Abstract
Biofilm formation is closely related to the pathogenetic processes of Klebsiella pneumoniae, which frequently causes infections in immunocompromised individuals. The immune system of astronauts is compromised in spaceflight. Accordingly, K. pneumoniae, which used to be isolated from orbiting spacecraft and astronauts, poses potential threats to the health of astronauts and mission security. Microgravity is a key environmental cue during spaceflight. Therefore, determining its effects on bacterial biofilm formation is necessary. In this study, K. pneumoniae ATCC BAA‐1705 was exposed to a simulated microgravity (SMG) environment. K. pneumoniae grown under SMG formed thicker biofilms compared with those under normal gravity (NG) control after 2 weeks of subculture. Two indicative dyes (i.e., Congo red and calcofluor) specifically binding to cellulose fibers and/or fimbriae were utilized to reconfirm the enhanced biofilm formation ability of K. pneumoniae grown under SMG. Further analysis showed that the biofilms formed by SMG‐treated K. pneumoniae were susceptible to cellulase digestion. Yeast cells mannose‐resistant agglutination by K. pneumoniae type 3 fimbriae was more obvious in the SMG group, which suggests that cellulose production and type 3 fimbriae expression in K. pneumoniae were both enhanced under the SMG condition. Transcriptomic analysis showed that 171 genes belonging to 15 functional categories were dysregulated in this organism exposed to the SMG conditions compared with those in the NG group, where the genes responsible for the type 3 fimbriae (mrkABCDF) and its regulator (mrkH) were upregulated.
Keywords: Biofilm, cellulose, Klebsiella pneumonia, simulated microgravity, type 3 fimbriae.
Introduction
Several opportunistic bacterial pathogens have been detected in samples from human space habitats and postflight astronauts (Castro et al. 2004; Novikova 2004). Microgravity is a key environmental factor in spaceflight, where microbes sense and respond to environmental stresses by modulating their gene expressions and altering their physiological and pathogenic processes (Wilson et al. 2007; Crabbe et al. 2011). Research resources in microgravity condition are extremely restricted because of the logistic reasons and safety considerations. To address this problem, high‐aspect ratio rotating‐wall vessels (HARVs) are extensively applied to investigate the physiological characteristics of microbes in a simulated microgravity (SMG) environment (Nickerson et al. 2000; Wilson et al. 2002a,b; Lynch et al. 2004, 2006; Crabbe et al. 2010; Lawal et al. 2010; Castro et al. 2011).
HARVs can create a low‐fluid shear condition after being filled with fluid medium and rotated at a setting speed, because air bubbles are completely removed and turbulent flow is minimized. The cells weight in the HARVs of the SMG group is offset with hydrodynamic forces (e.g., centrifugal, Coriolis, and shear components) and constantly suspended in a low‐fluid shear condition, which partially mimics the true microgravity environment. Several previous studies have adopted low‐shear modeled microgravity (LSMMG) to refer to the SMG condition in HARVs. Anaerobic culture is also avoided by oxygen diffusion through a gas‐permeable membrane in the back of HARVs during growth (Schwarz et al. 1992; Hammond and Hammond 2001).
Klebsiella pneumoniae is a ubiquitous opportunistic pathogen, commonly found in both clinical and nonclinical settings, and is associated with hospital‐acquired urinary, respiratory tract infections, bacteremia, and surgical wound infections, especially in immunocompromised individuals (Podschun and Ullmann 1998). This organism has been isolated from the equipment of on‐board spacecraft and from astronauts (Taylor 1974; Novikova 2004), and poses potential threats to the health of astronauts because their immune system is compromised in spaceflight (Taylor et al. 1996; Gueguinou et al. 2009). The biofilms alter the adaptation ability of bacterial cells to stressful environments, including antibiotic exposure and host immune responses, and might account for antibiotic treatment failure in chronic infection (Walters et al. 2003; Jefferson et al. 2005). The SMG effects on K. pneumoniae biofilm formation were examined in this study by using HARVs to continuously culture this organism under SMG and NG conditions.
Materials and Methods
Bacterial strains and growth conditions
The carbapenem‐resistant K. pneumoniae strain ATCC BAA‐1705 was clinically isolated from the urine of a male patient and utilized throughout this study. The bacterial strain was aerobically grown at 37°C in lysogeny broth (LB) or agar unless indicated otherwise. The test (SMG) and the control (NG) settings were established by growing bacterial cells for continuous cultivation in HARV bioreactors (Synthecon, Inc., Houston, Tex, USA). Figure 1A shows the SMG cultivation achieved by rotating the bioreactor with its axis perpendicular to gravity. NG cultivation was achieved with its axis parallel to gravity (Nickerson et al. 2000). Overnight cultures grown at 37°C with shaking at 200 rpm were inoculated at a dilution of 1:200 in the HARV bioreactors. Each bioreactor was completely filled with ~58 mL fresh LB medium. Air bubbles were carefully removed. After 24 h of incubation at 37°C in HARVs with a rotation of 25 rpm, both the SMG and NG bacterial cultures were diluted into new HARVs completely filled with the LB medium and then incubated at 37°C and 25 rpm for another 24 h. Experimental manipulations of bacterial inoculation in the HARV bioreactors were successively performed for 2 weeks. Bacterial cell numbers in the SMG and NG groups were counted through serial dilution in phosphate‐buffered saline (PBS) and plating on LB agar. The resulting cultures were subjected to the following assays.
Figure 1.
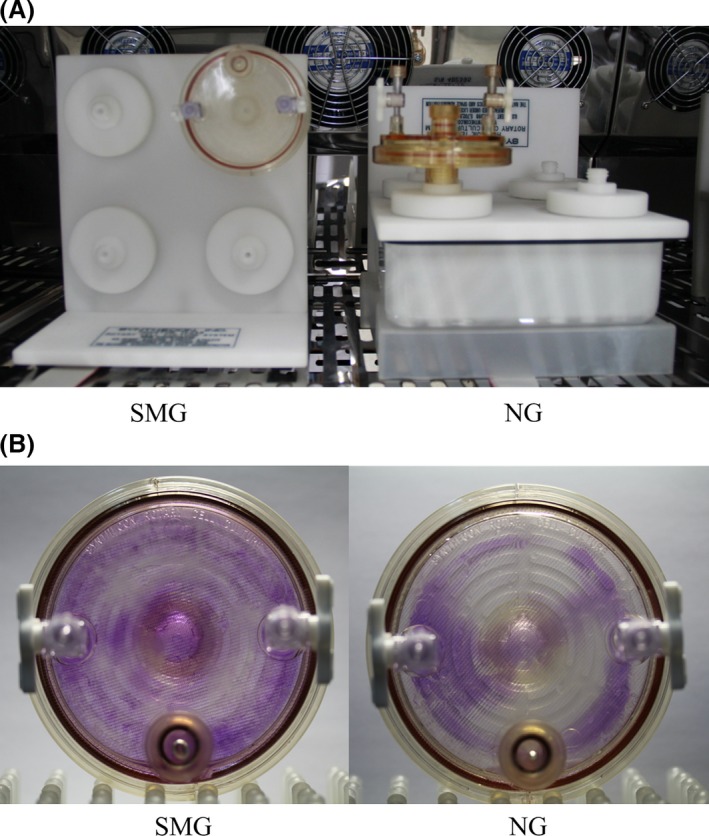
HARV bioreactors in the experimental setup. The bacterial cells in the HARV bioreactor are grown under the simulated microgravity (SMG) condition with its axis of rotation perpendicular to gravity or grown under the NG condition with its axis of rotation vertical to gravity when the medium is filled and the bubbles are removed (A). Both SMG and NG HARV bioreactors are stained with 0.1% crystal violet after 2‐week cultivation (B).
Crystal violet staining
Crystal violet staining was performed to evaluate the biofilm formation ability in K. pneumoniae after 2 weeks cultivation under SMG and NG conditions. The cultures in the HARV bioreactors were removed. The bioreactors were washed gently with deionized water and then stained with 0.1% crystal violet dye for 15 min at room temperature. Both the test and control cultures were separately diluted (1:100) in 5 mL LB medium in glass tubes and grown at 37°C and 200 rpm for 24 h. The planktonic bacteria were removed. Subsequently, each tube was washed three times with deionized water. The glass tubes were then stained with 0.1% crystal violet dye for 15 min at room temperature.
The biofilm formation in the SMG and NG groups was quantified by separately diluting both the test and control cultures (1:100) on a 24‐well plate. Each well contained 1 mL LB medium. The planktonic bacteria were removed after the 24‐well plate was incubated at 37°C and 200 rpm for 24 h. Each well was washed three times with deionized water. The 24‐well plate was then incubated at 80°C for 15 min to fix the biofilms. The adherent bacterial cells were stained with 0.1% crystal violet dye for 15 min and subsequently rinsed with deionized water. Bound crystal violet was solubilized with 2 mL dimethyl sulfoxide and quantified by measuring the optical density (OD) values at 570 nm. The results are presented as mean ± SD for three biological replicates.
Congo red‐based colony morphology
Ten microliters of 2‐week cultures under both NG and SMG conditions were spotted onto an LB agar plate containing 25 μg/mL Congo red. The spotted cultures were incubated at 37°C. The images of the spotted cultures were captured after 72 h.
Calcofluor staining
Five milliliters of K. pneumoniae cultures under both NG and SMG conditions were harvested and resuspended in 5 mL of PBS with 200 μg/mL calcofluor. The cells were recovered by centrifugation after 10 min of incubation at room temperature. The cells were then rinsed and resuspended with 5 mL PBS. Afterward, 10 μL of the resuspended mixture was dropped onto plastic plates. The fluorescent colony was visualized by exposure to UV light and imaged by using a UV light imaging system (Gel DocTM XR+, Bio‐Rad). For fluorescence quantification, 100 μL bacterial cell suspension was added to 96‐well black cell culture plates (Corning® Costar®). The fluorescence values were then measured with a spectrophotometer (SpectraMax M2, Molecular Devices) with 366 nm excitation and 525 nm emission. The results are indicated as mean ± SD calculated from three replicate samples.
Cellulase digestion of biofilms
The biofilm formation of the SMG and NG cultures was measured in 24‐well plate as described in the “Crystal violet staining” section. Each well was rinsed with deionized water after the planktonic bacteria were removed. Two milliliters of 50 mmol/L citrate buffer (pH 4.6) with 0.1% cellulase (Sigma Chemical Co., St. Louis, Mo, USA) were added to the wells, which were then incubated at 45°C for 48 h (Huertas, Zarate et al. 2014). The biofilms were stained with crystal violet and quantified through spectrometry.
Yeast cell agglutination assays
The PBS‐resuspended cultures in both SMG and NG groups were serially and doubly diluted in 24‐well plates to identify the yeast cell agglutination titers. Briefly, 100 μL of the PBS‐resuspended bacterial cells of each dilution degree in both the SMG and NG groups was applied to a polystyrene plate and mixed with 50 μL of 1% yeast cells (Sigma Aldrich) and 50 μL of 5% mannose (Sigma Aldrich). The polystyrene plate was rotated until agglutination was visible (Stahlhut, Struve et al. 2012). The logarithm means of the agglutination titers in both the SMG and NG groups were obtained from at least three experiments.
RNA‐seq‐based transcriptional analysis
The total RNAs of the bacterial cells incubated in the SMG and NG bioreactors were extracted with the PureLink™ RNA Mini Kit (Ambion, USA) according to the manufacturer's instructions. RNA quality was checked through agarose gel electrophoresis. The RNAs were then quantified with a spectrophotometer. The RNA samples were subsequently subjected to cDNA library construction and deep sequencing. The fragments per kilobase of transcript per million fragments mapped values were calculated to determine the expression levels of transcripts. The fold changes in the transcript levels between the SMG and NG groups were also calculated. A twofold change was defined as the arbitrary threshold of the differentially regulated genes.
Quantitative real‐time PCR
cDNA was synthesized by 7 μg of the total RNAs and 3 μg of random hexamer primers with the Superscript III reverse transcriptase (Invitrogen) for quantitative real‐time PCR (qRT‐PCR). All primer pairs were designed to produce amplicons with expected sizes of 100–200 nt using K. pneumoniae ATCC BAA‐1705 genomic DNA as the PCR template (Table 1). qRT‐PCR was performed in duplicate for each RNA sample using the LightCycler system (Roche, Switzerland) with cDNA as a template. The relative fold change of the target genes in the test and control RNA samples was determined. The 16S rRNA gene was used as an internal reference.
Table 1.
Primers for the target genes used in qRT‐PCR
| Gene name | Primers (forward/reverse, 5′–3′) |
|---|---|
| bcsA | GCTTGGCTGTCTGTGGG/GCGATGCGGATTTATGTG |
| mrkA | TTCTTCTCTCTGCAGCAATG/TACCGGAGACAGGTAAACGT |
| mrkB | ACCCGCTTTATTTATCCAGG/AAACGGGGTGGTAATGGTAT |
| mrkC | TGTGCTGCTTTCCGCCATTT/CGCCCTTTCCACTCGTCGTT |
| mrkD | GCGTCTCTCATCGCCAACGG/CACGATCTTCGCCGCAAAGC |
| mrkF | CCTCGGCGTGGGGTTTTGAG/GACTTCCGCCAGGCTGACCG |
| mrkH | TGGACTTTGCCGAGTT/ACCGCTATTGTCATGTTT |
| 16S rRNA | GAGCGGCGGACGGGTGAGTA/GGGCACATCTGATGGCATGA |
Statistical analysis
All the quantitative experiments were independently performed at least in triplicate. The data were analyzed with SPSS 20.0 (SPSS, Chicago, IL, USA), and Student's t‐test was utilized to determine the statistical significance (P < 0.05) between any two groups.
Results
Enhanced effect of SMG on K. pneumoniae biofilm formation
The biofilm formation phenotypes of both the SMG and NG cultures were analyzed after 2‐week continuous cultivation by observing the pellicles that formed and adhered to the gas‐permeable membrane at the back of the bioreactors and the glass tubes at the air–liquid interface. OD570 was also measured through crystal violet staining. The pellicles formed by K. pneumoniae grown under the SMG condition were thicker than those grown under the NG condition (Figs. 1B and 2A). The adhered pellicles were further quantified by the OD570 values of crystal violet staining. More than a twofold increase in the adhered dye was observed in K. pneumoniae under the SMG condition relative to the NG condition (Fig. 2B). The cell numbers in the SMG and NG groups were also counted by dilution in PBS and plating on LB agar for three times (~2.1 × 109 CFU/mL and 3.2 × 109 CFU/mL in the SMG and NG groups, respectively). This process confirmed that the thicker biofilm formation in the SMG group was not caused by more bacterial cell inoculation in the SMG group.
Figure 2.
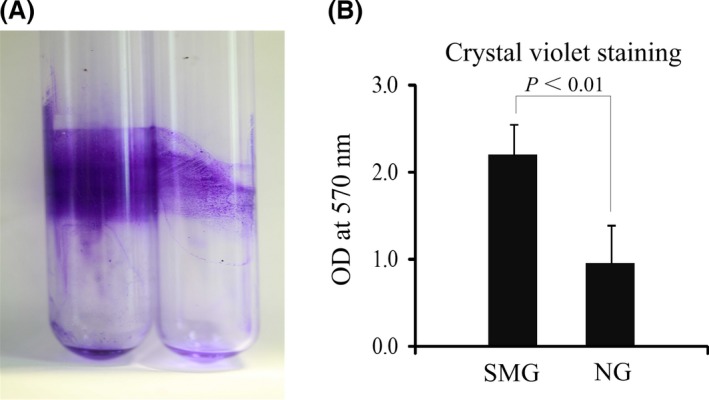
Quantification of biofilm formation by K. pneumoniae grown under SMG and NG conditions. The biofilms formed on the glass tubes are monitored by crystal violet staining (A) and quantified by measuring the OD 570 values (B). SMG, simulated microgravity.
Increased Congo red binding to the colony and yeast cell agglutination formed by K. pneumoniae grown under SMG
The major biofilm components were identified by spotting the bacterial culture on Congo red agar plates. The K. pneumoniae in the SMG group resulted in an RDAR (red, dry, and rough) colony, whereas the NG group remained white (Fig. 3A). The RDAR morphology is often associated with cellulose and fimbriae production characterized in Salmonella enteric serovar Typhimurium and Escherichia coli (Zogaj, Nimtz et al. 2001). The results of the different binding abilities of Congo red dye indicated that SMG might promote cellulose and fimbriae expression, which results in increased biofilm formation in K. pneumoniae.
Figure 3.
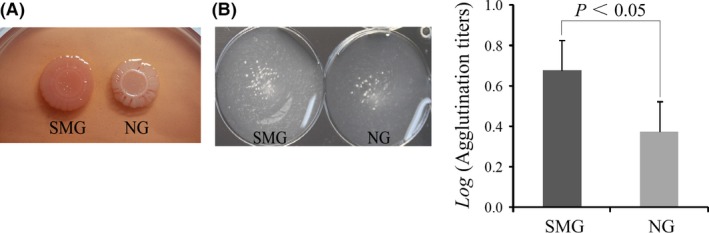
Colony morphology observation and type 3 fimbriae expression analysis. Colony morphology of the simulated microgravity (SMG)‐ and NG‐treated K. pneumoniae on Congo red agar (A). Klebsiella pneumoniae type 3 fimbriae can agglutinate yeast cells in a mannose‐resistant manner. Yeast cell agglutination (induced by undiluted bacterial cells) is observed on polystyrene plates for the SMG and NG groups. The agglutination titers of K. pneumoniae type 3 fimbriae in the SMG and NG groups are also compared by their logarithm means (B).
The agglutination assays also showed that yeast cell agglutination by K. pneumoniae type 3 fimbriae in a mannose‐resistant manner was more obvious in the SMG group than in the NG group. The geometric mean of the agglutination titers in the SMG group was 1:4.8, whereas that in the in the NG group was 1:1.7. And the logarithm mean of the agglutination titers in the SMG higher than that in the NG groups (Fig. 3B). This result indicates that a higher type 3 fimbriae expression exists in the SMG‐treated K. pneumoniae.
Cellulose as a major component of the biofilms formed by K. pneumoniae grown under SMG
Cellulose participates in the biofilm formation process in many bacteria (Romling 2002). The fluorescent dye, calcofluor, specifically binds to cellulose but not to curli fimbriae. The dye binds β‐1, 4 glucose cellulose linkages and causes a fluorescence strain phenotype under a long‐wavelength UV light source. Hence, calcofluor dye is widely used to analyze cellulose production in bacteria (Zogaj, Bokranz et al. 2003). The calcofluor fluorescence phenotype was detected to access cellulose production in K. pneumoniae under SMG and NG conditions. The fluorescence intensity of the SMG‐treated K. pneumoniae in the calcofluor agar was higher than that of the NG‐treated group. Quantitative results show that the fluorescence values at 525 nm of the SMG cultures are approximately twice those of the NG cultures (Fig. 4A). This result suggests that cellulose production increased after exposure to the SMG condition.
Figure 4.
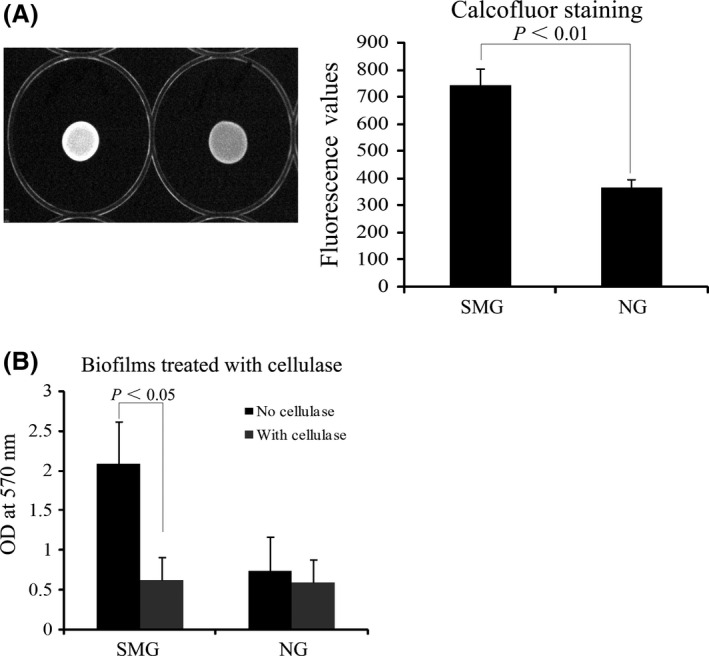
Cellulose production assay for K. pneumoniae cultured under SMG and NG conditions. Cellulose production is monitored by calcofluor staining (A). The biofilms formed in the wells of the 24‐well plate are quantified by crystal violet staining after digestion with cellulase (B). SMG, simulated microgravity.
The existence of cellulose in the biofilms was further confirmed by adding cellulase to digest the cellulose within the biofilms. A sharp decrease in the OD570 values was observed in the SMG group upon cellulase treatment, whereas no significant difference was found in the NG group under the same condition (Fig. 4B). Cellulose might be an important component of thick biofilms in SMG‐treated K. pneumoniae.
Increased transcriptional level of genes for type 3 fimbriae in SMG‐treated K. pneumoniae
RNA‐seq was utilized to monitor differences in the transcriptomic mRNA profile at a global scale between the SMG‐ and NG‐treated K. pneumoniae. A total of 171 genes were differentially expressed in the SMG group relative to the NG group (i.e., 89 upregulated genes and 82 downregulated genes) (see Data S1). The differentially regulated genes were then classified based on the KEGG pathway database. A total of 88 genes were categorized to 15 biochemical or signal transduction pathways. Approximately 64% (i.e., 56 genes) of the dysregulated genes belonged to five “metabolic pathway” categories (i.e., amino acid, carbohydrate, and energy metabolisms) (Table 2). A total of 14 dysregulated genes involved in mRNA translation were upregulated in K. pneumoniae grown under the SMG condition.
Table 2.
KEGG pathways of dysregulated genes in K. pneumoniae grown under SMG
| No. | Pathway | Upregulated genes | Downregulated genes | ||
|---|---|---|---|---|---|
| No. | Gene ID (KPBAA1705_) | No. | Gene ID (KPBAA1705_) | ||
| 1 | Amino acid metabolism | 5 | 06833, 14946, 14961, 17616, 25221 | 9 | 03301, 10183, 13245, 19907, 19912, 20820, 21421, 23663, 23678 |
| 2 | Carbohydrate metabolism | 2 | 16206, 25221 | 10 | 05906, 06221, 10902, 14095, 20210, 20820, 23023, 23323, 23663, 26174 |
| 3 | Cell growth and death | 0 | 1 | 25071 | |
| 4 | Energy metabolism | 7 | 05591, 05596, 13845, 13850, 13855, 13860, 25221 | 1 | 10183 |
| 5 | Glycan biosynthesis and metabolism | 3 | 14946, 14961, 14966 | 2 | 14095, 23023 |
| 6 | Virulence | 3 | 02841, 02891, 13830 | 0 | |
| 7 | Lipid metabolism | 2 | 14395, 25221 | 4 | 14095, 20820, 23023, 23663 |
| 8 | Membrane transport | 4 | 02836, 13820, 13830, 15096 | 4 | 02701, 05936, 24409, 26184 |
| 9 | Metabolism of cofactors and vitamins | 1 | 02401 | ||
| 10 | Metabolism of terpenoids and polyketides | 2 | 19092, 25221 | 1 | 23663 |
| 11 | Nucleotide metabolism | 1 | 00965 | 0 | |
| 12 | Replication and repair | 4 | 02586, 19637, 27259, 27454 | 0 | |
| 13 | Signal transduction | 2 | 13550, 13830 | 1 | 23323 |
| 14 | Translation | 14 | 21446, 26464, 26469, 26474, 26479, 26484, 26489,26494, 26499, 26504, 26509, 26514, 26554, 26559 | 0 | |
| 15 | Xenobiotics biodegradation and metabolism | 1 | 25221 | 3 | 03301, 23323, 23663 |
The biofilm formation ability of K. pneumoniae is determined by the intracellular levels of capsule, exopolysaccharides, and cell surface fimbriae (Jagnow and Clegg 2003; Schroll, Barken et al. 2010; Huertas, Zarate et al. 2014). Therefore, the expressions of the cellulose synthase‐encoding gene bcsA and type 3 fimbriae‐encoding genes mrkABCDF and its regulatory gene mrkH were selected for further analysis through RNA‐seq and qRT‐PCR. As expected, the mrkABCDF and mrkH gene expressions were upregulated in the SMG‐treated K. pneumoniae. However, the bcsA gene expression was unchanged at the transcriptional level (Fig. 5).
Figure 5.
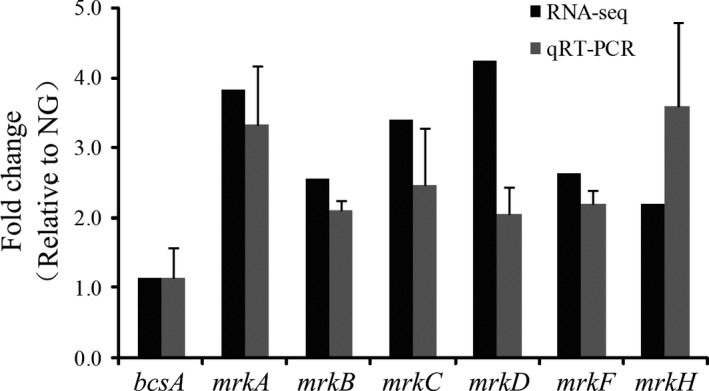
Expression validation of the biofilm‐associated genes by RNA‐seq (black) and qRT‐PCR (gray). Relative expression is indicated as fold changes ± standard derivation between the SMG‐ and NG‐treated K. pneumoniae. SMG, simulated microgravity.
Discussion
The bacterial cells in biofilms are embedded in extracellular polymeric substances (EPS) mostly produced by themselves, including exopolysaccharides, proteins, extracellular DNA, surfactants, lipids, and water (Flemming and Wingender 2010). Cellulose, an exopolysaccharide composed of β‐1, 4 glycosidic bond‐linked glucose units, has been identified as a significant component of EPS associated with biofilm formation in various bacteria (Zogaj, Nimtz et al. 2001; Spiers, Sm. et al. 2003). Extracellular structures (pili or fimbriae) in EPS are also required for biofilm formation in Enterobacteriaceae (Pratt and Kolter 1998). The altered biofilms in bacteria often correlate with the fluctuant binding ability of the Congo red dye and the fluorescence dye calcofluor. These two indicative dyes can specifically bind to cellulose and/or fimbriae. Thus, both dyes were utilized in this study to assess biofilm formation and ascertain the major components of thick biofilms formed by the SMG‐treated K. pneumoniae.
Cellulose biosynthesis in K. pneumoniae is silent under laboratory conditions (Zogaj, Bokranz et al. 2003). The present study showed that cellulose production was enhanced and mainly responsible for biofilm formation in K. pneumoniae grown under the SMG condition. However, no significant difference in the cellulose synthase‐encoding bcsA transcript abundance of K. pneumoniae was observed between the SMG and NG groups. Given the previous finding that cellulose synthesis is positively regulated by c‐di‐GMP via the allosteric effect on BcsA, the high cellulose production and the indistinctive value of bcsA transcript abundance in the SMG group may be attributed to the fact that bcsA expression is mainly regulated at the posttranslational level in the SMG condition (Morgan, Strumillo et al. 2013).
In K. pneumoniae, the type 3 fimbriae mediate the biofilm formation on biotic and abiotic surfaces (Murphy and Clegg 2012). Type 3 fimbriae are encoded by the mrkABCDF operon, mrkA, which encodes the major fimbria subunit; mrkB and mrkC, which encode a chaperone–usher system; mrkD, which encodes the fimbrial tip ahesin; and mrkF, which encodes an unknown function protein. The type 3 fimbriae expression is also regulated by the c‐di‐GMP molecule. The mrk gene cluster expression is activated at the transcriptional level via the c‐di‐GMP‐dependent regulatory effect on MrkH (Wilksch, Yang et al. 2011). The yeast cell agglutination assays and the transcriptomic analysis in this study indicated that the expression of K. pneumoniae type 3 fimbriae was upregulated in the SMG‐treated group.
Previous reports have shown that the expression of numerous genes is affected for bacterial cells in simulated microgravity (Wilson et al. 2002b; Crabbe et al. 2010). The transcriptomic analysis in this study showed that 171 genes belonging to 15 functional categories were dysregulated in K. pneumoniae of the SMG group. Among these dysregulated genes, 45 gene products were hypothetical proteins; the others were not closely related to K. pneumoniae biofilm formation, except for cellulose and the type 3 fimbriae‐encoding genes. The expression of type 1 fimbriae (encoding by the fimBEAICDFGHK gene cluster) subunit‐encoding gene fimA was also significantly upregulated in K. pneumoniae of the SMG group. But no other correlated gene upregulation or expression was identified in both groups, which indicates that type 1 fimbriae were not correctly expressed in the K. pneumoniae cultured in the HARVs.
Biofilm formation in bacteria was first investigated under microgravity in 2001 (McLean, Cassanto et al. 2001). Short‐term (24 h) exposure to microgravity affects the biofilm production in E. coli and Pseudomonas aeruginosa (Lynch et al. 2006; Crabbe, De Boever et al. 2008). Li et al. cultured K. pneumoniae ATCC BAA‐2146 in a semisolid medium for 15 days in spaceflight. Their results showed an enhanced K. pneumoniae biofilm formation ability after cultivation, which was consistent with our results obtained from culturing K. pneumoniae in the SMG environment (Jia Li 2014). However, the K. pneumoniae cultured in the SMG environment in Li et al.'s study showed a similar biofilm formation capacity with the strain cultured on the ground. Moreover, no gene closely related to biofilm formation was upregulated in spaceflight or SMG condition. The differences in bacteria strains and culture methods (culture medium and microgravity analogs) selected in the studies may be the cause of the different results.
Biofilm communities control the diffusion of nutrition components and shield bacteria from various environmental stress, which may cause the bacteria to colonize inside their hosts and induce infections (Donlan and J William 2002; López, Vlamakis et al. 2010). One of the main habitats of K. pneumoniae is the mucosal surfaces of mammals, which includes intestinal, respiratory, and urogenital mucosa. This organism encounters a low‐fluid shear environment in these habitats, which is achieved by the brush border microvilli of epithelial cells (Nickerson, Ott et al. 2004). Epidemiological studies have shown that the usual precondition for K. pneumoniae to induce nosocomial infections is its colonization of a patient's gastrointestinal (GI) tract (Mircˇeva, Žigon et al. 1980; De Champs, Sauvant et al. 1989). Thus, the microenvironment in the GI tract may provide a proper niche for K. pneumoniae colonization and survival. We believe that to a certain extent, several similarities exist between SMG environments and the mucosal surfaces of mammals. Therefore, the enhanced biofilm formation in the SMG condition might not only show the altered adaptation of K. pneumoniae to this special environment, but also provide insights into the mechanisms of K. pneumoniae colonization of the human mucosa.
In this study, HARVs are completely filled with LB medium, so low‐fluid shear condition was achieved in both the SMG and NG groups. Although low‐fluid shear condition may be important for K. pneumoniae biofilm formation, it is not the main cause of K. pneumoniae biofilm formation differences between the SMG and NG groups. Unlike bacterial cell sedimentation in the NG group, cells in the SMG group are constantly suspended by hydrodynamic forces during cultivation, which partially mimics the true microgravity environment and may benefit biofilm formation in K. pneumoniae.
HARVs are designed to have a large gas‐permeable membrane and a small thickness value, which guarantee oxygen permeability into the medium. Previous studies on E. coli have shown that sufficient oxygen is present for bacterial growth throughout HARVs (Lynch et al. 2006). However, in our study, K. pneumoniae biofilm formation on the membrane reduced the oxygen permeability and an oxygen concentration gradient might form in HARVs during the later stage of 24‐h cultivation, which might also be a factor that influenced the growth of K. pneumoniae.
Long‐term spaceflight missions of astronauts on space stations require urgent monitoring of the potential changes in bacterial physiology and virulence caused by exposure to prolonged microgravity. Given the safety and constraints of equipment and spaceflight time, the development of earth‐based microgravity analogs to conduct experiments on microbes is necessary. A detailed investigation of the SMG‐regulated genes in this bacterium is currently being undertaken in our laboratory. This investigation would deepen our understanding of the SMG effects on Klebsiella physiology and pathogenicity.
Conflict of Interest
None declared.
Supporting information
Data S1. The dysregulated genes of K. pneumoniae cultured in SMG environment.
Acknowledgments
This study was funded by the National Basic Research Program of China (2014CB744405), the National Key Program for Infectious Diseases of China (2016ZX10004215‐001‐002), and the National Natural Science Foundation of China (31430006).
MicrobiologyOpen 2016; 5(5): 793–801
Contributor Information
Ruifu Yang, Email: ruifuyang@gmail.com.
Yanping Han, Email: yanpinghan@gmail.com.
References
- Castro, V. A. , Thrasher A. N., Healy M., Ott C. M., and Pierson D. L.. 2004. Microbial characterization during the early habitation of the International Space Station. Microb. Ecol. 47:119–126. [DOI] [PubMed] [Google Scholar]
- Castro, S. L. , Nelman‐Gonzalez M., Nickerson C. A., and Ott C. M.. 2011. Induction of attachment‐independent biofilm formation and repression of Hfq expression by low‐fluid‐shear culture of Staphylococcus aureus . Appl. Environ. Microbiol. 77:6368–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe, A. , De Boever P., Van Houdt R., Moors H., M. Mergeay , and Cornelis P.. 2008. Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PA01. Environ. Microbiol. 10:2098–2110. [DOI] [PubMed] [Google Scholar]
- Crabbe, A. , Pycke B., Van Houdt R., Monsieurs P., Nickerson C., Leys N., et al. 2010. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol. 12:1545–1564. [DOI] [PubMed] [Google Scholar]
- Crabbe, A. , Schurr M. J., Monsieurs P., Morici L., Schurr J., Wilson J. W., et al. 2011. Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl. Environ. Microbiol. 77:1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Champs, C. , Sauvant M. P., Chanal C., Sirot D., Gazuy N., Malhuret R., et al. 1989. Prospective survey of colonization and infection caused by expanded‐spectrum‐beta‐lactamase‐producing members of the family Enterobacteriaceae in an intensive care unit. J. Clin. Microbiol. 27:2887–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan, R. M. , and William C. J.. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming, H. C. , and Wingender J.. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633. [DOI] [PubMed] [Google Scholar]
- Gueguinou, N. , Huin‐Schohn C., Bascove M., Bueb J.‐L., Tschirhart E., Legrand‐Frossi C., et al. 2009. Could spaceflight‐associated immune system weakening preclude the expansion of human presence beyond Earth's orbit? J. Leukoc. Biol. 86:1027–1038. [DOI] [PubMed] [Google Scholar]
- Hammond, T. G. , and Hammond J. M.. 2001. Optimized suspension culture: the rotating‐wall vessel. Am. J. Physiol. Renal. Physiol. 281:F12–F25. [DOI] [PubMed] [Google Scholar]
- Huertas, M. G. , Zarate L., Acosta I. C., Posada L., Cruz D. P., Lozano M., et al. 2014. Klebsiella pneumoniae yfiRNB operon affects biofilm formation, polysaccharide production and drug susceptibility. Microbiology 160:2595–2606. [DOI] [PubMed] [Google Scholar]
- Jagnow, J. , and Clegg S.. 2003. Klebsiella pneumoniae MrkD‐mediated biofilm formation on extracellular matrix‐ and collagen‐coated surfaces. Microbiology 149:2397–2405. [DOI] [PubMed] [Google Scholar]
- Jefferson, K. K. , Goldmann D. A., and Pier G. B.. 2005. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 49:2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Li, F. L. , Wang Qi, Ge Pupu, Woo Patrick C. Y., Yan Jinghua, Zhao Yanlin, et al. 2014. Genomic and transcriptomic analysis of NDM‐1 Klebsiella pneumoniae in spaceflight reveal mechanisms underlying environmental adaptability. Sci. Rep. 4:6216–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal, A. , Jejelowo O. A., and Rosenzweig J. A.. 2010. The effects of low‐shear mechanical stress on Yersinia pestis virulence. Astrobiology 10:881–888. [DOI] [PubMed] [Google Scholar]
- López, D. , Vlamakis H., and Kolter R.. 2010. Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, S. , Brodie E., and Matin A.. 2004. Role and regulation of sigma S in general resistance conferred by low‐shear simulated microgravity in Escherichia coli.” J. Bacteriol. 186: 8207–8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, S. V. , Mukundakrishnan K., Benoit M. R., Ayyaswamy P. S., and Matin A.. 2006. Escherichia coli biofilms formed under low‐shear modeled microgravity in a ground‐based system. Appl. Environ. Microbiol. 72:7701–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, R. J. , Cassanto J. M., Barnes M. B., and Koo J. H.. 2001. Bacterial biofilm formation under microgravity conditions. FEMS Microbiol. Lett. 195:115–119. [DOI] [PubMed] [Google Scholar]
- Mirčeva, A. , Žigon M., and Malavašič T.. 1980. Sequential outbreaks of infection due to Klebsiella pneumoniae in a neonatal intensive care unit: implication of a conjugative R plasmid. J. Infect. Dis. 142:106–112. [DOI] [PubMed] [Google Scholar]
- Morgan, J. L. , Strumillo J., and Zimmer J.. 2013. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C. N. , and Clegg S.. 2012. Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol. 7:991–1002. [DOI] [PubMed] [Google Scholar]
- Nickerson, C. A. , Ott C. M., Mister S. J., Morrow B. J., Burns‐Keliher L., and Pierson D. L.. 2000. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 68:3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, C. A. , Ott C. M., Wilson J. W., Ramamurthy R., and Pierson D. L.. 2004. Microbial Responses to Microgravity and Other Low‐Shear Environments. Microbiol. Mol. Biol. Rev. 68:345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova, N. D. 2004. Review of the knowledge of microbial contamination of the Russian manned spacecraft. Microb. Ecol. 47:127–132. [DOI] [PubMed] [Google Scholar]
- Podschun, R. , and Ullmann U.. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, L. A. , and Kolter R.. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285–293. [DOI] [PubMed] [Google Scholar]
- Romling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205–212. [DOI] [PubMed] [Google Scholar]
- Schroll, C. , Barken K. B., Krogfelt K. A., and Struve C.. 2010. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, R. P. , Goodwin T. J., and Wolf D. A.. 1992. Cell culture for three‐dimensional modeling in rotating‐wall vessels: an application of simulated microgravity. J. Tissue Cult. Methods 14:51–57. [DOI] [PubMed] [Google Scholar]
- Spiers, A. B. J. , Sm G., and Rainey P. B.. 2003. Biofilm formation at the air‐liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50:15–27. [DOI] [PubMed] [Google Scholar]
- Stahlhut, S. G. , Struve C., and Krogfelt K. A.. 2012. Klebsiella pneumoniae type 3 fimbriae agglutinate yeast in a mannose‐resistant manner. J. Med. Microbiol. 61:317–322. [DOI] [PubMed] [Google Scholar]
- Taylor, G. R . 1974. Recovery of medically important microorganisms from Apollo astronauts. Aerosp. Med. 45: 824–828. [PubMed] [Google Scholar]
- Taylor, G. R. , Konstantinova I., Sonnenfeld G., and Jennings R.. 1996. Changes in the immune system during and after spaceflight. Adv. Space Biol. Med. 6: 1–32. [DOI] [PubMed] [Google Scholar]
- Walters, M. C. 3rd , Roe F., Bugnicourt A., Franklin M. J., and Stewart P. S.. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilksch, J. J. , Yang J., Clements A., Gabbe J. L., Short K. R., Cao H., et al. 2011. MrkH, a novel c‐di‐GMP‐dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7:e1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J. W. , Ott C. M., Ramamurthy R., Porwollik S., McClelland M., Pierson D. L., et al. 2002a. Low‐Shear modeled microgravity alters the Salmonella enterica serovar Typhimurium stress response in an RpoS‐independent manner. Appl. Environ. Microbiol. 68:5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J. W. , Ramamurthy R., Porwollik S., McClelland M., Hammond T., Allen P., et al. 2002b. Microarray analysis identifies Salmonella genes belonging to the low‐shear modeled microgravity regulon. Proc. Natl. Acad. Sci. U. S. A. 99:13807–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J. W. , Ott C. M., Honer zu Bentrup K., Ramamurthy R., Quick L., Porwollik S., et al. 2007. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl. Acad. Sci. U. S. A. 104:16299–16304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj, X. , Nimtz M., Rohde M., Bokranz W., and Mling U. R.. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463. [DOI] [PubMed] [Google Scholar]
- Zogaj, X. , Bokranz W., Nimtz M., and Romling U.. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. The dysregulated genes of K. pneumoniae cultured in SMG environment.


