Abstract
Maturation defects occurring in adult tissue progenitor cells have the potential to contribute to tumor development; however, there is little experimental evidence implicating this cellular mechanism in the pathogenesis of solid tumors. Inhibitor of DNA-binding 2 (Id2) is a transcription factor known to regulate the proliferation and differentiation of primitive stem and progenitor cells. Id2 is derepressed in adult tissue neural stem cells (NSC) lacking the tumor suppressor Tp53 and modulates their proliferation. Constitutive expression of Id2 in differentiating NSCs resulted in maturation-resistant oligodendroglial precursor cells (OPC), a cell population implicated in the initiation of glioma. Mechanistically, Id2 overexpression was associated with inhibition of the Notch effector Hey1, a bHLH transcription factor that we here characterize as a direct transcriptional repressor of the oligodendroglial lineage determinant Olig2. Orthotopic inoculation of NSCs with enhanced Id2 expression into brains of mice engineered to express platelet-derived growth factor in the central nervous system resulted in glioma. These data implicate a mechanism of altered NSC differentiation in glioma development and characterize a novel mouse model that reflects key characteristics of the recently described proneural subtype of glioblastoma multiforme. Such findings support the emerging concept that the cellular and molecular characteristics of tumor cells are linked to the transformation of distinct subsets of adult tissue progenitors.
Introduction
Glioblastoma is an aggressive malignant primary brain tumor characterized by distinctive histopathologic features, including cellular heterogeneity, necrosis, and endothelial proliferation. These tumors are incurable by currently available therapies and frequently recur following an initial therapeutic response (1). Numerous studies have been conducted suggesting that glioblastoma arises from stem-like cells, including lineage-specific precursor cells (for review, refs. 2–4). Genomic-based classification has identified subtypes of glioblastoma, including classical, neural, proneural, and mesenchymal tumors defined by structural and expression-based genomic characteristics, treatment response, and patient outcomes (5). With this framework, transformation events affecting different precursor cell populations can be evaluated for their importance in giving rise to molecularly distinct subtypes of glioma.
Amplification of genomic DNA encoding PDGFRA, a plate-let-derived growth factor receptor, and high levels of PDGFRA expression are most commonly found in the proneural subset of glioma (5). Oligodendroglial precursor cells (OPC) are central nervous system (CNS) progenitor cells in which some glioma may arise (6). Because PDGF, a major mitogen for OPCs, and other molecular markers of OPCs are associated with proneural tumors (7), it is possible that this subset of glioma may arise from OPCs. Inhibitor of DNA-binding 2 (Id2) is expressed by OPCs and is important for determination of the oligodendroglial lineage (8). Forced expression of Id2 inhibits the terminal differentiation of oligodendrocyte marker 4 (O4)–positive rat optic nerve OPCs (9). Little is known about the specification of neural stem cells (NSC) into the oligodendroglial lineage and expansion of early OPCs, a progenitor cell type implicated in the initiation of glioma (6).
Id2 is directly repressed by p53 and in Tp53−/− NSCs isolated from the adult subventricular zone sustained overexpression of Id2 is required for proliferation (10). Here, we characterize the functional consequences of Id2 deregulation in adult brain tissue–derived NSCs during oligodendroglial differentiation and gliomagenesis. Our findings identify a novel mechanism by which oligodendroglial differentiation is mediated by Id2 and characterize a novel mouse model of PDGF-dependent gliomagenesis highly relevant to the study of proneural glioma.
Materials and Methods
Mouse procedures
Animal husbandry was performed in accordance with Dartmouth College guidelines under Institutional Animal Care and Use Committee–approved protocols. Descriptions of intracranial NSC injections and histologic analysis can be found in the Supplementary Materials. Intracranial NSC injections were made into entire litters of mice resulting from breedings designed to generate equal numbers of hPDGFB-positive (+) and hPDGFB-negative (−) animals previously reported by our laboratory (11). Innoculations were performed by injecting 2 µL of 4 × 104 NSCs/µL in sterile PBS into the ventricular region of the neonatal forebrain with a Hamilton syringe between postnatal day 0 and 3 as previously described (12). Genotyping was performed using standard techniques and verified in tail tissue obtained from experimental animals at time of death.
Recombinant DNA constructs
An Id2 retroviral expression vector was generated by BamHI digestion of a previously generated N-terminal flag tagged human Id2 into the pBMN–IRES–puromycin vector (10). The pBMN–IRES–puromycin vector was kindly provided by Dr. Michael Cole (Dartmouth College, Hanover, NH). The Olig2 luciferase reporter was kindly provided by Dr. David Gottlieb (Washington University, St Louis, MO). pBabe–puromycin–Hey1 plasmid was kindly provided by Dr. Tom Kadesch (University of Pennsylvania, Philadelphia, PA).
Cell culture, differentiation, and immunocytochemistry
NSCs were isolated from neonatal C57BL/6 mice (The Jackson Laboratory) or Id2−/− animals previously reported (13) and propagated in culture as neurospheres in media containing 20 ng/mL EGF (STEMCELL Technologies), 1× B27 without vitamin A (Invitrogen), and penicillin/streptomycin as previously described (14). NSC differentiation was induced by removal of mitogens and transfer of cultured neurospheres onto adherent substrates in Dulbecco's Modified Eagle Medium (DMEM)/F12 media containing 4% FBS, 2 mmol/L glutamine, and B27 with vitamin A. OPC enrichment was induced using media containing DMEM/F12, 1× N2 supplement (Invitrogen), 20ng/mL PDGF-BB (Peprotech), 20 ng/mL bFGF (basic fibroblast growth factor; Peprotech), 10 mmol/L forskolin (Sigma), and penicillin/streptomycin as previously described (15). For immunocytochemistry (ICC), cells were fixed briefly with 4% paraformaldehyde (PFA). For nuclear antigens, cells were permeabilized with 0.2% triton/PBS. Blocking reagents contained 1% BSA (bovine serum albumin) and 5% normal sera. ICC antibodies for single antigen immunostains included rabbit anti-Olig2 (1:400; Millipore; AB9610), mouse anti-O4 (1:200; Millipore; MAB345), and rabbit anti-NG2 (1:1,000; Millipore; AB5320). For dual labeling, we used mouse anti-Olig2 (Millipore; 1:500; 211F1.1), rabbit anti- PDGFRA (1:500; Cell Signaling Technology; #3164), and rabbit anti-Ng2 detailed above. Species directed fluorescent secondary antibodies were conjugated to Alexa-488 or Alexa-555 (1:1,000; Invitrogen). Nuclei were counterstained with Hoechst dye. Fluorescent images were captured under identical monochrome conditions and color composites were created using ImagePro software.
Flow cytometry
NSCs were rinsed and disrupted with accutase (STEMCELL Technologies), fixed using PFA, and permeabilized with 0.2% triton PBS. For single color labeling, cells were blocked with 1% BSA and 10% normal sera followed by incubation with rotation overnight in the presence of mouse monoclonal anti-Olig2 and rabbit anti-Ki67 antibodies detailed above. Fluorescent detection was accomplished using species-specific Alexa Fluor–conjugated secondary antibodies detailed above. Labeled cells were analyzed using a FACSCalibur flow cytometer and data were analyzed using the FlowJo analysis platform (Tree Star Inc.). For four-color labeling, accutase disrupted live cells were incubated with fluorophore-conjugated primary antibodies: anti-Ng2 biotin [eBioscience; 1:10, detected using streptavidin PE (phycoerythrin)-Cy7], anti-O4-PE (Miltenyi; 1:10), and anti-PDGFRα–APC (antigen-presenting cell; eBioscience; 1:10). Labeled cells were then fixed with formalin and permeabilized with 2% triton in PBS, and then incubated with fluorophore-conjugated anti-Olig2–Alexa-488 (Millipore; 1:1,000). Labeled cells were analyzed using a MACSQuant cytometer (Miltenyi) and data were digitally compensated using beads labeled with identical antibodies (OneComp; eBioscience). Data analysis was performed using the FlowJo analysis platform (Tree Star Inc.).
Luciferase reporter assays
Electroporation using the nucleofection (Lonza) method was used to introduce recombinant plasmid DNA into primary NSCs. Alternatively, NSCs were subject to adenoviral-assisted lipofection by incubating empty adenoviral capsids with LipoD293 liposome:DNA complexes (SignaGen Laboratories). In all experiments luciferase activity was analyzed using the Dual-Luciferase Assay (Promega) and quantified using an LMax II microplate lumenometer (Molecular Devices).
Real-time PCR
RNA was isolated using the RNeasy Extraction Kit (Qiagen) and subjected to on-column DNase digestion. cDNA was reverse transcribed using the iScript cDNA synthesis kit and quantified in real-time in the iQ Sybr Green PCR reaction mix (Bio-Rad). Relative fold change in gene expression was calculated using the ΔΔ−Ct method after normalization to Actb expression. Primer sequences used for quantitative PCR (qPCR) are available upon request.
Chromatin immunoprecipitation
Proliferating and differentiating NSCs were fixed in 1% formalin for 10 minutes before isolation of nuclear extracts. Nuclei were isolated with 20 mmol/L HEPES pH 7.6, 20% glycerol, 1.5 mmol/L MgCL2, 0.2 mmol/L EDTA, 0.1% Triton, 10 mmol/L NaCl, and dounce homogenized and pelleted at 2000 rpm. Protein:DNA complexes were immunoprecipitated with 5 µg of either anti-Hey1 (Millipore; AB5714) or isotype control antibodies overnight as previously described (16). Olig2 promoter fragments were detected using qPCR and calculated using the percent input DNA method (Invitrogen). Olig2 primer sequences were (sense) 5′-CAGCGGAGAGCCTCCGATAATTC-3′ and (antisense) 5′-GCTTTGGGGACAGGCGGTAGC-3′.
Histology and immunohistochemistry
Tissue fixation, histologic processing, and immunohistochemical analysis were performed as described previously (13). Briefly, brain tissues were perfusion fixed with 4% PFA or fixed by immersion in 4% PFA overnight. Tissues were embedded in paraffin and sectioned at 5 µmol/L using a microtome. Immunohistochemical staining was conducted at the Norris Cotton Cancer Center Research Pathology Shared Resource (Lebanon, NH) using standard techniques. Antibodies used for immunohistochemistry included rabbit polyclonal anti-GFAP (1:500; Millipore; AB5804), rabbit polyclonal anti-Ki67 (1:500; Novocastra; NCL-Ki67p), and rabbit polyclonal anti-Olig2 (1:400; Millipore; AB9610). Sections were counterstained with hematoxylin.
Results
Id2 expression promotes the accumulation of oligodendrocytic precursor cells
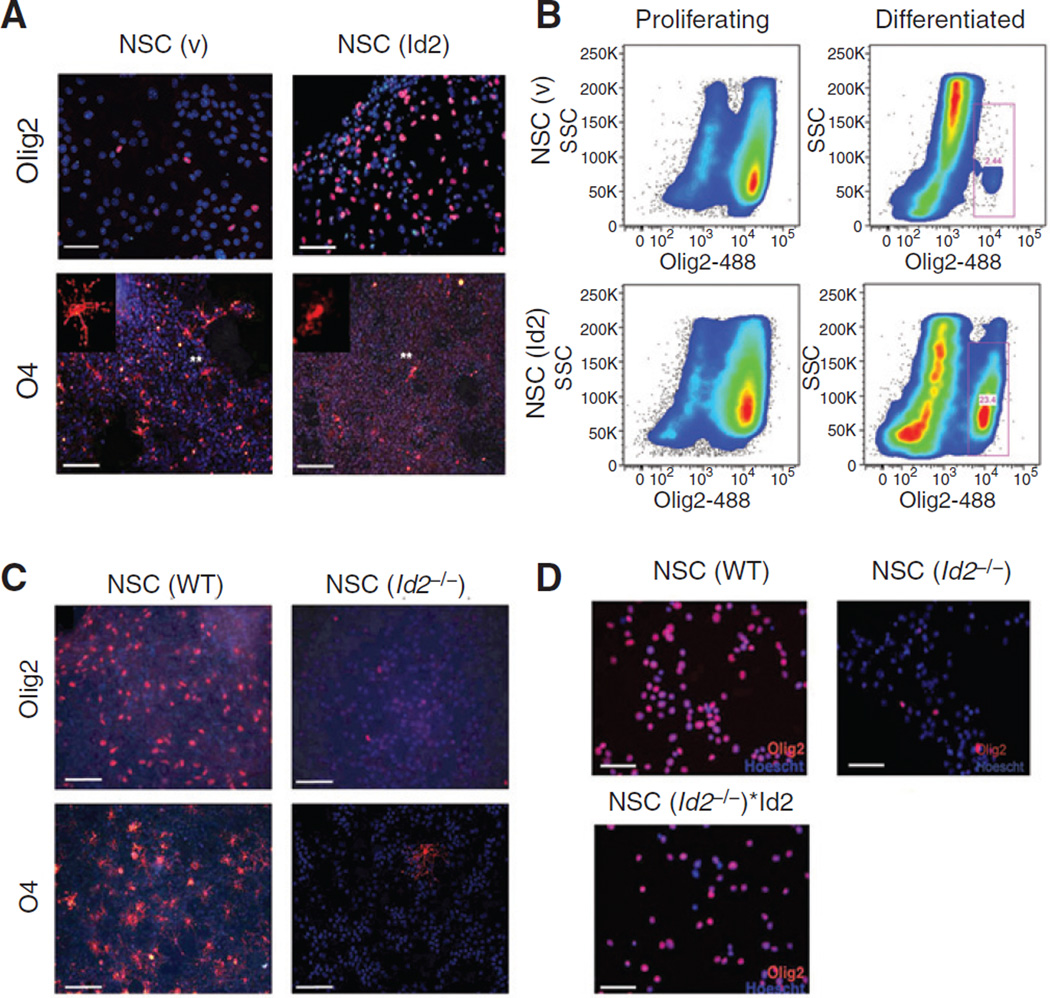
Because Id2 is derepressed in Tp53-inactivated NSCs (10) and known to promote the expansion of late-stage optic nerve OPCs (9), we examined the effect of deregulated Id2 expression on oligodendroglial differentiation of adult tissue–derived NSCs in vitro. After we differentiated NSCs modified by infection with a recombinant retrovirus to constitutively express Id2 [NSC (Id2)], we examined these cells for expression of oligodendrocyte differentiation markers. In these differentiated cultures, cells expressing O4, a marker of late-stage OPCs was readily detectable in differentiated NSC (v) cultures in all microscopic fields examined (7.7% ± 0.83%). In contrast, O4-positve cells in differentiated NSC (Id2) cultures were virtually absent, appearing as rare individual cells below the level of reliable quantification (<1%)(Fig. 1A). However, NSC (Id2) seemed to include a greater proportion of cells expressing the pan-oligodendroglial marker Olig2 than did NSCs modified by infection with only the parental retrovirus vector [NSC (v; Fig. 1A)]. This large Olig2+ population in differentiated NSC (Id2) cultures was validated using flow cytometry (Fig. 1B, red boxes).
Figure 1.
Id2 expression alters oligodendrocyte differentiation. A, Olig2 and O4immunofluorescence following 7 days of serum-induced differentiation of NSC (v) or NSC (Id2). Olig2 image scale bars, 100 µm; O4 image scale bars, 200 µm. Asterisks indicate the location of cells shown at higher magnification in the inset. B, flow cytometric analysis of NSC (v) and NSC (Id2) stained with anti-Olig2 antibody conjugated to Alexa-488 fluorophore. Gated cells (red boxes) represent Olig2 (+)-expressing cells following 7 days of differentiation. C, Olig2 and O4immunofluorescence in NSC(v) and NSC (Id2−/−) 7 days after differentiation. Olig2 image scale bars, 100 µm; O4image scale bars, 200 µm. D, Olig2 immunofluorescence in NSC (v), NSCId2−/− and NSC(Id2−/−)*Id2 7 days after differentiation. Scale bars, 100 µm. Nuclei are counterstained with Hoechst dye (blue).
We differentiated NSCs isolated from Id2−/− mice [NSC (Id2−/−)] and found dramatically decreased numbers of Olig2+ cells compared with differentiated NSC cultures from wildtype (WT) mice [NSC (WT); Fig. 1C]. We observed virtually no O4+ cells in differentiated NSC (Id2−/−) compared with easily identified O4+ oligodendrocyte precursors in differentiated NSC (WT; Fig. 1C). To determine whether Id2 expression was sufficient to restore an Olig2+ cell population to differentiating NSC (Id2−/−) cultures, we examined NSC (Id2−/−) in which Id2 expression had been restored by infection with an Id2-expressing recombinant retroviral vector, [NSC (Id2−/−)*Id2]. Expression of Id2 in NSC (Id2−/−) resulted in restoration of an Olig2+ population in differentiated cultures (Fig. 1D), providing additional evidence that Id2 expression regulates differentiation of NSCs into Olig2+ precursors (Fig. 1A–D).
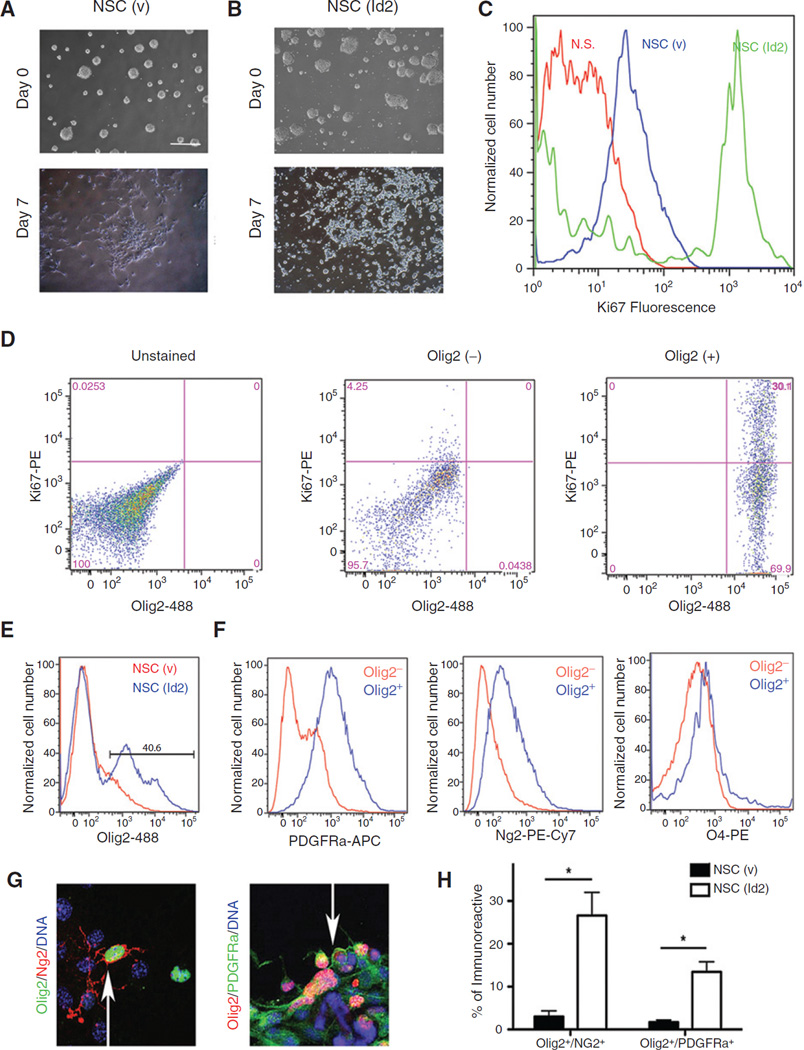
Changes in NSC (v) cultures following incubation under differentiation conditions were highly reproducible and included a transition from neurospheres to an adherent monolayer (Fig. 2A). In contrast, NSC (Id2) adhered minimally to the culture substrate and instead gave rise to increasing numbers of free floating spheres reminiscent of undifferentiated cells (Fig. 2B). We stained the differentiated cells examined in Fig. 2A and B with antibodies against Ki67, a marker of proliferative cells (Fig. 2C). Differentiated cultures of NSC (v) had few proliferative cells as indicated by a low level of Ki67 positivity. NSC (Id2) retained high levels of Ki67+ fluorescence following differentiation, indicating ongoing proliferative activity, which is a characteristic of OPCs (17–19). To determine whether these Ki67+ cells expressed Olig2, we used a dual labeling approach and found that a significant percentage of Olig2+ cells in NSC (Id2) cultures coexpressed Ki67, whereas Ki67 expression was not detected in Olig2− population (Fig. 2D). These observations were consistent with our findings that Olig2-expressing precursor cells were arrested in their maturation as a result of the constitutive expression of Id2 (Figs. 1, 2A–D).
Figure 2.
Id2 expression promotes the accumulation proliferative Olig2/PDGFRa/Ng2–positive cells. A and B, NSC (v; A) and NSC (Id2; B) before and after 7 days of differentiation. C, flow cytometric histograms of NSC (v; blue) and NSC (Id2; green) following 7 days of differentiation stained for Ki67 fluorescence and compared with unstained controls (N.S., no stain, red). D, flow cytometry analysis of differentiated NSC (Id2) dual-labeled for Olig2 and Ki67. Unstained cells (left) compared with Olig2− (middle) and Olig2+ (right) populations. E and F, NSC(v) and NSC(Id2) differentiated for 7 days and analyzed by four-color flow cytometry. The Olig2+ population observed in NSC (Id2) cultures (E, gated population) was compared with the remaining Olig2− population. Individual histograms represent fluorescent labeling of PDGFRa (APC), Ng2 (PE-Cy7), and O4 (PE) in the Olig2+ (blue) and Olig2− (red) populations (F). The representative experiment shown, N = 3. G, representative images of NSC (v) and NSC (Id2) differentiated for 7 days on coated glass slides and colabeled for either Olig2 and Ng2 or Olig2 and PDGFRa. Arrows, dual-labeled cells. H, total numbers of immunofluorescent, dual-labeled cells were counted in 10 randomly selected fields following 7 days of differentiation and expressed as a percentage of total cells per field. Data represent mean double-positive cell number of two experiments conducted in triplicate. Error bars ± SD. *, P < 0.05 determined by the t test.
We next interrogated the Olig2+ population in differentiated NSC (Id2) for the expression of additional OPC markers, including Pdgfra, Ng2, and O4 (19) using four-color labeling and flow cytometry. We again identified a population of Olig2+ cells present only in NSC (Id2) following differentiation (Fig. 2E, gated population). We examined further this Olig2+ population comparing it with the remaining Olig2− population. Olig2+ cells in differentiated cultures of NSC (Id2) were enriched for the expression of Pdgfra as well as Ng2 (Fig. 2F). Although very rare, O4+ cells could be detected in differentiated NSC (Id2) cultures (Fig. 1A); too few cells to quantitate were detected using flow cytometry, indicating that Id2 expression prevented the further maturation of early-stage OPCs (Fig. 2F). To substantiate these findings, we examined differentiated NSC (Id2) cultures for the coexpression of Olig2 with Pdgfra or Ng2. We found that these cultures contained increased numbers of both Olig2+/Ng2+ and Olig2+/Pdgfra+ cells (Fig. 2G and H). On the basis of the experiments described in Figs. 1 and 2, we concluded that under conditions that normally promote cell-cycle exit and differentiation, NSC (Id2) cultures instead retained proliferative cells (Fig. 2C) that express early OPC markers, including Olig2, Pdgfra, and Ng2 but not the late OPC marker, O4 (Fig. 2D–H).
Id2 promotes Olig2 expression by antagonizing the transcriptional repressor Hey1
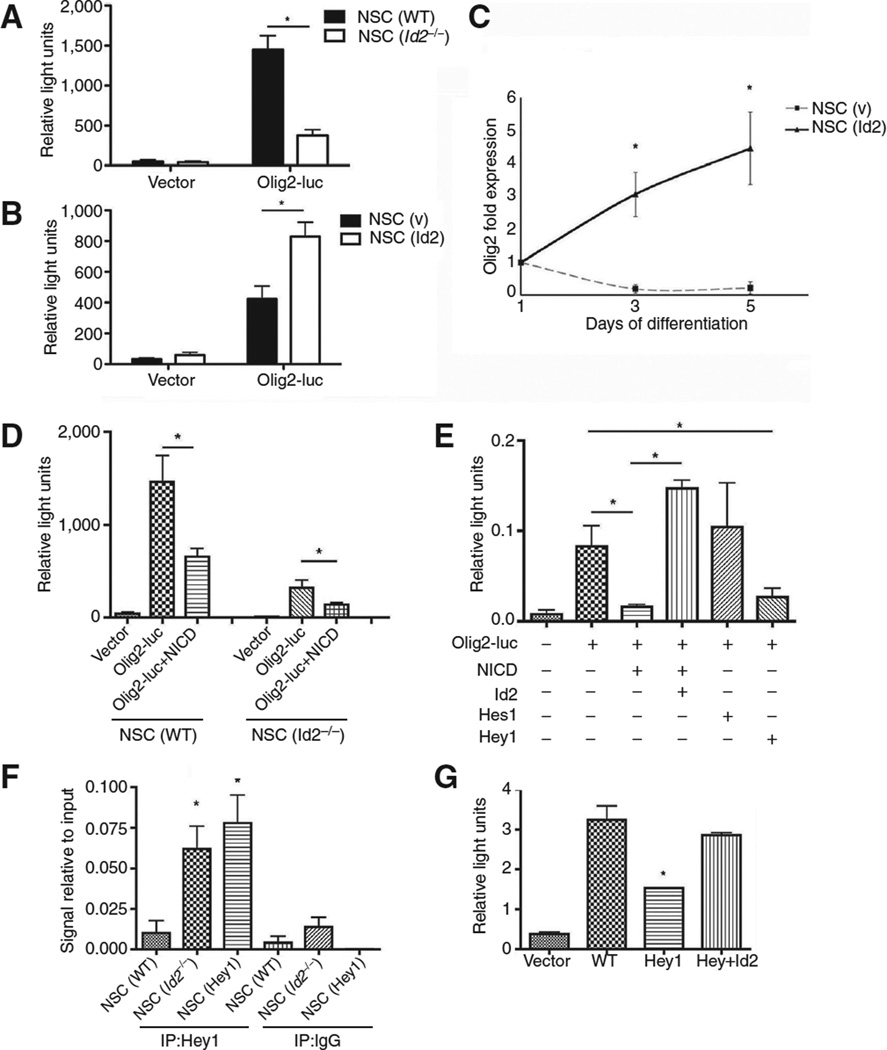
Olig2 and its role in glioma cell proliferation have been extensively characterized (20–22). Id2 clearly led to an expansion of the OPC population (Figs. 1 and 2), and so we sought to determine whether Id2 might enhance the expression of Olig2, which is important for oligodendrocyte specification (23). We used an Olig2 luciferase reporter driven by a 1.1-kb genomic fragment corresponding to the minimal Olig2 promoter region (24). Olig2 luciferase reporter activity was reduced in proliferating NSC (Id2−/−) as compared with NSC (WT; Fig. 3A), but increased in NSC (Id2; Fig. 3B). These findings were supported by qPCR analysis showing that NSC (Id2) cultures had higher levels of Olig2 mRNA than NSC (v) cultures differentiated under identical conditions (Fig. 3C).
Figure 3.
Id2-dependent transcriptional regulation of Olig2 involves Notch effector Hey1. A, Olig2 luciferase reporter activity in NSC (v) and NSC (Id2−/−); *, P < 0.01 determined by the t test. Data, mean ± SD of three independent experiments plated in triplicate and normalized to constitutively expressed Renilla luciferase. B, Olig2 luciferase reporter activity in NSC (v) and NSC (Id2); *, P < 0.01 analyzed using a t test. Data, mean ± SD of three independent experiments plated in triplicate normalized to constitutively expressed Renilla. C, Olig2 mRNA during differentiation time course in NSC (v) and NSC (Id2) determined by qPCR; *, P < 0.05 determined by the t test. Data, mean ± SD of two independent experiments PCR amplified in triplicate. D, Olig2 luciferase reporter activity in NSC(v) and NSC (Id2−/−) transfected with vector or NICD; *,P< 0.05. Data, mean±SD of three independent experiments plated in triplicate normalized to constitutively expressed Renilla. E, expression of the Olig2 luciferase reporter transfected with the indicated expression constructs; *, P < 0.05. Data, mean ± SD of three independent experiments plated in triplicate normalized to constitutively expressed Renilla. F, Hey1 ChIP from NSC (v), NSC (Id2−/−), or NSC (Hey1) 3 days after induction of differentiation. ChIP DNA fragments were PCR amplified with primers corresponding to the Olig2 promoter; *, P < 0.01. Data, mean ± SD of five independent experiments. G, Olig2 luciferase reporter expression from NSC (v), Hey1-expressing, or Id2- and Hey1-coexpressing NSCs 3 days after serum-induced differentiation; *, P < 0.01. Data, mean ± SD of two independent experiments plated in triplicate normalized to constitutively expressed Renilla.
Notch is a key regulator of nervous system development, and Notch activation inhibits oligodendrocyte differentiation (25). Notch effector basic helix-loop-helix (bHLH) proteins, including Hes1 and Hes5, repress oligodendrocyte lineage specification and maturation (8, 26) and are known to interact with Id proteins (27). Id2 typically functions as a transcriptional inhibitor because it lacks a basic DNA-binding domain and functions as a dominant-negative transcription factor, heterodimerizing with bHLH transcription factors and inhibiting their binding to DNA (27). We evaluated whether the Notch pathway contributed to the regulation of Olig2 expression in NSCs and sought to determine whether Id2 could affect that regulation. Transient expression of constitutively active Notch1 intracellular domain (NICD) repressed activity of the Olig2 reporter in both NSC (WT) and NSC (Id2−/−; Fig. 3D), whereas expression of Id2 with NICD was sufficient to inhibit NICD-mediated transcriptional repression (Fig. 3E). Transient expression of Hey1, a bHLH Notch signaling effector, but not Hes1, was sufficient to repress Olig2 promoter reporter activity in NSCs in the absence of NICD (Fig. 3F).
Evaluation of the Olig2 minimal promoter region revealed a series of three E-box promoter elements and one N-box binding element, known targets of Hey1 binding (29). To examine further the effect of Hey1 on Olig2 expression during differentiation, we prepared stable NSC cultures constitutively expressing Hey1 [NSC (Hey1)], and performed anti-Hey1 chromatin immunoprecipitation (ChIP) in these cells using primers that flanked the region noted above that was enriched for Hey1-binding sites. Hey1 avidly bound the Olig2 promoter in NSC (Id2−/−) as well as in NSCs (Hey1), but not in NSC (WT) cultures (Fig. 3F), most likely as the result of Id2 expression in these cells (13). To further validate that Hey1 was acting as a transcriptional repressor, we used the Olig2 promoter luciferase reporter assay and found that after 3 days of differentiation, a time point following the initiation of differentiation when we observed significant interaction of Hey1 with the Olig2 promoter in NSC (Hey1; Fig. 3F), the Olig2 reporter was repressed in NSC (Hey1) cultures. Transient expression of Id2 in these cells expressing Hey1 was sufficient to derepress Hey1 inhibition of the reporter (Fig. 3G).
NSCs expressing dysregulated Id2 are tumorigenic in a PDGF-rich microenvironment
Among the now recognized subclassifications of glioblastoma, the most commonly occurring primary brain tumor, tumors of the "proneural" subtype are characterized by TP53 mutations, high levels of OLIG2 expression, and PDGFRA mutation or expression at abnormally high levels (5, 7). These findings suggested to us an important role for PDGF/PDGFR in the proliferation of proneural glioma. Because high levels of Id2 result from inactivation of Tp53 and deregulated Id2 leads to the enrichment of Olig2+ OPCs in our differentiated NSC cultures (Figs. 1 and 2), we reasoned that these OPCs expressing high levels of Id2 may give rise to glioma-resembling tumors of the proneural subtype in a PDGF-rich microenvironment.
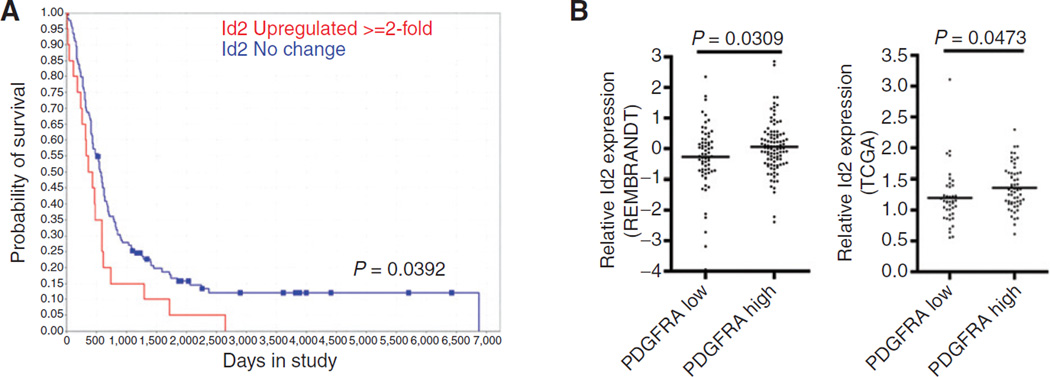
Before pursuing studies to examine this possibility, we sought evidence for ID2 being important in glioma biology. We interrogated the National Cancer Institute Repository for Molecular Brain Neoplasia Data (REMBRANDT; https://caintegrator. nci.nih.gov), a knowledgebase that combines gene expression, copy number, and clinical data from patients diagnosed with glioma. We found that patients with adult glioma (age ≥ 40 at onset) expressing high levels of ID2 mRNA (2-fold or more increase) had significantly decreased overall survival compared with patients whose tumors had normal levels of ID2 expression (Fig. 4A). We next compared ID2 mRNA expression in tumors with either a high or low level of PDGFRA expression. We defined high levels of expression in glioblastoma as having a z-score greater than 1.25 in the REMBRANDT database or an expression level greater than 2-fold above nontumor tissues in The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov). We found, in both databases, that increased ID2 mRNA expression was associated with a higher level of PDGFRA expression compared with tumors with lower levels of PDGFRA expression (Fig. 4B). These observations indicated that ID2 expression was associated with the decreased survival of patients with glioma, and are consistent with the notion that ID2 may contribute to the pathogenesis of tumors in which PDGFRA is most active.
Figure 4.
Role of Id2 and PDGF in gliomagenesis. A, Kaplan–Meier analysis of patients with glioma annotated in the REMBRANDT database binned into two groups of adult patients (40 years of age) representing those with Id2 expression levels equivalent to nontumor controls and Id2 levels greater than 2-fold above nontumor controls (P = 0.0392 determined by the log-rank test). B, relative Id2 mRNA expression in tumors annotated in the Rembrandt database (left; P = 0.0473) and the TCGA glioblastoma database (right; P = 0.0309) binned into two groups, PDGFRA high (expression Z-score ≥ 1 or expression 2-fold greater than normal) and PDGFRA low (expression Z-score ≥ 1 or expression 2-fold less than normal).
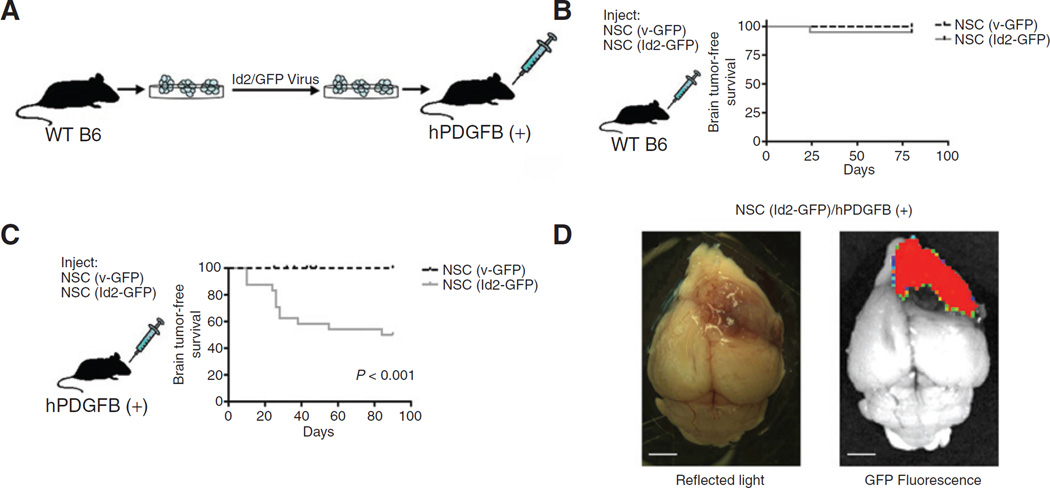
To evaluate the tumorigenicity of NSCs in which maturation along an oligodendrocytic lineage was enhanced by Id2 expression that also arrested these cells before their terminal differentiation (Figs. 1 and 2), we used a recombinant retrovirus encoding GFP to prepare cultures of modified NSCs constitutively expressing GFP that were designated NSC (v-GFP), or expressing GFP and Id2, NSC (Id2-GFP). We inoculated these cells orthotopically into the ventricular region of the forebrain of mice established in our laboratory to overexpress human PDGFB ligand in their CNS [hPDGFB (+); ref. 11], and WT B6 littermate control mice (Fig. 5A). Inoculation of NSC (v-GFP) or NSC (Id2-GFP) into WT B6 mice did not produce any pathologic changes (Fig. 5B); however, GFP+ cells could be detected readily in needle tracts, the subventricular zone, and eventually the olfactory bulb of asymptomatic mice (data not shown) confirming that injected cells survived engraftment and persisted over time in the adult CNS. Inoculation of NSC (Id2-GFP) into hPDGFB (+) animals, but not the inoculation of NSC (v-GFP), led to 50% of animals developing neurologic symptomatology, including head tilts, decreased mobility, and evidence of paralysis before 100 days of age (Fig. 5C). Postmortem examination of these animals frequently revealed large tumor masses and these masses had a high level of GFP fluorescence (Fig. 5D).
Figure 5.
PDGF-dependent gliomagenesis resulting from syngeneic engraftment of NSC (Id2). A, a schematic representation of orthotopic NSC engraftment procedure. B, Kaplan–Meier survival curve of WT mice orthotopically inoculated with NSC (v-GFP; n = 23) or NSC (Id2-GFP; n = 28) and sacrificed after developing severe neurologic symptoms. C, Kaplan–Meier survival curve of hPDGFB(+) mice orthotopically inoculated with NSC(v-GFP; n = 20) or NSC (Id2-GFP; n = 25), P < 0.001 determined using a log-rank (Mantel–Cox) test. D, the representative image of NSC (Id2-GFP)-induced tumor in an hPDGFB (+) mouse visualized using reflected light and GFP fluorescence. Scale bars, 2.5 mm.
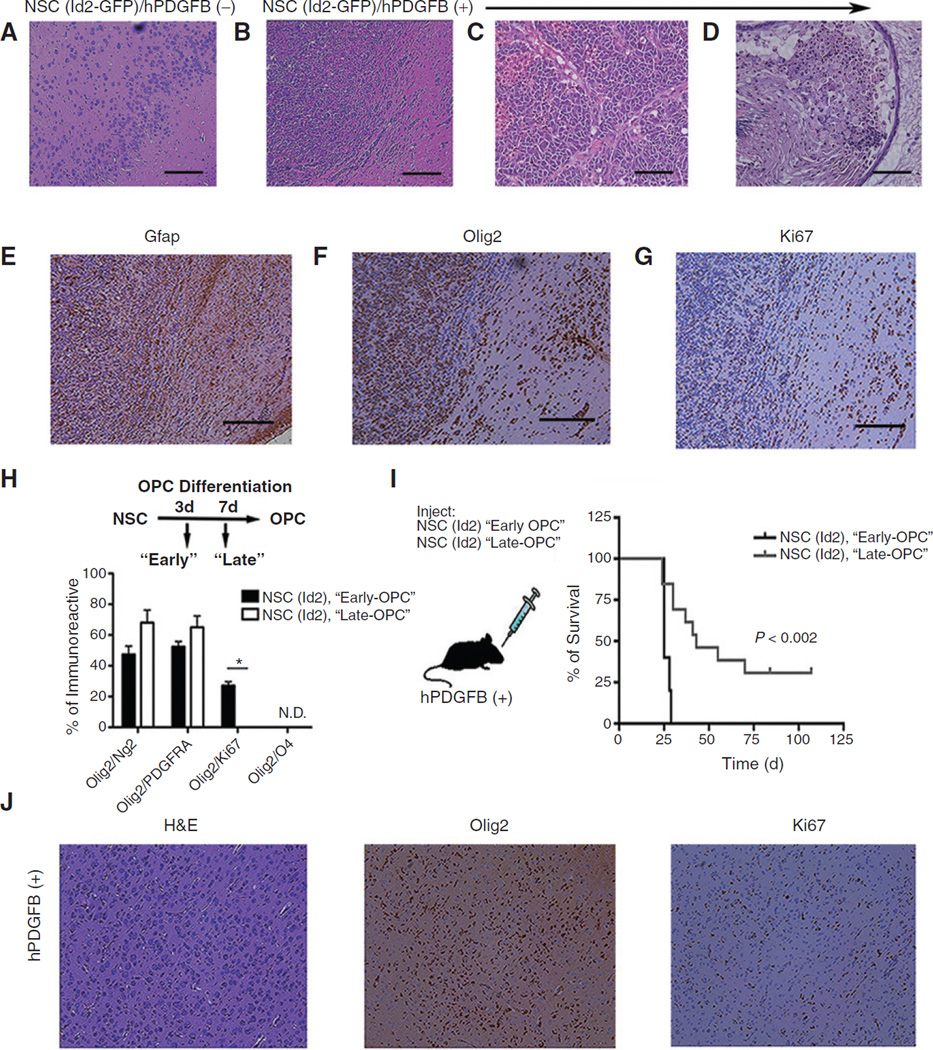
In these experiments no neurologic or other pathologic symptoms developed in hPDGFB (+) mice inoculated with NSC (v-GFP) and orWTB6 animals inoculated with either NSC (v-GFP) or NSC (Id2-GFP). We examined histopathologically brains from all animals that developed debilitating neurologic symptoms and manyWTB6 animals inoculated with NSC (Id2-GFP; Fig. 5C). As noted above, histologic examination ofWTB6 animals inoculated with NSC (Id2-GFP) revealed histologically normal tissues (Fig. 6A), although fluorescent cells were routinely found in several areas of the brain. In contrast, examination of hPDGFB (+) mice inoculated with NSC (Id2-GFP) revealed highly cellular masses with diffuse infiltrative borders (Fig. 6B). Histologic and cytologic characteristics of high-grade glioma, including high cell density; nuclear atypia; tortured, atypical blood vessels; hemorrhage (Fig. 6C); and areas of extensive cytologic variability typical of glioblastoma multiforme (Fig. 6D) were easily observed in these tumors. Immunohistochemical analyses of these tumor tissues revealed the expression of Gfap (Fig. 6E), Olig2 (Fig. 6F), and Ki67 (Fig. 6G) both within the dense masses and in tumor cells infiltrating normal tissue beyond the apparent tumor border.
Figure 6.
Histologic examination of glioma arising in hPDGFB (+) mice orthotopically inoculated with NSC (Id2-GFP). A and B, photomicrographs of hematoxylin and eosin (H&E) stained histologic sections showing the ventral forebrain of a WT animal (A) and an hPDGFB (+) age-matched littermate (B) orthotopically inoculated with NSC (Id2-GFP). Original magnification, ×4; scale bars, 200 µm. C and D, photomicrographs of H&E stained histologic sections of tumors obtained from hPDGFB (+) animals inoculated with NSC (Id2-GFP). Original magnification, ×20; scale bars, 100 µm. E–G, adjacent histologic sections to B immunostained for Gfap, Olig2, and Ki67 (brown) and counterstained with hematoxylin (blue); original magnification, ×4. H, quantification of Olig2/Ng2, Olig2/PDGFRa, and Olig2/Ki67 double-positive cells detected using immunostaining at 3 and 7 days following the initiation of OPC-directed differentiation. Data, mean ± SD from two independent experiments plated in triplicate; *, P < 0.05. I, Kaplan–Meier survival plot from NSC (Id2-GFP) inoculated 3 days after initiation of after OPC-directed differentiation compared with NSC (Id2-GFP) inoculated 7 days following differentiation. J, histologic sections taken from an hPDGFB (+) animal following inoculation NSC (Id2-GFP) differentiated for 3 days under OPC enrichment conditions. Sections are stained with H&E, Olig2, and Ki67 (brown). Original magnification, ×10; scale bars, 100 µm.
The occurrence of these tumors only in PDGF/PDGFR– expressing animals and their expression of Olig2 (Fig. 6F) mimicking the expression of these genes in proneural glioblastoma is consistent with data suggesting that Olig2+ glioblastoma arises in OPCs. These findings provide strong evidence for the malignant transformation of Id2 maturation– arrested NSCs, namely OPCs (Figs. 1 and 2), in the development of tumors dependent upon PDGF (Fig. 5). To examine further the importance of the OPC phenotype for tumor formation, we used defined culture conditions well characterized to enhance oligodendroglial differentiation of NSCs (15). Days 3 and 7 after the initiation of differentiation of NSC (Id2-GFP), we characterized these cells for oligodendroglial markers (Fig. 6H) and collected cells for orthotopic inoculation into hPDGFB (+) mice (Fig. 6I). Three days after the initiation of differentiation, these cultures developed cells coexpressing Olig2/Ng2, Olig2/Pdgfra, and Olig2/Ki67 (Fig. 6H). Longer periods in these culture conditions resulted in the loss of detectable Ki67 positivity, suggesting further differentiation along the oligodendroglial pathway (15). It is noteworthy, however, that maturation remained incomplete as evidenced by the absence of O4-expressing cells (Fig. 6H). Orthotopic inoculation into hPDGFB (+) mice of NSC (Id2-GFP) differentiated for 3 days caused rapid tumor onset and death (Fig. 6I). hPDGFB (+) mice inoculated with NSC (Id2-GFP) differentiated for 7 days developed a less penetrant tumor phenotype with a more protracted period of time, during which tumor developed and increased survival time (Fig. 6I). Pathologic examination of brains from mice orthotopically inoculated with NSC (Id2-GFP) differentiated for 3 days revealed disrupted normal histology resulting from extensive infiltration by Olig2+ and Ki67+ cells exhibiting small nuclei with a low nuclear to cytoplasmic ratio (Fig. 6J).
Discussion
The role of arrested differentiation of lineage-specific precursor cells in contributing to tumor development has been extensively examined in hematopoietic malignancies (30). This pathologic mechanism has been less well studied in solid tumors, perhaps because lineage-specific differentiation markers are not as well defined in solid tissues. The identification of proliferative precursor cells marked by lineage-specific patterns of gene expression in the mature mammalian brain (31) and the molecular characterization of distinct subtypes of glioblastoma (5, 32, 33) has enhanced the opportunity to identify cells of origin of glioma and to understand the biology underlying the transformation of such cells. The possibility that normal adult tissue NSCs (34) or precursor cells arising from them (6) are a cell of origin for brain tumors has been an active area of study. Our findings provide compelling evidence for a mechanism by which maturation-arrested OPCs become vulnerable to transformation and give rise to tumors resembling a clinically relevant subset of glioblastoma.
Our findings provide evidence that deregulation of Id2 in NSCs can lead to increased numbers of early OPC-like cells (Figs. 1 and 2), a target cell for the development of glioma (6). Olig2 is important not only as a marker of cells in the oligodendrocyte lineage, but also as a gene responsible for early differentiation of NSCs along the oligodendrocyte lineage (35), and proliferative expansion of neuronal precursors and glioma cells (36). Our model supports the work of others indicating that OPCs can be a cell of origin for glioma. In our study, we also observed that quiescent, maturation-arrested OPCs can give rise to glioma upon exposure to enhanced PDGF (Fig. 6H–J). We speculate that in a PDGF-rich brain microenvironment, maturation-arrested OPCs reenter the cell cycle in response to PDGF stimulation, resulting in tumor formation. This observation highlights the potential utility of our novel mouse model in characterizing the importance of an oncogenic microenvironment in promoting gliomagenic transformation in premalignant OPCs.
A predictable interpretation of our findings, that Id2 protein inhibits OPC maturation by directly blocking the activity of the bHLH transcription factor Olig2 (37), is confounded by the observation that differentiation NSC (Id2−/−) do not give rise to Olig2+ cells (Fig. 1C). Rather, Id2 is necessary for the expression of Olig2. Id2-mediated repression of Hey1 to enhance the expression of Olig2 indicates a dual role for Id2. The first of these in promoting early oligodendrocytic differentiation of NSCs into OPCs (Figs. 1 and 2) and the second, later in differentiation, inhibiting OPC maturation into mature oligodendrocytes (Fig. 1) by direct interaction with Olig2 as described by others (9, 37). Consistent with this model is our observation that immunoprecipitation of Id2 in undifferentiated NSCs did not result in the coprecipitation of Olig2, although Id2 and Olig2 did coimmunoprecipitate from NSCs undergoing differentiation (our unpublished observations). This dual role for Id2 in facilitating expansion of the OPC compartment by enhancing oligodendrocytic differentiation, while blocking later OPC maturation, may be of particular importance in situations in which Id2 is deregulated, for example, following the inactivation of Tp53 (10, 38).
In summary, we have developed a novel model of gliomagenesis that reveals a role for maturation arrest in the development of glioblastoma that reflect closely the proneural subtype of adult glioblastoma (5) and pediatric high-grade glioma (39). The proneural subtype is characterized by inactivation of TP53, activation of PDGFRA, and tumor cells that mimic an OPC phenotype (7). Our data suggest strongly that Id2, a gene known to be repressed by Tp53, mediates the biologic behavior of OPCs following Tp53 inactivation. This model can provide insights into PDGF-dependent glioma progression and may be a uniquely helpful system in which to evaluate the efficacy of OPC-targeted therapies.
Acknowledgments
The authors thank the expert assistance of Alison Young with animal experiments, Eric York and Rebecca O'Meara for assistance with histology, and Gary Ward, Alan Bergeron, Robert Grady, and Jacqueline Smith for assistance with flow cytometry. The authors also thank Dr. Christopher Dant and Tabatha Richardson for the helpful review during the writing of this article.
Grant Support
This research has been supported by the Jordan and Kyra Memorial Foundation (M.A. Israel), The Theodora B. Betz Foundation (M.A. Israel), an NINDS Ruth Kirschstein Predoctoral Fellowship (F31NS064634; B.R. Paolella), a Dartmouth SYNERGY Scholars Development Award (M.C. Havrda), and The Hitchcock Foundation (M.C. Havrda).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: M.C. Havrda, B.R. Paolella, K.S. Jering, M.A. Israel
Development of methodology: M.C. Havrda, B.R. Paolella, C. Ran, K.S. Jering, J.M. Sullivan, M.A. Israel
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): M.C. Havrda, B.R. Paolella, K.S. Jering, C.M. Wray, J.M. Sullivan, A. Nailor, Y. Hitoshi
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M.C. Havrda, B.R. Paolella, C. Ran, K.S. Jering, J.M. Sullivan, M.A. Israel
Writing, review, and/or revision of the manuscript: M.C. Havrda, B.R. Paolella, M.A. Israel
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): K.S. Jering, A. Nailor, M.A. Israel Study supervision: M.A. Israel
References
- 1.Havrda MC, Israel MA. In: The molecular basis of cancer. 3rd. Mendelsohn JHP, Israel MA, Gray JW, Thompson CB, editors. Philadelphia, PA: W.B. Saunders Co; 2008. [Google Scholar]
- 2.Zong H, Verhaak RG, Canoll P. The cellular origin for malignant glioma and prospects for clinical advancements. Expert Rev Mol Diagn. 2012;12:383–394. doi: 10.1586/erm.12.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 4.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 5.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C, Sage Jonathan C, Miller Michael R, Verhaak Roel GW, Hippenmeyer S, Vogel H, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Liu Y, Levine EM, Rao MS. Hes1 but not Hes5 regulates an astrocyte versus oligodendrocyte fate choice in glial restricted precursors. Dev Dyn. 2003;226:675–689. doi: 10.1002/dvdy.10278. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 10.Paolella BR, Havrda MC, Mantani A, Wray CM, Zhang Z, Israel MA, et al. p53 directly represses Id2 to inhibit the proliferation of neural progenitor cells. Stem Cells. 2011;29:1090–1101. doi: 10.1002/stem.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hitoshi Y, Harris BT, Liu H, Popko B, Israel MA. Spinal glioma: platelet-derived growth factor B-mediated oncogenesis in the spinal cord. Cancer Res. 2008;68:8507–8515. doi: 10.1158/0008-5472.CAN-08-1063. [DOI] [PubMed] [Google Scholar]
- 12.Lim DA, Fishell GJ, Alvarez-Buylla A. Postnatal mouse subventricular zone neuronal precursors can migrate and differentiate within multiple levels of the developing neuraxis. Proc Natl Acad Sci U S A. 1997;94:14832–14836. doi: 10.1073/pnas.94.26.14832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havrda MC, Harris BT, Mantani A, Ward NM, Paolella BR, Cuzon VC, et al. Id2 is required for specification of dopaminergic neurons during adult olfactory neurogenesis. J Neurosci. 2008;28:14074–14087. doi: 10.1523/JNEUROSCI.3188-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 15.Glaser T, Pollard SM, Smith A, Brüustle O. Tripotential differentiation of adherently expandable neural stem(NS) cells. PLoS ONE. 2007;2:e298. doi: 10.1371/journal.pone.0000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kintner C. Neurogenesis in embryos and in adult neural stem cells. J Neurosci. 2002;22:639–643. doi: 10.1523/JNEUROSCI.22-03-00639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori T, Wakabayashi T, Takamori Y, Kitaya K, Yamada H. Phenotype analysis and quantification of proliferating cells in the cortical gray matter of the adult rat. Acta Histochem Cytochem. 2009;42:1–8. doi: 10.1267/ahc.08037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horner PJ, Thallmair M, Gage FH. Defining the NG2-expressing cell of the adult CNS. J Neurocytol. 2002;31:469–480. doi: 10.1023/a:1025739630398. [DOI] [PubMed] [Google Scholar]
- 19.Levine JM, Reynolds R, Fawcett JW. The oligodendrocyte precursor cell in health and disease. Trends Neurosci. 2001;24:39–47. doi: 10.1016/s0166-2236(00)01691-x. [DOI] [PubMed] [Google Scholar]
- 20.Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 21.Mehta S, Huillard E, Kesari S, Maire Cecile L, Golebiowski D, Harrington Emily P, et al. The central nervous system-restricted transcription factor olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Meijer Dimphna H, Alberta John A, Mehta S, Kane Michael F, Tien A-C, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906–917. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Horrell SA, Delaney D, Gottlieb DI. Embryonic stem cells as a platform for analyzing neural gene transcription. Stem Cells. 2008;26:1841–1849. doi: 10.1634/stemcells.2007-0902. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, et al. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 26.Ogata T, Ueno T, Hoshikawa S, Ito J, Okazaki R, Hayakawa K, et al. Hes1 functions downstream of growth factors to maintain oligodendrocyte lineage cells in the early progenitor stage. NSC. 2011;176:132–141. doi: 10.1016/j.neuroscience.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 27.Bai G, Sheng N, Xie Z, Bian W, Yokota Y, Benezra R, et al. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 28.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 29.Fischer A, Gessler M. Delta Notch and then? protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–4596. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sell S. Leukemia: stem cells, maturation arrest, and differentiation therapy. Stem Cell Rev. 2005;1:197–205. doi: 10.1385/SCR:1:3:197. [DOI] [PubMed] [Google Scholar]
- 31.Johansson CB, Svensson M, Wallstedt L, Janson AM, Frisen J. Neural stem cells in the adult human brain. Exp Cell Res. 1999;253:733–736. doi: 10.1006/excr.1999.4678. [DOI] [PubMed] [Google Scholar]
- 32.Zong H, Verhaak RG, Canolk P. The cellular origin for malignant glioma and prospects for clinical advancements. Expert Rev Mol Diagn. 2012;12:383–394. doi: 10.1586/erm.12.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 36.Ligon K, Huillard E, Mehta S, Kesari S, Liu H, Alberta J, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samanta J. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- 38.Park HJ, Hong M, Bronson RT, Israel MA, Frankel WN, Yun K. Elevated Id2 expression results in precocious neural stem cell depletion and abnormal brain development. Stem Cells. 2013;31:1010–1021. doi: 10.1002/stem.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]