Abstract
The ubiquitous polyamine spermidine is indispensable for eukaryotic growth and cell proliferation. A conserved vital function of spermidine across eukaryotes is conferred by its aminobutyl group that is transferred to a single lysine in translation factor eIF5A to form the essential hypusine post-translational modification required for cellular translation. In direct contrast, although spermidine is absolutely essential for growth of alpha-proteobacterial plant pathogen Agrobacterium tumefaciens, we have found, by employing a suite of natural polyamines and synthetic methylated spermidine analogues together with spermidine biosynthetic mutants, that it is solely the 1,3-diaminopropane moiety of spermidine that is required for growth. Indeed, any polyamine containing an intact terminal 1,3-diaminopropane moiety can replace spermidine for growth, including the simple diamine 1,3-diaminopropane itself, a paradigm shift in understanding polyamine function in bacteria. We have identified for the first time a spermidine retroconversion activity in bacteria, producing diamine putrescine from triamine spermidine, however, exogenously-supplied tetraamine spermine is resistant to retroconversion. When spermidine levels are pharmacologically decreased, synthesis of spermine from spermidine is induced via the same biosynthetic enzymes, carboxyspermidine dehydrogenase and carboxyspermidine decarboxylase that produce spermidine from putrescine, the first identification of a spermine biosynthetic pathway in bacteria. This also suggests that spermidine represses spermine biosynthesis, but when spermidine levels decrease, it is then converted by carboxyspermidine dehydrogenase and decarboxylase enzymes to spermine, which is resistant to retroconversion and constitutes a sequestered pool of protected 1,3-diaminopropane modules required for growth. We also identify an efficient N-acetylspermidine deacetylase activity, indicative of a sophisticated bacterial polyamine homeostasis system.
The polyamine spermidine is essential for eukaryotic life, including growth, cell proliferation and development.1 Spermidine serves as an aminobutyl group donor for post-translational modification of translation factor eIF5A, which takes place on a specific lysine residue.2 The spermidine-derived aminobutyl modification of eIF5A and subsequent hydroxylation results in formation of a modified amino acid known as hypusine [Nε-(4-amino-2-hydroxybutyl)lysine]. Hypusine modification of eIF5A is essential for its function, and consequently for cell proliferation.3 Hypusinated eIF5A functions in the translation of mRNAs encoding proteins containing polyproline tracts, and without functional eIF5A, ribosomes arrest on the nascent polyproline stretches.4, 5 The aminobutyl moiety of spermidine is thus an essential structural and functional determinant of normal cellular translation in eukaryotes. In addition to hypusine formation, spermidine is the biosynthetic precursor of tetraamine spermine (N8-aminopropylspermidine). Biosynthesis of spermine has evolved independently in metazoa, flowering plants and yeasts, and biosynthesis of spermine isomer thermospermine has evolved in the archaeplastidia and diatoms, via distinct spermine and thermospermine synthases.6
In bacteria, the translation factor analogous to eIF5A is EFP, which in Escherichia coli is post-translationally modified by lysine and not by the aminobutyl moiety of spermidine.7 Furthermore, most bacteria do not synthesize the tetraamines spermine or thermospermine. Although spermidine is by far the most common triamine polyamine found in bacteria, its physiological role is obscure. It is presumed that its function is a consequence of its physicochemical attributes, i.e., it is positively charged at physiological pH. In E. coli, spermidine has been found to be electrostatically bound to negatively charged molecules, particularly RNA, DNA and phospholipids.8 It has been proposed that in E. coli, binding of spermidine to RNA influences translation of a subset of mRNAs described as the polyamine modulon, thereby affecting cellular physiology and growth.9 A completely polyamine-deficient strain of E. coli grows at about 50% the rate of the wild-type strain but nevertheless appears to grow indefinitely without polyamines.10
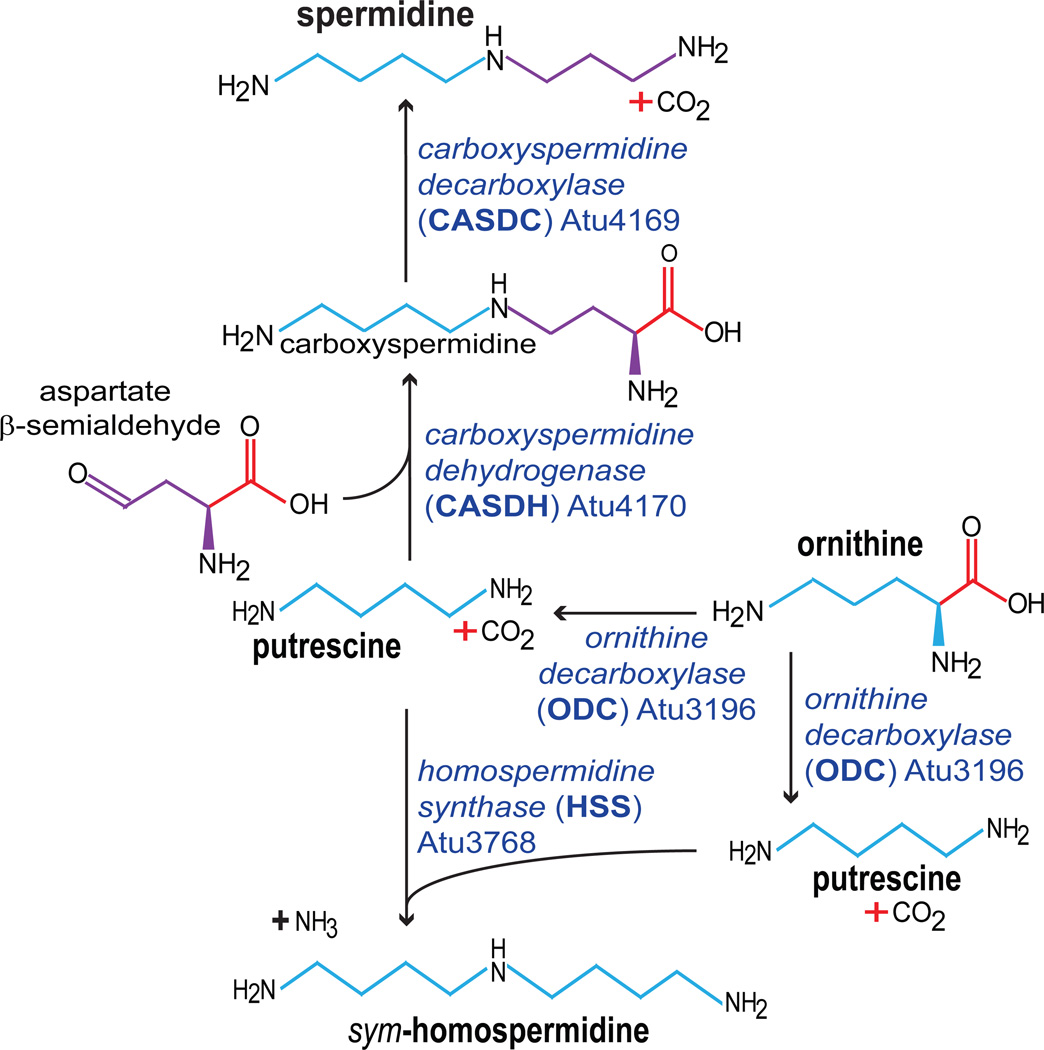
We have determined that spermidine is absolutely essential for growth of the α-proteobacterium Agrobacterium tumefaciens, a plant pathogen. Spermidine is synthesized from putrescine by the addition of an aminopropyl group derived from aspartate β-semialdehyde in A. tumefaciens, unlike eukaryotes and E. coli, which use decarboxylated S-adenosylmethionine (dcAdoMet) as an aminopropyl group donor (Figure 1). We now show, by employing a suite of natural polyamines and methylated spermidine analogues (Figure 2), that in contrast to eukaryotes, it is the aminopropyl side of the spermidine molecule that is essential for growth of A. tumefaciens, rather than the aminobutyl side, which is known to be essential in eukaryotes.11–13 Any polyamine or analogue containing an intact terminal 1,3-diaminopropane moiety was found to support growth, and polyamines lacking a 1,3-diaminopropane moiety were completely unable to support growth, including putrescine, cadaverine, homospermidine, carboxyspermidine, and spermidine analogues methylated on the aminopropyl side. Moreover, the simple diamine 1,3-diaminopropane alone is able to replace spermidine for growth of A. tumefaciens. We also show that the aspartate β-semialdehyde-dependent polyamine biosynthetic pathway is surprisingly plastic and is able to synthesize norspermidine from 1,3-diaminopropane, the triamine aminopropylcadaverine and tetraamine bis(aminopropyl)cadaverine or its isomer aminobutylnorspermidine from the diamine cadaverine. In addition, inhibition of the first step of polyamine biosynthesis, ornithine decarboxylase (ODC), results in accumulation of ornithine and induction of biosynthesis of the tetraamine spermine /thermospermine. We identify for the first time in bacteria the pathway for spermine biosynthesis. Finally, we have discovered an efficient retroconversion activity that produces putrescine from spermidine, and the activity of an N8-acetylspermidine deacetylase that releases spermidine from its acetylated form.
Figure 1.
Polyamine biosynthesis in A. tumefaciens C58. Homospermidine biosynthesis is repressed by spermidine and is not accumulated unless spermidine is depleted.
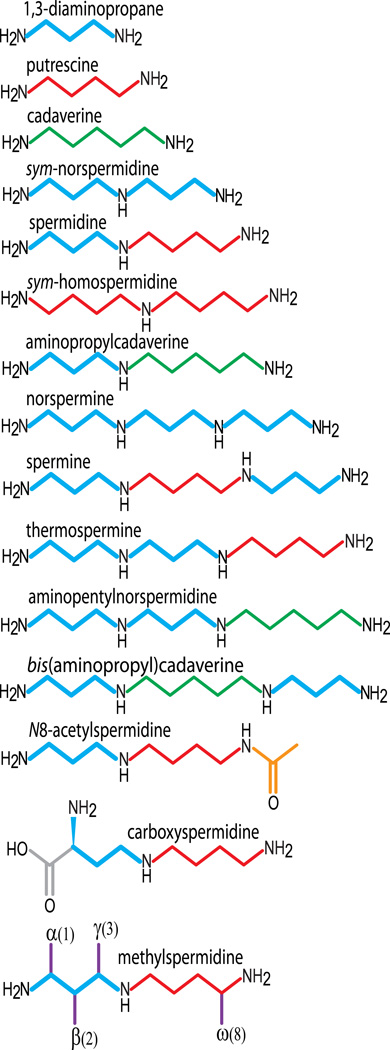
Figure 2.
Natural and synthetic polyamines used in this study. The 1,3-diaminopropane moiety in each molecule is shown in bold blue, putrescine (1,4-diaminobutane) in red and cadaverine in green (1,5-diaminopentane). Carboxyspermidine accumulates only in the ΔCASDC mutant. Each of the methylated carbon positions (in purple) of the methylated spermidine analogues is displayed on the same molecule.
RESULTS
Identification of a polyamine submolecular moiety essential for growth of A. tumefaciens C58
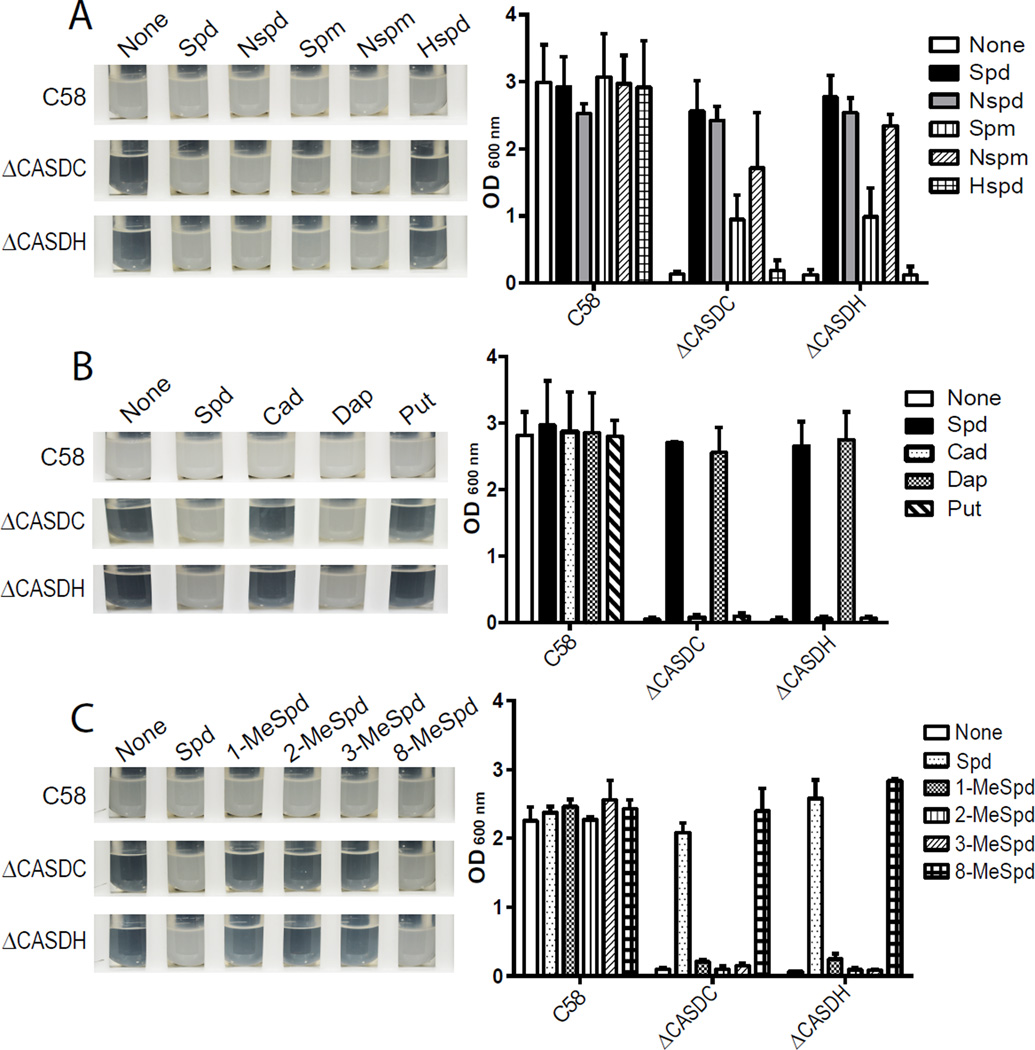
We sought to identify any functional subdomains of spermidine required for growth of A. tumefaciens spermidine auxotrophic strains. Compared to the C58 wild-type strain, the ΔCASDH (Atu4170) strain accumulates putrescine and homospermidine but lacks spermidine (Supporting Information Figure 1A); the ΔCASDC (Atu4169) strain accumulates carboxyspermidine, some putrescine and a small amount of homospermidine (Supporting Information Figure 2A). Neither strain is able to grow without provision of exogenous spermidine. First, we compared growth of ΔCASDH and ΔCASDC strains provided with the spermidine natural structural analogues sym-norspermidine and sym-homospermidine, which are one methylene group shorter and longer than spermidine, respectively (Figure 3A). Cells were grown initially in LB liquid growth medium, which naturally contains approximately 40 µM spermidine.14 They were then subcultured into chemically-defined growth medium (CDM) and subcultured a second time into CDM in order to dilute out by cell division any remaining spermidine from the LB medium. The CDM medium contained 100 µM of the indicated added polyamine. After two rounds of growth in CDM medium without polyamines, negligible growth was detected, confirming that spermidine biosynthesis is essential for growth in CDM, and provision of exogenous spermidine completely restored growth (Figure 3A). Cells grown with norspermidine grew to the same density as with spermidine, however, there was no growth with homospermidine. We also tested the natural tetraamines spermine and norspermine (Figure 2) and found that norspermine was as efficient as spermidine in restoring normal growth, and spermine consistently restored approximately 50% growth (Figure 3A). We then assessed the ability of the diamines 1,3-diaminopropane, putrescine (1,4-diaminobutane) and cadaverine (1,5-diaminopentane) to restore growth of the ΔCASDH and ΔCASDC strains (Figure 3B). Only 1,3-diaminopropane restored growth, and was as efficient as spermidine. A dosage response of growth to different concentrations of exogenous 1,3-diaminopropane was performed with the ΔCASDH and ΔCASDC strains in CDM. Complete restoration of growth was seen with 10 µM but not 1 µM 1,3-diaminopropane (Supporting Information Figure 3), whereas even 100 µM putrescine had no effect on restoration of growth.
Figure 3.
Growth of A. tumefaciens with different exogenous natural and synthetic polyamines. Cells were grown first in liquid LB and then subcultured twice in CDM to fully deplete through cell division the spermidine taken up from the LB medium. All exogenous polyamines were present in the growth medium at 100 µM. The results shown are for the second subculture, on the left are photographs of the cultures and on the right the corresponding OD600 nm. Data represent the means of triplicate cultures ± standard deviation. (A) Triamines and tetraamines: Spd, spermidine; Nspd, norspermidine; Spm, spermine; Nspm, norspermine; Hspd, homospermidine. (B) Diamines: Cad, cadaverine; Dap, 1,3-diaminopropane; Put, putrescine. (C) Methylated spermidine analogues: 1-MeSpd, 1-methylspermidine; 2-MeSpd, 2-methylspermidine; 3-MeSpd, 3-methylspermidine; 8-MeSpd, 8-methylspermidine. C58, A. tumefaciens wild-type parental strain.
The common feature of polyamines that could restore growth was the presence of a terminal 1,3-diaminopropane moiety and the common feature of polyamines that could not restore growth was the absence of a 1,3-diaminopropane moiety. It was formally possible that the exogenously provided tetraamines spermine and norspermine could be retroconverted to spermidine and norspermidine, respectively, by unknown polyamine oxidases. The polyamine content of all cultures presented in Figure 3 is shown in Supporting Information Figures 1 and 2 and there is no detectable retroconversion of the added tetraamines to generate spermidine or norspermidine. Furthermore, the ΔCASDC strain contains a large amount of endogenously produced carboxyspermidine, which does not restore growth in the absence of exogenous polyamines. The carboxyl group of carboxyspermidine is on the N1 position (Figure 2), i.e., on the aminopropyl side of the spermidine, so no intact 1,3-diaminopropane moiety is present in carboxyspermidine. In addition, the Δ4170 strain contains endogenously produced putrescine and homospermidine, but there is no growth of this strain in the absence of exogenously supplied polyamines. Homospermidine biosynthesis is induced only after spermidine depletion. These data confirm that neither endogenous nor exogenous putrescine or homospermidine support growth of A. tumefaciens. To further explore the importance of a terminal 1,3-diaminopropane moiety for growth of A. tumefaciens, we grew the ΔCASDH and ΔCASDC strains with 100 µM of a suite (Figure 2) of C-methylated spermidine analogues in CDM (Figure 3C). The 1-, 2- and 3-methylated analogues are methylated on the aminopropyl side of spermidine and thus do not possess an intact 1,3-diaminopropane moiety, whereas the 8-methylated analogue is methylated on the aminobutyl side and therefore contains an intact 1,3-diaminopropane moiety. Only the 8-methylated spermidine analogue restored growth (Figure 3C), confirming the essential role of a terminal 1,3-diaminopropane moiety in growth of A. tumefaciens.
Biosynthetic plasticity of the CASDH/CASDC polyamine pathway
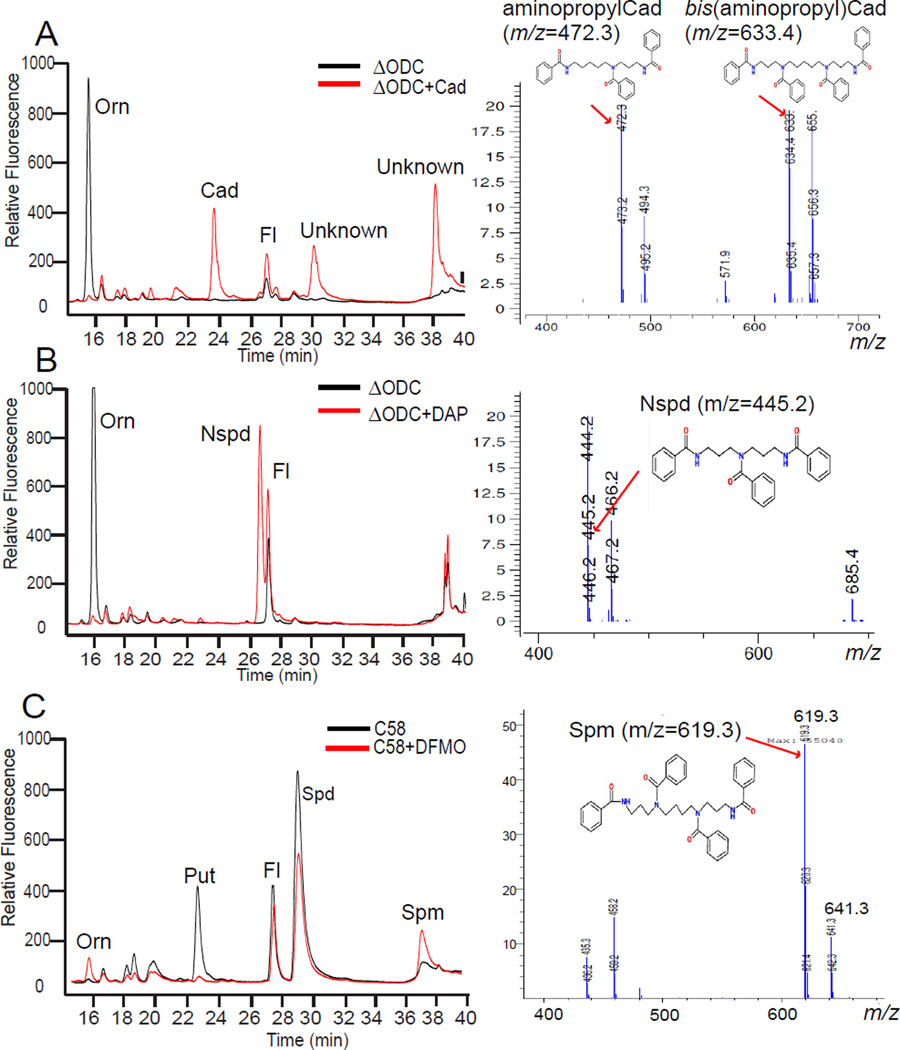
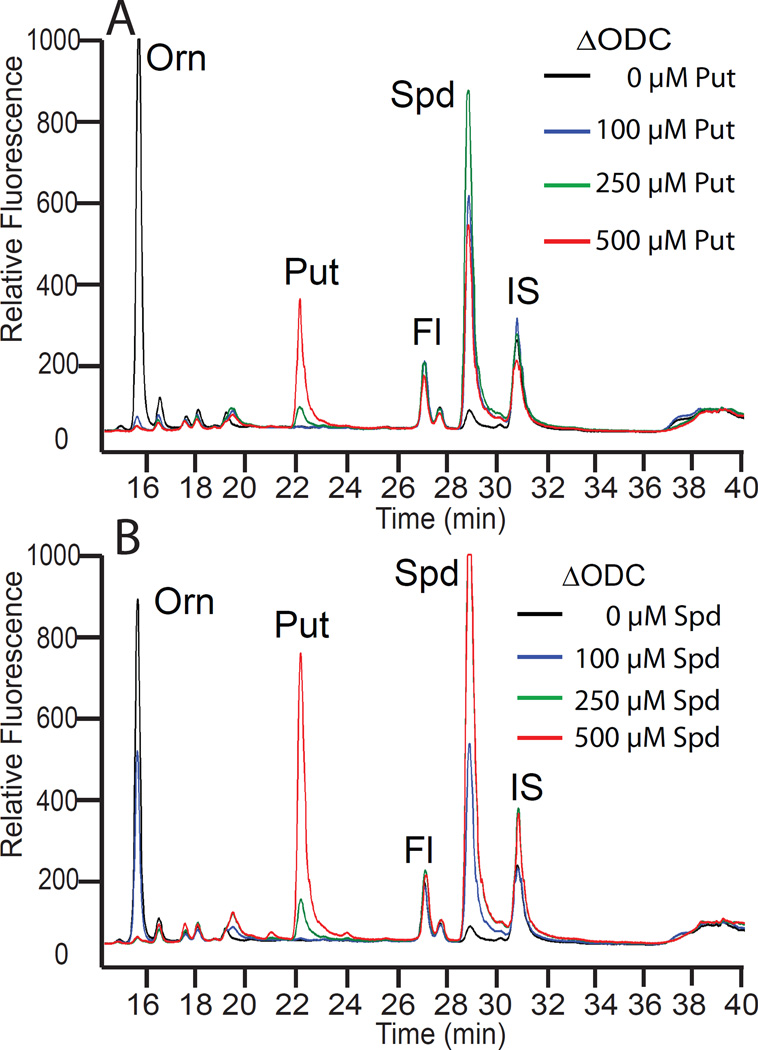
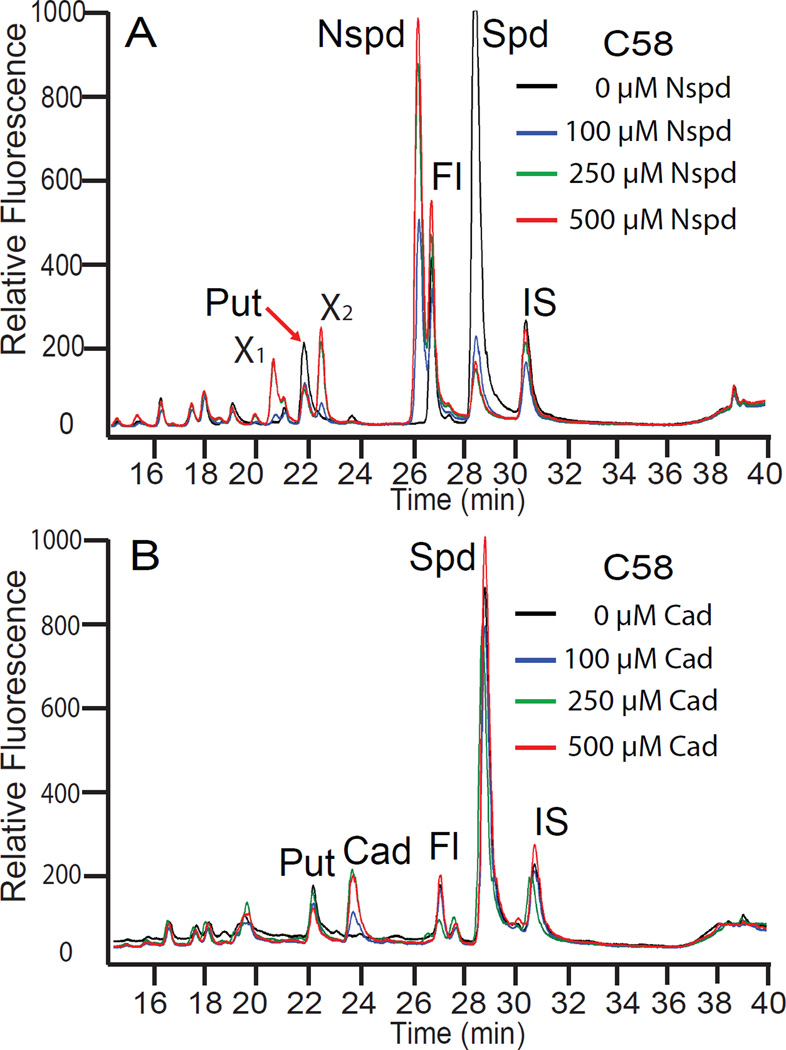
Disruption of the A. tumefaciens C58 ODC gene (Atu3196) eliminates putrescine and spermidine biosynthesis and causes a large accumulation of ornithine, which does not accumulate in the C58 wild-type strain (Figure 4). The ΔODC strain provides an opportunity to investigate the biosynthetic flexibility of the CASDH/CASDC pathway when supplied with exogenous diamines in place of putrescine. We grew the ΔODC strain in CDM with 100 µM exogenous cadaverine and observed a repression of the ornithine accumulation, the accumulation of cadaverine and appearance of two new peaks in the HPLC trace (Figure 4A). The simplest explanation for the new peaks, which did not correspond to any of the normal polyamine pure standards, was that the cadaverine had been aminopropylated to form the triamine aminopropylcadaverine (Figure 2) and this in turn had been aminopropylated to form the tetraamine bis(aminopropyl)cadaverine or its structural isomer aminopentylnorspermidine (Figure 2). A second aminopropylation of aminopropylcadaverine on the cadaverine moiety produces bis(aminopropyl)cadaverine, whereas if it is on the aminopropyl moiety, aminopentylnorspermidine is produced (Figure 2). Liquid chromatography-mass spectrometric (LC-MS) analysis of the benzoylated polyamines from cell extracts of the ΔODC strain grown with cadaverine detected peaks (Figure 4A) of m/z 472.3 and m/z 633.4, corresponding to the tribenzoylated aminopropylcadaverine and tetrabenzoylated bis(aminopropyl)cadaverine or aminopentylnorspermidine, respectively, and their sodium ion adducts (+22 Da) at m/z 494.3 and m/z 655.4. When the ΔODC strain was grown in CDM with 100 µM exogenous 1,3-diaminopropane, ornithine accumulation was repressed, a new peak of the triamine norspermidine appeared, and unlike with cadaverine, which accumulated to some extent as the diamine, all of the 1,3-diaminopropane appeared to have been converted to norspermidine (Figure 4B). No significant peak corresponding to aminopropylated norspermidine i.e., tetraamine norspermine (Figure 2) was produced by growth with 1,3-diaminopropane (Figure 4B), and this was confirmed by LC-MS, which detected a peak at m/z 445.2 corresponding to tribenzoylated norspermidine, but no peak for tetrabenzoylated norspermine (m/z 605.4) was detected.
Figure 4.
Polyamine biosynthetic plasticity in A. tumefaciens C58. The left panels are HPLC analyses and on the right are independent LC-MS mass spectra. (A) The ΔODC strain grown with or without 100 µM cadaverine (Cad). The unknown peak at 38.5 min in the HPLC trace is either the tetraamine bis(aminopropyl)cadaverine or its isomer N1-aminopentylnorspermidine, which are not distinguished by the LC-MS analysis. (B) The ΔODC strain grown with or without 100 µM 1,3-diaminopropane (DAP). The tetraamine norspermine is not formed. (C) The parental A. tumefaciens C58 wild-type strain grown with 10 mM DFMO (α-difluoromethylornithine), a highly specific suicide inhibitor of ornithine decarboxylase (ODC). The accumulation of tetraamine spermine (Spm) or its isomer thermospermine is confirmed by LC-MS. Sodium adducted forms of the polyamines are present as ions with a 22 Da higher m/z. Orn, ornithine; Fl, AccQ-Fluor tag.
The ODC protein of A. tumefaciens belongs to the alanine racemase fold,15 a different fold to the E. coli ODC, but homologous to the human ODC. We decided to compare the effect of ODC knockout with pharmacological inhibition of ODC enzymatic activity by using the highly specific ODC suicide inhibitor α-difluoromethylornithine (DFMO). Growth of the A. tumefaciens C58 wildtype strain in CDM containing 10 mM DFMO resulted in a small accumulation of ornithine, the depletion of putrescine, a reduction of spermidine content and the appearance of a new peak corresponding to the tetraamine spermine (Figure 4C), indicating that growth with DFMO resulted in the aminopropylation of spermidine to form spermine or its isomer thermospermine. If spermidine is aminopropylated on the aminobutyl moiety, spermine is produced, whereas if it occurs on the aminopropyl moiety, thermospermine is produced (Figure 2). The presence of spermine/thermospermine (Figure 2) was confirmed by LC-MS of the benzoylated cell extracts, which detected a peak at m/z 619.3, corresponding to the tetrabenzoylated spermine or thermospermine (Figure 4C).
Polyamine metabolic regulation
Accumulation of ornithine in the ΔODC strain could be due to arrest of flux from ornithine to putrescine and spermidine, or it could be due to alleviation of product feedback inhibition. To distinguish between these two possibilities, ΔODC cells were grown in CDM containing different concentrations of putrescine (Figure 5A) or spermidine (Figure 5B). Both exogenous putrescine and spermidine completely repressed ornithine accumulation at 100 µM in the ΔODC cells, strongly indicating that product feedback represses ornithine accumulation, rather than arrest of metabolic flux. An important observation with ΔODC cells, which cannot make putrescine, is that growth in CDM with exogenous spermidine results in the accumulation of high levels of putrescine (Figure 5B). Production of putrescine from spermidine indicates the presence of an active polyamine retroconversion pathway and the presence of an efficient spermidine oxidase/dehydrogenase. To determine whether the spermidine retroconversion pathway might represent a catabolic or regulatory function, we assessed whether 1 mM exogenous spermidine could be used as sole nitrogen or carbon source with A. tumefaciens C58 strain in liquid CDM, however, no growth was observed (results not shown). These results suggest that the spermidine retroconversion pathway is unlikely to be involved in a nutritional role.
Figure 5.
A spermidine retroconversion activity in A. tumefaciens. Polyamine content assessed by HPLC in the ΔODC strain after growth in CDM containing either (A) putrescine (Put) or (B) spermidine (Spd) at the indicated concentrations. After the second subculture (30°C), cell numbers were adjusted through OD600 nm equalization and, after centrifugation of equal cell number aliquots, the cell pellet was washed three times with PBS. Orn, ornithine; Fl, AccQ-Fluor tag; IS, internal standard (1,7-diaminoheptane).
As putrescine negatively regulates ornithine accumulation, we sought to determine whether spermidine negatively regulates its own production. Supplying exogenous spermidine to the C58 wild-type strain will mask any decrease in its biosynthesis, so we reasoned that since norspermidine can completely replace spermidine for growth, and can be distinguished from spermidine by HPLC, then norspermidine may be useful to detect negative feedback regulation of spermidine accumulation. Indeed, even 100 µM exogenous norspermidine had a marked repressive effect on spermidine accumulation (Figure 6A). In addition to repressing spermidine accumulation, provision of exogenous norspermidine caused the appearance of two new peaks (X1 and X2, Figure 6A) of unknown identity, which may represent catabolic or acetylated products. To assess potential negative feedback regulation of putrescine accumulation by putrescine, we used exogenous cadaverine as structural proxy to assess whether putrescine accumulation in the C58 strain was repressed by this diamine analogue. Exogenous cadaverine had no effect on putrescine accumulation and cadaverine appeared to be taken up inefficiently in the C58 wild-type strain (Figure 6B). The cadaverine that was taken up was not converted to a triamine or tetraamine in the C58 wild-type strain. These results suggest that putrescine does not repress its own biosynthesis, consistent with the large accumulation of putrescine in the ΔCASDH mutant (Supporting Information Figure 2A).
Figure 6.
Exogenous norspermidine represses spermidine accumulation. Polyamine content assessed by HPLC in the parental wild-type A. tumefaciens C58 strain. Cultures were grown as in Figure 5, in the presence of (A) exogenous norspermidine (Nspd) or (B) cadaverine (Cad) at the indicated concentrations. Put, putrescine; Spd, spermidine; Fl, AccQ-Fluor tag; IS, internal standard (1,7-diaminoheptane); X1, X2, unknown peaks.
An acetylspermidine deacetylase activity
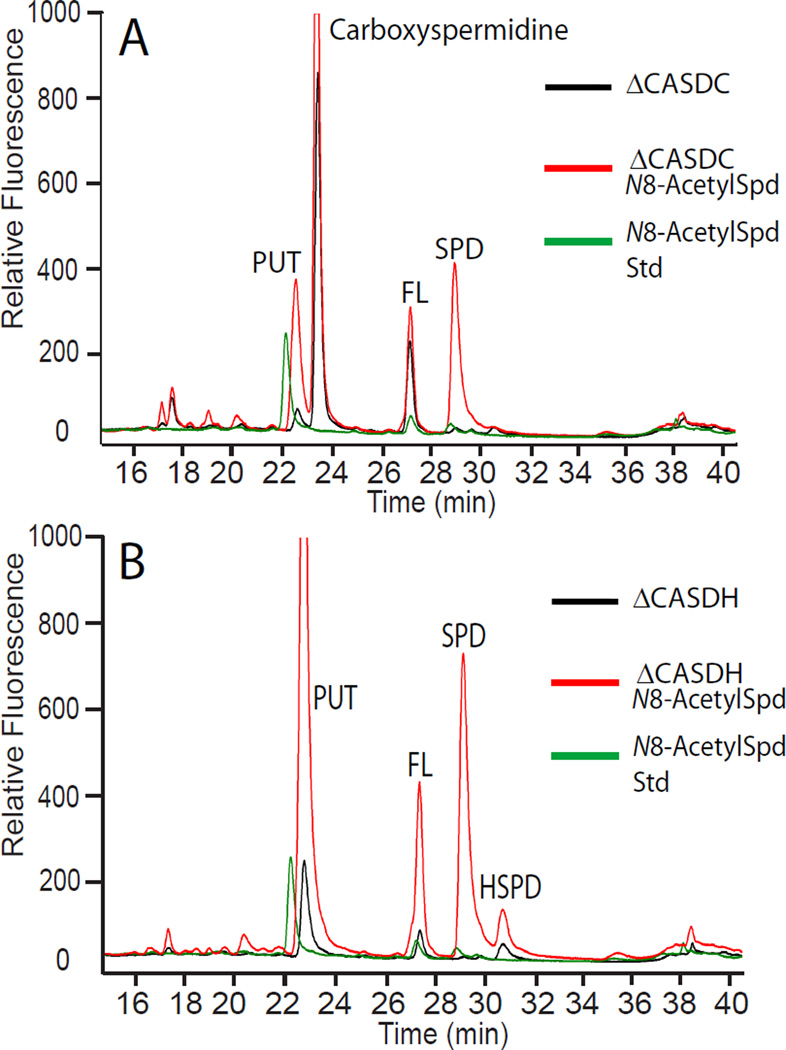
If a terminal 1,3-diaminopropane moiety is sufficient to restore growth to A. tumefaciens spermidine auxotrophs, then N8-acetylspermidine should be able to restore growth because the acetyl group is on the aminobutyl side of spermidine. Indeed, the ΔCASDH and ΔCASDC strains grow well in CDM with 100 µM exogenous N8-acetylspermidine (Supporting Information Figure 5). Analysis of the polyamine content of cells grown with and without 100 µM N8-acetylspermidine revealed that the pure chemical standard N8-acetylspermidine eluted very close to putrescine (Figure 7A,B). Surprisingly, a large peak of spermidine was seen to accumulate in both the ΔCASDH and ΔCASDC strains, indicating that the N8-acetylspermidine was subject to a deacetylase activity releasing spermidine. A zinc-dependent acetylpolyamine amidohydrolase/deacetylase (BAA01256) has been isolated from the α-proteobacterium Mycoplana ramosa.16 Both A. tumefaciens and M. ramosa belong to the Rhizobiales order of the α-Porteobacteria but A. tumefaciens is a member of the family Rhizobiaceae and M. ramosa belongs to the Brucellaceae. A homologue of the M. ramosa acetylpolyamine amidohydrolase is encoded in the A. tumefaciens C58 genome (Atu 0748, NP_353772) and exhibits 31% identity to the M. ramosa protein.
Figure 7.
An acetylspermidine deacetylase activity in A. tumefaciens. The ΔCASDC (A) and ΔCASDH (B) strains were grown as described for the ΔODC strain in Figure 5, with or without 100 µM exogenous N8-acetylspermidine. PUT, putrescine; SPD, spermidine; HSPD, homospermidine, FL, AccQ-Fluor tag; N8-acetylspermidine Std, sample of pure N8-acetylspermidine.
DISCUSSION
It is now clear that spermidine is absolutely essential for growth of A. tumefaciens but it is also evident that polyamines containing a terminal 1,3-diaminopropane moiety, including 1,3-diaminopropane itself, can fulfill the function of spermidine for growth. This is a very surprising finding and it is worth noting that in eukaryotes, the aminobutyl donor function of spermidine cannot be fulfilled by putrescine. The broader implication from these findings is that in A. tumefaciens spermidine may be less likely to function in growth by general binding of RNA, as its essential growth function can be replaced by 1,3-diaminopropane, which as a diamine has a much lower binding affinity for RNA.17 In addition, the related diamines putrescine and cadaverine cannot support growth, whereas their aminopropylated forms, spermidine and aminopropylcadaverine, which do contain a terminal 1,3-diaminopropane moiety, restore growth (Figure 3). It is not apparent whether 1,3-diaminopropane is the functional module, or the aminopropyl group contained within. The preference for 1,3-diaminopropane over putrescine or cadaverine in growth suggests that its function is part of a more specific interaction, electrostatic or covalent, rather than general electrostatic binding to polyanionic substrates. However, it is also possible that 1,3-diaminopropane, or at least the 1,3-diaminopropane moiety of spermidine, is the key structural feature for general electrostatic binding to nucleic acids in bacteria. It may be incorporated into another small but essential metabolite, it may be transferred as the intact diamine or an aminopropyl group to protein, lipid, nucleic acid or cell wall substrates. Alternatively, and perhaps in addition to its essential functions, it may act as a signal molecule. The biosynthesis of spermidine and spermine may be simply an indirect route to synthesize 1,3-diaminopropane modules. Unlike A. tumefaciens, certain bacteria such as Vibrio cholerae can directly synthesize 1,3-diaminopropane.18 It is clear that the ability of norspermine and spermine to replace spermidine is unlikely to be due to any catabolism of the tetraamines to triamines, eg., the retroconversion of spermine to spermidine. No catabolic products of the tetraamines were detectable by HPLC (Supporting Information Figures 1 and 2).
In eukaryotes, the synthesis of spermidine19 from putrescine, and spermine20 and thermospermine21 from spermidine are performed by distinct dcAdoMet-dependent aminopropyltransferases. Human and E. coli dcAdoMet-dependent spermidine synthases are highly specific, with limited if any ability to make spermine under physiological conditions, and in mammals a separate spermine synthase is required for spermine biosynthesis. In contrast to the dcAdoMet-dependent polyamine biosynthetic pathways, the aspartate β-semialdehyde-dependent pathway, consisting of the CASDH and CASDC enzymes, although first described by Tait nearly 40 years ago22 has received scant attention. The CASDH/CASDC pathway is apparently plastic, deletion of 1,3-diaminopropane biosynthesis in V. cholerae results in the abolition of norspermidine biosynthesis and induction of spermidine biosynthesis from putrescine.18 In Campylobacter jejuni, the pathway synthesizes only spermidine,23 since diamines other than putrescine are not produced. Our present study has uncovered a more extensive biosynthetic plasticity for the CASDH/CASDC pathway. The ΔODC strain allowed us to replace putrescine with 1,3-diaminopropane resulting in synthesis of norspermidine, and also with cadaverine, resulting in synthesis of aminopropylcadaverine and the corresponding tetraamine bis(aminopropyl)cadaverine or its isomer aminopentylnorspermidine. It is intriguing that norspermidine is not converted into a tetraamine, whereas aminopropylcadaverine does yield a tetraamine. Of relevance to this finding is the fact that inhibition of the putrescine biosynthetic enzyme ODC with DFMO causes a depletion of putrescine, a reduction of spermidine levels and the induction of spermine (or thermospermine) biosynthesis. One possible explanation for these differential tetraamine biosynthetic behaviours is that optimal spermidine levels may repress spermine/thermospermine biosynthesis by the CASDH/CASDC pathway through allosteric regulation of CASDH or by a more direct effect on the active site, such as competitive inhibition. There is an active spermidine retroconversion pathway to putrescine (Figure 5B), so if spermidine levels become limiting, the continued activity of the retroconversion pathway would further deplete spermidine. Derepression of tetraamine biosynthesis through lowered spermidine levels would allow diversion of spermidine into spermine, which does not participate in retroconversion (Supporting Information Figures 1D and 2D) and is still able to replace spermidine for growth. In the case of aminopropylcadaverine, the absence of spermidine would allow tetraamine biosynthesis from aminopropylcadaverine. The lack of tetraamine biosynthesis from norspermidine may be explained if norspermidine binds to the spermidine-regulated site that prevents tetraamine biosynthesis, and this site would not be bound by aminopropylcadaverine. Therefore, if spermidine levels decrease below optimal levels, the CASDH/CASDC pathway synthesizes spermine from spermidine, thereby sequestering the 1,3-diaminopropane moieties from retroconversion of spermidine that would produce putrescine and unstable 3-aminopropanaldehyde as products. Thus, even if spermidine levels fall, the low flux of the 1,3-diaminopropane moieties into spermine compensates for the decrease by protecting the 1,3-diaminopropane moiety from catabolism through retroconversion. Some indirect corroboration for this hypothesis comes from the related α-proteobacterium Paracoccus denitrificans, which synthesizes a spermidine-based triscatecholate siderophore parabactin,24 similar to agrobactin. When spermidine is diverted into parabactin biosynthesis, spermine is synthesized.24
A spermidine dehydrogenase activity was previously detected in the γ-proteobacteria Serratia marascens,25 Citrobacter freundii26 and Pseudomonas aeruginosa.27 This activity catabolizes spermidine to 1,3-diaminopropane and unstable 4-aminobutyraldehyde. The A. tumefaciens spermidine retroconversion activity we have identified is the first case of spermidine to putrescine retroconversion identified in bacteria. Conversion of spermidine to putrescine in A. tumefaciens eliminates the essential 1,3-diaminopropane moiety required for growth, thus the retroconversion activity functions to reduce the growth promoting role of spermidine. However, the growth function can be restored by re-aminopropylating the putrescine product to form spermidine again. Total spermidine levels may reflect the balance between biosynthesis and retroconversion, which provides an opportunity for a robust homeostatic circuit to control spermidine levels.
The α-Proteobacteria of which A. tumefaciens and P. denitrificans are members, are unusual in that they possess an ODC from the alanine racemase-fold, similar to eukaryotes but unlike most other bacteria.15 Regulation of ornithine levels by the downstream polyamines has not been investigated previously but in this study we have shown that putrescine represses ornithine accumulation, an example of negative feedback by a downstream metabolite. We also observed that spermidine, and especially norspermidine repressed carboxyspermidine accumulation in the ΔCASDC strain and caused an accumulation of putrescine. Spermidine and particularly norspermidine caused a repression of homospermidine accumulation in the ΔCASDH strain. Since the CASDH and homospermidine synthase enzymes are distantly related,28 they may be subject to similar polyamine feedback regulation.
Finally, the observation of an active spermidine deacetylase suggests that acetylation/deacetylation may be an additional mechanism to regulate spermidine levels in A. tumefaciens. As a plant pathogen, a spermidine deacetylase/amidohydrolase activity could be very relevant to the interaction of A. tumefaciens with plants. Spermidine and putrescine and often found at very high concentrations in plants in the form of hydroxycinnamic amides.29, 30 A spermidine amidohydrolase activity might liberate spermidine from these otherwise inaccessible forms if these compounds are taken up into the bacterial cell, and at the same time neutralize their potential antibacterial activity, as postulated for tyramine hydroxycinnamic amides.31
METHODS
Reagents
Natural polyamines were purchased from Sigma-Aldrich, sym-homospermidine and α-difluoromethyornithine (DFMO) were gifts from Patrick Woster (Medical University of South Carolina). Methylated spermidine analogues were synthesized as previously described.32, 33
Bacterial strains and growth conditions
Construction of in-frame markerless deletions of the odc (Atu3196), casdh (Atu4170) and casdc (Atu4169) are described in detail in the Supplementary Information. Strains of A. tumefaciens C58 and gene deletions were grown from −80°C glycerol stocks on LB solid agar containing relevant antibiotic for 2 days at 30°C and a single colony was used to inoculate LB liquid medium grown overnight at 30°C for 18 to 20 h on a shaking incubator with 220 rpm. After measuring OD600 nm, cells were washed three times with PBS and diluted to initial OD600 nm of 0.05 for subsequent growth in 2 ml chemically-defined growth medium,34 in a 14 ml plastic screw-top tube with or without 100 µM added polyamines, unless indicated otherwise. Second subcultures were grown using the same conditions. Minimal medium (CDM) was prepared from sterile stocks of 20× buffer and 20× salts solution supplemented with glucose.
Polyamine extraction
Bacterial cells were harvested by centrifugation (18,000 × g, 3 min) and washed in PBS three times. Cells were resuspended in MOPS lysis buffer (20 mM MOPS, pH 8.0, 10 mM NaCl, 4 mM MgCl2) and the bacterial cell suspension was subject to three cycles of freeze/thawing. Forty percent trichloroacetic acid was added to a final concentration of 10%. After centrifugation (18,000 × g, 5 min, 4°C), the supernatant was used for immediate analysis or stored at −20°C until needed.
Polyamine analysis by HPLC
Precolumn derivatization of 5 µl of polyamine sample was performed with AccQ-Fluor reagent labeling kit (Waters, cat. no. WAT052880). Reactions were performed in AccQ-fluor borate buffer in a total volume of 100 µl and heated to 55°C for 10 min. Labeled polyamines were separated by HPLC using a hydrolysate amino acid analysis column (AccQ-Tag Column, 60 Å, 4 µm, 3.6 mm × 150 mm, Waters) on a Beckman Coulter (System Gold) apparatus with fluorescence detection (excitation 248 nm; emission 398 nm, Prostar). Solvent A was 520 mM sodium acetic acid, 17 mM trimethylamine, 0.01% sodium azide, pH 4.65; Solvent B was 60% acetonitrile, 0.01% acetone at a flow rate of 1.0 ml/min and the elution was performed using the following linear gradient of elution buffer B: 2–6 min 0–20%, 6–11 min 20%, 11–27 min 20–50%, 27–34 min 50%, 34–37 min 50–100%, 37–40 min 100%, 40–50 min 0%.
Polyamine benzoylation for LC-MS analysis
Bacterial strains were grown in CDM with or without 100 µM exogenous polyamines. After centrifugation, cell pellets were processed using the same polyamine extraction procedure used for HPLC analysis except that 200 µl of lysis buffer was used and extracts were kept on ice for 5 min. Then in a glass vial 1 ml of 2 M NaOH was mixed with 200 µl of cell lysate containing polyamines, and 10 µl of benzoyl chloride (Sigma 259950-250) was added and vortexed for 1 min or until the benzoyl chloride was fully mixed. After 20 min incubation at room temperature, 2 ml of saturated NaCl was added and vortexed vigorously for 1 min. Two ml of diethyl ether was added to the mixture and vortexed 5 seconds and then centrifuged at 1500 × g for 5 min to separate the polyamine-containing diethyl ether layer from the aqueous layer. About 1 ml of the upper ether layer, which contains the benzoylated polyamines was carefully collected in a glass vial and evaporated completely in a chemical hood and kept at −20°C until further analysis. Benzoylated cell extracts were analyzed by LC-MS. The LC component was performed at a flow rate of 0.5 ml/min with a 50 µl injection volume and the isocratic elution starting with 20% and with a hold of 50% aqueous acetonitrile containing 0.1% formic acid over 10 min, and with a hold of 90% for the following 10 min. Cone voltage was 50 V and desolvation temp., was 50 °C. LC-MS analyses were performed on a liquid chromatograph Agilent 1100 series (flow rate of 0.5 ml/min) equipped with a Waters 2487 dual-wavelength absorbance detector (210 & 254 nm), column oven 30°C and with an Agilent Zorbax Eclipse XDB-C18 (4.6 × 150 mm, 5 µm column that was coupled to a Waters Quattro Micro API (atmospheric pressure ionization) mass spectrometer in positive mode with a scan range 100 to 640 m/z. MassLynx software (Waters) was used for data acquisition and processing.
Supplementary Material
Acknowledgments
AJM was supported by a UT Southwestern Medical Center seed grant, CF was supported by NIH grant GM080546, MK and AK (syntheses of methylated spermidine analogues) were supported by Russian Science Foundation grant 14-14-01099. We thank Meg Phillips (UT Southwestern) for constructive criticism of the manuscript.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available contains Supplementary Methods and Supporting Figures 1–4: This material is available free of charge via the Internet.
The authors declare no competing financial interests.
REFERENCES
- 1.Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–894. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. U S A. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishimura K, Lee SB, Park JH, Park MH. Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids. 2012;42:703–710. doi: 10.1007/s00726-011-0986-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol. Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen S, Leija C, Kinch L, Regmi S, Li Q, Grishin NV, Phillips MA. Deoxyhypusine Modification of Eukaryotic Translation Initiation Factor 5A (eIF5A) Is Essential for Trypanosoma brucei Growth and for Expression of Polyprolyl-containing Proteins. J. Biol. Chem. 2015;290:19987–19998. doi: 10.1074/jbc.M115.656785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pegg AE, Michael AJ. Spermine synthase. Cell. Mol. Life Sci. 2010;67:113–121. doi: 10.1007/s00018-009-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 2010;17:1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- 8.Igarashi K, Kashiwagi K. Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 2010;42:39–51. doi: 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Igarashi K, Kashiwagi K. Polyamine Modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J. Biochem. 2006;139:11–16. doi: 10.1093/jb/mvj020. [DOI] [PubMed] [Google Scholar]
- 10.Chattopadhyay MK, Tabor CW, Tabor H. Polyamines are not required for aerobic growth of Escherichia coli: preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J. Bacteriol. 2009;191:5549–5552. doi: 10.1128/JB.00381-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolff EC, Park MH, Folk JE. Cleavage of spermidine as the first step in deoxyhypusine synthesis. The role of NAD. J. Biol. Chem. 1990;265:4793–4799. [PubMed] [Google Scholar]
- 12.Jakus J, Wolff EC, Park MH, Folk JE. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. Effective inhibition by bis- and mono-guanylated diamines and polyamines. J. Biol. Chem. 1993;268:13151–13159. [PubMed] [Google Scholar]
- 13.Hyvonen MT, Keinanen TA, Khomutov M, Simonian A, Vepsalainen J, Park JH, Khomutov AR, Alhonen L, Park MH. Effects of novel C-methylated spermidine analogs on cell growth via hypusination of eukaryotic translation initiation factor 5A. Amino Acids. 2012;42:685–695. doi: 10.1007/s00726-011-0984-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGinnis MW, Parker ZM, Walter NE, Rutkovsky AC, Cartaya-Marin C, Karatan E. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol. Lett. 2009;299:166–174. doi: 10.1111/j.1574-6968.2009.01744.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Michael AJ, Martynowski D, Goldsmith EJ, Phillips MA. Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha-barrel fold basic amino acid decarboxylases. J. Biol. Chem. 2007;282:27115–27125. doi: 10.1074/jbc.M704066200. [DOI] [PubMed] [Google Scholar]
- 16.Sakurada K, Ohta T, Fujishiro K, Hasegawa M, Aisaka K. Acetylpolyamine amidohydrolase from Mycoplana ramosa: gene cloning and characterization of the metal-substituted enzyme. J. Bacteriol. 1996;178:5781–5786. doi: 10.1128/jb.178.19.5781-5786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lightfoot HL, Hall J. Endogenous polyamine function--the RNA perspective. Nucleic Acids Res. 2014;42:11275–11290. doi: 10.1093/nar/gku837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Sperandio V, Frantz DE, Longgood J, Camilli A, Phillips MA, Michael AJ. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 2009;284:9899–9907. doi: 10.1074/jbc.M900110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korolev S, Ikeguchi Y, Skarina T, Beasley S, Arrowsmith C, Edwards A, Joachimiak A, Pegg AE, Savchenko A. The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor. Nat. Struct. Biol. 2002;9:27–31. doi: 10.1038/nsb737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, Min J, Zeng H, McCloskey DE, Ikeguchi Y, Loppnau P, Michael AJ, Pegg AE, Plotnikov AN. Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism. J. Biol. Chem. 2008;283:16135–16146. doi: 10.1074/jbc.M710323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knott JM, Romer P, Sumper M. Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 2007;581:3081–3086. doi: 10.1016/j.febslet.2007.05.074. [DOI] [PubMed] [Google Scholar]
- 22.Tait GH. A new pathway for the biosynthesis of spermidine. Biochem. Soc. Trans. 1976;4:610–612. doi: 10.1042/bst0040610. [DOI] [PubMed] [Google Scholar]
- 23.Hanfrey CC, Pearson BM, Hazeldine S, Lee J, Gaskin DJ, Woster PM, Phillips MA, Michael AJ. Alternative spermidine biosynthetic route is critical for growth of Campylobacter jejuni and is the dominant polyamine pathway in human gut microbiota. J. Biol. Chem. 2011;286:43301–43312. doi: 10.1074/jbc.M111.307835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergeron RJ, Weimar WR. Increase in spermine content coordinated with siderophore production in Paracoccus denitrificans. J. Bacteriol. 1991;173:2238–2243. doi: 10.1128/jb.173.7.2238-2243.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabor CW, Kellogg PD. Identification of flavin adenine dinucleotide and heme in a homogeneous spermidine dehydrogenase from Serratia marcescens. J. Biol. Chem. 1970;245:5424–5433. [PubMed] [Google Scholar]
- 26.Hisano T, Murata K, Kimura A, Matsushita K, Toyama H, Adachi O. Characterization of membrane-bound spermidine dehydrogenase of Citrobacter freundii. Biosci. Biotechnol. Biochem. 1992;56:1916–1920. doi: 10.1271/bbb.56.1916. [DOI] [PubMed] [Google Scholar]
- 27.Dasu VV, Nakada Y, Ohnishi-Kameyama M, Kimura K, Itoh Y. Characterization and a role of Pseudomonas aeruginosa spermidine dehydrogenase in polyamine catabolism. Microbiology. 2006;152:2265–2272. doi: 10.1099/mic.0.28920-0. [DOI] [PubMed] [Google Scholar]
- 28.Shaw FL, Elliott KA, Kinch LN, Fuell C, Phillips MA, Michael AJ. Evolution and multifarious horizontal transfer of an alternative biosynthetic pathway for the alternative polyamine sym-homospermidine. J. Biol. Chem. 2010;285:14711–14723. doi: 10.1074/jbc.M110.107219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo J, Fuell C, Parr A, Hill L, Bailey P, Elliott K, Fairhurst SA, Martin C, Michael AJ. A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell. 2009;21:318–333. doi: 10.1105/tpc.108.063511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M. A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J. 2009;58:246–259. doi: 10.1111/j.1365-313X.2008.03773.x. [DOI] [PubMed] [Google Scholar]
- 31.Campos L, Lison P, Lopez-Gresa MP, Rodrigo I, Zacares L, Conejero V, Belles JM. Transgenic tomato plants overexpressing tyramine N-hydroxycinnamoyltransferase exhibit elevated hydroxycinnamic acid amide levels and enhanced resistance to Pseudomonas syringae. Mol. Plant Microbe Interact. 2014;27:1159–1169. doi: 10.1094/MPMI-04-14-0104-R. [DOI] [PubMed] [Google Scholar]
- 32.Khomutov AR, Weisell J, Khomutov MA, Grigorenko NA, Simonian AR, Hakkinen MR, Keinanen TA, Hyvonen MT, Alhonen L, Kochetkov SN, Vepsalainen J. Methylated polyamines as research tools. Methods Mol. Biol. 2011;720:449–461. doi: 10.1007/978-1-61779-034-8_29. [DOI] [PubMed] [Google Scholar]
- 33.Hyvonen MT, Keinanen TA, Khomutov M, Simonian A, Weisell J, Kochetkov SN, Vepsalainen J, Alhonen L, Khomutov AR. The use of novel C-methylated spermidine derivatives to investigate the regulation of polyamine metabolism. J. Med. Chem. 2011;54:4611–4618. doi: 10.1021/jm200293r. [DOI] [PubMed] [Google Scholar]
- 34.Tempe J, Petit A, Holsters M, Montagu M, Schell J. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: Possible relation to transformation in crown gall. Proc. Natl. Acad. Sci. U S A. 1977;74:2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.