Abstract
The leaves of Hibiscus sabdariffa L. have been used as traditional folk medicines for treating high blood pressure and fever. There are many accessions of H. sabdariffa L. throughout the world. To assess the chemical variations of 31 different accessions of H. sabdariffa L., fingerprinting analysis and quantitation of major flavonoids were performed by high‐performance liquid chromatography (HPLC). The HPLC method was validated for linearity, sensitivity, precision, repeatability and accuracy. A quadrupole‐time‐of‐flight mass spectrometry (Q‐TOF‐MS) was applied for the characterization of major compounds. A total of 9 compounds were identified, including 6 flavonoids and 3 phenolic acids. In the fingerprint analysis, similarity analysis (SA) and principal component analysis (PCA) were used to differentiate the 31 accessions of H. sabdariffa L. Based on the results of PCA and SA, the samples No. 15 and 19 appeared much different from the main group. The total content of five flavonoids varied greatly among different accessions, ranging from 3.35 to 23.30 mg/g. Rutin was found to be the dominant compound and the content of rutin could contribute to chemical variations among different accessions. This study was helpful to understand the chemical variations between different accessions of H. sabdariffa L., which could be used for quality control. © 2015 The Authors Biomedical Chromatography Published by John Wiley & Sons Ltd.
Keywords: Hibiscus sabdariffa L, flavonoids, fingerprints, principal components analysis (PCA), LC‐Q‐TOF‐MS
Abbreviations used
- ACN
acetonitrile
- HPLC
high performance liquid chromatography
- ICH
International Conference on Harmonisation
- LC‐Q‐TOF‐MS
liquid chromatography‐quadrupole‐time‐of‐flight mass spectrometry
- LOD
limit of detection
- LOQ
limit of quantification
- PA
peak area
- PCA
principal component analysis
- PC1
the first principal component
- PC2
the second principal component
- RSD
relative standard deviations
- RT
retention time
- SA
similarity analysis
- S/N
signal‐to‐noise ratio
- SUAREC
Southern University Agricultural Research and Extension Center
- USDA‐ARS
United States Department of Agriculture‐Agricultural Research Service
- UV
ultraviolet
- WHO
World Health Organization.
Introduction
Hibiscus sabdariffa L. (family: Malvaceae) is used for both food and traditional medicine (Da‐Costa‐Rocha et al., 2014). It is most popular for its calyces used as sour tea (Ali et al., 2005). In India, Africa and Mexico, infusions of the leaves or calyces are traditionally used for their diuretic, cholerectic, febrifugal and hypotensive effects (Kuo et al., 2012; Guardiola and Mach, 2014). Research has shown that H. sabdariffa L. leaves have multiple biological activities, such as antiatherosclerotic effect (Chen et al., 2013), anticancer activity (Lin et al., 2012), antioxidant and antihyperlipidemic activities (Ochani and D'Mello, 2009; Gosain et al., 2010; Sindi et al., 2014). Flavonoids and phenolic acids are considered as the major bioactive compounds in the leaves of H. sabdariffa L. (Chen et al., 2013).
Many accessions (samples of a crop variety collected at a specific location and time) of H. sabdariffa L. are widely cultivated in Africa, Asia, and America (Patel, 2014). The H. sabdariffa L. leaves from different countries and accessions could have different chemical constituents, which may result in the improper clinical usage under the same name. Most of the research about H. sabdariffa L. does not specify the origin of the variety and the crop site, making it difficult to make comparisons between the chemical profile and bioactivities of extracts obtained in different studies (Borrás‐Linares et al., 2015). Furthermore, the amount of bioactive compounds in H. sabdariffa L. leaves is an important aspect that influences their therapeutic effects. Therefore, to evaluate chemical variations of different accessions of H. sabdariffa L. is needed.
A strategy for clarifying the chemical variations of different accessions of H. sabdariffa L. consist of two aspects. One is the qualitative and quantitative analysis of several bioactive components (Jin et al., 2008). The other is chemical fingerprint analysis, which has been accepted by World Health Organization (WHO) (Kong et al., 2009; World Health Organization, 1991). At present, fingerprint analysis, based on multivariate statistical analysis, such as similarity analysis (SA) and principal component analysis (PCA), is widely applied to discriminate the medicinal plants (Tian et al., 2009; Xu et al., 2011), and fruits (Sârbu et al., 2012).
Flavonoids in edible and medicinal plants possess wide range of biochemical and pharmacological effects. Rutin, quercetin and its derivatives, and kaempferol and its derivatives are identified as major flavonoids in H. sabdariffa L. leaves (Zhen et al., 2016). The flavonoid content is an important factor in plant foods, which has been archived in the United States Department of Agriculture (USDA) database (U.S. Department of Agriculture, Agricultural Research Service, 2013). Therefore, flavonoids could be used as marker compounds to evaluate chemical consistency among 31 H. sabdariffa L. leaves. In our previous studies, high radical scavenging activity was observed in the leaves of H. sabdariffa L. The activity varied among different accessions (Wang et al., 2014). Therefore, this study was designed to profile the chemical fingerprint in the leaves of 31 H. sabdariffa L. accessions cultivated in United States. A Q‐TOF‐MS was carried out to identify the major fingerprint peaks. The chemical profiles of H. sabdariffa L. leaves were investigated by chemometric methods. Moreover, a reliable HPLC method was developed and validated for simultaneous determination of five flavonoids in the leaves of 31 H. sabdariffa L. accessions.
Materials and methods
Chemicals and reagents
HPLC‐grade acetonitrile (ACN), methanol, water and formic acid were obtained from Fisher Scientific (Fair Lawn, NJ, USA). Standards including quercetin (95% purity) and rutin (97% purity) were purchased from Acros Organics (Morris Plains, NJ, USA). Kaempferol (97% purity) was purchased from Sigma Aldrich (St. Louis, MO, USA). Kaempferol‐3‐o‐rutinoside (98% purity) and kaempferol‐3‐o‐glucoside (99% purity) were obtained from Indofine Chemicals (Hillsborough, NJ). Their chemical structures are shown in Fig. 1.
Figure 1.
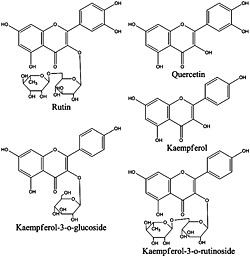
Structures of five investigated flavonoids.
Plant materials
A total of 31 accessions of H. sabdariffa L. were included in this study. Their sample identity numbers, country origins, and accession labels are listed in the Table 1. The first 25 accessions were obtained from United States Department of Agriculture‐Agricultural Research Service (USDA‐ARS) Plant Genetic Resources Conservation Unit in Griffin, Georgia and the additional 6 were collected by the Southern University Agricultural Research and Extension Center (SUAREC) Hibiscus Research Group.
Table 1.
Thirty‐one accessions of H. sabdariffa L.
| No. | Country (seed source) | Accession label |
|---|---|---|
| 1 | India | PIa‐180026 |
| 2 | Cuba | PI‐207920 |
| 3 | Bangladesh | PI‐256038 |
| 4 | Bangladesh | PI‐256039 |
| 5 | Poland | PI‐256041 |
| 6 | Cuba, La Habana | PI‐265319 |
| 7 | Sudan | PI‐267778 |
| 8 | Nigeria | PI‐268097 |
| 9 | Nigeria | PI‐268100 |
| 10 | Taiwan | PI‐273388 |
| 11 | Taiwan | PI‐273389 |
| 12 | Taiwan | PI‐273391 |
| 13 | South Africa, Transvaal | PI‐273459 |
| 14 | Nigeria | PI‐274245 |
| 15 | Senegal | PI‐275413 |
| 16 | Ghana | PI‐286316 |
| 17 | Ghana | PI‐286319 |
| 18 | Thailand | PI‐365477 |
| 19 | United States | PI‐468411 |
| 20 | United States, Georgia | PI‐468413 |
| 21 | Sudan | PI‐496717 |
| 22 | Sudan | PI‐496938 |
| 23 | Zambia | PI‐500725 |
| 24 | Zambia | PI‐500737 |
| 25 | South Africa | PI‐638933 |
| 26 | Jamaica | SUARECb‐1 |
| 27 | Liberia | SUAREC‐2 |
| 28 | Nigeria | SUAREC‐3 |
| 29 | Senegal | SUAREC‐4 |
| 30 | Thailand | SUAREC‐5 |
| 31 | South Africa | SUAREC‐6 |
PI, Plant Identification. The PI numbers were assigned by the USDA‐ARS.
SUAREC, the accessions collected by the Southern University Agricultural Research and Extension Center.
The seeds from all the accessions were germinated in the greenhouse in March 2012. On May 14, 2012, the seedlings were transplanted to the field in the Horticultural Farm at Southern University, Baton Rouge, Louisiana, USA. The leaves were collected in June 2012 and authenticated by Dr. Kit L. Chin (Southern University Agricultural Research and Extension Center). Leaf samples were oven‐dried and ground into fine powder.
Preparation of standard and sample solutions
Dried H. sabdariffa L. leaves powder (0.20 g) were accurately weighed and extracted with 70% aqueous methanol (20 mL) by ultrasonic‐assisted extraction (44 KHz, 70 W, Branson Ultrasonic Corporation, Danbury, CT, USA) for 30 min at room temperature(Chen et al., 2012; Zu et al., 2006). Each sample was prepared in triplicate for quantification of analytes. The sample solutions were filtered through 0.45‐µm membrane.
Stock solutions of the five analytes were prepared in methanol. The stock solution was diluted with 70% methanol to yield a series of appropriate concentrations. All the prepared samples were stored at ‐10°C till their analysis.
HPLC and Q‐TOF‐MS analysis conditions
HPLC analysis was carried out on an Agilent 1200 series equipped with a diode array detector (Agilent Technologies, USA). Chromatographic separation was performed on a C18 column (4.6 × 150 mm, 3.5 µm, Zorbax eclipse plus, Agilent USA) at 30°C with a guard column (4.6 × 12.5 mm, 5 µm, Zorbax eclipse plus). The mobile phase consisted of H2O (solvent A) and ACN (solvent B) with 0.1% (v/v) formic acid, respectively. The gradient program was as follows: 0‐19 min, 10‐46% B; 19‐20 min, 46‐10% B. A post‐run equilibrium time of 3 min was used for all samples. The flow rate was set at 1 mL/min with ultraviolet (UV) detection at 350 nm. The T splitter gave a flow rate of 0.25 mL/min toward the MS detector.
Mass spectrometry was performed using an Agilent 6540 Q‐TOF‐MS system (Agilent Corp., USA) equipped with a jet stream ESI interface. The TOF/MS system was operated in both negative and positive ion modes. Mass spectra were recorded over the mass range m/z 50‐1100. The mass analysis conditions were set as follows: drying gas (N2) flow rate, 10 L/min; nebulizer, 40 psi; drying gas temperature, 350°C; sheath gas temperature, 350°C; capillary voltage, 3500 V; fragmentor voltage, 120 V.
Method validation for the determination of five flavonoids
The linearity, precision (inter‐day and intra‐day), repeatability, and recovery were carried out to validate the HPLC method according to ICH guideline (Validation of Analytical Procedures: Text and Methodology Q2 (R1), 2005)
Six working solutions were analyzed for the construction of calibration curves. Linearity was evaluated by the calculation of a regression line by the method of least squares. The limits of detection (LOD) and quantification (LOQ) were estimated experimentally by injecting a series of dilute solutions with known concentrations until the signal‐to‐noise ratio (S/N) for the standards reached a 3:1 ratio for LOD and 10:1 for LOQ, respectively. Intra‐ and inter‐day variations were applied to determine the precision of the developed method. The repeatability of the proposed HPLC method was studied at three levels (0.10 g, 0.20 g, 0.25 g) of the sample No.15 (Senegal). The samples of each level were extracted and analyzed triplicates. The accuracy of the method was tested by detecting the recovery, which was evaluated by adding three concentration levels (low, middle and high) of standard solutions into certain amount (0.10 g) of sample No.15 (Senegal) (Peng et al., 2013). The samples were extracted and analyzed for quantitative analysis as the developed method mentioned above.
Data analysis
Accurate mass data were recorded and processed by MassHunter B.04.00 software (Agilent Technologies, USA). Principal components analysis (PCA) and similarity analysis (SA) were performed to analyze the 31 samples of H. sabdariffa L. based on the common characteristic peaks. PCA and SA were calculated and generated using a professional software named “ChemPattern 1.0.1.0” (fingerprint chromatography processing software, ChemMind Technologies (Beijing) Co., Ltd., China). All the data were pretreated including data normalization and chromatograms alignment before PCA and SA.
Results and discussion
Optimization of sample extraction and chromatographic conditions
According to the previous studies, methanol‐water (70:30) has a higher efficiency in extracting flavonoids than ethanol‐water (70:30) (Chen et al., 2012). Ultrasonic extraction is considered a simpler and more effective method for extraction of flavonoids in the plant leaves. Moreover, ultrasonication for 30 min gave a similar result as the soxhlet for 240 min (Zu et al., 2006). The ultrasonic extraction method in this study was also optimized including extraction times (30 min, 45 min and 60 min), extraction temperatures (room temperature, 35°C and 45°C) and times of extraction (once and twice). The peak areas of the five flavonoids were used as a marker for evaluation of extraction efficiency. There were no significant differences in the peak areas of the five flavonoids among different extraction conditions as described above. Therefore, the simple and convenient extraction method was selected as follows: solvent, 70% methanol; extraction temperature, room temperature; ultrasonic extraction time, 30 min.
HPLC conditions including chromatographic column, mobile phase and detection wavelength were optimized. The best results were obtained using an Agilent zorbax eclipse plus C18 (150 × 4.6 mm i.d., 3.5 µm) column at 30°C, with gradient elution of 0.1 % aqueous formic acid and ACN with 0.1% formic acid (v/v) as the mobile phase. According to the peak area of principal peaks, 350 nm was chosen as the detection wavelength. Under the optimized conditions, the separation of five marker compounds can be easily achieved in 20 min. Typical HPLC‐DAD chromatograms are shown in Fig. 2A.
Figure 2.
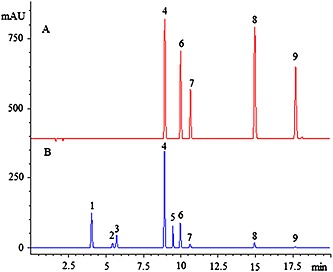
Chromatograms of mixed standards (A) including: rutin (4), kaempferol‐3‐o‐rutinoside (6), kaempferol‐3‐o‐glucoside (7), quercetin (8), kaempferol (9), and (B) common pattern of HPLC fingerprint of 31 samples of H. sabdariffa L.
HPLC fingerprint analysis
The fingerprints of 31 samples of H. sabdariffa L. were obtained under the optimal HPLC conditions and are shown in Fig. 3. Peaks that existed in all the samples were assigned as “common pattern”, which was generated based on all chromatograms by the professional software of ChemPattern 1.0.1.0 (Fig. 2B).
Figure 3.
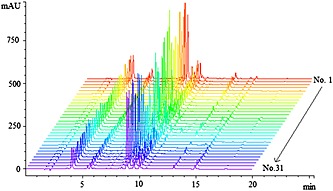
The HPLC fingerprints of the 31 H. sabdariffa L. samples at wavelength 350 nm.
HPLC fingerprint method precision and reproducibility were evaluated by the analysis of five runs of the same sample solution and five replicates from the same sample, respectively. The relative standard deviations (RSD) of retention time (RT) and peak area (PA) of 9 characteristic peaks in the precision test were found in the range of 0.15‐3.27%, whereas in the reproducibility test the RSDs of RT and PA were also below 0.75 and 4.26%, respectively. The stability of sample solution was evaluated at different time points (0, 2, 4, 8, 12 and 24 h), and the RSDs of RT and PA were less than 0.32 and 3.90%, respectively. All of these results indicated that the HPLC fingerprint method was reliable.
Identification of major compounds in H. sabdariffa L.
The structural identification of 9 characteristic peaks was performed by the LC‐Q‐TOF‐MS. Both negative and positive ion modes were used because they provided more information about chemical structure. Some phenolic acids, flavonoids and ascorbic acid have been reported in the leaves of H. sabdariffa L. (Rodriguez‐Medina et al., 2009; Wang et al., 2014; Kumar et al., 2015). In this study, the nine characteristic peaks were identified by comparison of their retention time and accurate MS with those of reference standards. The MS data are shown in Table 2. Of these 9 compounds identified, the 5 flavonoids (rutin, kaempferol‐3‐o‐rutinoside, kaempferol‐3‐o‐glucoside, quercetin and kaempferol) were chosen as the marked components.
Table 2.
Identification of chemical constituents in H. sabdariffa L. by LC‐Q‐TOF‐MS
| No. | RT (min) | [M + H]+ | Main fragments | Measured m/z | Calculated m/z | Error (ppm) | Formula | Identification* |
|---|---|---|---|---|---|---|---|---|
| 1. | 4.05 | 355.1025 | 163.0390 | 354.0952 | 354.0951 | ‐0.33 | C16H18O9 | Neochlorogenic acid |
| 2. | 5.45 | 355.1020 | 163.0390 | 354.0947 | 354.0951 | 1.14 | C16H18O9 | Chlorogenic acid |
| 3. | 5.71 | 355.1029 | 163.0391 | 354.0956 | 354.0951 | ‐1.46 | C16H18O9 | Cryptochlorogenic acid |
| 4 | 8.90 | 611.1607 | 465.1025, 303.0466 | 610.1526 | 610.1534 | 1.36 | C27H30O16 | Rutin |
| 5 | 9.50 | 465.1033 | 303.0498 | 464.0960 | 464.0955 | ‐1.1 | C21H20O12 | Isoquercitin |
| 6 | 10.01 | 595.1663 | 449.1075, 287.0553 | 594.1590 | 594.1585 | ‐0.86 | C27H30O15 | Kaempferol‐3‐o‐rutinoside |
| 7 | 10.66 | 449.1070 | 287.0541 | 448.0997 | 448.1006 | 1.88 | C21H20O11 | Kaempferol‐3‐o‐glucoside |
| 8 | 14.96 | 303.0495 | 158.0028 | 302.0422 | 302.0427 | 1.39 | C15H10O7 | Quercetin |
| 9 | 17.67 | 287.0549 | 158.0029 | 286.0476 | 286.0477 | 0.46 | C15H10O6 | Kaempferol |
All the identified compounds were compared with standard compounds.
Method validation for the determination of five flavonoids
The linear ranges, regression equations, LODs and LOQs of the five analytes were detected using the developed HPLC method. As shown in Table 3, the calibration curves of the analytes showed good linearity (r 2 ≥ 0.9996) with given concentration ranges. LOD and LOQ values were less than 0.10 µg/mL and 0.35 µg/mL, respectively, and showed the adequate sensitivity of the proposed method. The intra‐ and inter‐day variations (RSD) of five analytes peak areas were less than 0.67% and 0.90%, while retention time were less than 0.26% and 0.26%, respectively (Table 4). The average recoveries obtained in this study ranged from 86.80% to 103.50% (Table 5). The repeatability of the developed method was evaluated at three levels (Table 6). The results showed that the repeatability (RSD, n = 3) was less than 1.05% (0.10 g), 0.81% (0.20 g), 1.29% (0.25 g), respectively. Therefore, the proposed HPLC method could be considered accurate for quantitative determination of the five investigated compounds.
Table 3.
Linearity, LOD and LOQ in the determination of analytes
| Compounds | Linear range (µg/mL) | Regression equationa | Correlation coefficient r 2 | LOD (µg/mL) | LOQ (µg/mL) |
|---|---|---|---|---|---|
| Rutin | 2.65‐340.00 | y = 14.832x + 1.400 | 0.9996 | 0.10 | 0.35 |
| Kaempferol‐3‐o‐rutinoside | 4.00‐122.00 | y = 17.225x‐17.691 | 0.9997 | 0.10 | 0.32 |
| Kaempferol‐3‐o‐glucoside | 0.20‐50.00 | y = 18.385x‐0.512 | 1.0000 | 0.07 | 0.20 |
| Quercetin | 0.40‐20.00 | y = 25.426x‐0.847 | 0.9999 | 0.09 | 0.28 |
| Kaempferol | 0.20‐15.75 | y = 35.377x‐0.172 | 0.9998 | 0.07 | 0.20 |
y is the peak area; x is the concentration (µg/mL).
Table 4.
Intra‐ and interday precision of the investigated analytes
| Analytes | Concentration Level (ug/mL) | Intra‐day (RSD, %, n = 6) | Inter‐day (RSD, %, n = 3) | ||
|---|---|---|---|---|---|
| Retention time | Peak area | Retention time | Peak area | ||
| Rutin | 3.4 | 0.26 | 0.18 | 0.17 | 0.21 |
| 34.0 | 0.09 | 0.09 | 0.19 | 0.09 | |
| 170.0 | 0.06 | 0.07 | 0.26 | 0.32 | |
| Kaempferol‐3‐o‐rutinoside | 2.0 | 0.26 | 0.29 | 0.11 | 0.24 |
| 20.0 | 0.07 | 0.11 | 0.18 | 0.06 | |
| 100.0 | 0.06 | 0.06 | 0.23 | 0.32 | |
| Kaempferol‐3‐o‐glucoside | 1.0 | 0.25 | 0.30 | 0.09 | 0.16 |
| 10.0 | 0.07 | 0.10 | 0.18 | 0.05 | |
| 50.0 | 0.06 | 0.06 | 0.23 | 0.32 | |
| Quercetin | 2.0 | 0.20 | 0.64 | 0.26 | 0.90 |
| 20.0 | 0.08 | 0.12 | 0.13 | 0.45 | |
| 100.0 | 0.03 | 0.08 | 0.18 | 0.40 | |
| Kaempferol | 1.0 | 0.17 | 0.67 | 0.03 | 0.37 |
| 10.0 | 0.07 | 0.11 | 0.12 | 0.09 | |
| 50.0 | 0.04 | 0.06 | 0.16 | 0.27 | |
Table 5.
Recoveries for the assay of five compounds in H. sabdariffa L. leaves
| Analytes | Original amount (µg) | Spiked (µg) | Amount founda (µg) | Recoveryb (%) | RSDc (%) |
|---|---|---|---|---|---|
| Rutin | 683.05 | 347.70 | 1036.64 | 101.69 | 2.59 |
| 683.05 | 695.40 | 1402.76 | 103.50 | 0.70 | |
| 683.05 | 1043.10 | 1752.19 | 102.50 | 1.50 | |
| Kaempferol‐3‐o‐rutinoside | 888.67 | 940.00 | 1801.70 | 97.13 | 3.72 |
| 888.67 | 1880.00 | 2696.76 | 96.18 | 1.94 | |
| 888.67 | 2820.00 | 3336.39 | 86.80 | 1.49 | |
| Kaempferol‐3‐o‐glucoside | 194.46 | 220.00 | 410.85 | 98.36 | 5.04 |
| 194.46 | 440.00 | 632.84 | 99.63 | 1.76 | |
| 194.46 | 660.00 | 774.56 | 87.89 | 1.94 | |
| Quercetin | 35.33 | 31.10 | 66.44 | 100.03 | 1.68 |
| 35.33 | 62.20 | 96.67 | 98.62 | 5.70 | |
| 35.33 | 93.30 | 128.97 | 100.36 | 1.58 | |
| Kaempferol | 22.37 | 13.02 | 34.75 | 95.08 | 1.67 |
| 22.37 | 26.04 | 46.82 | 93.89 | 3.01 | |
| 22.37 | 39.06 | 59.16 | 94.19 | 0.70 |
The data was present as average of three determination.
Recovery (%) = (amount found‐original amount)/spiked amount × 100.
RSD (%) = (recovery SD/mean) × 100.
Table 6.
Repeatability of five investigated analytes
| Analytes | Repeatability (RSD, %, n = 3) | ||
|---|---|---|---|
| Level I (0.1 g of sample) | Level II (0.2 g of sample) | Level III (0.25 g of sample) | |
| Rutin | 0.58 | 0.81 | 0.46 |
| Kaempferol‐3‐o‐rutinoside | 0.84 | 0.65 | 0.41 |
| Kaempferol‐3‐o‐glucoside | 0.80 | 0.73 | 0.30 |
| Quercetin | 0.54 | 0.26 | 1.29 |
| Kaempferol | 1.05 | 0.54 | 0.53 |
Quantitative analysis of five analytes in 31 H. sabdariffa L. accessions
The developed HPLC method was successfully applied to the simultaneous quantification of the five marker compounds in 31 accessions of H. sabdariffa L. The results are shown in Table 7. The quantitative analysis results showed that the content ranges (mg/g) were 0.47‐19.16 (rutin), 0.66‐8.65 (kaempferol‐3‐o‐rutinoside), 0.18‐1.94 (kaempferol‐3‐o‐glucoside), 0.18‐0.82 (quercetin), and 0.03‐0.22 (kaempferol), respectively. The total content of the five flavonoids showed great variations among different accessions, ranging from 3.35 to 23.30 mg/g. Among the tested samples, the sample No. 2 from Cuba had the highest contents of five flavonoids, while the sample No. 19 from USA had the lowest total amount (Table 7).
Table 7.
Contents of the investigated compounds in the leaves of H. sabdariffa L.
| No.a | Content, (mg/g, mean ± SD, n = 3) | |||||
|---|---|---|---|---|---|---|
| Rutin | Kaempferol‐3‐o‐rutinoside | Kaempferol‐3‐o‐glucoside | Quercetin | Kaempferol | Totalb | |
| 1 | 12.864 ± 0.230 | 2.860 ± 0.053 | 0.254 ± 0.029 | 0.333 ± 0.015 | 0.050 ± 0.001 | 16.361 |
| 2 | 18.810 ± 0.249 | 3.646 ± 0.067 | 0.355 ± 0.003 | 0.433 ± 0.005 | 0.062 ± 0.001 | 23.304 |
| 3 | 14.845 ± 0.174 | 3.448 ± 0.055 | 0.333 ± 0.003 | 0.277 ± 0.005 | 0.040 ± 0.001 | 18.943 |
| 4 | 11.182 ± 0.123 | 2.495 ± 0.023 | 0.228 ± 0.019 | 0.181 ± 0.017 | 0.027 ± 0.002 | 14.112 |
| 5 | 15.821 ± 0.467 | 4.069 ± 0.129 | 0.336 ± 0.029 | 0.305 ± 0.021 | 0.048 ± 0.003 | 20.579 |
| 6 | 9.988 ± 0.130 | 2.526 ± 0.040 | 0.270 ± 0.006 | 0.200 ± 0.005 | 0.032 ± 0.000 | 13.017 |
| 7 | 11.882 ± 0.165 | 2.809 ± 0.045 | 0.291 ± 0.009 | 0.224 ± 0.000 | 0.031 ± 0.000 | 15.237 |
| 8 | 9.820 ± 0.273 | 2.828 ± 0.082 | 0.344 ± 0.008 | 0.489 ± 0.011 | 0.083 ± 0.001 | 13.565 |
| 9 | 11.889 ± 0.201 | 2.277 ± 0.063 | 0.275 ± 0.002 | 0.471 ± 0.007 | 0.061 ± 0.001 | 14.974 |
| 10 | 19.164 ± 0.262 | 2.642 ± 0.043 | 0.269 ± 0.005 | 0.370 ± 0.009 | 0.038 ± 0.001 | 22.483 |
| 11 | 15.098 ± 0.510 | 2.654 ± 0.067 | 0.223 ± 0.010 | 0.380 ± 0.013 | 0.050 ± 0.001 | 18.406 |
| 12 | 9.765 ± 0.115 | 2.310 ± 0.016 | 0.208 ± 0.019 | 0.219 ± 0.008 | 0.032 ± 0.000 | 12.534 |
| 13 | 15.173 ± 0.441 | 2.177 ± 0.065 | 0.189 ± 0.003 | 0.394 ± 0.005 | 0.042 ± 0.000 | 17.976 |
| 14 | 12.259 ± 0.243 | 2.713 ± 0.052 | 0.354 ± 0.016 | 0.389 ± 0.015 | 0.057 ± 0.000 | 15.772 |
| 15 | 6.758 ± 0.020 | 8.654 ± 0.057 | 1.944 ± 0.014 | 0.347 ± 0.001 | 0.217 ± 0.001 | 17.920 |
| 16 | 11.075 ± 0.187 | 3.105 ± 0.056 | 0.713 ± 0.013 | 0.494 ± 0.013 | 0.077 ± 0.003 | 15.463 |
| 17 | 14.711 ± 0.295 | 2.844 ± 0.056 | 0.375 ± 0.011 | 0.397 ± 0.006 | 0.053 ± 0.001 | 18.381 |
| 18 | 10.490 ± 0.410 | 2.572 ± 0.103 | 0.255 ± 0.014 | 0.276 ± 0.015 | 0.043 ± 0.001 | 13.637 |
| 19 | 0.465 ± 0.002 | 0.661 ± 0.008 | 1.458 ± 0.012 | 0.693 ± 0.010 | 0.072 ± 0.001 | 3.349 |
| 20 | 10.089 ± 0.224 | 2.333 ± 0.041 | 0.206 ± 0.009 | 0.319 ± 0.008 | 0.052 ± 0.002 | 12.999 |
| 21 | 6.783 ± 0.258 | 1.721 ± 0.057 | 0.776 ± 0.032 | 0.426 ± 0.018 | 0.064 ± 0.002 | 9.770 |
| 22 | 7.050 ± 0.086 | 1.703 ± 0.049 | 0.773 ± 0.014 | 0.430 ± 0.008 | 0.062 ± 0.000 | 10.019 |
| 23 | 11.934 ± 0.125 | 2.553 ± 0.027 | 0.314 ± 0.022 | 0.371 ± 0.014 | 0.058 ± 0.001 | 15.229 |
| 24 | 14.123 ± 0.185 | 3.488 ± 0.027 | 0.478 ± 0.005 | 0.437 ± 0.002 | 0.079 ± 0.001 | 18.605 |
| 25 | 15.309 ± 0.202 | 2.202 ± 0.051 | 0.190 ± 0.020 | 0.563 ± 0.013 | 0.059 ± 0.001 | 18.322 |
| 26 | 15.979 ± 0.707 | 2.318 ± 0.057 | 0.203 ± 0.026 | 0.636 ± 0.018 | 0.066 ± 0.003 | 19.203 |
| 27 | 12.689 ± 0.358 | 1.881 ± 0.058 | 0.177 ± 0.002 | 0.508 ± 0.012 | 0.055 ± 0.002 | 15.309 |
| 28 | 16.067 ± 0.238 | 2.228 ± 0.052 | 0.232 ± 0.009 | 0.522 ± 0.012 | 0.051 ± 0.001 | 19.100 |
| 29 | 13.731 ± 0.266 | 1.870 ± 0.049 | 0.202 ± 0.016 | 0.520 ± 0.012 | 0.050 ± 0.000 | 16.374 |
| 30 | 15.151 ± 0.252 | 2.035 ± 0.054 | 0.199 ± 0.023 | 0.822 ± 0.008 | 0.079 ± 0.001 | 18.286 |
| 31 | 15.218 ± 0.082 | 2.241 ± 0.017 | 0.226 ± 0.014 | 0.544 ± 0.008 | 0.057 ± 0.001 | 18.286 |
| Meanc | 12.457 | 2.705 | 0.408 | 0.418 | 0.059 | 16.049 |
| Min | 0.465 | 0.661 | 0.177 | 0.181 | 0.027 | 3.349 |
| Max | 19.164 | 8.654 | 1.944 | 0.822 | 0.217 | 23.304 |
Sample No. corresponds to Table 1.
Total represents the sum of the individual selected five flavonoids.
Mean, Min and Max were obtained based on the contents of 31 samples.
Based on the previous studies, it is found that both the total flavonoid content and antioxidant activity of H. sabdariffa L. leaves were higher than those of H. sabdariffa L. flowers (Chen et al., 2013; Mohd‐Esa et al., 2010). Variations in antioxidant activity may be due to different phenolic, flavonoid and ascorbic acid contents (Kumar et al., 2015). Therefore the total antioxidant activities of samples could vary greatly among different accessions of H. sabdariffa L.
Principal component analysis (PCA)
To evaluate the variations among the 31 accessions, PCA was performed. The data of chromatographic fingerprints were imported into the ChemPattern 1.0.1.0 software. The score plot of the first two principal components (PC1‐PC2) is shown in Fig. 4.
Figure 4.
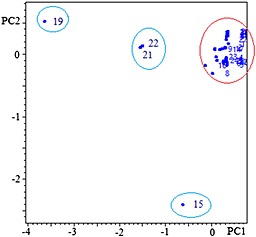
PC1‐PC2 score plot of 31 samples. The labels of the samples refer to Table 1.
The first two principal components (PC1‐PC2) were accounted for 85.9% of the total variance of the samples. The PCA analysis showed that the differences observed in the H. sabdariffa L. accessions were derived from the concentration of rutin. PC1, which explained 63.4% of the variance, was positively correlated with rutin content. For PC2 significant variables were flavonoid content and kaempferol‐3‐o‐rutinoside content.
PCA analysis revealed clearly the relationships among the tested samples. As shown in Fig. 4, the two samples No. 21 and 22 were clustered in one group, which were from Sudan. Except the sample No. 15 from Senegal and No. 19 from USA, the other samples can be clustered in one main group. The main group may be considered as rutin‐rich chemotype, which contains much more rutin than others. The results also indicated that samples No.15 and 19 produced greater variations in their chemical compositions and content. Because all the leaf samples were obtained from the accessions grown in the same place and under the same cultivation conditions, such variations may result from the inherent variability of the accessions. No. 19 could be a low quality accession in terms of its major flavonoid contents.
Similarity analysis (SA)
Similarity analysis is a conventional method describing the similarity among the fingerprints. In this study, cosine similarity algorism was applied for the similarity analysis. By comparison with the fingerprint common pattern of H. sabdariffa L., the similarity index of 31 samples was not less than 0.983 except those of samples No. 15, 19, 21 and 22 (Fig. 5).
Figure 5.
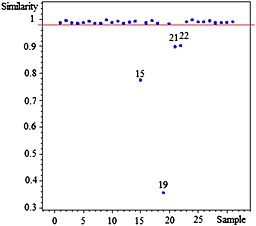
Similarity evaluation of HPLC fingerprint for H. sabdariffa L. Different samples (No.1‐31) are listed in Table 1.
As shown in Fig. 5, taking 0.98 as the threshold, the samples with the correlation coefficients above it should be clustered to a group, which has been properly proved by the previous results from PCA. The different similarity values of samples means the different internal quality of these samples. Therefore, the developed HPLC fingerprint common pattern could be as a quality assessment model for classifying H. sabdariffa L. accessions.
Generally, the climatic and edaphic conditions are important environmental factors that can affect chemical composition of the leaf samples. In order to eliminate the environmental effect, we collected the seeds from 31 accessions and germinated and grew them in the same environment. The leaves were collected at the same time for quantitative and chemical fingerprint analyses. We assume the chemical variations among the accessions could be attributed to the variations of the seed sources where these individual accessions may have adapted to their original climatic and edaphic conditions. Therefore, the resulting genetic variations could be one of the key factors affecting the contents of bioactive compounds and quality of H. sabdariffa L. Many of these accessions could also be called ecotypes of the species. This study is a first comprehensive evaluation of the chemical variations among 31 H. sabdariffa L. accessions based on the combination of quantitative and chromatographic fingerprint analyses. Because of the tremendous potential of utilizing the leaves of this species for food, nutrition, and medicine, as well as value added products for human health, this research would be helpful for the quality control of H. sabdariffa L. in the future.
Conclusion
A strategy for clarifying the chemical variations of different accessions of H. sabdariffa L. was developed. Firstly, chemical fingerprint analysis was performed based on the multivariate analysis methods (PCA and SA). Secondly, a simple HPLC method was developed and validated for simultaneously quantitative analysis of major flavonoids in 31 accessions of H. sabdariffa L. The samples were clustered in one group except four accessions (No.15, 19, 21 and 22). The chemical differences could be due to the inherent variability of the accessions. The proposed HPLC and fingerprint method were validated and proved to be reliable. The chemical fingerprint analysis is helpful to clarify the relationship among different accessions of H. sabdariffa L. and useful for quality control.
Acknowledgments
The authors would like to acknowledge the financial support from the Project of the State Forestry Administration of China (2014‐4‐33) and USDA‐NIFA grants #2008‐38814‐04772 and 2012‐38821‐20092, and the Central Public‐Interest Scientific Institution Basal Research Fund, China (No.1632014009). We are also grateful to Mr. Run‐tao Tian for his technical assistance.
Wang, J. , Cao, X. , Ferchaud, V. , Qi, Y. , Jiang, H. , Tang, F. , Yue, Y. , and Chin, K. L. (2016) Variations in chemical fingerprints and major flavonoid contents from the leaves of thirty‐one accessions of Hibiscus sabdariffa L.. Biomed. Chromatogr., 30: 880–887. doi: 10.1002/bmc.3623.
References
- Ali BH, Al Wabel N and Blunden G. Phytochemical, pharmacological and toxicological aspects of Hibiscus sabdariffa L.: A review. Phytotherapy Research 2005; 19: 369–375. [DOI] [PubMed] [Google Scholar]
- Borrás‐Linares I, Fernández‐Arroyo S, Arráez‐Roman D, Palmeros‐Suárez PA, Del Val‐Díaz R, Andrade‐Gonzáles I, Fernández‐Gutiérrez A, Gómez‐Leyva JF and Segura‐Carretero A. Characterization of phenolic compounds, anthocyanidin, antioxidant and antimicrobial activity of 25 varieties of Mexican Roselle (Hibiscus sabdariffa). Industrial Crops and Products 2015; 69: 385–394. [Google Scholar]
- Chen JH, Wang CJ, Wang CP, Sheu JY, Lin CL and Lin HH. Hibiscus sabdariffa leaf polyphenolic extract inhibits LDL oxidation and foam cell formation involving up‐regulation of LXRα/ABCA1 pathway. Food Chemistry 2013; 141: 397–406. [DOI] [PubMed] [Google Scholar]
- Chen S, Wu BH, Fang JB, Liu YL, Zhang HH, Fang LC, Guan L and Li SH. Analysis of flavonoids from lotus (Nelumbo nucifera) leaves using high performance liquid chromatography/photodiode array detector tandem electrospray ionization mass spectrometry and an extraction method optimized by orthogonal design. Journal of Chromatography A 2012; 1227: 145–153. [DOI] [PubMed] [Google Scholar]
- Da‐Costa‐Rocha I, Bonnlaender B, Sievers H, Pischel I and Heinrich M. Hibiscus sabdariffa L. ‐ A phytochemical and pharmacological review. Food Chemistry 2014; 165: 424–443. [DOI] [PubMed] [Google Scholar]
- Gosain S, Ircchiaya R, Sharma PC, Thareja S, Kalra A, Deep A and Bhardwaj TR. Hypolipidemic effect of ethanolic extract from the leaves of Hibiscus sabdariffa L. in hyperlipidemic rats. Acta Poloniae Pharmaceutica 2010; 67: 179–184. [PubMed] [Google Scholar]
- Guardiola S and Mach N. Therapeutic potential of Hibiscus sabdariffa: a review of the scientific evidence. Endocrinología y Nutrición 2014; 61: 274–295. [DOI] [PubMed] [Google Scholar]
- Jin XF, Lu YH, Wei DZ and Wang ZT. Chemical fingerprint and quantitative analysis of Salvia plebeia R.Br. by high‐performance liquid chromatography. Journal of Pharmaceutical and Biomedical Analysis 2008; 48: 100–104. [DOI] [PubMed] [Google Scholar]
- Kong WJ, Zhao YL, Xiao XH, Jin C and Li ZL. Quantitative and chemical fingerprint analysis for quality control of rhizoma Coptidischinensis based on UPLC‐PAD combined with chemometrics methods. Phytomedicine 2009; 16: 950–959. [DOI] [PubMed] [Google Scholar]
- Kumar SS, Manoj P, Shetty NP and Giridhar P. Effect of different drying methods on chlorophyll, ascorbic acid and antioxidant compounds retention of leaves of Hibiscus sabdariffa L. Journal of the Science of Food and Agriculture 2015; 9: 1812–1820. [DOI] [PubMed] [Google Scholar]
- Kuo C, Kao E, Chan K, Lee H, Huang T and Wang C. Hibiscus sabdariffa L. extracts reduce serum uric acid levels in oxonate‐induced rats. Journal of Functional Foods 2012; 4: 375–381. [Google Scholar]
- Lin H, Chan K, Sheu J, Hsuan S, Wang C and Chen J. Hibiscus sabdariffa leaf induces apoptosis of human prostate cancer cells in vitro and in vivo . Food Chemistry 2012; 132: 880–891. [Google Scholar]
- Mohd‐Esa N, Hern FS, Ismail A and Yee CL. Antioxidant activity in different parts of roselle (Hibiscus sabdariffa L.) extracts and potential exploitation of the seeds. Food Chemistry 2010; 122: 1055–1060. [Google Scholar]
- Ochani PC and D'Mello P. Antioxidant and antihyperlipidemic activity of Hibiscus sabdariffa Linn. leaves and calyces extracts in rats. Indian Journal of Experimental Biology 2009; 47: 276–282. [PubMed] [Google Scholar]
- Patel S. Hibiscus sabdariffa: An ideal yet under‐exploited candidate for nutraceutical applications. Biomedicine and Preventive Nutrition 2014; 4: 23–27. [Google Scholar]
- Peng B, Qiao CF, Zhao J, Huang WH, Hu DJ, Liu HG and Li SP. Simultaneous determination of flavonoids, isochlorogenic acids and triterpenoids in Ilex hainanensis using high performance liquid chromatography coupled with diode array and evaporative light scattering detection. Molecules 2013; 18: 2934–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Medina IC, Beltran‐Debon R, Molina VM, Alonso‐Villaverde C, Joven J, Menendez JA, Segura‐Carretero A and Fernandez‐Gutierrez A. Direct characterization of aqueous extract of Hibiscus sabdariffa using HPLC with diode array detection coupled to ESI and ion trap MS. Journal of Separation Science 2009; 32: 3441–3448. [DOI] [PubMed] [Google Scholar]
- Sârbu C, Naşcu‐Briciu RD, Kot‐Wasik A, Gorinstein S, Wasik A and Namieśnik J. Classification and fingerprinting of kiwi and pomelo fruits by multivariate analysis of chromatographic and spectroscopic data. Food Chemistry 2012; 130: 994–1002. [Google Scholar]
- Sindi HA, Marshall LJ and Morgan MR. Comparative chemical and biochemical analysis of extracts of Hibiscus sabdariffa . Food Chemistry 2014; 164: 23–29. [DOI] [PubMed] [Google Scholar]
- Tian RT, Xie PS and Liu HP. Evaluation of traditional Chinese herbal medicine: Chaihu (Bupleuri Radix) by both high‐performance liquid chromatographic and high‐performance thin‐layer chromatographic fingerprint and chemometric analysis. Journal of Chromatography A 2009; 1216: 2150–2155. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Agriculture, Agricultural Research Service . 2013. USDA Database for the Flavonoid Content of Selected Foods Release 3.1. Available online: https://www.ars.usda.gov/Services/docs.htm?docid=6231 (accessed on 15 July 2015).
- Validation of Analytical Procedures: Text and Methodology Q2 (R1) . 2005. ICH Harmonised Tripartite Guideline, International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use. Available online: http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Quality/Q2_R1/Step4/Q2_R1__Guideline.pdf (accessed on 24 November 2014).
- Wang J, Cao X, Jiang H, Qi Y, Chin KL and Yue Y. Antioxidant activity of leaf extracts from different Hibiscus sabdariffa accessions and simultaneous determination five major antioxidant compounds by LC‐Q‐TOF‐MS. Molecules 2014; 19: 21226–21238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Guidelines for the assessment of herbal medicines, 1991. http://apps.who.int/medicinedocs/en/d/Jh1813e/3.html#Jh1813e.3.1 (accessed on 3 April 2015).
- Xu X, Jiang J, Liang Y, Li X, Yi L and Cheng J. Chromatographic fingerprint analysis of Fructus Aurantii Immaturus by HPLC‐DAD and chemometric methods. Journal of Central South University of Technology 2011; 18: 353–360. [Google Scholar]
- Zhen J, Villani TS, Guo Y, Qi Y, Chin K, Pan M, Ho C, Simon JE and Wu Q. Phytochemistry, antioxidant capacity, total phenolic content and anti‐inflammatory activity of Hibiscus sabdariffa leaves. Food Chemistry 2016; 190: 673–680. [DOI] [PubMed] [Google Scholar]
- Zu Y, Li C, Fu Y and Zhao C. Simultaneous determination of catechin, rutin, quercetin kaempferol and isorhamnetin in the extract of sea buckthorn (Hippophae rhamnoides L.) leaves by RP‐HPLC with DAD. Journal of Pharmaceutical and Biomedical Analysis 2006; 41: 714–719. [DOI] [PubMed] [Google Scholar]


