SUMMARY
PRDM16 is a transcription cofactor that plays critical roles in development of brown adipose tissue (BAT) as well as maintenance of adult hematopoietic and neural stem cells. Here we report that PRDM16 is a histone H3 K4 methyltransferase on chromatin. Mutation in the N-terminal PR-domain of PRDM16 completely abolishes the intrinsic enzymatic activity of PRDM16. We show that the methyltransferase activity of PRDM16 is required for specific suppression of MLL leukemogenesis both in vitro and in vivo. Mechanistic studies show that PRDM16 directly activates the SNAG family transcription factor GFI1b, which in turn down regulates the HOXA gene cluster. Knockdown GFI1b represses PRDM16-mediated tumor suppression while GFI1b overexpression mimics PRDM16 overexpression. In further support of the tumor suppressor function of PRDM16, silencing PRDM16 by DNA methylation is concomitant with MLL-AF9 induced leukemic transformation. Taken together, our study reveals a previously uncharacterized function of PRDM16 that depends on its PR-domain activity.
INTRODUCTION
PRDM16 (also known as MEL, PFM13) encodes a protein of 1,247 amino acids and is comprised of an N-terminal PR domain (PRD1-BF1 and RIZ1 homologous) and several zinc fingers at C-terminus (Figure 1A) (Fog et al., 2012). PRDM16 was initially characterized as an essential factor that regulates the cell-fate switch between brown adipose tissue (BAT) and skeletal myoblasts (Kajimura et al., 2010; Seale et al., 2011). Recent studies show that PRDM16 is highly enriched in multiple adult stem cell lineages and plays important roles in regulating stem cell homeostasis (Aguilo et al., 2011; Chuikov et al., 2010). PRDM16 is also able to suppress stem cell deficiency induced by Bmi1 deletion (Chuikov et al., 2010). Loss-of-function of PRDM16 leads to multi-lineage defects including hematopoiesis, neurogenesis and palotogenesis in vivo (Bjork et al., 2010; Deneault et al., 2009; Endo et al., 2012; Horn et al., 2011).
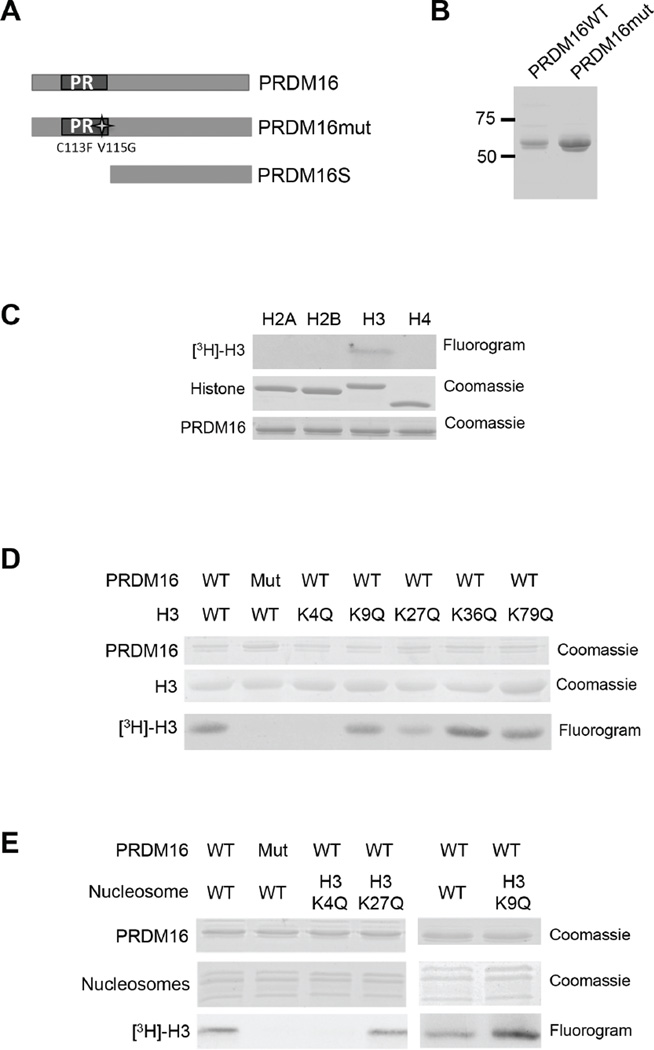
Figure 1. PRDM16 specifically methylates histone H3 K4 on nucleosomes.
(See also Figure S1). A. Schematic representation of PRDM16, PRDM16mut and PRDM16S. The mutations in PRDM16mut are shown on bottom of the PR domain. B. Coomassie-stained SDS-PAGE gel of the His-tagged PR-domains of PRDM16 and PRDM16mut purified from insect cells. C, D, E. In vitro HMT assays using free recombinant histones (C), recombinant wild type histone H3 or H3 mutants with individual lysine to glutamine mutations (D), or recombinant nucleosomes (E) as the substrates. The fluorograms for [3H]-H3 methylation as well as coomassie gels for loading controls were included as indicated on right.
PRDM16 resides in chromosome 1p36, which is frequently deleted or rearranged in multiple human cancers (Martinez-Climent et al., 2003; Mochizuki et al., 2000; Morishita, 2007; White et al., 2005). At least two protein isoforms are encoded by PRDM16: the full-length PRDM16 and the shorter isoform PRDM16S (also known as MELS) that lacks the PR domain (Nishikata et al., 2003). These two PRDM16 isoforms are transcribed from distinct transcription start sites (TSS) and are subject to differential regulation by DNA methylation (Yoshida et al., 2004). In particular, TSS of PRDM16, but not PRDM16S, resides in large regions of CpG islands (CGIs). PRDM16 silencing by DNA hypermethylation has been described in several cancers (Morishita, 2007; Tan et al., 2014). Consistent with differential regulation in cancer, aberrant expression of PRDM16S has been described in leukemia (Du et al., 2005b; Mochizuki et al., 2000; Nishikata et al., 2003; Xiao et al., 2006). Since genetic loss of function study in mouse models often simultaneously disrupts both PRDM16 isoforms, the specific function of PRDM16, especially its PR-domain, in cancer remains unclear.
PRDM16 is a transcription co-activator that acts in concert with transcription factors such as PPAR-γ (peroxisome proliferator-activated receptor), PPAR-β, PGC-1α as well as CEBP-β (CCAAT/enhancer-binding protein) in promoting expression of thermogenic genes in vivo (Kajimura et al., 2009; Seale et al., 2008; Seale et al., 2007). PRDM16 also directly interacts with the MED1 subunit of the Mediator complex and enhances thyroid hormone receptor (TR)-driven transcription in a Mediator-dependent manner (Iida et al., 2015). These functions involve direct interactions between PRDM16 C-terminal zinc fingers and transcription cofactors. It remains unclear whether the N-terminal PR-domain plays a role in transcription regulation. The PR domain of PRDM16 is a variant of the SET (Suv39, E(z) and Trithorax) domain that is commonly found in histone lysine methyltransferases (HKMT) (Jenuwein and Allis, 2001). It was reported that the PR domain of PRDM16 has intrinsic HKMT activity and is able to mono-methylate a 6-mer H3 peptide that harbors histone H3 lysine (K) 9 (H3K9) in vitro (Pinheiro et al., 2012). However, it was not determined whether PRDM16 is able to methylate other lysine residues on histones and whether it has substrate specificity on nucleosomes (Pinheiro et al., 2012). Furthermore, the function of the intrinsic methyltransferase activity of PRDM16 was not directly established in cells (Pinheiro et al., 2012).
By rigorous biochemical analyses, we demonstrate that PRDM16 is a highly specific histone methyltransferase that target histone H3 lysine 4 (H3K4) on nucleosomes. Mutating H3K4 to glutamine (Q) completely abolishes PRDM16 dependent histone methylation in vitro. Furthermore, the HKMT activity of PRDM16 is essential for suppressing mixed lineage leukemia (MLL). Overexpression or depletion of PRDM16 significantly alters MLL progression in vivo. Mechanistic studies reveal an important regulatory network involving PRDM16, the SNAG family transcription factor GFI1b and HOXA genes that is regulated by PRDM16 activity. Taken together, our studies provide insights into the sequence of events that lead to MLL by uncovering a PR-domain dependent tumor suppression function for PRDM16.
RESULTS
PRDM16 is a specific H3 K4 methyltransferase on chromatin
In order to establish the substrate specificity of PRDM16 on chromatin, we purified the PR/SET domains of PRDM16 and a PRDM16 mutant (PRDM16mut) carrying two amino acid mutations (C113F and V115G) using the insect cell expression system (Figure 1A and 1B). The PR-domain mutations did not affect thermostability of PRDM16 (Supplemental Figure 1A), suggesting minimal perturbation of the protein structure. The methyltransferase activity of PRDM16 was first tested on free histones. PRDM16 specifically methylates histone H3 but not other core histones (Figure 1C). To explore the substrate specificity, we generated histone mutants, in which lysine at positions 4, 9, 27, 36, or 79 was each mutated to glutamine (Q). We found that wild-type PRDM16 failed to methylate H3K4Q while it was able to methylate H3 that contained other lysine mutation (i.e. K9Q, K27Q, K36Q and K79Q) in vitro (Figure 1D). As the control, PRDM16mut showed no HKMT activity under the same condition, confirming that H3K4 methylation was due to the intrinsic activity of the PR-domain (Figure 1D). To examine PRDM16 specificity on nucleosomes, we used the reconstituted nucleosomes that contained wild type H3, H3K4Q, H3K9Q or H3K27Q mutant as the substrate. As shown in Figure 1E, PRDM16 could not methylate H3K4Q-containing nucleosomes whereas it had robust activity on nucleosomes comprised of H3K9Q or H3K27Q. Both mass spectrometry (Supplemental Figure 1B) and immunoblot (Supplemental Figure 1C) showed that PRDM16 was able to mono- and di-methylate H3K4 in nucleosomes. No tri-methylation of H3K4 or methylation of other lysine residues of H3 was detected by mass spec (Supplemental Figure 1B and data not shown). Taken together, we conclude that the PR domain of PRDM16 has intrinsic HKMT activity that is specific for nucleosomal H3 K4 in vitro.
PRDM16 methyltransferase activity is essential to suppress MLL-AF9 leukemogenesis
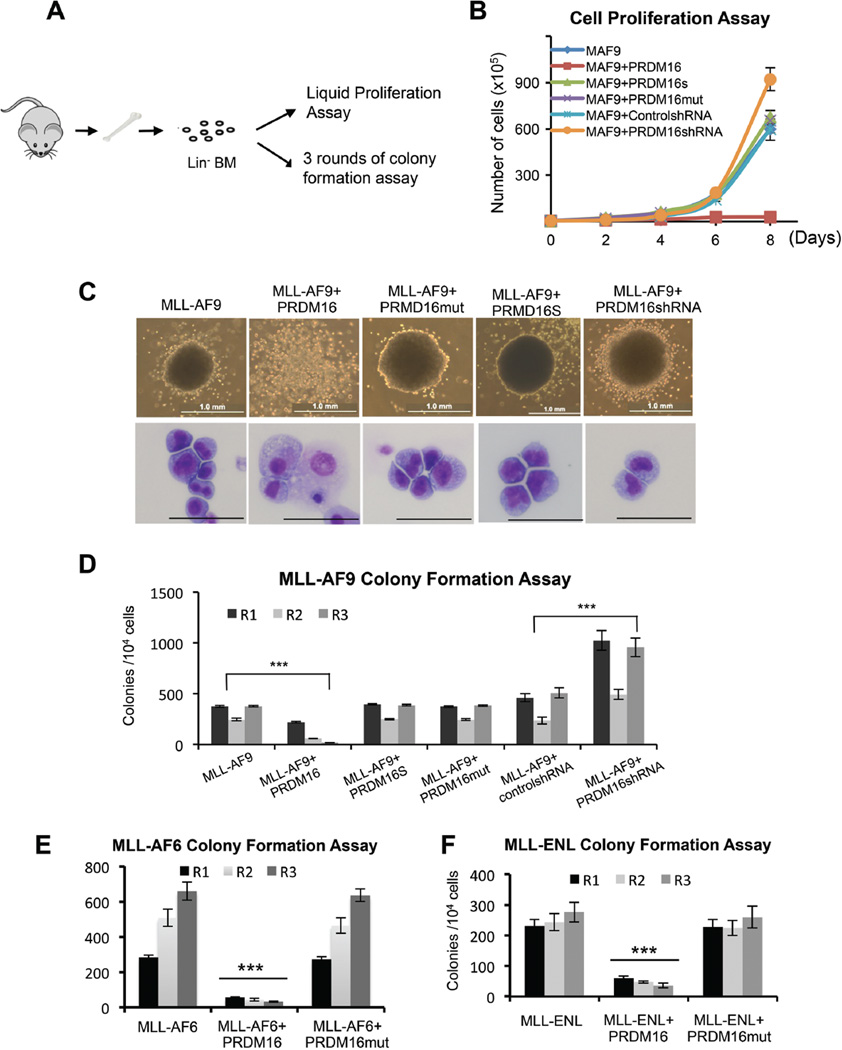
Since PRDM16 is a direct target of mixed lineage leukemia protein (MLL1, also called MLL, ALL1, HTRX and KMT2A) (Artinger et al., 2013), we wondered whether PRDM16 and more specifically, its PR domain, played a role in MLL-rearranged leukemia. We co-transduced Lin− murine bone marrow (BM) cells with retroviruses expressing Prdm16, inactive Prdm16mut or the PR-less Prdm16S isoform together with MLL-AF9 (Supplemental Figure 1D). The proliferations of co-transduced MLL-AF9+Prdm16, MLL-AF9+Prdm16mut or MLL-AF9+Prdm16S cells were measured by the liquid proliferation and myeloid colony formation assays (Figure 2A). As shown in Figure 2B and 2C, overexpression of Prdm16 completely inhibited MLL-AF9-induced leukemic transformation. It reduced cell proliferation and clonogenicity of MLL-AF9 cells. No compact colonies on the methylcellulose medium were observed for MLL-AF9+Prdm16 cells beyond second plating (Figure 2C and 2D). Furthermore, Wright-Giemsa staining showed that MLL-AF9+Prdm16 co-transduced cells largely differentiated into monocyte-like cells. (Figure 2C, bottom panel). In contrast, MLL-AF9, MLL-AF9+Prdm16mut and MLL-AF9+Prdm16S cells formed compact colonies in serial plating and the transformed cells remained undifferentiated by Wright-Giemsa staining (Figure 2C and 2D). To confirm that endogenous Prdm16 inhibits MLL-AF9 leukemogenesis, we co-transduced MLL-AF9 with either control shRNA or Prdm16 shRNA (Supplemental Figure 1E). We found that Prdm16 knockdown significantly enhanced clonogenicity of the MLL-AF9-transformed cells in serial plating (Two-way ANOVA analysis, p<0.0001, Figure 2D) and increased cell proliferation in liquid culture (Figure 2B). These results indicate that PRDM16 suppresses MLL-AF9-mediated leukemic transformation in vitro.
Figure 2. PRDM16 methyltransferase activity inhibits MLL-AF9 transformation in vitro.
(See also Figure S1, S2). A. Schematic for the in vitro proliferation and colony formation experiments. B. Cell proliferation assay for MLL-AF9 (MAF9) with or without overexpression of PRDM16, PRDM16mut, PRDM16S as well as MLL-AF9 with or without PRDM16 shRNA mediated depletion. Error bars indicate standard deviation (SD) from duplicates. The results were repeated at least three times. C. Representative colonies (top) and Wright-Giemas-stained cells (bottom) from the tertiary plating were shown. Scan bar: 50µm. Genes used in co-transduction were indicated on top. D. Myeloid colony formation assay for co-transduced bone marrow cells as indicated on bottom. Colony counts were summarized from primary, secondary and tertiary plating on methycellulose medium in the presence of IL3, IL6, SCF and GM-CSF. Error bars indicate SD from duplicates. The results were repeated at least three times. (E, F). Myeloid colony formation assay for co-transduced bone marrow cells as indicated on bottom. Means and standard deviations (as error bars) were derived from at least three experiments. For (D–F), ***, p<0.0001, two-way ANOVA test.
PRDM16 specifically inhibits the MLL leukemic transformation
To determine whether PRDM16 suppresses leukemic transformation by other oncogenes, we performed similar studies in which primary BM cells were transformed by additional MLL fusion genes (i.e. MLL-AF6 and MLL-ENL) as well as non-MLL oncogene E2A-HLF. Interestingly, we found that Prdm16, but not Prdm16mut, specifically inhibits leukemic transformation by MLL fusion genes. It reduced myeloid colony formation on methylcellulose and decreased cell proliferation in liquid culture (Figure 2E and 2F and Supplemental Figure 1F). Interestingly, overexpression of Prdm16, Prdm16S or Prdm16mut had indistinguishable effects when co-transduced with E2A-HLF (Supplemental Figure 1G). They did not affect cell proliferation in liquid culture (Supplemental Figure 2A), colony formation in serial plating (Supplemental Figure 2B) or maintenance of leukemic blasts (Supplemental Figure 2C). Together, these results suggest that PRDM16 specifically inhibits MLL fusion mediated leukemogenesis in vitro and this function requires the PR domain activity.
PRDM16 alters MLL leukemia progression in vivo
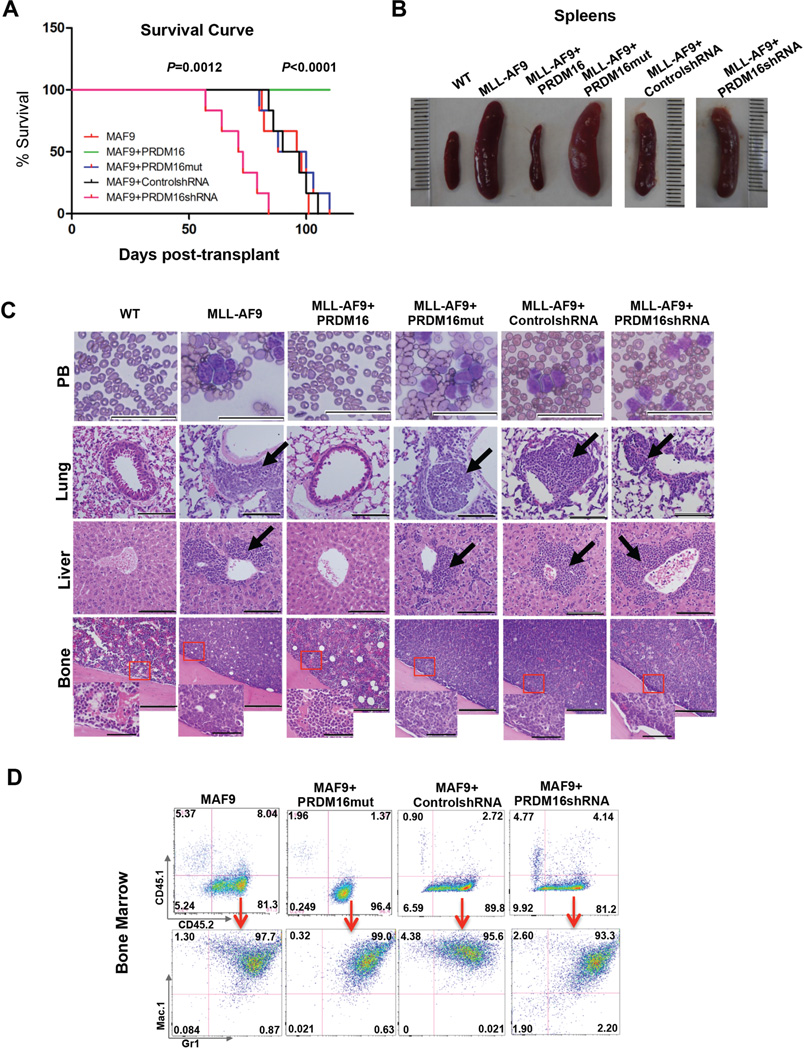
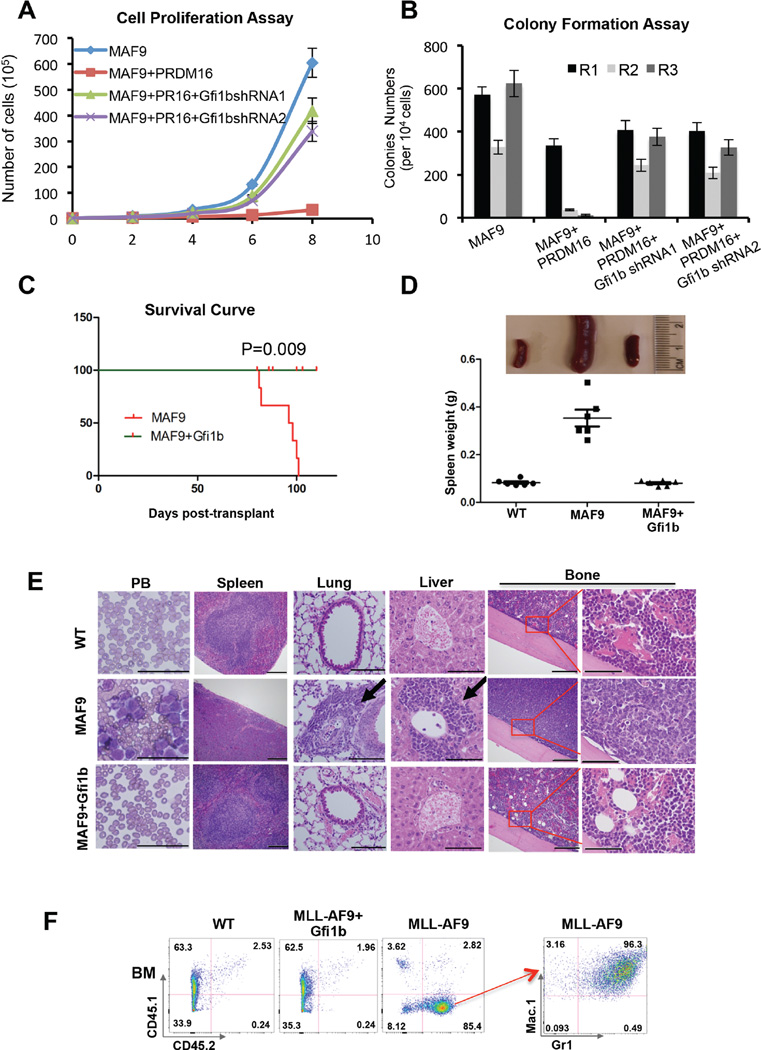
To further explore the role of PRDM16 and its HMT activity in MLL leukemogenesis, we isolated BM cells from C57BL/6 mice (CD45.2+), and co-transduced them with MLL-AF9+vector control, MLL-AF9+Prdm16 or MLL-AF9+Prdm16mut, respectively. About 1x105 cells were competitively transplanted into the lethally irradiated B6.SJL mice (CD45.1+), together with supporting CD45.1+ cells (Muntean et al., 2010). The engrafted mice, six in each cohort, were monitored for acute myeloid leukemia (AML). Flow cytometric analyses at 48-hour post-transplantation showed similar number of engrafted CD45.2+ BM cells in recipient mice (Supplemental Figure 2D). Importantly, mice receiving MLL-AF9+vector control or MLL-AF9+Prdm16mut developed acute myeloid leukemia (AML) about 80 days post transplantation (Figure 3A). They showed obvious emaciation and significant weight loss (Supplemental Figure 2E). All mice receiving MLL-AF9+vector control or MLL-AF9+Prdm16mut cells died before 110 days with splenomegaly and hepatomegaly (Figure 3B and Supplemental Figure 2E). Leukemic blasts were detected in peripheral blood and infiltrated into bone, spleen, liver and lung of these mice (Figure 3C and Supplemental Figure 2F). In contrast, none of the MLL-AF9+Prdm16 recipient mice developed AML by the end point of the study (Figure 3A, Log-rank (Mantel-Cox) test, p<0.0001). No notable defects in peripheral blood and hematopoietic organs in MLL-AF9+Prdm16 recipient mice were observed (Figure 3B and 3C). Flow cytometric analysis of isolated BM cells and splenocytes from each cohort supported the histologic findings. Significant amplification of CD45.2+ cells was found in BM and spleens of MLL-AF9 and MLL-AF9+Prdm16mut recipient mice. Majority of these cells expressed Mac1high and Gr-1high surface makers, as expected for AML (Muntean et al., 2010) (Figure 3D and Supplemental Figure 2G). In contrast, very few CD45.2+ cells were found in MLL-AF9+Prdm16 recipients, consistent with a disease-free phenotype. These results suggest an essential role of PRDM16 HMT activity in suppressing MLL-AF9 leukemogenesis in vivo.
Figure 3. PRDM16 represses MLL-AF9 leukemia in vivo.
(See also Figure S2). A. Kaplan-Meier survival curves for recipient mice engrafted with cells as indicated (n=6). Mantel-Cox test was performed to obtain p values for overexpression (p<0.0001) and knockdown (p=0.0012) experiments, respectively. B. The representative images of the spleens from recipient mice at the end point of the study. C. Representative Wright-Giemsa staining of peripheral blood (PB) smear (Scan bar: 50µm) and H&E staining of lung (Scan bar: 100µm), liver (Scan bar: 100µm) and bone (Scan bar: 200um and 50µm) as indicated on right. Co-transduced genes were indicated on top. D. Representative flow cytometry analyses of bone marrow cells isolated for each group of recipient mice as indicated on top. Top row: antibodies against CD45.1 and CD45.2 surface markers to separate supporter cells and donor cells. Bottom row: antibodies against myeloid surface markers Mac-1 and Gr-1 to identify leukemic cell population. Percentage of cells for each immnophenotype was indicated in each quadrant.
In contrast to Prdm16 overexpression, depletion of endogenous Prdm16 during MLL-AF9 transduction greatly accelerated MLL leukemogenesis. We co-transduced CD45.2+ BM cells with retroviruses expressing MLL-AF9 and Prdm16 shRNAs or control shRNAs and transplanted the cells into CD45.1+ recipient mice. As shown in Figure 3A, all mice receiving cells expressing MLL-AF9+Prdm16 shRNA developed AML. Importantly, onset of AML in these mice was ~50 days post transplantation, which was significantly earlier than mice that were grafted with MLL-AF9-transduced cells alone. All mice receiving MLL-AF9+Prdm16 shRNA cells died before 84 days (Figure 3A, Log-rank (Mantel-Cox) test, p=0.0012). Histologic studies showed that these mice had typical AML phenotypes that were indistinguishable from MLL-AF9 leukemia (Figure 3B–3D and Supplemental Figure 2E–2G). Given the much shortened disease latency for mice grafted with MLL-AF9+Prdm16shRNA cells, our results signify that PRDM16 alters the trajectory of MLL leukemogenesis in vivo and that down regulation of endogenous PRDM16 is probably one of the rate-limiting steps for the onset of MLL leukemia.
PRDM16 suppresses leukemic transformation by repressing the HoxA gene cluster
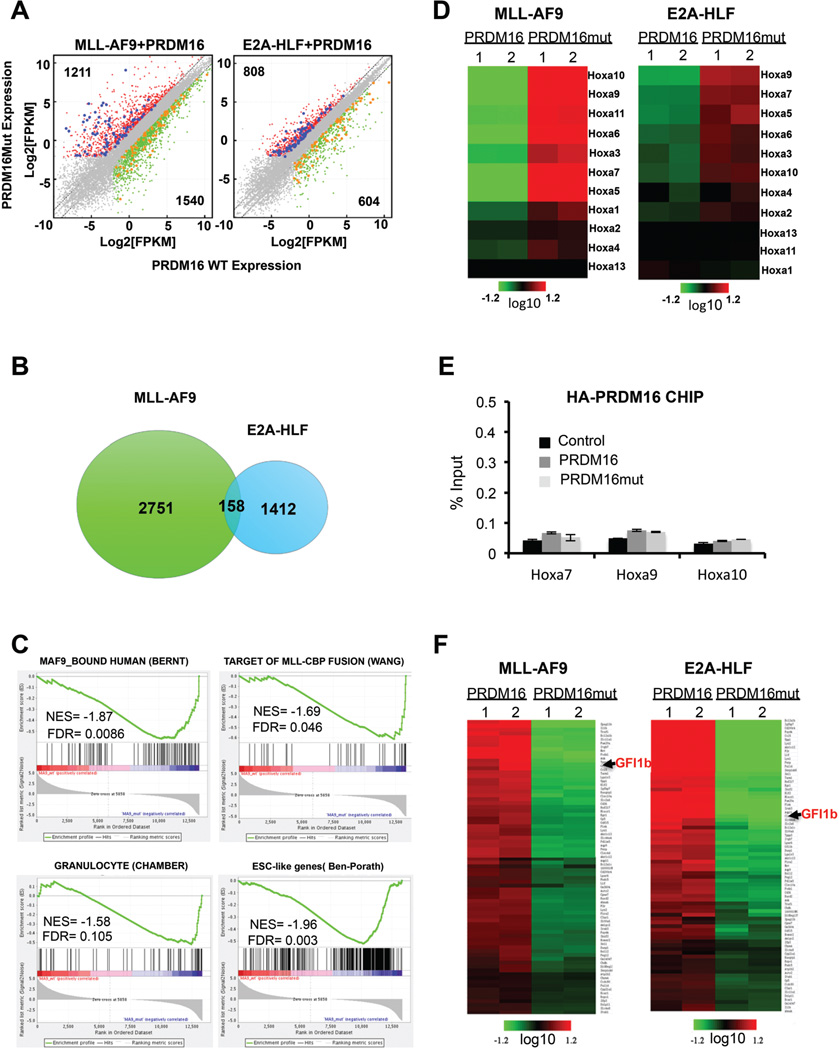
To better understand the mechanism by which PRDM16 HKMT activity suppresses MLL leukemogenesis, we performed Illumina-based RNA-sequencing (RNA-seq). To identify the PRDM16 activity-dependent transcriptome, we focused on genes that showed differential expression between MLL-AF9+Prdm16 and MLL-AF9+Prdm16mut-transduced cells. A comparison of transcriptomes from these two groups revealed a total of 2,751 genes that showed more than two-fold differential expression (False discovery rate (FDR) <=0.05) (Figure 4A, 4B and Supplemental Table S1). 1,540 genes were down regulated (green) and 1,211 genes were up regulated (red) in MLL-AF9+Prdm16mut-transduced cells (Figure 4A and 4B). The global changes in gene expression suggest that PRDM16 HMT activity is important for transcriptome-wide regulation in MLL-AF9-transduced cells. Gene ontology (GO) analyses showed that Prdm16 regulates broad biological processes including immune responses, signal transduction, hematopoietic differentiation and metabolic pathways via its methyltransferase activity (Supplemental Figure 3A and Supplemental Table S2). Importantly, gene set enrichment analyses (GSEA) showed significant negative correlations of PRDM16 transcriptome with gene signatures associated with MLL leukemic cells (Bernt et al., 2011; Wang et al., 2005), granulocytes (Chambers et al., 2007) and embryonic stem cells gene (Ben-Porath et al., 2008) (Figure 4C). These results demonstrate that PRDM16 HMT activity shapes the Lin− BM cell transcriptome during MLL-AF9 mediated-leukemogenesis.
Figure 4. PRDM16 regulates broad transcriptome in MLL-AF9 and E2A-HLF cells.
(See also Figure S3 and Table S1–3). A. Left, scatter plot for transcripts in MLL-AF9+Prdm16mut down regulated (X-axis) and MLL-AF9+Prdm16mut up regulated (Y-axis). Right, scatter plot for transcripts in E2A-HLF+Prdm16 down regulated (X-axis) and E2A-HLF+PRDM16mut up regulated (Y-axis). Transcripts levels were presented as log2 FPKM (fragments per kilobase of transcript per Million mapped reads). Genes that have > 2 fold differences in expression as well as <=0.05 FDR corrected p-value in each cell type are represented by red and green dots for up and down regulated genes, respectively. Grey, genes with no expression change in either cell type. Blue dots, genes expressed higher in both MLL-AF9+Prdm16mut and E2A-HLF/Prdm16mut co-transduced cells. Orange dots, genes expressed higher in both MLL-AF9+Prdm16 and E2A-HLF+Prdm16 co-transduced cells. B. Venn diagram for the differentially expressed genes upon inactivation of PRDM16 in co-transduced MLL-AF9 or E2A-HLF cells. C. Gene set enrichment analysis (GSEA) on genes regulated by PRDM16 methyltransferase activity in MLL-AF9 co-transduced cells. NES: normalized enrichment score. FDR: false discovery rate. References see text. D. Heat map for Hox A genes (indicated on right) in MLL-AF9 and E2A-HLF cells as indicated on top. Color bar indicates scale of log2 fold change after centering of expression values. Duplicate RNA-seq data sets were used. E. ChIP for exogenous PRDM16 at Hox A genes (X-axis). Anti-HA antibody is used. Signals for each experiment were normalized to 1% input. Means and standard deviations (as error bars) from at least three independent experiments were presented. F. Heat map for genes that have lower expression in PRDM16mut+MLL-AF9 and PRDM16mut+E2A-HLF cells as indicated on top. Color bar indicates scale of log2 fold change after centering of expression values.
To determine whether the PRDM16-associated gene expression pattern in MLL-AF9 mediated-leukemogenesis is distinct from that of non-MLL leukemia, we co-transduced Lin− BM cells with E2A-HLF+Prdm16 and E2A-HLF+Prdm16mut constructs and performed similar RNA-seq experiments. Interestingly, while we found no differences in clonogenicity and proliferation between E2A-HLF+Prdm16 and E2A-HLF+Prdm16mut-transduced cells during E2A-HLF transformation (Supplement Figure 1), we found that PRDM16 inactivation led to transcriptome-wide changes. Specifically, in our comparison of the transcriptomes of E2A-HLF+Prdm16 and E2A-HLF+Prdm16mut-transduced cells, we found at least two-fold difference in 1,412 genes, with 808 genes (red) up regulated and 604 genes (green) down regulated (Figure 4A). A comparison of these genes with those altered in MLL-AF9 cells co-transduced with Prdm16 and Prdm16mut revealed only a small set of genes (~158) was regulated by the PR-domain activity in both MLL-AF9 and E2A-HLF co-transduced cells (Figure 4B). Among them, 77 genes were down regulated and 82 genes were up regulated upon PRDM16 inactivation in both cells (Supplemental Table S3). A closer examination of the 82 genes that were commonly up regulated upon Prdm16 inactivation during MLL-AF9 and E2A-HLF-mediated transformation showed that almost all HoxA cluster genes were significantly up regulated (Figure 4D). The RNA-seq results were confirmed by real-time RT-PCR (Supplemental Figure 3B–3D). These results suggest that PRDM16 mediated Hox A repression is probably a general regulatory mechanism that is independent of specific oncogene context. Consistent with this notion, PRDM16 mediated repression of Hox A genes was also observed in normal BM cells transduced with PRDM16 (Supplemental Figure 3E). PRDM16 inactivation did not affect expression of other Hox clusters (e.g. Hox B) (Supplemental Figure 3F). We speculate that the specific inhibition of MLL leukemia by PRDM16 is probably due to the reliance of MLL on Hox A overexpression (see discussion).
Gfi1b is a key mediator of PRDM16 regulation of Hox A genes
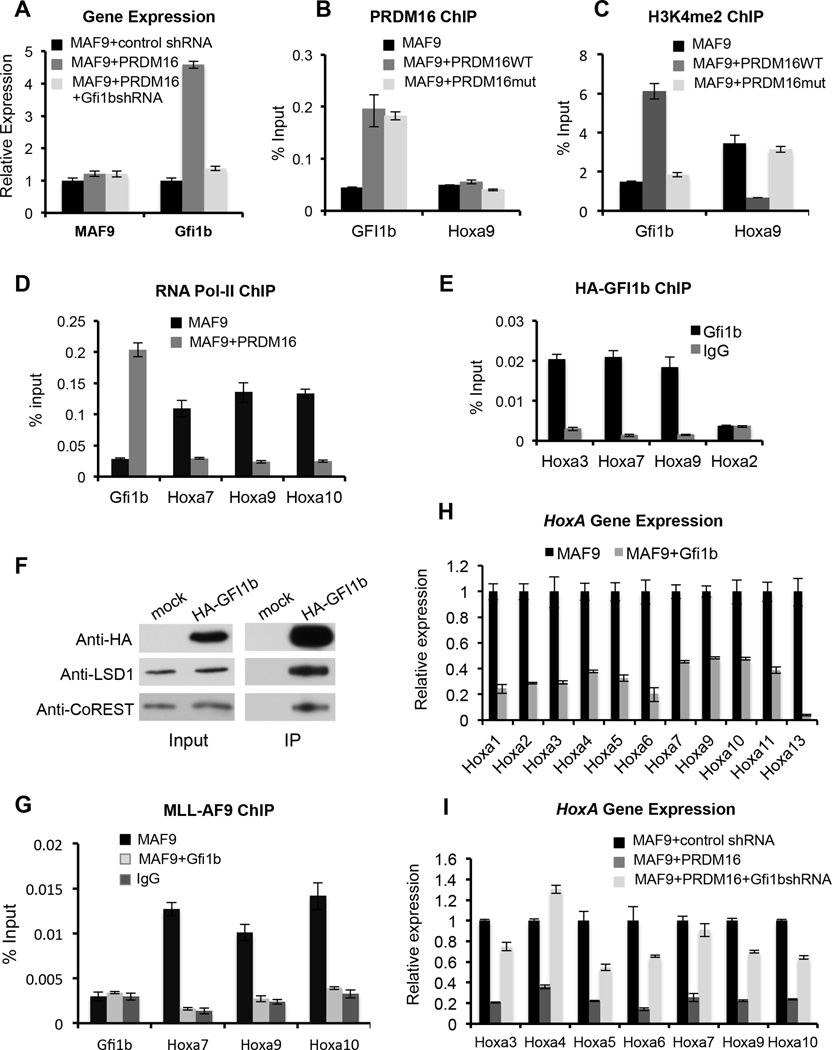
To determine whether PRDM16 directly repressed HoxA gene cluster, we performed chromatin immunoprecipitation (ChIP) experiment for the affinity tagged-exogenous PRDM16. We could not find enrichment of PRDM16 at any HoxA gene (Figure 4E and data not shown). This result suggests that PRDM16 probably represses HoxA genes indirectly. To identify the PRDM16 direct target(s) that mediate HoxA repression, we focused on the 77 PRDM16 targets that were down regulated upon co-transduction of Prdm16mut in both MLL-AF9 and E2A-HLF cells (Figure 4F and Supplemental Table S3). These down regulated genes included several important hematopoietic transcription factors (e.g. KLF2, ETS1 and GFI1b) implicated in HSPC function. Among them, GFI1b appeared as a potentially appealing candidate target of PRDM16. GFI1b belongs to the GFI subfamily of the SNAG zinc finger containing transcription factors restricted to hematopoietic stem cells (Chiang and Ayyanathan, 2013; Saleque et al., 2007; van der Meer et al., 2010). The homolog of GFI1b, GFI1, has been reported to repress gene expression by recruiting the CoREST complex (Chowdhury et al., 2013; Saleque et al., 2007). We first confirmed specific up regulation of GFI1b upon overexpression of wild type PRDM16 (Figure 5A). ChIP experiments showed that both PRDM16 and PRDM16mut directly bound to the Gfi1b promoter, but not the control Hoxa9 locus, in MLL-AF9 cells (Figure 5B). Importantly, an increase of H3K4me2 at the Gfi1b promoter was detected upon binding of PRDM16, but not PRDM16mut (Figure 5C). Concomitant increase in RNA polymerase II (Pol-II) binding was also observed at the Gfi1b promoter upon PRDM16 binding (Figure 5D). These results support a direct role of the PR-domain activity in Gfi1b activation.
Figure 5. Gfi1b is the key intermediate in PRDM16 regulation of Hox A genes.
(See also Figure S4, S5). A. Real-time PCR for MLL-AF9 and Gfi1b with or without PRDM16 overexpression and Gfi1b knock-down. Gene expression was normalized against GAPDH and presented as fold change against the level in MLL-AF9/control shRNA cells, which is arbitrarily set at 1. (B-E and G). ChIP experiments using antibodies as indicated on top. Signals for each experiment were normalized to 1% input. B, E, anti-HA antibody was used to detect exogenous PRDM16 or GFI1b. G, anti-Flag antibody was used to detect FLAG-MLL-AF9. F) Immunoprecipitation of exogenous HA-GFI1b using anti-HA antibody in MLL-AF9+Gfi1b cells. Antibodies were indicated on left. (H). Real-time PCR for Hox A genes in MLL-AF9 cells with or without Gfi1b overexpression. (I). Real-time PCR for Hox A genes in MLL-AF9 or MLL-AF9+PRDM16 cells treated with control or Gfi1b shRNAs as indicated. For H and I, gene expression was normalized against GAPDH and presented as fold change against the level in MLL-AF9, which is arbitrarily set at 1. For A–G and I, means and standard deviations (as error bars) from at least three independent experiments were presented.
Since the GFI1b homolog, GFI1, has been reported to repress Hox A gene expression by recruiting the CoREST complex (Chowdhury et al., 2013; Saleque et al., 2007), we tested whether GFI1b also binds to Hox A loci and recruits the CoREST/LSD1 complex. Indeed, GFI1b bound to three consensus sequences near Hoxa3, Hoxa7 and Hoxa9 (Figure 5E). The binding of GFI1b correlated with decrease in H3K4me2 and Pol-II binding at these loci (Supplemental Figure 4A and Figure 5D). Consistent with previous studies (Chowdhury et al., 2013; Saleque et al., 2007), immunoprecipitation using HA-tagged GFI1b showed that GFI1b physically interacted with the CoREST/LSD1 complex (Figure 5F) and GFI1b-depended recruitment of LSD1 and CoREST was observed in MLL-AF9+Gfi1b cells (Supplemental Figure 4B and 4C). In corroboration of GFI1b as a key mediator of PRDM16 function, overexpression of PRDM16 also led to recruitment of LSD1 and CoREST to Hox A cluster, albeit indirectly (Supplemental Figure 4D and 4E). Importantly, overexpression of -PRDM16 led to increase in the heterochromatin marks such as H3K9me1 and H3K9me2 (Supplemental Figure 4F and 4G). It also led to reduced chromatin binding of MLL-AF9 protein at Hox A genes, consistent with reduced chromatin accessibility (Figure 5G). These changes led to repression of all Hox A gene expression (Figure 5H).
Gfi1b mimics PRDM16 in suppressing MLL-AF9 leukemic transformation
To further establish that GFI1b functions downstream of PRDM16 in suppressing MLL leukemia, we knocked down endogenous Gfi1b in MLL-AF9+Prdm16 co-transduced cells by two independent shRNAs (Supplemental Figure 5A). Gfi1b depletion in MLL-AF9+Prdm16 co-transduced cells led to re-activation of Hox A genes that were repressed by PRDM16 overexpression (Figure 5I). Consistent with gene expression changes, Gfi1b depletion rescued the inhibitory effects of PRDM16 overexpression on MLL-AF9 leukemic transformation (Figure 6A and 6B) without affecting the PRDM16 protein level (Supplemental Figure 5A, bottom panel). Cell proliferation and clonogenicity of GFI1b-depleted MLL-AF9+Prdm16 cells were similar to those of MLL-AF9 cells (Figure 6A and 6B). On the contrary, overexpression of Gfi1b significantly inhibited leukemogenesis when co-transduced with MLL-AF9 in vitro (Supplemental Figure 5B–E). Consistent with in vitro studies, transplantation of MLL-AF9+Gfi1b co-transduced BM cells into lethally irradiated recipients did not give rise to AML in vivo (Figure 6C) despite similar engraftment efficiency as the MLL-AF9 cells (Supplemental Figure 6A). All mice survived beyond the end point of the study (115 days) (Figure 6C) without detectable hematological abnormalities (Figure 6D–6F and Supplemental Figure 6B–6D).
Figure 6. Gfi1b overexpression inhibits MLL-AF9 leukemogenesis.
(See also Figure S6). A. Liquid cell proliferation assays. Cell number (Y-axis) is counted every two days (X-axis). Error bars indicate SD from duplicates. The results were repeated at least three times. B. Myeloid colony formation assay. Colony counts from primary (R1), secondary (R2), and tertiary (R3) plating were summarized for each co-transduction as indicated on bottom. Means and SD (error bars) from duplicates were presented. The results were repeated at least three times. C. Kaplan-Meier survival curve of cohorts of recipient mice (n=6). p-value was calculated using the Mantel-Cox test. D. Top, representative image of spleens from the recipient mice. Bottom, the distribution of the spleen weight for each cohort. The MLL-AF9 cohort was the same as shown in Figure 3 since the experiments were performed at the same time. E. Wright-Giemsa staining of peripheral blood (PB) smear and histology of organs (H&E staining) of recipient mice (as indicated on top) at the end point of the study. Scan bars: 50µm for PB, 200µm for spleen, 100µm for lung and liver, 200µm and 50µm for bones. F. Representative flow cytometry analysis of BM cells in each cohort. Antibodies included CD45.1 vs. CD45.2 (left panels) and Mac-1 vs. Gr-1 (right panel) as indicated. Percentage of cell population was indicated in each quadrant.
Silencing endogenous PRDM16 accompanies MLL-AF9 leukemic transformation
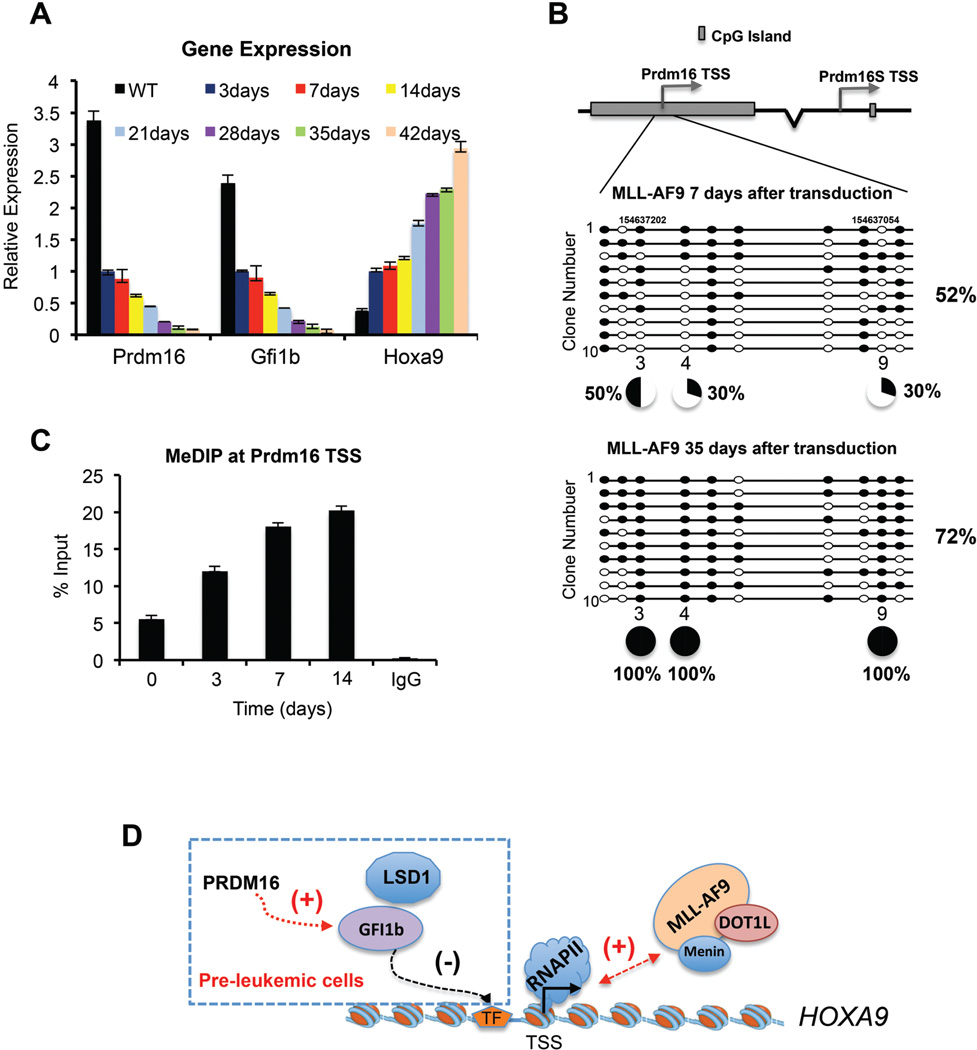
Hitherto, we demonstrated that PRDM16 played essential roles in suppressing MLL leukemic transformation via its HKMT activity. These results raised the question of whether silencing endogenous Prdm16 is a prerequisite for MLL-AF9-mediated leukemic transformation, especially given the relatively long latency of MLL leukemia (Ayton and Cleary, 2001; Krivtsov and Armstrong, 2007). Interestingly, we found a gradual reduction of Prdm16 transcripts following MLL-AF9 transduction; at 35 days, these transcripts were barely detectable (Figure 7A). Concomitantly, a gradual decrease of Gfi1b and an increase of Hoxa9 expression were found in these cells (Figure 7A). Next we examined whether Prdm16 silencing was due to DNA methylation since the TSS of Prdm16 resides within a large CpG island (CGI) (Figure 7B). We measured DNA methylation after 7 or 35 days of MLL-AF9 transduction by bisulfite sequencing. We found that out of 10 tested CpGs, three of them (i.e. 3, 4 and 9) showed dramatic increase of DNA methylation. While these CpGs were methylated at ~30–50% at early stage of transformation, they were fully methylated in completely transformed MLL-AF9 leukemia cells, consistent with silencing of Prdm16 in these cells (Figure 7B). Dynamic change in DNA methylation during the course of leukemic transformation was further confirmed by 5-methylcytosine immunoprecipitation (MeDIP) at the Prdm16 TSS (Figure 7C). Importantly, the regulation of Prdm16 by DNA methylation was reversible. When we treated the MLL-AF9-transduced cells with 50nM DNA methyltransferase (DNMT1) inhibitor decitabine (Nie et al., 2014), a dose-dependent reactivation of endogenous Prdm16 (Supplemental Figure 7A) and significant decrease of DNA methylation at Prdm16 CGIs were observed (Supplemental Figure 7B). Consistently, we found that MLL-AF9 leukemia cells showed increased sensitivity to decitabine (GI50 44.6nM) as compared to E2A-HLF leukemia cells (GI50 82.14nM, Supplemental Figure 7C). Decitabine induced both apoptosis and differentiation of the MLL-AF9 leukemia cells (Supplemental Figure 7D and 7E). Interestingly, knockdown Prdm16 in the MLL-AF9 leukemia cells compromised the effects of decitabine (GI50 65.06nM, Supplemental Figure 7C), suggesting that decitabine blocks MLL leukemia, at least in part, by reactivating endogenous Prdm16.
Figure 7. PRDM16 is regulated by DNA methylation.
(See also Figure S7). A. Real-time PCR for Prdm16, Gfi1b and Hoxa9 gene expression in pre-leukemic MLL-AF9 cells. Gene expression was normalized against GAPDH and presented as fold change against their respective levels in cells 3-day after transduction, which was arbitrarily set at 1. B. Top, schematic of CGIs at PRDM16 and PRDM16S. Bottom, bisulfite-sequencing results for 10 clones in each experimental group as indicated on top. Percentage of total methylated CpG sites was indicated on right and percentage of methylation at selected CpG sites were indicated on bottom. C. MeDIP was performed at different time after MLL-AF9 transduction as indicated on bottom. DNA corresponding to TSS of PRDM16 was amplified by real-time PCR. Signals for IP were normalized to 1% input. Means and standard deviations (as error bars) from at least three independent experiments were presented. D. Schematic for the PRDM16 mediated regulation in pre-leukemic cells. See text for detail.
DISCUSSION
Here we report that PRDM16 is a highly specific H3 K4 methyltransferase on chromatin and its intrinsic activity is essential for suppressing MLL1-rearranged acute leukemia. The tumor suppression function of PRDM16 is mediated by the SNAG family transcription factor GFI1b, which negatively regulates the Hox A gene cluster through recruiting the LSD1/CoREST complex. Overexpression of PRDM16 or GFI1b inhibits initiation of MLL leukemogenesis while knockdown PRDM16 significantly accelerates the disease trajectory of MLL leukemia in vivo. We further demonstrate that silencing endogenous PRDM16 by DNA methylation accompanies leukemic transformation induced by MLL-AF9.
The PR domain of PRDM16 is required for tumor suppression in MLL
PRDM16 is one of the 17 PRDM gene family proteins in human. A previous study reported that PRDM16 was an H3K9me1 HKMT, based on an in vitro HKMT assay using a 6-mer H3 peptide with H3K9 as the only lysine residue (Pinheiro et al., 2012). Our study here shows that PRDM16 is a highly specific HKMT for histone H3 K4 methylation. Mutation of H3 K4 completely abolished PRDM16 activity on nucleosomes in vitro and PRDM16 binding at the GFI1b promoter leads to increase of H3K4me2, but not H3K9me, in cells (Figure 5). These results argue that PRDM16 is a H3K4 specific methyltransferase on chromatin. We would like to point out that PRDM16 has extremely low activity on short peptides (i.e. 6–20aa) as compared to the nucleosome substrate (data not shown), which we have used for the mass spec and immunoblot studies. We also would like to point out that PRDM16 seems to have more robust mono-methylation activity in vitro, although we cannot rule out different detection efficiency of H3K4me1 and H3K4me2 peptides by mass spec due to propionylation of H3K4me1.
Importantly, we show that PRDM16 specifically suppresses MLL leukemogenesis via its PR-domain function. Since the PR domain mutation does not affect the thermostability of PRDM16 (Supplemental Figure 1A) or its interaction with known protein partners (e.g. PGC1 and MED1) (data not shown), it is likely that the intrinsic activity of the PR domain is specifically required for tumor suppression function of PRDM16. Several previous studies have shown that overexpression of PR-less PRDM16S isoform, but not PRDM16, is leukemogenic: 1) leukemogenic translocation in AMLs mostly involves PRDM16S, but not full length PRDM16 (Quentin et al., 2011; Shimizu et al., 2000); 2) PRDM16S is selectively overexpressed in adult T cell leukemia (Yoshida et al., 2004); 3) aberrant expression of PRDM16S by retroviral insertion promotes immortalization of murine bone marrow progenitors and blocks granulocytic differentiation (Du et al., 2005a; Nishikata et al., 2003); 4) overexpression of Prdm16S in p53 null bone marrow cells induces AML with full penetrance (Shing et al., 2007); and 5) overexpression of both Prdm16S and Hoxb4 led to myeloid expansion and leukemia (Yu et al., 2014). These studies suggest that loss of the PR-domain is compatible with leukemogenesis. Interestingly, our studies show that overexpression of Prdm16S or Prdm16mut in MLL leukemia does not affect cell proliferation (Figure 2), suggesting that loss of PR-domain or its activity is not sufficient to promote leukemic transformation in this context. Future studies on the context-dependent functions of PRDM16 and PRDM16S are necessary to further delineate the role of PRDM16 and PRDM16S in leukemia in future.
PRDM16 depletion shortens the latency of MLL in vivo
MLL1 rearranged leukemia has long latency, leading to the hypothesis of a ‘two-hit’ model (Ayton and Cleary, 2001; Krivtsov and Armstrong, 2007). This model implies that in addition to the balanced MLL1 translocation, subsequent genetic or epigenetic alterations are needed for leukemic transformation. However, the evolutionary trajectory that leads from the initial lesion to the eventual development of leukemia is not well understood and epigenetic alterations concurrent with HOXA9 overexpression are not clear. Our studies here show that PRDM16 plays a critical and specific role in suppressing MLL disease progression. Concurrent knockdown of PRDM16 during MLL-AF9 transduction significantly shortens disease latency while overexpression of PRDM16 blocks leukemogenesis (Figure 3). Although we cannot rule out that MLL inhibition by PRDM16 overexpression is due to increased latency beyond end point of the study, the significant anti-correlation between PRDM16 level and onset of MLL is striking. This result is also consistent with down regulation of PRDM16 in MLL patients (Hazourli et al., 2006).
Several recent genomic studies show that genetic and epigenetic heterogeneity within the pre-leukemic HSPC translates into variegated diagnostic and prognostic signatures (Corces-Zimmerman et al., 2014; Shlush et al., 2014). We speculate that epigenetic silencing of PRDM16 in pre-LSCs by local stochastic DNA methylation (Gruber and Wu, 2014) likely renders initial selective advantage for some pre-leukemic stem cells (pre-LSCs) by de-repressing HOXA genes. Unchecked HOXA expression that is essential for MLL maintenance (Chen et al., 2008), in turn, drives successive waves of clonal selection and expansion of LSCs that eventually leads to fully developed leukemia. Consistent with this model, gradual silencing of PRDM16 and concomitant increase of HOXA9 are observed upon MLL-AF9 transformation (Figure 7A). Furthermore, enforced expression or depletion of PRDM16 results in significantly altered disease progression, probably by blocking or accelerating these processes respectively. Since the pre-LSC provides a silent reservoir for the formation of LSCs in fully transformed leukemia, future delineation of pre-leukemic genetic and epigenetic events in MLL will facilitate the development of lasting cures for the disease by rationally eradiating all pre-LSC populations.
PRDM16 indirectly regulates HOXA cluster genes
Our study shows that PRDM16 is a master regulator of transcription in the pre-leukemic HSPC. Inactivating PRDM16 methyltransferase activity induces global changes in transcriptome of both MLL-AF9 and E2A-HLF cells. Importantly, PRDM16 regulates multiple pleiotropic transcription factors such as ETS1 (Findlay et al., 2013), KLF2 (Novodvorsky and Chico, 2014) and GFI1b. These results are consistent with previous studies that PRDM16 is required for HSPC homeostasis (Deneault et al., 2009) and overexpression of Prdm16 leads to increased HSC numbers and activity in vivo (Chuikov et al., 2010). More importantly, PRDM16 plays an important role in controlling the level of HOXA genes in hematopoietic cells. The regulation of HOXA genes by PRDM16 is consistent with a previous study that overexpression of PRDM16 could partially rescue Bmi1-deficiency in HSC (Chuikov et al., 2010), which also represses HOXA genes (Bracken et al., 2006). Deregulation of HOXA genes such as HOXA9 by either enforced overexpression or chromosomal rearrangements is sufficient to drive leukemic transformation (Argiropoulos and Humphries, 2007). In fact, elevated HOXA9 expression is reported in over 50% of AML patients and is also associated with myeloproliferative disorders (Owens and Hawley, 2002). Therefore, maintaining HOXA9 expression at appropriate level is important for balancing HSPC proliferation and malignant transformation. We show that PRDM16 plays a key role to repress HOXA gene expression in HSPC by activating transcription repressor GFI1b, which directly binds to HOXA loci and recruits histone demethylase LSD1 (Chowdhury et al., 2013; Saleque et al., 2007). Since PRDM16 is a direct target of MLL1 (Artinger et al., 2013), it is possible that PRDM16-dependent HOXA9 repression completes a feedback regulatory loop that prevents the unchecked up regulation of HOXA9 by MLL1 in normal HSPC. Given recent progresses in therapeutic targeting factors that up regulates HOXA9 in MLL (Rao and Dou, 2015), it will be interesting to explore whether reactivating endogenous HOXA9 repressive pathways (e.g. PRDM16) also has therapeutic value in future.
EXPERIMENTAL PROCEDURE
PRDM16 expression and purification
His-tagged PR domains of PRDM16 (9–517aa) were expressed from Sf9 insect cells. The recombinant proteins were purified by Ni-NAT magnetic agarose matrix (QIAGEN).
In vitro HMT assay
Preparation of recombinant histones and nucleosomes and in vitro HMT assay were performed as previously described (Wu et al., 2013). Details see Supplemental Method.
Flow Cytometry analysis
Cells from peripheral blood, BM, or spleen were harvested for immunophenotypic analysis. Analyses were performed on LSRII Files and analyzed by FlowJo (TreeStar) software.
DNA Methylation Analysis
MLL-AF9 stable leukemia cells (~2X106) were collected for genomic DNA extraction after 72 hours Decitabine treatment (Abcam ab1200842). DNA bisulfite conversion was performed using Methylation-Gold™ Kit (ZYMO RESEARCH D5005). Regions for detection were amplified by PCR and cloned into the TOPO vector (Invitrogen). The sequencing was done by DNA Sequencing Core facility at University of Michigan.
Retroviral Transduction and Myeloid Colony Formation Assay
The retroviral vectors were transduced to the BM cell as previously described (Muntean et al., 2010). Retroviruses were collected after 48 or 72 hours and transduce BM cells by spinoculation with 90 minutes at 3000rpm. After retroviral transduction, cells were selected for 3 days before plating in methylcelllose medium (M3234, STEMCELL Technologies) with 10ng/ml IL-3, 10ng/ml IL-6, 100ng/ml SCF, 10ng/ml GM-CSF. After three rounds of plating, Plates were scanned and the clone numbers were accounted. Cells harvested at the end of the experiment were cytospin and stained with Hema 3 Stain Kit (Thermo Fisher Scientific).
Murine Bone Marrow (BM) Transformation Assays
Six to eight week old C57BL/6 mice were treated with 4 mg/mouse 5-fluorouacil before BM cell isolation. BM cells were isolated using the EasySep® Mouse Hematopoietic Progenitor Cell Enrichment Kit (STEMCELL Technologies). Lin− BM cells were isolated from 6~8-week-old C57BL/6 mice and transduced with retroviruses. After selection by 1mg/ml G418 or 1.5µg/ml puromycin for 4 days, the cells were counted and injected through the tail vein into cohorts of lethal irradiated (900 rads) B6.SJL mice. Donor and supporting cells (from B6.SJL mice), 1X105 each, were injected into each mouse. Recipient mice were checked daily for leukemia development. Tissues from mice at the end of the study were fixed with 10% formalin, embedded and subject to histology studies (Tan et al., 2011). All animal experiments in this study were approved by the University of Michigan Committee on Use and Care of Animal and Unit for Laboratory Animal Medicine (ULAM).
CHIP Assay
The CHIP assay was performed as previously described (Dou et al., 2005). Antibody information can be found in Supplemental Information.
RNA-sequencing experiment
The RNA was extracted using Trizol reagent (Ambion) and further purified by RNeasy Mini kit (QIAGEN) following manufacture’s protocol. 10ng of total RNAs from each sample were used for preparation of lllumina sequencing library. RNA sequencing was performed on Illumina HiSeq2000 at University of Michigan DNA sequencing core facility. Details for data analyses are shown in Supplemental Information.
Supplementary Material
Acknowledgments
The work are supported by NIGMS (GM082856), American Cancer Society (ACS) and Leukemia and Lymphoma Society Scholar grants to YD, by NIH R01 CA151425 grant to JLH and by NIGMS (GM110174) and the Leukemia and Lymphoma Society Dr. Robert Arceci Scholar Award to BAG. We are grateful to Drs. Jiandie Lin for the PRDM16 cDNA, Patrick Seale for the anti-PRDM16 antibody and Mark Schlissel for GFI1b cDNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
B.Z designed, performed all experiments and prepared the manuscript. J.W performed RNA-seq analyses. P.M. helped with murine leukemia models. J.X. and Y.S helped with RNA-seq data presentation. S.L helped with the thermo shift assay. N.B, Q.G and B.A.G performed the mass spec experiments, J.L.H provided supervision for J.W and P.M. R.R helped with manuscript writing. Y.D helped with the experimental design and wrote the manuscript.
Accession Number
RNA-seq data sets are deposited with accession number GSE64218.
REFERENCE
- Aguilo F, Avagyan S, Labar A, Sevilla A, Lee DF, Kumar P, Lemischka IR, Zhou BY, Snoeck HW. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood. 2011;117:5057–5066. doi: 10.1182/blood-2010-08-300145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- Artinger EL, Mishra BP, Zaffuto KM, Li BE, Chung EK, Moore AW, Chen Y, Cheng C, Ernst P. An MLL-dependent network sustains hematopoiesis. Proc Natl Acad Sci U S A. 2013;110:12000–12005. doi: 10.1073/pnas.1301278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet. 2010;19:774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kumar AR, Hudson WA, Li Q, Wu B, Staggs RA, Lund EA, Sam TN, Kersey JH. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13:432–440. doi: 10.1016/j.ccr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Ayyanathan K. Snail/Gfi-1 (SNAG) family zinc finger proteins in transcription regulation, chromatin dynamics, cell signaling, development, and disease. Cytokine Growth Factor Rev. 2013;24:123–131. doi: 10.1016/j.cytogfr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury AH, Ramroop JR, Upadhyay G, Sengupta A, Andrzejczyk A, Saleque S. Differential transcriptional regulation of meis1 by Gfi1b and its co-factors LSD1 and CoREST. PloS one. 2013;8:e53666. doi: 10.1371/journal.pone.0053666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol. 2010;12:999–1006. doi: 10.1038/ncb2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneault E, Cellot S, Faubert A, Laverdure JP, Frechette M, Chagraoui J, Mayotte N, Sauvageau M, Ting SB, Sauvageau G. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137:369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005a;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Spence SE, Jenkins NA, Copeland NG. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood. 2005b;106:2498–2505. doi: 10.1182/blood-2004-12-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Karim MR, Taniguchi H, Krejci A, Kinameri E, Siebert M, Ito K, Bray SJ, Moore AW. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat Neurosci. 2012;15:224–233. doi: 10.1038/nn.2998. [DOI] [PubMed] [Google Scholar]
- Findlay VJ, LaRue AC, Turner DP, Watson PM, Watson DK. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv Cancer Res. 2013;119:1–61. doi: 10.1016/B978-0-12-407190-2.00001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fog CK, Galli GG, Lund AH. PRDM proteins: important players in differentiation and disease. Bioessays. 2012;34:50–60. doi: 10.1002/bies.201100107. [DOI] [PubMed] [Google Scholar]
- Gruber M, Wu CJ. Evolving understanding of the CLL genome. Semin Hematol. 2014;51:177–187. doi: 10.1053/j.seminhematol.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazourli S, Chagnon P, Sauvageau M, Fetni R, Busque L, Hebert J. Overexpression of PRDM16 in the presence and absence of the RUNX1/PRDM16 fusion gene in myeloid leukemias. Genes Chromosomes Cancer. 2006;45:1072–1076. doi: 10.1002/gcc.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn KH, Warner DR, Pisano M, Greene RM. PRDM16 expression in the developing mouse embryo. Acta Histochem. 2011;113:150–155. doi: 10.1016/j.acthis.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S, Chen W, Nakadai T, Ohkuma Y, Roeder RG. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes & development. 2015;29:308–321. doi: 10.1101/gad.252809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Martinez-Climent JA, Alizadeh AA, Segraves R, Blesa D, Rubio-Moscardo F, Albertson DG, Garcia-Conde J, Dyer MJ, Levy R, Pinkel D, et al. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood. 2003;101:3109–3117. doi: 10.1182/blood-2002-07-2119. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- Morishita K. Leukemogenesis of the EVI1/MEL1 gene family. Int J Hematol. 2007;85:279–286. doi: 10.1532/IJH97.06174. [DOI] [PubMed] [Google Scholar]
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, Liu L, Li X, Han W. Decitabine, a new star in epigenetic therapy: the clinical application and biological mechanism in solid tumors. Cancer Lett. 2014;354:12–20. doi: 10.1016/j.canlet.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N, Nakamura T, Morishita K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood. 2003;102:3323–3332. doi: 10.1182/blood-2002-12-3944. [DOI] [PubMed] [Google Scholar]
- Novodvorsky P, Chico TJ. The role of the transcription factor KLF2 in vascular development and disease. Prog Mol Biol Transl Sci. 2014;124:155–188. doi: 10.1016/B978-0-12-386930-2.00007-0. [DOI] [PubMed] [Google Scholar]
- Owens BM, Hawley RG. HOX and non-HOX homeobox genes in leukemic hematopoiesis. Stem Cells. 2002;20:364–379. doi: 10.1634/stemcells.20-5-364. [DOI] [PubMed] [Google Scholar]
- Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, Mittler G, Genoud C, Goyama S, Kurokawa M, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150:948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- Quentin S, Cuccuini W, Ceccaldi R, Nibourel O, Pondarre C, Pages MP, Vasquez N, Dubois d’Enghien C, Larghero J, Peffault de Latour R, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117:e161–e170. doi: 10.1182/blood-2010-09-308726. [DOI] [PubMed] [Google Scholar]
- Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Suzukawa K, Kodera T, Nagasawa T, Abe T, Taniwaki M, Yagasaki F, Tanaka H, Fujisawa S, Johansson B, et al. Identification of breakpoint cluster regions at 1p36.3 and 3q21 in hematologic malignancies with t(1;3)(p36;q21) Genes Chromosomes Cancer. 2000;27:229–238. doi: 10.1002/(sici)1098-2264(200003)27:3<229::aid-gcc2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Shing DC, Trubia M, Marchesi F, Radaelli E, Belloni E, Tapinassi C, Scanziani E, Mecucci C, Crescenzi B, Lahortiga I, et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J Clin Invest. 2007;117:3696–3707. doi: 10.1172/JCI32390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SX, Hu RC, Liu JJ, Tan YL, Liu WE. Methylation of PRDM2, PRDM5 and PRDM16 genes in lung cancer cells. International journal of clinical and experimental pathology. 2014;7:2305–2311. [PMC free article] [PubMed] [Google Scholar]
- van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. 2010;24:1834–1843. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- Wang J, Iwasaki H, Krivtsov A, Febbo PG, Thorner AR, Ernst P, Anastasiadou E, Kutok JL, Kogan SC, Zinkel SS, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. Embo J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M, Winter C, Gregory SG, Hogarty MD, Maris JM, et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- Wu L, Lee SY, Zhou B, Nguyen UT, Muir TW, Tan S, Dou Y. ASH2L regulates ubiquitylation signaling to MLL: trans-regulation of H3 K4 methylation in higher eukaryotes. Mol Cell. 2013;49:1108–1120. doi: 10.1016/j.molcel.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Zhang M, Liu X, Zhang Y, Yang L, Hao Y. MEL1S, not MEL1, is overexpressed in myelodysplastic syndromes patients with t(1;3)(p36;q21) Leuk Res. 2006;30:332–334. doi: 10.1016/j.leukres.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Nosaka K, Yasunaga J, Nishikata I, Morishita K, Matsuoka M. Aberrant expression of the MEL1S gene identified in association with hypomethylation in adult T-cell leukemia cells. Blood. 2004;103:2753–2760. doi: 10.1182/blood-2003-07-2482. [DOI] [PubMed] [Google Scholar]
- Yu H, Neale G, Zhang H, Lee HM, Ma Z, Zhou S, Forget BG, Sorrentino BP. Downregulation of Prdm16 mRNA is a specific antileukemic mechanism during HOXB4-mediated HSC expansion in vivo. Blood. 2014;124:1737–1747. doi: 10.1182/blood-2013-10-534735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.