Abstract
Research using twins has found that much of the variability in externalizing phenotypes – including alcohol and drug use, impulsive personality traits, risky sex and property crime – is explained by genetic factors. Nevertheless, identification of specific genes and variants associated with these traits has proven to be difficult, likely because individual differences in externalizing are explained by many genes of small individual effect. Moreover, twin research indicates that heritable variance in externalizing behaviors is mostly shared across the externalizing spectrum rather than specific to any behavior. We use a longitudinal, “deep phenotyping” approach to model a general externalizing factor reflecting persistent engagement in a variety of socially problematic behaviors measured at eleven assessment occasions spanning early adulthood (ages 18 to 28). In an ancestrally homogenous sample of non-Hispanic Whites (N = 337), we then tested for enrichment of associations between the persistent externalizing factor and a set of 3,281 polymorphisms within 104 genes that were previously identified as associated with alcohol-use behaviors. Next we tested for enrichment among domain-specific factors (e.g., property crime) composed of residual variance not accounted for by the common factor. Significance was determined relative to bootstrapped empirical thresholds derived from permutations of phenotypic data. Results indicated significant enrichment of genetic associations for persistent externalizing, but not for domain-specific factors. Consistent with twin research findings, these results suggest that genetic variants are broadly associated with externalizing behaviors rather than unique to specific behaviors.
General Scientific Summary
This study shows that variation in 104 genes is associated with socially problematic “externalizing” behavior, including substance misuse, property crime, risky sex, and aspects of impulsive personality. Importantly, this association was with the common variation across these behaviors rather than with the variation unique to any given behavior. The manuscript demonstrates a potentially advantageous technique for relating sets of hypothesized genes to complex traits or behaviors.
Keywords: externalizing behavior, enrichment analysis, problem behavior, genetic polymorphisms, impulsive traits, deviant behavior
Genetic influences account for substantial variation in externalizing behaviors, a constellation of risky and deviant behaviors that includes hazardous alcohol use, tobacco use, illicit drug use, and theft. Twin studies, which estimate heritability by comparing identical twins to fraternal twins, have found that 30% to 70% of the variance in substance use disorders (Agrawal & Lynskey, 2008), 30% to 40% of the variance in antisocial behavior (Viding, Larsson, & Jones, 2008), and about 33% of the variance in risky sex (Zietsch, Verweij, Bailey, Wright, & Martin, 2010) is due to genes. Personality traits that are associated with externalizing behavior, such as impulsivity and sensation seeking (Cooper, Wood, Orcutt, & Albino, 2003; Jentsch et al., 2014; Krueger et al., 2002), are also heritable (Harden, Quinn, & Tucker-Drob, 2012).
Moreover, researchers have long understood that externalizing behaviors commonly co-occur, and likely share a common etiology. Problem behavior theory posits a single common factor that explains the high correlations between rates of various “problem” behaviors (Jessor & Jessor, 1977). For example, a single common factor model adequately captures the correlations between drunkenness, tobacco use, marijuana use, delinquent or antisocial behaviors and sexual precociousness (Donovan & Jessor, 1985; Donovan, Jessor, & Costa, 1988). Support for a single externalizing factor has also been found more recently by other authors (e.g., Caspi et al., 2014; Cooper et al., 2003; Krueger et al., 2002).
Furthermore, analyses using twin samples have demonstrated that, not only are externalizing behaviors phenotypically correlated, they also share a substantial common genetic etiology (Kendler, Myers, & Prescott, 2007; Krueger et al., 2002; Xian et al., 2008). In other words, the same genetic risks that predispose individuals toward hazardous drinking also predispose them toward illicit drug use (Kendler et al., 2007; Palmer et al., 2012), non-substance-related criminal behaviors (Krueger et al., 2002), and impulsive personality traits (Harden et al., 2012; Krueger et al., 2002). Notably, the genetic signal for a general factor representing the tendency to engage in a variety of externalizing behaviors is stronger (estimated as high as 80%) than for any one behavior considered in isolation (Krueger et al., 2002).
Value of Psychometric Approaches for Genetic Research
Although genes are unquestionably important for understanding individual differences in externalizing behaviors, the specific genetic architecture is unknown. Despite some major strides in elaborating the genetic basis of psychiatric illnesses and complex behaviors with polygenic causes, determining which specific genes and variants are involved remains largely elusive (Geschwind & Flint, 2015). How can it be that most of the substantial heritable variation in externalizing phenotypes appears “missing” (Manolio et al., 2009)? Current scientific consensus is that behavioral and psychiatric phenotypes, including externalizing behaviors, are influenced by a very large number of genes, each of which explains perhaps less than 1% of the total variability in a particular phenotype (C. Chabris, Lee, Cesarini, Benjamin, & Laibson, 2015; C. F. Chabris et al., 2013). Moving forward, psychology must grapple with the implications of a “many genes of small effect” model of complex behaviors and psychiatric diseases, including the enormous sample sizes necessary for a robust genome-wide association study (e.g., Rietveld et al., 2013) and the improbability of a large effect size for any single genetic variant (Dick et al., 2015).
Sophisticated quantitative modeling and measurement of behavioral and psychiatric phenotypes is a unique strength of psychological science, providing the potential to contribute to our understanding of the genetic underpinnings of complex behavior. As such, taking a broader view of the etiology of externalizing behavior and related traits using factor model approaches may lead to unique insights. Indeed, analyses of simulated data have demonstrated that using latent factor models to measure phenotypes (as opposed to manifest variables, such as dichotomous clinical diagnoses or symptom-count sum scores) generally increases power to detect genetic associations (van der Sluis, Verhage, Posthuma, & Dolan, 2010).
Enrichment Analyses
Given massive polygeneticity of complex behavior, one method for testing hypotheses about the genetic underpinnings of behavior (including samples with moderate sample sizes) is an enrichment analysis (Aliev et al., 2015). Like a candidate gene approach, an enrichment analysis examines associations with a circumscribed and hypothesis-driven set of genetic variants, rather than testing associations with every marker across the genome. Enrichment is detected if there are more significant associations between the genetic variants and the phenotype of interest than would be expected by chance. Still, unlike a well-powered genome-wide association study (which may require sample sizes in the tens or hundreds of thousands), an enrichment analysis cannot specify which variants within a set are driving an enrichment effect. That is, enrichment indicates that there is a genetic signal from a circumscribed set of genetic variants, but does not identify which specific associations are “true” effects. Results from an enrichment analysis can thus be conceptualized as intermediate to results from twin studies (which estimate the entirety of the genetic variance but are silent about which genetic variants are involved) and results from GWAS (which account for only small fractions of total phenotypic variability but can identify specific variants).
Recently, investigators from the Collaborative Studies on Genetics of Alcoholism (COGA) conducted an enrichment analysis of single nucleotide polymorphisms (SNPs) in a set of over 100 genes, which were selected because of evidence that they were associated with alcohol outcomes in either GWAS or candidate gene analyses (Aliev et al., 2015). Consistent with results from previous twin research (e.g., Krueger et al., 2002), Aliev and colleagues found significant enrichment of associations not only with alcohol dependence symptoms, but also with antisocial behavior symptoms, conduct disorder symptoms, and sensation seeking personality. As these outcomes were tested individually as separate observed measures, the enrichment results observed for these outcomes could be interpreted in one of two ways. First, these genetic variants could influence biological processes that underlie all of the different symptoms. For example, an individual with genetic variants that predisposed them to greater reward sensitivity may be more sensation seeking, more likely to use drugs, and more likely to have break rules to obtain desired ends. Alternatively, the gene set could be heterogeneous, with some variants affecting only alcohol use, some affecting only personality, and some affecting rule-breaking. In this case, one might detect significant enrichment of associations for all externalizing phenotypes, but in this case the genetic influences are operating through processes unique to each phenotype rather than on their overlap. The former interpretation is consistent with results from twin research, which have found evidence for strong genetic influences on a general factor of externalizing. However, for the genetic variants examined by Aliev et al. (2014), these alternative models of genetic influence have not yet been directly tested.
Goals of the Current Paper
The goal of the current paper, then, is to test an enrichment of associations between the gene set used by Aliev et al. (2014) and externalizing phenotypes, modeled using a latent general factor and a series of residual behavior-specific (e.g., alcohol use, sensation seeking personality) factors, in an independent sample. Importantly, our sample was not selected for alcohol use disorder, as was the COGA sample, broadening the generalizability of the findings. Our “deep phenotyping” approach (Robinson, 2012) captures persistent externalizing behavior and traits across time. Participants were followed prospectively from late adolescence through early adulthood over eleven assessment waves (from about age 18 to 28), and they were measured on a variety of externalizing phenotypes, including hazardous alcohol use, tobacco smoking, marijuana use, risky sex, property crime, and impulsive/sensation-seeking personality traits. Like other studies of the externalizing spectrum (Latendresse et al., 2015; Salvatore et al., 2015), we used latent factor analyses to model variance common across behaviors, and, by using 10 waves of longitudinal data, variance common across time. Therefore, individuals who scored highly on our key phenotype of interest – a general externalizing factor – have been persistently involved in an array of socially problematic and disinhibited behaviors or traits across emerging adulthood. After establishing a well-fitting model of persistent externalizing behavior, we then used the derived factor scores to test for enrichment in an ancestrally homogenous sample of non-Hispanic Whites who had high-quality genetic information (N = 337).
Only two previous studies have used composite measures derived from factor models of externalizing phenotypes to test for association with individual genetic variants. First, Latendresse and colleagues tested for associations between a general problem behavior factor and a limited number of variants found within three candidate genes (CHRM2, GABRA2, and OPRM1) in a sample of African American adolescents (Latendresse et al., 2015). Although these authors implemented sophisticated techniques for modeling the broad problem behavior or externalizing phenotype, testing against only three genes likely limited the potential phenotypic variance explained, as compared to a more polygenic approach. At the other polygenic extreme, Salvatore and colleagues (2015) demonstrated the utility of a genome-wide polygenic risk score approach in relation to a composite score derived from a principal components analysis of symptoms of externalizing disorders. In this kind of analysis, researchers first conduct a genome wide association study (GWAS) between all available SNPs across the genome and phenotypes in a discovery sample. Then, beta weights for each SNP are used in a separate target sample to compute an additive “risk score” for all individuals given their specific alleles. These authors did indeed find a significant relation between polygenic risk scores and externalizing in an independent target sample. Nevertheless, this approach is genome-wide, and therefore agnostic about the sources of association, and it does not leverage the hypothesis-driven selection of candidate genes that have emerged from the extant literature. The approach used in the current study, therefore, may provide more stable estimates of genetic risk than candidate gene studies, while still proving some specificity regarding target genes compared to a genome-wide polygenic risk score approach.
Combining a factor analytic approach with an enrichment analysis of an independently developed gene set is a novel method for examining the extent to which genes associated with alcohol outcomes also associate with externalizing more generally. This approach provides advantages both in terms of strong phenotypic measurement and by providing a degree of genetic specificity not afforded by some genome-wide approaches by relying on prior evidence. In addition, by using a bi-factor model with residual domain-specific factors (Latendresse et al., 2015), we can test if enrichment effects are present for specific behaviors over and above the common variance shared across domains. Our hypothesis is that the enrichment will primarily be evident for a general externalizing factor, rather than for residual variance specific to certain behaviors, given the strong genetic signal for common variance identified in twin studies (Krueger et al., 2002).
Method
Participants
We recruited participants from an entering freshman class at a large public Southwestern university beginning in 2004. A subset of those invited to participate (N = 6,391) indicted potential interest and met inclusion criteria of being first-time students between the ages of 17 and 19 years old, unmarried, and providing valid contact information (N = 4,832, 75.6%). A group of eligible participants (N = 3,046) were randomized to complete a series of repeated surveys, with the first given during the summer after the end of high school and the remaining surveys given over six years. The final sample included in the externalizing factor model analysis comprises those who provided informed consent and completed the first longitudinal survey (N = 2,245). A majority of respondents in the full survey sample were female (N = 1,345, 59.9%). The flow of participants from initial invitation and recruitment in 2004 to the provision of saliva samples and the most recent survey data in summer 2014 is presented in Supplemental Figure S1.
Longitudinal Design and Sample Collection
Web-based survey data were gathered at eleven assessment waves. The first eight waves were conducted twice per year (fall and spring), whereas Waves 9 and 10 occurred one year after the previous assessment as described previously (Ashenhurst, Harden, Corbin, & Fromme, 2015). A targeted sample of respondents (N = 1,060) were invited to complete a Wave 11 (W11) survey and to provide salivary DNA five years after W10. Only a subset of the original W1–W10 sample was invited for this next phase of the study due to constraints on the possible number of individuals who could be feasibly genotyped due to budgetary constraints. Criteria for invitation to the W11 survey included a) permission to re-contact, b) completion of W1 and at least one other survey. To date, 601 individuals have provided saliva samples.
In order to avoid any spurious results due to population stratification (Cardon & Palmer, 2003), the target sample for these analyses was comprised of non-Hispanic White individuals only. Self-reported White individuals who provided DNA and passed quality control procedures were the largest racial segment (N = 341, 56%) of those who provided DNA. Genomic principal components analysis (see Genotyping Procedures) within this group identified four ancestral outliers who were removed from analysis (sigma > 6.0), resulting in a final sample size for enrichment analysis of N = 337. A comparison of these participants versus non-Hispanic White participants who did not provide DNA and W11 survey data or pass quality control procedures is reported in the Results.
Respondents were compensated $30 for completion of the W1 survey, $20 for completing the fall surveys (Waves 2, 4, 6), $25 for the spring surveys (Waves 3, 5, 7), and $40 for the remaining Waves 8–10. Respondents received $30 for survey completion if DNA was provided, and $20 for completing Wave 11 without providing a DNA sample. The local university Institutional Review Board approved all study surveys and procedures.
Demographics
Basic demographic measures gathered at Wave 1 (full survey sample) included: biological sex (coded 0 = female [59.9%], 1 = male) and Race (White [53.9%], Asian, [18%], Histpanic/Latino(a) [15.2%], African American [4.1%], Other/Multiethnic [7.1%], missing or not provided [1.6%]). A small number of participants who provided DNA did not provide self-report ethnicity at Wave 1, but did at later waves. The total available pool of non-Hispanic Whites included in the externalizing phenotype model was N = 1223.
Measurement of Externalizing Phenotypes
Hazardous Alcohol Use
Three measures captured hazardous alcohol use: binge drinking frequency, times “drunk,” and driving after drinking. These three indicators were chosen as they assess patterns of excess use, aspects of subjective intoxication, and alcohol-related antisocial behavior, respectively. The definition of a binge episode was consistent with NIAAA guidelines at study onset (NIAAA, 2004). Respondents were asked to provide a free response to the open-ended question, “During the past three months, how many times did you have [five (men)/four (women)] drinks at a sitting?” Respondents were also asked to freely provide an integer response to determine the number of times drunk by the question, “During the past 3 months, how many TIMES did you get drunk (not just a little high) on alcohol?” Driving after drinking was captured by two questions: “During the past three months, how many times did you… [drive after having 1–3 alcoholic beverages/drive after having 4 or more alcoholic beverages?”. The available responses were: 0 = 0 times, 1 = 1 time, 2 = 2 times, 3 = 3–5 times, 4 = 6–10 times, 5 = 11–20 times, 6 = >20 times. The two drinking and driving items were summed to create a single variable.
Cannabis Use
One item assessed cannabis use: “During the last 3 months, how many times did you smoke marijuana?” The available responses were: 0 = 0 times, 1 = 1 time, 2 = 2 times, 3 = 3–5 times, 4 = 6–10 times, 5 = 11–20 times, 6 = >20 times.
Property Crime
Two items captured incidence of property crime, “During the last 3 months, how many times did you… [destroy property/steal something]?” The available responses were: 0 = 0 times, 1 = 1 time, 2 = 2 times, 3 = 3–5 times, 4 = 6–10 times, 5 = 11–20 times, 6 = >20 times.
Tobacco Use
Respondents were asked, “How often did you use tobacco during the last three months?” Available responses were coded: 0 = never, 1 = occasionally, 2 = weekly but not daily, 3 = daily.
Risky Sex
Scores were summed across two items at each wave: “During the past 3 months, how many times did you have sex without protection against STDs and pregnancy with an [exclusive/non-exclusive] dating partner?” We included both exclusive and non-exclusive unprotected sex because being in a reportedly exclusive relationship is not necessarily protective against sexually transmitted infections or unintended pregnancy. These two questions had the following choices: 0 = 0 times, 1 = 1 time, 2 = 2 times, 3 = 3–5 times, 4 = 6–10 times, 5 = 11–20 times, 6 = >20 times.
Personality Scales
Impulsivity and sensation seeking were assessed at only Waves 1 and 8–11, thus data at intervening waves were not available. The two domains of personality were taken from the Impulsivity (8 item) and Sensation Seeking (11 item) subscales of the Zuckerman-Kuhlmen Personality Questionnaire (Zuckerman, Kuhlman, Joireman, Teta, & Kraft, 1993). Factor analytic work has demonstrated that there are distinct facets of impulsivity including: urgency, lack of planning, and lack of perseverance (Stautz & Cooper, 2013; Whiteside & Lynam, 2001). The items provided on the Zuckerman-Kuhlman inventory most closely resemble the lack of planning facet rather than the broader construct of impulsivity. As such, scores on this measure will be referred to as “ZK Impulsivity” to avoid the implication that this measure provides comprehensive coverage of impulsivity domains. Examples of items for each scale include: ZK Impulsivity, “I usually think about what I am going to do before doing it,” (reverse scored) and Sensation Seeking, “I like doing things just for the thrill of it.” Each item was scored dichotomously and reversed scored where appropriate, with respondents endorsing either 0 = false or 1 = true.
Data Management for Survey Data
In order to appropriately model the externalizing behavior variables (which were not normally distributed), to reduce the computational burden required for model fitting and to obtain standard fit indices, the following measures were re-scored as dichotomous variables such that 0 = no behavior, 1 = at least one incidence of behavior: binge drinking, times drunk, drinking before driving, sex without protection, and indicators of property crime. The following variables remained continuous or quasi-continuous: tobacco use and personality measures. Next, indicators within a specific domain were summed to create a manifest variable used in model fitting. For example, the three dichotomized alcohol variables were summed into a single ordinal categorical variable such that 0 = no behaviors, and 3 = endorsing all three behaviors. A sum score was also generated for indicators of property crime. See Supplement for descriptive summary statistics for all variables at all waves (Table S1), and a large bivariate correlation matrix between all manifest variables in the measurement model (Supplemental Data file).
Genetic Data
Lab Procedures
Two mL of saliva were collected in Oragene-Discover (Oragene™, DNAgenotek, Ottawa, Ontario, Canada) kits distributed to participants through the mail. When capped, the collection vessel released a room-temperature stabilizing lysis buffer. DNA extraction and purification was conducted at the Institute for Behavior Genetics at the University of Colorado, Boulder. The DNA was prepared from 500 μl of the Oragene™ solution with the Beckman-Coulter DNAdvance (Brea, CA) system according to the manufacturer’s protocol, with the final elution volume being 150 μl. Samples were diluted 1:20 in TE and the DNA was quantified using Picogreen fluorescence (Invitrogen, ThermoFisher, Grand Island, NY). Samples were standardized to 50 ng DNA/μl for chip genotyping.
Purified and diluted samples were sent to the Neuroscience Genomics Core at the University of California, Los Angeles, for single nucleotide polymorphism (SNP) genotyping assay. Samples were run on an Illumina BeadLab platform using an Illumina Infinium PsychArray BeadChip array (San Diego, CA). This chip includes 265,000 tag-SNPs across the genome, with about 50,000 markers associated with common psychiatric disorders. Chips were scanned on an Illumina iScan confocal laser, with genotype calls performed using manufacture’s parameters in GenomeStudio (Illumina, v 2011.1, genotyping module v1.9.5). Data was converted to PLINK (v1.90b3v) format for subsequent analyses (Purcell et al., 2007).
Quality Control
Of the samples sent for DNA extraction (N = 601), a subset yielded insufficient concentrations of DNA for further processing (N = 28), or suffered from poor amplification (N = 8). Furthermore, three randomly selected samples were not assayed in order to run full plates only. Thus, the total sample with available genetic information was 93.5% (N = 562) of the total collection sample.
We then followed quality control procedures recommended for the chip-based genomic data (Anderson et al., 2010; Turner et al., 2011). Each of the six plates contained a standard sample, and seven samples were run as duplicates on separate plates. Identity by descent (IBD) analysis conducted in PLINK (Purcell et al., 2007) showed high quality duplication of genotyping results (pi-hat = 1.0 for all duplicate pairs and standard pairs). Sex check analysis of X chromosome heterozygosity identified 6 individuals with sexes inconsistent with their self-reported sexes, likely an indication of a plating error. Inbreeding coefficients and IBD analyses identified four samples that were likely contaminated, and one that was erroneously duplicated. After removing these samples, no sample pairs showed IBD relatedness values greater than 0.1875 (Anderson et al., 2010), and all self-reported sexes were consistent with X chromosome heterozygosity. One further sample was removed from analyses due to low genotyping efficiency (genome-wide missingness greater than 10%).
Genomic Principal Components Analysis
We then used EIGENSTRAT (Price et al., 2006) in order to address any residual population stratification and to identify ancestral outliers. Genomic principal components were extracted within the self-reported non-Hispanic White sample. Default parameters were used in accordance with recommendations for this procedure (Turner et al., 2011), and data was linkage disequilibrium pruned (r2 < 0.5) prior to extraction in order to reduce computational burden. Iterative extraction identified four individuals who were ancestral outliers relative to the rest of the self-reported White sample (sigma > 6.0); these four individuals were removed, resulting in a final analysis sample after all quality control procedures of N = 337.
Gene Set for Analyses
The genes selected by COGA authors as being associated with alcohol phenotypes (Aliev et al., 2015) were used in the analyses. Nine of these genes were not represented by SNPs on the chip array we used for genotyping, prohibiting their inclusion in the analysis. In total, 104 genes were included. Gene locations and ranges were gathered using GRCH37.p13 coordinates from the UCSC Table Browser. These gene location ranges (and the genes that could not be included in analyses) are provided in a downloadable excel file (Supplemental Results). All available SNPs (3,281) within these coordinate ranges that passed SNP-level quality control filters (minor allele frequency > 5%, genotype missing by SNP < 2%) were included in analysis. The list of SNPs included in each analysis and their corresponding individual p-values are provided as Supplemental Results (to be found online). One SNP deviated from Hardy-Weinberg equilibrium just beyond the p < 5 × 10−5 level, which commonly occurs in studies using genome-wide data (Anderson et al., 2010; Turner et al., 2011). All other SNPs were consistent with Hardy-Weinberg equilibrium above this value. Hardy-Weinberg Statistics for every SNP tested are provided in the supplemental materials.
Data Analytic Plan
Factor Model of Problem Behavior
Individual behaviors were modeled as indicators of an Externalizing factor at each individual wave; these wave-specific factors, in turn, loaded on a higher-order Persistent Externalizing factor (Figure 1). Loadings for individual behaviors (e.g., hazardous alcohol use) on the wave-specific Externalizing factors were constrained to be equal a priori across all waves.1 Residual covariances among measures of the same behavior at different waves were modeled with a series of domain-specific factors (Alcohol Use, Cannabis Use, Property Crime, Tobacco Use, Risky Sex, and Sensation Seeking/ZK Impulsivity). Factor models were estimated using Mplus version 7.31 (Muthén & Muthén, Los Angeles, CA), and model fit was evaluated using root mean square error of approximation (RMSEA), with values less than 0.05 indicating good fit (Steiger, 1990). We also used the Bentler Comparative Fit Index (CFI) and Tucker-Lewis Index (TLI) indices, which are sensitive to model fit as well as parsimony. Values of CFI and TLI vary between 0 and 1 with excellent values being greater than 0.95 (Hu & Bentler, 1999). Factor scores for the Persistent Externalizing factor and the six domain-specific factors were then estimated using Mplus, and these factor score estimates were used as the phenotypes in all subsequent analyses.
Figure 1.
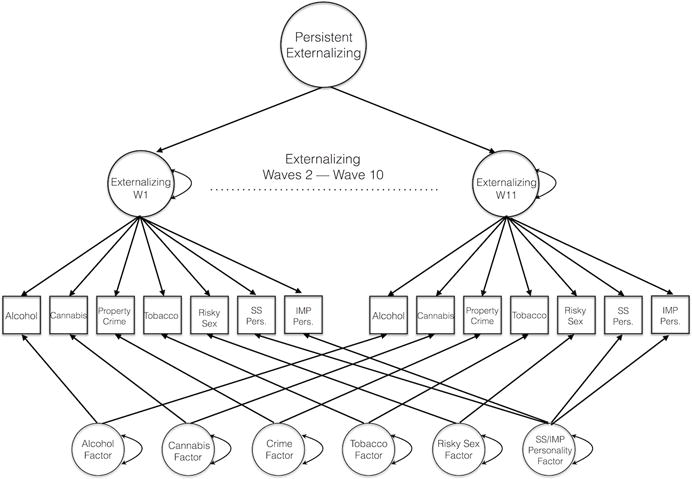
Factor Model of Externalizing Behaviors and Traits. The phenotypes for the enrichment analyses were estimated factor scores from a multi-level model of eleven waves of data capturing (a) variance common across all behaviors and across time (Persistent Externalizing), and (b) variance unique to particular domains (e.g., Alcohol, Cannabis) across time. Thus, a high score on the Persistent Externalizing factor indicated continued engagement in many of the behaviors over the span of emerging adulthood.
Enrichment Analysis
Enrichment analyses were conducted separately for each factor score phenotype (Persistent Externalizing and six domain-specific factors) following the enrichment methods of Aliev and colleagues (2015) in their series of analyses. We first used PLINK to estimate the association between each individual SNP and the phenotype (factor scores), controlling for biological sex and genomic principal components (PC). We included only two of the top ten PC eigenvectors as covariates in these association analyses, as only these two were significantly correlated (p < 0.05) with any of the measures under examination (Turner et al., 2011). All analyses were conducted three times at three different significance thresholds for determining a SNP association hit, p = 0.05, 0.01, and 0.001 levels. An association with a p-value above a given significance threshold (e.g., p < .05) was considered a “hit.”
We then considered whether this number of significant hits exceeded the number of hits that one could expect solely from chance. Given, for example, a p-value threshold of p = .05 and N = 1000 SNPs, one would expect N × p = 50 “hits” by chance, if each test were independent (an assumption to which we will return shortly). This expectation of 50 hits is actually the mean of a distribution of expected outcomes, where the standard deviation of that distribution is . Consequently, the number of observed hits can be converted to a Z score, and a one-tailed Z test can provide what is essentially a p-value for the set of p-values; in other words, it is a determination of the probability of observing this number of hits if there were no true association between the phenotype and the SNPs in this gene set.
The calculations described in the previous paragraph assume that each test of association is independent from the others. However, SNPs within a gene (or even in different genes) can be correlated with one another (i.e., in linkage disequilibrium [LD]). Consequently, in order to increase our confidence in the thresholds for determining enrichment, our primary test for significance was a stringent permutation test that determined an empirical threshold for the number of hits that could be expected from chance. Using RStudio (v0.98.994), individual phenotype data were randomly permuted and paired with preserved genomic data 1000 times for each phenotype among the target sample. We shuffled the phenotypes but preserved the SNPs under examination in order to preserve the LD structure of the genotypes in our target sample. Association analyses (using the SNP-level significance thresholds of 0.05, 0.01, and 0.001) were performed in PLINK (v1.90b3v) on all 1000 permuted data sets for each phenotype, and Z scores were calculated for the shuffled data in the same manner as the observed data. An empirical threshold was drawn at the 95th percentile highest-ranked Z score (an empirical α = 0.05) among the 1000 permuted sets for each phenotype, and we concluded that there was a significant enrichment of associations if the number of observed hits met or exceeded this empirical threshold. Empirical thresholds are provided in Table 2. As is evident in the Results, the permutation analyses provided a rather conservative test of enrichment.
Table 2.
Enrichment Analysis in a non-Hispanic White Sample
| Significance Threshold for “Hit” | Observed Data
|
95th Percentile Empirical Threshold
|
|||||
|---|---|---|---|---|---|---|---|
| Hits | Z-score | p value | Hits | Z-score | p value | ||
| p < 0.05 | Externalizing* | 200 | 2.89 | 0.0019 | 199 | 2.81 | 0.0025 |
| Alcohol* | 204 | 3.21 | 0.0007 | 199 | 2.81 | 0.0025 | |
| Cannabis | 151 | 201 | 2.97 | 0.0015 | |||
| Property Crime* | 203 | 3.13 | 0.0009 | 199 | 2.81 | 0.0025 | |
| Tobacco | 134 | 200 | 2.89 | 0.0019 | |||
| Risky Sex | 161 | 198 | 2.73 | 0.0032 | |||
| SS/IMP Personality | 147 | 198 | 2.73 | 0.0032 | |||
|
| |||||||
| p < 0.01 | Externalizing* | 54 | 3.72 | 0.0001 | 48 | 2.67 | 0.0038 |
| Alcohol | 41 | 1.44 | 0.0748 | 49 | 2.85 | 0.0022 | |
| Cannabis | 37 | 0.74 | 0.2300 | 48 | 2.67 | 0.0038 | |
| Property Crime | 42 | 1.62 | 0.0530 | 49 | 2.85 | 0.0022 | |
| Tobacco | 22 | 48 | 2.67 | 0.0038 | |||
| Risky Sex | 32 | 46 | 2.32 | 0.0102 | |||
| SS/IMP Personality | 22 | 50 | 3.02 | 0.0013 | |||
|
| |||||||
| p < 0 .001 | Externalizing* | 8 | 2.61 | 0.0045 | 8 | 2.61 | 0.0045 |
| Alcohol | 5 | 0.95 | 0.1708 | 8 | 2.61 | 0.0045 | |
| Cannabis | 1 | 8 | 2.61 | 0.0045 | |||
| Property Crime* | 8 | 2.61 | 0.0045 | 8 | 2.61 | 0.0045 | |
| Tobacco | 1 | 8 | 2.61 | 0.0045 | |||
| Risky Sex | 2 | 8 | 2.61 | 0.0045 | |||
| SS/IMP Personality | 2 | 8 | 2.61 | 0.0045 | |||
Note. Number of significant association hits at three p-value levels compared to bootstrapped empirical significance thresholds in the non-Hispanic White sample. P values provided for the observed data are from a one-tailed Z-test; inferential statistics are not present for constructs that did not at least meet the ‘expected’ number of hits, and would therefore have negative Z-scores. SS is sensation seeking and IMP is ZK Impulsive personality.
Bolded factors are significant at the empirically derived significance threshold.
Results
Sample Characteristics
Respondent retention by Wave 10 (five years after study commencement) was 62.7% of the Wave 1 sample, and 56% of those targeted for re-recruitment (26.8% of the original sample) completed surveys for Wave 11. Our factor models used the robust weighted least squares means and variance adjusted estimator (WLSMV) to account for missing data, and most of those who provided DNA and were included in the enrichment analyses also completed W11 survey data (92.9%). Nonetheless, we tested for sample differences between those with Wave 11 (W11) data available versus those without W11 data. There was a higher proportion of women with (64.9%) than without W11 data (58.0%), χ2(1) = 8.63, p < 0.01. Further, those who provided data at W11 had significantly higher factor scores in terms of persistent externalizing, F(1,2244) = 5.0, p < 0.05, d = 0.11, alcohol involvement, F(1,2244) = 8.93, p < 0.01, d = 0.14, and marijuana use, F(1,2244) = 9.42, p <0.01, d = 0.14. All other factor scores means were not different between those missing versus those providing data at Wave 11 (ps > 0.05).
We next compared the characteristics of those included in the genetic enrichment analysis to those who did not provide DNA or were dropped from analysis for quality control issues among non-Hispanic Whites only (total pool of non-Hispanic Whites included in the factor model N = 1223, non-Hispanic Whites without genetic data N = 886). The proportion of females in the analysis sample (64.4%) was significantly greater than in the non-analysis sample (56.9%), χ2(1) = 5.69, p = 0.017. Among the phenotypes examined (the factor scores), only marijuana use, F(1,1221) = 9.61, p < 0.01, d = 0.19, significantly differed between those who were (M = 0.19, SD = 0.57) and were not (M = 0.09, SD = 0.48) included in enrichment analyses (all other ps > 0.05).
Factor Model of Externalizing Phenotypes
The initial externalizing factor model (illustrated in Figure 1) demonstrated excellent fit: RMSEA = 0.020, CFI = 0.952, TLI = 0.950, χ2(1987) = 3719.97, p < 0.001. Standardized factor loadings for all paths in the model are presented in Table 1. We next tested if measurement invariance held across biological sex using multigroup modeling features of Mplus. A model that allowed for all loadings to be free by sex (but retained the constraint of equality over time) had slightly better fit indices (RMSEA = 0.019, CFI = 0.951, TLI = 0.949, χ2(4017) = 5643.899, p < 0.001) than a model constraining all loadings to be equal across the sexes (RMSEA = 0.021, CFI = 0.937, TLI = 0.936, χ2(4089) = 6187.347, p < 0.001). Although this appeared to be a significant decrement in model fit when using a chi-square difference test, Δχ2(72) = 307.918, p < 0.001, this form of comparative model testing can be sensitive to relatively trivial differences in model fit when the sample size is large. Thus, we re-scaled χ2 differences to an RMSEA metric (Hildebrandt, Wilhelm, & Robitzsch, 2009), whereby values greater than 0.05 on the resulting Index of Root Deterioration per Restriction (RDR) suggest that the change in model fit is significant. The resulting value (RDR = 0.038), indicated that we could constrain loadings to be equal by sex, and this model (the initial model) was used to derive all factor scores for enrichment testing.
Table 1.
Standardized Loadings from Externalizing Factor Model
| Externalizing Factor by Wave | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Alcohol | Cannabis | Property Crime | Tobacco | Risky Sex | SS | IMP | ||
| W1 | 0.59 | 0.81 | 0.20 | 0.54 | 0.51 | 0.48 | 0.30 | |
| W2 | 0.70 | 0.88 | 0.24 | 0.55 | 0.55 | – | – | |
| W3 | 0.65 | 0.85 | 0.25 | 0.52 | 0.53 | – | – | |
| W4 | 0.67 | 0.91 | 0.25 | 0.53 | 0.57 | – | – | |
| W5 | 0.66 | 0.90 | 0.27 | 0.56 | 0.56 | – | – | |
| W6 | 0.62 | 0.85 | 0.28 | 0.51 | 0.53 | – | – | |
| W7 | 0.60 | 0.83 | 0.30 | 0.52 | 0.52 | – | – | |
| W8 | 0.57 | 0.80 | 0.32 | 0.51 | 0.50 | 0.42 | 0.29 | |
| W9 | 0.53 | 0.73 | 0.35 | 0.48 | 0.46 | 0.38 | 0.27 | |
| W10 | 0.52 | 0.72 | 0.37 | 0.48 | 0.45 | 0.37 | 0.26 | |
| W11 | 0.50 | 0.63 | 0.32 | 0.45 | 0.39 | 0.37 | 0.27 | |
| Persistent Externalizing | Residual Alcohol | Residual Cannabis | Residual Property Crime | Residual Tobacco | Residual Risky Sex | Residual SS/IMP | Residual SS/IMP | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Externalizing | Alcohol | Cannabis | Property Crime | Tobacco | Risky Sex | SS | IMP | |
| W1 | 0.86 | 0.32 | 0.27 | 0.36 | 0.50 | 0.53 | 0.44 | 0.47 |
| W2 | 0.91 | 0.34 | 0.25 | 0.52 | 0.60 | 0.53 | – | – |
| W3 | 0.95 | 0.43 | 0.30 | 0.54 | 0.62 | 0.52 | – | – |
| W4 | 0.94 | 0.51 | 0.29 | 0.59 | 0.69 | 0.59 | – | – |
| W5 | 0.94 | 0.53 | 0.33 | 0.51 | 0.64 | 0.59 | – | – |
| W6 | 0.93 | 0.58 | 0.42 | 0.60 | 0.69 | 0.56 | – | – |
| W7 | 0.92 | 0.60 | 0.48 | 0.58 | 0.67 | 0.62 | – | – |
| W8 | 0.91 | 0.54 | 0.50 | 0.43 | 0.70 | 0.50 | 0.65 | 0.64 |
| W9 | 0.88 | 0.54 | 0.65 | 0.29 | 0.63 | 0.49 | 0.69 | 0.67 |
| W10 | 0.84 | 0.51 | 0.64 | 0.22 | 0.57 | 0.42 | 0.70 | 0.70 |
| W11 | 0.75 | 0.43 | 0.54 | 0.29 | 0.55 | 0.26 | 0.59 | 0.58 |
Note. Standardized loadings on factors from bi-factor model of externalizing traits (shown in Figure 1). Italicized names are indicator variables for each of the bolded factor names. SS is sensation seeking and IMP is ZK Impulsive personality. The upper portion of the table presents the loadings for each indicator on a wave-specific Externalizing factor from Wave 1 (W1) to W11; these loadings (unstandardized, Table S#) were constrained to be equal across time. The lower portion presents 1) the loadings of these wave-specific factors onto the higher-order Persistent Externalizing factor, capturing the common variance of these behaviors across time, and 2) wave-specific loadings onto each of the residual domain-specific factors. Domain-specific factors were composed of residual variance not accounted for by the common factor. All loadings were significant at p < 0.001.
Although measurement invariance held across the sexes, we tested if the mean factor scores differed by sex. As expected, males had significantly higher factor scores than females for Persistent Externalizing, F(1,2243) = 18.16, p < 0.001, Property Crime, F(1,2243) = 102.75, p < 0.001, Tobacco, F(1,2243) = 13.30, p < 0.001, Risky Sex, F(1,2243) = 6.09, p < 0.001, and IMP/SS Personality, F(1,2243) = 71.72, p < 0.001. There were no significant sex effects on the Alcohol, F(1,2243) = 3.24, p = 0.072, or Cannabis factors, F(1,2243) = 2.72, p = 0.099. Sex was included as a covariate in all subsequent genetic association analyses.
Enrichment Analyses
The number of significant hits at the p < 0.05, p < 0.01, and p < 0.001 levels are presented in Table 2. We concluded that there was significant enrichment of associations when the number of hits met or exceeded the empirical threshold determined from the permutation analyses, our primary criterion for significant enrichment. There was significant enrichment of associations for Persistent Externalizing, regardless of the p-value thresholds used to define a SNP association “hit” (p = .05, .01, and .001). Moreover, the Alcohol and Property Crime factors showed enrichment at the p < 0.05 level, as did Property Crime at the p < 0.001 level. The five genes containing the strongest SNP hits of association with the Persistent Externalizing factor (8 hits with p < 0.001) were GRID2, GABBR2, CSMD1, LINGO2, and MARCH1. Next, we determined if any individual SNP in any set of tests was significantly associated with factor scores. No single SNP was significant after correction for multiple comparisons using a false discovery rate correction (Benjamini & Hochberg, 1995).
In order to confirm that the enrichment signal is not dependent on the domain with the strongest standardized loadings onto the wave-specific externalizing factors (cannabis, Table 1) or on the domain of primary interest to the consortium that assembled the gene set (alcohol), we performed a series of sensitivity tests by removing each of these two measures from the analysis. We re-fit the hierarchical factor model with either all cannabis-related measures removed or all alcohol indicators removed, and re-ran the enrichment analyses on these new sets of factor score estimates. Despite the absence of either cannabis or alcohol measures, the enrichment signal persisted at or above the permutation threshold (Supplemental Tables S2–S3). Thus, it is clear that this signal does not depend on the presence of either cannabis or alcohol measures in the model.
Finally, we sought to determine the proportion of variance (R2) in persistent externalizing explained by variants within the SNP set. In order to achieve a total R2 across the SNPs tested, we used a polygenic risk score approach with five-fold within-sample validation following recommended methods (Vrieze, McGue, Miller, Hicks, & Iacono, 2013). In order to avoid gross overfitting that occurs when the same data are used as both discovery and target samples, the data were first split into five nearly equally sized groups (N = 337, subgroup Ns = 67, with one random group having N = 68). Next, we conducted linear association analyses in PLINK (exactly as done in the full sample analysis) five times, each time using 4/5th of the total data set (i.e., leaving out one sub-sample). The group not included in analysis then became the target sample for computing polygenic risk scores. As such, each participant was included in a ‘discovery’ sample four times, and was in the target sample once. Importantly, no individual was included in both the discovery and target samples in any analysis workflow.
We used the PRSice package (Euesden, Lewis, & O’Reilly, 2015) in order to determine the p-value thresholds for including SNPs in the polygenic score that would optimize R2 in each of the five target samples. The p-value lower bound was 0.0, the upper bound was 0.5, and models were run at increments of 0.01. There was a narrow range of optimal thresholds (p range = 0.02 to 0.07, M = 0.036, SD = 0.021). We used this mean p-value as the inclusion threshold for computing polygenic risk scores for each target sample. We then re-merged the five target samples and regressed the Persistent Externalizing factor scores on polygenic risk scores. There was a significant linear association (β = 0.16, p < 0.01, R2 = 0.026). In other words, about 2.6% of the variance in the Persistent Externalizing factor was accounted for by polygenic risk across a relatively small number of SNPs present in the hypothesized gene set.
Discussion
This study tested for an enrichment of associations between SNPs in an independently assembled set of genes previously associated with alcohol outcomes (Aliev et al., 2015) and factor scores from a multivariate, hierarchical model of externalizing behaviors. Overall, the results indicated that, within an ancestrally homogenous sample (non-Hispanic White individuals), there was significant enrichment of associations between genetic polymorphisms previously identified as relevant for alcohol outcomes and a common factor underlying a broad spectrum of externalizing behaviors and impulsive/sensation seeking personality traits across time. Our “deep phenotyping” approach (Robinson, 2012) provides an important complement to many genetic association studies, which, because of their sample size burdens, more typically use cross-sectional or retrospective measures of single clinical categories. More generally, this approach illustrates how psychological science can uniquely contribute to understanding the genetic underpinnings of complex behavior through multivariate analyses of behavioral and psychiatric phenotypes.
A Common Factor Model of Externalizing Behavior and Personality
A single, higher-order factor captured persistent involvement in externalizing behaviors and personality across emerging adulthood (ages 18 to 28). We tested for measurement invariance by biological sex and found that the model could be constrained to be equal across the sexes. Importantly, our model is largely consistent with a recent factor analytic examination of externalizing behavior in a genetic context (Latendresse et al., 2015). We extended the best-fitting model design (a bi-factor model) used by Latendresse and colleagues into a longitudinal framework, and included measures of hazardous alcohol use, cannabis use, property crime, tobacco use, and risky sex, as well as individual differences in sensation seeking and impulsive personality traits. Our phenotypic approach is also similar to that of an examination of polygenic risk for externalizing symptomology as captured by the first principal component extracted from a composite of clinical symptomology for externalizing disorders (Salvatore et al., 2015). However, our structured model allows for estimation of both common variance across the dimensions and unique domain-specific residual variances.
Including both clinically relevant behaviors in combination with disinhibited personality traits in a common factor model of externalizing represents a broader approach than used previously in models of problem behavior (Donovan & Jessor, 1985; Donovan et al., 1988). This approach complements the previous finding that enrichment of associations with this gene set extends beyond clinical symptomology into individual differences in these personality traits (Aliev et al., 2015). That is, in addition to identifying enrichment in a series of separate analyses for alcohol and drug dependence symptoms, antisocial behavior symptoms, and conduct disorder symptoms, Aliev and colleagues reported the strongest enrichment effect for sensations seeking personality. We believe it is important to consider both behavior and personality traits when examining the externalizing spectrum, given the strong phenotypic associations between sensation-seeking/impulsive personality facets and manifestations of externalizing behavior (e.g., problematic alcohol use; Dick et al., 2010; Hittner & Swickert, 2006) and previous evidence from twin studies that much of the genetic influence on antisocial behavior is shared with disinhibited personality traits (Harden et al., 2012; Mann et al., under review). Of note, the indicators with the largest (standardized) loadings to the general factor were substance use behaviors (cannabis, alcohol, tobacco), followed by risky sex, sensation seeking, ZK Impulsivity, and finally property crime (Table 1).
An Enrichment of Associations Implies Shared Genetic Etiology
Within a non-Hispanic White sample, this gene set (see Supplement for list of genes) showed an enrichment of associations with a Persistent Externalizing factor beyond a conservative, empirically derived, significance threshold. Importantly, the enrichment effect was more robust for this general factor than for any domain-specific residual factor, and the enrichment signal survived removal of the measure with the highest loading (cannabis, Table 1) in a re-estimated model. The only two residual factors for which some enrichment was detected were alcohol and property crime. These results are consistent with the hypothesis that externalizing behaviors share a substantial degree of genetic etiology (Krueger et al., 2002), and that genetic risks for alcohol use disorders are associated not only with alcohol-related outcomes, but with the broader construct of externalizing behavior. Lastly, we demonstrated that the total variance captured by this narrow set of 104 genes was 2.6%, which is nearly half the variance accounted for by genome-wide polygenic risk approach in another recent study (6%, Salvatore et al., 2015).
The five genes containing the strongest SNP hits of association with the Persistent Externalizing factor (8 hits with p < 0.001) were GRID2, GABBR2, CSMD1, LINGO2, and MARCH1. No individual SNP remained significant after correcting for multiple comparisons, and enrichment effects pertain to the set of SNPs as a whole rather than any particular SNP. Nevertheless, it is worth noting how polymorphisms in these 5 genes performed in Aliev et al.’s (2015) previous study this gene set and individual externalizing phenotypes. They found no significant associations between any phenotype and LINGO2, and one association between MARCH1 and extraverted personality. Furthermore, CSMD1 had significant hits for sensation seeking, and GRID2 (which encodes a glutamate receptor subunit) had hits with respect to both conduct disorder symptoms and sensation seeking. Lastly, among these five genes, GABRR2 (which encodes a GABA receptor subunit) showed the most hits in the COGA sample, with hits for antisocial personality symptoms, conduct disorder symptoms, extraversion and sensation seeking. Thus, across both studies, GABRR2 was the one of the most promising genetic signals to emerge.
Furthermore, all of these genes have shown some degree of association with externalizing phenotypes or neuropsychiatric disorders in other studies, although the strength of the evidence varies. Of note, one SNP in CSMD1 showed genome-wide significance in a large GWAS study of cannabis dependence (Sherva et al., 2016), and SNPs within CSMD1 were associated with tobacco use cessation in a clinical trial (Uhl et al., 2008). Similarly GABRR2 was associated with alcohol dependence in a sample (N = 1000+) with family history of alcohol use disorders (Xuei et al., 2010). Three genes, CSMD1, GRID2 (Greenwood et al., 2016), and GABRR2 (Wang, Liu, & Aragam, 2010) have also been associated with schizophrenia or bipolar disorder in moderately large samples (N = 1000+). Potentially related to behavioral self-control or reward processes, LINGO2 is associated with body mass or obesity (Rask-Andersen, Almén, Lind, & Schiöth, 2015), as is MARCH1 (Lee et al., 2016). Lastly, there is preliminary evidence of association between variants in GABRR2 and general cognitive ability (Ma et al., 2016) in a moderately sized (N = 987) Han Chinese sample. The associations with other, non-externalizing forms of psychopathology and with cognitive ability serve as reminder that genetic influences may operate even more broadly, through a general vulnerability to psychopathology, sometimes referred to as the p-factor (Caspi et al., 2014; Pettersson, Larsson, & Lichtenstein, 2016).
It is important to note that this is not a definitive set of genes regulating these behaviors, as much of the putative remaining polygenic variance remains unaccounted for. Indeed, a consortium specifically interested in alcohol use disorders assembled this specific set. These genes nominated by COGA authors (Aliev et al., 2015) did not emerge exclusively from more rigorous GWAS studies (criterion p < 0.00001), but also included genes with putative evidence from candidate gene studies (criterion p < 0.05). Nevertheless, our results also demonstrate that this set of genes is associated with externalizing behavior and traits in a general sample, rather than one selected for familial risk for alcohol use disorders, as was the COGA sample. Our results provide evidence that these genes may confer risk across the broad spectrum of individual differences in the general population rather than being limited to those with genetic risk for specific clinical manifestations of externalizing. Further work by other consortia, continued GWAS in larger samples, data from analyses using polygenic risk scores (e.g., Salvatore et al., 2015) and bioinformatics work will be necessary to prune this set of potential false positives, and to add additional candidates that may be related to the broad range of externalizing problems rather than to alcohol-related outcomes specifically.
The shared phenotypic variability and shared genetic etiology for externalizing behavior and traits in the general population implies that risk for exhibiting these behaviors may also share underlying neurobiology. This “cross-cutting” conceptualization is consistent with recent efforts of the U.S. National Institute of Mental Health to move away from the historical diagnostic categories of mental illness and towards dimensional constructs of psychopathology (Sanislow et al., 2010). The purpose of this Research Domain Criterion (RDoC) approach is to harmonize clinical research with neuroscience approaches in order to identify both psychological and biological mechanisms that underlie the traits shared across many clinical disorders, including externalizing (Patrick et al., 2013). Given the large number of genes included in the set analyzed here, we can only speculate about which neural systems may be impacted by variation within these genes. Likely candidate systems include those involved in self-regulation, motivation, affect, and reward processing, including the prefrontal cortex, striatum, and limbic systems (Aron, 2011; Bechara, 2005; Coccaro, Sripada, Yanowitch, & Phan, 2011; Jentsch et al., 2014; Jentsch & Taylor, 1999; Patrick et al., 2013; Verdejo-García & Bechara, 2009). Future work in integrative neuroscience including both human and animal model-based approaches may yield paths from genetic variation in this set to functional differences in the brain, and finally to behavior.
Improving Genetic Signal Detection for Complex Traits
Researchers interested in the genetic causes and correlates of complex phenotypes like externalizing behaviors face a clear quandary in how to optimally bridge the gap between individual SNPs and behavior given the clearly massive polygenicity (C. Chabris et al., 2015; C. F. Chabris et al., 2013) and the unavoidable measurement error when using diagnostic indicators or single manifest scales as outcomes. We believe that the present approach provides advantages on both sides of the genotype-to-phenotype equation, particularly in samples of moderate size. Capturing the common variance across a range of related behaviors and traits both reduces measurement error, and in the present analysis may more accurately represent the broader externalizing dimension that cuts across many psychological disorders and conditions of interest. Indeed, factor analytic approaches to phenotypic measurement may ultimately increase the power to detect meaningful and reliable genetic associations, as evidenced by analyses of simulated data (van der Sluis et al., 2010) and in recent analyses of depression phenotypes (Laurin, Hottenga, Willemsen, Boomsma, & Lubke, 2015).
Our method also provides advantages for genetic discovery by leveraging the posterior-probabilities of the extant GWAS and candidate gene literature. Although an enrichment analysis cannot be used to identify specific SNPs or genes that may regulate a trait (similar to a genome-wide polygenic risk approach), a confirmed enrichment signal can indicate that a much more narrow and hypothesis-driven set of genes (e.g., 104 in a set versus 20,000+ in a genome-wide polygenic risk model) is associated with a trait of interest. Indeed, our narrow set may represent half of the previously identified polygenic risk for externalizing (Salvatore et al., 2015). Thus, our approach strengthens both the assessment of the phenotype, in addition to refining and narrowing the genes under study, while also being feasable in samples smaller than tens of thousands of people.
Limitations and Conclusions
Our results must be weighed with respect to the study’s strengths and limitations. One important limitation of our analyses is the population sampled – entering college students at a competitive university. Individuals who exhibited more extreme externalizing behavior – thus disrupting academic performance in adolescence – were less likely to be present in our sample. However, our results independently extend previous results with this gene set found in the COGA sample, which was specifically selected for family history of alcohol use disorders. The consistency of results across samples with different potential sources of selection bias increases confidence in the robustness of the enrichment findings. Next, the set of SNPs available for analyses were limited to those that were present on the specific chip platform used in the current study and that passed quality control filters. Thus, nine genes from the original set could not be included, nor could potential rare causal variants not present on the chip. The personality measures were only available at five of the eleven waves and by the final wave, data was missing on a majority of participants, meaning that the information contributing to the full model was incomplete; nevertheless, we used robust methods for handling missing data. Lastly, our analyses did not include individuals from ancestral backgrounds other than non-Hispanic Whites, because we sought to eliminate population stratification confounds, limiting the generalizability of these findings to the broader population.
These results provide further evidence that variation in this set of 104 genes is associated with Persistent Externalizing within the non-Hispanic White population, a demonstration of our advantageous approach to bridging the gap between genetic variation and complex behavior. Bioinformatics approaches could be used to ascertain if these genes operate within a confined network, or if multiple gene networks, perhaps serving distinct neural systems, are present within this set of genes. Identifying these networks and neural systems may yield greater insight into the origins of broad externalizing traits and behavior, which may be fruitful for understanding the shared biological origins of a number of clinical disorders on the externalizing spectrum.
Supplementary Material
Acknowledgments
This research was supported by NIAAA grants 5T32 AA7471-28 (Ashenhurst), R01-AA013967 and R01-AA020637 (Fromme). We thank Emily Wilhite, Elise Marino, Dr. Andy Smolen, Peter Piliere, and Dr. Elliot Tucker-Drob for their assistance with data collection and statistical methods.
Footnotes
All listed authors contributed significantly to this manuscript in terms of study design, analysis, and writing. All authors approved the final manuscript.
The authors declare no conflicts of interest pertaining to the data or analysis presented herein.
These hypotheses and results were presented at the annual Research Society on Alcoholism meeting in New Orleans, LA in 2016.
One conceptual question, which is outside the scope of the current paper, is the extent to which externalizing in young adulthood shows heterotypic continuity, i.e., whether the behavioral manifestations of an underlying externalizing liability change over the course of development. This could be modeled by testing how the pattern of factor loadings changes over the waves. If, however, the pattern of factor loadings on the wave-specific Externalizing factors (and thus the residual variances for the specific behaviors) were allowed to vary across waves, this introduces the possibility that the domain-specific factors would be confounded with developmental differences. Heterotypic continuity and developmental changes in the magnitude and specificity of genetic influence remain interesting questions for future research.
Contributor Information
James R. Ashenhurst, Psychology Department, University of Texas at Austin, Austin, Texas, USA
K. Paige Harden, Psychology Department, University of Texas at Austin, Austin, Texas, USA.
William R. Corbin, Psychology Department, Arizona State University, Tempe, Arizona, USA
Kim Fromme, Psychology Department, University of Texas at Austin, Austin, Texas, USA.
References
- Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction. 2008;103(7):1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- Aliev F, Wetherill RR, Bierut L, Bucholz KK, Edenberg H, Foroud T, Investigators, C Genes associated with alcohol outcomes show enrichment of effects with broad externalizing and impulsivity phenotypes in an independent sample. Journal of Studies on Alcohol and Drugs. 2015;76(1):38–46. [PMC free article] [PubMed] [Google Scholar]
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nature Protocols. 2010;5(9):1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AR. From reactive to proactive and selective control: developing a richer model for stopping inappropriate responses. Biological Psychiatry. 2011;69(12):e55–68. doi: 10.1016/j.biopsych.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenhurst JR, Harden KP, Corbin WR, Fromme K. Trajectories of Binge Drinking and Personality Change Across Emerging Adulthood. Psychology of Addictive Behaviors. 2015;29(4):978–991. doi: 10.1037/adb0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara A. Decision making, impulse control and loss of willpower to resist drugs: a neurocognitive perspective. Nature Neuroscience. 2005;8(11):1458–1463. doi: 10.1038/nn1584. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. The Lancet. 2003;361(9357):598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Caspi A, Houts RM, Belsky DW, Goldman-Mellor SJ, Harrington H, Israel S, Moffitt TE. The p Factor: One General Psychopathology Factor in the Structure of Psychiatric Disorders? Clinical Psychological Science. 2014;2(2):119–137. doi: 10.1177/2167702613497473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabris C, Lee J, Cesarini D, Benjamin D, Laibson D. The Fourth Law of Behavior Genetics. Current Directions in Psychological Science. 2015;24(4):302–312. doi: 10.1177/0963721415580430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabris CF, Lee JJ, Benjamin DJ, Beauchamp JP, Glaeser EL, Borst G, Laibson DI. Why it is hard to find genes associated with social science traits: theoretical and empirical considerations. American Journal of Public Health. 2013;103(Suppl 1):S152–166. doi: 10.2105/AJPH.2013.301327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro EF, Sripada CS, Yanowitch RN, Phan KL. Corticolimbic function in impulsive aggressive behavior. Biological Psychiatry. 2011;69(12):1153–1159. doi: 10.1016/j.biopsych.2011.02.032. [DOI] [PubMed] [Google Scholar]
- Cooper ML, Wood PK, Orcutt HK, Albino A. Personality and the predisposition to engage in risky or problem behaviors during adolescence. Journal of Personality and Social Psychology. 2003;84(2):390–410. doi: 10.1037//0022-3514.84.2.390. [DOI] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Keller MC, Adkins A, Aliev F, Monroe S, Sher KJ. Candidate gene-environment interaction research: reflections and recommendations. Perspectives on Psychological Science. 2015;10(1):37–59. doi: 10.1177/1745691614556682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Smith G, Olausson P, Mitchell SH, Leeman RF, O’Malley SS, Sher K. Understanding the construct of impulsivity and its relationship to alcohol use disorders. Addiction Biology. 2010;15(2):217–226. doi: 10.1111/j.1369-1600.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan JE, Jessor R. Structure of problem behavior in adolescence and young adulthood. Journal of Consulting and Clinical Psychology. 1985;53(6):890–904. doi: 10.1037//0022-006x.53.6.890. [DOI] [PubMed] [Google Scholar]
- Donovan JE, Jessor R, Costa FM. Syndrome of problem behavior in adolescence: a replication. Journal of Consulting and Clinical Psychology. 1988;56(5):762–765. doi: 10.1037//0022-006x.56.5.762. [DOI] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349(6255):1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Calkins ME, Freedman R, Green MF, Gur RE, Braff DL. Genetic assessment of additional endophenotypes from the Consortium on the Genetics of Schizophrenia Family Study. Schizophr Res. 2016;170(1):30–40. doi: 10.1016/j.schres.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden KP, Quinn PD, Tucker-Drob EM. Genetically influenced change in sensation seeking drives the rise of delinquent behavior during adolescence. Developmental Science. 2012;15(1):150–163. doi: 10.1111/j.1467-7687.2011.01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt A, Wilhelm O, Robitzsch A. Complementary and competing factor analytic approaches for the investigation of measurement invariance. Review of Psychology. 2009;16(2):87–102. [Google Scholar]
- Hittner JB, Swickert R. Sensation seeking and alcohol use: a meta-analytic review. Addictive Behaviors. 2006;31(8):1383–1401. doi: 10.1016/j.addbeh.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Hu Lt, Bentler PM. Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives. Structural Equation Modeling. 1999;6(1):1–55. [Google Scholar]
- Jentsch JD, Ashenhurst JR, Cervantes MC, Groman SM, James AS, Pennington ZT. Dissecting impulsivity and its relationships to drug addictions. Annals of the New York Academy of Sciences. 2014;1327(1):1–26. doi: 10.1111/nyas.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch JD, Taylor JR. Impulsivity resulting from frontostriatal dysfunction in drug abuse: implications for the control of behavior by reward-related stimuli. Psychopharmacology. 1999;146(4):373–390. doi: 10.1007/pl00005483. [DOI] [PubMed] [Google Scholar]
- Jessor R, Jessor SL. Problem behavior and psychosocial development: a longitudinal study of youth. New York: Academic Press; 1977. [Google Scholar]
- Kendler KS, Myers J, Prescott CA. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Archives of General Psychiatry. 2007;64(11):1313–1320. doi: 10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: modeling the externalizing spectrum. Journal of Abnormal Psychology. 2002;111(3):411–424. [PubMed] [Google Scholar]
- Latendresse SJ, Henry DB, Aggen SH, Byck GR, Ashbeck AW, Bolland JM, Dick DM. Dimensionality and Genetic Correlates of Problem Behavior in Low-Income African American Adolescents. Journal of Clinical Child & Adolescent Psychology. 2015:1–16. doi: 10.1080/15374416.2015.1070353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin CA, Hottenga JJ, Willemsen G, Boomsma DI, Lubke GH. Genetic analyses benefit from using less heterogeneous phenotypes: an illustration with the hospital anxiety and depression scale (HADS) Genet Epidemiol. 2015;39(4):317–324. doi: 10.1002/gepi.21897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kwon DY, Kim MS, Choi CR, Park MY, Kim AJ. Genome-wide association study for the interaction between BMR and BMI in obese Korean women including overweight. Nutr Res Pract. 2016;10(1):115–124. doi: 10.4162/nrp.2016.10.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Niu B, Shi Z, Li J, Wang J, Zhang F, Zhang K. Genetic Polymorphism of GABRR2 Modulates Individuals’ General Cognitive Ability in Healthy Chinese Han People. Cell Mol Neurobiol. 2016 doi: 10.1007/s10571-016-0347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann FD, Engelhardt L, Briley DA, Grotzinger AD, Patterson MW, Harden KP. Sensation seeking and impulsive traits as personality endophenotypes for antisocial behavior: Results from two independent samples. doi: 10.1016/j.paid.2016.09.018. (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIAAA. National Institute of Alcohol Abuse and Alcoholism Council approves definition of binge drinking. 2004 Retrieved from http://pubs.niaaa.nih.gov/publications/Newsletter/winter2004/Newsletter_Number3.pdf.
- Palmer RH, Button TM, Rhee SH, Corley RP, Young SE, Stallings MC, Hewitt JK. Genetic etiology of the common liability to drug dependence: evidence of common and specific mechanisms for DSM-IV dependence symptoms. Drug and Alcohol Dependence. 2012;123(Suppl 1):S24–32. doi: 10.1016/j.drugalcdep.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick CJ, Venables NC, Yancey JR, Hicks BM, Nelson LD, Kramer MD. A construct-network approach to bridging diagnostic and physiological domains: application to assessment of externalizing psychopathology. Journal of Abnormal Psychology. 2013;122(3):902–916. doi: 10.1037/a0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson E, Larsson H, Lichtenstein P. Common psychiatric disorders share the same genetic origin: a multivariate sibling study of the Swedish population. Mol Psychiatry. 2016;21(5):717–721. doi: 10.1038/mp.2015.116. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, Sham P. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81 doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Andersen M, Almén MS, Lind L, Schiöth HB. Association of the LINGO2-related SNP rs10968576 with body mass in a cohort of elderly Swedes. Mol Genet Genomics. 2015;290(4):1485–1491. doi: 10.1007/s00438-015-1009-7. [DOI] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, Study LC. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340(6139):1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN. Deep Phenotyping for Precision Medicine. Human Mutation. 2012;33(5):777–780. doi: 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- Salvatore JE, Aliev F, Bucholz K, Agrawal A, Hesselbrock V, Hesselbrock M, Dick DM. Polygenic risk for externalizing disorders: Gene-by-development and gene-by-environment effects in adolescents and young adults. Clinical Psychological Science. 2015;3(2):189–201. doi: 10.1177/2167702614534211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanislow CA, Pine DS, Quinn KJ, Kozak MJ, Garvey MA, Heinssen RK, Cuthbert BN. Developing constructs for psychopathology research: research domain criteria. Journal of Abnormal Psychology. 2010;119(4):631–639. doi: 10.1037/a0020909. [DOI] [PubMed] [Google Scholar]
- Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A, Gelernter J. Genome-wide Association Study of Cannabis Dependence Severity, Novel Risk Variants, and Shared Genetic Risks. JAMA Psychiatry. 2016;73(5):472–480. doi: 10.1001/jamapsychiatry.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stautz K, Cooper A. Impulsivity-related personality traits and adolescent alcohol use: a meta-analytic review. Clinical Psychology Review. 2013;33(4):574–592. doi: 10.1016/j.cpr.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Steiger JH. Structural model evaluation and modification: An interval estimation approach. Multivariate Behavioral Research. 1990;25(2):173–180. doi: 10.1207/s15327906mbr2502_4. [DOI] [PubMed] [Google Scholar]
- Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, Ritchie MD. Quality control procedures for genome-wide association studies. Current Protocols in Human Genetics. 2011 doi: 10.1002/0471142905.hg0119s68. Chapter 1, Unit1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65(6):683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis S, Verhage M, Posthuma D, Dolan CV. Phenotypic complexity, measurement bias, and poor phenotypic resolution contribute to the missing heritability problem in genetic association studies. PLoS One. 2010;5(11):e13929. doi: 10.1371/journal.pone.0013929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdejo-García A, Bechara A. A somatic marker theory of addiction. Neuropharmacology. 2009;56(Suppl 1):48–62. doi: 10.1016/j.neuropharm.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viding E, Larsson H, Jones AP. Quantitative genetic studies of antisocial behaviour. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363(1503):2519–2527. doi: 10.1098/rstb.2008.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, McGue M, Miller MB, Hicks BM, Iacono WG. Three mutually informative ways to understand the genetic relationships among behavioral disinhibition, alcohol use, drug use, nicotine use/dependence, and their co-occurrence: twin biometry, GCTA, and genome-wide scoring. Behav Genet. 2013;43(2):97–107. doi: 10.1007/s10519-013-9584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu XF, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124(1–3):192–199. doi: 10.1016/j.schres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Whiteside SP, Lynam DR. The five factor model and impulsivity: Using a structural model of personality to understand impulsivity. Personality and Individual Differences. 2001;30(4):669–689. [Google Scholar]
- Xian H, Scherrer JF, Grant JD, Eisen SA, True WR, Jacob T, Bucholz KK. Genetic and environmental contributions to nicotine, alcohol and cannabis dependence in male twins. Addiction. 2008;103(8):1391–1398. doi: 10.1111/j.1360-0443.2008.02243.x. [DOI] [PubMed] [Google Scholar]
- Xuei X, Flury-Wetherill L, Dick D, Goate A, Tischfield J, Nurnberger J, Edenberg HJ. GABRR1 and GABRR2, encoding the GABA-A receptor subunits rho1 and rho2, are associated with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):418–427. doi: 10.1002/ajmg.b.30995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietsch BP, Verweij KJ, Bailey JM, Wright MJ, Martin NG. Genetic and environmental influences on risky sexual behaviour and its relationship with personality. Behavior Genetics. 2010;40(1):12–21. doi: 10.1007/s10519-009-9300-1. [DOI] [PubMed] [Google Scholar]
- Zuckerman M, Kuhlman DM, Joireman J, Teta P, Kraft M. A comparison of three structural models for personality: The Big Three, the Big Five, and the Alternative Five. Journal of Personality and Social Psychology. 1993;65(4):757–768. doi: 10.1037/0022-3514.65.4.757. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


