Abstract
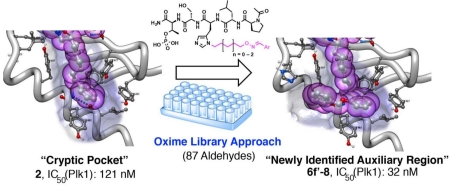
By a process involving initial screening of a set of 87 aldehydes using an oxime ligation-based strategy, we were able to achieve a several-fold affinity enhancement over one of the most potent previously known polo-like kinase 1 (Plk1) polo-box domain (PBD) binding inhibitors. This improved binding may result by accessing a newly identified auxiliary region proximal to a key hydrophobic cryptic pocket on the surface of the protein. Our findings could have general applicability to the design of PBD-binding antagonists.
Keywords: Plk1 polo-box domain, Ligand optimization, Oxime ligation, Cryptic binding pocket
Graphical abstract
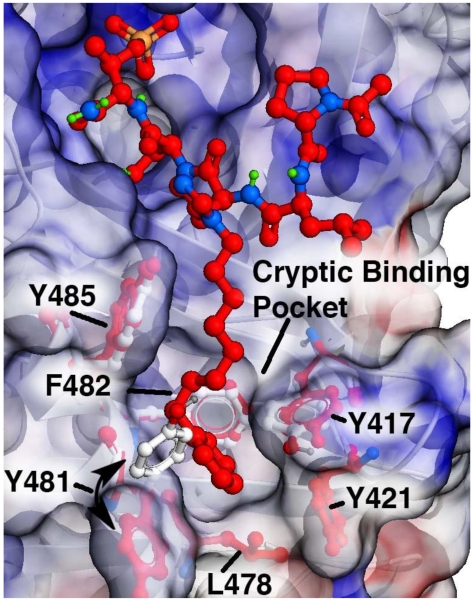
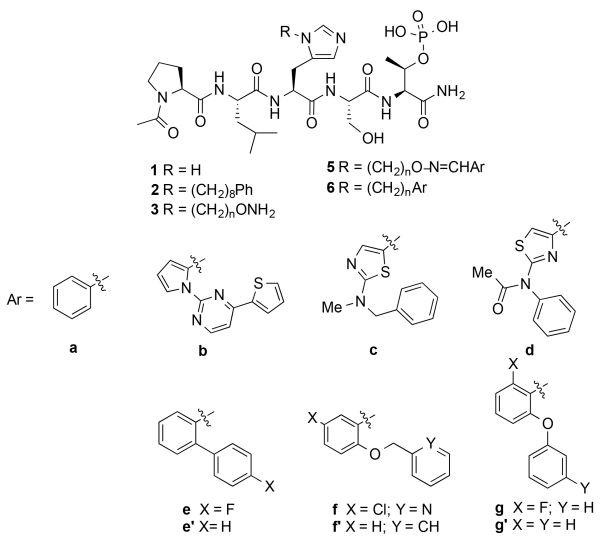
The serine/threonine specific polo-like kinase 1 (Plk1) is an important cell cycle regulator that has been defined as a molecular target for anti-cancer therapy development.1-3 Plk1 requires the coordinated actions of both an N-terminal catalytic kinase domain (KD) and a C-terminal polo-box domain (PBD), which engages in protein-protein interactions (PPIs) with phosphoserine (pS) and phosphothreonine (pT)-containing sequences. PBD-directed antagonists may afford attractive alternatives to more classical KD inhibitors for down-regulating Plk1 activity. The Plk1 PBD is remarkable by its inclusion of a hydrophobic “cryptic binding pocket” formed by Y417, Y421, Y481, F482, Y485 and L478, which is revealed by a more than 100° rotation of the Y481 side chain in the presence of ligands capable of accessing the pocket.4 Many efforts to develop PBD-binding antagonists have utilized peptides based on the region of the polo-box domain interacting protein 1 (PBIP1) proximal to the phosphorylated pT78 residue.5 Using the PBIP1 pT78-derived sequence, FDPPLHSpTA, Sledz et al. have shown that the N-terminal Phe residue can access this pocket,6 and that replacing the Phe residue with a variety of arylpropyl amides can improve binding affinities.7 Independently, we have found by tethering alkylphenyl groups from different positions on the shorter sequence PLHSpT (1), that we can occupy the cryptic binding pocket and achieve up to three-orders-of-magnitude enhancement in PBD-binding affinity.8-10 Of particular note, we were able to reach the pocket from the His residue using peptides of the form PLH*SpT (2), where H* indicates the presence of a -(CH2)8Ph group on the His N3 (π) nitrogen [ie, the His-[N(π)-(CH2)8Ph] (Figure 1).8,11 This is significant for developing reduced-size binding antagonists, since this residue is at the pT-2 position, which is immediately adjacent to the “SpT” minimal recognition motif.12
Figure 1.
Plk1 PBD semi-transparent electrostatic surface for bound ligand 2 (carbons in red) with underlying protein ribbon in white. Residues forming the “cryptic binding pocket” are identified with side chains shown in white for the parent “Ac-Pro-Leu-His-Ser-pThr-amide (1) and in red (for peptide 2). The cryptic binding pocket is accessed by ligand-induced 115° rotation of Y481. Structure is from PDB accession code 3RQ7.8
It is unlikely that the N(π)-(CH2)8Ph moiety provides maximal interaction from the pT-2 position. Unfortunately, the reagent N-Fmoc-His-[N(π)-(CH2)8Ph]-OH,13 which is currently used to incorporate H* residues into peptides, requires a lengthy synthesis.8,9,11,14-18 This has made difficult a direct examination of different functionality at the N(π)-position. In our current work we explore binding motifs originating from the His N(π)-position using a “tethered fragment” methodology that employs oxime ligation.19 By synthesizing an initial set of parent peptides (3) having terminal aminooxy groups tethered at various distances from this location and then reacting each member of the set with several aldehydes (4, see Supporting Information Table S1), we were able to interrogate interactions within the binding pocket using variety of different groups (5) (Figure 2). Once we had identified preferred binding motifs in this fashion, we replaced oxime linkages with methylene chains (6). The goal of this work was make linker variations that improve binding of the N(π)-side chain, and ultimately, this allowed us to achieve several-fold enhancement in binding affinities relative to the parent peptide (2). This is noteworthy, since 2 is one of the most potent Plk1 PBD-binding ligands known. The improved binding may result by accessing a newly identified auxiliary region proximal to the cryptic pocket.
Figure 2.
Structures of peptides discussed in the text.
We prepared a series of reagents of the form, N-Fmoc-His-[N(π)–X]-OH, where “X” indicates –(CH2)n–O–NHBoc and n is from 4 to 6, respectively (see Supporting Information). We then used these in solid-phase peptide synthesis to construct a corresponding set of free aminooxy-containing peptides, PLH†SpT, where H† indicates N(π)–(CH2)n–O–NH2 (3, Figure 2 and Supporting Information). We reacted each HPLC-purified peptide with a series of 87 aldehydes to yield libraries of oximes, PLH‡SpT, where H‡ indicates N(π)–(CH2)n–O–N=CH–Ar (5, Figure 2 and Supporting Information). By including the oxime functionality, the resulting peptides exhibited total overall tether lengths (including both methylene and oxime components) of from 7 to 9 units, as contrasted with a length of 8 units for the original parent peptide 2, whose tether is composed exclusively of methylene units.
An advantage of oxime ligation-based diversification is that reaction products can be subjected to direct biological evaluation without purification.19 We examined the Plk1 PBD-binding affinities of the constructs using an ELISA-based competition assay, which measured their ability to compete with an immobilized phosphopeptide, PMQSpTPLN, for the binding of either isolated PBD or full-length Plk1. We screened the full panel of oxime products by determining percent inhibition of isolated PBD at either 200 nM or 300 nM concentrations (Table S1). A total of 25 oximes showed greater than 60% inhibition at 300 nM, with longer tethers (5, n = 5 or 6) tending to be more potent than the shortest tether (n = 4). Several of the oximes demonstrated greater than 70% at 300 nM. A pattern emerged, in which higher inhibitory potency appeared to be associated with oximes having two aryl rings arranged in an angular orientation. This is exemplified by oximes derived from 5-membered heteroaryl 2-carboxaldehydes having phenyl-containing substituents at either their 1- or 5-positions (for example, 5b – 5d) as well as benzaldehydes with phenyl-containing substituents at the 2-position (for example, 5e – 5g) (Figure 2). We further evaluated the most active oximes (5b – 5g) from this initial screen for the inhibition of isolated Plk1 PBD and found that their IC50 values ranged from 91 nM to 244 nM. In this assay the IC50 values of 1 and 2 were 21 μM and 25 nM, respectively (Figure 2 and Supporting Information Figure S2).
Next, we determined IC50 values for a subset of oxime-containing peptides using the full-length Plk1 competitive ELISA binding assay (Table 1 and Supporting Information Figure S3). In this assay the original PBIP1-derived pentapeptide showed very low affinity (1, IC50 > 300,000 nM), while introducing the N(π)-(CH2)8Ph group onto the His residue resulted in greater than 1,000-fold enhancement of binding affinity (2, IC50 = 121 nM) (Table 1). These values are approximately 10-fold higher than our previously reported ELISA IC50 values for the same peptides.8,13,14 We attribute this to differences in binding affinities of the immobilized competitor peptides used for these assays. We used oxime-containing peptide 5a-5 as a reference (the second digit indicates the number of methylene units in the tether), since its phenyl group is tethered eight units from the His residue (based on five methylenes and the three members of the oxime linker) similar to the eight methylenes in the tether chain of the parent peptide 2. The inhibitory potency of 5a-5 was found to be lower than 2 (IC50 = 9 μM). When we extended the tether length of 5a-5 to nine units by adding one methylene, the affinity improved (5a-6, IC50 = 5 μM). Because the original screening data indicated that arranging aryl rings in an angular fashion was beneficial (Table S1), we examined phenyl oximes with a phenyl ether at the 2-position. With a five-methylene tether, we found that inhibitory potency increased relative to the unsubstituted parent (5g-5, IC50 = 1.6 μM as compared to 5a-5). Extending the tether length from eight to nine units by adding one additional methylene further enhanced inhibitory potency for this type of oxime (5g-6, IC50 = 0.7 μM) (Table 1).
Table 1.
Inhibitory potencies of peptide using a full-length Plk1-based ELISA competitive assay.a
| No. | IC50 (nM)b | Linker Length |
|---|---|---|
| 1 | >300,000 | - |
| 2 | 121 ± 9 | 8 |
| 5a-5 c | 9,000 ± 680 | 8 |
| 5a-6 c | 5,050 ± 130 | 9 |
| 5g-5 c | 1,550 ± 30 | 8 |
| 5g-6 c | 670 ± 400 | 9 |
| 6e’-8 | 43 ± 7 | 8 |
| 6f’-8 | 32 ± 4 | 8 |
| 6g’-6 | 139 ± 4 | 6 |
| 6g’-8 | 52 ± 7 | 8 |
| 6g-8 | 66 ± 8 | 8 |
| 6g-9 | 139 ± 16 | 9 |
Assays were conducted as described in the Supporting Information;
Values represent average ± SEM from three independent experiments and fit using nonlinear regression in GraphPad Prism 6 (curves are shown in the Supporting Information);
Freshly prepared oxime peptide solutions.
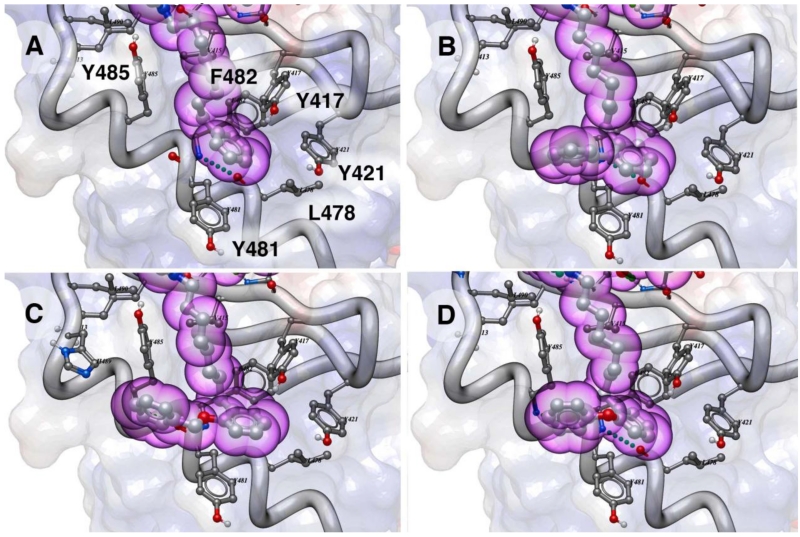
As exemplified by comparing the inhibitory potencies of 5a-5 as with 2, it was evident that replacing the oxime moiety by methylene groups could have additional beneficial effects. Therefore, for a select subset of oximes identified as presenting good geometries, we prepared peptide variants having tethers consisting entirely of methylene units (6). In order to prepare these peptides, we first synthesized the key reagents, N-Fmoc-His-[N(π)–X]-OH, in which “X” indicates –(CH2)n–Aryl and then used these in solid-phase protocols similar to those reported for the synthesis of 2 (full synthetic details are presented in the Supporting Information).11,14 We determined the IC50 values for these peptides using the full-length Plk1 competitive ELISA binding assay (Table 1). Analogs having phenyl ether substituents at the 2-position with tether chains of six or nine methylenes (6g’-6 and 6g-9, IC50 = 139 nM, respectively) exhibited potencies approximately equivalent to 2 (IC50 = 121 nM). However, by setting their tether lengths to the same as in 2 (eight methylenes), a significant increase in affinities were observed (6g’-8, IC50 = 52 nM and 6g-8, IC50 = 66 nM) (Table 1). Further increases in affinity were achieved either by removing the ether oxygen (6e-8, IC50 = 43 nM) or by converting the phenyl ether to a benzyl ether (6f’-8, IC50 = 32 nM). Parent peptide 2 is one of the highest affinity Plk1 PBD-binding antagonists known. In order to gain an understanding of the potential structural basis for the enhanced binding of the 2-aryl-containing analogs, we performed docking studies using a receptor defined by the Plk1 PBD-bound peptide 2 (PDB accession code 3rq7) (Figure 3). In addition to the primary cryptic pocket formed by Y417, Y421, Y481, F482 and Y485 and L478, a contiguous auxiliary binding region was observed that ran between the side chains of Y481 and Y485 and extended along the αB helix, overlaying N484 and terminating at H489. Access to this region was made possible by rotation of the Y481 side chain, which had been critical for revealing the original primary cryptic pocket.8 While the main-chain phenyl rings in 6e’-8, 6f’-8 and 6g’-8 remained within the primary pocket similar to the parent peptide 2, the 2-aryl-containing substituents extended into this auxiliary binding region (Figure 3). This is the first time that interactions of this nature have been reported.
Figure 3.
Docking of ligands in the Plk1 PBD showing interactions of alkyl-His side chains (purple spheres) in the cryptic binding pocket as described in the Supporting Information. Electrostatic surface, protein ribbon and sidechains forming the binding pocket are shown: (A) Ligand 2; (B) Ligand 6e’-8; (C) Ligand 6f’-8 and (D) Ligand 6g’-8.
In conclusion, by screening of a set of 87 aldehydes using an oxime ligation-based strategy, we arrived at 2-aryl-containing substituents (6e’ – 6g’), which exhibit up to four-fold higher affinities relative to parent peptide 2. This is noteworthy, since peptide 2 is already one of the highest affinity Plk1 PBD-binding ligands yet reported. More broadly, this process has led to our recognition of a potential auxiliary region proximal to previously identified hydrophobic cryptic pocket. These findings represent a fundamental advancement in understanding potential Plk1 PBD-binding interactions. Accessing this region may provide a general strategy for enhancing Plk1 PBD-binding interactions of a variety of ligands.
Supplementary Material
Acknowledgements
Work was supported by the NIH Intramural Program, Center for Cancer Research National Cancer Institute, National Insitutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data associated with this article can be found in the online version. This includes procedures for chemical synthesis and compound characterization; oxime ligation; Plk1 PBD-binding ELISA assays and generated data; and molecular modeling.
References
- 1.Strebhardt K. Nat. Rev. Drug Discov. 2010;9:643. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- 2.Lee KS, Burke TR, Park J-E, Bang JK, Lee E. Trends Pharmacol. Sci. 2015;36:858. doi: 10.1016/j.tips.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archambault V, Lepine G, Kachaner D. Oncogene. 2015;34:4799. doi: 10.1038/onc.2014.451. [DOI] [PubMed] [Google Scholar]
- 4.Tan YS, Sledz P, Lang S, Stubbs CJ, Spring DR, Abell C, Best RB. Angew. Chem., Int. Ed. 2012;51:10078. doi: 10.1002/anie.201205676. S10078/10071-S10078/10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun S-M, Moulaei T, Lim D, Bang JK, Park J-E, Shenoy SR, Liu F, Kang YH, Liao C, Soung N-K, Lee S, Yoon D-Y, Lim Y, Lee D-H, Otaka A, Appella E, McMahon JB, Nicklaus MC, Burke TR, Jr., Yaffe MB, Wlodawer A, Lee KS. Nat. Struct. Mol. Biol. 2009;16:876. doi: 10.1038/nsmb.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sledz P, Stubbs CJ, Lang S, Yang Y-Q, McKenzie GJ, Venkitaraman AR, Hyvoenen M, Abell C. Angew. Chem., Int. Ed. 2011;50:4003. doi: 10.1002/anie.201008019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sledz P, Lang S, Stubbs CJ, Abell C. Angew. Chem. Int. Ed. Engl. 2012;51:7680. doi: 10.1002/anie.201202660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Park J-E, Qian W-J, Lim D, Graber M, Berg T, Yaffe MB, Lee KS, Burke TR., Jr. Nat. Chem. Biol. 2011;7:595. doi: 10.1038/nchembio.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F, Park J-E, Qian W-J, Lim D, Scharow A, Berg T, Yaffe MB, Lee KS, Burke TR. ACS Chem. Biol. 2012;7:805. doi: 10.1021/cb200469a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu F, Park J-E, Qian W-J, Lim D, Scharow A, Berg T, Yaffe MB, Lee KS, Burke TR. ChemBioChem. 2012;13:1291. doi: 10.1002/cbic.201200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian W-J, Park J-E, Lee KS, Burke TR., Jr. Bioor. Med. Chem. Lett. 2012;22:7306. doi: 10.1016/j.bmcl.2012.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. Cell. 2003;115:83. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 13.Qian W, Liu F, Burke TR. J. Org. Chem. 2011;76:8885. doi: 10.1021/jo201599c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian W-J, Park J-E, Liu F, Lee KS, Burke TR., Jr. Bioorg. Med. Chem. 2013;21:3996. doi: 10.1016/j.bmc.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murugan RN, Park J-E, Lim D, Ahn M, Cheong C, Kwon T, Nam K-Y, Choi SH, Kim BY, Yoon D-Y, Yaffe MB, Yu D-Y, Lee KS, Bang JK. Bioorg. Med. Chem. 2013;21:2623. doi: 10.1016/j.bmc.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn M, Han Y-H, Park J-E, Kim S, Lee WC, Lee SJ, Gunasekaran P, Cheong C, Shin SY, Sr., Kim H-Y, Ryu EK, Murugan RN, Kim N-H, Bang JK. J. Med. Chem. 2015;58:294. doi: 10.1021/jm501147g. [DOI] [PubMed] [Google Scholar]
- 17.Qian W-J, Burke TR., Jr. Org. Biomol. Chem. 2015;13:4221–4225. doi: 10.1039/c5ob00171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian W-J, Park J-E, Grant R, Lai CC, Kelley JA, Yaffe MB, Lee KS, Burke TR., Jr. Biopolymers Pept. Sci. 2015;104:663. doi: 10.1002/bip.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahta M, Liu F, Kim S-E, Stephen AG, Fisher RJ, Burke TR., Jr. Nat. Protocols. 2012;7:686. doi: 10.1038/nprot.2012.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.