEGF-A and additional angiogenesis-related gene expression and blood vessel count in xenograft model of TNBC. (a) Growth curve and rate in TNBC xenografts treated with JQ1 or untreated (UT). Linear regression followed by Student t-test, n=5, *P<0.05. (b) Gene expression in TNBC xenografts treated with JQ1, assessed by qRT–PCR analysis of RNA extracted from tumours. Student t-test compared with UT, n=3, *P<0.05, **P<0.01. (c) Representative CD31 immunostaining in TNBC xenografts. Non-parametric Mann–Whitney test, n=3 shCTL and n=5 JQ1-treated. Xenografts were grown using HCC1806 cells in 6–7-week-old female CD1 nude mice.
Despite the downregulation effect observed in VEGF-A expression, angiogenesis is a broad phenomenon involving multiple molecular players.7, 14, 15 We thus investigated the expression of additional angiogenesis-related genes and observed JQ1-treated xenografts have lower expression of Tie2 (P<0.05, n=3) and NRP (P<0.05, n=3) and higher expression of EFNB2/ephrinB2 (P<0.05, n=3) (Figure 6b).
Interestingly, LDH-A (P<0.01, n=3), BNIP3 (P<0.05, n=3), PFKFB4 (P<0.01, n=3) and TMEM45A (P<0.05, n=3) (Figure 4a, Supplementary Figure S4 and Figure 6b) were unaltered or even downregulated by JQ1 (and other BETi) in cell culture and upregulated in xenografts.
We investigated the role of JQ1 in angiogenesis due to the consistent downregulation of VEGF-A found in cell lines (Figure 4a, Supplementary Figure S4) and the xenografts (Figure 6b) along with the reduction of the angiogenesis pathway expression (Figure 1c, Supplementary Figure S1B and Supplementary Tables S2 and S3). We observed that JQ1-treated xenografts had lower immunostaining of the blood vessel marker CD31 (P<0.05), thus indicating an anti-angiogenic effect of JQ1 (Figure 6c).
Discussion
Hypoxia represents a key target for the development of therapies in cancer.3, 32 Hypoxia induces a transcriptomic shift largely dependent on HIF,33 and there is evidence for HIF dependence upon epigenetic regulation in response to hypoxia.5, 6 We demonstrate an epigenetic approach to modulate the tumour response to hypoxia and reduce growth in TNBC. JQ1 modulated the expression of 44% of hypoxia-responsive genes in MDA-MB-231 TNBC cell lines, of which two-thirds were downregulated. More specifically, JQ1 downregulated the expression of the major regulators of hypoxic pH regulation and angiogenesis, CA9 and VEGF-A, in TNBC cell lines and xenografts. We observed that in hypoxic conditions, there was an increased histone acetylation at, and BRD4 binding to, the CA9 and VEGF promoters, suggesting an explanation for JQ1 effectiveness in this context. It is possible that BRD2 and/or BRD3 may also be important in the regulation of CA9 and VEGF. Analysis of published data sets26, 31 identified that many of the hypoxic JQ1-regulated genes have BRD4 binding and that 14% of the genes simultaneously upregulated by hypoxia, downregulated by JQ1 and bound by BRD4 are direct targets of either HIF-1α or HIF-2α these included VEGFA and CA9. Finally, as JQ1 prevents HIF binding to the CA9 promoter but not all HIF-regulated transcription, the data suggest that some HIF targets are BET-dependent.
JQ1 consistently downregulated CA9 in in vitro and in vivo models. Hypoxic tumours develop in an acidic microenvironment, owing to increased production of metabolic acids and poor vascularization.34 CA9 is highly induced in hypoxia, where it allows adaptation to this environment maintaining a more neutral intracellular pH.35, 36 Increased CA9 expression is a marker of poor prognosis in breast cancer and is more common in TNBC than other breast cancer subtypes.36 CA9 inhibition reduces tumour growth and metastasis.35, 36, 37
JQ1 also consistently demonstrated an anti-angiogenic effect, as it reduced the expression of the angiogenic pathway, the key angiogenic inducer VEGF-A and blood vessel count. JQ1-treated xenografts showed lower levels of Tie2 and NRP, involved in vascular stabilization and branching and promotion of arterial growth.38 Conversely, there was a higher expression of EFNB2/ephrinB2 (Figure 6b), described as a regulator of arterial/venous specialization and vessel branching.38 Collectively, this indicates that JQ1 could impair the early steps of angiogenesis, a major hallmark of cancer.
Many studies showed an anti-tumoural effect of BETi,19, 21, 22, 24 and recently it was found that JQ1 shows a typical behaviour of anti-angiogenic agents and, in fact, JQ1 can reduce tumour vascularization by suppressing VEGF stimulation.39 Angiogenesis is upregulated by hypoxia and supports tumour progression.7 Anti-angiogenic therapy is a major cancer treatment strategy used to treat eight solid tumour types. However, this strategy was found to induce hypoxia in around 50% of patients.7 Hypoxic adaptation enables resistance to anti-angiogenic therapy and may in part explain why the promise of anti-angiogenic therapy in breast cancer has not been fulfilled. We have shown that combined inhibition of VEGF and CA9 act at least additively and in some examples synergistically to reduce tumour growth.35
Anti-angiogenic therapies can lead to metabolic adaptation.7, 35 JQ1 treatment increased the expression of LDHA and PFKFB4 in xenografts, but not in cell cultures. LDHA was reported to be downregulated by JQ1 in ovarian cancer,40 may be because of its regulation by MYC.41 Whereas JQ1 reduced the expression of oxidative phosphorylation, pentose phosphate pathway, TCA gene data sets but not glycolysis in TNBC cell line MDA-MB-231, in the ER+ cell line MCF-7, JQ1 increased TCA. This may be due to differences in the metabolic requirements of these subtypes of breast cancer. Thus, we might expect that a co-treatment with an anti-glycolytic or pro-OXSPHOS drug (such as metformin) could lead to a synergistic effect and be a promising therapy, especially in TNBC.
We also highlight the impact of BET inhibition on wider hypoxic gene expression. The hypoxic regions of tumours are resistant to other therapies, therefore we propose that utilizing BET inhibitors to target the hypoxic tumour cells in combination with additional chemotherapy or radiotherapy may provide better responses. Combining hypoxia targeting with radiotherapy or chemotherapy has been shown previously to provide a greater therapeutic response. For example, targeting hypoxia-regulated genes including CA9, one of the JQ1-regulated genes, increases sensitivity to radiotherapy and chemotherapy.37, 42
Stem cell characteristics comprise another important hallmark of cancer and epigenetic regulation has an important role in this. BRD4 has been proposed as a marker for self-renewal43 and JQ1 can downregulate genes involved in this process in human cord-derived mesenchymal stem cells.44 Stem cells are maintained in an undifferentiated state through expression of the core transcriptional factors Nanog, Oct4 and Sox2. BRD4 is required for Nanog expression and JQ1 inhibits this inducing rapid differentiation of murine embryonic stem cells45, 46 as well as significantly downregulating Oct4 and SOX2.46 BET inhibition or BRD4 depletion reduces the expression of pluripotent genes and shifts cellular fate.45, 46 Collectively, these data show that BRD4 is critical for the maintenance of pluripotency and maintaining stem cell fate, while inhibition of BET proteins enhances differentiation.
Initially, studies described JQ1 effects as MYC-dependent.16, 18, 47 Although some studies reported MYC expression predicts JQ1 sensitivity, our results indicate other mechanisms are relevant; as JQ1 reduced tumour cell growth both in MYC-amplified (MCF-7 and HCC1806) and MYC-non-amplified cell lines (MDA-MB-231, Cal51 and SUM159). Our data provide further evidence for the context dependence of BETi.
Several studies show that BETi have broader MYC-independent effects.19, 48, 49 JQ1 impairs the recruitment of multiple TFs to their targets by physical disruption, for example, between the BRD4 and the N-terminal domain of the androgen receptor.19, 21 Thus, JQ1 acts by blocking BET protein ability to bind to chromatin, which in turn prevents TF recruitment, possibly including HIF. This is in agreement with our observation of reduced HIF binding to the CA9 promoter region in response to JQ1. Other important hypoxia-regulated genes demonstrated a similar pattern of downregulation by JQ1 in this study. These include CXCR7 and LOX. CXCR7 is a G protein-coupled receptor upregulated in breast cancer associated with worst outcome that mediates angiogenesis and metastasis.50 LOX is also upregulated in breast cancer and confers a poor prognosis, where it enables angiogenesis51 and disrupts bone homeostasis providing a favourable environment for metastatic cells from hypoxic ER− breast cancer.52 Taken together, this led us to the original suggestion that HIF targets can be divided into BET-dependent and BET-independent.
The SWI/SNF chromatin remodelling complex was the first epigenetic factor demonstrated to regulate the response to hypoxia.5, 53 This complex makes DNA accessible to other factors, especially through its ATPase subunits BRM and BRG1.54 In breast cancer, BRG1 and BRM are overexpressed in most primary breast cancers and are needed for in vivo tumour formation and TNBC cell line proliferation.55 SWI/SNF can either directly regulate the expression of HIF-1α and HIF-2α or regulate the expression of hypoxia-responsive genes, including CA9.5 The CA9 promoter nucleosome is BRG1-depedently remodelled in response to hypoxia.5 JQ1 does not bind to BRM or BRG1,16 and there is no current knowledge regarding interactions between BET proteins and SWI/SNF complex. While JQ1 prevents acetylated histones from being ‘read', SWI-SNF can promote deacetylation.54 Both interact with MYC.16, 18, 47, 54 Finally, just as CA9 was modulated by both factors, LDHA was not. Future studies should address to what extent the set of genes affected by these factors overlap. It might be the case that BET proteins and SWI/SNF complex interact at some level forming an enhanceosome and only some HIF targets are epigenetically regulated, rather than being BET- or SWI/SNF-dependent.
In conclusion, we showed that BETi impairs tumour response to hypoxia, targeting multiple pathways such as angiogenesis and pH control. Our findings alter the understanding of tumour response to hypoxia and identify a new avenue for epigenetic therapy to target the hypoxic tumour microenvironment. Furthermore, these results have a clear impact on the interpretation of the results from current clinical trials and future clinical use of drugs that inhibit the BET proteins in solid tumours.
Materials and Methods
Cell culture
Cells were maintained in DMEM+10% FBS at 5% CO2, 37 °C. Hypoxic incubations: 0.1%O2 in INVIVO2400 workstation (Baker Ruskinn, Sanford, ME, USA). Cell number was measured by Sulforhodamine B assay56 or CyQUANT (Molecular Probes, Waltham, MA, USA) following the manufacturer's instructions. Cell lines were purchased from ATCC (Manassas, VA, USA; MDA-MB-231, HCC1806, MCF-7), Creative Bioarray (Shirley, NY, USA) (CAL51) and Asterand (Royston, UK) (SUM159); these have stringent quality control for cell authenticity incorporating short tandem repeat profiling. Cells were regularly tested for mycoplasma. Stefen Knapp provided JQ1 (University of Oxford, UK) and Daniel Ebner provided I-BET151 and I-BET762 (University of Oxford, UK).
Spheroid culture
Cells were seeded in round-bottomed plates (Corning, Corning, NY, USA) with Matrigel (BD Bioscience, Franklin Lakes, NJ, USA)-supplemented media, aggregated by centrifugation (2000 r.p.m./10 min). Treatment was daily renewed. Pictures were taken 3 days/week with an inverted microscope (EVOS xl Core, AMG, Waltham, MA, USA) (n=3).
Gene expression array and ChIP-Seq data analysis
Illumina whole genome gene expression was performed (n=3 per group). Biotin-labelled aRNA was hybridized, according to the manufacturer's instruction (Illumina Inc., San Diego, CA, USA; #11286340), to high-density Illumina Human oligonucleotide arrays Human HT-12_V4_0_R1_15002873_B. Data are available at Array Express (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-4604/ E-MTAB-4604). Fluorescence emissions were detected using iScanner and data were extracted using BeadStudio v2011.1 Software (Illumina Inc) and imported to GeneSpring GX 12.1 (Agilent Technologies, Inc., Santa Clara, CA, USA); Illumina microarray data were pre-processed, normalized and differential expression analysed using R package LIMMA (v3.22.4). Significantly DEG were regarded as those with false discovery rate (Benjamini–Hochberg) corrected P value cutoff of <0.01. Analyses were performed using R (v3.1.2).
Functional analysis was carried out on DEG to identify statistically overrepresented ontologies using Database for Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov/). DEG fulfilled the criteria: FC log2⩾1 and P<0.05.
The prognostic value of DEG was generated using The Cancer Genome Atlas (TCGA) (more information available at http://cancergenome.nih.gov/).
Genecodis28, 29 (http://genecodis.cnb.csic.es/) was used to investigate pathway alterations. Hypergeometric test was used (false discovery rate=0.05). The list of genes comprising these pathways were obtained from KEGG (http://www.genome.jp/kegg/), GO or the study by Buffa et al.27 (Supplementary Table S3). The Hypoxic Response (HyR) network was designed in STRING (http://string-db.org/) containing the selected pathways (Figure 1b).
Gene Set Enrichment Analysis (v2.1) was used to evaluate pathway enrichment57 (Supplementary Table S3). ViaComplex (v1.0)58 was used to generate representative landscape images of these results (Figure 1c and Supplementary Figure S1B).
Publicly available MCF-7 ChIP-Seq data (GEO dataset GSE55923) was analysed using MACS2.59 Sequence alignments were performed using Bowtie v1 (parameters: -v 2 -m 1 -3 1 -S 2) against human genome assembly hg19. Peaks were called using MACS2 (parameters: —bdg —nomodel —extsize 283.67 —gsize 2.7e9 —pvalue 1e-3). Peaks were annotated with gene names (UCSC Hg19 gene annotations version: September 2014) if they overlapped within 10 000 bp upstream (strand-specific) or within gene body. For comparisons, each BRD4 replicate was independently compared with the same MCF-7 Input control. Peaks of three MCF-7 BRD4 replicates were compared for pairwise correlation using bigWigCorrelate. Correlation results suggested only modest correlation (rho 0.4-0.55), therefore replicates were deemed unsuitable for pooling.
Real-time PCR (qPCR)
qPCR was performed (n=3) as described previously.35 Primers sequences are available in Supplementary Table S4.
Immunoblotting
Immunoblotting was performed as described previously (n=3)35 with primary antibodies listed in Supplementary Table S7. Bands were quantified using ImageJ.
Xenograft studies
Mice were housed at BMS, University of Oxford, UK, and procedures were carried out under a Home Office licence (PPL30/2771). Female Crl:NU-Foxn1nu mice (Charles River, Oxford, UK) (6–7-week-old) were injected orthotopically into the mammary fat pad with 2.5 × 106 HCC1806 cells in 1:1 serum-free medium and Matrigel (BD Bioscience) (n=5 animals per group).
Tumour growth was measured with calipers by experienced technicians blinded to the experimental hypothesis and after three out of five animals reached 150 mm3, animals received JQ1 or vehicle (10% DMSO, 10% hydroxypropyl beta cyclodextrin) intraperitoneally at 50 mg/kg daily. Animals were randomly grouped at injection and one group was treated with JQ1 whilst the other was untreated (mean tumour sizes at the treatment starts were; shCTL, 120 mm3 and JQ1-treated, 137 mm3). When tumours reached 1.44 cm3, the mice were killed by cervical dislocation.
Immunohistochemistry
Immunohistochemistry was carried out as previously described (n=3 untreated, n=5 JQ1-treated).35 Slides were submitted to antigen retrieval, and the primary antibodies listed in Supplementary Table S7. Slides were incubated with secondary antibody (Dako, Cambridgeshire, UK) and DAB (Dako) and counterstained with haematoxylin solution (Sigma-Aldrich Corp., St Louis, MO, USA). Secondary-only control staining was performed routinely. ImageJ colour deconvolution was used for quantification.35
Gene silencing by RNA interference
Transfections of siRNA duplexes targeting BET proteins (BRD2–4 and BRDT) or a scramble control (ON-TARGETplus SMARTpool) were performed in Optimem (Invitrogen, Waltham, MA, USA), using Oligofectamine (Invitrogen) (n=3).
ChIP assay
ChIP assay was performed for antibodies listed in Supplementary Table S7 using the EZ-ChIP Chromatin Immunoprecipitation Kit (#17-371, Millipore, Billerica, MA, USA) or ChIP-IT Express Enzymatic (Active Motif) according to the manufacturer's instructions. DNA isolated from ChIP was quantified by qPCR using CA9 or VEGF promoter primers (Supplementary Table S4) (n=3).
Statistical analysis
Statistical analysis and graphs were performed using GraphPad Prism v6.0 (GraphPad, La Jolla, CA, USA). Results are plotted as mean values with standard deviation. Statistical tests and the number of repeats are described in the figure legends. Student's t-test was used for two sample analyses and normal distributions were assumed, otherwise the non-parametric Mann–Whitney test was used. Analysis of variance was used for >2 sample analyses. No samples or experimental repeats were excluded from analyses. For the in vivo experiment to detect a FC of 2 at alpha (P value)= 0.05, five animals in each group gave a 75% power of detection. No statistical methods were used for the samples size selection of other experiments.
Acknowledgments
This work was funded by grants from Cancer Research UK (ALH), Breast Cancer Research Foundation (ALH), Breast Cancer Now (AM), Oxford NIHR Biomedical Research Centre, and the CRUK Oxford Centre (ALH). Leonardo Lisboa da Motta has been awarded a scholarship from the CAPES Foundation/Brazil to conduct this study.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014; 13: 337–356. [DOI] [PubMed] [Google Scholar]
- Chaidos A, Caputo V, Karadimitris A. Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: emerging preclinical and clinical evidence. Ther Adv Hematol 2015; 6: 128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim Biophys Acta 2015; 1863: 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 2008; 13: 472–482. [DOI] [PubMed] [Google Scholar]
- Sena JA, Wang LY, Hu CJ. BRG1 and BRM chromatin-remodeling complexes regulate the hypoxia response by acting as coactivators for a subset of hypoxia-inducible transcription factor target genes. Mol Cell Biol 2013; 33: 3849–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YP, Wu KJ. Epigenetic regulation of hypoxia-responsive gene expression: focusing on chromatin and DNA modifications. Int J Cancer 2014; 134: 249–256. [DOI] [PubMed] [Google Scholar]
- McIntyre A, Harris AL. Metabolic and hypoxic adaptation to anti-angiogenic therapy: a target for induced essentiality. EMBO Mol Med 2015; 7: 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C, Langdon SP, Mullen P, Harris AL, Harrison DJ, Supuran CT et al. New strategies for targeting the hypoxic tumour microenvironment in breast cancer. Cancer Treat Rev 2013; 39: 171–179. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 2007; 12: 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzymski T, Milani M, Singleton DC, Harris AL. Role of ATF4 in regulation of autophagy and resistance to drugs and hypoxia. Cell Cycle 2009; 8: 3838–3847. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Gianni L. Hallmarks of triple negative breast cancer emerging at last? Cell Res 2014; 24: 904–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EY, Yan M, Campo L, Han C, Takano E, Turley H et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br J Cancer 2009; 100: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumler I, Christiansen OG, Nielsen DL. A systematic review of bevacizumab efficacy in breast cancer. Cancer Treat Rev 2014; 40: 960–973. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011; 473: 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothey A, Galanis E. Targeting angiogenesis: progress with anti-VEGF treatment with large molecules. Nat Rev Clin Oncol 2009; 6: 507–518. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O et al. Selective inhibition of BET bromodomains. Nature 2010; 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. The N Engl J Med 2012; 367: 647–657. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi JW, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Z, Shea MJ, Creighton CJ, Coarfa C, Hilsenbeck SG et al. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res 2014; 24: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V, Vaidyanathan M, Janardhanam VA, Bradner JE. Pre-clinical analysis of changes in intra-cellular biochemistry of glioblastoma multiforme (GBM) cells due to c-Myc silencing. Cell Mol Neurobiol 2014; 34: 1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014; 510: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA 2012; 109: 19408–19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA 2011; 108: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Qi J, Bradner JE, Said JW, Doan NB, Forscher C et al. Synergistic effect of JQ1 and rapamycin for treatment of human osteosarcoma. Int J Cancer 2015; 136: 2055–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YJ, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med 2014; 20: 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 2011; 117: e207–e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffa FM, Harris AL, West CM, Miller CJ. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene (vol 102, pg 428, 2010). Br J Cancer 2010; 103: 1136–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JJ, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschow MLS, Boettcher K, Kelm J. High-content analysis of biomarker intensity and distribution in 3D microtissues. Nat Methods 2012; 9. [Google Scholar]
- Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep 2014; 8: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer 2011; 11: 393–410. [DOI] [PubMed] [Google Scholar]
- Parks SK, Chiche J, Pouyssegur J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat Rev Cancer 2013; 13: 611–623. [DOI] [PubMed] [Google Scholar]
- Swietach P, Patiar S, Supuran CT, Harris AL, Vaughan-Jones RD. The role of carbonic anhydrase 9 in regulating extracellular and intracellular pH in three-dimensional tumor cell growths. J Biol Chem 2009; 284: 20299–20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre A, Patiar S, Wigfield S, Li JL, Ledaki I, Turley H et al. Carbonic anhydrase IX promotes tumor growth and necrosis in vivo and inhibition enhances anti-VEGF therapy. Clin Cancer Res 2012; 18: 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, McDonald PC, Oloumi A, Chia S, Ostlund C, Ahmadi A. Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res 2011; 71: 3364–3376. [DOI] [PubMed] [Google Scholar]
- Dubois L, Peeters S, Lieuwes NG, Geusens N, Thiry A, Wigfield S et al. Specific inhibition of carbonic anhydrase IX activity enhances the in vivo therapeutic effect of tumor irradiation. Radiother Oncol 2011; 99: 424–431. [DOI] [PubMed] [Google Scholar]
- Jain RK. Molecular regulation of vessel maturation. Nat Med 2003; 9: 685–693. [DOI] [PubMed] [Google Scholar]
- Bid HK, Phelps DA, Xiao L, Guttridge DC, Lin J, London C et al. The bromodomain BET inhibitor JQ1 suppresses tumor angiogenesis in models of childhood sarcoma. Mol Cancer Ther e-pub ahead of print 23 February 2016. [DOI] [PMC free article] [PubMed]
- Qiu HF, Jackson AL, Kilgore JE, Zhong Y, Chang LL, Gehrig PA et al. JQ1 suppresses tumor growth through downregulating LDHA in ovarian cancer. Oncotarget 2015; 6: 6915–6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 2009; 15: 6479–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieling RG, Parker CA, De Costa LA, Robertson N, Harris AL, Stratford IJ et al. Inhibition of carbonic anhydrase activity modifies the toxicity of doxorubicin and melphalan in tumour cells in vitro. J Enzyme Inhib Med Chem 2013; 28: 360–369. [DOI] [PubMed] [Google Scholar]
- Gonzales-Cope M, Sidoli S, Bhanu NV, Won KJ, Garcia BA. Histone H4 acetylation and the epigenetic reader Brd4 are critical regulators of pluripotency in embryonic stem cells. BMC Genomics 2016; 17: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghamdi S, Khan I, Beeravolu N, McKee C, Thibodeau B, Wilson G et al. BET protein inhibitor JQ1 inhibits growth and modulates WNT signaling in mesenchymal stem cells. Stem Cell Res Ther 2016; 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Pinto HB, Kamikawa YF, Donohoe ME. The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression. Stem Cell Rep 2015; 4: 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne GA, Stewart HJS, Dickson J, Knapp S, Ramsahoye B, Chevassut T. Nanog requires BRD4 to maintain murine embryonic stem cell pluripotency and is suppressed by bromodomain inhibitor JQ1 together with Lefty1. Stem Cells Dev 2015; 24: 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res 2014; 20: 912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Biarnes MC, Clarke R, Jordan VC. Inhibition of BET proteins impairs estrogen-mediated growth and transcription in breast cancers by pausing RNA polymerase advancement. Breast Cancer Res Treat 2015; 150: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratikopoulos EE, Dendy M, Szabolcs M, Khaykin AJ, Lefebvre C, Zhou MM et al. Kinase and BET inhibitors together clamp inhibition of PI3K signaling and overcome resistance to therapy. Cancer Cell 2015; 27: 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani NA, Nasser MW, Ahirwar DK, Zhao HL, Miao ZH, Shilo K et al. C-X-C motif chemokine 12/C-X-C chemokine receptor type 7 signaling regulates breast cancer growth and metastasis by modulating the tumor microenvironment. Breast Cancer Res 2014; 16: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AM, Bird D, Welti JC, Gourlaouen M, Lang G, Murray GI et al. Lysyl oxidase plays a critical role in endothelial cell stimulation to drive tumor angiogenesis. Cancer Res 2013; 73: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TR, Rumney RM, Schoof EM, Perryman L, Hoye AM, Agrawal A et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature 2015; 522: 106–U279. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang F, Zhang RX, Beischlag TV, Muchardt C, Yaniv M, Hankinson O. Roles of Brahma and Brahma/SWI2-related gene 1 in hypoxic induction of the erythropoietin gene. J Biol Chem 2004; 279: 46733–46741. [DOI] [PubMed] [Google Scholar]
- Halliday GM, Bock VL, Moloney FJ, Lyons JG. SWI/SNF: a chromatin-remodelling complex with a role in carcinogenesis. Int J Biochem Cell Biol 2009; 41: 725–728. [DOI] [PubMed] [Google Scholar]
- Wu Q, Madany P, Akech J, Dobson JR, Douthwright S, Browne G et al. The SWI/SNF ATPases are required for triple negative breast cancer cell proliferation. J Cell Physiol 2015; 230: 2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 2006; 1: 1112–1116. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MA, Filho JL, Dalmolin RJ, Sinigaglia M, Moreira JC, Mombach JC et al. ViaComplex: software for landscape analysis of gene expression networks in genomic context. Bioinformatics 2009; 25: 1468–1469. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.