Abstract
A bioactive compound was purified from the culture medium of a new strain of Lactococcus BSN307 by solvent extraction followed by chromatographic techniques. This bioactive compound was identified to belong to phenazine class of compounds by MS, NMR and FTIR. The phenazine compound showed antifungal activity against Aspergillus niger, Penicillium chrysogenum as well as Fusarium oxysporum by disc diffusion assay in addition to antioxidant potential as demonstrated by DPPH scavenging assay. The compound demonstrated selective cytotoxicity against cancer cell lines HeLa and MCF-7 where IC50 was achieved with 20 and 24 µg/mL respectively. At the same time no cytotoxicity was occurred in normal H9c2 cells. The bioactive found to be inhibitory to both leucine and proline aminopeptidases and thus revealed its potential as metalloenzyme inhibitor. This study, for the first time reports the production of phenazine class of compounds by lactic acid bacteria.
Keywords: Anticancer, Antifungal, Bioactive, Phenazine, Lactococcus
Introduction
Lactic acid bacteria (LAB) are thoroughly studied to genome level for their production of therapeutic compounds including antimicrobials. Phenazines which are nitrogen containing heterocyclic compounds that differ in their properties based on the type and position of functional groups are extensively studied because of their wide range antibiotic properties [1]. Members of the phylum Proteobacteria and Actinobacteria, most importantly Pseudomonas and Streptomyces are the major producers of phenazine compounds and are not yet reported to be produced by any member of phylum Firmicutes.
Here, we report the purification, identification and biological activity evaluation of a phenazine compound produced by a new strain of Lactococcus BSN307 (DSMZ 100577, MCC 2824). This novel LAB strain was closely related to Lactococcus garvieae as identified by 16S rRNA gene (KM261818) sequencing but showed phenotypic, chemotaxonomic and molecular level differences and stands separate from the type strain of L. garvieae T (data not provided). This compound showed antifungal, antioxidant and anticancer properties along with aminopeptidase enzyme inhibition potential. We have previously reported the identification of organic acids and a bioactive phenolic compound in the cell free supernatant of the same LAB strain [2, 3]. The advantage of LAB metabolites having both antifungal and antioxidant properties is that they can be developed as food preservatives with dietary antioxidant properties once incorporated into food. As far as we know, this is the preliminary report about the production of phenazine class of compound by LAB or any member of phylum Firmicutes as well as the aminopeptidase inhibitory potential of any LAB metabolite.
Materials and Methods
Microorganisms, Cell Lines and Culture Conditions
Growth medium used for Lactococcus BSN307 was de Man Rogosa Sharpe (MRS) with culture condition (Himedia, India) at 30 °C for 24 h and maintained for longer storage at −80 °C in MRS broth with 20 % (v/v) glycerol. The fungal test strains, Fusarium oxysporum (KACC 42109), Aspergillus niger (KACC 42589), Penicillium chrysogenum (NII 08137), Fusarium moniliforme (KACC 08141) were cultivated on potato dextrose agar (Himedia, India) at 30 °C. Mammalian cell lines, H9c2 (myoblast cell line), HeLa (cervical adenocarcinoma cell line) and MCF-7 (breast carcinoma cell line) were collected from ATCC (American Type Culture Collection).
Purification and Identification of Bioactive
MRS medium (10 L) was inoculated (5 % (v/v)) with 24 h old culture and incubated at 30 °C for 48 h under static condition. Extraction and purification of the bioactive compound was performed as described before [3]. In order to identify, the pure compound was subjected to 1H nuclear magnetic resonance (NMR) spectroscopy using Bruker Avance II 500 spectrometer (USA). Mass spectrometry (Thermo Orbitrap, USA) using electrospray (ESI) ionization method was used to determine molecular mass of the compound. The sample was prepared in acetonitrile and 0.1 N formic acid was used as the mobile phase. Fourier transform infrared spectroscopy (FTIR) spectrum was obtained with PerkinElmer model Spectrum 100 (USA). Antifungal activity of the pure compound (400 μg/disc) was confirmed against F. oxysporum (KACC 42109) by disc diffusion assay as described in “Antifungal Activity Assay of the Pure Compound” section.
Antifungal Activity Assay of the Pure Compound
Broad spectrum antifungal activity of the pure compound (400 μg/disc) was determined against A. niger (KACC 42589), F. moniliforme (KACC 08141) and P. chrysogenum (NII 08137) by disc diffusion method on PDA plates swabbed with individual fungal suspension (1 × 104 spores/mL). The compound was prepared in 50 % acetonitrile and the same was kept as control. The plates were checked after 4 days of incubation at 30 °C for visible zones of inhibition.
Assay of Free Radical Scavenging
The antioxidant activity of compound was calculated following the method of Shimada et al. [4] which is based on the scavenging of DPPH (1,1-diphenyl-2-picrylhydrazyl) radicals. The sample (2 mg/mL) was prepared in methanol and assay was performed as described before [3].
Cytotoxicity to Mammalian Cell Lines
MTT assay, previously described by Mosmann [5] was followed to check the cytotoxicity of the compound. Cell line maintenance and incubation was followed according to Varsha et al. [3]. Cytotoxicity (%) and IC50 (effective concentration of drug resulting in 50 % of maximal toxicity) were calculated as follows:
Aminopeptidase Inhibitory Activity
Aminopeptidase Production
Leucine aminopeptidase (LAP) was produced by Streptomyces gedanensis (IFO 13427) and proline aminopeptidase (PAP) was produced by S. lavendulae (ATCC 14162). The production medium for PAP was YEME (yeast extract malt extract) and for LAP was AP3 medium [(g/L): sucrose; 10, KH2PO4; 2, MgSO4. 7H2O; 1, NaCl; 15, Na2CO3; 0.5, soybean powder; 5 and tween 80; 1.5, pH 7] [6]. Individual 250 mL Erlenmeyer flasks with 50 mL of the media were inoculated with 1 % (v/v) of 24 h old inoculums of S. gedanensis and S. lavendulae separately and incubated for 5 days at 30 °C and 200 rpm. The cell free supernatants were collected after centrifugation at 8000×g and used as enzyme.
Aminopeptidase Assay
Aminopeptidase assay was performed according to the protocol described by Nandan et al. [7]. The reaction mixture contained 2.5 mM leucine p-nitroanilide (pNA) or proline p-nitroanilide as substrate, 50 mM Tris HCl buffer (pH 8.5), 50 μL enzyme sample (leucine or proline aminopeptidase) and 50 μL of either B1 fraction (250 mg/mL) or purified 2, 4 DTBP or phenazine compound (10 mg/mL). The assay mixture was incubated at 37 °C for 10 min. The bright yellow color produced due to the release of pNA by aminopeptidase enzyme activity was estimated spectrophotometrically by absorbance at 405 nm (Tecan NanoQuant, Switzerland). Enzyme activity (One IU) was defined as the amount of enzyme that hydrolyses 1 μM of paranitroanilide substrate. When the inhibitor is added, a decrease in color due to inhibition or decrease in activity of enzyme happens which was estimated as explained before.
Statistical Analysis
The experiments were done in triplicates and the results calculated as mean values ± standard deviation. One-way ANOVA and Dunnet’s test were carried out to test the differences between test and control groups (p < 0.05). Statistical analyses were performed using the Minitab statistical package v. 17 (Minitab Inc., USA).
Results and Discussion
Purification and Characterization
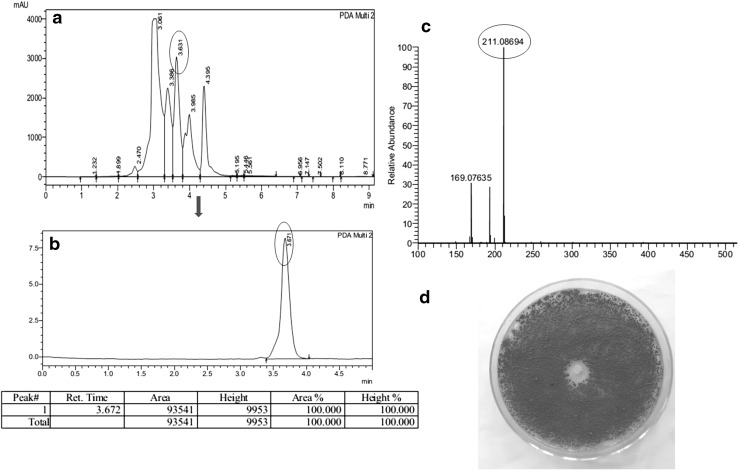
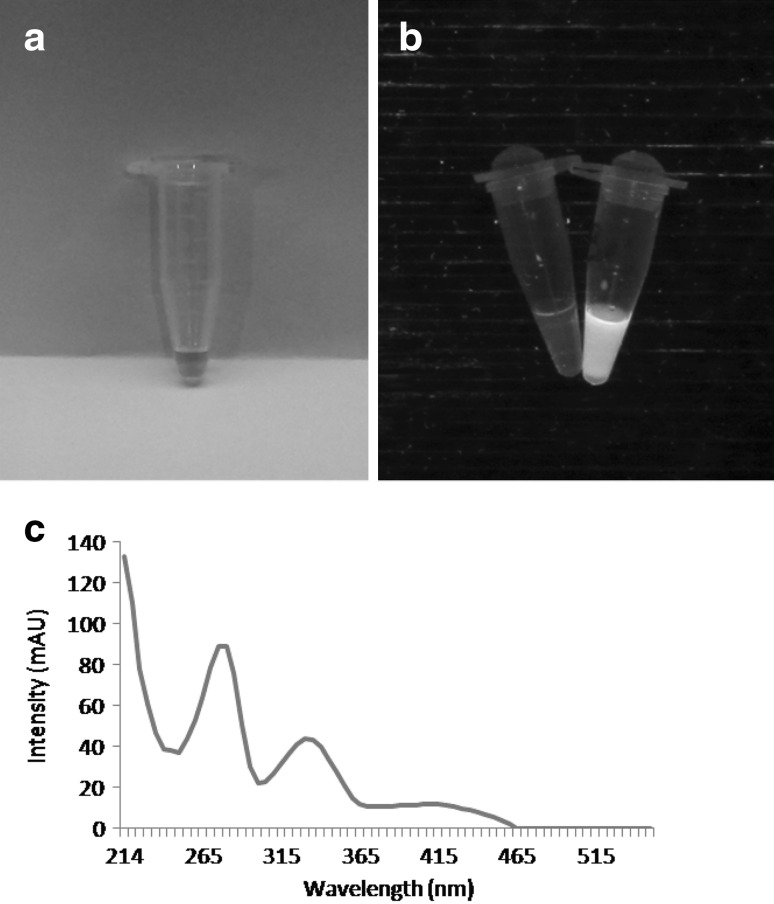
The bioactive compound with antifungal activity was purified and when subjected to HPLC analysis revealed a single peak (Fig. 1b). In LC–MS the compound generated a protonated molecular ion at m/z 211 in the positive ion mode and mass was calculated to be 210 Da (Fig. 1c). The full UV spectrum of this yellow colored (Fig. 2a), fluorescent (Fig. 2b), active material showed a of 214, 278 and 332 nm (Fig. 2c) which belongs to phenazine class of compounds. The structural resemblance of phenazines to anthracene and flavin compounds creates uncertainty in interpretation when only UV–visible spectra considered for identification of phenazine and hence IR spectra of the compound were also looked into. The IR spectra of the compound showed a peak at 3329 cm−1 indicating the presence of N–H stretching along with N–H bending observed at 749 cm−1. Presence of carbonyl group was identified by peak at 1710 cm−1. Aromatic ring of the phenazine structure were identified by the peaks at 2858–2956 cm−1. In the NMR spectra the multiple peaks around 7.1 to 8.4 ppm indicated the presence of aromatic region of Phenazine compound. Singlet peak at 4.12 ppm suggest the presence of methyl group. Thus from the UV, NMR and IR data the compound appears to be phenazine class of bioactive. The final yield of phenazine (≥90 % purity) was 2 mg/L. Using purified phenazine a clearing zone of 1.5 cm against F. oxysporum (Fig. 1d) was obtained.
Fig. 1.
Purification and antifungal activity of phenazine compound. HPLC chromatogram of B1 fraction (a), purified phenazine compound (b), MS chromatogram of phenazine (c) and antifungal activity of pure compound against F. oxysporum (d)
Fig. 2.
Pure phenazine compound (a) showing fluorescence (b) along with UV absorbance spectrum (c)
Phenazine production is based on phz gene expressions. Enzymes, PhzE, PhzD, PhzF, PhzB, and PhzG convert chorismic acid to phenazine-1,6-dicarboxylic acid (PDC) and phenazine-1-carboxylic acid (PCA). PDC and PCA then act as “core” phenazines that strain-specific enzymes convert to other phenazine derivatives [8]. From complete genomes of lactococcal strains deposited in NCBI database, it was found that they possess the genes required for production of phenazines but the enzymes and biosynthetic pathway are yet to be characterized.
Antifungal Activity Assay
Pure phenazine compound (400 µg/disc) produced a clear zone of 1.7 cm against F. moniliforme and 1.4 cm against P. chrysogenum. Minimum inhibitory concentration against A. niger was 400 µg/disc where it produced a clearing zone of 0.7 cm. Three different genera of fungi were selected to study the antifungal activity and found that all of them are sensitive to the compound.
The capacity of phenazine compounds to inhibit the growth of pathogens has been attributed to the ability to generate reactive oxygen species (ROS) and oxidative stress in organisms [9]. Phenazines are inhibitory to the growth of bacteria and fungi because of their ability to undergo cellular redox cycling in the presence of oxygen and reducing agents that result in buildup of toxic superoxide and hydrogen peroxide [10].
Assay of Free Radical Scavenging and Cytotoxicity
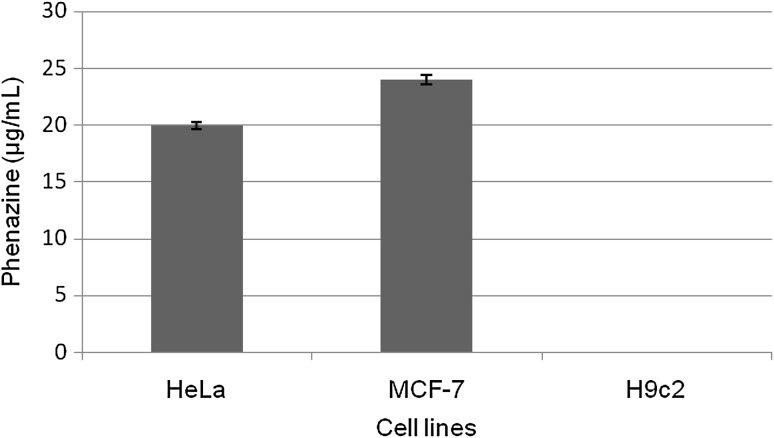
400 µg of phenazine compound caused 77 % of free radical scavenging. Apart from antioxidant activity, this compound showed selective cytotoxicity in cancer cells. 20 and 24 µg/mL phenazine brought abount IC50 in HeLa and MCF-7 cell lines respectively (Fig. 3). At the same time no cytotoxicity occurred in normal H9c2 cells and remained unaffected even when the concentration of compound was increased to 60 µg/mL.
Fig. 3.
IC50 of phenazine compound in µg/mL against different cell lines (p < 0.05)
Benthocyanin B and its congener benthocyanin C, phenazine derivatives isolated from Psueodomonas punicolar are free radical scavengers and inhibit free radical induced lipid peroxidation in rat liver microsomes [11]. Apart from the excellent antifungal activity, the antioxidant activity assays revealed the potential of this compound to act as reducing agent and natural antioxidant with protective effect against oxidative damage.
Phenazines are associated with anti-tumor potential. Actively respiring cells, like cancer cells are more vulnerable to respiratory intervention and ROS generation by phenazines [12]. Phenazines also interfere with the activities of topoisomerase I and II in eukaryotic cells, and cancer cells because of high levels of both topoisomerases are more susceptible to this interference [1]. Development of anti-cancer drugs based on phenazine derivatives is a current topic of research. Saphenamycin from S. canaries and S. antibioticus, showed IC50 values of 0.15 µg/mL in L5178Y and of 0.6 µg/mL in L1210 mouse leukaemia cell lines [13] and IC50 of 0.6 µg/mL in CCRF/CEM T cell leukaemia cells [12, 14]. Apart from this, saphenamycin displays life-extending impact on mice with leukaemia cell implants. 250 µg/mL mouse/day × 10 days slightly (p > 0.05) prolonged by 19 and 20 % the survival of mice intra-peritoneally implanted with the L1210 leukaemia and the Ehrlich ascite carcinoma, respectively [13, 15]. A recent investigation revealed the selective cytotoxicity of Phenazine-1-carboxamide (PCN) against lung (A549) and breast (MDA-MB-231) cancer cell lines. It was also demonstrated that the cause of cytotoxicity is apoptosis as evidenced by morphological characteristics, loss of mitochondrial membrane potential and caspase-3 activation. The in silico docking studies aiming Bcl-2 family proteins revealed the binding of PCN with the BH3 domain and hence the possible inhibition of anti-apoptotic proteins that led to apoptosis [16]. PCA is reported to have anticancer activity against the human skin melanoma cell line SK-MEL-2 in addition to the UV-B protecting activity as calculated by spectrophotometry. Erythrocyte haemolysis assay showed that PCA is non-toxic up to concentration of 100 ppm [17]. Production of phenazines by LAB explains many of the reasons behind the therapeutic potential of probiotics based on LAB.
Aminopeptidase Inhibitory Activity
The enzyme activity calculated for LAP was 4.1 ± 0.04 IU/mL and 1 ± 0.08 IU/mL for PAP. Phenazine prevented 67.7 % activity of LAP and 94 % activity of PAP. Gilpin et al. [18] reported that phenazine compounds produced by Streptomyces possess metalloenzyme inhibitory activity. The mechanism of how phenazines cause aminopeptidase inhibition is not studied thoroughly and much data are not available about the same.
Amino peptidase inhibition disrupts protein turnover leading to an accumulation of peptides and a reduction in the cellular free amino acid content, which has profound effect on cell survival and proliferation especially in myeloma cells which are more reliant on protein production and unfolded protein response [19]. Clinical trials of cancer therapies in combination with aminopeptidase inhibitors are in progress. Bestatin is an example of aminopeptidase inhibitor used to treat acute and chronic acute myeloid leukemia under the trademark Ubenimex in Japan [20]. It is possible that a bioactive that is inhibitory to Streptomyces aminopeptidase can be inhibitory to eukaryotic aminopeptidases too. Both human and Streptomyces LAP belong to the M28 family that have two zinc atoms in their active site. The double -zinc coordinated active centre carries structural similarity among LAP from different sources [21]. Amastatin that mimics the LAP catalytic transition state inhibits eukaryotic aminopeptidases and is also inhibitory to LAP of S. hygroscopicus [22].
Conclusion
This work reveals the production of phenazine class of compounds by lactic acid bacteria. Antifungal, antioxidant and anti-cancer properties of this compound disclose the role in the probiotic and therapeutic potential of LAB. Selective cytotoxicity against cancer cells without affecting normal cells and aminopeptidase inhibitory activity of the strain BSN307 would be of particular interest in development of personal medicines. Since LAB are normal part of our daily diet, their intake can bring along therapeutic effects considering they are source of natural antioxidants and may be particularly useful when long term treatment is required.
Acknowledgments
K. K. Varsha greatly acknowledges the Research Fellowship from Council of Scientific and Industrial Research, India. KMN acknowledges the financial support from CSIR Net work project (CSC0133).
Compliance with Ethical Standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Pierson Iii LS, Pierson EA. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86:1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varsha KK, Priya S, Devendra L, Nampoothiri KM. Control of spoilage fungi by protective lactic acid bacteria displaying probiotic properties. Appl Biochem Biotechnol. 2014;172:3402–3413. doi: 10.1007/s12010-014-0779-4. [DOI] [PubMed] [Google Scholar]
- 3.Varsha KK, Devendra L, Shilpa G, Priya S, Pandey A, Nampoothiri KM. 2,4-Di-tert-butyl phenol as the antifungal, antioxidant bioactive purified from a newly isolated Lactococcus sp. Int J Food Microbiol. 2015;211:44–50. doi: 10.1016/j.ijfoodmicro.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 4.Shimada K, Fujikawa K, Yahara K, Nakamura T. Antioxidative properties of xanthan on the autoxidation of soybean oil in cyclodextrin emulsion. J Agric Food Chem. 1992;40:945–948. doi: 10.1021/jf00018a005. [DOI] [Google Scholar]
- 5.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 6.Rahulan R, Nampoothiri KM, Szakacs G, Nagy V, Pandey A. Statistical optimization of l-leucine amino peptidase production from Streptomyces gedanensis IFO 13427 under submerged fermentation using response surface methodology. Biochem Eng J. 2009;43:64–71. doi: 10.1016/j.bej.2008.08.011. [DOI] [Google Scholar]
- 7.Nandan A, Nampoothiri KM. Extracellular proline aminopeptidase production by Streptomyces lavendulae ATCC14162 under solid-state fermentation. J Sci Ind Res. 2013;72:591–595. [Google Scholar]
- 8.Blankenfeldt W (2013) The biosynthesis of phenazines. In: Chincholkar SB, Thomashow L (eds) Microbial phenazines. Springer, Berlin, pp 1–17. doi:10.1007/978-3-642-40573-0_1
- 9.Price-Whelan A, Dietrich LEP, Newman DK. Rethinking secondary metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 10.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinya K, Furihata K, Teshima Y, Hayakawa Y, Seto H. Benthocyanins B and C, new free radical scavengers from Streptomyces prunicolor. J Org Chem. 1993;58:4170–4172. doi: 10.1021/jo00067a069. [DOI] [Google Scholar]
- 12.Laursen JB, Nielsen J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem Rev. 2004;104:1663–1686. doi: 10.1021/cr020473j. [DOI] [PubMed] [Google Scholar]
- 13.Kitahara M, Nakamura H, Matsuda Y, Hamada M, Naganawa H, Maeda K, Umezawa H, Iitaka Y. Saphenamycin, a novel antibiotic from a strain of Streptomyces. J Antibiot. 1982;35:1412–1414. doi: 10.7164/antibiotics.35.1412. [DOI] [PubMed] [Google Scholar]
- 14.Geiger A, Keller-Schierlein W, Brandl M, Zahner H. Metabolites of microorganisms. 247. Phenazines from Streptomyces antibioticus, strain TUE 2706. J Antibiot. 1988;41:1542–1551. doi: 10.7164/antibiotics.41.1542. [DOI] [PubMed] [Google Scholar]
- 15.Cimmino A, Evidente A, Mathieu V, Andolfi A, Lefranc F, Kornienko A, Kiss R. Phenazines and cancer. Nat Prod Rep. 2012;29:487–501. doi: 10.1039/c2np00079b. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy RK, Veena V, Naik PR, Lakshmi P, Krishna R, Sudharani S, Sakthivel N. Phenazine-1-carboxamide (PCN) from Pseudomonas sp. strain PUP6 selectively induced apoptosis in lung (A549) and breast (MDA MB-231) cancer cells by inhibition of antiapoptotic Bcl-2 family proteins. Apoptosis. 2015;20:858–868. doi: 10.1007/s10495-015-1118-0. [DOI] [PubMed] [Google Scholar]
- 17.Patil S, Paradeshi J, Chaudhari B. Anti-melanoma and UV-B protective effect of microbial pigment produced by marine Pseudomonas aeruginosa GS-33. Nat Prod Res. 2016 doi: 10.1080/14786419.2016.1154057. [DOI] [PubMed] [Google Scholar]
- 18.Gilpin ML, Fulston M, Payne D, Cramp R, Hood I. Isolation and structure determination of two novel phenazines from a Streptomyces with inhibitory activity against metallo-enzymes, including metallo-. BETA.-lactamase. J Antibiot. 1995;48:1081–1085. doi: 10.7164/antibiotics.48.1081. [DOI] [PubMed] [Google Scholar]
- 19.Davenport EL, Moore HE, Dunlop AS, Sharp SY, Workman P, Morgan GJ, Davies FE. Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood. 2007;110:2641–2649. doi: 10.1182/blood-2006-11-053728. [DOI] [PubMed] [Google Scholar]
- 20.Ichinose Y, Genka K, Koike T, Kato H, Watanabe Y, Mori T, Iioka S, Sakuma A, Ohta M. Randomized double-blind placebo-controlled trial of bestatin in patients with resected stage I squamous-cell lung carcinoma. J Natl Cancer Inst. 2003;95:605–610. doi: 10.1093/jnci/95.8.605. [DOI] [PubMed] [Google Scholar]
- 21.Arima J, Uesugi Y, Uraji M, Yatsushiro S, Tsuboi S, Iwabuchi M, Hatanaka T. Modulation of Streptomyces leucine aminopeptidase by calcium identification and functional analysis of key residues in activation and stabilization by calcium. J Biol Chem. 2006;281:5885–5894. doi: 10.1074/jbc.M509025200. [DOI] [PubMed] [Google Scholar]
- 22.Liew SM, Tay ST, Puthucheary SD. Enzymatic and molecular characterisation of leucine aminopeptidase of Burkholderia pseudomallei. BMC Microbiol. 2013;13:110. doi: 10.1186/1471-2180-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]