Abstract
Background
Urea transporters (UTs) are important in urine concentration and in urea recycling, and UT-B has been implicated in both. In kidney, UT-B was originally localized to outer medullary descending vasa recta, and more recently detected in inner medullary descending vasa recta. Endogenously produced microRNAs (miRs) bind to the 3’UTR of genes and generally inhibit their translation, thus playing a pivotal role gene regulation.
Methods
Mice were dehydrated for 24h then sacrificed. Inner and outer medullas were analyzed by PCR and qPCR for microRNA expression and western blotting for protein abundance.
Results
MicroRNA seq analysis of mouse inner medullas showed a 40% increase in microRNA-200c in dehydrated mice compared with controls. An in silico analysis of the targets for miR-200c revealed that microRNA-200c could directly target the gene for UT-B. PCR confirmed that miR-200c is up-regulated in the inner medullas of dehydrated mice while western blot showed that UT-B protein abundance was down-regulated in the same portion of the kidney. However in the outer medulla, miR-200c was reduced and UT-B protein was increased in dehydrated mice.
Conclusions
This is the first indication that UT-B protein and miR-200c may each be differentially regulated by dehydration within the kidney outer and inner medulla. The inverse correlation between the direction of change in miR-200c and UT-B protein abundance in both the inner and outer medulla suggests that miR-200c may be associated with the change in UT-B protein in these two portions of the kidney medulla.
Keywords: Urine concentration, miR, qPCR, dehydration
Introduction
Urea plays an important role in the urinary concentration system. Urea has a low lipid solubility so it crosses cell membranes very slowly by passive diffusion. Physiologic studies demonstrate that highly efficient transmembrane urea transporters (UTs) mediate intrarenal urea recycling [1–3]. There are two types of urea transporters; UT-A and UT-B. Both are members of the solute carrier 14 (SLC14) family and found in the kidney. UT-A1 and UT-A3 are located in the inner medulla and UT-A2 is located in the outer medulla. UT-B was originally localized to outer medullary descending vasa recta, but was recently reported in inner medullary descending vasa recta [4, 5].
MiRs are short non-coding RNAs found endogenously in all mammals. In recent years, many studies show that miRs play crucial roles in gene regulation (reviewed in [6, 7]). With the growing knowledge of the roles of miRs in animal models, it is becoming clear that miRs are important in the development and physiology of multiple organs, including the kidney (reviewed in [8–10]. In the present study, we analyzed predicted targets according to the algorithms of TargetScan [11], PITA [12], and miRanda [13] to identify potential targets of miR-200c. Although some data demonstrate that specific miRs have different expression profiles in renal cortex and medulla [10, 14], the difference in expression and functional role of miR-200c in different sections of the kidney medulla has not previously been reported. Further, the impact of this difference on the regulation of UT-B expression in inner medulla and outer medulla has yet to be elucidated.
Most previous studies of UT-B have examined the protein in whole medulla or even whole kidney, so differential changes in UT-B in different parts of the kidney, or even the medulla, may have been missed. In this study, UT-B protein abundance and miR-200c levels in inner medulla and outer medulla were compared. In the inner medulla there was an increase in miR-200c but a decrease in UT-B protein abundance; whereas in outer medulla miR-200c decreased but UT-B protein abundance increased.
Methods
Animal preparation
All animal protocols and procedures were approved by the Emory University Institutional Animal Care and Use Committee and conducted according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Male C57bl/6J mice (8 weeks old) were used. Control mice received standard mouse chow (Teklad diet # 5001) and water ad libitum. Dehydrated mice were provided free access to mouse chow but their access to water was restricted. Water was removed for 12 hours, reinstated for 15 minutes, and removed for an additional 12 hours. Following the 24 hour dehydration period, the animals were euthanized by cervical dislocation, and mouse kidneys were dissected into inner medulla (IM) and outer medulla (OM). Tissue to be used for RNA was either placed directly into Tri Reagent for RNA isolation, or flash frozen in liquid nitrogen for later analysis. Tissue to be used for protein analysis was placed directly into isolation buffer and processed as described under Western Blot below.
MiR seq
Control and dehydrated mouse inner medulla were harvested and sent to the Emory miR sequencing core at the Yerkes National Center for Primate Research for analysis. Qualitative and quantitative analysis of the total RNA was performed using the Thermo Nanodrop 2000 and Agilent 2100 Bioanalyzer respectively. Small RNA libraries were prepared using the SeqMatic tailormix miRNA sample preparation kit (SeqMatic. Union City, CA, USA) as per the manufacturer’s instructions. The libraries were further quantified on Qubit® 2.0 Fluorometer (Life Technologies, Grand Island, NY, USA) using the High Sensitivity dsDNA assay. Libraries from all the samples were multiplexed and run in a single lane of Illumina v3 flowcell. PhiX was used as an internal control on each lane to monitor the error statistics. Cluster generation was performed on the v3 flowcell on the Illumina cBot. The clustered flowcell was sequenced on the Illumina HiSeq1000 system as a 100-cycle single read multiplexed run. The GenCore uses the software platform PARTEK Genomics Suite for performing statistical analyses. They employ the Benjamini-Hochberg False Discovery Rate (FDR) method for adjusting p-values. We defined differential expression at this step as a change of at least 10% (either increase or decrease) in dehydrated mice versus the controls.
Quantitative real-time PCR (qPCR)
Each sample contained the IM or OM from both kidneys from a single mouse. The tissues were frozen in liquid nitrogen immediately (within 2 minute of cervical dislocation) and stored until RNA extraction. The RNA quality was assessed by PCR and agarose gel electrophoresis to verify band integrity. MicroRNA (miR-200c) quantification was then accomplished by qPCR. Reverse transcription and quantitative PCR (q-PCR): Total RNA was extracted using Tri-Reagent (Molecular Research Inc., Cincinnati, OH). RNA concentrations were measured by spectrophotometry (Nano Drop Lite, Thermo Scientific, Wilmington, DE). To remove contaminating DNA, samples were treated with recombinant RNase-free DNase I (Thermo Fisher Scientific, West Palm Beach, FL). Total RNA was reverse transcribed to cDNA using the miRCURY LNA™ Universal cDNA Synthesis kit (Exiqon INC., Woburn, MA). The primers were custom designed by Exiqon. Real-time qPCR was performed on CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using the miRCURY LNA microRNA PCR SYBR Green master mix (Exiqon INC). The following cycle parameters were used: 95°C for 10 minutes and 40 cycles at 95°C for 10 seconds and 60°C for 60 seconds. The quantification cycle (Cq) values was defined as the cycle at which the curvature of the amplification curve is maximal and was automatically detected by the CFX Manager software (BioRad). The primer sequence (EXIQON) is shown in Table 1. MiR200c expression was standardized to U6 gene and expression was calculated as the difference between the threshold values of the two miRs (ΔΔcq). Melting curve analysis was routinely performed to verify the specificity of the reaction [15].
Table 1.
PCR primer sequence
| Product name | Product No. | Target sequence 5’-3’ | Product size,nt |
|---|---|---|---|
| Has-miR-3p | 204482 | UAAUACUGCCGGGUAAUGAUGGA | 22 |
Western blot analysis
Mouse IM and OM were placed into ice-cold protein isolation buffer (10 mM triethanolamine, 250 mM sucrose, pH 7.6, 1 μg/ml leupeptin, and 2 mg/ml PMSF) and homogenized manually using glass/glass tissue grinders. SDS (10%) was added to the total cell lysate to a final concentration of 1% SDS [16, 17]. Insoluble material was removed by centrifugation at 16,000 ×g for 10 minutes and the protein concentration in the supernatant fraction was determined by a modified Lowry method (Bio-Rad DC protein assay reagent, Bio-Rad, Richmond, CA). Detergent solubilized proteins (20 μg/lane) were size separated by SDS-PAGE using 10% polyacrylamide gels, then transferred to polyvinylidene difluoride membranes (Immobilon, Millipore, Bedford, MA) as described previously [16, 17]. Blots were blocked with 5% nonfat dry milk in Tris-buffered saline (TBS: 20 mM Tris HCl, 0.5 M NaCl, pH 7.5) at room temperature for 1 hour, then incubated with our primary polyclonal antibody to UT-B overnight at 4°C. Blots were washed three times in TBS with 0.5% Tween-20 (TBS/Tween) and then incubated with Alexa Fluor 680-linked anti-rabbit IgG (Molecular Probes, Eugene, OR). Blots were washed three times with TBS/Tween, and then the bound secondary antibody was visualized using infrared detection with the LICOR Odyssey protein analysis system (Licor Bioscience, Lincoln, NE). Total protein staining of the blots using Ponceau S was used to determine protein loading. Blots were stained with Ponceau S, then scanned and a grayscale tiff generated. Using Image J, a central horizontal section equal to about 25% of the total lane length was evaluated for pixel density. Specific band densities were divided by total loading protein band densities to normalize the western blot band densities.
Statistical analysis
All data are expressed as means ± SE and n is the number of mice per group. Differences between each dehydrated group and control group were tested by unpaired, two-tailed Student’s t-test. The criterion for statistical significance is p < 0.05.
Results
miR-200c changes in dehydrated mouse inner medulla
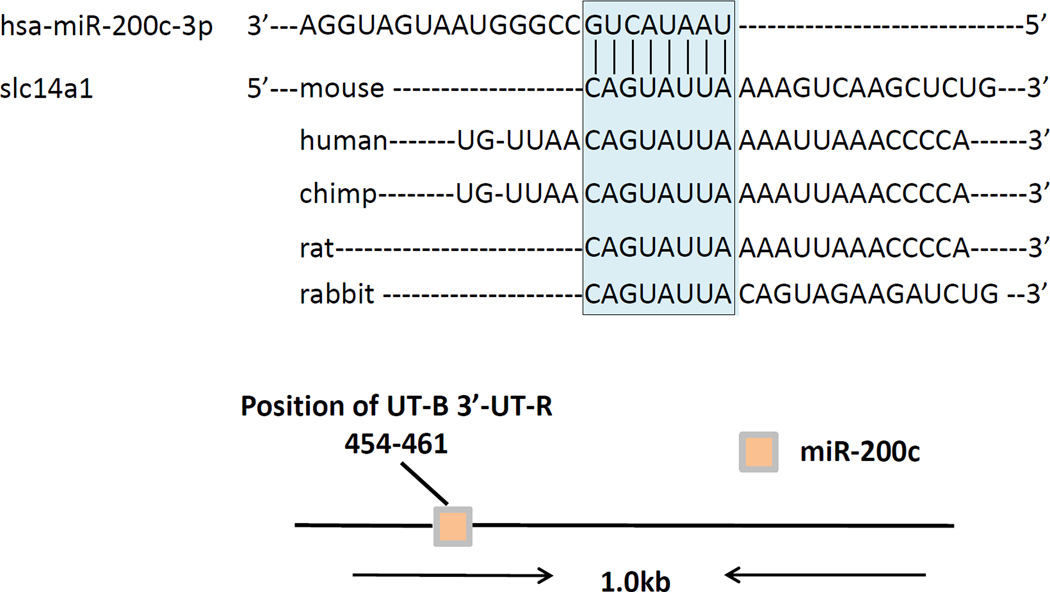
We performed a microRNA seq analysis of inner medullas from dehydrated and control mice using the Emory microRNA seq core facility. We found that miR-200c was increased by 42% (1.4 fold) in dehydrated versus control mice (P=0.007). Next, we used Targetscan (www.targetscan.org) to assess the potential targets of miR-200c. We found that UT-B was a direct target of miR-200c with one section of complementary sequence in the UT-B 3’UTR (Figure 1).
Figure 1.
miR-200c directly targets slc14a1 (UT-B). Top: Shown is an RNA sequence for UT-B from different animals. The sequence contains an identical complementary sequence to one in miR-200c, shown at the top. Bottom: a diagram of the gene structure showing the approximate location of the target sequence for binding of miR-200c to UT-B.
Differential changes in miRNA-200c in inner medulla and outer medulla during dehydration
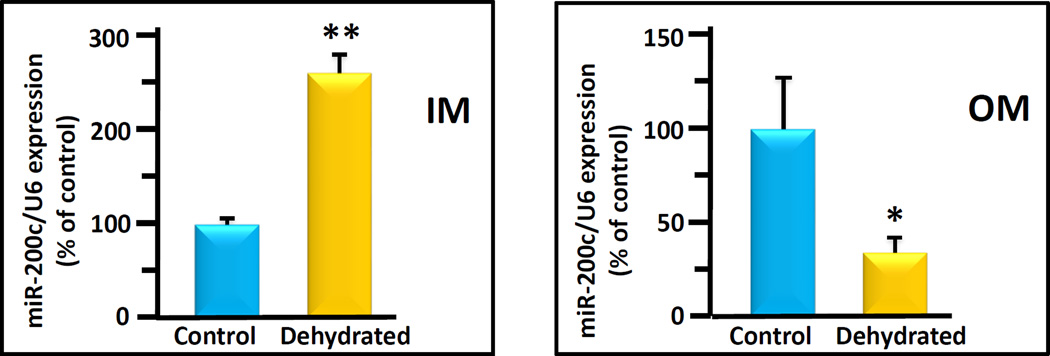
To confirm the microRNA seq response using an alternate analysis, we performed qPCR of inner medulla from control or dehydrated mice. We also performed qPCR on the outer medulla of the same mice. . The combined results from 12 animals / group are shown in Figure 2. In the IM, the miR-200c level of the dehydrated group was significantly increased by 182 ± 33% above control levels (100 ± 5%). In contrast, in the OM, the level of miR-200c of the dehydrated group was significantly decreased by 32± 11% compared to the control group (100 ± 27, Figure 2).
Figure 2.
Inner medulla and outer medulla miR-200c expression in control (black bars) and dehydrated (white bars) mice. The bar graphs show average qPCR results from 10 animals per bar/condition combined from 3 separate experiments. Data is presented as % of control, with control designated as 100%. All data were normalized to U6 expression level. Statistical analysis was performed using Student’s 2 tailed t-test. Error bars represent standard error (S.E.). (** p< 0.01, * p<0.05, dehydrated vs. control).
Differential changes in UT-B protein in inner medulla and outer medulla during dehydration
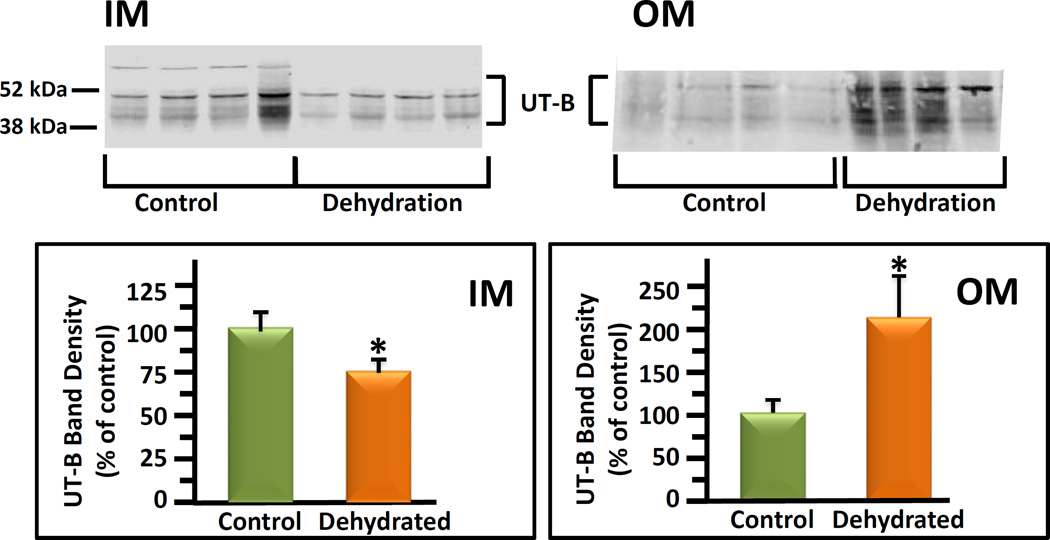
Control and 24 hour dehydrated mice were killed and kidneys dissected into inner medulla and outer medulla. Tissues were prepared for protein analysis as described above. UT-B protein was detected by Western blot. UT-B is a glycosylated protein and is seen on blots as a multiple group band between 41 and 54 kDa [18]. These bands included a heavy band just below 52 kDa and lighter bands below that are glycosylated variants of the UT-B protein. The brackets denote the section of the gel that was used for densitometry analysis. There was a band apparent at 97 kDa (not shown) but this has previously been shown to be non-specific [19]. UT-B protein abundance in the IM dehydrated mice was 75 ± 8% of the level in control mice (100 ± 9%, Figure 3). In contrast, UT-B protein abundance was dramatically increased by 112% over controls in the OM of dehydrated mice (212 ± 52% dehydrated vs 100 ± 17% control mice, Figure 3).
Figure 3.
UT-B protein abundances in inner medulla (IM) and outer medulla (OM) of control mice compared to dehydrated mice. Top: representative western blots of IM (left data set) and OM (right data set) proteins. Left lanes, control mice; right lanes, dehydrated mice. Each lane was tissue from a separate animal. UT-B bands are identified by a bracket. Bottom: combined data bar graph showing the average band density for control and dehydrated groups. The average band densities were calculated using the UT-B band density normalized to the total protein loading control density per sample determined by Ponceau S staining of the same membrane. Error bars show standard error. N=15 animals per group from 3 separate experiments. The y-axis is density in arbitrary units. Values are means ± s.e. (* p< 0.05, dehydrated vs. control).
Discussion
The major result of this study is that both miR-200c expression and UT-B protein abundance change in opposite directions in the inner and outer medulla following dehydration. The inverse association between miR-200c and UT-B levels in distinct sections of the kidney medulla suggests that these changes may be related. However, the present findings show an association and do not establish cause and effect. In addition, the opposite changes in UT-B protein abundance in the two portions of the kidney medulla emphasize the need to examine each region of the kidney separately to avoid missing these changes.
MiRs belong to a class of small, endogenous non-coding RNAs of 21–25 nucleotides in length [20–23]. They play an important negative regulatory role at the post-transcriptional level by targeting specific mRNAs for degradation or translation repression [24–26]. In recent years, the study of miRs has become an important research focus for understanding the regulation of renal development, physiology to pathology. Previous studies have linked the miR-200 family with kidney diseases [27–29]. MiR-200c is a member of the miR-200 family that includes miR-141, miR-200a, miR-200b, miR-200c, and miR-429 [30]. Several articles show that miR-200c regulates epithelial to mesenchymal transition (EMT) and endothelial mesenchymal transition (EndMT) by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2 in kidney fibrosis and cancer [31–35]. Additional EMT-related targets of the miR-200 family that contribute to progression of kidney fibrosis have been identified: miR141 inhibits TGF-β2 [36] and miR200a suppresses E-catenin [37].
In our initial miR seq study, miR-200c was increased by 42% (1.4 fold, P=0.007). We chose to study miR-200c further because it had a theoretical target, UT-B, which could be involved in urine concentration. Our first goal was to perform qPCR experiments to confirm that miR-200c was changed in the dehydrated animals by a second method (in addition to the miR seq approach). The qPCR results showed that the level of miR-200c in the inner medulla was significantly increased by dehydration, consistent with the miR seq result.
UT-B is an important transporter for controlling urea movement and tissue osmolality leading to urine concentration. Urine concentration is a key component for the regulation of sodium and water excretion so that the body can maintain a nearly constant blood plasma osmolality and sodium concentration. Urea is the major form of nitrogen metabolism in most mammals. Urea is the most abundant solute contributor to the osmotic gradient in kidney inner medulla [38–40]. It has low lipid solubility and a corresponding low permeability through artificial lipid bilayers [41]. It crosses cell membranes very slowly by passive diffusion [40]. UT-B is expressed primarily in the red blood cells and descending vasa recta [5, 42]. Vasa recta for the countercurrent exchange system are chiefly responsible for medullary urea accumulation in the urinary concentration process. Transgenic mice lacking UT-B show decreased urinary urea and increased plasma urea concentrations. Yang et al. have verified that UT-B selectively transports urea [43]. Although there are many reports focusing on the relationship between UT-B and urine concentration, the mechanism is not clear.
In our study, the UT-B abundance was decreased in the IM of dehydrated mice. This suggested that miR-200c might be involved in the urine concentration mechanism by altering the levels of UT-B in the IM. In contrast, we found that UT-B expression was increased in dehydrated mouse OM. The increase in UT-B in the OM has been reported previously by Lim et al., where they looked at thick sections of kidneys from control, dehydrated and water loaded rats, stained with antibody to UT-B [44]. We cannot exclude possibility that some of the increase in UT-B could reflect the presence of increased erythrocytes in outer medullary tissue from the dehydrated animals and this was not apparent in the thick sections in the Lim manuscript. Lim et al. did not show results of UT-B in the inner medulla [44]. We used qPCR analysis to probe miR-200c level in the OM of dehydrated mice to see if it was changing in the same direction in OM as in IM. Unlike the IM, miR-200c is decreased in OM of dehydrated mice relative to control animals. The inverse correlation between the direction of change in miR-200c and UT-B protein abundance in both the inner and outer medulla suggests that miR-200c may be associated with the change in UT-B protein in these two portions of the kidney medulla.
UT-B is thought to play a physiological role in intrarenal urea recycling [1–3]. One recycling pathway is urea reabsorption from the IMCD into the descending vasa recta. We speculate that the increase in UT-B protein in the OM, along with the decrease in UT-B protein in the IM during dehydration, could result in more urea recycling into the OM descending vasa recta, rather than into the IM portion. This may allow the recycled urea to be improve the efficacy of countercurrent exchange, and hence urine concentration.
Several reports [10, 14] revealed differences in some miR expression profiles between renal cortex and medulla. Most of the studies of the kidney were based on the whole kidney, cortex, whole medulla, or just inner medulla. Our study shows that this is probably not sufficient to understand the different function of distinct kidney sections. MiRs have not previously been reported to be connected with regulation of UTs. Future studies will be needed to assess whether the changes in miR-200c and UT-B are simply an association or whether miR-200c regulates UT-B abundance.
Acknowledgments
This work was supported by NIH grants DK41707 and DK89828.
Thanks to Gregory Tharp and Steve Bosinger at the Yerkes National Primate Center RNA sequencing core for help with interpretation of the microRNA seq data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: No conflicts of interest are declared by the authors.
References
- 1.Bagnasco SM. Gene structure of urea transporters. AmJPhysiolRenal Physiol. 2003;284(1):F3–F10. doi: 10.1152/ajprenal.00260.2002. [DOI] [PubMed] [Google Scholar]
- 2.Sands JM. Mammalian urea transporters. AnnuRevPhysiol. 2003;65:543–566. doi: 10.1146/annurev.physiol.65.092101.142638. [DOI] [PubMed] [Google Scholar]
- 3.Trinh-Trang-Tan MM, Bankir L. Integrated function of urea transporters in the mammalian kidney. Exp Nephrol. 1998;6(6):471–479. doi: 10.1159/000020559. [DOI] [PubMed] [Google Scholar]
- 4.Klein JD, Blount MA, Sands JM. Urea transport in the kidney. Compr Physiol. 2011;1(2):699–729. doi: 10.1002/cphy.c100030. [DOI] [PubMed] [Google Scholar]
- 5.Yang B, Verkman AS. Analysis of double knockout mice lacking aquaporin-1 and urea transporter UT-B. JBiolChem. 2002;277(39):36782–36786. doi: 10.1074/jbc.M206948200. [DOI] [PubMed] [Google Scholar]
- 6.Friedman LM, Avraham KB. MicroRNAs and epigenetic regulation in the mammalian inner ear: implications for deafness. Mammalian genome : official journal of the International Mammalian Genome Society. 2009;20(9–10):581–603. doi: 10.1007/s00335-009-9230-5. [DOI] [PubMed] [Google Scholar]
- 7.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA (New York, NY) 2005;11(3):241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Koo S, White N, et al. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic acids research. 2004;32(22):e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Kertesz M, Iovino N, Unnerstall U, et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 13.John B, Enright AJ, Aravin A, et al. Human MicroRNA targets. PLoS biology. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian Z, Greene AS, Pietrusz JL, et al. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome research. 2008;18(3):404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Klein JD, Gunn RB, Roberts BR, et al. Down-regulation of urea transporters in the renal inner medulla of lithium-fed rats. Kidney Int. 2002;61(3):995–1002. doi: 10.1046/j.1523-1755.2002.00210.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim D-U, Klein JD, Racine S, et al. Urea may regulate urea transporter protein abundance during osmotic diuresis. AmJPhysiolRenal Physiol. 2005;288(1):F188–F197. doi: 10.1152/ajprenal.00200.2004. [DOI] [PubMed] [Google Scholar]
- 18.Timmer RT, Klein JD, Bagnasco SM, et al. Localization of the urea transporter UT-B protein in human and rat erythrocytes and tissues. AmJPhysiolCell Physiol. 2001;281(4):C1318–C1325. doi: 10.1152/ajpcell.2001.281.4.C1318. [DOI] [PubMed] [Google Scholar]
- 19.Klein JD, Sands JM, Qian L, et al. Upregulation of urea transporter UT-A2 and water channels AQP2 and AQP3 in mice lacking urea transporter UT-B. JAmSocNephrol. 2004;15(5):1161–1167. doi: 10.1097/01.asn.0000125617.19799.72. [DOI] [PubMed] [Google Scholar]
- 20.Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14(10):1036–1040. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 21.Ruvkun G. The perfect storm of tiny RNAs. Nat Med. 2008;14(10):1041–1045. doi: 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- 22.Sontheimer EJ, Carthew RW. Silence from within: endogenous siRNAs and miRNAs. Cell. 2005;122(1):9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 23.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature reviews Molecular cell biology. 2008;9(3):219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 24.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 25.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 26.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 27.Chung AC, Lan HY. MicroRNAs in renal fibrosis. Front Physiol. 2015;6:50. doi: 10.3389/fphys.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv LL, Cao YH, Ni HF, et al. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am J Physiol Renal Physiol. 2013;305(8):F1220–F1227. doi: 10.1152/ajprenal.00148.2013. [DOI] [PubMed] [Google Scholar]
- 29.Patel V, Hajarnis S, Williams D, et al. MicroRNAs regulate renal tubule maturation through modulation of Pkd1. J Am Soc Nephrol. 2012;23(12):1941–1948. doi: 10.1681/ASN.2012030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhlmann S, Zhang JD, Schwager A, et al. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29(30):4297–4306. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- 31.Bracken CP, Gregory PA, Kolesnikoff N, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 32.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 33.Hurteau GJ, Carlson JA, Spivack SD, et al. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67(17):7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 34.Korpal M, Lee ES, Hu G, et al. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of biological chemistry. 2008;283(22):14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer biology & therapy. 2010;10(3):219–222. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO reports. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia H, Ng SS, Jiang S, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 2010;391(1):535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 38.Hediger MA, Smith CP, You GF, et al. Structure, regulation and physiological roles of urea transporters. Kidney Int. 1996;49(6):1615–1623. doi: 10.1038/ki.1996.235. [DOI] [PubMed] [Google Scholar]
- 39.Knepper MA, Roch-Ramel F. Pathways of urea transport in the mammalian kidney. Kidney Int. 1987;31:629–633. doi: 10.1038/ki.1987.44. [DOI] [PubMed] [Google Scholar]
- 40.Sands JM, Timmer RT, Gunn RB. Urea transporters in kidney and erythrocytes. AmJPhysiol. 1997;273(3):F321–F339. doi: 10.1152/ajprenal.1997.273.3.F321. [DOI] [PubMed] [Google Scholar]
- 41.Galluci E, Micelli S, Lippe C. Non-electrolyte permeability across thin lipid membranes. ArchIntPhysiolBiochim. 1971;79:881–887. doi: 10.3109/13813457109104847. [DOI] [PubMed] [Google Scholar]
- 42.Klein JD, Blount MA, Sands JM. Urea transport in the kidney. Compr Physiol. 2011;1(2):699–729. doi: 10.1002/cphy.c100030. [DOI] [PubMed] [Google Scholar]
- 43.Yang B, Bankir L, Gillespie A, et al. Urea-selective concentrating defect in transgenic mice lacking urea transporter UT-B. JBiolChem. 2002;277:10633–10637. doi: 10.1074/jbc.M200207200. [DOI] [PubMed] [Google Scholar]
- 44.Lim S-W, Han K-H, Jung J-Y, et al. Ultrastructural localization of UT-A and UT-B in rat kidneys with different hydration status. AmJPhysiolRegulIntegrCompPhysiol. 2006;290(2):R479–R492. doi: 10.1152/ajpregu.00512.2005. [DOI] [PubMed] [Google Scholar]