In this review article, the history and pathophysiology of mismatch repair, the process of testing for mismatch repair deficiency and microsatellite instability, and the role of immunotherapy in a number of tumor types, including those that are highly sensitive to PD-1 blockade, are discussed.
Keywords: Microsatellite instability, DNA mismatch repair, Immunotherapy, Colonic neoplasms, Colorectal neoplasms, Hereditary nonpolyposis
Abstract
More than 1.6 million new cases of cancer will be diagnosed in the U.S. in 2016, resulting in more than 500,000 deaths. Although chemotherapy has been the mainstay of treatment in advanced cancers, immunotherapy development, particularly with PD-1 inhibitors, has changed the face of treatment for a number of tumor types. One example is the subset of tumors characterized by mismatch repair deficiency and microsatellite instability that are highly sensitive to PD-1 blockade. Hereditary forms of cancer have been noted for more than a century, but the molecular changes underlying mismatch repair-deficient tumors and subsequent microsatellite unstable tumors was not known until the early 1990s. In this review article, we discuss the history and pathophysiology of mismatch repair, the process of testing for mismatch repair deficiency and microsatellite instability, and the role of immunotherapy in this subset of cancers.
Implications for Practice:
Mismatch repair deficiency has contributed to our understanding of carcinogenesis for the past 2 decades and now identifies a subgroup of traditionally chemotherapy-insensitive solid tumors as sensitive to PD-1 blockade. This article seeks to educate oncologists regarding the nature of mismatch repair deficiency, its impact in multiple tumor types, and its implications for predicting the responsiveness of solid tumors to immune checkpoint blockade.
Introduction
Although the success of numerous checkpoint inhibitors has changed the therapeutic paradigm for several cancers, many tumors remain highly resistant to immunotherapy. A recent study has demonstrated that a well-known subpopulation of patients with solid tumors has since achieved excellent and durable responses to these new therapies [1]. These tumors demonstrate alterations in the mismatch repair (MMR) pathway that leads to high levels of microsatellite instability (MSI-H). This clinical entity is most familiar as the molecular etiology of Lynch syndrome, which was characterized more than 20 years ago. In this review, we discuss a brief history of MMR deficiency, the diagnostic tools for detecting deficient MMR (dMMR) and MSI-H, and the prognostic and predictive value they play in cancer care, including recent studies investigating the therapeutic potential of treating dMMR/MSI with checkpoint inhibitors.
History
In the early 1900s, Dr. Aldred Warthin first traced the pedigree of Family G, who were plagued with early-onset uterine and gastrointestinal tumors. However, it was not until the mid-1960s that Dr. Henry Lynch revisited the genealogy and cataloged a cancer family syndrome afflicting two large families with a propensity for early tumor development [2]. Over the subsequent years, similar pedigrees were noted worldwide, each with slight differences in cancer predispositions but characterized by high rates of nonpolyposis colon, endometrial, and gastric cancers. By the mid-1980s, the terms Lynch syndrome and hereditary non-polyposis colon cancer (HNPCC) were coined, but identifying those at risk remained difficult without a molecular etiology [3]. In the spring of 1993, several groups (Aaltonen et al., Ionov et al., Peltomaki et al., and Thibodeau et al. [4–7]) published findings that these tumors demonstrated high rates of mutations, particularly in regions of short tandem repeats (named microsatellites), a phenomenon referred to as replication error or MSI [8]. These changes showed uncanny similarity to known Escherichia coli strains with mutations in the proteins mutS and mutL, which coordinate DNA MMR. Over the next several years, the human homologs were sequenced and identified, thereby beginning our understanding of the molecular basis of Lynch syndrome and a novel form of carcinogenesis [6, 9–19].
Molecular Basis of MMR Deficiency
Alterations in four key human proteins in the DNA MMR pathway—MLH1, MSH2, MSH6, and PMS2—were identified in 1993–1996 as the predominant causative germline mutations responsible for Lynch syndrome [9, 13–15, 19–21]. During normal DNA replication with proficient MMR (pMMR), small DNA mismatch errors are initially detected and bound by MSH2/MSH6 and MSH2/MSH3 heterodimers. MLH1/PMS2 heterodimers are subsequently recruited for excision and resynthesis of a new, corrected strand. Throughout the genome are, more than 100,000 areas of short tandem repeat sequences named microsatellites (mono-, di-, tri, or tetranucleotide repeats) that are particularly prone to “slippage” during replication and thus rely more heavily on the MMR system for repair. Deficiency in MMR through qualitative or quantitative protein abnormalities leads to accelerated accumulation of genetic errors (i.e., mutations) at these microsatellites and subsequent diffuse MSI. The damage to the MMR process leads to additive mutations throughout the genome, leading to a “hypermutator” phenotype. Although most microsatellites are located in noncoding regions of the genome, ill-placed mutations can cause frameshift mutations, which can lead to subsequent inactivation, constitutive activation, or truncated/nonfunctional proteins necessary for DNA repair, apoptosis, cell growth, and epigenetics. Commonly targeted regions include the promoter regions of MMR genes, TGFBRII, IGFRII, BAX, Caspase5, BCL10, and PTEN [22]. Accumulation of mutations of these key proteins that control DNA repair, cell signaling, and apoptosis thus facilitates carcinogenesis.
Lynch syndrome arises from germline mutations in the MMR pathway, and affected patients are prone to a number of tumor types, mainly nonpolyposis colon cancer and endometrial cancer. Other mutations in these proteins produce different clinical syndromes, with autosomal dominant MLH1 and MSH2 mutations and reduced-penetrance MSH6 and PMS2 mutations. MSH2 variants have higher rates of extracolonic cancers, whereas MSH6 variants demonstrate development of tumors later in the lifetime [23]. More than 1,500 variants of Lynch syndrome mutations have been identified, comprising mainly deletions in MLH1 (50%) and MSH2 (39%) and less frequently in MSH6 (7%) or PMS2 [24, 25]. Inherited epigenetic modifications, such as the 3′-end deletions in the EPCAM gene, have also been identified that lead to inactivation of the adjacent MSH2 gene [26].
Although dMMR can lead to Lynch syndrome, sporadic somatic mutations in the MMR pathway can also be found in tumors unrelated to hereditary syndromes. Sporadic dMMR more often results from epigenetic changes, in particular methylation of the MLH1 promoter, which leads to subsequent silencing of MLH1. This methylation may be sporadic or associated with a CpG island methylator phenotype (CIMP), which is rarely seen in Lynch syndrome patients. Additional studies suggest that alterations in microRNA pathways, particularly miR-155 and miR-21, can also lead to suppression of mismatch repair proteins, with subsequent MSI-H [27–29].
Although MMR deficiency and MSI-H pathways are linked in our understanding of the pathogenesis of these tumors, there are both familial and sporadic cases in which MSI-H and dMMR are not definitively seen together. dMMR resulting from MSH6 mutations results in lower rates of MSI and thus may not reach the criteria for defining MSI-H tumors; in contrast, MSI-H-positive tumors occasionally do not reveal a known underlying protein loss, potentially reflecting heretofore unidentified proteins in the MMR pathway [30, 31]. Additionally, low rates of MSI (MSI-L) are noted in many tumors, particularly in regions of di- or trinucleotide repeats [32]. In some studies, these tumors appear to have an increased association with KRAS mutations compared with microsatellite-stable (MSS) disease [32], but overall MSI-L tumors show few genetic differences from MSS disease in colorectal cancer (CRC) lines and appear to have little meaningful clinical impact on CRC patients [33, 34]. Results showing the similarity of MSI-L/MSS tumors have also been seen in endometrial and gastric cancers [27, 35]. These factors suggest that low levels of baseline mutability may occur in tumors even without clinical significance or basis in dMMR. This has pushed the classification MSS/MSI-L tumors into a single category, in contrast to MSI-H tumors. This change in classification has led to discrepancies in results between studies that use different definitions of MSI to investigate its use in prognostication and prediction. In addition, older studies include testing of tetranucleotide repeats (elevated microsatellite instability at selected tetranucleotide repeats, or EMAST), which appear to derive from a different molecular driver than traditional MMR and may hold different clinical significance than mono- or dinucleotide repeats; these are not included in newer definitions of MSI-H [36].
Diagnosis of dMMR and MSI
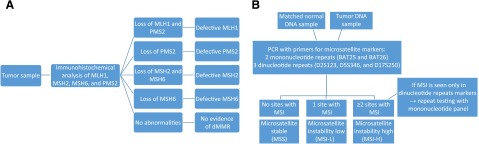
Two forms of testing are commonly used in screening patients and tumors for a deficiency in MMR or MSI: immunohistochemical staining (IHC) for altered proteins and polymerase chain reaction (PCR) testing for MSI. IHC works by staining slides of tumor samples to evaluate for the presence of the four known MMR proteins (Fig. 1A). IHC is inexpensive and readily available at most institutions, allowing for widely disseminated use; however, it may miss abnormalities caused by untested proteins or mutations that lead to qualitative, but not quantitative, changes in the tested proteins. For example, more than 30% of MLH1 mutations are missense mutations and therefore would not alter protein expression. Additionally, staining can be heterogeneous throughout tumor samples, and scoring may not be readily reproducible [37]. However, the sensitivity for detection of dMMR is increased when all four MMR proteins are tested. Because of the heterodimer structure of MLH1/PMS2 and MSH2/MSH6, when MLH1 or MSH2 is functionally lost, the heterodimer becomes unstable and PMS2 or MSH6, respectively, is degraded (although the inverse is not seen). IHC sensitivity can be improved when combined with testing for MLH1 promoter methylation status, and BRAF V600E mutations can usually help identify sporadic tumors in CRC patients. In those patients who have histories and IHC consistent with a germline protein loss, IHC also allows for subsequent screening for Lynch syndrome in the proband and relatives.
Figure 1.
Test schemas for mismatch repair deficiency with IHC staining (A) and MSI with PCR (B) per NCI guidelines.
Abbreviations: dMMR, deficient mismatch repair; IHC, immunohistochemical; MSI, microsatellite instability; NCI, National Cancer Institute; PCR, polymerase chain reaction.
MSI testing by PCR is considered the gold standard and directly compares the length of repeats at microsatellites to the patient’s germline tissue. This provides a more objective measure of functional dMMR activity, with better reproducibility between testing centers, and allows for identification of abnormalities even in the setting of nontruncating protein mutations in the four tested MMR proteins, as well as identification of MSI caused by other defects that result in dMMR [38]. However, PCR-based testing is more labor-intensive and requires comparison with nontumor tissue [37]. In 1998, the National Cancer Institute (NCI) proposed a standardized microsatellite panel for CRC testing that includes five microsatellite markers: two mononucleotide repeats (BAT25 and BAT26) and three dinucleotide repeats (D2S123, D5S346, and D17S250). Tumors with MSI at more than 30%–40% of sites (two or more of five sites) are considered MSI-H, those with less than 30%–40% (one of five) mutations are considered MSI-L, and those without instability are MSS (Fig. 1B). Because of higher mutability at dinucleotide repeats, further testing is advised if only dinucleotide repeats show instability [39] (Fig. 1B). However, there are many microsatellites throughout the genome, and investigators have argued that using only the NCI panel is controversial because of of inclusion of dinucleotide repeats and unclear applicability to all tumor types [40, 41]. Even for CRC, the updated NCI guidelines suggest that a pentaplex of quasimonomorphic mononucleotide repeats may have improved sensitivity and specificity over the NCI panel [21]. This lack of standardization has led to significant heterogeneity in historical studies, with researchers testing between one and hundreds of microsatellites, whereas testing of a single site can lead to decreased sensitivity, and testing of excess sites can lead to poor specificity or a MSI-L phenotype [33]. PCR may additionally be less effective at identifying MSH6 abnormalities, leading to lower rates of MSI [30].
Both IHC and PCR are sensitive and specific for dMMR and MSI, and the two tests show high concordance (>95%) across a number of tumor types [42–44]. Because of the limitations discussed above, however, they will likely continue to remain complementary to one another, with PCR determining functional abnormalities and IHC identifying underlying protein abnormalities [37]. Multiple next-generation sequencing platforms are being explored that may improve on speed and the ability to identify MSI across the whole genome by identifying high-mutational-load tumors, with several platforms already showing high concordance with traditional PCR testing [45–47].
The Amsterdam and Bethesda Criteria
Current testing guidelines for dMMR or MSI are based on the original Amsterdam and Bethesda criteria, which sought to identify families at high risk for Lynch syndrome. More recent recommendations now take into consideration the prognostic and predictive value of dMMR and suggest a need for potential expansion of the pool of patients who undergo testing.
The Amsterdam II criteria (1999) require that a patient (a) has at least three relatives with Lynch-associated tumors, (b) one of whom is a first-degree relative of the other two, with (c) two successive generations affected and (d) at least one relative diagnosed before age 50; (e) familial adenomatous polyposis should be excluded [48]. In contrast, the revised Bethesda guidelines (2004) advise testing in (a) diagnosis at <50 years old of synchronous or metachronous CRC or other HNPCC-associated tumors (including colorectal, endometrial, gastric, ovarian, pancreas, ureteral and renal pelvic, biliary, brain, or small bowel tumors or sebaceous gland adenomas or keratoachanthomas); (b) diagnosis at <60 years old of CRC with MSI-H histology (Crohn’s-like lymphocytic reaction, presence of tumor-infiltrating lymphocytes, mucinous/signet-ring differentiation, or medullary growth pattern); (c) diagnosis of CRC in one or more first-degree relatives with an HNPCC-related tumor, one of which was diagnosed at <50 years old; and (d) diagnosis of CRC in two or more first- or second-degree relatives with HNPCC-related tumors [39].
However, as the role of dMMR in nonhereditary tumors affects prognostic and predictive indices for various tumor types, suggestions for broadening testing has increased. At this time, per NCCN guidelines, MSI testing should be considered in tumors from all colon cancer patients <70 years of age or those with stage II colon cancers, even without a family history of cancer, and most recently all those diagnosed with metastatic colon cancer [49], as well as in those with endometrial cancer diagnosed in younger patients (<50 years old) or with significant family history. Some institutions are now undertaking universal MSI testing for all newly diagnosed endometrial tumors and CRC, to increase identification of Lynch syndrome patients [50].
Frequency of MSI Across Tumor Types
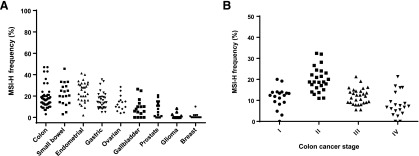
In part because of the lack of standardization of testing, reports on MSI-H frequency in the literature have varied greatly (Fig. 2A), but repeatedly demonstrate higher percentages in endometrial, gastric, and colon cancer, although there is large variability even between tumor subtypes or populations [51–105]. In endometrial cancer, the rates of MSI-H vary from 40% to 50% in endometrioid tumors to 2% in serous types. Gastric cancer rates also differ in Asian countries (8%–12%) in contrast to Western nations (20%–25%), suggesting different pathways of carcinogenesis [106]. Stage II colon cancers show higher rates of MSI-H than more advanced tumors (Fig. 2B), and rectal cancers and other left-sided colon cancers are noted to have lower frequencies of MSI-H tumors [7].
Figure 2.
MSI-H frequency. (A): Colon, small bowel, endometrial, gastric ovarian, gallbladder, prostate, glioma, and breast cancer based on MSI-H frequency rates in populations reported in the literature. (B): Colon cancer MSI-H frequency by stage.
Abbreviations: MSI-H, high-frequency microsatellite instability.
Prognosis in Tumors With Mismatch Repair Deficiency
Although dMMR deficiency with MSI-H is seen in a number of tumor histologies with or without corresponding Lynch syndrome, the clinical presentation varies depending on tumor type.
Colon Cancer
Colon cancer with MSI-H is often identified by pathologists as presentation with larger, more proximal tumors, poor differentiation, increased mucin secretion, increased lymphocytic infiltrates, and signet ring morphology [107]. Despite some negative prognostic characteristics, overall localized MSI-H colon cancers have better overall survival (OS) than MSS counterparts [108, 109]. However, patients with metastatic MSI-H disease have significantly poorer survival compared with MSI-L/MSS disease, which may be related to the high rates of BRAF mutations in sporadic MSI-H tumors [110]. Comparisons of metastatic dMMR and pMMR CRC of a small group at our institution demonstrated comparably short durations of treatment on prior therapies and before enrollment in trials, suggesting similar development of treatment resistance [1, 111, 112]. Sporadic MSI-H tumors are also commonly associated with CIMP, which is rarely seen in Lynch syndrome patients [113]. The Cancer Genome Atlas (TCGA) analyzed 276 CRC samples, with 97 undergoing whole-genome sequencing. Of the tumors found to be hypermutated (16%) in the TCGA analysis, only 75% were MSI-H, and these were highly associated with concurrent mutations in multiple tumor suppressors and oncogenes, including APC, TGFBRII, MSH3, and MSH6 [20].
Gastric Cancer
MSI-H gastric cancer is more frequently identified in older female patients, with tumors presenting in the distal stomach, of intestinal and mucinous type, with lower incidence of lymph node metastases and p53 abnormalities [114]. MSI-H tumors also show improved prognosis: pooled data from 24 studies with 712 cases of MSI-H showed an OS advantage of 0.76 compared with MSS/MSI-L cases [115]. These sporadic tumors, similar to colon cancer, are most frequently associated with loss of hMLH1 in the context of CIMP [116]. TCGA subdivided tumors into EBV+ tumors, genomically stable tumors, tumors with chromosomal instability, and tumors with MSI (22%) [51]. Both EBV+ and MSI tumors exhibited CIMP, but in distinct profiles, with EBV+ tumors lacking hMLH1 promoter hypermethylation [117]. Notably, these MSI tumors also included increased alterations in major histocompatibility complex (MHC) class I genes, including B2M and HLA-B, as well as recurrent amplifications in PIK3CA, ERBB3, ERBB2, and EGFR and frequent inactivation of ARID1A (83% of MSI tumors), which regulates chromatin structure [118]. Unlike MSI-H colon cancers, MSI-H gastric tumors were not associated with BRAF V600E mutations [119]. Similar outcomes were seen in Asian Cancer Research Group whole-genome sequencing [120].
Endometrial Cancer
Endometrial cancers are commonly classified into type I endometrioid tumors, 30%–40% of which demonstrate MSI-H, versus type II serous or clear-cell tumors that have a clinical presentation similar to ovarian cancers and less often express MSI-H, and then only in the setting of underlying Lynch syndrome [90]. Sporadic MSI-H endometrial tumors are more often associated with MLH1 deficiency and present with higher-grade tumors and more lymphovascular invasion than Lynch syndrome and MSS sporadic tumors [121]. Although multiple trials in endometrial cancer have shown that MSI-H is associated with poorer prognosis and acts as a negative prognostic factor in early-stage endometrial cancers, a meta-analysis of 23 trials showed no definitive detrimental effect on disease-free survival or OS [90, 122]. A TCGA analysis recently subdivided endometrial cancers into four molecular categories: POLE ultramutated, MSI, copy number-low, and copy number-high [84]. Most MSI-H tumors were secondary to MLH1 promoter hypermethylation, associated frequently with KRAS and ARID5B mutations (same family as ARID1A), as well as low PTEN expression that correlated with alterations in the PI3K pathway. p53 mutations were seen infrequently, and POLE mutations, which affect DNA polymerase proofreading, were present in approximately 5% of MSI-H disease and only in tumors without hypermethylation of MLH1 [123].
Ovarian Cancer
Ovarian tumors are seen in increased numbers in Lynch syndrome cohorts and are often noted to include a higher frequency of nonserous subtypes and earlier-stage presentation, with small studies showing improved prognosis [124–126].
Other Tumors
Although the risk of cancer development in Lynch syndrome patients is increased over the general population for a number of tumor types (colon, endometrial, gastric, ovarian, hepatobiliary, upper urinary tract, small bowel, central nervous system [Turcot syndrome], sebaceous [Muir-Torre syndrome], and prostate cancer), MSI-H tumors in sporadic malignancies of these types is rare [23, 127]. Breast cancer is not considered a part of Lynch syndrome; some studies suggest a fourfold risk in Lynch syndrome carriers, while others show no difference from the general population [128]. Sporadic breast cancer tumors with dMMR are rare (<2% in 316 breast cancers, including 226 triple-negative breast cancer, and may be associated with ER/PR-negative tumors; however, variations in earlier studies may be due to testing for MSI using tetra- and trinucleotide repeats [71, 129]. The inclusion of prostate cancer as a part of Lynch syndrome is controversial, although multiple studies demonstrate a near twofold increase in prostate cancer in carriers of Lynch syndrome mutations, with one meta-analysis finding 73% of those tumors being dMMR [130, 131]. Most studies evaluating MSI-H in sporadic prostate cancer have been small and inconclusive, but rates of MSI-H are thought to be low. Glioblastomas arise in a rare subset of Lynch syndrome patients with Turcot syndrome. A few small studies suggest that MSI-H may be associated with lower-grade astrocytomas; however, studies of MSI in central nervous system tumors remain limited [59]. Few recent trials have looked into the incidence of MSI in leukemias, but older studies suggest that de novo leukemias and myelodysplasia rarely demonstrate MSI-H, whereas the rate is increased by up to 33% in some secondary leukemias (including treatment-related leukemias and leukemias arising from myelodysplasia) [132, 133].
Chemotherapy Response in Mismatch Repair-Deficient Tumors
Because of the differences in underlying mutational drivers and prognosis, MSI-H status is hypothesized to predict responses to therapy. Carethers et al. first studied CRC cell lines with dMMR and found that they had limited response to 5-fluorouracil (5-FU) in contrast to MSS cell lines [134]. It is theorized that an intact MMR system is necessary to identify the DNA damage by 5-FU to halt cell growth.
Colon Cancer
The mainstay of treatment for early-stage colon cancer involves surgical resection and adjuvant chemotherapy for stage III tumors and high-risk stage II tumors. Initial studies in 2000 showed dramatic improvements in 5-year OS, up to 90%, when patients with Duke stage C MSI tumors were treated with adjuvant chemotherapy [135, 136]. However, those publications were believed to be limited by their retrospective strategies, inclusion of mainly younger patients with MSI, and use of only a single site (BAT26) in MSI testing. Prospective trials have since shown a detrimental effect of adjuvant 5-FU monotherapy in patients with MSI-H disease [137] or no difference in disease outcome after 5-FU therapy, although MSI-H maintains superior survival over MSS disease regardless of treatment [138, 139]. Subpopulation analysis suggests that subsets of stage II/III MSI-H CRC with concurrent TGFBRII mutations may benefit from adjuvant 5-FU [140, 141]. Several meta-analyses of stage II–III CRC were unable to demonstrate a statistically significant benefit for adjuvant 5-FU-based therapy for MSI-H disease, and so controversy remains over exactly how MSI status should be used in clinical decision making [8, 100, 101, 142–144], although the NCCN advises against adjuvant 5-FU in MSI-H stage II colon cancer patients [49, 145, 146]. In contrast, a retrospective Association des Gastro-Entérologues Oncologues (AGEO) study suggests that addition of oxaliplatin to adjuvant therapy in dMMR stage III patients may overcome the detrimental effect of 5-FU monotherapy [147]. In metastatic disease, response to chemotherapy is less clear, likely because of the smaller relative percentage of MSI-H cases. A retrospective review of 55 patients showed no improvement in prognosis in patients with metastatic MSI-H CRC with or without chemotherapy [111]. Additionally, a pooled analysis of COIN, CAIRO, CAIRO2, and FOCUS trials suggested inferior prognosis with dMMR colon cancers, in part driven by association with BRAF mutations [148].
Gastric Cancer
The benefit of adjuvant therapy after curative resection in gastric cancer was not clear until 2001 with INT-0016, and the subsequent MAGIC, ACTS-GC, and CLASSIC trials [149]. Like colon cancer, however, MSI-H tumors do not appear to show the same clear OS benefit after adjuvant chemotherapy as MSI-L/MSS tumors, with some trials suggesting worse outcomes with chemotherapy. A retrospective analysis of 1,276 Korean patients demonstrated a hazard ratio of 0.49 for MSI-H versus MSI-L/MSS without chemotherapy, which decreased to 1.16 when adjuvant therapy was provided [57]. MSI and MMR testing are not part of the NCCN guidelines for determining treatment at this time.
Immunogenicity of Tumors With Mismatch Repair Deficiency
It has long been recognized that the infiltration of lymphocytes within tumors (tumor-infiltrating lymphocytes [TILs]) represents a positive prognostic factor in many cancer types [150, 151]. In particular, certain lymphocyte subsets (CD45R0+, a memory T cell marker) appear to play important roles in limiting tumor dissemination and ultimately improving survival [152]. MSI-H tumors are associated with increased TIL density [153, 154], although the rationale for this has not always been understood. They are associated with an abundance of CD3+CD8+ cytotoxic TILs, suggesting that these tumors trigger an immune response in the host [155]. Tumor cells interact with the immune system in the tumor microenvironment, resulting in cancer immunoediting, which is described in three phases: elimination, equilibrium, and escape. The attempted elimination of tumor cells by T lymphocytes leads to residual immune-resistant tumor cells [156]. This is followed by a period of immunologic pressure on these tumor cells (equilibrium), which ultimately results in immune escape, where the downregulation of major histocompatibility complex molecules occurs. The end result is immune tolerance, where tumor antigens prevent rejection by the host immune system by creating unsuitable conditions in the tumor microenvironment that prevent appropriate immune responses [157]. This process of immunoediting is regulated through a series of checkpoint receptors, including CTLA-4, programmed death 1 pathway (including PD-1 and its ligand PD-L1), and LAG3. Inhibitors of these pathways (including US Food and Drug Administration-approved ipilimumab, nivolumab, and pembrolizumab) interrupt this pathway, leading to stimulation of activated T-cells and antitumor immunity [158].
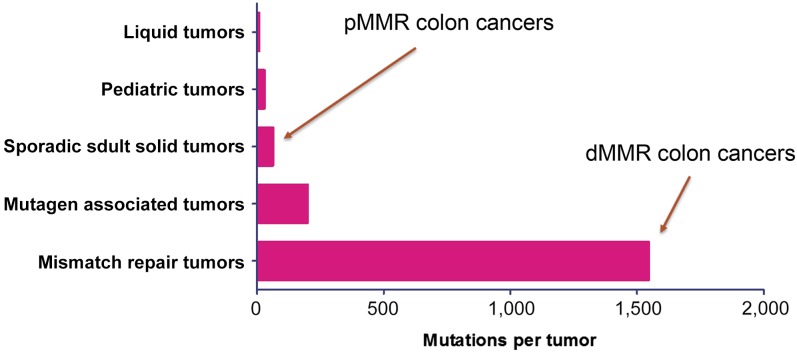
MSI-H tumors are thought to possess higher TIL densities relative to MSS tumors because of their higher mutational load. Next-generation sequencing studies have shown that MSI-H tumors typically harbor more than 1,000 coding somatic mutations per tumor cell genome compared with the 50 to 100 somatic mutations found in MSS tumors [159] (Fig. 3). dMMR tumors result in frameshifting mutations within coding sequences, often resulting in functionally inactive proteins [160, 161]. These abnormal peptides have the potential to be presented through the tumor’s MHC I to cytotoxic T-lymphocytes (CTLs) as neoantigens, resulting in the increased TIL density synonymous with MSI-H tumors.
Figure 3.
Total somatic mutations per tumor identified by comparison exome sequencing of tumor and matched normal DNA according to tumor type [181].
Abbreviations: dMMR, deficient mismatch repair; pMMR, proficient mismatch repair.
MSI-H tumors are thought to possess higher TIL densities relative to MSS tumors because of their higher mutational load. Next-generation sequencing studies have shown that MSI-H tumors typically harbor more than 1,000 coding somatic mutations per tumor cell genome compared with the 50 to 100 somatic mutations found in MSS tumors.
The MSI-H CRC tumors identified in the TCGA exhibited a strong expression of a group of tightly coregulated immune-related genes (coordinate immune response cluster) [162]. This cluster predominantly consists of T-helper 1 (Th1) genes but also contains class I and II genes and genes relating to immune checkpoints, innate immune recognition, and T cell activation. This cluster confirmed previous findings of the presence of a strong density of Th1 T cells in MSI-H tumors relative to MSS tumors and higher expression of chemokines CCL5, CXCL9, and CXCL10, which are associated with the Th1 response [163]. The Th1 response is particularly antitumorigenic because of the role of interferon (IFN)-γ in tumor surveillance [164].
Looking specifically at T cell subsets within CRC tumors, Llosa et al. showed that MSI-H tumors displayed a high degree of infiltration with CD8+ CTLs, activated Th1 cells characterized by the production of IFN-γ (the canonical Th1 cytokine) and T-bet, the Th1 transcription factor [165]. Conversely, MSI-H and MSS tumors have similar expression of interleukin (IL)-13 and IL-14 (Th2 response) and FOXP3 (Treg-associated gene), indicating that MSI-H tumors have selective Th1 and CTL infiltration. There is also evidence of increased macrophage density in MSI-H tumors compared with MSS CRC [166].
MSI-H tumors also express genes encoding checkpoint receptors at significantly higher levels than MSS tumors, including PD-1, CTLA-4, and LAG-3 [165]. Both PD-1 expression in TILs and PD-L1 expression on tumor cells differ in MSI-H compared with MSS tumors. [167] TILs from MSI-H tumors have 77% PD-1 expression compared with 39% in MSS tumors. MSI-H tumors have 32% PD-L1 expression compared with 13% in MSS tumors. The expression of checkpoint receptors is induced by IFN-γ, which may represent adaptive resistance to an active Th1/CTL microenvironment induced by MSI-H [168]. Modulation of IFN-γ receptor (IFN-γRα) expression on tumor cells can result in tumor immunoediting, acquiring a stem cell-like phenotype and the development of resistance to CTL-mediated death [169–171]. The extent of PD-L1 expression is modest compared with other classically immunosensitive tumors (renal cell cancer, non-small-cell lung cancer) [172]. Instead, MSI-H tumors are infiltrated with PD-L1-expressing myeloid cells (CD115−CD14+CD33+CD11c+), which produce IFN-γ, suggesting that these cells have an important role in the PD-1 pathway interactions in the MSI tumor microenvironment [165]. Because of the heterogeneity in immune gene expression between MSI-H and MSS tumors, efforts have been made to establish a scoring system that can predict clinical outcomes independently of MMR status. The immunoscore, based on the immunohistochemical quantification of cytotoxic and memory T cells at the tumor’s core and invasive margin, has been shown to be predictive of survival in stages I–III CRC [166]. A high score (I3/I4) indicates high densities of TILs and is also associated with overexpression of PD-1. A scoring system has the potential to predict subsets of patients with MSI-H and MSS tumors likely to benefit from immune checkpoint therapy.
Immune Checkpoint Inhibition in Tumors With Mismatch Repair Deficiency
In mid-2015, we published results of a phase 2 trial demonstrating the benefit of PD-1 inhibitors for patients with MSI-H tumors [1]. Three cohorts of patients were recruited: cohort A, patients with dMMR CRC (n = 11); cohort B, pMMR CRC (n = 21); and cohort C, dMMR non-CRC cancers (n = 9, including ampullary/cholangiocarcinoma, endometrial, small bowel, and gastric tumors). All patients with CRC had previously received at least two lines of therapy, and all patients in cohort C had received at least one prior regimen. Patients received 10 mg/kg pembrolizumab every 14 days. All patients had heavily pretreated advanced cancer, with the CRC population receiving a median of four prior chemotherapy regimens and with non-CRC patients receiving a median of two prior regimens. In general, patients with dMMR tumors were younger than those with pMMR tumors, and 31.7% had germline mutations or known Lynch syndrome. Patients were considered evaluable if they underwent radiographic scanning at 12 weeks. The primary endpoints were immune-related objective response rate and immune-related progression-free survival at 20 weeks.
Both PD-1 expression in TILs and PD-L1 expression on tumor cells differ in MSI-H compared with MSS tumors. TILs from MSI-H tumors have 77% PD-1 expression compared with 39% in MSS tumors. MSI-H tumors have 32% PD-L1 expression compared with 13% in MSS tumors.
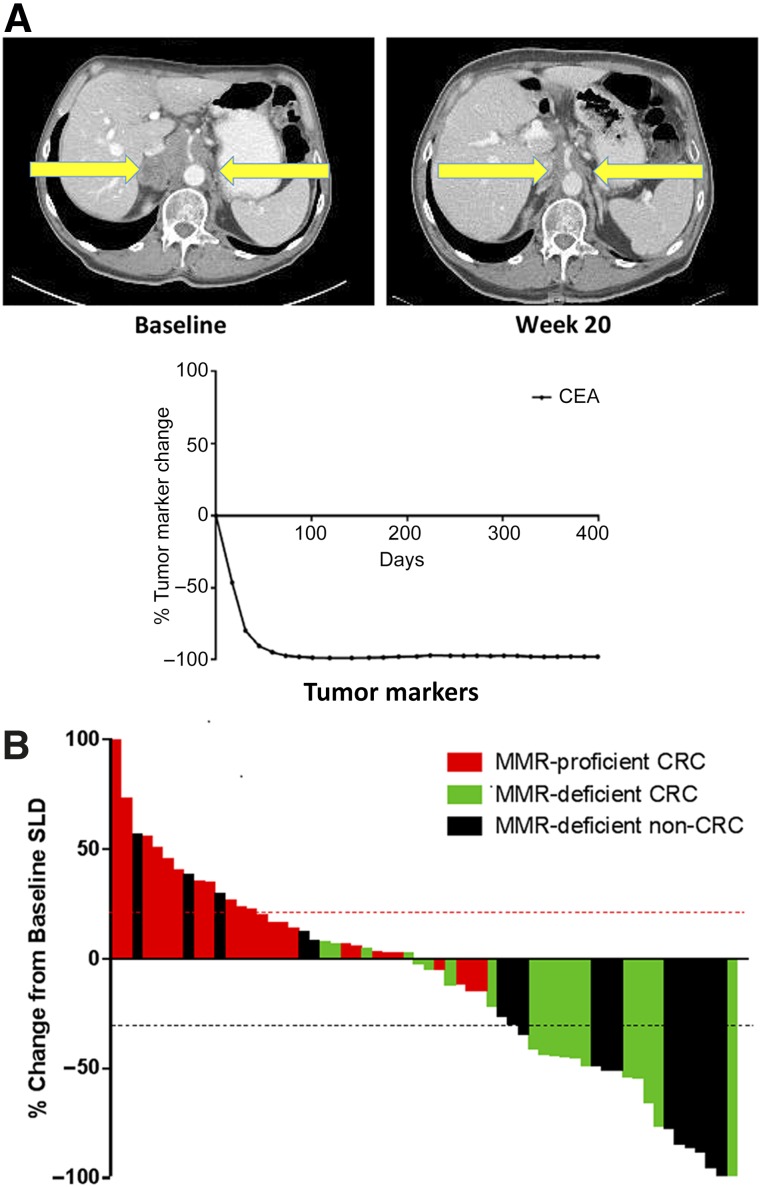
Upon treatment, virtually all patients with dMMR had an immediate clinical and biomarker (CEA or CA19-9) response; a representative patient is shown in Figure 4. In cohorts A, B, and C, immune-related objective response rates were 40%, 0%, and 71%, respectively, and immune-related progression-free survival rates were 78%, 11%, and 57% (Fig. 4).
Figure 4.
Responses to anti-PD-1 therapy in tumors with dMMR. (A): Single patient response after 20 weeks of anti-PD-1 therapy with complete response of retroperitoneal lymphadenopathy. (B): CEA response to checkpoint inhibition in the same patient with durable response over time. (C): Radiographic response to pembrolizumab as represented by the largest percentage change of SLD based on RECIST. Each bar represents one patient.
Abbreviations: CEA, carcinoembryonic antigen; CRC, colorectal cancer; dMMR, deficient mismatch repair; MMR, mismatch repair; anti-PD-1, antibodies to programmed cell death protein 1; RECIST, Response Evaluation Criteria in Solid Tumors; SLD, sum of longest diameters.
IHC of MSI-H tumors showed greater densities of CD8+ TILs and PD-L1 expression (not seen in any pMMR tumors), which were associated with a trend toward objective response and stable disease, although this did not reach statistical significance. Whole-exome sequences also showed an average 1,782 somatic mutations and 578 potential mutation-associated neoantigens per dMMR tumor, in contrast to 73 mutations and 21 neoantigens per tumor in pMMR cancers. Those in cohorts A and C with objective partial or complete responses have yet to reach median progression-free survival as of 1 year after publication; this is in contrast to cohort B, for whom median PFS was only 2.2 months (95% confidence interval, 1.4–2.8 months) [173, 174]. On November 2, 2015, the Food and Drug Administration granted breakthrough therapy designation for pembrolizumab in MSI-H advanced CRCs. Future studies are ongoing in MSI-H colon cancer in the first metastatic setting, as well in other tumor types with dMMR [175]. These studies will not be limited to metastatic or treatment-refractory disease, but will likely move to first-line therapy compared with standard-of-care treatment. In addition, trials evaluating this approach in the neoadjuvant and adjuvant setting are in development, especially in high-risk disease. If checkpoint blockade in this population of patients achieves curative results in the metastatic setting, then one could envision a time when checkpoint blockade could even replace surgery for this subgroup of patients.
Where Do Tumors With Mismatch Repair Deficiency Fit Into the Checkpoint Inhibition Paradigm?
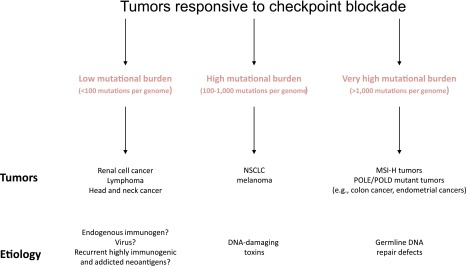
The human studies of PD-1 blockade in MSI-H tumors, although encouraging, will need to be validated in a larger cohort of patients. Nonetheless, the role of somatic cancer mutations in priming the immune system is becoming more evident, and data from prospective clinical trials is more clearly defining the association between mutations and response to checkpoint blockade. This trial in MSI-H solid tumors is the first prospective demonstration that mutation burden may influence response to PD-1 inhibitors. Retrospective studies in melanoma and non-small-cell lung cancer have also demonstrated the importance of mutational burden and response to checkpoint blockade [176, 177]. However, other tumor types, such as renal cell carcinomas, have a relatively low mutational burden and yet still show significant activity to checkpoint inhibitors [178–180]. These observations, summarized in Figure 5, suggest that different mechanisms of tumorigenesis may be related to tumors responsiveness to checkpoint blockade.
Figure 5.
Schematic of anti-PD-1-responsive tumors.
Abbreviations: MSI-H, high-frequency microsatellite instability; NSCLC, non-small-cell lung cancer.
The checkpoint blockade data in pMMR and dMMR tumors suggest that clinically there are three groups of tumors that respond differently to immunotherapy: (1) tumors that are not immunogenic and will likely never respond to any immunotherapy (MSS tumors that rapidly progressed on immunotherapy), (2) tumors that are highly immunogenic (MSI-H tumors that briskly respond to single-agent immunotherapy), and (3) tumors that are borderline immunogenic (MSS tumors that develop stable disease with immunotherapy). It is the borderline immunogenic tumors that will likely need combination therapy for a significant durable response. Strategies for combination therapy—radiation, small molecules, or even other immunomodulatory agents—will likely dominate drug development in the next few years. Equally as important will be understanding how to molecularly define these subgroups before initiating therapy and identifying a host of novel predictive biomarkers that modulate immune-therapeutic response. Whether these predictors of response and resistance are tumor cell intrinsic (i.e., somatic mutations) or extrinsic (tumor stroma) remains to be seen but will certainly be an active area of research.
Conclusion
Current guidelines suggest dMMR/MSI testing in select groups of patients with colon and endometrial cancer. The data summarized in this review suggest that dMMR/MSI testing will go beyond testing for heritable risk and likely be used to dictate therapy. In fact, this testing will likely not be limited to colon or endometrial cancer, but may be beneficial for any cancer in which dMMR has been reported. If the data continue to show such durable responses of MSI-H tumors to PD-1 blockade, MSI status may be a pan-cancer biomarker predictive for response to immunotherapy that is independent of tumor histology and fully dependent on a tumor’s genetic composition.
Author Contributions
Conception/Design: Valerie Lee, Adrian Murphy, Dung T. Le, Luis A. Diaz, Jr.
Provision of study material or patients: Valerie Lee, Adrian Murphy, Dung T. Le, Luis A. Diaz, Jr.
Collection and/or assembly of data: Valerie Lee, Adrian Murphy, Dung T. Le, Luis A. Diaz, Jr.
Data analysis and interpretation: Valerie Lee, Adrian Murphy, Dung T. Le
Manuscript writing: Valerie Lee, Adrian Murphy, Dung T. Le, Luis A. Diaz, Jr.
Final approval of manuscript: Valerie Lee, Adrian Murphy, Dung T. Le, Luis A. Diaz, Jr.
Disclosures
Luis A. Diaz, Jr.: Merck, PGDx (C/A), PGDx (E), Papgene, PGDx (OI), PapGene, PGDx, Inostics (IP); Dung T. Le: Merck, Bristol-Myers Squibb, Aduro Biotech (RF), Merck (H). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch HT, Snyder CL, Shaw TG, et al. Milestones of Lynch syndrome: 1895-2015. Nat Rev Cancer. 2015;15:181–194. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 3.Boland CR, Lynch HT. The history of Lynch syndrome. Fam Cancer. 2013;12:145–157. doi: 10.1007/s10689-013-9637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aaltonen LA, Peltomäki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 5.Ionov Y, Peinado MA, Malkhosyan S, et al. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 6.Peltomäki P, Aaltonen LA, Sistonen P, et al. Genetic mapping of a locus predisposing to human colorectal cancer. Science. 1993;260:810–812. doi: 10.1126/science.8484120. [DOI] [PubMed] [Google Scholar]
- 7.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 8.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 10.Leach FS, Nicolaides NC, Papadopoulos N, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 11.Parsons R, Li GM, Longley MJ, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 12.Lindblom A, Tannergård P, Werelius B, et al. Genetic mapping of a second locus predisposing to hereditary non-polyposis colon cancer. Nat Genet. 1993;5:279–282. doi: 10.1038/ng1193-279. [DOI] [PubMed] [Google Scholar]
- 13.Bronner CE, Baker SM, Morrison PT, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 14.Papadopoulos N, Nicolaides NC, Wei YF, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 15.Nicolaides NC, Papadopoulos N, Liu B, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 16.da Costa LT, Liu B, el-Deiry W, et al. Polymerase delta variants in RER colorectal tumours. Nat Genet. 1995;9:10–11. doi: 10.1038/ng0195-10. [DOI] [PubMed] [Google Scholar]
- 17.Palombo F, Gallinari P, Iaccarino I, et al. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 18.Papadopoulos N, Nicolaides NC, Liu B, et al. Mutations of GTBP in genetically unstable cells. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 19.Miyaki M, Konishi M, Tanaka K, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 22.Woerner SM, Benner A, Sutter C, et al. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative real common target genes. Oncogene. 2003;22:2226–2235. doi: 10.1038/sj.onc.1206421. [DOI] [PubMed] [Google Scholar]
- 23.Kohlmann W, Gruber SB. Lynch syndrome. In: Pagon RA, Adam MP, Ardinger HH et al, eds. GeneReviews. Seattle, WA: University of Washington, Seattle; 1993. [Google Scholar]
- 24.Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition—Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woods MO, Williams P, Careen A, et al. A new variant database for mismatch repair genes associated with Lynch syndrome. Hum Mutat. 2007;28:669–673. doi: 10.1002/humu.20502. [DOI] [PubMed] [Google Scholar]
- 26.Ligtenberg MJ, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto H, Imai K. Microsatellite instability: An update. Arch Toxicol. 2015;89:899–921. doi: 10.1007/s00204-015-1474-0. [DOI] [PubMed] [Google Scholar]
- 28.Valeri N, Gasparini P, Fabbri M, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci USA. 2010;107:21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, Berends MJ, Mensink RG, et al. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet. 1999;65:1291–1298. doi: 10.1086/302612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carethers JM, Stoffel EM. Lynch syndrome and Lynch syndrome mimics: The growing complex landscape of hereditary colon cancer. World J Gastroenterol. 2015;21:9253–9261. doi: 10.3748/wjg.v21.i31.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perucho M. Correspondence re: C.R. Boland et al., A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res., 58: 5248-5257, 1998. Cancer Res. 1999;59:249–256. [PubMed] [Google Scholar]
- 33.Laiho P, Launonen V, Lahermo P, et al. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62:1166–1170. [PubMed] [Google Scholar]
- 34.Wu CW, Chen GD, Jiang KC, et al. A genome-wide study of microsatellite instability in advanced gastric carcinoma. Cancer. 2001;92:92–101. doi: 10.1002/1097-0142(20010701)92:1<92::aid-cncr1296>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 35.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carethers JM, Koi M, Tseng-Rogenski SS. EMAST is a form of microsatellite instability that is initiated by inflammation and modulates colorectal cancer progression. Genes (Basel) 2015;6:185–205. doi: 10.3390/genes6020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing. J Mol Diagn. 2008;10:301–307. doi: 10.2353/jmoldx.2008.080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sood AK, Holmes R, Hendrix MJ, et al. Application of the National Cancer Institute international criteria for determination of microsatellite instability in ovarian cancer. Cancer Res. 2001;61:4371–4374. [PubMed] [Google Scholar]
- 41.Laghi L, Bianchi P, Malesci A. Differences and evolution of the methods for the assessment of microsatellite instability. Oncogene. 2008;27:6313–6321. doi: 10.1038/onc.2008.217. [DOI] [PubMed] [Google Scholar]
- 42.McConechy MK, Talhouk A, Li-Chang HH, et al. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecol Oncol. 2015;137:306–310. doi: 10.1016/j.ygyno.2015.01.541. [DOI] [PubMed] [Google Scholar]
- 43.Bartley AN, Luthra R, Saraiya DS, et al. Identification of cancer patients with Lynch syndrome: clinically significant discordances and problems in tissue-based mismatch repair testing. Cancer Prev Res (Phila) 2012;5:320–327. doi: 10.1158/1940-6207.CAPR-11-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Li J. Era of universal testing of microsatellite instability in colorectal cancer. World J Gastrointest Oncol. 2013;5:12–19. doi: 10.4251/wjgo.v5.i2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tafe LJ. Targeted next-generation sequencing for hereditary cancer syndromes: A focus on Lynch syndrome and associated endometrial cancer. J Mol Diagn. 2015;17:472–482. doi: 10.1016/j.jmoldx.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Stadler ZK, Battaglin F, Middha S, et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J Clin Oncol. 2016;34:2141–2147. doi: 10.1200/JCO.2015.65.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salipante SJ, Scroggins SM, Hampel HL, et al. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 48.Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 49.National Comprehensive Cancer Network. NCCN Guidelines: Colon Cancer, Version 2.2016. Available at https://www.nccn.org/professionals/physician_gls/pdf/colon_blocks.pdf. Accessed April 2, 2016.
- 50.Heald B, Plesec T, Liu X, et al. Implementation of universal microsatellite instability and immunohistochemistry screening for diagnosing lynch syndrome in a large academic medical center. J Clin Oncol. 2013;31:1336–1340. doi: 10.1200/JCO.2012.45.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corso G, Velho S, Paredes J, et al. Oncogenic mutations in gastric cancer with microsatellite instability. Eur J Cancer. 2011;47:443–451. doi: 10.1016/j.ejca.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 53.Zhu L, Li Z, Wang Y, et al. Microsatellite instability and survival in gastric cancer: A systematic review and meta-analysis. Mol Clin Oncol. 2015;3:699–705. doi: 10.3892/mco.2015.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Czopek J, Bialas M, Rudzki Z, et al. The relationship between gastric cancer cells circulating in the blood and microsatellite instability positive gastric carcinomas. Aliment Pharmacol Ther. 2002;16(Suppl 2):128–136. doi: 10.1046/j.1365-2036.16.s2.5.x. [DOI] [PubMed] [Google Scholar]
- 55.Oki E, Kakeji Y, Zhao Y, et al. Chemosensitivity and survival in gastric cancer patients with microsatellite instability. Ann Surg Oncol. 2009;16:2510–2515. doi: 10.1245/s10434-009-0580-8. [DOI] [PubMed] [Google Scholar]
- 56.Seo JY, Jin EH, Jo HJ, et al. Clinicopathologic and molecular features associated with patient age in gastric cancer. World J Gastroenterol. 2015;21:6905–6913. doi: 10.3748/wjg.v21.i22.6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim SY, Choi YY, An JY, et al. The benefit of microsatellite instability is attenuated by chemotherapy in stage II and stage III gastric cancer: Results from a large cohort with subgroup analyses. Int J Cancer. 2015;137:819–825. doi: 10.1002/ijc.29449. [DOI] [PubMed] [Google Scholar]
- 58.Williams AS, Huang WY. The analysis of microsatellite instability in extracolonic gastrointestinal malignancy. Pathology. 2013;45:540–552. doi: 10.1097/PAT.0b013e3283653307. [DOI] [PubMed] [Google Scholar]
- 59.Rodríguez-Hernández I, Garcia JL, Santos-Briz A, et al. Integrated analysis of mismatch repair system in malignant astrocytomas. PLoS One. 2013;8:e76401. doi: 10.1371/journal.pone.0076401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Viana-Pereira M, Lee A, Popov S, et al. Microsatellite instability in pediatric high grade glioma is associated with genomic profile and differential target gene inactivation. PLoS One. 2011;6:e20588. doi: 10.1371/journal.pone.0020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pollack IF, Hamilton RL, Sobol RW, et al. Mismatch repair deficiency is an uncommon mechanism of alkylator resistance in pediatric malignant gliomas: A report from the Children’s Oncology Group. Pediatr Blood Cancer. 2010;55:1066–1071. doi: 10.1002/pbc.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viana-Pereira M, Almeida I, Sousa S, et al. Analysis of microsatellite instability in medulloblastoma. Neuro-oncol. 2009;11:458–467. doi: 10.1215/15228517-2008-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maxwell JA, Johnson SP, McLendon RE, et al. Mismatch repair deficiency does not mediate clinical resistance to temozolomide in malignant glioma. Clin Cancer Res. 2008;14:4859–4868. doi: 10.1158/1078-0432.CCR-07-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vladimirova V, Denkhaus D, Soerensen N, et al. Low level of microsatellite instability in paediatric malignant astrocytomas. Neuropathol Appl Neurobiol. 2008;34:547–554. doi: 10.1111/j.1365-2990.2007.00919.x. [DOI] [PubMed] [Google Scholar]
- 65.Eckert A, Kloor M, Giersch A, et al. Microsatellite instability in pediatric and adult high-grade gliomas. Brain Pathol. 2007;17:146–150. doi: 10.1111/j.1750-3639.2007.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinez R, Schackert HK, Appelt H, et al. Low-level microsatellite instability phenotype in sporadic glioblastoma multiforme. J Cancer Res Clin Oncol. 2005;131:87–93. doi: 10.1007/s00432-004-0592-5. [DOI] [PubMed] [Google Scholar]
- 67.Alonso M, Hamelin R, Kim M, et al. Microsatellite instability occurs in distinct subtypes of pediatric but not adult central nervous system tumors. Cancer Res. 2001;61:2124–2128. [PubMed] [Google Scholar]
- 68.Leung SY, Chan TL, Chung LP, et al. Microsatellite instability and mutation of DNA mismatch repair genes in gliomas. Am J Pathol. 1998;153:1181–1188. doi: 10.1016/S0002-9440(10)65662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siah SP, Quinn DM, Bennett GD, et al. Microsatellite instability markers in breast cancer: A review and study showing MSI was not detected at ‘BAT 25’ and ‘BAT 26’ microsatellite markers in early-onset breast cancer. Breast Cancer Res Treat. 2000;60:135–142. doi: 10.1023/a:1006315315060. [DOI] [PubMed] [Google Scholar]
- 70.Sarrió D, Moreno-Bueno G, Hardisson D, et al. Epigenetic and genetic alterations of APC and CDH1 genes in lobular breast cancer: Relationships with abnormal E-cadherin and catenin expression and microsatellite instability. Int J Cancer. 2003;106:208–215. doi: 10.1002/ijc.11197. [DOI] [PubMed] [Google Scholar]
- 71.Wen YH, Brogi E, Zeng Z, et al. DNA mismatch repair deficiency in breast carcinoma: A pilot study of triple-negative and non-triple-negative tumors. Am J Surg Pathol. 2012;36:1700–1708. doi: 10.1097/PAS.0b013e3182627787. [DOI] [PubMed] [Google Scholar]
- 72.Lacroix-Triki M, Lambros MB, Geyer FC, et al. Absence of microsatellite instability in mucinous carcinomas of the breast. Int J Clin Exp Pathol. 2010;4:22–31. [PMC free article] [PubMed] [Google Scholar]
- 73.Adem C, Soderberg CL, Cunningham JM, et al. Microsatellite instability in hereditary and sporadic breast cancers. Int J Cancer. 2003;107:580–582. doi: 10.1002/ijc.11442. [DOI] [PubMed] [Google Scholar]
- 74.Seitz S, Wassmuth P, Plaschke J, et al. Identification of microsatellite instability and mismatch repair gene mutations in breast cancer cell lines. Genes Chromosomes Cancer. 2003;37:29–35. doi: 10.1002/gcc.10196. [DOI] [PubMed] [Google Scholar]
- 75.Demokan S, Muslumanoglu M, Yazici H, et al. Investigation of microsatellite instability in Turkish breast cancer patients. Pathol Oncol Res. 2002;8:138–141. doi: 10.1007/BF03033724. [DOI] [PubMed] [Google Scholar]
- 76.Huang HN, Lin MC, Tseng LH, et al. Ovarian and endometrial endometrioid adenocarcinomas have distinct profiles of microsatellite instability, PTEN expression, and ARID1A expression. Histopathology. 2015;66:517–528. doi: 10.1111/his.12543. [DOI] [PubMed] [Google Scholar]
- 77.Shilpa V, Bhagat R, Premalata CS, et al. Microsatellite instability, promoter methylation and protein expression of the DNA mismatch repair genes in epithelial ovarian cancer. Genomics. 2014;104:257–263. doi: 10.1016/j.ygeno.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 78.Lee JH, Cragun D, Thompson Z, et al. Association between IHC and MSI testing to identify mismatch repair-deficient patients with ovarian cancer. Genet Test Mol Biomarkers. 2014;18:229–235. doi: 10.1089/gtmb.2013.0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chui MH, Gilks CB, Cooper K, et al. Identifying Lynch syndrome in patients with ovarian carcinoma: the significance of tumor subtype. Adv Anat Pathol. 2013;20:378–386. doi: 10.1097/PAP.0b013e3182a92cf8. [DOI] [PubMed] [Google Scholar]
- 80.Caliman LP, Tavares RL, Piedade JB, et al. Evaluation of microsatellite instability in women with epithelial ovarian cancer. Oncol Lett. 2012;4:556–560. doi: 10.3892/ol.2012.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aysal A, Karnezis A, Medhi I, et al. Ovarian endometrioid adenocarcinoma: Incidence and clinical significance of the morphologic and immunohistochemical markers of mismatch repair protein defects and tumor microsatellite instability. Am J Surg Pathol. 2012;36:163–172. doi: 10.1097/PAS.0b013e31823bc434. [DOI] [PubMed] [Google Scholar]
- 82.Murphy MA, Wentzensen N. Frequency of mismatch repair deficiency in ovarian cancer: A systematic review. Int J Cancer. 2011;129:1914–1922. doi: 10.1002/ijc.25835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pal T, Akbari MR, Sun P, et al. Frequency of mutations in mismatch repair genes in a population-based study of women with ovarian cancer. Br J Cancer. 2012;107:1783–1790. doi: 10.1038/bjc.2012.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McConechy MK, Ding J, Cheang MC, et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol. 2012;228:20–30. doi: 10.1002/path.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thoury A, Descatoire V, Kotelevets L, et al. Evidence for different expression profiles for c-Met, EGFR, PTEN and the mTOR pathway in low and high grade endometrial carcinomas in a cohort of consecutive women. Occurrence of PIK3CA and K-Ras mutations and microsatellite instability. Histol Histopathol. 2014;29:1455–1466. doi: 10.14670/HH-29.1455. [DOI] [PubMed] [Google Scholar]
- 87.Ruiz I, Martín-Arruti M, Lopez-Lopez E, et al. Lack of association between deficient mismatch repair expression and outcome in endometrial carcinomas of the endometrioid type. Gynecol Oncol. 2014;134:20–23. doi: 10.1016/j.ygyno.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 88.Egoavil C, Alenda C, Castillejo A, et al. Prevalence of Lynch syndrome among patients with newly diagnosed endometrial cancers. PLoS One. 2013;8:e79737. doi: 10.1371/journal.pone.0079737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nelson GS, Pink A, Lee S, et al. MMR deficiency is common in high-grade endometrioid carcinomas and is associated with an unfavorable outcome. Gynecol Oncol. 2013;131:309–314. doi: 10.1016/j.ygyno.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 90.Diaz-Padilla I, Romero N, Amir E, et al. Mismatch repair status and clinical outcome in endometrial cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2013;88:154–167. doi: 10.1016/j.critrevonc.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 91.Stelloo E, Bosse T, Nout RA, et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod Pathol. 2015;28:836–844. doi: 10.1038/modpathol.2015.43. [DOI] [PubMed] [Google Scholar]
- 92.Peterson LM, Kipp BR, Halling KC, et al. Molecular characterization of endometrial cancer: a correlative study assessing microsatellite instability, MLH1 hypermethylation, DNA mismatch repair protein expression, and PTEN, PIK3CA, KRAS, and BRAF mutation analysis. Int J Gynecol Pathol. 2012;31:195–205. doi: 10.1097/PGP.0b013e318231fc51. [DOI] [PubMed] [Google Scholar]
- 93.Steinbakk A, Malpica A, Slewa A, et al. High frequency microsatellite instability has a prognostic value in endometrial endometrioid adenocarcinoma, but only in FIGO stage 1 cases. Anal Cell Pathol (Amst) 2010;33:245–255. doi: 10.3233/ACP-CLO-2010-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mackay HJ, Gallinger S, Tsao MS, et al. Prognostic value of microsatellite instability (MSI) and PTEN expression in women with endometrial cancer: Results from studies of the NCIC Clinical Trials Group (NCIC CTG) Eur J Cancer. 2010;46:1365–1373. doi: 10.1016/j.ejca.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 95.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Azzouzi AR, Catto JW, Rehman I, et al. Clinically localised prostate cancer is microsatellite stable. BJU Int. 2007;99:1031–1035. doi: 10.1111/j.1464-410X.2006.06723.x. [DOI] [PubMed] [Google Scholar]
- 97.Burger M, Denzinger S, Hammerschmied CG, et al. Elevated microsatellite alterations at selected tetranucleotides (EMAST) and mismatch repair gene expression in prostate cancer. J Mol Med (Berl) 2006;84:833–841. doi: 10.1007/s00109-006-0074-0. [DOI] [PubMed] [Google Scholar]
- 98.Rohrbach H, Haas CJ, Baretton GB, et al. Microsatellite instability and loss of heterozygosity in prostatic carcinomas: Comparison of primary tumors, and of corresponding recurrences after androgen-deprivation therapy and lymph-node metastases. Prostate. 1999;40:20–27. doi: 10.1002/(sici)1097-0045(19990615)40:1<20::aid-pros3>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 99.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 100.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 101.Guastadisegni C, Colafranceschi M, Ottini L, et al. Microsatellite instability as a marker of prognosis and response to therapy: A meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–2798. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 102.Moy AP, Shahid M, Ferrone CR, et al. Microsatellite instability in gallbladder carcinoma. Virchows Arch. 2015;466:393–402. doi: 10.1007/s00428-015-1720-0. [DOI] [PubMed] [Google Scholar]
- 103.Sessa F, Furlan D, Zampatti C, et al. Prognostic factors for ampullary adenocarcinomas: Tumor stage, tumor histology, tumor location, immunohistochemistry and microsatellite instability. Virchows Arch. 2007;451:649–657. doi: 10.1007/s00428-007-0444-1. [DOI] [PubMed] [Google Scholar]
- 104.Saetta AA, Gigelou F, Papanastasiou PI, et al. High-level microsatellite instability is not involved in gallbladder carcinogenesis. Exp Mol Pathol. 2006;80:67–71. doi: 10.1016/j.yexmp.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 105.Kidd M, Eick G, Shapiro MD, et al. Microsatellite instability and gene mutations in transforming growth factor-beta type II receptor are absent in small bowel carcinoid tumors. Cancer. 2005;103:229–236. doi: 10.1002/cncr.20750. [DOI] [PubMed] [Google Scholar]
- 106.An JY, Kim H, Cheong JH, et al. Microsatellite instability in sporadic gastric cancer: Its prognostic role and guidance for 5-FU based chemotherapy after R0 resection. Int J Cancer. 2012;131:505–511. doi: 10.1002/ijc.26399. [DOI] [PubMed] [Google Scholar]
- 107.Karahan B, Argon A, Yıldırım M, et al. Relationship between MLH-1, MSH-2, PMS-2,MSH-6 expression and clinicopathological features in colorectal cancer. Int J Clin Exp Pathol. 2015;8:4044–4053. [PMC free article] [PubMed] [Google Scholar]
- 108.Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer. 2005;41:2060–2070. doi: 10.1016/j.ejca.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 109.Gryfe R, Kim H, Hsieh ET, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 110.Tran B, Kopetz S, Tie J, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–4632. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goldstein J, Tran B, Ensor J, et al. Multicenter retrospective analysis of metastatic colorectal cancer (CRC) with high-level microsatellite instability (MSI-H) Ann Oncol. 2014;25:1032–1038. doi: 10.1093/annonc/mdu100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Koopman M, Kortman GA, Mekenkamp L, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100:266–273. doi: 10.1038/sj.bjc.6604867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang L, Cunningham JM, Winters JL, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res. 2003;63:5209–5212. [PubMed] [Google Scholar]
- 114.Yamamoto H, Perez-Piteira J, Yoshida T, et al. Gastric cancers of the microsatellite mutator phenotype display characteristic genetic and clinical features. Gastroenterology. 1999;116:1348–1357. doi: 10.1016/s0016-5085(99)70499-3. [DOI] [PubMed] [Google Scholar]
- 115.Choi YY, Bae JM, An JY, et al. Is microsatellite instability a prognostic marker in gastric cancer? A systematic review with meta-analysis. J Surg Oncol. 2014;110:129–135. doi: 10.1002/jso.23618. [DOI] [PubMed] [Google Scholar]
- 116.Falchetti M, Saieva C, Lupi R, et al. Gastric cancer with high-level microsatellite instability: Target gene mutations, clinicopathologic features, and long-term survival. Hum Pathol. 2008;39:925–932. doi: 10.1016/j.humpath.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 117.Matsusaka K, Kaneda A, Nagae G, et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187–7197. doi: 10.1158/0008-5472.CAN-11-1349. [DOI] [PubMed] [Google Scholar]
- 118.Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 119.Sunakawa Y, Lenz HJ. Molecular classification of gastric adenocarcinoma: Translating new insights from the cancer genome atlas research network. Curr Treat Options Oncol. 2015;16:17. doi: 10.1007/s11864-015-0331-y. [DOI] [PubMed] [Google Scholar]
- 120.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 121.Broaddus RR, Lynch HT, Chen LM, et al. Pathologic features of endometrial carcinoma associated with HNPCC: A comparison with sporadic endometrial carcinoma. Cancer. 2006;106:87–94. doi: 10.1002/cncr.21560. [DOI] [PubMed] [Google Scholar]
- 122.Nout RA, Bosse T, Creutzberg CL, et al. Improved risk assessment of endometrial cancer by combined analysis of MSI, PI3K-AKT, Wnt/β-catenin and P53 pathway activation. Gynecol Oncol. 2012;126:466–473. doi: 10.1016/j.ygyno.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 123.Billingsley CC, Cohn DE, Mutch DG, et al. Polymerase E (POLE) mutations in endometrial cancer: Clinical outcomes and implications for Lynch syndrome testing. Cancer. 2015;121:386–394. doi: 10.1002/cncr.29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pal T, Permuth-Wey J, Kumar A, et al. Systematic review and meta-analysis of ovarian cancers: Estimation of microsatellite-high frequency and characterization of mismatch repair deficient tumor histology. Clin Cancer Res. 2008;14:6847–6854. doi: 10.1158/1078-0432.CCR-08-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Segev Y, Pal T, Rosen B, et al. Risk factors for ovarian cancers with and without microsatellite instability. Int J Gynecol Cancer. 2014;24:664–669. doi: 10.1097/IGC.0000000000000134. [DOI] [PubMed] [Google Scholar]
- 126.Xiao X, Melton DW, Gourley C. Mismatch repair deficiency in ovarian cancer—Molecular characteristics and clinical implications. Gynecol Oncol. 2014;132:506–512. doi: 10.1016/j.ygyno.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 127.National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal 2015. Available at https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf. Accessed April 2, 2016.
- 128.Buerki N, Gautier L, Kovac M, et al. Evidence for breast cancer as an integral part of Lynch syndrome. Genes Chromosomes Cancer. 2012;51:83–91. doi: 10.1002/gcc.20935. [DOI] [PubMed] [Google Scholar]
- 129.Murata H, Khattar NH, Kang Y, et al. Genetic and epigenetic modification of mismatch repair genes hMSH2 and hMLH1 in sporadic breast cancer with microsatellite instability. Oncogene. 2002;21:5696–5703. doi: 10.1038/sj.onc.1205683. [DOI] [PubMed] [Google Scholar]
- 130.Raymond VM, Mukherjee B, Wang F, et al. Elevated risk of prostate cancer among men with Lynch syndrome. J Clin Oncol. 2013;31:1713–1718. doi: 10.1200/JCO.2012.44.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ryan S, Jenkins MA, Win AK. Risk of prostate cancer in Lynch syndrome: A systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014;23:437–449. doi: 10.1158/1055-9965.EPI-13-1165. [DOI] [PubMed] [Google Scholar]
- 132.Casorelli I, Bossa C, Bignami M. DNA damage and repair in human cancer: Molecular mechanisms and contribution to therapy-related leukemias. Int J Environ Res Public Health. 2012;9:2636–2657. doi: 10.3390/ijerph9082636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Das-Gupta EP, Seedhouse CH, Russell NH. Microsatellite instability occurs in defined subsets of patients with acute myeloblastic leukaemia. Br J Haematol. 2001;114:307–312. doi: 10.1046/j.1365-2141.2001.02920.x. [DOI] [PubMed] [Google Scholar]
- 134.Carethers JM, Chauhan DP, Fink D, et al. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hemminki A, Mecklin JP, Järvinen H, et al. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–928. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- 136.Elsaleh H, Joseph D, Grieu F, et al. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 137.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Klingbiel D, Saridaki Z, Roth AD, et al. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: Results of the PETACC-3 trial. Ann Oncol. 2015;26:126–132. doi: 10.1093/annonc/mdu499. [DOI] [PubMed] [Google Scholar]
- 139.Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: A National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 140.Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Collura A, Lagrange A, Svrcek M, et al. Patients with colorectal tumors with microsatellite instability and large deletions in HSP110 T17 have improved response to 5-fluorouracil-based chemotherapy. Gastroenterology. 2014;146:401–411. doi: 10.1053/j.gastro.2013.10.054. [DOI] [PubMed] [Google Scholar]
- 142.Jover R, Nguyen TP, Pérez-Carbonell L, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140:1174–1181. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Webber EM, Kauffman TL, O’Connor E, et al. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer. 2015;15:156. doi: 10.1186/s12885-015-1093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: Selecting optimal treatments for individual patients. J Clin Oncol. 2015;33:1787–1796. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- 146.Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16:30. doi: 10.1007/s11864-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tougeron D, Mouillet G, Trouilloud I, et al. Efficacy of adjuvant chemotherapy in colon cancer with microsatellite instability: A large multicenter AGEO study. J Natl Cancer Inst. 2016;108:djv438. doi: 10.1093/jnci/djv438. [DOI] [PubMed] [Google Scholar]
- 148.Venderbosch S, Nagtegaal ID, Maughan TS, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: A pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014;20:5322–5330. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Brooks GA, Enzinger PC, Fuchs CS. Adjuvant therapy for gastric cancer: Revisiting the past to clarify the future. J Clin Oncol. 2012;30:2297–2299. doi: 10.1200/JCO.2012.42.4069. [DOI] [PubMed] [Google Scholar]
- 150.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 151.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pagès F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 153.Kim H, Jen J, Vogelstein B, et al. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- 154.Alexander J, Watanabe T, Wu TT, et al. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158:527–535. doi: 10.1016/S0002-9440(10)63994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dolcetti R, Viel A, Doglioni C, et al. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am J Pathol. 1999;154:1805–1813. doi: 10.1016/S0002-9440(10)65436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology. 2007;121:1–14. doi: 10.1111/j.1365-2567.2007.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stewart TJ, Abrams SI. How tumours escape mass destruction. Oncogene. 2008;27:5894–5903. doi: 10.1038/onc.2008.268. [DOI] [PubMed] [Google Scholar]
- 158.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Timmermann B, Kerick M, Roehr C, et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS One. 2010;5:e15661. doi: 10.1371/journal.pone.0015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jung B, Doctolero RT, Tajima A, et al. Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology. 2004;126:654–659. doi: 10.1053/j.gastro.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 161.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 162.Lal N, Beggs AD, Willcox BE, et al. An immunogenomic stratification of colorectal cancer: Implications for development of targeted immunotherapy. OncoImmunology. 2015;4:e976052. doi: 10.4161/2162402X.2014.976052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boissière-Michot F, Lazennec G, Frugier H, et al. Characterization of an adaptive immune response in microsatellite-instable colorectal cancer. OncoImmunology. 2014;3:e29256. doi: 10.4161/onci.29256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kaplan DH, Shankaran V, Dighe AS, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 167.Gatalica Z, Snyder C, Maney T, et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev. 2014;23:2965–2970. doi: 10.1158/1055-9965.EPI-14-0654. [DOI] [PubMed] [Google Scholar]
- 168.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 170.Kmieciak M, Payne KK, Wang XY, et al. IFN-γ Rα is a key determinant of CD8+ T cell-mediated tumor elimination or tumor escape and relapse in FVB mouse. PLoS One. 2013;8:e82544. doi: 10.1371/journal.pone.0082544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hallermalm K, Seki K, De Geer A, et al. Modulation of the tumor cell phenotype by IFN-gamma results in resistance of uveal melanoma cells to granule-mediated lysis by cytotoxic lymphocytes. J Immunol. 2008;180:3766–3774. doi: 10.4049/jimmunol.180.6.3766. [DOI] [PubMed] [Google Scholar]
- 172.Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Le DT, Uram JN, Wang H et al. PD-1 blockade in mismatch repair deficient non-colorectal gastrointestinal cancers. J Clin Oncol 2016;34(suppl 4S):195a.
- 174.Le DT, Uram JN, Wang H et al. Programmed death-1 blockade in mismatch repair deficient colorectal cancer. J Clin Oncol 2016;34(suppl):103a.
- 175.Merck Sharp & Dohme Corp. Study of Pembrolizumab (MK-3475) vs Standard Therapy in Participants With Microsatellite Instability-High (MSI-H) or Mismatch Repair Deficient (dMMR) Stage IV Colorectal Carcinoma (MK-3475-177/KEYNOTE-177). NLM Identifier: NCT02563002. Available at https://clinicaltrials.gov/ct2/show/NCT02563002. Accessed March 25, 2016.
- 176.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 180.Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]