Abstract
Non-viral, biomaterial-mediated gene delivery has the potential to treat many diseases, but is limited by low efficacy. Elucidating the bottlenecks of plasmid mass transfer can enable an improved understanding of biomaterial structure-function relationships, leading to next-generation rationally designed non-viral gene delivery vectors. As proof of principle, we transfected human primary glioblastoma cells using a poly(beta-amino ester) complexed with eGFP plasmid DNA. The polyplexes transfected 70.6 ± 0.6% of the cells with 101 ± 3% viability. The amount of DNA within the cytoplasm, nuclear envelope, and nuclei was assessed at multiple time points using fluorescent dye conjugated plasmid up to 24 hours post-transfection using a quantitative multi-well plate-based flow cytometry assay. Conversion to plasmid counts and degradation kinetics were accounted for via quantitative PCR (plasmid degradation rate constants were determined to be 0.62 hr−1 and 0.084 hr−1 for fast and slow phases respectively). Quantitative cellular uptake, nuclear association, and nuclear uptake rate constants were determined by using a four-compartment first order mass-action model. The rate limiting step for these poly(beta-amino ester)/DNA polyplex nanoparticles was determined to be cellular uptake (7.5×10−4 hr−1) and only 0.1% of the added dose was taken up by the human brain cancer cells, whereas 12% of internalized DNA successfully entered the nucleus (the rate of nuclear internalization of nuclear associated plasmid was 1.1 hr−1). We describe an efficient new method for assessing cellular and nuclear uptake rates of non-viral gene delivery nanoparticles using flow cytometry to improve understanding and design of polymeric gene delivery nanoparticles.
Keywords: gene delivery, nanoparticle, polymer, computational modeling, brain cancer
Graphical abstract
1. Introduction
Gene therapy has the potential to treat inheritable diseases such as cystic fibrosis [1], Duchenne muscular dystrophy [2, 3], and hemophilia [4] as well as acquired diseases such as cancer [5, 6]. In general, viruses are highly efficient in delivering nucleic acid with a multiplicity of infection as low as one viral particle per cell [7], but are immunogenic and can cause insertional mutagenesis [8]. Non-viral methods, although generally less efficient at delivering nucleic acids than viral methods, are easier to scale-up from a manufacturing standpoint, are easily chemically modified for optimization of function, and can have low toxicity and host immune response [9, 10].
Although high-throughput synthesis and screening methods can enable the evaluation of a large library of biomaterials for non-viral gene delivery [11], an improved understanding of polymeric structure-function relationships could enable improved rational design of next-generation non-viral gene delivery vectors. Quantifying plasmid cellular and nuclear uptake rates and differences can elucidate salient functional differences between biomaterial structures and can reveal bottlenecks associated with specific vectors. PCR is a well-established method to quantify the number of plasmids within the cell or nucleus [12, 13]. However, lysing cells and nuclei, isolating and purifying DNA, and then subsequently running PCR on the samples is a time-consuming process that is less amenable to scale up for semi-high throughput analysis of multiple time points and multiple structures.
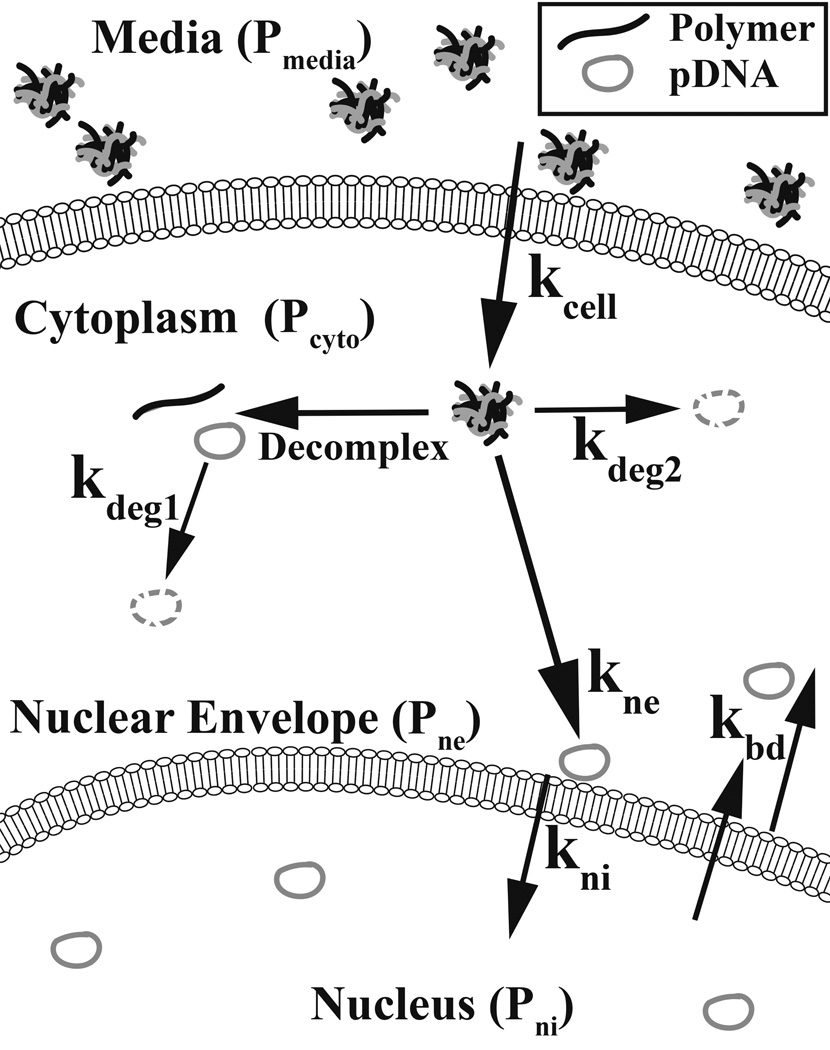
This work describes a new approach to quantify the number of plasmids within the cell’s cytoplasm (Pcyto), associated with nuclei (Pne), and internalized by nuclei (Pni) based on flow cytometry. Through the use of mathematical fitting to a system of differential equations and a four-compartment model, this method also allows the quantitative determination of cellular and nuclear uptake rate constants. We synthesized a newly discovered and leading non-viral polymeric vector, poly(beta-amino ester) (PBAE) B4-S4-E7 or 447 [14, 15] to transfect human primary glioblastoma cells and subsequently evaluated its intracellular delivery properties using this flow cytometry-based method. This technique can be utilized to quantify the rate constants associated with cellular entry (kcell), nuclear envelope association (kne), and nuclear internalization (kni) for a given non-viral vector (Scheme 1). This tool could be used to further elucidate biomaterial structure-function relationships of non-viral gene delivery vectors [16, 17] and quantitative characterization could enable improved design of next generation polymer vectors [18].
Scheme 1. Depiction of plasmid transfer rates between compartments of interest.
The four compartments of interest are: the media (Pmedia), the cytoplasm (Pcyto), nuclear envelope-associated (Pne), and nuclei-internalized (Pni). The associated transfer rate constants between these four compartments are kcell, kne, and kni respectively. kbd is the rate to the cytoplasm from the nuclear envelope and from within the nucleus. The fast and slow degradation rate constants are kdeg1 and kdeg2 which are hypothesized to be associated with decomplexed and complexed plasmids, respectively.
2. Materials and methods
2.1 Polymer synthesis
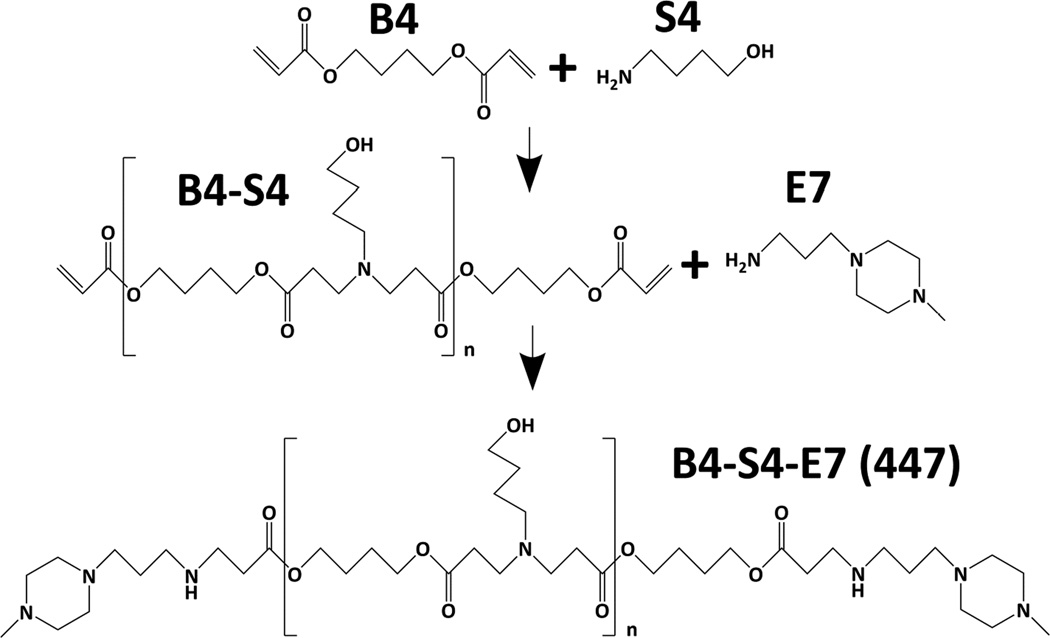
Polymer B4-S4-E7 (447) was synthesized via a Michael addition reaction (Scheme 2) by adding 4-amino-1-butanol (S4) neat to 1,4-butanediol diacrylate (B4) in a 1.2:1 B4:S4 monomeric ratio, immediately vortexed, and placed in a 90°C oven for 24 hours in the dark using a stir bar greater than 100 RPM, resulting in a B4-S4 acrylate-terminated base polymer [14]. The B4-S4 polymer was then solvated to 166.7 mg/mL using anhydrous tetrahydrofuran (THF) and subsequently endcapped using a 0.5 M solution of 1-(3-aminopropyl)-4-methylpiperazine, or E7, solvated in anhydrous THF at room temperature for 1 hour at a final concentration of 100 mg/mL (Scheme 2). The polymer was then precipitated using anhydrous ethyl ether using two centrifugation washing steps at 4000 RPM for approximately 4 minutes. After decanting the ether the second time, the polymer was placed in a vacuum chamber for at least 24 hours in the dark. The polymer was then stored neat at −20°C until the fractionation process.
Scheme 2. Reaction scheme of polymer B4-S4-E7 (447).
1,4-butanediol diacrylate (B4) was polymerized with 4-amino-1-butanol (S4) neat at 90°C for 24 hours and subsequently end-capped with 1-(3-aminopropyl)-4-methylpiperazine (E7) in THF at room temperature for 1 hour.
2.2 Polymer fractionation
Gel permeation chromatography (GPC) (Waters Corp.) was used to obtain a similar number- and weight-average molecular weight (Mn and Mw, respectively) of polymer 447 used previously which had high transfection efficacy in glioblastoma cells [14]. The GPC setup included an autosampler, a refractive index detector, a Styragel column (WAT025861), and an autofractionator (Waters Corporation). The GPC diluent flowed at 1 mL/min and constituted 94% THF, 5% dimethyl sulfoxide (DMSO), 1% piperidine and a few 100 mg of butylated hydroxytoluene to inhibit free radical formation. A two minute time fraction was chosen based on the calibration curve for the targeted molecular weight. After collection, the polymer was again precipitated and vacuum dried as previously described [14]. The polymer was then solvated in anhydrous DMSO to 100 mg/mL and frozen in 20 µL aliquots at −20°C until use. Each aliquot used for further study underwent one freeze-thaw cycle only. Post-fractionation, the Mn and Mw was quantitated via GPC against polystyrene standards. 1H Nuclear magnetic resonance spectroscopy (1H NMR; Bruker) was also used to quantify the absolute molecular weight of polymer 447. 1H NMR in CDCl3 was used to measure 447 polymer molecular weight by comparing the ratio of area between peaks for the endcap secondary amine hydrogen at 0.81 ppm and the hydrogens of the α-carbons of the B repeat units at 4.07 ppm.
2.3 Cy3-plasmid DNA conjugation
A Mirus Label IT® Tracker™ Cy™ 3 Kit was used to conjugate Cy3 to enhanced Green Fluorescent Protein (eGFP-N1; 4733 base pairs (bp)) plasmids. In brief, 75 µg plasmid DNA (pDNA; 1 µg/µL), 60 µL of Mirus Buffer A and 415 µL of ultrapure distilled water (DNase and RNase free) was added to 50 µL of the Cy3 dye reagent and incubated for at least 3 hours; at each hour the vials were briefly centrifuged to ensure all reagents were homogeneously reacting. Subsequently, 60 µL of 3 M sodium acetate (NaAc) and 1880 µL of ice cold 190 proof ethanol (EtOH) were added and the solution was kept at −20°C for at least 30 additional minutes. After which, the sample was centrifuged at 4°C for 15 minutes at 13 krcf, the supernatant was discarded and the pellet was then resuspended in 2 mL of 70% EtOH at room temperature. The sample was again centrifuged and the supernatant was discarded and the pellet was resuspended in 300 µg (1 mg/mL) of Cy3-unconjugated eGFP pDNA (1 µg:4 µg conjugated:unconjugated). Prior to adding the unconjugated pDNA the cap was left open and covered with aluminum to protect from light to allow the EtOH to evaporate for approximately 10 minutes. The sample was then characterized via a NanoDrop 2000 Spectrophotometer (Thermo Scientific). The absorbance of the DNA was corrected for Cy3-spectral overlap using the following equation (obtained from Mirus): Abase = A260 nm − 0.08A550 nm. The nucleotide:dye ratio (N:D) was calculated using Eq. 1 (obtained from Mirus):
| Eq.1 |
The eGFP-N1 plasmid contained a 77 bp Simian virus-40 (SV40) DNA-targeted sequence (DTS): 5’-AACCAGCTGT GGAATGTGTG TCAGTTAGGG TGTGGAAAGT CCCCAGGCTC CCCAGCAGGC AGAAGTATGC AAAGCAT-3’. The sequence binds to a transcription factor which then undergoes a morphological change, revealing its nuclear localization signal (NLS) [19]. The DNA and its indirectly associated NLS can then be transported into the nucleus.
2.4 Cy3 pH-sensitivity
Various uptake pathways such as macropinocytosis, and caveolae- and clathrin-mediated endocytosis lead to the lysosomal degradation pathway, referred to as the proton sponge effect [20]. The cargo must have the ability to buffer the endosome to escape without degradation [20]. Because the pH of the Cy3-conjugated pDNA during the delivery process may range anywhere from approximately 5 to 7.4, the pH-sensitivity of Cy3 was assessed using a fluorescence plate reader (excitation and emission of 550 and 570 nm, respectively; Synergy2, Gen5) by using equal molar concentrations of the Cy3-eGFP pDNA in 150 mM sodium acetate (pH 5.2) and 150 mM phosphate buffered saline (PBS; pH 7.4) in triplicate. A Student’s t-test was used to assess significance (α = 0.05).
2.5 Cell culture
The glioblastoma cells used for transfection were derived from human primary brain cancer [21, 22] and were cultured in a 5% CO2 atmosphere at 37°C, using DMEM:Ham’s F12 (1:1) (Invitrogen) and was supplemented with 10% fetal bovine serum and 1× antibiotic-antimycotic (Invitrogen).
2.6 Polyplex formation and cell incubation
Polymer 447 at a working concentration of 100 mg/mL in anhydrous DMSO was diluted to 1.35 mg/mL which was used for preparing the 30 weight/weight (w/w; mass of polymer to mass of DNA) in 25 mM NaAc (buffered at a pH of 5.2) and mixed in a 1:1 v/v ratio with 45 µg/mL of plasmid DNA which was diluted from 1 mg/mL (originally in water) using 25 mM NaAc. Uptake studies utilized Cy3-conjugated pDNA, whereas qPCR and expression studies used Cy3-unconjugated pDNA, as the presence of Cy3 within such close proximity to the pDNA would inhibit primer annealing or the transcription and translation processes. Polyplex diameter and concentration were assessed in PBS via nanoparticle tracking analysis in triplicate via a NanoSight NS500 (Malvern Instruments, Ltd.). Zeta potential via dynamic light scattering was quantified using two independently prepared samples diluted to 700 µL in ultra-pure distilled water (DNase- and RNase-free) Malvern Instruments, Ltd.). 24-well plates (2.0 µm2) were seeded with 93,750 glioblastoma cells (625 µL) 24 hours prior to transfection. 125 µL (2813 ng of pDNA) was delivered to each well. Separate 24-well plates for each time point were used to ensure that the cells containing Cy3-pDNA at later time points did not have more exposure to light, ensuring photobleaching was not a confounding factor.
Gel electrophoresis was used to visualize whether the polymer and DNA would dissociate with the addition of heparin. Polyplexes of 447 and DNA were formed at 30 w/w in 25 mM NaAc at a DNA concentration of 16 µg/mL. Heparin sulfate was diluted in PBS in a serial dilution then 20 µL were mixed in a 1:1 ratio with the nanoparticles to assess its ability to disrupt 447 interaction with DNA. Thirty-percent glycerol (8 µL) was added to the 40 µL of nanoparticles and heparin as a loading buffer and 15 µL of each sample were loaded in a 1% agarose gel. The gel contained 100 ng DNA and 3000 ng of 447 per well.
2.7 Washing extracellular and extranuclear pDNA
To ensure the heparin washing was sufficient to remove uninternalized plasmid pDNA from the cells (n=3) and nuclei (n≥2), the 24-well plates were placed at 4°C to inhibit ATP-dependent uptake mechanisms. Any fluorescence above the untreated groups associated with the cells or nuclei was thus attributed to unwashed plasmids on the phospholipid bilayer membrane of the cells or nuclei.
To assess how well cells were washed, polymer 447-pDNA polyplexes were incubated with the cells for 1 hour using 30 w/w (and 60 w/w which would likely be more difficult to wash). After the incubation period, the cells were either not washed or quickly washed with gentle trituration using heparin twice (1 mL; 50 µg/mL of heparin in PBS) and subsequently washed with 1 mL of PBS. Heparin is a highly anionic macromolecule used to destabilize the uninternalized polyplexes which frees the pDNA, causing the pDNA to be easily removed. The washed cells were immediately trypsinized after the PBS wash, quenched using 2% fetal bovine serum (FBS) in PBS, and then used for flow cytometry.
The nuclei were also challenged similarly at 4 °C for 1 hour. Similar to the nuclei isolation process, one heparin wash was used (350 µL at 50 µg/mL) which was followed by centrifugation (600 rcf for 6 minutes). Furthermore, to more closely assimilate the intracellular condition, rather than polyplexes, naked pDNA was used to challenge the nuclei as the plasmid in contact with the nuclei would likely have little or even no polymer ionically complexed at that stage due to degradation and dissociation from charged intracellular molecules [23–25]. The amount of plasmids per nuclei (1×105 plasmids/nuclei) used to assess the thoroughness of the heparin washing procedure was chosen to be exceedingly higher than previously reported values for plasmids found in the nucleus using non-viral methods [12, 13].
For cellular and nuclear uptake studies all samples were washed at either the time point of interest up to two hours (0 (aspirated nearly simultaneously), 1, and 2 hours) or at two hours for all time points thereafter (4, 7, 18, and 24 hours). Cells were washed before nuclei isolation at the time point of interest. Unwashed and washed nuclei were then used for flow cytometry to assess the amount of plasmids either associated (extranuclear (Pne) + intranuclear (Pni)) or internal to the nuclei (Pni), respectively. pDNA which is loosely associated with nuclei would likely not retain association with the nuclei throughout the nuclei isolation process.
2.8 Nuclei isolation
Nuclei isolation was accomplished using an isolation kit (Nuc101; Sigma Aldrich). At each time point of interest after transfection the cells were placed on ice and 1 mL of an ice cold solution of heparin sodium salt from porcine intestinal mucosa (Sigma-Aldrich H3393) at 50 µg/mL in PBS was quickly used to wash the cells twice with gentle trituration. Subsequently, 150 µL of ice cold lysis buffer (Nuc101; Sigma Aldrich) was placed on the cells and the cells were immediately scraped thoroughly and transferred to a 15 mL conical tube on ice. The cells were then homogenized using a round-bottom glass tube while on ice and were by volume approximately 1/3 froth at completion and sat on ice for an additional 5 minutes. The samples were then transferred to 0.5 mL microcentrifuge tubes and centrifuged at 4°C for 5 minutes at 500 rcf. The supernatant was carefully aspirated as to not agitate the pellet. 30 µL of ice cold lysis buffer was used to resuspend the pellet, followed by vortexing for 1–2 seconds. 120 µL of ice cold lysis buffer was then added to the samples and vortexed for a fraction of a second. The samples were placed on ice for another 5 minutes. The centrifugation and resuspension were completed twice and the samples were filtered into a new 0.5 mL microcentrifuge tube with a 40 µm nylon filter. The time points were divided into two tubes. 350 µL of ice cold 50 µg/mL heparin was added to half of the divided tubes and vortexed briefly. Samples were then centrifuged for 6 minutes at 600 rcf at 4°C, the tubes were carefully aspirated, and the pellets were resuspended in 30% glycerol (3:7 v:v glycerol: ultrapure distilled water), and frozen at −80°C until further use for either flow cytometry analysis or genomic DNA (gDNA)/pDNA isolation and purification.
The nuclei isolation efficiency was calculated via a hemocytometer by dividing the number of cells used for the nuclei isolation process by the number of 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI)-stained nuclei isolated and multiplying by 100 (n=3).
To confirm nuclei purity, stably expressing eGFP(+) glioblastoma cells [15] underwent the same culturing, transfection, and nuclear isolation process and were assessed via flow cytometry (n=2). The singlet nuclei population was identified using FSC-H and SSC-H. The percent of eGFP(+) cells and nuclei were quantified by multiplying the ratio of the number of eGFP(+) cells to singlets by 100. Furthermore, DAPI stained nuclei with a combined brightfield image for qualitative purposes were taken to show the purity of the nuclei.
2.9 Cellular and nuclear uptake
Cellular and nuclear uptake were assessed in triplicate at 0, 1, 2, 4, 7, 18, and 24 hours. To prepare the cells (n=3) for flow cytometry, the cells were washed as previously discussed. After the PBS washing step, 150 µL of warm 0.05% Trypsin-ethylenediaminetetraacetic acid (EDTA) was added. After incubating at 37°C for approximately 5 minutes, 850 µL of 2% Trypsin in PBS was added to each well to quench the reaction. Each well was transferred to a 1.5 mL microcentrifuge tube and placed on ice before analysis. To prepare the nuclei (n=3) for flow cytometry, nuclei were taken from the −80°C freezer, thawed and diluted to approximately 150 µL using 2% FBS in PBS. Flow cytometry using a BD Accuri™ C6 Cytometer at a flow rate of 66 µL/min (note FL2-A = function of flow rate) was then used to identify the singlet populations and to assess the presence and intensity of Cy3. All gating and analysis was accomplished using FloJo. The singlet populations of nuclei and cells were identified using FSC-H and SSC-H; the amount of Cy3 per cell or nucleus was quantified using the geometric mean of FL2-A of the singlet population and normalized to an untreated group. Flow cytometry was used to measure the fluorescence of each cell (plasmids within the cytoplasm (Pcyto + Pne + Pni), unwashed nuclei (Pne+Pni), and washed nuclei (Pni)).
2.10 Plasmid and genomic DNA isolation and purification
The cells at their respective time points after transfection with unconjugated pDNA were prepared in triplicate for gDNA/pDNA isolation and purification by washing with heparin as previously described, trypsinizing, quenching with 2% FBS, and then doing a centrifugation washing step in a 1.5 mL tube to remove the trypsin. After the supernatant was trypsinized the samples were resuspended in 30 µL of PBS. The samples (7 time points in triplicate and the untreated group) were then incubated in a lysis buffer which constituted 0.5% sodium dodecyl sulfate (SDS), 100 µg/ml proteinase K, and 20 µg/ml DNase-free RNase [6]. The samples were placed in an oven at 50°C while shaking at 200 rpm for 4 hours. Tris-EDTA-saturated phenol was used to do a 1× volume extraction where an appropriate amount of phenol was added and the emulsion was vortexed briefly. To separate the layers, the samples were placed in a quickspin apparatus. The aqueous top layer was placed in a new vial and a 1× volume extraction with 25:24:1 phenol/chloroform/isoamyl alcohol was completed twice. The same procedure from the previous extraction was followed and the aqueous top layer was collected. The samples were set on ice and washed with ice-cold water-saturated ether twice. Water-saturated ether was prepared by mixing ultrapure distilled, RNase-/DNase-free water and ether in a 1:1 volume ratio. After the water-saturated ether was added, the samples were briefly vortexed and spun to separate the layers. The top ether layer was aspirated and the aqueous bottom layer was set on ice. To evaporate the remaining ether in the solution, the vials were left open for ~5 minutes. DNA was precipitated using 3× volumes of ice cold 95% EtOH at a final concentration of 0.3 M NaAc. The samples were incubated for at least 30 minutes at −20°C, and then centrifuged at 21 krcf at 4°C for 15 minutes. The supernatants were then removed and the pellets were resuspended in 3× volumes of 70% EtOH at room temperature. The same centrifugation steps were followed, and the supernatant was aspirated. The samples were allowed to dry by leaving the caps open for ~15 minutes. Ultrapure distilled, RNase-/DNase-free water was used to resuspend the samples to 30 µL before storing at −80°C. After storage, the concentrations of the isolated and purified gDNA and pDNA at each of the respective time points were assessed via UV-vis spectroscopy. The 260 nm/280 nm absorbance ratios were also assessed (n=24) as an indicator for nucleic acid purity. Aromatic amino acids absorb at the 280 nm wavelength which can used as an indicator of protein contamination.
2.11 Cy3 fluorescence to plasmid number conversion for cells
The normalized geometric mean (NGM) of the samples were calculated using flow cytometry by dividing FL2-A (Cy3) by the untreated FL2-A. Because there may be slight variations in the intensity of the flow cytometry from day to day, the samples were normalized to their own untreated samples at each time point. When there is no Cy3 present in the samples the NGM is 1. Since we are interested in the increase in the fluorescence in time above the untreated group, NGMSample – (NGMUnt = 1) versus time was plotted thus Cy3’s fluorescence began at 0 at the 0 hour time point. NGMSample – NGMUnt is the equivalence of the relative-fold fluorescence of Cy3 above the untreated. We use the term NGM to refer to . The other values needed to convert Cy3 fluorescence to plasmid number included the bulk fluorescence (550/570 nm excitation/emission) measurement of the samples via a fluorescence plate reader (FluorPR) and the associated number of cells as quantified with a hemocytometer (# cellsHemocytometer). The plasmid number (# Plasmids) contained within all of the cells was calculated using a calibration curve (n = 4; # Plasmids = m * FluorPR + b). Eq. 2 was used to calculate how much fluorescence (above the untreated group) is associated per plasmid according to flow cytometry:
| Eq.2 |
In order to be above the bulk fluorescence background 3 wells of a 24-well plate were combined in order to have sufficient Cy3 signal (n=3; 9 wells total). The value was measured (n=3) multiple times at multiple time points to ensure it was constant.
2.12 Cy3 fluorescence to plasmid number conversion for nuclei
The fluorescence signal per nucleus is weaker than the fluorescence signal per cell because there are significantly fewer plasmids per nucleus than plasmids per cell. We scaled the value to be applicable for nuclei by multiplying by the relative fluorescence (flow cytometry) of untreated cells and nuclei (which was confirmed with qPCR and a Student’s t-test) using Eq. 3:
| Eq.3 |
For each time point for either the cells or nuclei the fluorescence by flow cytometry was converted to the number of plasmids using Eq. 4:
| Eq.4 |
2.13 Intracellular plasmid DNA degradation kinetics
Cy3 is not an intercalating dye and its fluorescence intensity does not change if a plasmid’s structural integrity is compromised because Cy3 is conjugated to the pDNA via a reactive alkyl group to heteroatoms within the plasmid. Because quantifying fluorescence of Cy3-pDNA does not necessarily correspond to intact plasmids, qPCR was used in order to correct for pDNA degradation in time.
After the isolated and purified gDNA/pDNA samples from the glioblastoma cells (n=3) were quantified via NanoDrop, each sample was diluted to 1 µg/mL in ultrapure distilled water (DNase and RNase free). Each qPCR well contained 6 ng of the purified gDNA/pDNA, 14 µL of SYBR® Green PCR Master Mix (2× concentration; Life Technologies), and 2 µL of the reverse and forward primers for eGFP and human β-cytoskeleton actin (β-actin), totaling 20 µL. β-actin is a housekeeping gene and was used to compare the relative amounts of eGFP present. The eGFP and β-actin primer amplicons were 95 and 250 nucleotides in length, respectively. The eGFP and β-actin sequences were as follows: forward eGFP primer: AGGGCATCGACTTCAAGG (55.6% GC; Tm: 54.7°C); reverse eGFP primer: CTACGTCTATATCATGGCCG (50% GC: %Tm: 52.1°C); forward β-actin primer: CATGTACGTTGCTATCCAGGC (52.4% GC; Tm: 60.8°C); reverse β-actin primer: CTCCTTAATGTCACGCACGA (50.0% GC; Tm: 60.2°C). The thermal cycling conditions for qPCR were set at 95°C for the first 10 minutes to fully denature the template and activate the enzymes. Immediately thereafter, a temperature cycle was repeated between 95°C and 60°C for 15 seconds and 1 minute, respectively, 40 times.
PCR was performed on a dilution series for each gene of interest in order to calculate the efficiency of replication (ER) and was calculated using the cycle threshold value (CT) according to Eq. 5:
| Eq.5 |
We ensured the ERs for the eGFP and the β-actin amplicon amplifications were between 0.95–1.05 which allowed 2−ΔΔCT to be a valid quantification of the relative amounts of genes present [26]. The ER was calculated for the linear range in this plot and the unknown samples’ CT values were within this same linear CT range. ΔΔCT was calculated using Eq. 6:
| Eq.6 |
Because β-actin is a single copy gene, there are two copies per cell. By multiplying 2−ΔΔCT by 2, the total number of plasmids per cell can be obtained [6]. Thus: PlasmidqPCR = 2(2−ΔΔCT).
Once qPCR was completed, melt curves of the qPCR products were examined for quality control; furthermore, the qPCR products were assessed for off-target amplification and primer dimers using gel electrophoresis (4% agarose; 1µg/mL ethidium bromide) and a DNA ladder. The structural integrity of the plasmids was taken into account (PlasmidCorr) by multiplying PlasmidPR by the ratio of intact plasmids (of the 447 polymer in the glioblastoma cells) at each time point, using Eq. 7:
| Eq.7 |
The polymer chosen for this poof of principle paper has a PlasmidCorr value equal to the PlasmidsqPCR 447 value. If additional polymer structures were compared to assess structure-function relationships at gene delivery, the ratios at each time point could be used without repeating qPCR. The corrected plasmid values for the cytoplasm, the extranuclear space, and the intranuclear space will be referred throughout the paper as Pcyto, Pne, and Pni, respectively. Each plasmid compartment (Pcyto, Pne and Pni) was assumed to have the same degradation kinetics. For the investigation of other polymers, the polymer-pDNA binding strength would also likely need to be relatively similar for the degradation kinetics to be comparable [14, 27].
The intracellular plasmid degradation time constants were calculated from using exponential decay fittings from 2 to 24 hours, which were the time points after the polymer/DNA particles were removed from the media. We investigated fittings with both mono- and bi-exponential terms, which were accomplished using the MATLAB function nlinfit.
2.14 Expression efficacy and relative metabolic activity
Fluorescence microscopy (10× magnification, 200 ms eGFP exposure time) and flow cytometry (66 µL/min) were performed to assess expression 48-hours post transfection (n=3). The relative metabolic activity (viability) was assessed using a CellTiter 96® AQueous One assay at an absorbance of 490 nm using a UV-Vis spectrophotometer (Synergy2, Gen5) (n=3).
2.15 Modeling via first order mass-action kinetics
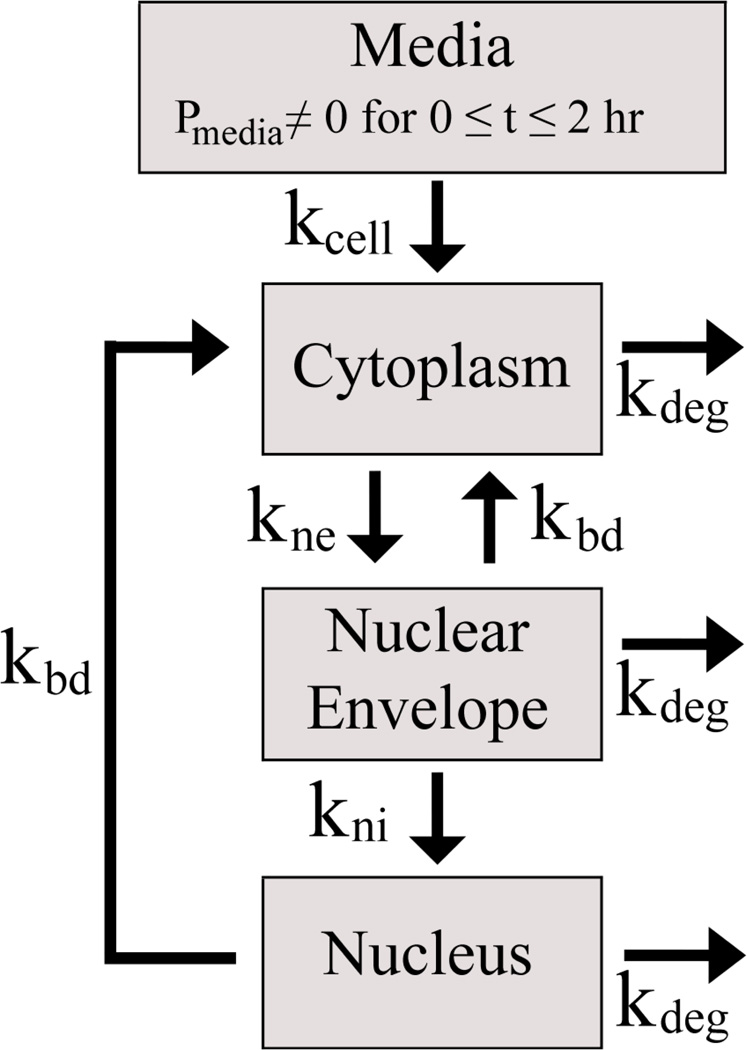
The set of first order mass-action differential equations used in this work composes a simplified model where the experimental data could be sufficiently recapitulated using a minimum number of fitted variables and without requiring additional parameters from the literature that dealt with different experimental systems [28]. The model included 4 compartments from 0 to 2 hours: the media, cytoplasm, nuclear envelope (ne; tightly-associated extranuclear space), and nucleus (ni; intranuclear space) (Scheme 3) and 3 compartments after 2 hours because the plasmid number in the media drops to zero at the completion of the polyplex-cell incubation time period.
Scheme 3. Four-compartmental model depicting the direction of plasmid transfer.
The number of plasmids in the media, cytoplasm, associated with the nuclear envelope, and internal to the nucleus are represented by Pmedia, Pcyto, Pne, and Pni, respectively. The rate contributing term transferring plasmids from the media into the cytoplasm, kcell Pmedia, in the set of differential equations included a Heaviside or unit step function (𝔄) which was able to bring the non-zero term to zero at 2 hours by multiplying kcell Pmedia by 𝔄[−(t − 2)]; where t = time (hours).
The rate constants for plasmids transferring from the media to the cytoplasm, the cytoplasm to the nuclear envelope, and the nuclear envelope to the nucleus are represented as kcell, kne, and kni, respectively (Scheme 3). kbd is a backflow rate term from the nuclear envelope and the nucleus back to the cytoplasm. The backflow encompasses plasmid transport that could be due to active plasmid transport from the nucleus or from nuclear envelope breakdown. For simplification, we used the same kdeg term for all intracellular compartments. The differential equation set used was as follows (Eq. 8–10):
| Eq.8 |
| Eq.9 |
| Eq.10 |
MATLAB was used to fit the experimental data by minimizing the sum of the residuals squared (SRS). MATLAB’s ODE23s solver was used to assess the model plasmid values (Pcyto, Pne, Pni) through all the experimental time points. All four rate constants were optimized such that their values did not change with further iterations of the model. The fitting optimization process was repeated 20 times with different random initial values for the four rate constants each time and convergence to minima were obtained.
2.16 Statistics
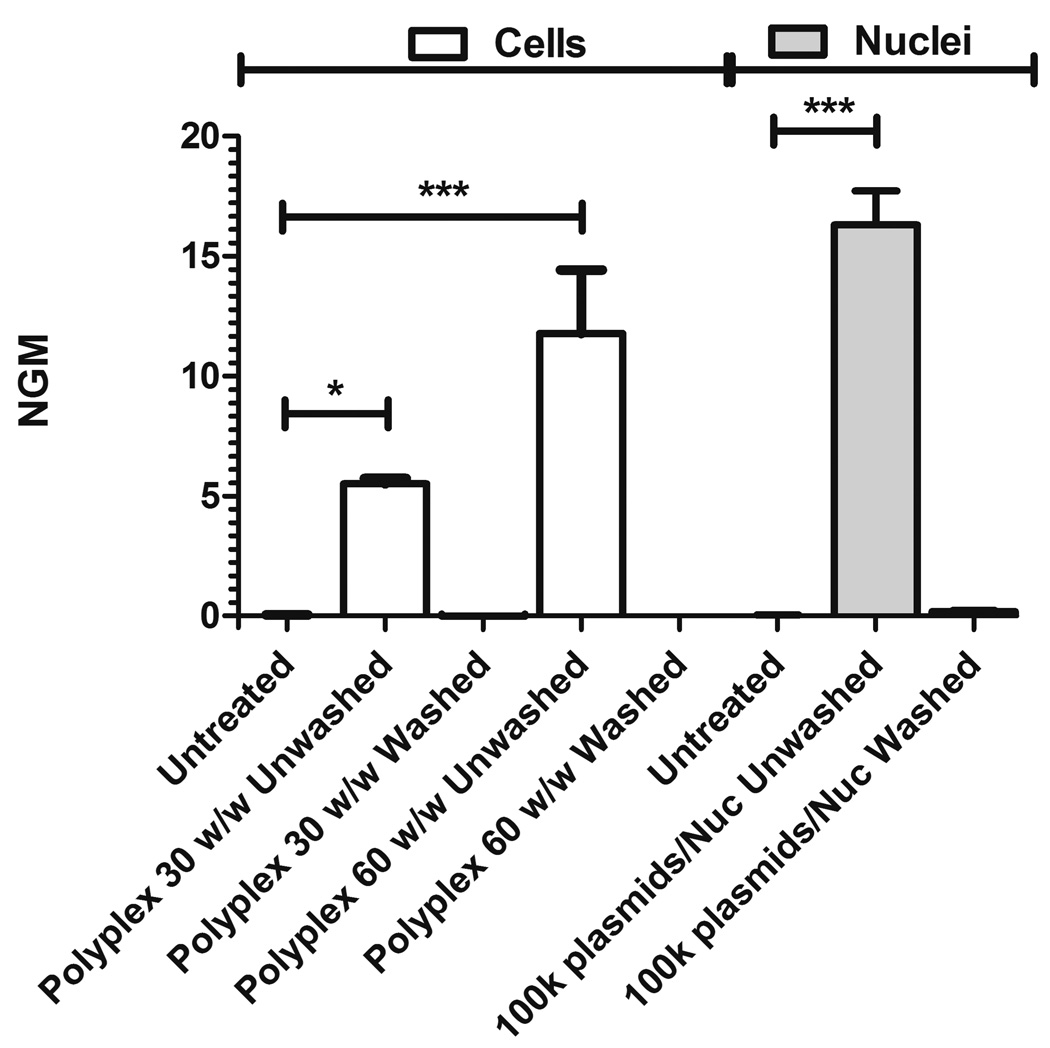
All errors reported are standard errors of the mean and are of independently prepared samples. Sample numbers are reported in their respective method sections. A Student’s t-test was performed for Figure S1 (n=3) and Figure S2 (n=2). Two independent one-way ANOVAs were performed for cells (n=3) and nuclei (n≥2) for Figure 2.
Figure 2. Heparin washing procedure successfully removes all bound plasmid.
Following an incubation for 1 hour at 4°C, 50 µg/mL heparin in PBS was used to eliminate all associated DNA. Cy3-labeled plasmid is removed from both cells (with added polyplex) and nuclei (with added plasmids). NGM indicated normalized geometric mean fluorescence (unitless).
3. Results
3.1 Polymer synthesis, DNA conjugation, and particle formation
Polymer 447, 1-(3-aminopropyl)-4-methylpiperazine end-terminated poly(1,4-butanediol diacrylate-co-4-amino-1-butanol), was successfully synthesized by Michael addition. Post-endcapping, the polymer 447’s Mn and Mw according to GPC were 6.7 kDa and 8.5 kDa, respectively. A polydispersity index (PDI) close to unity of 1.30 was obtained. The molecular weight according to 1H NMR was 7.6 kDa (Figure S3). The mean repeat unit (MW = 287.36 g/mol) was determined to be 25.5 using the ratio of area between peaks for the endcap secondary amine hydrogen at 0.81 ppm and the hydrogens of the α-carbons of the B repeat units at 4.07 ppm. The peak and satellites at 2.59 ppm were due to DMSO contamination following isolation of the polymer from a DMSO stock (Figure S3).
eGFP pDNA was successfully labeled with Cy3 dye so that it could be tracked intracellularly. The number of nucleotides per dye immediately post-conjugation and before dilution into unconjugated pDNA was approximately 35 nucleotides/dye. The Cy3 fluorescence at pH 7.4 (150 mM PBS) and 5.2 (150 mM NaAc) were not different (p-value = 0.63) as shown in Figure S1, demonstrating the local pH environment within the cell would not affect the fluorescence readings. Polymer/DNA polyplex nanoparticles were formed by self-assembly between the cationic polymer and the anionic DNA in 25 mM NaAc buffer. A mass ratio of 30 polymer weight: DNA weight (w/w) was used as the standard dose for particle formation. The diameter and zeta potential of the polyplexes was 98 ± 7 nm, 2.6 ± 0.3 mV, respectively. Heparin sulfate was used in a serial dilution to dissociate polyplexes and this disassociation was characterized by gel electrophoresis. Heparin was found to be disruptive of the polyplexes at a 1:1 mass ratio of heparin:PBAE (3125 ng heparin) (Figure S4). It should be noted that multiple washing instances were used to sufficiently wash uninternalized plasmid from extracellular or extranuclear membranes.
The polyplex concentration was (4.3 ± 0.7)×1010 particles/well. Based on the pDNA dosage delivered per well (2813 ng) and the molecular weight of the plasmid (3.12×106 g/mol), there were 5.42×1011 plasmids per well. This suggests there were 13 ± 2 plasmids on average per polyplex particle. At the time of seeding there were approximately 93,750 cells, which suggests that on average there were 5.78×106 plasmids available per cell and (4.6 ± 0.7)×105 polyplexes available per cell.
3.2 Cellular and nuclear uptake
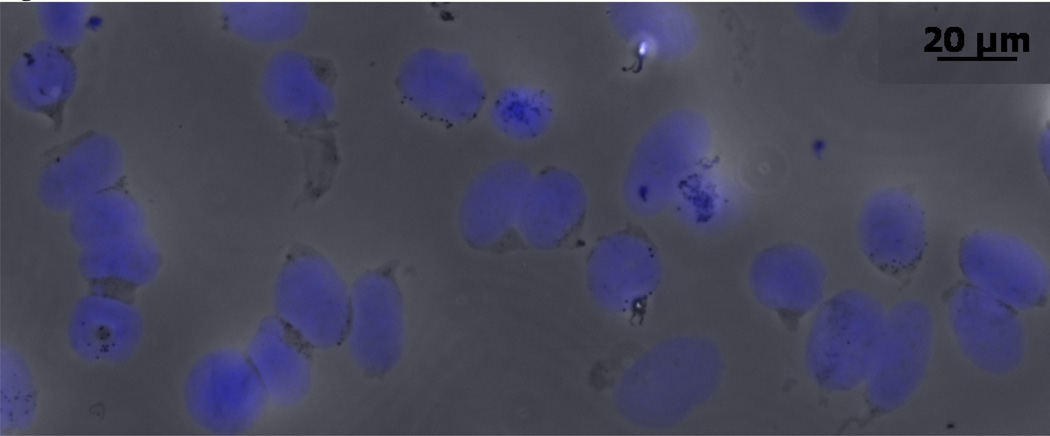
Following the nuclei separation steps described in the methods, the isolated nuclei were approximately 20 µm in diameter. The efficiency of the nuclei isolation process was 25 ± 5%. Figure 1 shows isolated nuclei stained with DAPI. To confirm the purity of the isolation step in separating nuclei from intact cells, the purity of gated nuclei from eGFP-stably expressing cells (79%) according to flow cytometry was 99.6% as shown in Figure S2 (p-value = 0.02) which is more than a 99% drop in eGFP.
Figure 1. Isolated nuclei.
Isolated nuclei are obtained and stained with DAPI following the described nuclei isolation protocol.
The heparin wash regimen was sufficient in removing the pDNA from both cells and nuclei. Washing steps were conducted, as described in the methods, at 4°C to prevent endocytosis from occurring, as it is an ATP-dependent process. In all cases, for both polyplex mass ratios (the standard 30 w/w and a higher 60 w/w ratio designed to be more difficult to wash away), and for both cells and nuclei (100k plasmids/nucleus), the washing process fully removed all bound plasmids (Figure 2).
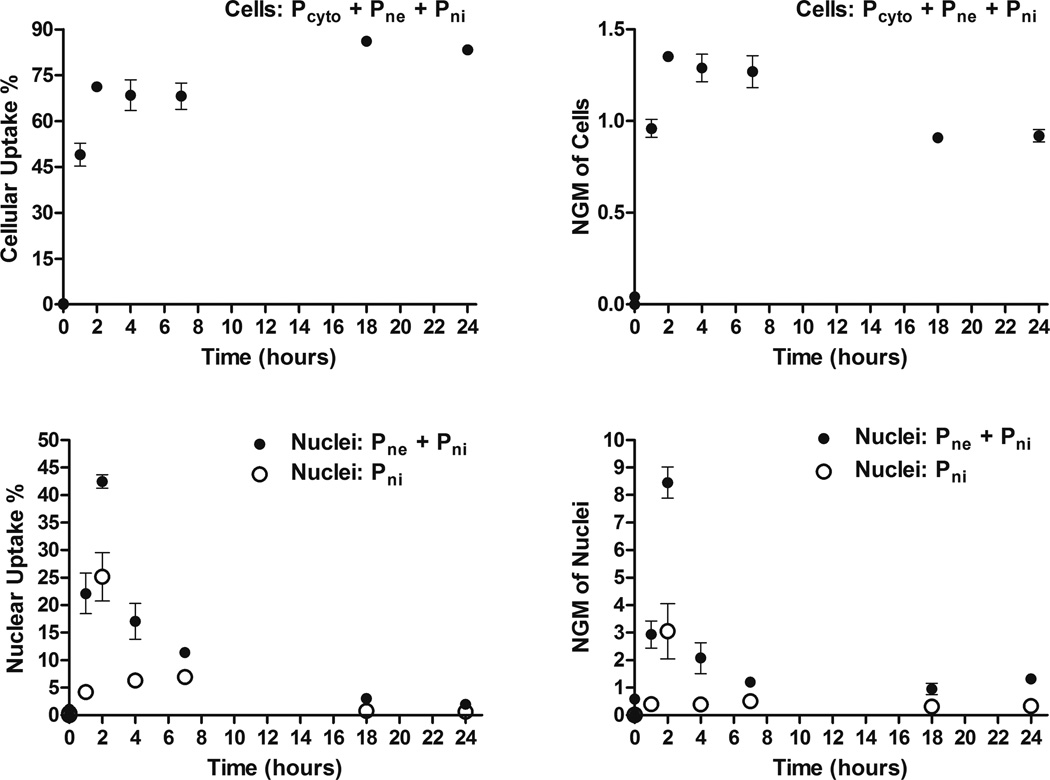
As expected, the isolated nuclei are associated with a lower FSC-H signal by flow cytometry than the cells which can be seen in the ungated FSC-H vs SSC-H dot plot (Figure S5; top) and in the FSC-H histogram of gated singlet cells and nuclei (Figure S5; bottom). The 0 hour time point for the cells and nuclei were as expected in that the fluorescence values were near the untreated groups’ fluorescence values. As an example, a dot plot of FL1-A and FL2-A for the untreated, and 0 and 2 hour time points for cells are shown in Figure S6. This can also be observed for cells and nuclei by the 0 hour time points’ closeness in proximity to the origin in Figure 3.
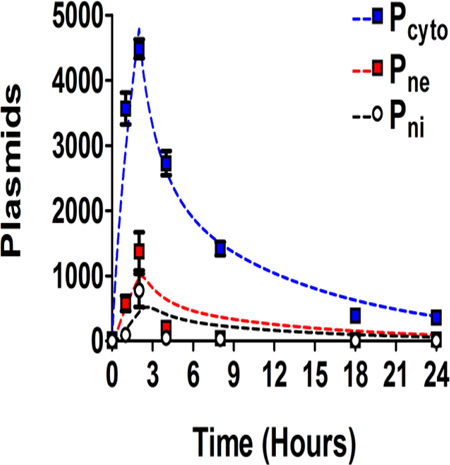
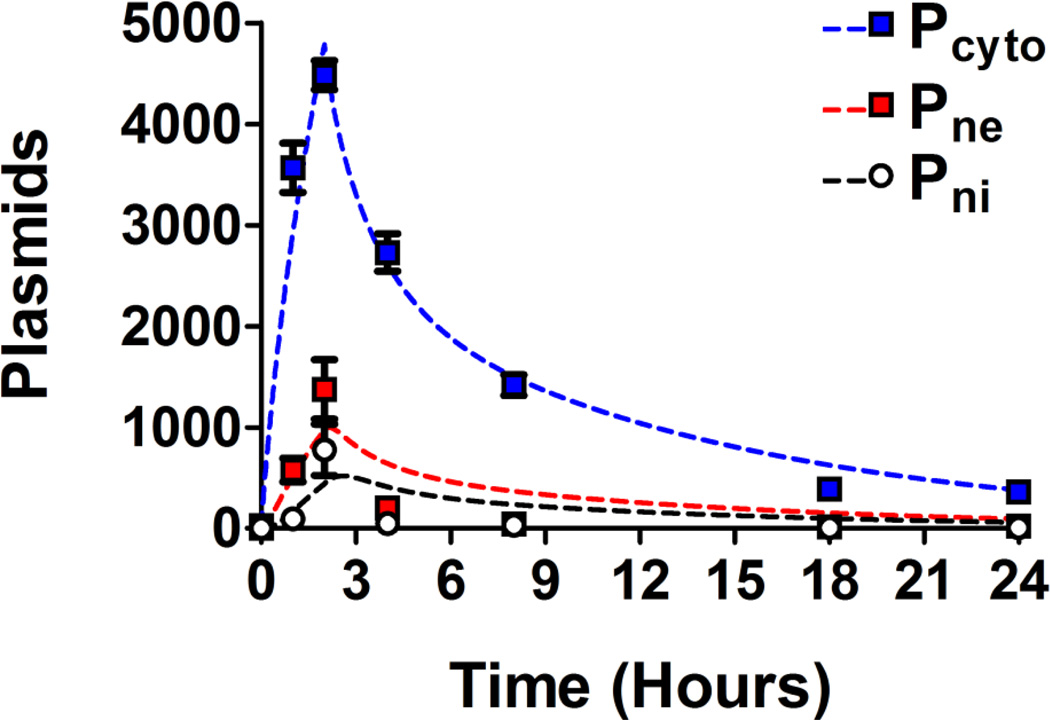
Figure 3. Cellular and nuclear uptake of PBAE 447 polyplexes in human brain cancer cells.
Percentages (left) and NGM (right) of Cy3-labeled DNA for both cells (top) and nuclei (bottom). Pcyto are plasmids which are in the cytoplasm, Pne are plasmids which are nucleus-associated, and Pni are plasmids which are intranuclear.
The percent fluorescently positive time profiles for Cy3-pDNA in cells (top left) and nuclei (bottom left) are shown in Figure 3. The Cy3(+) percent for cells increased from zero to a relatively constant value of ~75% at 2 hours (top left), with the 18 and 24 hour time points being ~80%; the Cy3(+) percent value for nuclei (bottom left) reached a maximum at 2 hours. The NGM fluorescence increased from 0 to a maximum at 2 hours for both cells (top right) and nuclei (bottom right) (Figure 3).
The concentrations of the gDNA and pDNA isolated and purified from the cells for qPCR was 16 ± 2 µg/mL. The 260 nm/280 nm absorbance ratio was 2.03 ± 0.08. A calibration curve (Figure S7) was used to convert the bulk fluorescence on the plate reader to a plasmid number. The and values were (1.8 ± 0.2)×10−4 per plasmid and (3.5 ± 0.4)×10−3 per plasmid, respectively. Confirming the scaling of the value, a Student’s t-test at the 2 hour time point between the PlasmidqPCR and Pni values showed no statistical difference (p-value = 0.25). The slope (m) was equal to 1.90×105 plasmids and the y-intercept (O) was 5.65×107 plasmids.
3.3 Intracellular plasmid DNA degradation kinetics
The replication efficiencies of the eGFP and β-actin primers were 0.97, and 0.95, respectively. The eGFP and β-actin qPCR products were the same lengths as the amplicon lengths; 95 and 250 bp, respectively, according to gel electrophoresis (Figure S8). No primer dimers or off-target amplification was observed on the gel. Figure S9 shows the plasmid numbers over time in the cells (Pcyto + Pne + Pni) based on PlasmidPR and PlasmidqPCR, which separated further in time as intracellular nucleases degraded the exogenous pDNA (but did not affect the plasmid labeling dye). The similarity of the PlasmidPR and PlasmidqPCR values for the initial time points further validate the conversion from fluorescence to plasmid number. The PlasmidPR and plasmid values accounting for degradation for both cells and nuclei are shown in Figure S10 on the top and bottom, respectively.
The PlasmidqPCR values in time from 2 to 24 hours are shown in Figure S11 with mono-(dotted line) and bi-exponential degradation (solid line) fittings. The bi-exponential degradation fits the experimental data more stringently, suggesting there are likely two terms involved in the degradation process. For simplicity, we initially used the mono-exponential kdeg value of 0.350 hr−1 in the model and the plasmid values reached zero at a much earlier time point after the 2 hour peak than the experimental data suggested which prompted us to explore the bi-exponential degradation approach for kdeg. The bi-exponential degradation fit, which better matches the experimental data, assumes there are two different rates at which internalized plasmid can be degraded. We hypothesize that the faster rate corresponds to free DNA plasmid and that the slower rate corresponds to DNA plasmid bound with polymer to form a polyplex nanoparticle. The kdeg incorporating the two exponential degradation terms in the set of differential equations was calculated using Supplemental derivation 1, where A, B, kdeg1, and kdeg2 were obtained from the bi-exponential fitting (Figure S11; Plasmids (Pcyto + Pne + Pni) = Ae−kdeg1t + Be−kdeg2t) and is as follows (Eq. 11):
| Eq.11 |
The values for kdeg1 and kdeg2 are 0.62 hr−1 and 0.084 hr−1 respectively. Of the available plasmids initially dosed per well, the cells had uptaken just 0.1% over the 2 hr incubation. This demonstrates that the first step in the plasmid mass transport process, cellular uptake, is quite inefficient with these polyplex nanoparticles.
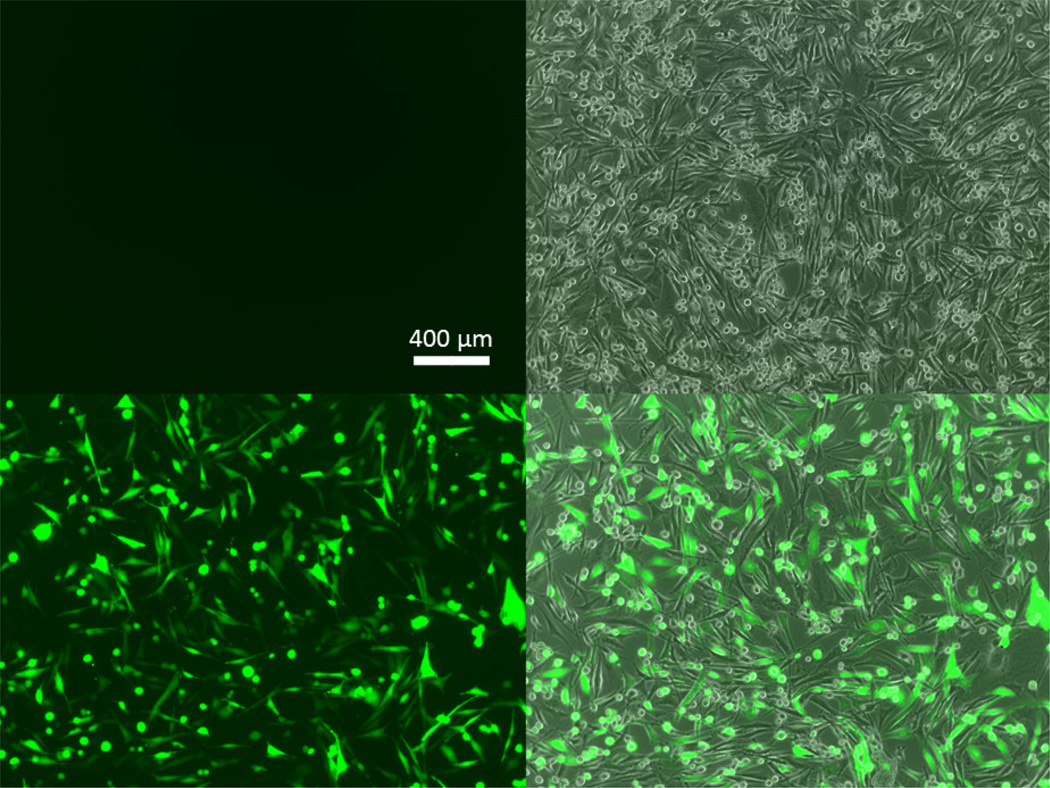
Ultimately, the exogenous gene expression efficacy of 447 30 w/w polyplexes (2813 ng; 2 hour polyplex incubation) 48 hours post transfection was 70.6 ± 0.6% (Figure 4; bottom). Although, it was possible to use a higher dosage (i.e., 3750 ng/well) which resulted in higher transfection levels (92 ± 0.5%), we opted to use a more moderate dosage and w/w value which also resulted in no apparent cytotoxicity. The relative metabolic activity of the selected 447 polyplex formulation and dosage and was 101 ± 3% compared to the untreated group (100%). As these polymeric nanoparticles have high transfection efficacy and low cytotoxicity, they are an important class of biomaterials to study for non-viral gene delivery. Successful fitting optimization was obtained using MATLAB (Figure S12). The values for the four kinetic constants, kcell, kne, kni, and kbd, fitted over the experimental time points were 7.5×10−4, 0.72, 1.1, and 1.8 hr−1, respectively. The minima of the sum of the residuals squared, SRS, between the experimental data and the bi-exponential model are shown in Figure S13 and demonstrate clear minima. The experimental data in conjunction with the bi-exponential degradation model is shown in Figure 5 and demonstrated close fitting. The experimental data in conjunction with the alternative mono-exponential degradation models is shown Figure S14 (with fitted kinetic constant values of kcell, kne, kni, and kbd as 7.9×10−4, 0.87, 1.3, and 2.1 hr−1, respectively).
Figure 4. GFP transfection of human brain cancer cells with PBAE 447 polyplexes.
Fluorescence microscopy images of human primary glioblastoma cells showing the eGFP channel (left) and the eGFP and phase contrast channels combined (right) for untreated cells (top) and for 447 30 w/w polyplex transfected cells (bottom) at 48 hours post transfection.
Figure 5. Fitting of the four-compartment first order mass-action model to the experimental data.
The first order mass-action model is represented as dotted lines (bi-exponential degradation) and the experimental data as data points. Curves are shown for plasmid number within the cell (Pcyto), plasmid number associated with the nucleus (Pne), and plasmid number within the nucleus (Pni).
4. Discussion
This study has demonstrated a new approach to quantify steps in the intracellular delivery of non-viral gene delivery materials. A new flow cytometry-based technique is presented as well as a four-compartment model that captures key features of intracellular delivery. The technique is applied to a new leading biomaterial, polymer 447 or 1-(3-aminopropyl)-4-methylpiperazine end-terminated poly(1,4-butanediol diacrylate-co-4-amino-1-butanol), for non-viral gene delivery to better understand its mechanistic advantages and limitations. The molecular weight of polymer 447 was controlled through the use of a GPC fractionation process. The obtained PDI of 1.30 is similar to previously reported PDIs of fractionated polymer [14]. Because Cy3 is not pH-sensitive in the range of interest (pH 5.2–7.4), it is suitable to use as a probe through the different compartments of our model. The model includes an extracellular compartment (the media), an intracellular compartment (the cytoplasm), and then differentiates between nuclear-associated plasmid/polyplex and plasmid internalized to the nucleus. Polyplex/plasmid in endosomes is treated the same as plasmid out of endosomes as they have both not yet reached the nucleus. In this manner, kne, represents a combined rate of multiple intracellular steps, whereas kcell and kni are the rates of crossing the specific membranes of the cell and nucleus respectively.
The washing experiments successfully demonstrated the feasibility of removing extracellular and extranuclear plasmids from the surface of the cells and nuclei, which would have otherwise contributed false-positively to the fluorescence. Following our approach, separate sampling can be taken from each of the four compartments over time without requiring specialized equipment.
The percentage of cells with a detectable amount of labeled plasmid by flow cytometry increased significantly from 0 to 2 hours, while polyplexes remained in the media. There was also a slight increase after 2 hours in the percentage of positive cells which may be due to differential cell proliferation of the cells, with those preparing to enter mitosis having an increased endocytosis rate, and greater internalization of polyplexes [29]. This could result in a greater proportion of cells with a detectable amount of Cy3-pDNA after the polyplex incubation time period. There was a decrease in the labeled DNA’s average fluorescence intensity per cell after the incubation time period. The decrease in the fluorescence was likely due to dilution from cell division and also, to some degree, exocytosis.
The mono-exponential degradation value for kdeg of 0.350 hr−1 was similar to Varga, et al.’s reported value of 5×10−3 min−1 or 0.3 hr−1 [30]. Usage of the bi-exponential degradation allowed for a better fit for the first order mass-action kinetic model which suggests that there may be two different plasmid states as has been reported previously in literature: 1) an unbound or free plasmid state where the plasmids are more susceptible to nuclease activity and 2) the plasmid-polymer bound state, where the plasmids are better protected from nuclease activity [27]. The polymer degradation or unpacking of the plasmid cargo would lead to more degradation in time accounting for the greater differences between the PlasmidPR and PlasmidqPCR values in time [27].
The calculated degradation rate constants are particular to these glioblastoma cells and should be re-evaluated for other cell types that utilize this quantitative model as plasmid degradation rate constants can vary by cell type. If the binding affinity [31] between the polymer types of interest vary substantially it may also be beneficial to assess the degradation rate even within the same cell type [14] as this could change the proportion of DNA in an unbound state vs. a polymer-bound state, affecting DNA degradation rate. Furthermore, if the Cy3 density conjugated to pDNA varies, re-calibrating the plasmid conversion would also be necessary. The cellular and nuclear uptake rates observed are similar to other kinetic rates previously reported in literature [28, 30, 32, 33] through other approaches, which further validates the flow cytometry approach developed and used.
An extension of this high-throughput fluorescence-based analysis of polyplex uptake and transport could be incorporation of Förster Resonance Energy Transfer (FRET) [34]. Such studies could reveal unpacking dynamics of the polyplexes and DNA release in addition to nuclear uptake. For the current uptake studies, cell populations with asynchronous cell cycle stages were used because the literature has shown that cell cycle stage synchronization processes themselves can affect multiple cellular processes, confounding interpretation of results [35]. An asynchronous cell population also more closely resembles the in vivo environment.
Intriguingly, approximately 25% of the cells did not uptake plasmids sufficiently to be Cy3(+) and ~30% of cells did not transcribe and translate enough protein to be considered eGFP(+) according to flow cytometry. Only 0.1% of the pDNA delivered in the media was successfully taken up by the cells during the 2 hr incubation. This high number of non-internalized plasmids for non-viral gene delivery highlights the need for more efficient delivery systems at the cellular level. The PBAE 447 polymer is effective at transfecting the human glioblastoma cells, but is not efficient at doing so. Of the 0.1% of the plasmids that enter the cells, 12% of this internalized plasmid successfully enters the nucleus. Thus, the rate-limiting barrier for polymer 447 is at the cellular uptake level. These results point to increasing cellular uptake as an important design criteria for creating more efficient gene delivery polyplexes. Incorporating cell-specific uptake modalities, such as a targeting ligand, may improve the cellular uptake efficiency and also control transfection to specific targeted cells [36].
5. Conclusions
A flow cytometry-based method was developed and utilized to measure intracellular rate constants of a non-viral gene delivery material. The gene delivery of PBAE 447/DNA polyplexes to human primary glioblastoma cells was used as a model biomaterial transfection system. A four-compartment first order mass-action kinetics model was found to be sufficient at capturing the trends found in the experimental transport data without needing to incorporate any additional information extrapolated from the literature. Quantitative rate constants were identified for the intracellular transport of PBAE 447/DNA polyplexes, including DNA degradation rate from polyplexes, cellular uptake rate, and nuclear uptake rate. This approach and model could be utilized to more easily differentiate polyplex intracellular delivery in different cell types as a function of time as well as delineate the activity of structurally different polymers at transfection. This model is flexible to also incorporate further features to increase complexity and describe additional intracellular steps.
Supplementary Material
Statement of Significance.
In this work, a quantitative high throughput flow cytometry-based assay and computational modeling approach was developed for assessing cellular and nuclear uptake rates of non-viral gene delivery nanoparticles. This method is significant as it can be used to elucidate structure-function relationships of gene delivery nanoparticles and improve their efficiency. This method was applied to a particular type of biodegradable polymer, a poly(beta-amino ester), that transfected human brain cancer cells with high efficacy and without cytotoxicity. A four-compartment first order mass-action kinetics model was found to model the experimental transport data well without requiring external fitting parameters. Quantitative rate constants were identified for the intracellular transport, including DNA degradation rate from polyplexes, cellular uptake rate, and nuclear uptake rate, with cellular uptake identified as the rate-limiting step.
Acknowledgments
CJB thanks the NSF for a GRF: DGE-0707427. This work was also supported in part by the National Institutes of Health (R01EB016721 and a Core Grant to the Wilmer Institute, P30-EY001865).
Abbreviations
- 447
B4-S4-E7
- bp
base pair (nucleotides)
- CT
cycle threshold
- DAPI
2-(4-amidinophenyl)-1H-indole-6-carboxamidine
- DMSO
dimethyl sulfoxide
- DTS
DNA-targeted sequence
- EDTA
ethylenediaminetetraacetic acid
- eGFP
enhanced green fluorescent protein
- EtOH
ethanol
- ER
efficiency of replication
- FBS
fetal bovine serum
- FluorPR
Fluorescence according to the plate reader
- FRET
Förster resonance energy transfer
- gDNA
genomic DNA
- GPC
gel permeation chromatography
- kbd
rate constant of plasmids being recycled from either the nuclear envelope or being internal to the nucleus to the cytoplasm
- kcell
rate constant of plasmids into the cell
- kdeg1
fast degradation constant of plasmid DNA
- kdeg2
slow degradation constant of plasmid
- kne
rate constant of plasmid onto the nuclear envelope
- kni
rate constant of plasmids entering nucleus
- Mn
number-average molecular weight
- Mw
weight-average molecular weight
- N:D
nucleotide to dye ratio
- NaAc
sodium acetate
- NGM
normalized geometric mean
- NLS
nuclear localization signal
- PBAE
poly(beta-amino ester)
- NMR
nuclear magnetic resonance
- Pcyto
plasmid number within the cytoplasm
- PDI
polydispersity index
- pDNA
plasmid DNA
- Plasmidcorr
corrected plasmid number
- PlasmidPR
plasmid number according to the plate reader calibration
- PlasmidqPCR
plasmid number according to qPCR
- Pne
plasmid number on the nuclear envelope
- Pni
plasmid number within the nucleus
- SDS
sodium dodecyl sulfate
- SRS
sum of the residuals squared
- SV40
simian virus-40
- THF
tetrahydrofuran
- 𝔄
Heaviside function
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alton EW, Middleton PG, Caplen NJ, Smith SN, Steel DM, Munkonge FM, Jeffery PK, Geddes DM, Hart SL, Williamson R, et al. Non-invasive liposome-mediated gene delivery can correct the ion transport defect in cystic fibrosis mutant mice. Nat. Genet. 1993;5:135–142. doi: 10.1038/ng1093-135. [DOI] [PubMed] [Google Scholar]
- 2.Bishop CJ, Kim J, Green JJ. Biomolecule Delivery to Engineer the Cellular Microenvironment for Regenerative Medicine. Ann. Biomed. Eng. 2014;42:1557–1572. doi: 10.1007/s10439-013-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filareto A, Parker S, Darabi R, Borges L, Iacovino M, Schaaf T, Mayerhofer T, Chamberlain JS, Ervasti JM, McIvor RS, Kyba M, Perlingeiro RCR. An ex vivo gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nat. Commun. 2013;4:1549. doi: 10.1038/ncomms2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nathwani AC, Tuddenham EGD, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O'Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CYC, Kay MA, Zhou JF, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. Adenovirus-Associated Virus Vector-Mediated Gene Transfer in Hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy AM, Rabkin SD. Current status of gene therapy for brain tumors. Transl. Res. 2013;161:339–354. doi: 10.1016/j.trsl.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohyeldin A, Chiocca EA. Gene and Viral Therapy for Glioblastoma A Review of Clinical Trials and Future Directions. Cancer J. 2012;18:82–88. doi: 10.1097/PPO.0b013e3182458b13. [DOI] [PubMed] [Google Scholar]
- 7.Toyoda E, Doi R, Kami K, Mori T, Ito D, Koizumi M, Kida A, Nagai K, Ito T, Masui T, Wada M, Tagawa M, Uemoto S. Midkine promoter-based conditionally replicative adenovirus therapy for midkine-expressing human pancreatic cancer. J. Exp. Clin. Cancer Res. 2008;27:30. doi: 10.1186/1756-9966-27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, Gilmour KC, Adams S, Thornhill SI, Parsley KL, Staal FJT, Gale RE, Linch DC, Bayford J, Brown L, Quaye M, Kinnon C, Ancliff P, Webb DK, Schmidt M, von Kalle C, Gaspar HB, Thrasher AJ. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou JB, Liu J, Cheng CJ, Patel TR, Weller CE, Piepmeier JM, Jiang ZZ, Saltzman WM. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat. Mater. 2012;11:82–90. doi: 10.1038/nmat3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeney M, Ong SG, Padilla A, Yao ZY, Goodman S, Wu JC, Yang F. Development of Poly(beta-amino ester)-Based Biodegradable Nanoparticles for Nonviral Delivery of Minicircle DNA. Acs Nano. 2013;7:7241–7250. doi: 10.1021/nn402657d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmueli RB, Sunshine JC, Xu ZH, Duh EJ, Green JJ. Gene delivery nanoparticles specific for human microvasculature and macrovasculature. Nanomedicine-Nanotechnology Biology and Medicine. 2012;8:1200–1207. doi: 10.1016/j.nano.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carapuca E, Azzoni AR, Prazeres DMF, Monteiro GA, Mergulhao FJM. Time-course determination of plasmid content in eukaryotic and prokaryotic cells using Real-Time PCR. Mol. Biotechnol. 2007;37:120–126. doi: 10.1007/s12033-007-0007-3. [DOI] [PubMed] [Google Scholar]
- 13.Cohen RN, van der Aa MAEM, Macaraeg N, Lee AP, Szoka FC. Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J. Control. Release. 2009;135:166–174. doi: 10.1016/j.jconrel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop CJ, Ketola TM, Tzeng SY, Sunshine JC, Urtti A, Lemmetyinen H, Vuorimaa-Laukkanen E, Yliperttula M, Green JJ. The Effect and Role of Carbon Atoms in Poly(beta-amino ester)s for DNA Binding and Gene Delivery. J. Am. Chem. Soc. 2013;135:6951–6957. doi: 10.1021/ja4002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop CJ, Tzeng SY, Green JJ. Degradable polymer-coated gold nanoparticles for co-delivery of DNA and siRNA. Acta Biomater. 2015;11:393–403. doi: 10.1016/j.actbio.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol Ther. 2005;11:426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Millili PG, Selekman JA, Blocker KM, Johnson DA, Naik UP, Sullivan MO. Structural and Functional Consequences of Poly(ethylene glycol) Inclusion on DNA Condensation For Gene Delivery. Microsc. Res. Tech. 2010;73:866–877. doi: 10.1002/jemt.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop CJ, Abubaker-Sharif B, Guiriba T, Tzeng SY, Green JJ. Gene delivery polymer structure-function relationships elucidated via principal component analysis. Chem. Commun. 2015;51:12134–12137. doi: 10.1039/c5cc04417k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacramento CB, Moraes JZ, Denapolis PMA, Han SW. Gene expression promoted by the SV40 DNA targeting sequence and the hypoxia-responsive element under normoxia and hypoxia. Braz. J. Med. Biol. Res. 2010;43:722–727. doi: 10.1590/s0100-879x2010007500064. [DOI] [PubMed] [Google Scholar]
- 20.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - Polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero-Cazares H, Chaichana KL, Quinones-Hinojosa A. Neurosphere Culture and Human Organotypic Model to Evaluate Brain Tumor Stem Cells. Cancer Stem Cells: Methods and Protocols. 2009;568:73–83. doi: 10.1007/978-1-59745-280-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on poly(beta-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32:5402–5410. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 24.Sunshine JC, Peng DY, Green JJ. Uptake and Transfection with Polymeric Nanoparticles Are Dependent on Polymer End-Group Structure, but Largely Independent of Nanoparticle Physical and Chemical Properties. Mol. Pharm. 2012;9:3375–3383. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4:438–446. doi: 10.1006/mthe.2001.0475. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Mullen PM, Lollo CP, Phan QC, Amini A, Banaszczyk MG, Fabrycki JM, Wu DP, Carlo AT, Pezzoli P, Coffin CC, Carlo DJ. Strength of conjugate binding to plasmid DNA affects degradation rate and expression level in vivo. BBA-Gen. Subjects. 2000;1523:103–110. doi: 10.1016/s0304-4165(00)00104-5. [DOI] [PubMed] [Google Scholar]
- 28.Banks GA, Roselli RJ, Chen R, Giorgio TD. A model for the analysis of nonviral gene therapy. Gene Ther. 2003;10:1766–1775. doi: 10.1038/sj.gt.3302076. [DOI] [PubMed] [Google Scholar]
- 29.Raucher D, Sheetz MP. Membrane expansion increases endocytosis rate during mitosis. J. Cell Biol. 1999;144:497–506. doi: 10.1083/jcb.144.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12:1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 31.Ketola TM, Hanzlikova M, Leppanen L, Ravina M, Bishop CJ, Green JJ, Urtti A, Lemmetyinen H, Yliperttula M, Vuorimaa-Laukkanen E. Independent versus Cooperative Binding in Polyethylenimine-DNA and Poly(L-lysine)-DNA Polyplexes. Journal of Physical Chemistry B. 2013;117:10405–10413. doi: 10.1021/jp404812a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James MB, Giorgio TD. Nuclear-associated plasmid, but not cell-associated plasmid, is correlated with transgene expression in cultured mammalian cells. Mol. Ther. 2000;1:339–346. doi: 10.1006/mthe.2000.0054. [DOI] [PubMed] [Google Scholar]
- 33.Zhou JY, Yockman JW, Kim SW, Kern SE. Intracellular kinetics of non-viral gene delivery using polyethylenimine carriers. Pharm. Res. 2007;24:1079–1087. doi: 10.1007/s11095-006-9229-5. [DOI] [PubMed] [Google Scholar]
- 34.Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH, Leong KW. Simultaneous Non-invasive Analysis of DNA Condensation and Stability by Two-step QD-FRET. Nano Today. 2009;4:125–134. doi: 10.1016/j.nantod.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper S. Rethinking synchronization of mammalian cells for cell cycle analysis. Cell. Mol. Life Sci. 2003;60:1099–1106. doi: 10.1007/s00018-003-2253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunshine JC, Bishop CJ, Green JJ. Advances in polymeric and inorganic vectors for nonviral nucleic acid delivery. Ther. Deliv. 2011;2:493–521. doi: 10.4155/tde.11.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.