Abstract
Cell activity is coordinated by dynamic interactions with the extracellular matrix, often through stimuli-mediated spatiotemporal stiffening and softening. Dynamic changes in mechanics occur in vivo through enzymatic or chemical means, processes which are challenging to reconstruct in cell culture materials. Here we present a magnetoactive hydrogel material formed by embedding magnetic particles in a hydrogel matrix whereby elasticity can be modulated reversibly by attenuation of a magnetic field. We show orders of magnitude change in elasticity using low magnetic fields and demonstrate reversibility of stiffening with simple permanent magnets. The broad applicability of this technique is demonstrated with two therapeutically relevant bioactivities in mesenchymal stem cells: secretion of proangiogenic molecules, and dynamic control of osteogenesis. The ability to reversibly stiffen cell culture materials across the full spectrum of soft tissue mechanics, using simple materials and commercially available permanent magnets, will make this approach viable for a broad range of laboratory environments.
Keywords: Tissue engineering, hydrogels, magnetic materials, biomedical applications, stimuli responsive materials
1. Introduction
Cells adapt to and respond to their local microenvironment in a context dependent fashion, that depends on spatiotemporal control of biophysical and biochemical properties[1]. The chemistry[2], structure[3] and mechanics[4,5] of the extracellular matrix (ECM) all influence cellular activity. This sensitivity to the environment enables cells to maintain ECM homeostasis by responding to external changes[6] and allows highly versatile stem cells to take on multiple roles in different niches[7]. This complexity, however, is significantly challenging to deconstruct with in vitro approaches when trying to decipher the properties that facilitate a specific activity or outcome[8]. Thus, the development of engineered model matrices is imperative for the study of ECM properties that guide cellular behavior.
Mechanotransduction, whereby cells sense their mechanical environment and process this information into biochemical signals, is a complex process[9] that occurs ubiquitously[10] and has severe consequences when disrupted[11]. Adult mesenchymal stem cells (MSCs), due to their relevance to regenerative medicine, have been studied extensively to investigate their responsiveness to mechanical properties of the ECM. MSC fate decisions can be driven by mechanical factors such as substrate stiffness[12,13] and cell geometry[14–16]. Furthermore, the MSC secretome can be influenced by such factors, which may be used to optimize their pro-angiogenic therapeutic capabilities[17,18]. Interestingly, in several of these studies[12,19,20], using physiological conditions gave optimal results.
Contrary to the polymeric cell culture substrates used in most of these studies, cellular environments are not static. In fact, the ECM is constantly changing in normal[21] and diseased[22,23] tissue. Several reports have shown that, in addition to sensing their current environment, stem cells are affected by or ‘remember’ their mechanical history[24–26]. In addition, although great care is usually given to the temporal regulation of chemical factors used in several protocols (during somatic cell reprogramming[27] for instance), there is no reason to assume that mechanical regulation is not just as important. In fact, a recent study utilizing tunable polymeric materials to study the effects of parameters such as anisotropic topgraphic cues[28] or stress relaxation[29] reveal highly dynamic cellular responses. Therefore, there is a need for in-vitro cell culture platforms with externally tunable mechanical properties in order to study the temporal effect of these parameters[30].
There have been several innovative strategies for making tunable stiffness systems. For example, pH[31], DNA strands[32] and calcium ion concentration[33] have been used to reversibly change substrate stiffness. However, these factors may affect cellular signaling and stiffness changes occur over different time scales. Another strategy is to alter the structure of the hydrogel using multi-step crosslinking[34] (for stiffening) or controlled degradation[35] (for softening) using various methods. For example, Kloxin et al. used photodegradable hydrogels to tune the gel microenvironment through visible light irradiation[36]. Although these methods work well for one-directional changes in elasticity, they cause irreversible changes in gel structure and do not offer reversibility. Recently, Rosales et al.[37] used an azobenzene based reversibly photo-switchable PEG hydrogel. However these gels can thermally relax and only show a modest change in modulus. Reversibility and the ability to modulate stiffness in a controlled manner are important to study continuous, temporally modulated changes that occur in vivo such as during development, homeostasis or disease (For example, fibrosis or wound healing)[38] or in vitro to ascertain how long it takes for changes in cell behavior to become permanent (mechanical dosing). MSCs, for instance, show irreversible changes in localization of transcriptional co-activator Yes-associated protein (YAP) in the nucleus by 10 days of culture in stiff conditions but these changes could be reversed by switching the environment before 10 days[25].
In this paper we adapt magnetorheological gels[39] and elastomers[40,41] into a magnetically tunable hydrogel platform for cell culture. We modify carbonyl iron (CI) particles for incorporation into polyacrylamide hydrogels and we demonstrate over two orders of magnitude shift in hydrogel compliance in response to magnetic fields. The gel stiffness can be easily and reversibly changed using permanent magnets, obviating the need for complex instrumentation, and thus making this technique amenable to virtually any research laboratory. Using mesenchymal stem cells as a model adult stem cell with therapeutic potential we show how magnetic fields modulate cell spreading and cytoskeletal tension, which impacts secretion of pro-angiogenic molecules and the propensity to undergo osteogenesis. The simplicity in which hydrogel mechanical properties can be modulated in situ will make this tool useful for a wide variety of applications, where temporal control over the biophysical microenvironment is desired.
2. Results
2.1. Gel concept and fabrication
We used polyacrylamide (PA) as the base polymeric hydrogel for our system, because of PA’s flexibility as a cell culture platform with tunable elasticity within physiological stiffness ranges[42]. Furthermore, PA has been previously used in multiple studies with MSCs[12,16,17,43], which provides a wealth of preliminary data for MSC behavior on soft and stiff PA substrates. These gels are formed through radical addition polymerization (Figure 1a) as described previously[42].
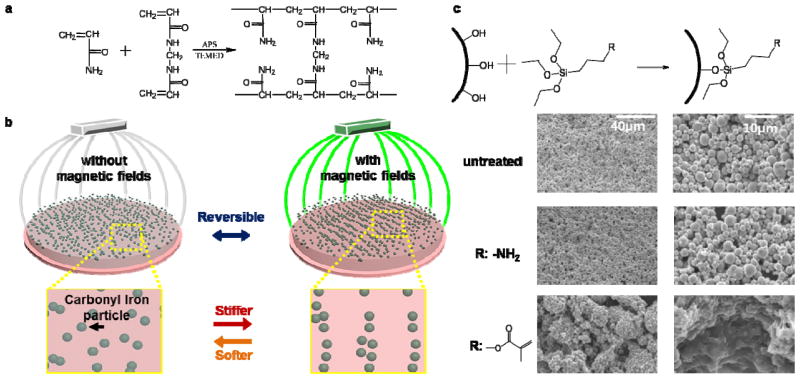
Figure 1. Carbonyl iron polyacrylamide composite hydrogel formation and stiffening.
(a) Formation of polyacrylamide via radical polymerization from acrylamide and bis-acrylamide monomers. (b) Schematic of magnetoactive hydrogel system formed by incorporation of carbonyl iron (CI) particles in a polyacrylamide matrix. Subjecting these gels to magnetic fields causes alignment of the particles, stiffening the hydrogels35. (c) Silane modification of CI particles (top) Silane chemistry can be used to modularly modify the surface of CI particles. (bottom) SEM images of dried PA hydrogels with incorporated CI particles that are untreated, treated with an amine-terminated silanes or treated with a methacrylate terminated silane. The latter shows covalent incorporation into the hydrogel and aggregation behavior.
In order to add magnetic tunability to our hydrogels, we adapted the approach of Mitsumata et al. to incorporate carbonyl iron (CI) particles in carrageenan hydrogels[39] (Figure 1b). In order to make the particles more stable in cell culture conditions, and allow functionalization of the particles, we used silane chemistry to modify the surface with different functional groups (Figure 1c, top) This has the added benefit of allowing modular modifications to the system such as covalent incorporation of moieties to the particles or the chemical crosslinking of particles in the hydrogel network. To demonstrate the efficacy of this treatment, we used two different silanes: aminopropyl triethoxysilane (APTES) to passivate the particles to stabilize them for long term cell culture, and 3-(Trimethoxysilyl)propyl-methacrylate (TMSPM) to enable covalent incorporation into PA gels. FTIR analysis shows a change in the spectrum upon treatment with APTES and TMSPM with characteristic peaks showing chemical conjugation at ~1080 cm−1, attributed to open chain siloxane groups, with the TMSPM treated particles showing characteristic peaks at ~1638 cm−1 (C=C) and 1720 cm−1 (C=O)[44] (Figure S1). Upon incorporation of the modified particles into the hydrogels, the gels were dried and imaged using SEM to see whether there was any visible effect on the structure (Figure 1c, bottom). The amine-terminated particles, while they appear similar to untreated particles in SEM, demonstrated a significant improvement in stability under prolonged cell culture conditions, where gels made with untreated particles dissolve within 10 days (Figure S2). The methacylate-terminated particles, on the other hand, show different characteristics to both amine terminated and untreated particles. SEM images show gel residues attached to the particles indicating covalent attachment of the hydrogel to the particles during gelation. An unforeseen consequence of the treatment, however, was aggregation of the particles into ‘clumps’, presumably due to heterogeneous polymerization between monomer and particles. For all particle treatments, repeated application and removal of magnetic fields does lead to some limited leaching of particles from the hydrogel, however leached particles sediment to the bottom of well plates and do not further impact cell behavior. APTES modified particles were used for the remainder of the study unless otherwise noted.
2.2. Mechanical characterization of hydrogels
Shear rheometry was used to characterize the hydrogel’s mechanical properties and their response to magnetic fields using a magnetorheological setup (Figure S3a). The gel composite is a viscoelastic solid as observed by creep compliance and oscillatory shear measurements (Figures S3b, S3c), with linear viscoelastic equilibrium compliance J≈0.01 Pa−1 (modulus G≈0.1 kPa). Figure S3b shows the strain amplitude dependence of the gel composite viscoelasticity at a frequency of 1 rad s−1; the sweep range was chosen to avoid a nonlinear response that may irreversibly affect the sample. We observed almost constant storage modulus and a slowly rising loss modulus between 0.1% and 10% strain. This agrees with previous data for polyacrylamide[45]. We use a strain amplitude of 1% for performing all further linear viscoelastic measurements on the gel. Linear viscoelastic frequency sweep measurements (Figure S3c) indicate minimal frequency dependence. Further information at longer timescales is obtained from a creep compliance test (Figure S3d). At short times, inertio-elastic oscillations are observed, due to the well-known effect of sample elasticity coupling to instrument rotational inertia. At longer times beyond 100s, the compliance J(t) is nearly constant, indicating solid-like behavior at these timescales.
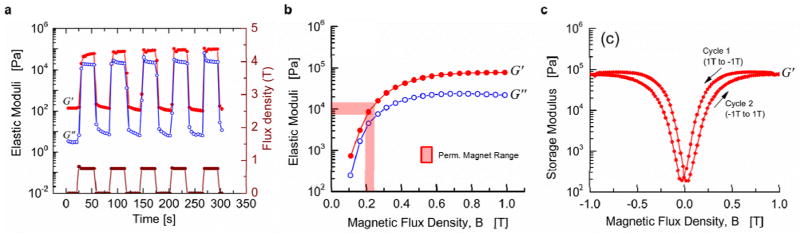
The gel composite dramatically stiffens in response to magnetic field, as shown in Figure 2. The reproducibility of the magnetic field effect is shown in Figure 2a, as observed by cycling the magnetic field between 0 and 0.75T several times. Each full cycle was 1 minute (30 seconds at 0T and 30 seconds at 0.75T). The storage modulus G′ at 0T ranged between 0.1–0.14 kPa and at 0.75T stiffened to 60–90 kPa. The gel recovered its elastic modulus at 0T with each cycle, indicating no irreversible disruption of the polymer network. The modulus at 0.75T increased marginally with each cycle, but this is insignificant in the context of the relative change in moduli from 0T to 0.75T. Importantly, this range of mechanical behavior corresponds to nearly the entire range of physiological stiffness observed in biology, from brain tissue to collagenous bone[12]. Thus, a single hydrogel can cover the entire spectrum of physiological elasticity, dynamically, and on a single gel surface.
Figure 2. Magnetic field dependent elasticity of PAC-CI gels.
(a) Pulsed magnetic field from 0T to 0.75 T showing three orders of magnitude change to elasticity. (b) A magnetic field ramp showing a continuous rise in the modulus with field strength, with the highlighted region representing magnetic flux density achieved with permanent magnets. (c) Elastic hysteresis is observed between back and forth ramps from −1T to 1T.
The sensitivity to continuously modulated magnetic field strength is shown in Figure 2b, with a magnetic field ramp from 0.1T to 1T. The elasticity (G′) transitioned smoothly and continuously as the magnetic field was raised, starting to level off and saturate around 0.8T. Magnetic saturation is a well-known effect in magneto-responsive materials[46]. Hysteresis effects are also apparent when the magnetic field is cycled through positive and negative values (Figure 2c), with cyclic ramps from 1T to −1 T. Between cycles, the gel appeared to attain identical values of G′ at 0T, but the approach to this value depended on the direction of the field, showing signs of hysteresis in the mechanical response. When compared with cycle 1 (1T to 0T), the gel elasticity in cycle 2 is larger in the first half of the cycle (−1T to 0T), but smaller in the second half (0T to 1T). We attribute this to the magnetic hysteresis of the particles, which may consequently result in the particle network maintaining its configuration from a previous cycle (cycle 1, 0T to −1T), and rearranging to a different configuration in cycle 2 when the field was being ramped up from 0T to 1T.
Next, we checked the particle fraction dependence of the composite (Figure S4). At B=0T, the linear elastic modulus (G′) shows no frequency dependence for all considered concentrations and the loss modulus (G″) shows weak frequency dependence before being affected by instrument rotational inertia, shown as a limit line in the figure[47]. Up to a volume fraction of 30% in the composite, both moduli increase with increasing particle concentration, beyond which the composite elasticity drops with particle inclusion, and the gel degrades. The magnetic field dependent mechanical properties of each resulting composite are outlined (Figure S4b), and serve as a good reference for the choice of 30% particle volume fraction in the gel, which represents the maximum change in (G′).
To determine the effect of the surface functionality on treated/untreated CI particles in the composite, we studied the magnetic field dependent mechanical properties of untreated and amine-treated particles in the polyacrylamide gel (Figure S5). The linear viscoelastic properties of the composite show no significant difference when compared to the properties of the composite with untreated particles. Both moduli show strong magnetic field dependence (Figure S5b) and increase ~100X as the field is ramped from 0T to 1T. The same material is then exposed to five cycles of a pulsed magnetic field from 0T to 0.75 T (Figure S5c). The linear viscoelastic moduli are reversible between cycles 2–5 and show ~200X increase in moduli. A larger relative change in moduli here is attributed to a faster rate of change of magnetic field, in contrast to a slow and gradual growth from 0T to 1T in the ramps shown in Figure S5b. A study with methacrylate treated particles in the gel was also attempted, but consistently resulted in a heterogeneous composite that showed noticeable phase separation at the volume fractions of interest, ϕ=30%.
As a control with particles alone, we formulated a suspension of amine-treated carbonyl particles in a silicone oil-grease medium (Figure S6). This medium has a low yield stress of ~2 Pa which inhibits particle sedimentation for ~24 hours[48]. The storage modulus and loss modulus increase with increasing particle volume fraction. We show this volume fraction dependence with a characteristic shear modulus G0≡G′ (ω=1 rad s−1), both with and without magnetic field (Figure S6b). The suspension shows magnetic field dependent linear viscoelasticity that scales as G0 ~ ϕ2.2, an exponent that is slightly larger than the earlier observed scaling of 1.7[49].
While tunability across the entire range of elasticity is desirable, the equipment used to impose a variable magnetic field is expensive, requires electrical power, and is cumbersome in cell culture environments. An alternate solution is to use permanent magnets which can provide a constant magnetic field, which can be changed by varying the distance between magnet and gel. We used permanent rare earth magnets attached to a well plate cover (Figure 3, inset), upon which the hydrogel samples with cells in well plates can be placed. By measuring the magnetic field strength using a Hall probe, we found the field to be ~0.2–0.25T which corresponds to a storage modulus in the range ~8–15 kPa. (Figure 2b) Therefore, with this setup, we can ‘switch’ or ‘oscillate’ the modulus between 0.1–0.14 Pa (no magnet) and 8–15 kPa (with magnet). Henceforth these will be given the designations ‘soft’ and ‘stiff’, respectively.
Figure 3. Stiffening hydrogels enhances secretion of pro-angiogenic factors.
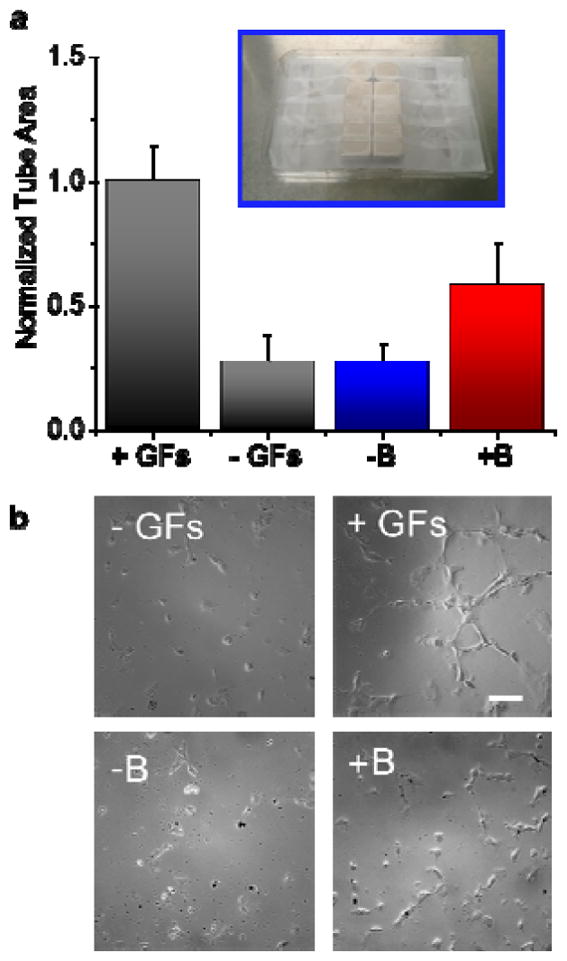
hMVEC tube formation on matrigel is quantified as a function of secreted factors in the conditioned medium. Conditioned medium is obtained from MSCs cultured on magnetoactive hydrogels cultured with and without a magnetic field. EGM-2 media is used as a positive control and unconditioned DMEM is used as a negative control. (below) representative images of tube formation on the positive, with or without magnetic field conditions (scale bar:100μm) (Inset) Permanent rare earth magnets taped to the underside of a well plate cover. Well plates with samples inside them can be placed on top of this well plate cover to introduce a magnetic field to substrates. Error bars represent standard error.
2.3. Modulation of substrate stiffness guides secretion of pro-angiogenic molecules, cell spreading and differentiation
Previously we demonstrated how increasing stiffness of fibronectin conjugated hydrogels will enhance MSC pro-angiogenic potential[17]. First, to confirm protein incorporation onto the magnetoactive gel surfaces we used fluorescently tagged fibronectin and confirmed increased fluorescence signal from the hydrogels after addition (Figure S7a). Next, we micropatterned fibronectin on our gel surface using a polydimethylsiloxane (PDMS) stamp fabricated through photolithography to present oval features in relief. After addition of MSCs we observe preferential adhesion to the micropatterned regions (Figure S7b). We also tested the effect of magnetic fields on protein arrangement on the gel surface (Figure S8). Fluorescence analysis indicates negligible changes in the uniformity; however, direct gel contact with the magnet leads to the appearance of fluorescent lines suggesting some adhesion to the particles within the gel that becomes apparent after alignment in a magnetic field.
We cultured MSCs on our tunable hydrogel for two days and performed a tube formation assay (hMVECs on matrigel as described previously[17]) using the conditioned medium from MSCs cultured on the surfaces with (+B) or without (−B) a magnetic field. We also used EGM-2 growth media as a positive control and unconditioned DMEM as a negative control. Quantifying the tube area from the different media (Figure 3) shows almost double the tube formation for MSCs cultured on stiff vs soft surfaces. This trend agrees with our expectations for MSCs on soft or stiff substrates and demonstrates the potential for using dynamic magnetoactive materials to guide angiogenesis when MSCs are a therapeutic agent.
To further investigate how changing the elasticity in-vitro will influence adherent cells, we used one of the fastest ‘cellular indicators’ of substrate elasticity: cell spread area. Cell spread area generally increases with substrate stiffness and the changes happen relatively quickly. We cultured MSCs on soft and stiff conditions for 4 hours and then we switched a subset of the MSCs from stiff to soft and cultured them for a further 4 hours (Figure 4a). Overall, MSCs on soft substrates had an average area of about 750 μm2 while those on stiff substrates had an average area of ~1,300 μm2 (Figure 4b). Cells cultured on stiff-to-soft substrates reverted to an area of ~900 μm2, just above that from soft substrates. Interestingly, some cells showed more prominent actin stress fibers when cultured on stiff vs soft substrates which persisted for 4 hours (Figure 4a, bottom). Taken together, this shows that reversible changes in substrate stiffness can lead to reversible changes in cellular spreading.
Figure 4. Modulating hydrogel stiffness in-vitro reversibly affects MSC spread area.
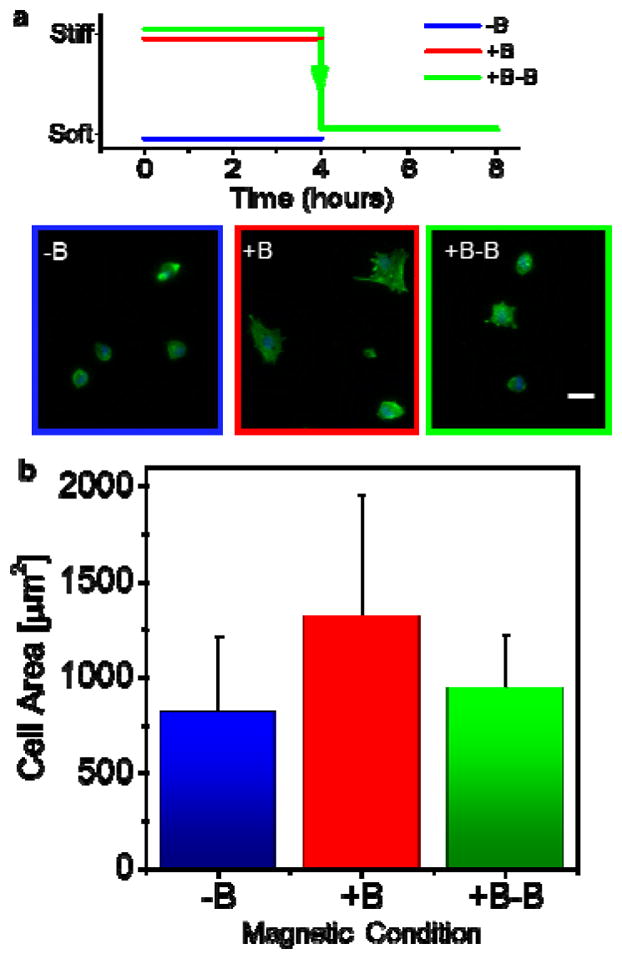
(a) (top) cells were cultured with (+B) or without (−B) a magnetic field or cultured for 4 hours on a magnetic field and then then the field was switched off (+B−B). (bottom) Representative images of MSCs cultured on these substrates at these different conditions (Scale bar: 50μm). (b) Quantification of average MSC area.
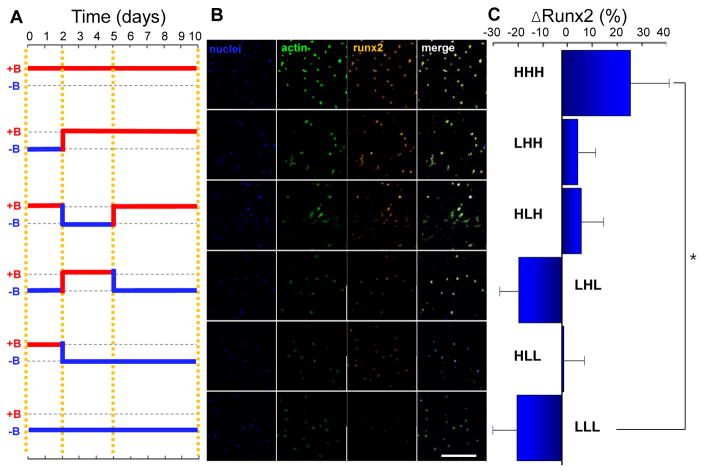
MSC osteogenesis is another phenomenon that has been studied extensively on hydrogels[12,17,50]. Generally, going to stiffer substrates increases osteogenic marker expression. Runx2 is an important transcription factor involved in regulating lineage specification, and is the most common marker used to classify early osteogenesis. Several reports have demonstrated a ‘memory effect’ where the properties of a previous microenvironment influences cell state to a degree that lineage-specific activity remains apparent[25,26,34]. Guvendiren et al. show a dependence of Runx2 expression when gels are switched from soft-to-stiff at different time points, with gels switched earlier showing increased Runx2 expression[34]. We performed a similar experiment with MSCs cultured on soft, stiff or switched from soft-to-stiff. First, cells were seeded and allowed to attach for 1 day at one stiffness, then the stiffness was changed at day 2, and again at day 5 (Figure 5a, left). We then imaged and quantified Runx2 expression at 10 days (Figure 5b). The experiment was performed in this way in order to see the relative effects of stiffness changes during cell attachment and spreading (up to day 2) and in the early (up to day 5) and late stages (day 5–10) of our study of early osteogenic lineage specification. We used 6 different variations labeled with H for high stiffness and L for low stiffness (e.g. HLH indicates high stiffness for 1 day, low stiffness for 4 days and high stiffness for the remaining 5 days). To highlight relative differences between samples, the data is shown as deviation of Runx2 expresssion from the average of all samples at day 10. Analysis of Runx2 expression indicates that initial stiffening plays a significant role in guiding the final differentiation state. Furthermore, stiffeness condition during the latter stages of culture (last 5 days) corresponded with the largest increase in Runx2 intensity. Surprisingly, intermediate stiffening (between days 2 and 5) did not exert a significant influence on Runx2 expression.
Figure 5. Dynamic stiffneing guides the extent of osteogenesis in mesenchymal stem cells.
(a) Magnetization profile for dynamic stiffening in culture. (b) Representative images of DAPI, Phalloidin and Runx2 of MSCs cultured for 10 days at various magnetic field profiles, with (+B) or without (−B) magnetic fields (scale bar: 500um). (c) Percent change of Runx2 expression (compared to average of all samples at day 10) in MSCs cultured for 10 days in bi-potential osteogenic/adipogenic medium at different magnetic field profiles from the overall average. * P<0.05.
Interestingly, after 10 days the average cell area was similar between the soft and stiff conditions (Figure S9a). This result is somewhat surprising, and may be related to cells adapting a preferred shape after differentation, or through remodelling their microenvironment and the hydrogel properties. Nevertheless, looking at cell area vs. Runx2 expression for a random sample of 200 cells (Figure S9b) we observed that for virtually any particular range of areas, Runx2 expression was higher in cells on stiff matrices when compared to soft.
3. Discussion
We have demonstrated a magnetically tunable hydrogel system, using functionalized carbonyl iron particles in a polyacrylamide matrix, which shows a several-fold change in elasticity when subjected to magnetic fields. The CI particles reinforce the gel at higher magnetic fields, increasing elasticity reversibly. The CI particles can be modified by flexible silane chemistries for conjugation to the polymer network or other moieties. Using a magnetic field as a stimulus has the added benefit of not affecting cellular cycle and growth which has been shown for fields up to 10T[51].
Mechanical characterization shows possible modulation of storage modulus between ~0.1kPa to ~80 kPa reversibly. This accessible range of elasticities can cover most of the physiologically relevant tissue stiffness giving this platform wide applicability as a model system for studying mechanical effects on different cellular systems in-vitro.
We show how simple and inexpensive permanent magnets can be used to dynamically stiffen hydrogels for the investigation of several cellular activities that are influenced by mechanical properties. Although this approach sacrifices the ability to continuously tune the elasticity, it allows switching between soft and stiff conditions and retains the reversibility. It is important to consider that, with particle alignment, there are some changes in protein distribution and changes in surface topography and alignment may also occur that modulate cell behavior or adhesion[40]. MSCs cultured on soft substrates show nearly a 2-fold increase in spread area when a magnet is applied. The influence on cell area is reversible and the cell area is reduced after removal of the magnet; curiously however, stained actin remains brighter in cells cultured on magnetically treated gels compared to cells cultured on soft gels without an applied field. This may be related to residual “stiffening” effects caused by hysteresis we observed when magnetic fields are removed.
Using this simplified system, we show that the pro-angiogenic potential of MSCs increase when they are cultured on magnetically-stiffened substrates, which agrees with our previous observations on static polyacrylamide hydrogels. In addition to pro-angiogenic secretion, we show how the degree of MSC osteogenesis can be dynamically modulated by simply adding or removing a magnet below the culture plate. After 10 days exposure to differentiation promoting media, MSCs cultured on magnetoactive hydrogels display susceptibility to the temporal dynamics of stiffening. MSCs initially seeded on a stiff matrix appear predisposed to osteogenesis, while initial seeding on soft matrices appears to discourage lineage commitment. Interestingly, stiffening during intermediate times in our experimet (days 2–5) did not enhance osteogenesis compared to MSCs cultured on soft gels for the entire experiment. Stiffening at later timepoints (days 5–10) exerted a larger influence on osteogenesic marker expression at 10 days. Notably, since we are evaluating Runx2 expression through immunofluorescent staining, accumulation of protein with time plays a role, giving extra weight to total time spent at each stiffness condition, perhaps explaining why the HHH condition shows higher protein expression than the LHH or HLH conditions. Overall, we speculate that mechanotransduction during early stages of culture are important for initiation of osteogenic signaling. This is consistent with previous reports of early mechanical signals promoting a susceptibility to the osteogenesis program[25].
4. Conclusion
The dynamic modulation of stem cell activity using magnetic fields demonstrates the potential of this system for studying temporal regulation of ECM mechanical properties in physiological and pathological contexts, adding tunable stiffness to other applications of magnetoactive hydrogels in tissue engineering such as on-demand drug and cell delivery[52] and modulation of surface roughness and topography[53]. To our knowledge, there has only been one other study where magnetoactive hydrogels have been used in cell culture[40], and then with PDMS elastomer. Our study here facilitates bridging the gap between the need for tunable hydrogels for cell culture and magnetoactive systems to study dynamic microenvironments. Some examples of dynamic microenvironments observed in vivo include gastrulation[54], branching morphogenesis[55], cardiovascular development and function[56], and pathophysiological processes such as fibrosis and cancer[22,57]. Although the use of permanent magnets is convenient, more advanced magnetic accessories will be necessary to capture subtle changes underlying many biological processes. Nevertheless, this simple technique for studying the effect of dynamic temporal modulation of substrate mechanics on cell activity, that is flexible enough to be used in many different hydrogel platforms, may find broad applicability for cell biology studies and for ‘priming’ cells to an appropriate state for therapy.
5. Experimental Section
All materials were bought from Sigma Aldrich unless otherwise noted.
5.1. Carbonyl Iron (CI) particle modification and hydrogel preparation
CI particles (grade EW) were generously provided by QED technologies. The particles were are either amino functionalized using aminopropyl trimethoxysilane or methacrylate functionalized using 3-(Trimethoxysilyl)propyl-methacrylate. The treatment is performed by incubating the particles in the desired silane dissolved in 90% ethanol solution overnight under shaking. Modified CI particles are washed thoroughly with DI water at least 4 times before use.
For gel preparation, a pre-polymer solution mixture of acrylamide and bis-acrylamide (Fisher Scientific) is mixed according to the desired crosslinking density (here we use 3% acrylamide and 0.06% bis-acrylamide) and degassed under nitrogen gas for 10 minutes. 18 mm glass coverslips are cleaned by sonication under ethanol for 15 minutes followed by sonication in DI water for 15 minutes. Coverslips are activated for gel attachment by treatment with 0.5% solution of APTES for 3 minutes followed by thorough washing with DI water 3 times. Coverslips are then treated with 0.5% glutaraldehyde solution for 30 minutes and washed with DI water. A hydrophobic microscope slide is prepared by treatment with Rain-X (SOPUS). The desired mixture of CI particles by volume is prepared in pre-polymer solution. Gelation is initiated by addition of 0.1% ammonium persulfate and 0.1% tetramethylethylenediamine (TEMED) and vortexing the solution. 20 μl of solution is pipetted onto the hydrophobic microscope slide and the activated coverslip is placed face down on the drop. The gel is left to solidify for ~20 minutes and is then detached from the hydrophobic slide. Gels are washed at least 3 times with DI water to remove any particles not incorporated during gelation.
For protein incorporation, fibronectin (from human plasma) at 50μg ml−1 is incubated on the surface of PDMS stamps for 30 minutes. Then, air is used to blow excess solution from the surface of the PDMS stamps and fibronectin is transferred to the surface of the gel by stamping. Gels are washed several times in DI water and stored in 12 well plates until cell culture.
5.2. Scanning electron microscopy (SEM)
SEM images of dried PA hydrogels with modified CI particles were acquired using a JEOL 6060-LV scanning electron microscope under high vacuum. A thin layer of gold was sputtered onto the surfaces to ensure electrical conductivity of the samples. Images were taken at either 1,000x or 3,500x.
5.3. Fourier transform infrared spectroscopy
FTIR was performed on a Spectrum 100 (Perkin Elmer) machine in transmittance mode. Spectra were taken at each point between 450 to 4000 cm−1on samples in dichloromethane on a potassium bromide salt plate. Baseline correction and normalization were performed on spectra.
5.4. Mechanical characterization
Dynamic shear measurements were performed on a rotational rheometer (combined-motor-transducer, DHR-3, TA Instruments) with a Magneto-Rheology (MR) setup for uniform and controlled application of magnetic fields from −1 T to +1 T (experimental setup shown in Figure S2a). Disks of samples of 1 mm thickness and 20 mm diameter were prepared, and measurements were made with a non-magnetic 20 mm diameter parallel plate fixture. An electro-magnetic coil beneath the sample imposed magnetic field lines orthogonal to the plate surface, and a hall probe under the bottom fixed plate gave real time measurement of the external field strength during tests. An upper yoke surrounded the upper geometry to draw field lines orthogonal to the plate surfaces. Tests were run at a constant temperature of 37°C, maintained by a closed-loop-control fluid circulator through the bottom MR fixture. For experiments performed in the presence of a magnetic field, oscillations were run at a frequency of 1 rad s−1 and shear strain amplitude of 1% (in the linear viscoelastic regime).
5.5. Cell culture
Human MSCs were purchased and tested for purity from Lonza and were positive for CD105, CD166, CD29, and CD44 and negative for CD14, CD34, and CD45 by flow cytometry. Human microvascular endothelial cells (HMVECs) were purchased from Cell Systems. Endothelial growth media-2 (EGM-2)–supplemented media was purchased from Lonza. The use of human cell lines in this work was reviewed and approved by the University of Illinois at Urbana-Champaign Biological Safety Institutional Review Board.
MSCs were cultured in low glucose DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. HMVECs were cultured in EGM-2 on attachment factor (Life Technologies). Media was changed every 3–4 days and cells were passaged at ~80% confluence. Cells were seeded on surfaces at passages 3–7.
For differentiation experiments, MSCs were cultured for 10 days in mixed (1:1) bipotential adipogenic/osteogenic differentiation media (Lonza) as per the manufacturer’s instructions.
5.6. Tube formation assay
Vascularization assays were performed with MSC conditioned media as described previously[17]. Briefly, growth-factor-reduced basement membrane (matrigel, Trevigen) was coated on the bottom surfaces of a 48-well plate and gelled at 37°C for 30 minutes. HMVECs were seeded onto the matrigel at ~15,000 cells/well in serum and growth factor free media (EBM-2, Lonza) and conditioned media from MSCs cultured at different conditions was added. After 8 hours, tube formation was imaged using a Rebel DSLR camera (Cannon) and tube area quantified using imageJ (NIH).
5.7. Immunostaining
Surfaces were rinsed twice with phosphate buffered saline (PBS) and were then fixed with 4% paraformaldehyde for 20 minutes. Surfaces were permeabilized for 30 minutes in 0.1% Triton X-100 and then blocked in 1% bovine serum albumin (BSA) for 1 hour. Runx2 was stained with a primary rabbit-anti-Runx2 (ABCAM) overnight at 4°C and secondary 555-Alexa fluor goat anti-rabbit antibody (1:200) (Invitrogen). Actin and nuclei were stained by Alexa-Fluor 488-phalloidin (1:200) and 4,6-Diamidino-2-phenylindole (DAPI; 1:5000), respectively. Secondary staining was performed for 20 minutes at 37°C.
5.8. Fluorescence imaging and data analysis
Immunostained cells were imaged using a Zeiss Axiovert inverted fluorescence microscope (Carl Zeiss) or an IN Cell Analyzer 2000 (General Electric). Cell area was measured from phalloidin staining of the actin cytoskeleton using ImageJ and nuclear Runx2 intensity was measured using DAPI as a mask for nuclei. Runx2 intensity is reported as Nuclear intensity minus cytoplasmic intensity. Statistical significance was determined using two-tailed p-values from unpaired t-test for comparing two groups. Error bars represent standard error.
Supplementary Material
Acknowledgments
This work was supported by the National Heart Lung and Blood Institute of the National Institutes of Health, grant number HL121757
Footnotes
Author contributions
A.A., N.B., R.E. and K.K. conceived the ideas and designed the experiments. A.A., J.L., and N.B. conducted the experiments and A.A., J.L., N.B., R.E. and K.A.K analyzed the data. A.A., J.L.,N.B., R.E., and K.K. interpreted the data and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Amr A. Abdeen, Department of Materials Science and Engineering, University of Illinois at Urbana-Champaign Urbana, Illinois 61801 (USA)
Junmin Lee, Department of Materials Science and Engineering, University of Illinois at Urbana-Champaign Urbana, Illinois 61801 (USA).
N. Ashwin Bharadwaj, Department of Mechanical Science and Engineering, University of Illinois at Urbana-Champaign Urbana, Illinois 61801 (USA).
Prof. Randy H. Ewoldt, Department of Mechanical Science and Engineering, University of Illinois at Urbana-Champaign Urbana, Illinois 61801 (USA)
Prof. Kristopher A. Kilian, Email: kakilian@illinois.edu, Department of Materials Science and Engineering, University of Illinois at Urbana-Champaign Urbana, Illinois 61801 (USA)
References
- 1.Lukashev ME, Werb Z. Trends Cell Biol. 1998;8:437. doi: 10.1016/s0962-8924(98)01362-2. [DOI] [PubMed] [Google Scholar]
- 2.Mouw JK, Ou G, Weaver VM. Nat Rev Mol Cell Biol. 2014;15:771. doi: 10.1038/nrm3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7:265. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 4.Mammoto A, Mammoto T, Ingber DE. J Cell Sci. 2012;125:3061. doi: 10.1242/jcs.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 6.Humphrey JD, Dufresne ER, Schwartz MA. Nat Publ Gr. 2014;15:802. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watt FM, Huck WTS. Nat Rev Mol Cell Biol. 2013;14:467. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 8.Rosso F, Giordano A, Barbarisi M, Barbarisi A. J Cell Physiol. 2004;199:174. doi: 10.1002/jcp.10471. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman BD, Grashoff C, Schwartz Ma. Nature. 2011;475:316. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolahi KS, Mofrad MRK. Wiley Interdiscip Rev Syst Biol Med. 2010;2:625. doi: 10.1002/wsbm.79. [DOI] [PubMed] [Google Scholar]
- 11.Jaalouk DE, Lammerding J. Nat Rev Mol Cell Biol. 2009;10:63. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 13.Lee J, Abdeen AA, Zhang D, Kilian KA. Biomaterials. 2013;34:8140. doi: 10.1016/j.biomaterials.2013.07.074. [DOI] [PubMed] [Google Scholar]
- 14.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Proc Natl Acad Sci. 2010;107:4872. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Abdeen AA, Tang X, Saif TA, Kilian KA. Biomaterials. 2015;69:174. doi: 10.1016/j.biomaterials.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Abdeen AA, Kim AS, Kilian KA. ACS Biomater Sci Eng. 2015 doi: 10.1021/ab500003s. 150306103621001. [DOI] [PubMed] [Google Scholar]
- 17.Abdeen AA, Weiss JB, Lee J, Kilian KA. Tissue Eng Part A. 2014;20:2737. doi: 10.1089/ten.tea.2013.0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seib FP, Prewitz M, Werner C, Bornhäuser M. Biochem Biophys Res Commun. 2009;389:663. doi: 10.1016/j.bbrc.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 19.Engler AJ, Griffin MA, Sen S, Bönnemann CG, Sweeney HL, Discher DE. J Cell Biol. 2004;166:877. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Biophys J. 2008;95:4426. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daley WP, Peters SB, Larsen M. J Cell Sci. 2008;121:255. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 22.Cox TR, Erler JT. Dis Model Mech. 2011;4:165. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu P, Weaver VM, Werb Z. J Cell Biol. 2012;196:395. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Science. 2010;329:1078. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C, Tibbitt MW, Basta L, Anseth KS. Nat Mater. 2014;13:645. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Abdeen AA, Kilian KA. Sci Rep. 2014;4:5188. doi: 10.1038/srep05188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaeta X, Xie Y, Lowry WE. Nat Cell Biol. 2013;15:725. doi: 10.1038/ncb2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mengsteab PY, Uto K, Smith AST, Frankel S, Fisher E, Nawas Z, Macadangdang J, Ebara M, Kim DH. Biomaterials. 2016;86:1. doi: 10.1016/j.biomaterials.2016.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H, Lippens E, Duda GN, Mooney DJ. Nat Mater. 2015 doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdick JA, Murphy WL. Nat Commun. 2012;3:1269. doi: 10.1038/ncomms2271. [DOI] [PubMed] [Google Scholar]
- 31.Yoshikawa HY, Rossetti FF, Kaufmann S, Kaindl T, Madsen J, Engel U, Lewis AL, Armes SP, Tanaka M. J Am Chem Soc. 2011;133:1367. doi: 10.1021/ja1060615. [DOI] [PubMed] [Google Scholar]
- 32.Jiang FX, Yurke B, Schloss RS, Firestein BL, Langrana NA. Biomaterials. 2010;31:1199. doi: 10.1016/j.biomaterials.2009.10.050. [DOI] [PubMed] [Google Scholar]
- 33.Gillette BM, Jensen JA, Wang M, Tchao J, Sia SK. Adv Mater. 2010;22:686. doi: 10.1002/adma.200902265. [DOI] [PubMed] [Google Scholar]
- 34.Guvendiren M, Burdick JA. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 35.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kloxin AM, Tibbitt MW, Anseth KS. Nat Protoc. 2010;5:1867. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosales AM, Mabry KM, Nehls EM, Anseth KS. Biomacromolecules. 2015 doi: 10.1021/bm501710e. 150210101436008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosales AM, Anseth KS. Nat Publ Gr. 2016;1:1. [Google Scholar]
- 39.Mitsumata T, Honda A, Kanazawa H, Kawai M. J Phys Chem B. 2012;116:12341. doi: 10.1021/jp3049372. [DOI] [PubMed] [Google Scholar]
- 40.Mayer M, Rabindranath R, Börner J, Hörner E, Bentz A, Salgado J, Han H, Böse H, Probst J, Shamonin M, Monkman GJ, Schlunck G. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitsumata T, Ohori S, Honda A, Kawai M. Soft Matter. 2013;9:904. [Google Scholar]
- 42.Tse JR, Engler AJ. Curr Protoc Cell Biol. 2010;Chapter 10(Unit 10.16) doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 43.Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Cohen Stuart Ma, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WTS. Nat Mater. 2012;11:642. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 44.Lin-Vien D, Colthup NB, Fateley WG, Grasselli JG. Handb Infrared Raman Charact Freq Org Mol. 1991:251. [Google Scholar]
- 45.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nature. 2005;435:191. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 46.de Vicente J, Klingenberg DJ, Hidalgo-Alvarez R. Soft Matter. 2011;7:3701. [Google Scholar]
- 47.Ewoldt RH, Johnston MT, Caretta LM. Complex Fluids in Biological Systems. In: Spagnolie SE, editor. Biological and Medical Physics, Biomedical Engineering. Springer New York; New York, NY: 2015. pp. 207–241. [Google Scholar]
- 48.Park JH, Kwon MH, Park OO. Korean J Chem Eng. 2001;18:580. [Google Scholar]
- 49.Claracq J, Sarrazin J, Montfort JP. Rheol Acta. 2004;43:38. [Google Scholar]
- 50.Rowlands AS, George PA, Cooper-White JJ. Am J Physiol Cell Physiol. 2008;295:1037. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 51.Miyakoshi J. Prog Biophys Mol Biol. 2005;87:213. doi: 10.1016/j.pbiomolbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Zhao X, Kim J, Cezar Ca, Huebsch N, Lee K, Bouhadir K, Mooney DJ. Proc Natl Acad Sci. 2011;108:67. doi: 10.1073/pnas.1007862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiang JD, Wen JH, Del Álamo JC, Engler AJ. J Biomed Mater Res - Part A. 2013;101A:2313. doi: 10.1002/jbm.a.34543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Latimer A, Jessen JR. Matrix Biol. 2010;29:89. doi: 10.1016/j.matbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Kim HY, Nelson CM. Organogenesis. 2012;8:56. doi: 10.4161/org.19813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rienks M, Papageorgiou AP, Frangogiannis NG, Heymans S. Circ Res. 2014;114:872. doi: 10.1161/CIRCRESAHA.114.302533. [DOI] [PubMed] [Google Scholar]
- 57.Bonnans C, Chou J, Werb Z. Nat Rev Mol Cell Biol. 2014;15:786. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.