Abstract
This study reports the identification of ionising radiation tolerant bacteria from a high elevation arid region of central Tibet. Nineteen isolates were isolated from soil exposed to ionising radiation at doses from 0 to 15 kGy. Isolates were phylogenetically characterised using 16S rRNA gene sequences. Most isolates comprised taxa from the Actinobacteria, Cyanobacteria, Firmicutes and proteobacteria and these survived doses up to 5 kGy. The Firmicutes and Deinococci also survived doses up to 10 kGy, and the highest dose of 15 kGy was survived only by the Deinococci. No altitude-related pattern was discernible within the range 4638–5240 m, instead culturable bacterial estimates for irradiated soil were strongly influenced by the abundance of Deinococci. We conclude that the relatively high UV exposure in Tibet has contributed to the high diversity of radiation tolerant soil bacteria. In addition, the strong association between desiccation-tolerance and radiation tolerance pathways suggests the arid environment may also have selected in favour of radiation tolerant taxa.
Keywords: Actinobacteria, Cyanobacteria, Deinococcus, Firmicutes, Proteobacteria, Radiation, Tibet
The cellular mechanism for desiccation tolerance [1] in microorganisms also confers radiation tolerance [2]. Several studies have shown that arid environments support radiation-tolerant bacteria and some are able to withstand relatively high doses of ionising radiation [3–5]. We hypothesized that high elevation arid region of central Tibet may have selected for microorganisms with tolerance to ionising radiation [6–8]. Here we report the isolation and phylogenetic identification of ionising radiation tolerant bacteria from Tibetan soils.
Surface soil (upper 20 mm) was sampled aseptically during May 2006 at four altitude-defined locations in central Tibet: 4638 m (32°5′1.44″N 84°7′55.02″E), 4810 m (29°27′42.00″N 86°8′37.32″E), 5240 m (31°33′57.30″N 85°11′9.48″E), 5520 m (32°5′1.44″N 84°7′55.02″E). A control sample of sub-tropical soil from Hong Kong (approx. 200 m altitude) was used as a control and yielded no viable bacteria after all radiation treatments. Soils were irradiated in glass vials using a 60Co source at the following doses: 0, 2.7, 5, 5.0. 10.0 15.0 kGy (Isotron plc, Swindon, UK). After irradiation soils were serially diluted and incubated on nutrient agar (Oxoid, Hampshire, UK) at 20 °C. The number of colony forming units (CFU) per gram of soil was determined after 5 days of incubation. Colonies representative of each morphology observed were then characterised by phylogenetic analysis of the 16S rRNA gene. Extraction of DNA and PCR amplification using primers 8F (AGA GTT TGA TCC TGG CTC AG) and 1391R (GAC GGG CGG TGT GTR CA) was conducted as previously described [9]. Sequencing was conducted using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems, California, USA) and performed on an Applied Biosystems 3730 Genetic Analyzer. Sequences were edited using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and then aligned using Clustal W (http://www.genome.jp/tools/clustalw/). Approximate phylogentic affiliations were then determined by BLAST searches of the NCBI GenBank database (http://www.ncbi.nim.nic.gov).
Phylogenetic analysis were conducted using PAUP* 4.0b8 [10]. Model sequence of evolution that best fit the data by hierarchical likelihood ratio test (hLRTs) was first generated using MODELTEST v.3.06 program [11]. Based on this model, maximum likelihood trees were generated with a starting tree obtained by stepwise addition, sequence addition taken as-is and tree bisection-reconnection (TBR) branch swapping. Gaps were treated as missing data. Nonparametric bootstrap support [12, 13] was calculated for each internal node with a simple stepwise sequence addition, TBR branch swapping and 1000 replicates. Bayseian posterior probablities were also calculated. Phylogenetic trees were drawn using TREEVIEW [14]. Sequences were deposited in the NCBI GenBank database under accession numbers (HQ144151-HQ144169).
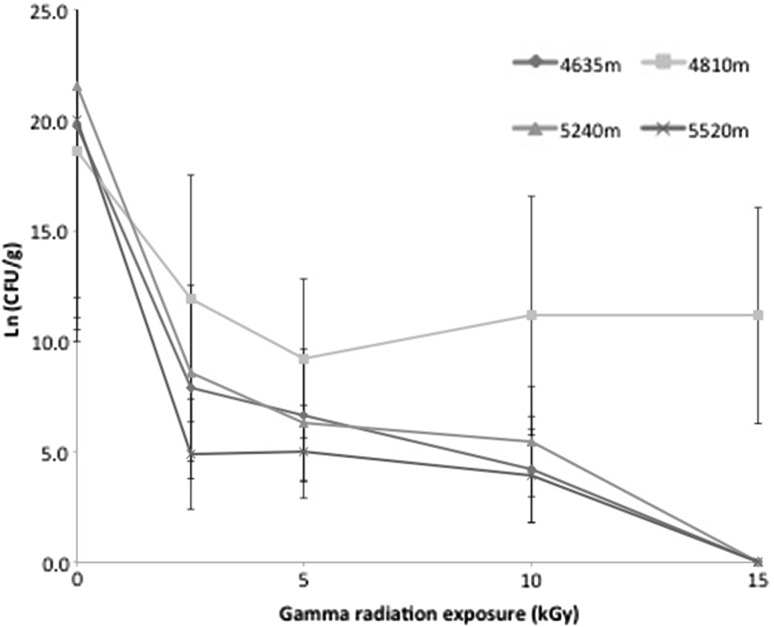
The total cultivable bacterial cell count for non-irradiated soils ranged from 1.2 × 108 to 2.32 × 109 CFU/g. A dose–response curve for irradiated soil samples revealed that viability decreased with increasing ionising radiation dose (Fig. 1). At a dose of 15 kGy samples from 4638, 5240 and 5520 m were effectively sterilised, but soil from 4810 m maintained a recoverable population of CFU/g at 7.21 × 104. We explain this in terms of the relatively high abundance of isolates from the known radiation-tolerant genus Deinococcus [1] recovered from this location. A total of 19 ionizing radiation tolerant isolates were cultivated from soil samples. Phylogenetic analysis revealed that isolates were from five bacterial phyla, comprising Actinobacteria, Cyanobacteria, Deinococcus-Thermus, Firmicutes, Proteobacteria.
Fig. 1.
Dose-response curve for bacterial abundance in soils with increasing exposure to ionising radiation. Error bars illustrate standard deviation for the mean of five replicates
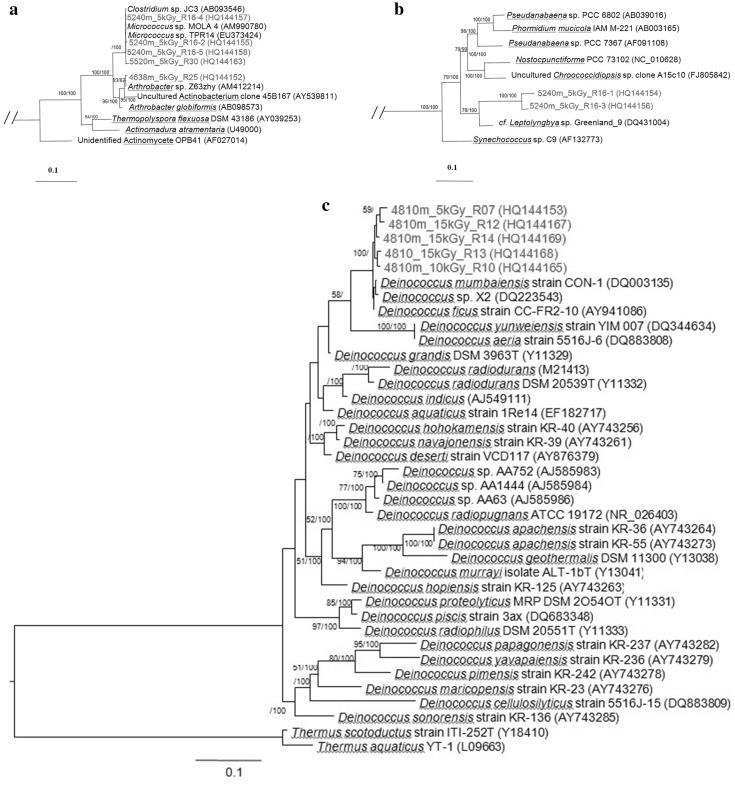
Four Actinobacteria taxa were isolated from 4638, 5240 and 5520 m soil samples that was exposed to 5 kGy of gamma radiation. The 16S rRNA gene analysis showed that they were closely affiliated to Arthrobacter/Rubrobacter and Micrococcus sp (Fig. 2a). The Actinobacteria are a high GC, gram-positive phylum found as a major component in desert soils [15] and Rubrobacter species are known to be radiotolerant [16].
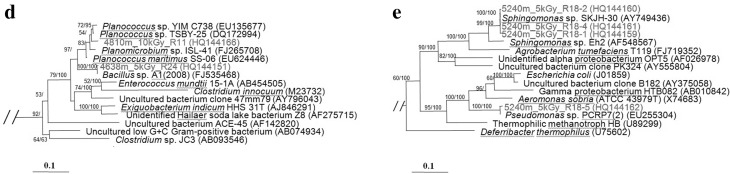
Fig. 2.
Phylogenetic identity of radiation tolerant bacteria isolated from high altitude soils in Tibet. Trees were created after Maximum Likelihood analysis of near-full length 16S rRNA gene sequences. Partial trees are shown but bootstrap (first number) and Bayesian posterior probabilities (second number) were generated from the full tree. Scale bars represent nucleotide change per position. a Actinobacteria, b Cyanobacteria, c Deinococci, d Firmicutes, e Proteobacteria
Two oscillatorian cyanobacteria were recovered from 5240 m samples exposed to 5 kGy of radiation (Fig. 2b). The molecular phylogeny for cyanobacteria is highly ambiguous although these taxa affiliated most closely with a Leptolyngbya taxon. This cyanobacterium has been implicated in forming extracellular polymeric substances which would help bind soil together and help in forming desert biotic soil crust [17]. The ability to produce this polymer has been linked directly to desiccation tolerance in cyanobacteria [18]. Cyanobacteria dominate soil biota in the most extreme desert habitats [15]. Cyanobacteria can tolerate desiccation as well as high doses of ionizing radiation [4]. Cyanobacteria also have been studied extensively for the ability to tolerate UV radiation [19].
The isolates surviving the highest dose (15 kGy) of ionising radiation were all phylogenetically affiliated with the genus Deinococcus within the Deinococcus/Thermus group (Fig. 2c). This genus comprises several species with extremely high tolerance to UV radiation and ionising radiation due to a highly effective cellular repair mechanism [1]. At least 29 species are recognized [5] and the genus has been encountered in several other arid environments [7, 9, 20, 21], as well as other extreme Tibetan environments such as geothermal pools [22–24].
Three Firmicutes taxa were isolated from 4638 to 4810 m (Fig. 2d). Sample R-09 and R-11 were closely affiliated with Planomicrobium/Planococcus sp. Sample R-24 is closely affiliated with Bacillus sp. Bacilli form resistant endospores and occur in extreme arid environments [3]. In this study, Planococcus and Planomicrobium taxa survived doses up to 10 kGy. Planococcus was first described as radiation tolerant in 2005 [5], whereas Planomicrobium has not previously been reported as radiation tolerant.
Isolates from 5240 m were identified as belonging to the alpha- and gamma- sub-phyla of proteobacteria after being exposed to 5 kGy of gamma radiation (Fig. 2e). Previous study has indicated proteobacteria may be a highly abundant group in extreme desert environments [25]. Here we encountered several radio-tolerant taxa including Methylobacterium and Chelatococcus [5]. A single Sphingomonas-like alpha proteobacterium encountered in this study. The family Sphingomonadaceae was reportedly resistant to high doses of radiation of up to 15 kGy [5]. The Gammaproteobacteria clone was closely affiliated with Pseudomonas-like taxa from high altitude soil [26]. Pseudomonas was previously identified as very susceptible to low-intensity ionizing radiation [27].
Overall this study has identified high altitude arid soils support diverse radio-tolerant bacteria from five phyla. In view of the enhanced UV exposure at high altitude Tibetan locations [6, 7, 23, 24] and the strong association between desiccation-tolerance pathways and radiation tolerance [1] it is likely that the relatively high UV environment at high altitude combined with the periodic desiccation imposed by the arid environment have selected for this trait. Since other Indian environments have also yielded radiation-tolerant bacteria [28, 29] we speculate that this continent may prove to be a biodiversity hotspot for radiation-tolerant taxa.
Contributor Information
Donnabella C. Lacap-Bugler, Email: dlacapbu@aut.ac.nz
Stephen B. Pointing, Email: steve.pointing@aut.ac.nz
References
- 1.Daly MJ. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat Rev Micro. 2009;7:237–245. doi: 10.1038/nrmicro2073. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell MM, Bornman JF, Ballare CL, et al. Terrestrial ecosystems, increased solar ultraviolet radiation, and interactions with other climate change factors. Photochem Photobiol Sci. 2007;6:252–266. doi: 10.1039/b700019g. [DOI] [PubMed] [Google Scholar]
- 3.Chanal A, Chapon V, Benzerara K, et al. The desert of Tataouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ Microbiol. 2006;8:514–525. doi: 10.1111/j.1462-2920.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- 4.Billi D, Friedmann EI, Hofer KG, et al. Ionizing-radiation resistance in the desiccation-tolerant Cyanobacterium Chroococcidiopsis. Appl Environ Microbiol. 2000;66:1489–1492. doi: 10.1128/AEM.66.4.1489-1492.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rainey FA, Ray K, Ferreira M, et al. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl Environ Microbiol. 2005;71:5225–5235. doi: 10.1128/AEM.71.9.5225-5235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong FKY, Lacap DC, Lau MCY, et al. Endolithic microbial colonization of limestone in a high-altitude arid environment. Microb Ecol. 2010;59:689–699. doi: 10.1007/s00248-009-9607-8. [DOI] [PubMed] [Google Scholar]
- 7.Wong FKY, Lacap DC, Lau MCY, et al. Hypolithic microbial community of quartz pavement in the high-altitude tundra of central Tibet. Microb Ecol. 2010;60:730–739. doi: 10.1007/s00248-010-9653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahl J, Lau MCY, Smith GJD, et al. Ancient origins determine global biogeography of hot and cold desert cyanobacteria. Nat Commun. 2011;2:163. doi: 10.1038/ncomms1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pointing SB, Chan Y, Lacap DC, et al. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci. 2009;106:19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swofford DL (2003) PAUP* phylogenetic analysis using parsimony (*and Other Methods) Version 4. http://www.sinauer.com/paup-phylogenetic-analysis-using-parsimony-and-other-methods-4-0-beta.html
- 11.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J (1989) PHYLIP—phylogeny inference package (Version 3.2). Cladistics 5:164–166. http://evolution.genetics.washington.edu/phylip.html
- 13.Sander C, Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 14.Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 15.Pointing SB, Belnap J. Microbial colonization and controls in dryland systems. Nat Rev Micro. 2012;10:551–562. doi: 10.1038/nrmicro2831. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira AC, Nobre MF, Moore E, et al. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles. 1999;3:235–238. doi: 10.1007/s007920050121. [DOI] [PubMed] [Google Scholar]
- 17.Mazor G, Kidron GJ, Vonshak A, Abeliovich A. The role of cyanobacterial exopolysaccharides in structuring desert microbial crusts. FEMS Microbiol Ecol. 1996;21:121–130. doi: 10.1111/j.1574-6941.1996.tb00339.x. [DOI] [Google Scholar]
- 18.Billi D, Potts M. Life and death of dried prokaryotes. Res Microbiol. 2002;153:7–12. doi: 10.1016/S0923-2508(01)01279-7. [DOI] [PubMed] [Google Scholar]
- 19.Castenholz RW, Garcia-Pichel F (2002) Cyanobacterial responses to UV-radiation. In: Ecol cyanobacteria. Springer, pp 591–611
- 20.Warren-Rhodes KA, Rhodes KL, Pointing SB, et al. Hypolithic cyanobacteria, dry limit of photosynthesis, and microbial ecology in the hyperarid Atacama Desert. Microb Ecol. 2006;52:389–398. doi: 10.1007/s00248-006-9055-7. [DOI] [PubMed] [Google Scholar]
- 21.Pointing SB, Warren-Rhodes KA, Lacap DC, et al. Hypolithic community shifts occur as a result of liquid water availability along environmental gradients in China’s hot and cold hyperarid deserts. Environ Microbiol. 2007;9:414–424. doi: 10.1111/j.1462-2920.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 22.Lau CY, Aitchison JC, Pointing SB. Early colonization of thermal niches in a silica-depositing hot spring in central Tibet. Geobiology. 2008;6:136–146. doi: 10.1111/j.1472-4669.2007.00124.x. [DOI] [PubMed] [Google Scholar]
- 23.Jing H, Lacap DC, Lau CY, Pointing SB. Community phylogenetic diversity of cyanobacterial mats associated with geothermal springs along a tropical intertidal gradient. Extremophiles. 2006;10:159–163. doi: 10.1007/s00792-005-0477-9. [DOI] [PubMed] [Google Scholar]
- 24.Lau CY, Jing H, Aitchison JC, Pointing SB. Highly diverse community structure in a remote central Tibetan geothermal spring does not display monotonic variation to thermal stress. FEMS Microbiol Ecol. 2006;57:80–91. doi: 10.1111/j.1574-6941.2006.00104.x. [DOI] [PubMed] [Google Scholar]
- 25.Connon S, Lester E, Shafaat H, Al E. Bacterial diversity in hyperarid Atacama Desert soils. J Geophys Res Biogeosci. 2007;112:G04S17. doi: 10.1029/2006JG000311. [DOI] [Google Scholar]
- 26.Selvakumar G, Joshi P, Mishra P, et al. Mountain aspect influences the genetic clustering of psychrotolerant phosphate solubilising pseudomonads in the Uttarakhand Himalaya. Curr Microbiol. 2009;59:432–438. doi: 10.1007/s00284-009-9456-1. [DOI] [PubMed] [Google Scholar]
- 27.Ito H, Lizuka I. Taxonomic studies on a radio-resistant pseudomonas part XII. Studies on the microorganisms of cereal grain. Agric Biol Chem. 1971;35:1566–1571. doi: 10.1080/00021369.1971.10859995. [DOI] [Google Scholar]
- 28.Suresh K, Reddy GSN, Sengupta S, Shivaji S. Deinococcus indicus sp. nov., an arsenic-resistant bacterium from an aquifer in West Bengal. India. Int J Syst Evol Microbiol. 2004;54:457–461. doi: 10.1099/ijs.0.02758-0. [DOI] [PubMed] [Google Scholar]
- 29.Shashidhar R, Bandekar JR. Deinococcus mumbaiensis sp. nov., a radiation-resistant pleomorphic bacterium isolated from Mumbai, India. FEMS Microbiol Lett. 2006;254:275–280. doi: 10.1111/j.1574-6968.2005.00033.x. [DOI] [PubMed] [Google Scholar]