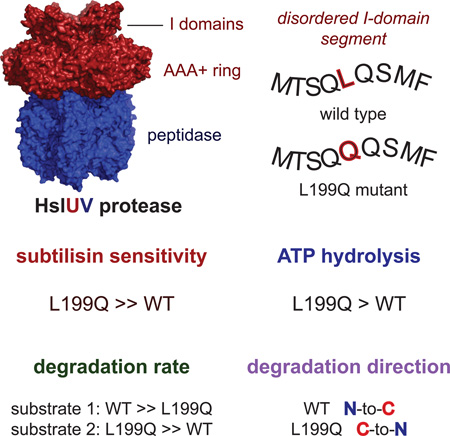
Summary
The I domain of HslU sits above the AAA+ ring and forms a funnel-like entry to the axial pore, where protein substrates are engaged, unfolded, and translocated into HslV for degradation. The L199Q I-domain substitution, which was originally reported as a loss-of-function mutation, resides in a segment that appears to adopt multiple conformations as electron density is not observed in HslU and HslUV crystal structures. The L199Q sequence change does not alter the structure of the AAA+ ring or its interactions with HslV but increases I-domain susceptibility to limited endoproteolysis. Notably, the L199Q mutation increases the rate of ATP-hydrolysis substantially, results in slower degradation of some proteins but faster degradation of other substrates, and markedly changes the preference of HslUV for initiating degradation at the N-terminus or C-terminus of model substrates. Thus, a structurally dynamic region of the I domain plays a key role in controlling protein degradation by HslUV.
Keywords: AAA+ protease, ATP-dependent degradation, allosteric control, HslUV protease
Graphical abstract
eTOC Blurb
Baytshtok et al. demonstrate that a mutation in a highly dynamic region of the intermediate domain of the AAA+ HslU hexamer plays a key role in controlling both the rate and direction of ATP-fueled degradation by the HslUV protease.
Introduction
Proteases of the AAA+ superfamily play critical roles in protein quality control, in signaling, in control of cell division, and in regulating transcriptional responses to stress in all organisms (Ogura and Wilkinson, 2001; Striebel et al., 2009; Sauer and Baker, 2011). Escherichia coli has five AAA+ proteases: HslUV, ClpXP, ClpAP, FtsH, and Lon. Despite differences in structure and substrate specificity, these proteases share a common mechanism in which a hexameric AAA+ ring harnesses the energy of ATP binding and hydrolysis to recognize, unfold, and translocate substrates through an axial pore and into the degradation chamber of the peptidase. Most bacterial proteins are targeted for degradation via short terminal peptide sequences, known as degrons. For example, the seven N-terminal residues of the Arc protein are required for degradation by HslUV (Sundar et al., 2012).
The HslUV protease consists of the hexameric AAA+ unfoldase HslU and the dodecameric HslV peptidase and is involved in the degradation of substrates during heat shock, the DNA-damage response, and other stresses (Rohrwild et al., 1996; Kanemori et al., 1997; 1999; Seong et al., 1999; Wu et al., 1999; Bochtler et al., 2000; Sousa et al., 2000; Kuo et al., 2004). In model substrates, both N- and C-terminal sequences can affect the kinetics of degradation by HslUV, suggesting that multi-degron recognition by different binding sites in the HslU hexamer regulates substrate selection (Kwon et al., 2004; Burton et al., 2005; Koodathingal et al., 2009; Lien et al., 2009; Sundar et al., 2010; 2012). HslUV can initiate degradation of appropriately tagged fusion proteins from either terminus, but it is unclear if degron position alone is responsible for controlling the directionality of degradation (Sundar et al., 2012; Kwon et al., 2003, Koodathingal et al., 2009). Interestingly, HslUV appears to be a considerably more powerful unfoldase in the N→C than in the C→N direction (Koodathingal et al., 2009), suggesting that directionality may also be an important factor in substrate degradation in vivo.
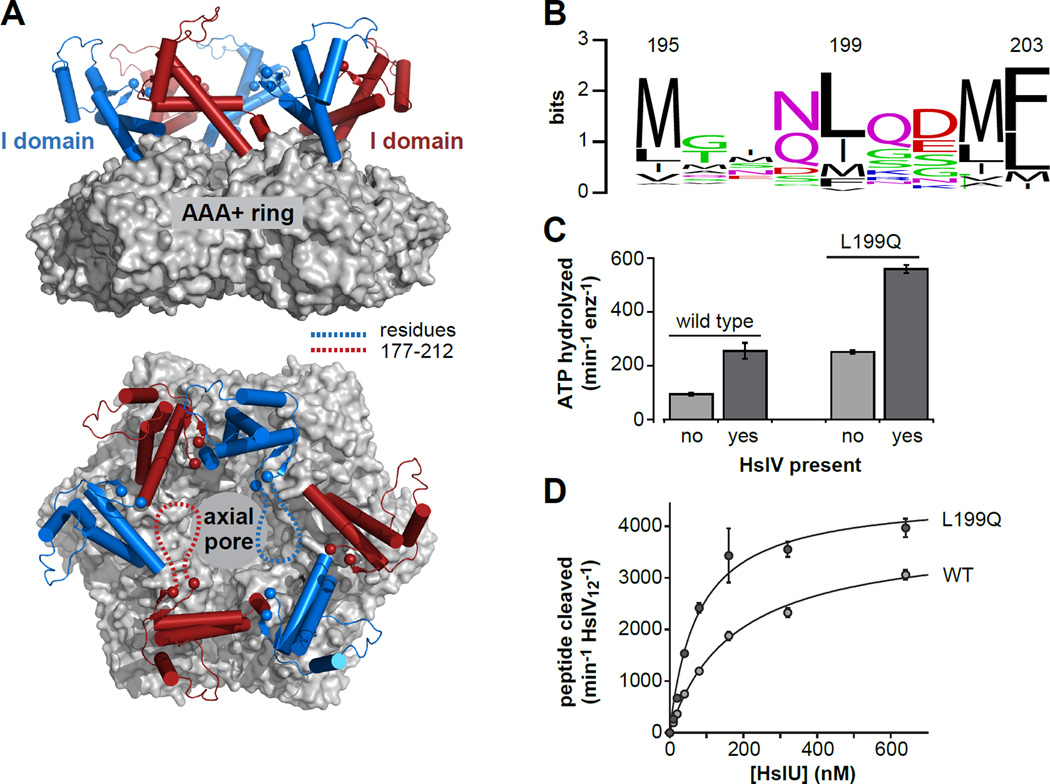
HslU contains a unique auxiliary domain, called the intermediate or I domain, that emerges from the top of the AAA+ ring following the Walker-A ATPase motif and GYVG translocation loop of each subunit and reconnects with the ring just before the Walker-B motif (Bochtler et al., 2000; Sousa et al., 2000). Deletion of this domain severely depresses ATP hydrolysis and prevents substrate degradation by HslUV (Kwon et al., 2003; Sundar et al., 2012). In HslU and HslUV crystal structures, electron density for I-domain residues 177–212 is absent or extremely poor (Bochtler et al., 2000; Sousa et al., 2000; Wang et al., 2001a), indicating that these amino acids adopt multiple conformations with respect to the crystal lattice. Based on flanking regions, the 177–212 segment appears to be positioned near the top of a funnel-like cavity leading to the axial translocation pore of HslU (Fig. 1A). Indeed, deleting these residues or replacing them with a linker results in reduced substrate binding and alters the coupling between substrate binding and ATP hydrolysis (Sundar et al., 2012). A Leu199 →Gln mutation in this region abolishes HslUV degradation of several substrates in vivo (Lien et al., 2009), but the mechanism by which it affects HslUV function is unknown. Here, we characterize the L199QHslU and L199QHslUV enzymes and show that the structured regions of the mutant and wild-type enzymes and their interactions with HslV are essentially identical. Compared to WTHslUV, however, L199QHslUV has substantially elevated ATP-hydrolysis, degrades some substrates more rapidly, and preferentially degrades model substrates from the opposite terminus. Thus, a structurally dynamic region of the I-domain modulates substrate specificity, regulates ATP hydrolysis, and controls the direction of substrate degradation.
Figure 1. The HslU hexamer and initial characterization of L199QHslU.
(A) Views of a wild-type HslU hexamer (pdb 1do0; Bochtler et al., 2000) from the side and from above the axial pore. The hexameric AAA+ ring is shown in surface representation and colored light grey. The I domains are shown in cartoon representation and colored blue or red. The α-carbons of residues 176 and 213 are shown as spheres. The intervening residues (177–212) are not observed in this or other HslU structures, and are schematically depicted as dashed lines in two I domains in the bottom panel. (B) WebLogo representation (Crooks et al., 2004) of sequence conservation for HslU residue 199 and flanking amino acids. (C) The rate of hydrolysis of 5 mM ATP at 37 °C was assayed for L199QHslU and WTHslU (300 nM each) in the absence or presence of HslV (900 nM) using a coupled assay (Nørby, 1988). (D) Increasing concentrations of L199QHslU or WTHslU were titrated against HslV12 (5 nM) in the presence of 5 mM ATP at 25 °C and the rate of cleavage of a Z-GGL-AMC tripeptide (200 nM) was monitored by increased fluorescence. The lines are fits to a hyperbolic binding equation. For L199QHslUV, half-maximal activation was observed at a HslU hexamer concentration of 78 ± 14 nM and maximal activity was 4580 ± 262 peptides cleaved min−1 HslV12−1. For WTHslUV, these values were 176 ± 16 nM and 3820 ± 137 peptides cleaved min−1 HslV12−1, respectively. In panels C and D, values are averages of 3 independent replicates ± 1 SD.
Results
L199Q enhances ATP hydrolysis and HslV binding
In a multiple sequence alignment of 783 HslU orthologs (POA6H5 from http://gremlin.bakerlab.org/ecoli.php), the position corresponding to residue 199 in E. coli HslU is a mid-sized hydrophobic residue, typically Leu, Ile, or Met (Fig. 1B). As the Leu199 →Gln mutation prevents HslUV degradation of several substrates in E. coli (Lien et al., 2009), Leu199 appears to play an important role in HslU structure and/or function. We purified the E. coli L199Q HslU mutant with an N-terminal H6 tag (called L199QHslU hereafter) and assayed rates of ATP hydrolysis for this enzyme and wild type (WTHslU) in the presence and absence of the HslV peptidase. Notably, L199QHslU hydrolyzed ATP at a rate ∼2.5-fold faster than WTHslU, and HslV substantially enhanced the ATP-hydrolysis rate for both the mutant and wild-type enzymes (Fig. 1C). We titrated increasing concentrations of L199QHslU or WTHslU against a fixed concentration of HslV, assayed changes in the rate of cleavage of a tripeptide that HslV alone degrades poorly, and fit the resulting curves to obtain apparent binding constants (Fig. 1D). L199QHslU bound HslV more tightly (78 nM) than WTHslU (180 nM) and also supported higher maximal peptidase activity. In combination, these results suggest that the degradation defect associated with L199QHslU in vivo does not arise from impaired HslV binding or from defects in ATP hydrolysis in the L199QHslUV complex.
L199QHslUV and WTHslUV have similar structures
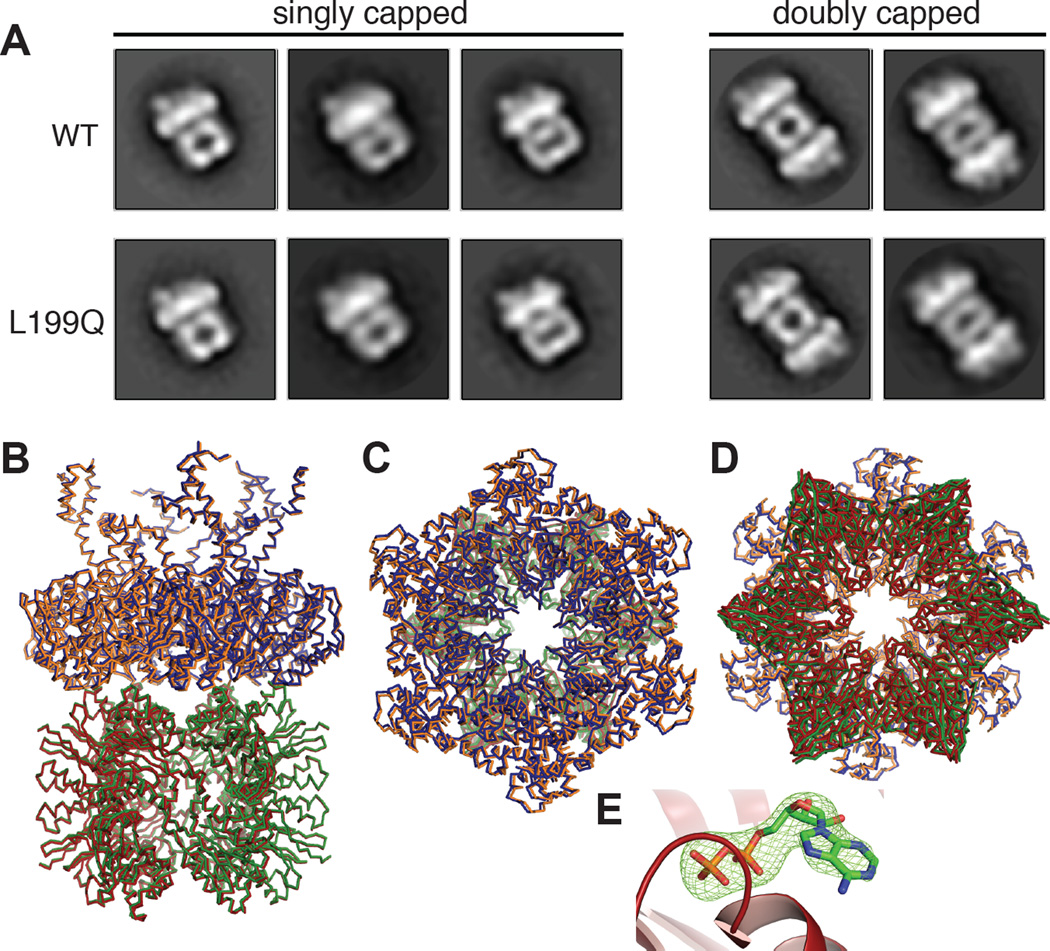
We examined ATPγS-bound L199QHslUV and WTHslUV by negative-stain electron microscopy. In these experiments, most images were side views of singly capped (HslU6•HslV12) or doubly capped (HslU6•HslV12•HslU6) complexes. As shown in Fig. 2A, two-dimensional class averages of L199QHslUV and WTHslUV were extremely similar. We also crystallized the L199QHslUV complex in the presence of ATP and a high concentration of ammonium sulfate at 4 °C. Surprisingly, the unit-cell dimensions and space group of this crystal, which diffracted to 3.3 Å resolution, were almost identical to those of a crystal of WTHslUV grown in the presence of dATP from a low-salt buffer at 20 °C (1G4A.pdb; Wang et al., 2001a; Table 1). After phasing by molecular replacement, we refined the L199QHslUV structure to a model with excellent geometry and good R and Rfree values given the moderate resolution (Table 1). We also re-refined the 1G4A structure to improve the statistics and geometry (Table 1). For both the mutant and wild-type structures, crystallographic symmetry generated an HslU hexamer and an associated HslV dodecamer, which were nearly identical in the mutant and wild-type complexes (Fig. 2B–D; RMSD 0.62 Å for 4270 common Cα atoms). The main differences were in the I domain, where the maximum RMSD was 2.4 Å, but the wild-type I domains in some HslU structures differ by substantially larger values. Electron density for the I-domain segment containing residue 199 and flanking sequences was absent in both L199QHslU and WTHslU. These results establish that L199QHslU and WTHslU adopt the same basic hexameric structure and interact with HslV in the same fashion in the crystal lattice. In the active site of each L199QHslU subunit, a simulated-annealing omit map showed electron density for ADP but no density for the γ-phosphate of ATP (Fig. 2E), suggesting that ATP hydrolysis occurred during crystallization.
Figure 2. L199QHslUV structures.
(A) Two-dimensional class averages of singly- and doubly-capped WTHslUV and L199QHslUV complexes based upon negative-stain electron microscopy. HslV is likely to appear wider in some views than others depending if the view is normal to a face or edge of the double-hexameric ring. (B) Superposition of side views of the L199QHslUV and WTHslUV crystal structures shown in ribbon representation. The L199QHslU and WTHslU hexamers are shown in blue and orange, respectively. The HslV dodecamer is shown in green for the L199QHslUV structure and in red for the WTHslUV structure. (C) Same superposition viewed from the top of the HslU hexamer. (D) Same superposition viewed from the bottom of the HslV dodecamer. (E) ADP (stick representation) in the active site of chain E of L199QHslU (cartoon representation). The green mesh is electron density (3.5 σ) from a FO-FC simulated annealing omit map in which the ADP was absent.
Table 1.
Crystallographic scaling and refinement statistics
| 5JI2 (L199Q) | 5JI3 (1G4A re- refined) |
1G4A | |
|---|---|---|---|
| Space group | P321 | P321 | P321 |
| Unit cell (Å) | a = b = 170.01 c = 163.39 |
a = b = 167.0 c= 161.32 |
a = b = 167.0 c= 161.32 |
| Resolution (Å) | 3.3 | 3.0 | 3.0 |
| Rsym | 14.2 (1.59) | 13.1 (*) | 13.1 (*) |
| Rpim | 0.045 (0.51) | * | * |
| # reflections | 41,171 | 46,551 | 46,551 |
| Completeness (%) | 100 (100) | 85.7 (80.6) | 85.7 (80.6) |
| Rwork/ | 0.236 (0.317) | 0.222 (0.322) | 0.257 (*) |
| Rfree | 0.281 (0.367) | 0.253 (0.370) | 0.294 (*) |
| MolProbity score (percentile) | 100 | 100 | 12 |
| Clash score | 2.7 | 1.9 | 72 |
| Poor rotamers (%) | 0.17 | 0.17 | 16.9 |
| Ramachandran outliers (%) | 0 | 0 | 13.1 |
| Ramachandran favored | 98.6 | 98.8 | 64.8 |
| Bad bonds / angles | 0 / 0 | 0 / 0 | 0 / 9 |
| Cβ deviations | 0 | 0 | 0 |
Values in parentheses are for the highest resolution shell of data.
Values not reported in 1G4A pdb entry.
Rsym = ShSj |Ij(h) - <I(h)>| / ShSj <I(h)>, where Ij(h) is the jth reflection of index h and <I(h)> is the average intensity of all observations of I(h).
Rpim= = Σh[1/(N-1)]1/2Σj |Ij(h) - <I(h)>| / ΣhΣj <I(h)>, where N is the redundancy of the reflection with index h
Rwork = Sh |Fobs(h) - Fcalc(h)| | / Sh |Fobs(h)|, calculated over ∼95% of the data in the working set.
Rfree equivalent to Rwork except calculated over ∼5% of the data in the test set.
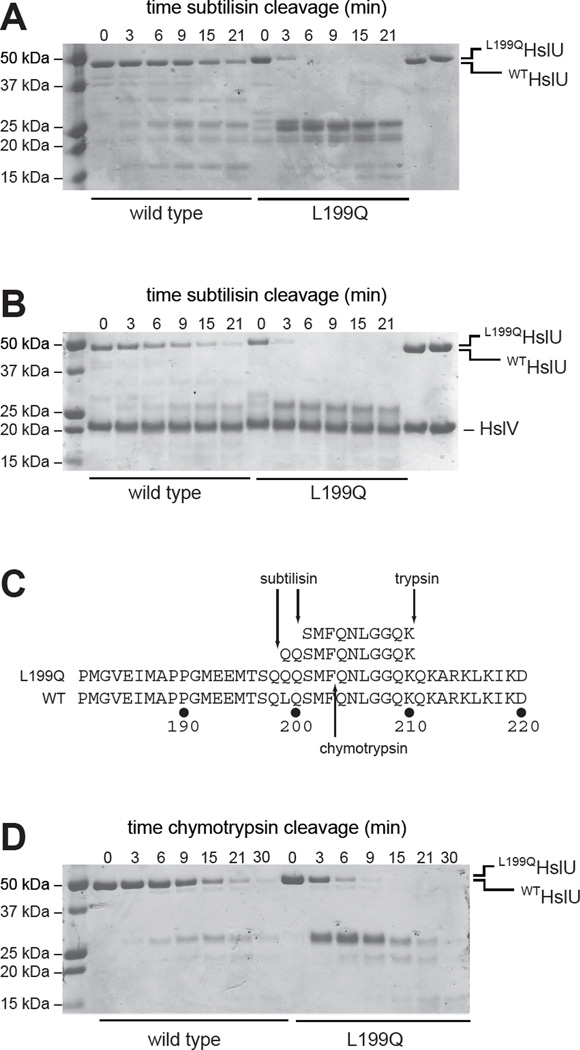
L199QHslU is more sensitive to limited proteolysis
To probe potential differences in the structurally dynamic regions of the mutant and wild-type I domains, we performed limited proteolysis time courses monitored by SDS-PAGE. In the presence of ATP, subtilisin cleaved L199QHslU substantially faster than WTHslU both in the absence and presence of HslV (Fig. 3A, 3B). Subtilisin cleavage of WTHslU and L199QHslU performed in the presence of ADP or absence of nucleotide gave results similar to the ATP experiments (not shown). Whether HslV was present or not, the early major products of L199QHslU cleavage had molecular weights of ∼25 kDa, suggesting cleavage within the I domain, which includes residues 110–240 in the intact 443-residue enzyme. We isolated major proteolytic product(s) of L199QHslU cleavage by subtilisin by anion-exchange chromatography, cleaved with trypsin, and analyzed the peptides by LC MS/MS sequencing. This analysis identified peptides corresponding to L199QHslU residues 199–210 and 201–210, whose N-terminal sequences are inconsistent with generation by tryptic cleavage (Fig. 3C). Thus, subtilisin appears to cleave L199QHslU after Gln198 and Gln200, which flank the site of the L199Q mutation. Chymotrypsin also cleaved L199QHslU more rapidly than WTHslU (Fig. 3D). As the cleavage specificities of chymotrypsin and subtilisin differ, these results suggest that the L199Q mutation affects limited proteolysis by altering the local structure or dynamics of the I domain rather than by introducing new cleavage sites. MALDI-TOF mass spectrometry of the major L199QHslU chymotryptic products gave masses close to those expected for one fragment extending from the N-terminus to Phe203 (expected 23,906 Da; observed 23,858 Da) and second fragment extending from Gln204 to the C-terminus (expected 26,618 Da; observed 26,593 Da). To confirm that chymotryptic cleavage of L199QHslU occurred between Phe203 and Gln204, we excised the top band of the doublet near 25 kDa and subjected it to sequential Edman degradation. The resulting sequence, Gln-Asn-Leu-Gly-Gly-Gln, corresponded to HslU residues 204–209.
Figure 3. Limited proteolysis.
(A) Kinetics of cleavage of WTHslU and L199QHslU (500 nM each) by subtilisin (11 nM) at 37 °C in the presence of 5 mM ATP, assayed by SDS-PAGE and staining with Coomassie Blue. The rightmost two lanes show the HslU proteins in the absence of subtilisin. (B) Kinetics of cleavage of WTHslUV and L199QHslUV (500 nM HslU; 1000 nM HslV) by subtilisin (11 nM) at 37 °C in the presence of 5 mM ATP. The rightmost two lanes show the HslU and HslV proteins in the absence of subtilisin. (C) Peptide sequences obtained by LC MS/MS following tryptic digestion of the major subtilisin cleavage products of L199QHslU aligned with residues 181–220 of L199QHslU and WTHslU. The major site of chymotryptic cleavage of intact L199QHslU is also shown. (D) Kinetics of cleavage of WTHslU and L199QHslU (500 nM each) by chymotrypsin (60 nM) at 37 °C in the presence of 5 mM ATP.
L199QHslUV is an active protease
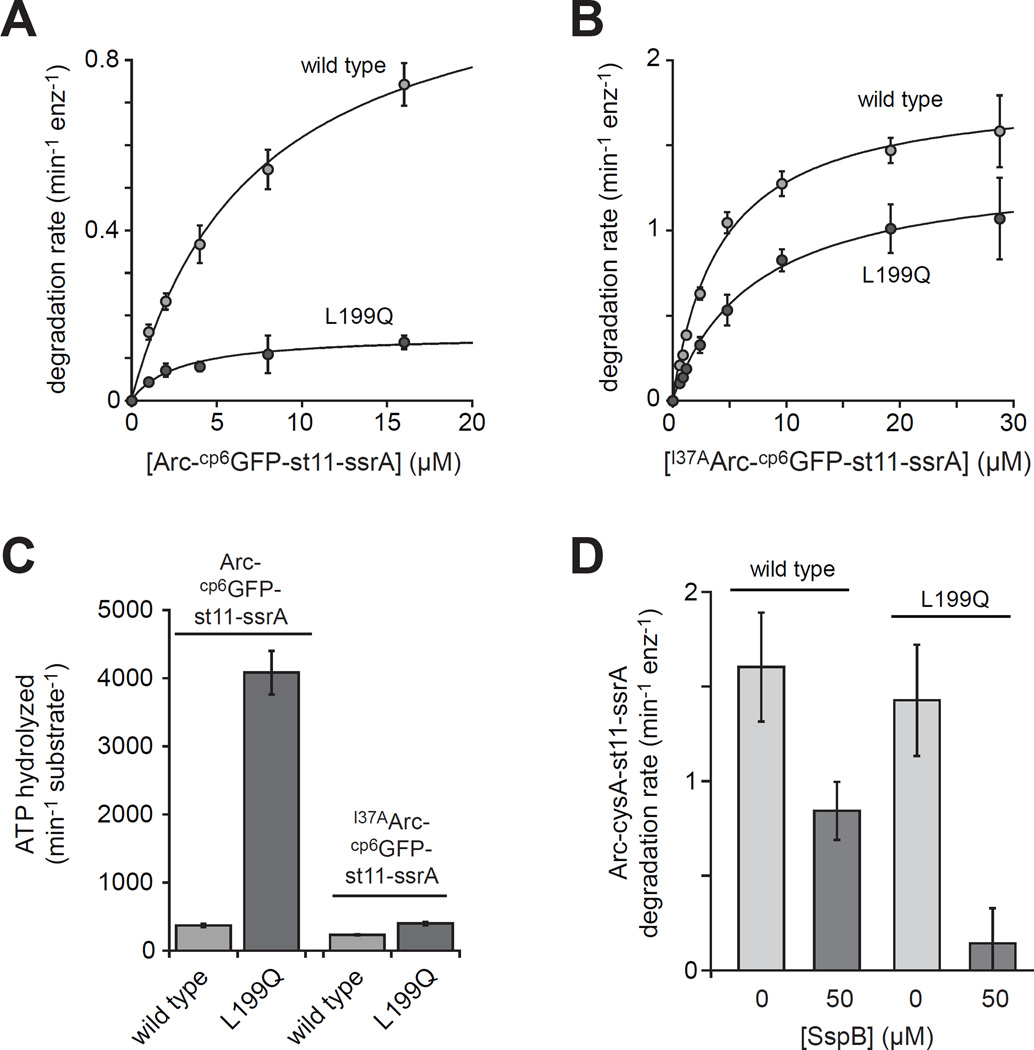
To probe degradation activity, we initially used a circularly permuted variant of superfolder GFP (cp6GFP) with a C-terminal st11-ssrA sequence and an N-terminal Arc sequence (Milla et al., 1993; Nager et al., 2011). Arc is normally dimeric and is a good HslUV substrate (Bowie et al., 1989; Burton et al., 2005). Steady-state rates of degradation of different concentrations of Arc-cp6GFP-st11-ssrA by L199QHslUV and WTHslUV substrate were quantified by loss of native fluorescence of the cp6GFP module and fit to the Michaelis-Menten equation to determine KM and Vmax values (Fig. 4A; Table 2). Compared to WTHslUV, Vmax was ∼7-fold slower for L199QHslUV and the catalytic efficiency (Vmax/KM) was ∼3-fold lower. For both enzymes, degradation was ATP dependent and deletion of the Arc portion of the substrate prevented degradation (not shown). We also assayed degradation of I37AArc-cp6GFP-st11-ssrA (Fig. 4B; Table 2), in which the I37A mutation results in natively unfolded monomeric Arc (Milla et al., 1994). For this substrate, Vmax values were similar for both enzymes, and the wild-type catalytic efficiency was about twice that of the mutant. Thus, the degree of impairment of the mutant protease depends on the substrate.
Figure 4. Degradation of substrates with different N-terminal signals.
(A) Degradation of Arc-cp6GFP-st11-ssrA by WTHslUV or L199QHslUV. The lines are non-linear least-squares fits to the Michaelis-Menten equation (fitted parameters in Table 2). Values are averages of 3 independent replicates ± 1 SD. (B) Degradation of I37AArc-cp6GFP-st11-ssrA by WTHslUV and L199QHslUV (fitted parameters in Table 2). (C) The energetic efficiency of substrate degradation was determined by dividing the ATPase rate in the presence of near saturating substrate by the maximal degradation rate. Rates of ATP hydrolysis by WTHslUV and L199QHslUV were determined in the presence of Arc-cp6GFP-st11-ssrA (60 µM) or I37AArc-cp6GFP-st11-ssrA (40 µM). Neither substrate significantly altered the ATPase rate of L199QHslUV, but both stimulated the ATPase rate of WTHslUV ∼1.5-fold. (D) Rates of degradation of Arc-cysA-st11-ssrA (40 µM) by WTHslUV or L199QHslUV in the absence or presence of monomeric SspB (50 µM). In all panels, experiments were performed at 37 °C in the presence of 5 mM ATP using 300 nM HslU and 900 nM HslV.
Table 2.
Steady-state kinetic parameters for degradation. Numbers in parenthesis are errors of non-linear least-squares fitting to the Michaelis-Menten equation for KM and Vmax and are propagated errors for Vmax/KM. n.d. – not determined.
| WTHslUV | L199QHslUV | |||||
|---|---|---|---|---|---|---|
| substrate | KM | Vmax | Vmax/KM | KM | Vmax | Vmax/KM |
| µM | min−1 enz−1 |
min−1 enz−1 µM−1 |
µM | min−1 enz−1 | min−1 enz−1 µM−1 |
|
| Arc-cp6GFP-st11-ssrA | 7.3 (1.0) | 1.1 (.07) | 0.15 (0.02) | 2.8 (1.0) | 0.15 (0.02) | 0.05 (0.02) |
|
I37AArc-cp6GFP-st11- ssrA |
4.3 (0.41) | 1.8 (0.06) | 0.43 (0.043) | 7.2 (1.3) | 1.4 (0.09) | 0.19 (0.04) |
| Arc-Cys-st11-sul20 | 11 (2.9) | 7.3 (0.88) | 0.68 (0.2) | 8.2 (2.0) | 12 (1.0) | 1.5 (0.38) |
| Arc-Cys-st11-ssrA | 18 (3.0) | 4.4 (0.28) | 0.24 (0.04) | 67 (14) | 5.3 (0.68) | 0.08 (0.02) |
| cp7GFP-sul20 | n.d. | n.d. | 0.0003 | 16 (3.9) | 0.39 (0.04) | 0.025 (.007) |
We determined rates of ATP hydrolysis in the presence of near saturating concentrations of Arc-cp6GFP-st11-ssrA or I37AArc-cp6GFP-st11-ssrA to calculate the number of ATPs hydrolyzed per molecule of substrate degraded (Fig. 4C). For dimeric Arc-cp6GFP-st11-ssrA, the energetic cost of L199QHslUV degradation was ∼11-fold higher than the wild-type value, suggesting that the mutant enzyme requires a substantially greater average number of power strokes to successfully degrade this substrate. For monomeric I37AArc-cp6GFP-st11-ssrA, by contrast, L199QHslUV was only 1.7-fold less efficient than WTHslUV. Thus, the N-terminal degron plays a role in determining the magnitude of energetic defect for L199QHslUV degradation.
C-terminal sequences appended to Arc can also affect HslUV degradation (Kwon et al., 2004; Sundar et al., 2010; 2012). We constructed Arc-cys-st11-ssrA and Arc-cys-st11-sul20 substrates, where sul20 corresponds to the C-terminal 20 residues of E. coli SulA (Gur and Sauer, 2009). We labeled the cysteines in these substrates with an Alexa-488 dye (designated cysA), and used changes in autoquenching to determine KM and Vmax values for L199QHslUV and WTHslUV degradation (Table 2). For Arc-cysA-st11-ssrA degradation, L199QHslUV had a slightly higher Vmax than WTHslUV, but WTHslUV had a 3-fold higher catalytic efficiency because of a substantially lower KM. Addition of a monomeric SspB variant (McGinness et al., 2007), which binds specifically to the ssrA tag, reduced the rate of WTHslUV degradation of Arc-cysA-st11-ssrA by ∼2-fold but almost completely eliminated degradation by L199QHslUV (Fig. 4D). Thus, the C-terminal region of Arc-cysA-st11-ssrA seems to be more important for degradation by L199QHslUV. Interestingly, for degradation of Arc-cysA-st11-sul20, L199QHslUV had a higher Vmax and a higher catalytic efficiency than WTHslUV (Table 2). Thus, the kinetic defects associated with L199QHslUV degradation are suppressed when sul20 is the targeting degron.
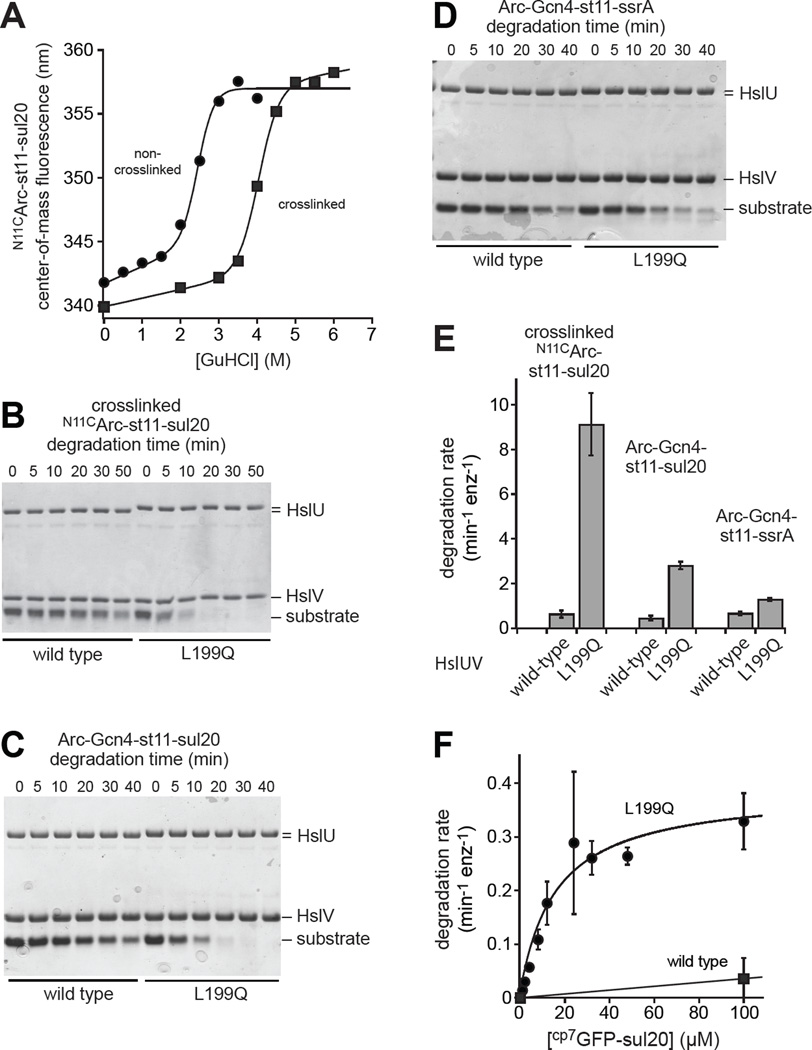
Because native Arc dimers are relatively unstable thermodynamically and kinetically (Bowie and Sauer, 1989; Milla and Sauer, 1994), we tested whether the sul20 degron could target more stable substrates for preferential L199QHslUV degradation. The Asn11 side chains in the Arc dimer are close in space, and a Cys11-Cys11 disulfide dramatically stabilizes the dimer (Robinson and Sauer, 2000). We constructed N11CArc-st11-sul20 and crosslinked the Cys11 side chains with a bismaleimide, resulting in marked stabilization to GuHCl denaturation (Fig. 5A). Strikingly, as assayed by SDS-PAGE, L199QHslUV degraded crosslinked NC11Arc-st11-sul20 ∼10-fold faster than did WTHslUV (Fig. 5B). Several additional experiments also show that L199QHslUV has robust unfolding activity. Fusion of Arc to a Gcn4 coiled-coil peptide increases the thermodynamic stability of both proteins by ∼4.5 kcal/mol (Baytshtok et al., 2015). L199QHslUV degraded Arc-Gcn4-st11-sul20 ∼5-fold faster than WTHslUV (Fig. 5C), but this difference was less than 2-fold for Arc-Gcn4-st11-ssrA (Fig. 5D). To confirm that degradation by L199QHslUV was faster than degradation by WTHslUV for these three substrates, we performed two additional independent replicates for the experiments shown in Fig. 5B–5D, quantified degradation by densitometry of the gels, fit each kinetic curve to a single exponential, and determined average rates and errors (Fig. 5E). We also found that a cp7 variant of superfolder GFP-sul20 (Nager et al., 2011; Wohlever et al., 2013) was degraded far more rapidly by L199QHslUV than by WTHslUV (Fig. 5F). Collectively, these results show that the sul20 degron enhances degradation by L199QHslUV to a greater extent than degradation by WTHslUV, and support a model in which C-terminal sequences play a major role in directing L199QHslUV degradation.
Figure 5. Degradation of substrates differing in structure and stability.
(A) Stability to GuHCl denaturation of N11CArc-st11-sul20, before and after crosslinking with 1,6-bismaleimidoethane, monitored by changes in tryptophan fluorescence. (B) Degradation monitored by SDS-PAGE of crosslinked N11CArc-st11-sul20 (20 µM) by WTHslUV or L199QHslUV (300 nM HslU; 500 nM HslV). (C) Degradation monitored by SDS-PAGE of Arc-Gcn4-st11-sul20 (15 µM) by WTHslUV or L199QHslUV (500 nM HslU; 1000 nM HslV). (D) Degradation monitored by SDS-PAGE of Arc-Gcn4-st11-ssrA (15 µM) by WTHslUV or L199QHslUV (500 nM HslU; 1000 nM HslV). (E) Rates of degradation of crosslinked N11CArc-st11-sul20, Arc-Gcn4-st11-sul20, and Arc-Gcn4-st11-ssrA by WTHslUV and L199QHslUV. Rates were determined by single-exponential fits of substrate remaining versus time determined by densitometry of the gels in panels B-E plus two additional independent replicates for each substrate. The values plotted are averages of the three replicates ± SD. (F) Michaelis-Menten plot for degradation of cp7GFP-sul20 by WTHslUV or L199QHslUV (500 nM HslU; 1000 nM HslV). Values are averages (n=3) ± SD. Fitted parameters for L199QHslUV are listed in Table 2. For WTHslUV, the line is a linear function and KM and Vmax were not determined. All degradation assays were performed at 37 °C in the presence of 5 mM ATP.
L199Q changes the preferred direction of degradation
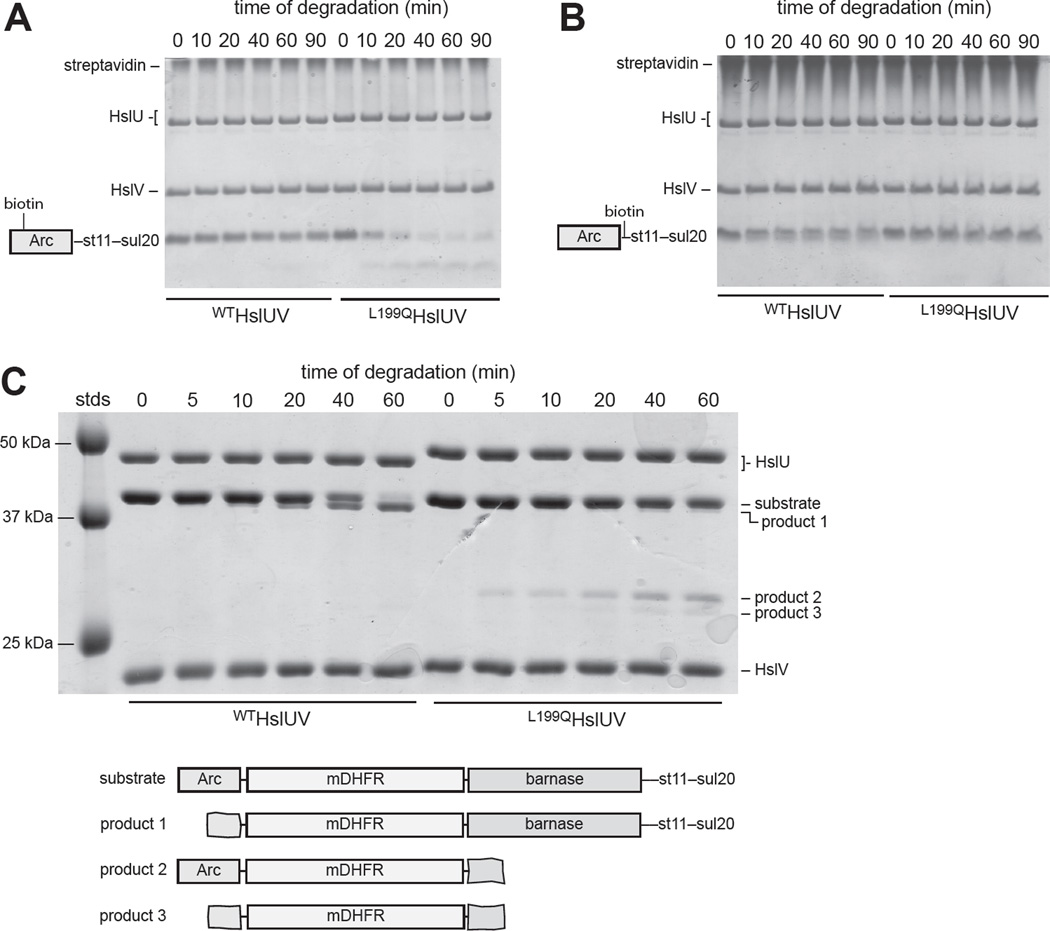
Many of our results could be explained if the L199Q mutation alters the preferred direction of HslUV degradation. To test this model, we used substrates for which degradation from either protein terminus was possible. N11CArc-st11-sul20 contains a single cysteine near the N-terminus, and Arc-cys-st11-sul20 contains a single cysteine near the C-terminus. We labeled the cysteines in these substrates with biotin-maleimide and added streptavidin to bind the biotin. As assayed by SDS-PAGE, the substrate with streptavidin bound near the N-terminus was degraded substantially faster by L199QHslUV than by WTHslUV (Fig. 6A). By contrast, the substrate with streptavidin bound near the C-terminus was degraded ∼2-fold faster by WTHslUV than by L199QHslUV (Fig. 6B).
Figure 6. Degradation directionality monitored by SDS-PAGE.
(A) WTHslUV and L199QHslUV degradation of biotinylated NC11Arc-st11-sul20 (15 µM) in the presence of 25 µM streptavidin (tetramer equivalents). (B) WTHslUV and L199QHslUV degradation of biotinylated Arc-cys-st11-sul20 (15 µM) in the presence of 25 µM streptavidin (tetramer equivalents). (C) WTHslUV and L199QHslUV degradation of Arc-mDHFR-barnase-st11-sul20 (5 µM) in the presence of 100 µM methotrexate. In all panels, the concentrations of HslU and HslV were 500 nM and 1000 nM, respectively and ATP was present at 5 mM. Assays were done at 37 °C in panels A and B and at 30 °C in panel C.
We also assayed degradation of Arc-DHFR-barnase-st11-sul20 in the presence of methotrexate, which stabilizes the DHFR domain and slows or blocks its degradation (Lee et al., 2001; Koodathingal et al., 2009). Strikingly, we observed accumulation of different truncation products for WTHslUV and L199QHslUV, supporting opposite directional preferences (Fig. 6C). The major product of WTHslUV degradation (product 1) was only a few kDa smaller than the full-length substrate (∼44 kDa), as expected if WTHslUV initiates degradation from the N-terminus, degrades part of Arc (∼6 kDa), and stalls upon encountering DHFR. By contrast, the major product of L199QHslUV degradation (product 2) was ∼30 kDa, a size expected if L199QHslUV initiates degradation from the C-terminus, degrades most of barnase (∼16 kDa), and stalls upon encountering DHFR. A minor and slightly smaller band (product 3) also accumulated during L199QHslUV degradation, which probably represents degradation from both the C-terminus and N-terminus, leaving a product slightly larger than DHFR. Likewise, a very faint band of the same size was observed during WTHslUV degradation, suggesting that the wild-type enzyme can also degrade this substrate from the C-terminus, albeit very inefficiently. Thus, although both the wild-type and mutant enzymes can initiate degradation of Arc-DHFR-barnase-st11-sul20 from either end, WTHslUV preferentially degrades this substrate from the N-terminus, whereas L199QHslUV preferentially degrades it from the C-terminus.
Discussion
The L199Q substitution in the I domain of HslU was initially reported as a loss-of-function mutation (Lien et al., 2009), and we do find that L199QHslUV is a less-active protease for some protein substrates. More importantly, however, this point mutation elevates the rate of ATP hydrolysis, results in much faster HslUV proteolysis of some protein substrates, and changes whether degradation is preferentially initiated at the substrate N- or C-terminus. These dramatic functional changes are not associated with major structural changes in the HslUV complex. For example, the L199Q mutation does not detectably change the structure of the hexameric AAA+ ring of HslU, which is responsible for ATP-fueled substrate unfolding and translocation. Similarly, the L199Q mutation does not disrupt the interaction of HslU with the HslV peptidase. Moreover, electron density for the portion of the I domain in which the L199Q mutation resides is absent in both the wild type and L199Q structures. Below, we discuss how a mutation in a segment of the I domain that is structurally dynamic might regulate ATP hydrolysis and protein degradation.
The position corresponding to Leu199 is a hydrophobic residue in HslU orthologs (Fig. 1B), and the Jpred4 homology-based secondary-structure algorithm (Drozdetskiy et al., 2015) strongly predicts that it is part of an amphipathic α-helix, extending from residues 193–204. Because this region can be proteolytically cleaved in WTHslU but becomes substantially more susceptible to cleavage in L199QHslU, we propose that this helix transiently unravels in the wild-type enzyme but is largely unfolded in the mutant because the L199Q substitution disrupts hydrophobic interactions required for helix stability. We refer to the 193–204 region as a “gating” helix, although the argument that follows is not dependent upon specific structural details. Based upon the flanking well-ordered regions of the I domain, the gating helix would fill part of the lumen of a funnel-shaped cavity above the axial pore of the AAA+ ring, and any protein substrate would need to traverse this funnel to reach the axial pore of HslU (Fig. 1A).
ATP-hydrolysis drives power strokes in the AAA+ ring of HslU and the related ring of ClpX that involve movements of rigid-body units, comprising a large AAA+ domain and a neighboring small AAA+ domain, with respect to each other (Wang et al., 2001b; Glynn et al., 2009; 2012). If the gating helices contribute to interactions between neighboring I domains that need to be broken to allow a power stroke, then the L199Q mutation could accelerate ATP hydrolysis simply by lowering the energy barrier for this rate-limiting conformational change. Alternatively, part of the gating helix might suppress ATP hydrolysis more directly by interacting with the AAA+ ring. Again, mutational disruption of such interactions could plausibly be responsible for increasing the ATP-hydrolysis rate of L199QHslU.
One striking property of L199QHslUV is that it prefers to begin degradation at the C-terminus of the model substrates tested, whereas WTHslUV prefers to initiate degradation at the N-terminus of these substrates. Because the mechanical stability of protein substrates can depend on the direction of pulling by a AAA+ enzyme (Lee et al., 2001; Kenniston et al., 2004; Gur et al., 2012), this change in directionality combined with faster ATP hydrolysis probably accounts for our finding that L199QHslUV degrades some substrates faster than WTHslUV. Similarly, for substrates easier to unfold from the N-terminus, L199QHslUV would be expected to be a poorer protease than WTHslUV. Although structures of wild-type or L199QHslU in complex with substrates may ultimately be needed to understand this change in directionality, we suggest that mutational disruption of the gating helices might affect directionality by reducing binding to certain degrons and/or improving binding to other degrons. For example, the native gating helix in WTHslU might preferentially bind C-terminal sequences of an Arc substrate, allowing the N-terminal residues to bind in the axial pore. Disruption of the gating helix in L199QHslU might then allow either the C-terminus or N-terminus of the substrate to be engaged by the pore. This model would result in faster L199QHslU degradation for substrates that are easier to unfold from the C-terminus than the N-terminus. Sites that are unmasked by disruption of the gating helices could also alter binding of N-terminal or C-terminal substrate sequences. For example, a binding or engagement site for the sul20 tag in the HslU pore might be unmasked in L199QHslU, explaining why L199QHslUV robustly degrades sul20-tagged substrates. Both types of models are consistent with our observation that WTHslUV and L199QHslUV can each degrade the same substrates, albeit often with very different efficiencies. Indeed, the binding of certain substrates might preferentially stabilize or destabilize the gating helices, altering how these proteins are engaged and degraded by HslU.
Although our models can account for changes in ATPase and degradation properties of L199QHslUV in vitro, they do not explain why this mutation prevents degradation of SulA in vivo (Lien et al., 2009). Indeed, we find that L199QHslUV degrades proteins bearing the C-terminal 20-residues of SulA faster than WTHslUV does at saturating substrate concentrations and with a higher second-order rate constant (Vmax/KM) at low substrate concentrations. In E. coli, SulA binds to FtsZ (Jones and Holland, 1985), and HslU may need to recognize both the N- and C-termini of SulA for efficient extraction of SulA from the complex and subsequent degradation. Alternatively, L199QHslU may be expressed at somewhat lower levels in vivo, competitively engage other substrates more efficiently, thereby slowing SulA degradation, or expose hydrophobic residues that result in the recruitment of cellular chaperones, thereby blocking substrate binding and engagement in the cell.
Methods
Cloning, expression, and protein purification
Unless noted, standard polymerase-chain-reaction techniques were used for cloning and site-directed mutagenesis. Genes encoding wild-type HslU or L199QHslU with N-terminal H6 tags were expressed from pET12b vectors (Novagen). The pET21b vector (Novagen) was used to express Arc, followed by a cysteine, followed by one of three tags: H6, st11-ssrA (H6KNQHDAANDENYALAA), or st11-sul20 (H6KNQHDASSHATRQLSGLKIHSNLYH). A pET21b plasmid expressing I37AArc-cp6GFP-st11-ssrA was a gift from Jiejin Chen (MIT); a related vector expressing Arc-cp6GFP-st11-ssrA was generated by reversing the I37A mutation. NC11Arc-st11-sul20 contains a glycine between residues Met1 and Lys2 of wild-type Arc, has a slightly different st11 tag (H6KNQHE), and was expressed from a pACYC-Duet plasmid constructed by Matthew Wohlever (MIT).
H6-tagged variants of HslU6 and HslV12 were overexpressed in E. coli strain X90 (λDE3) slyD::kan hslUV::tet and purified by Ni++-NTA affinity, ion-exchange, and gel-filtration chromatography using minor modifications of a published procedure (Sundar et al., 2010). Cell pellets were resuspended and lysed in buffer A (50 mM Tris, pH 7.5, 300 mM NaCl, 20 mM imidazole, 0.5 mM DTT, 0.5 mM EDTA), and 1.5 µL benzonase (250 units/µL, Sigma) per liter of culture was added. Lysates were cleared by centrifugation, polyethyleneimine was added to the supernatant to a final concentration of 0.1% (v/v), and precipitated material was removed by centrifugation. Proteins were eluted from Ni++-NTA with buffer B (50 mM Tris, pH 7.5, 300 mM NaCl, 250 mM imidazole, 0.5 mM DTT, 0.5 mM EDTA). The Ni++-NTA eluate was diluted with buffer C (50 mM Tris, pH 7.5, 10% (v/v) glycerol, 1 mM EDTA, 1 mM DTT), loaded onto a MonoQ 10/100 GL column (GE Healthcare), and eluted with a linear gradient from 0.15 to 0.5 M NaCl in buffer C. Appropriate fractions from the monoQ column were pooled, concentrated, and loaded onto a HiLoad Superdex 200 16/60 column (GE Healthcare) equilibrated in buffer D (50 mM Tris, pH 7.5, 300 mM NaCl, 10% (v/v) glycerol, 1 mM EDTA, 1 mM DTT). Fractions from this column were monitored by SDS-PAGE, and those containing HslU or HslV (> 95% purity) were pooled, concentrated, aliquoted, and frozen at –80 °C until use. HslU concentrations were calculated for hexamer equivalents; HslV concentrations were calculated for dodecamer equivalents.
Plasmids expressing Arc-cys-st11-ssrA, Arc-cys-st11-sul20, or NC11Arc-st11-sul20 were transformed into E. coli strain X90 (λDE3) slyD::kan hslUV::tet, grown to an OD600 of 0.6–0.8 at 37 °C, 1 mM IPTG was added to induce protein expression, and growth was continued for 4 h at room temperature. Cells were resuspended, lysed, and the Arc substrates were purified by Ni++-NTA affinity chromatography as described above for the HslU purification. For further purification, Arc-cys-st11-ssrA was diluted with an appropriate volume of buffer C to an ionic strength equivalent to 100 mM NaCl, loaded onto a Source 15S column (GE Healthcare), and eluted with a linear gradient from 0.1 to 0.5 M NaCl in buffer C. Appropriate fractions were pooled, exchanged into buffer D, concentrated, and frozen at –80 °C. For additional purification, Arc-cys-st11-sul20 or NC11Arc-st11-sul20 was chromatographed on a HiLoad Superdex 75 16/60 column (GE Healthcare) equilibrated in buffer D. Appropriate fractions were pooled, diluted with 0.66 volumes of buffer C, loaded onto the Source 15S column, and eluted with a linear gradient from 0.2 to 0.6 M NaCl in buffer C. Arc-GFP-st11-ssrA and I37AArc-GFP-st11-ssrA were purified in the same manner as Arc-cys-st11-ssrA except an additional gel-filtration step using a HiLoad Superdex 200 16/60 equilibrated in buffer D was performed. Arc-Gcn4-st11-ssrA was purified as described (Baytshtok et al., 2015). Arc-Gcn4-st11-sul20 was purified as described for Arc-Gcn4-st11-ssrA. Monomeric, H6-tagged SspB contained the L6R, R9E, Y12Q, L13K, A16E, F17K, W20E, and D23K mutations (McGinness et al., 2007) and was purified as described for Arc-cys-st11-ssrA except a monoQ 10/100 GL column was used for the ion-exchange step.
Electron microscopy
HslUV complexes were assembled by mixing wild-type or L199Q HslU (1 µM), HslV (0.5 µM), and freshly prepared ATPγS (5 mM) in 20 mM HEPES, pH 7.5, 5 mM MgCl2, 500 mM NaCl, 10% glycerol (v/v), and 0.032% NP40. After 15 min at room temperature, the mixture was diluted 10-times in EM buffer (20 mM HEPES, pH 7.5, 5 mM MgCl2, 500 mM NaCl and 5 mM ATPγS) and immediately applied on formvar/carbon grids, and stained with 0.7% uranyl formate. Electron micrographs of single particles were recorded with a 2k×2k Gatan CCD using a FEI Tecnai T12 instrument at 120 kev with a magnification of 67000×. Relion1.3 (Scheres, 2012a; 2012b) was used for single-particle picking, particle extraction, and reference-free 2D class averages. CTFFIND4 (Rohou and Grigorieff, 2015) was used to estimate contrast transfer functions. For WTHslUV, 10235 particles from 25 micrographs were picked initially, and 5200 particles were used to generate representative 2D class averages. For L199QHslUV, 6164 particles from 26 micrographs were picked initially, and 3313 particles were used to generate representative 2D class averages.
Crystallization and structure determination
H6-tagged L199QHslU and HslV were exchanged into buffer X (15 mM Tris, pH 7.5, 100 mM NaCl, 20 mM MgCl2, 0.2 mM EDTA) using Micro Bio-Spin 6 (Bio-Rad) columns and mixed with Arc-st11-ssrA and 5 mM ATP to a final concentration of 25 µM L199QHslU (7.5 mg/mL), 40 µM HslV (9.7 mg/mL), and 60 µM Arc-st11-ssrA (0.5 mg/mL). Hanging drops containing 0.5 µL of this mixture and 0.5 µL of the mother liquor (100 mM Bis-Tris, pH 5.5, 1.8 M ammonium sulfate) were incubated at 4 °C for crystallization. Large hexagonal crystals appeared within 1–2 days. Diffraction data for the L199QHslUV crystal were collected at the Advanced Photon Source (APS) beamline 24-ID-E at 100 K (wavelength 0.9792 Å) and were processed using HKL2000 (Otwinowski and Minor, 1997). The structure was solved by molecular replacement using PHASER (McCoy et al., 2007) and the 1G4A HslUV structure (Wang et al., 2001a) as a search model. Structure factors for re-refinement of 1G4A were obtained from the Protein Data Bank. All refinement was performed using PHENIX with Cartesian non-crystallographic restraints for identical chains in the asymmetric unit (Adams et al., 2010), model building was carried out using COOT (Emsley and Cowtan, 2010), and MolProbity was used to assess model geometry (Chen et al., 2010).
Fluorescent labeling of Arc-cys variants
Prior to labeling, a G25 column (GE Healthcare) was used to exchange Arc-cys-st11-ssrA into buffer E (50 mM Tris, pH 7.5, 300 mM NaCl, 1 mM EDTA) or to exchange Arc-cys-st11-sul20 into buffer F (50 mM Tris, pH 7.5, 300 mM NaCl, 5% (v/v) glycerol, 0.032% (v/v) Igepal CA-630, 1 mM EDTA). Alexa488-C5-maleimide (Thermo Fisher) was added to the buffer-exchanged samples in a 4:1 molar ratio of fluorophore to protein (in monomer equivalents), and the reaction was incubated in the dark for 2 h at room temperature. The reaction was quenched by addition of 5 mM DTT, and the labeled product was sequentially chromatographed on a PD-10 desalting column (GE Healthcare) and a HiLoad Superdex 75 16/60 column, both equilibrated in buffer D. Appropriate Superdex-75 fractions were pooled, concentrated, and the final concentration was determined by the Bradford assay (Bio-Rad) using unlabeled Arc-cys variants as standards.
Crosslinked NC11Arc-st11-sul20 dimer
NC11Arc-st11-sul20 was exchanged into buffer F using sequential Micro Bio-Spin 6 columns. Bismaleimidoethane (BMOE, Thermo Scientific) was added to a concentration of 1 mM to the sample, the reaction was incubated for 2 h at room temperature, 5 mM DTT was added as a quencher, and the protein was exchanged into buffer D using a Micro Bio-Spin 6 column. SDS-PAGE showed that the crosslinking reaction was >98% complete. For stability measurements, samples of non-crosslinked or BMOE-crosslinked NC11Arc-st11-sul20 (5 µM each) were incubated with increasing concentrations of guanidine hydrochloride in buffer G (50 mM Tris, pH 7.5, 300 mM NaCl, 1 mM EDTA, 5 mM DTT). After 20 h at room temperature, tryptophan fluorescence was excited at 295 nm, the emission spectra from 300 to 400 nm emission was taken using a PTI QM-2000-4SE spectrofluorimeter, and the center of fluorescence spectral mass was calculated to measure unfolding (Robinson and Sauer, 2000).
Biotinylation
NC11Arc-st11-sul20 and Arc-cys-st11-sul20 were exchanged into buffer F using sequential Micro Bio-Spin 6 columns, EZ-link maleimide-PEG2-biotin (Thermo Scientific) was added to a final concentration of 2 mM, and the reaction was incubated at room temperature for 1.5 h before quenching with 10 mM dithiothreitol. Biotinylated proteins were then exchanged into buffer D using sequential Micro Bio-Spin 6 columns.
Biochemical assays
Assays were performed in PD buffer (25 mM HEPES, pH 7.5, 5 mM KCl, 20 mM MgCl2, 10% glycerol, 0.032% Igepal CA-630) unless noted. ATPase rates at 37 °C were measured using a Spectramax M5 plate reader (Molecular Devices) by monitoring changes in absorbance at 340 nm using an NADH-coupled assay (Nørby, 1988). Cleavage of Z-Gly-Gly-Leu-AMC (200 µM; Santa Cruz Biotechnology, Inc.) by HslV (5 nM) was measured at 25 °C in the presence of 5 mM ATP and varying concentrations of HslU using a Spectramax M5 plate reader (excitation 380 nm; emission 455 nm).
Unless noted, degradation experiments monitored by SDS-PAGE were performed at 37 °C with 5 mM ATP and an ATP-regeneration system (16 mM creatine phosphate, 10 µg/mL creatine kinase). Degradation of GFP variants tagged with Arc, I37AArc, or sul20 was monitored by loss of fluorescence (excitation 467 nm; emission 511 nm). Degradation of Arc-cysA-st11-ssrA and Arc-cysA-st11-sul20 variants labeled with Alexa488 was monitored by increase in fluorescence (excitation 480 nm; emission 520 nm) on a Spectramax M5 plate reader at 37 °C in the presence of 5mM ATP and an ATP-regeneration system. When degradation of Arc-cysA variants did not go to completion, trypsin was added to a final concentration of 40 µg/mL to obtain the final fluorescence values. Rates for degradation of the Arc-cysA variants were calculated by dividing the initial linear slope by the total amplitude of the fluorescence change, and multiplying this quotient by the concentration of substrate. Degradation of biotinylated NC11Arc-st11-sul20 or Arc-cys-st11-sul20 (15 µM each) was performed in the presence of streptavidin tetramer (25 µM; Sigma, salt-free). At different times, aliquots were taken, added to SDS-PAGE loading buffer containing 10 mM d-biotin, and heated at 80 °C for 5 min before gel electrophoresis. Degradation of Arc-mDHFR-barnase-st11-sul20 was performed at 30 °C in the presence of 100 µM methotrexate.
Limited proteolysis experiments were performed at 37 °C in the presence of 5 mM ATP, 5 mM ADP, or the absence of nucleotide. HslU (0.5 µM) was pre-incubated with ATP in the presence or absence of HslV (1 µM) for 30 s and 11 nM subtilisin (Carlsberg variant; Sigma Aldrich) or 60 nM chymotrypsin (TLCK-treated, Worthington Biochemical Corporation) was added. Aliquots were taken at different times and mixed with SDS-PAGE loading buffer to quench the reaction. To purify subtilisin cleavage products for LC MS/MS, the reaction was quenched after 6 min by the addition of 1 mM phenylmethylsulfonyl fluoride (PMSF), diluted with buffer H (50 mM Tris, pH 7.5, 7 M urea, 1 mM DTT, 1 mM EDTA), loaded onto a source 15Q column (GE Healthcare), and eluted with a linear gradient from 0.1 to 1.0 M NaCl in buffer H. A peak containing the major fragments was run on SDS-PAGE, the ∼25 kDa bands were extracted from the gel and digested with trypsin, and LC MS/MS analysis was performed by the MIT Koch Institute Biopolymers and Proteomics Core Facility. To purify chymotrypsin cleavage products, the reaction was quenched after 6 min by addition of 1 mM PMSF, H6-tagged proteins and associated partners were bound to Ni++-NTA affinity in buffer A, and proteins eluted with buffer B were subjected to MALDI-TOF analysis by the MIT Koch Institute Biopolymers and Proteomics Core Facility. For sequential Edman degradation, L199QHslU was incubated with chymotrypsin for 6 min, reaction products were separated by SDS-PAGE, and the top band of a doublet running near 25 kDa was excised from the gel and submitted to the Tufts Core Facility for sequencing.
Highlights.
The L199Q mutation in HslUV alters the preferred direction of substrate degradation
EM and crystallography reveal very similar L199QHslUV and WTHslUV structures
L199QHslUV degrades certain protein substrates substantially faster than WTHslUV
L199Q increases the susceptibility of the I domain to limited proteolysis
Acknowledgments
We thank M. Bochtler, J. Chen, S.C. Harrison, A. Olivares, K. Schmitz, S. Sundar, and M. Wohlever for materials, advice, help, and use of equipment. Supported by NIH grant AI-16892. T.A.B. is an employee of the Howard Hughes Medical Institute. Studies at the North East Collaborative Access Team 24-ID-E beam line of the Advanced Photon Source were supported by the NIH National Institute of General Medical Sciences Grant P41 GM103403 and the US Department of Energy under Contract DE-AC02-06CH11357. EM studies were supported by an NIH program project grant (GM-62580) to S.C. Harrison.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers. The PDB accession numbers for the structures reported in this paper are 5JI2 and 5JI3.
Author contributions. V.B., T.A.B., and R.T.S. planned experiments. V.B. performed all biochemical studies. X.F. performed EM studies. V.B., R.A.G, and R.T.S. performed crystallographic studies. V.B. and R.T.S. wrote the manuscript with input from other authors.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baytshtok V, Baker TA, Sauer RT. Assaying the kinetics of protein denaturation catalyzed by AAA+ unfolding machines and proteases. Proc. Natl. Acad. Sci. USA. 2015;112:5377–5382. doi: 10.1073/pnas.1505881112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HslU and the ATP-dependent protease HslU-HslV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- Burton RE, Baker TA, Sauer RT. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat. Struct. Mol. Biol. 2005;12:245–251. doi: 10.1038/nsmb898. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Crystal structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Nager AR, Baker TA, Sauer RT. Dynamic and static components power unfolding in topologically closed rings of a AAA+ proteolytic machine. Nat. Struct. Mol. Biol. 2012;19:616–622. doi: 10.1038/nsmb.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Sauer RT. Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proc. Natl. Acad. Sci. USA. 2009;106:18503–8508. doi: 10.1073/pnas.0910392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Vishkautsan M, Sauer RT. Protein unfolding and degradation by the AAA+ Lon protease. Protein Sci. 2012;21:268–278. doi: 10.1002/pro.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Holland IB. Role of the SulB (FtsZ) protein in division inhibition during the SOS response in Escherichia coli: FtsZ stabilizes the inhibitor SulA in maxicells. Proc. Natl. Acad. Sci. USA. 1985;82:6045–6049. doi: 10.1073/pnas.82.18.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemori M, Nishihara K, Yanagi H, Yura T. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of sigma32 and abnormal proteins in Escherichia coli. J. Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemori M, Yanagi H, Yura T. Marked instability of the sigma(32) heat shock transcription factor at high temperature. Implications for heat shock regulation. J. Biol. Chem. 1999;274:22002–22007. doi: 10.1074/jbc.274.31.22002. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J. Struct. Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J. Biol. Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MS, Chen KP, Wu WF. Regulation of RcsA by the ClpYQ (HslUV) protease in Escherichia coli. Microbiology. 2004;150:437–446. doi: 10.1099/mic.0.26446-0. [DOI] [PubMed] [Google Scholar]
- Kwon AR, Kessler BM, Overkleeft HS, McKay DB. Structure and reactivity of an asymmetric complex between HslV and I-domain deleted HslU, a prokaryotic homolog of the eukaryotic proteasome. J. Mol. Biol. 2003;330:185–195. doi: 10.1016/s0022-2836(03)00580-1. [DOI] [PubMed] [Google Scholar]
- Kwon AR, Trame CB, McKay DB. Kinetics of protein substrate degradation by HslUV. J. Struct, Biol. 2004;146:141–147. doi: 10.1016/j.jsb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Lien HY, Shy RS, Peng SS, Wu YL, Weng YT, Chen HH, Su PC, Ng WF, Chen YC, Chang PY, Wu WF. Characterization of the Escherichia coli ClpY (HslU) substrate recognition site in the ClpYQ (HslUV) protease using the yeast two-hybrid system. J. Bacteriol. 2009;191:4218–4231. doi: 10.1128/JB.00089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Bolon DN, Kaganovich M, Baker TA, Sauer RT. Altered tethering of the SspB adaptor to the ClpXP protease causes changes in substrate delivery. J. Biol. Chem. 2007;282:11465–11473. doi: 10.1074/jbc.M610671200. [DOI] [PubMed] [Google Scholar]
- Milla ME, Sauer RT. P22 Arc repressor: folding kinetics of a single-domain dimeric protein. Biochemistry. 1994;33:1125–1133. doi: 10.1021/bi00171a011. [DOI] [PubMed] [Google Scholar]
- Milla ME, Brown BM, Sauer RT. P22 Arc repressor: enhanced expression of unstable mutants by addition of polar C-terminal sequences. Protein Sci. 1993;2:2198–2205. doi: 10.1002/pro.5560021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla ME, Brown BM, Sauer RT. Protein stability effects of a complete set of alanine substitutions in Arc repressor. Nat. Struct. Biol. 1994;1:518–523. doi: 10.1038/nsb0894-518. [DOI] [PubMed] [Google Scholar]
- Nager AR, Baker TA, Sauer RT. Stepwise unfolding of a β-barrel protein by the AAA+ ClpXP protease. J. Mol. Biol. 2011;413:4–16. doi: 10.1016/j.jmb.2011.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; Erratum J. Mol. Biol. 425:1241–1243. [Google Scholar]
- Nørby JG. Coupled assay of Na+, K+-ATPase activity. Methods Enz. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure—diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enz. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Robinson CR, Sauer RT. Striking stabilization of Arc repressor by an engineered disulfide bond. Biochemistry. 2000;39:12494–12502. doi: 10.1021/bi001484e. [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrwild M, Coux O, Huang HC, Moerschell RP, Yoo SJ, Seol JH, Chung CH, Goldberg AL. HslV-HslU: A novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc. Natl. Acad. Sci. USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Scheres SH. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012a;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012b;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong IS, Oh JY, Yoo SJ, Seol JH, Chung CH. ATP-dependent degradation of SulA, a cell division inhibitor, by the HslVU protease in Escherichia coli. FEBS Lett. 1999;456:211–214. doi: 10.1016/s0014-5793(99)00935-7. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Striebel F, Kress W, Weber-Ban E. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr. Opin. Struct. Biol. 2009;19:209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Sundar S, Baker TA, Sauer RT. The I domain of the AAA+ HslUV protease coordinates substrate binding, ATP hydrolysis, and protein degradation. Protein Sci. 2012;21:188–198. doi: 10.1002/pro.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar S, McGinness KE, Baker TA, Sauer RT. Multiple sequence signals direct recognition and degradation of protein substrates by the AAA+ protease HslUV. J. Mol. Biol. 2010;403:420–429. doi: 10.1016/j.jmb.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001a;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HslU. Structure. 2001b;9:1107–1016. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- Wohlever ML, Nager AR, Baker TA, Sauer RT. Engineering fluorescent protein substrates for the AAA+ Lon protease. Protein Eng. Des. Sel. 2013;26:299–305. doi: 10.1093/protein/gzs105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WF, Zhou Y, Gottesman S. Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J. Bacteriol. 1999;181:3681–3687. doi: 10.1128/jb.181.12.3681-3687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]