Abstract
Phylogenetic networks are rooted, labelled directed acyclic graphswhich are commonly used to represent reticulate evolution. There is a close relationship between phylogenetic networks and multi-labelled trees (MUL-trees). Indeed, any phylogenetic network N can be “unfolded” to obtain a MUL-tree U(N) and, conversely, a MUL-tree T can in certain circumstances be “folded” to obtain aphylogenetic network F(T) that exhibits T. In this paper, we study properties of the operations U and F in more detail. In particular, we introduce the class of stable networks, phylogenetic networks N for which F(U(N)) is isomorphic to N, characterise such networks, and show that they are related to the well-known class of tree-sibling networks. We also explore how the concept of displaying a tree in a network N can be related to displaying the tree in the MUL-tree U(N). To do this, we develop aphylogenetic analogue of graph fibrations. This allows us to view U(N) as the analogue of the universal cover of a digraph, and to establish a close connection between displaying trees in U(N) and reconciling phylogenetic trees with networks.
Keywords: Phylogenetic networks, Multi-labelled trees, Graph fibrations, Tree and network reconciliation, Universal cover of a digraph
Introduction
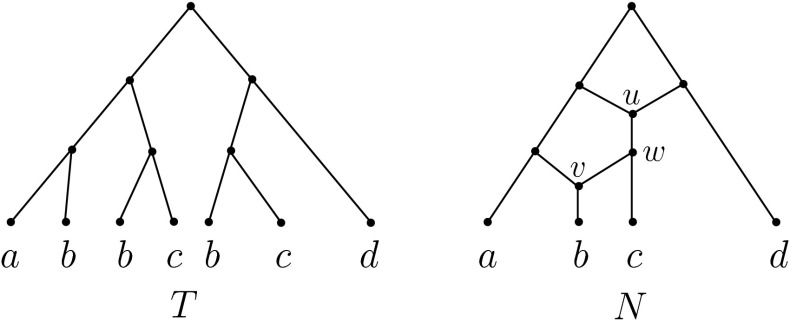
Phylogenetic networks are rooted, directed acyclic graphs whose leaves are labelled by some set of species (see Sect. 2 for precise definitions of the concepts that we introduce in this section). Such networks are used by biologists to represent the evolution of species that have undergone reticulate events such as hybridization and there is much recent work on these structures (cf.e.g. Gusfield 2014; Huson et al. 2010). In Huber and Moulton (2006) a close relationship is described between phylogenetic networks and multi-labelled trees (MUL-trees), leaf-labelled trees where more than one leaf may have the same label. Essentially, it is shown that it is always possible to “unfold” a phylogenetic network N to obtain a MUL-tree U(N) and that, conversely, a MUL-tree T can under certain conditions be “folded” to obtain a phylogenetic network F(T) that exhibits T. We illustrate these operations in Fig. 1 (see Sect. 3 for more details). The tree T is a MUL-tree, and the network N is obtained by first inserting vertices into T and then folding up the resulting tree by identifying vertices in T to obtain F(T); the unfolding U(N) of N (which is essentially obtained by reversing this process) is precisely T.
Fig. 1.
A MUL-tree T and its folding
Applications of the operations F and U include the construction of evolutionary histories of polyploids in terms of phylogenetic networks (Lott et al. 2009; Marcussen et al. 2015). In particular, polyploid organisms contain several copies of a genome, and if a tree is constructed from these genomes (or specific genes in these genomes) a MUL-tree can be obtained by labelling each leaf by the species that has the corresponding genome. By folding this MUL-tree a representation of the evolution of the species can then be obtained (in terms of a phylogenetic network), from the evolutionary history of the genomes. In this representation, vertices in the network with indegree two represent hybridisation events, where two parent species have produced a child which has the combined set of genomes of both of its parents.
In this paper, we study properties of the F and U operations in some detail and, in the process, show that they have some interesting connections with other areas such as gene tree/species network reconciliation (Wu and Zhang 2011; Zhang et al. 2011) and the theory of graph fibrations (Boldi and Vigna 2002). To do this, we begin by reviewing the concepts of MUL-trees and phylogenetic networks in the next section, and present some general properties of the folding and unfolding operations in Sect. 3. We then consider the interrelationship between the folding and unfolding operations.
More specifically, although it is always the case that U(F(T)) is isomorphic to T for any MUL-tree T (Huber and Moulton 2006), the same situation does not apply if the U and F operations are applied in the opposite order to some network as there are networks N for which F(U(N)) is not isomorphic to N (we give an example shortly in Fig. 3). Therefore, it is of interest to understand the networks N for which F(U(N)) is isomorphic to N. We call these stable networks. In Sect. 4, we present a characterization for stable phylogenetic networks (see Theorem 1). Using this result we are then able to show that the well-known class of binary, tree-sibling networks as defined in Cardona et al. (2009) are stable (see Corollary 1). We expect that stable networks could be of interest as they can provide a canonical representative for the set of all networks that display a particular MUL-tree (cf. Pardi and Scornavacca 2015 for choosing canonical representatives of networks that display a set of trees).
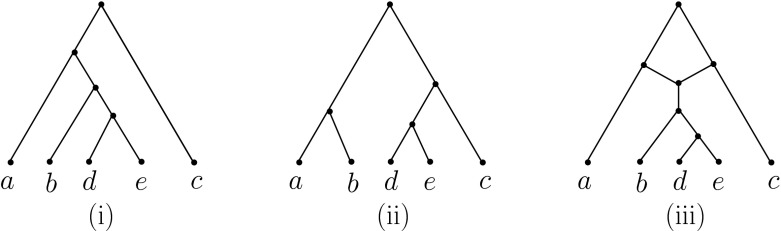
Fig. 3.
(i) A phylogenetic network N, (ii) U(N), and (iii) the X-network F(U(N)). Clearly, N and F(U(N)) are not isomorphic
In Sect. 5, we show that the unfolding and folding operations are closely related to concepts that arise in the theory of graph fibrations (cf. Boldi and Vigna 2002 for a review of this area). In particular, we define the concept of a folding map between a MUL-tree and a phylogenetic network. As one consequence, we show that the unfolding of a network can be considered as a phylogenetic analogue of the universal cover of a digraph. This allows us to provide an alternative characterisation for stable networks (Corollary 3). It is worth noting that an alternative framework for considering maps between phylogenetic networks is developed in Willson (2012).
We then focus on the problem of displaying trees in networks. In Sect. 6, we demonstrate that it is NP-complete to decide whether or not a phylogenetic tree is displayed by a stable network (Theorem 5). This is of interest since in Kanj et al. (2008) it is shown that it is NP-complete to decide if a tree is displayed by a network, but in Iersel et al. (2010) it is shown that this problem is polynomial for certain special classes of networks (such as normal and tree-child networks).
Finally, in Sect. 7, we define and study a new way in which a tree may be displayed in a network: We say that a phylogenetic tree is weakly displayed by a phylogenetic network N if it is displayed by the MUL-tree U(N). Using the concepts developed in Sect. 5, we provide a characterization for when a tree is weakly displayed by a network in terms of a special type of tree reconciliation (Theorem 6). This characterisation allows us to show that, in contrast to displaying a tree, it is possible to decide in polynomial time whether or not a phylogenetic tree is weakly displayed by a phylogenetic network having the same leaf-set (Corollary 4).
Definitions
Throughout this paper, we let X denote a finite set of size at least two. In addition, all graphs that we consider are connected.
Rooted DAGs
Suppose G is a directed acyclic graph with vertex set V(G), arc set A(G) and a single root . For an arc in G, we denote the head v of a by , and its tail u by . We say that two arcs a and of G are parallel if and hold. If v is a vertex of G, then the in-degree of v, denoted by indeg(v), is the number of incoming arcs of v, and the out-degree of v, denoted by outdeg(v), is the number of outgoing arcs of v. We say that a vertex is below a vertex if there exists a directed path from w to v in G. We call a vertex v of G a reticulation vertex of G, if and holds. We call v a tree vertex of G if and either holds, in which case we call v a leaf of G, or . We denote the set of leaves of G by L(G) and the set of interior vertices v of G, that is, v is neither the root nor a leaf of G, by .
MUL-trees
We say that a multi-set M is a multi-set on X if the set resulting from M by ignoring the multiplicities of the elements in M is X. Following Huber and Moulton (2006), we define a pseudo multi-labelled tree on X, or a pseudo MUL-tree on X for short, to be a pair consisting of a rooted directed tree T together with a labelling map from X into the set of non-empty subsets of the leaf set of T such that
-
(i)
for all distinct , and
-
(ii)
for every leaf there exists some with .
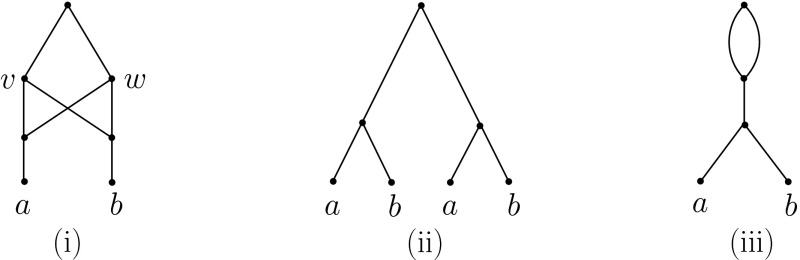
In addition, we call a MUL-tree on X if the tree T does not have any vertices with in-degree one and out-degree one. If the map is clear from the context then we will write T rather than and if the set X is of no relevance to the discussion then we will call T a (pseudo) MUL-tree rather than a (pseudo) MUL-tree on X. For example, the tree T in Fig. 1 is a MUL-tree on and in Fig. 2 is a pseudo MUL-tree on X that is not a MUL-tree on X. We say that two (pseudo) MUL-trees and on X are isomorphic if there is a digraph isomorphism such that, for all and , we have if and only if .
Fig. 2.
A sequence of MUL-trees and the intermediate pseudo MUL-tree used by the folding operation F for the MUL-tree T in Fig. 1. Tree is obtained from by considering the maximal inextendible subMUL-tree with root . That is, we have and is constructed from by first deleting the subMUL-tree , then removing the arc , and finally suppressing vertex . Similarly, is obtained from by considering the maximal inextendible subMUL-tree on and incident with vertex
Suppose T is a pseudo MUL-tree. For v a non-root vertex of T, we denote by T(v) the connected subgraph of T that contains v obtained by deleting the incoming arc of v. Clearly T(v) is a pseudo MUL-tree. We call a pseudo MUL-tree a pseudo subMUL-tree of T if there exists a non-root vertex v of T such that T(v) and are isomorphic. For T a MUL-tree we say that a subMUL-tree of T is inextendible if there exist distinct vertices v and of T such that and T(v) and are isomorphic. Loosely speaking, a subMUL-tree of T is inextendible if T contains more than one copy of that subMUL-tree. We say that a subMUL-tree of T is maximal inextendible if is inextendible and any other inextendible subMUL-tree of T that contains as a subMUL-tree is isomorphic with . Note that although the definition of inextendible used in this paper is slightly different from the one in (Huber and Moulton 2006), the maximal inextendible subMUL-trees coincide under both definitions.
To illustrate these definitions consider for example the MUL-tree T and its folding depicted in Fig. 1. The three leaves labelled b are all inextendible subtrees of T and so are the two leaves labelled c. Each one of two subtrees of T of length two (ignoring the directions of the arcs of T) that have leaf label set is maximal inextendible.
Phylogenetic networks
An X -network N is a rooted directed acyclic graph, in which parallel arcs are allowed,
such that
-
(i)
there exists a unique root of N that has in-degree zero and out-degree at least two,
-
(ii)
every vertex of N except the root is either a reticulation vertex or a tree vertex,
-
(iii)
there exists no vertex of in-degree one and out-degree one, and
-
(iv)
the set L(N) of leaves of N is X.
A phylogenetic network ( on X) is an X-network that does not have parallel arcs. A phylogenetic tree on X is a phylogenetic network on X that has no reticulation vertices. We say that a phylogenetic network N is binary if the degree of every reticulation vertex and every non-leaf tree-vertex is three and . Finally, we say that two X-networks N and are isomorphic if there exists a bijection such that for all vertices the number of arcs in N with head u and tail v equals the number of arcs in with head and tail , and is the identity on X.
Folding and unfolding
In this section, we recall the unfolding and folding operations mentioned in the introduction that were first proposed in Huber and Moulton (2006) (see also Huber et al. 2012 for the binary case).
We first describe the unfolding operation U which constructs a pseudo MUL-tree from an X-network N as follows:
the vertices of are the directed paths in N that start at ,
there is an arc from vertex in to vertex in if and only if holds for some arc a in N (i.e. is the path in N that extends the path in N by the arc a), and
the vertices in that start at and end at some x in X are labelled by x.
The MUL-tree obtained by suppressing all in-degree one and out-degree one vertices in , if there are any, is denoted by U(N). In Huber and Moulton (2006) it is shown that U(N) is indeed a MUL-tree.
We denote for all vertices v of an directed graph G as in Sect. 2.1 the set of children of v by ch(v) and say that an X-network N exhibits a MUL-tree T if the MUL-trees U(N) and T are isomorphic. In particular, any X-network N exhibits the MUL-tree U(N). Note that there exist MUL-trees T for which there is no phylogenetic network that exhibits T (for example, the binary MUL-tree with two leaves both labelled by the same element).
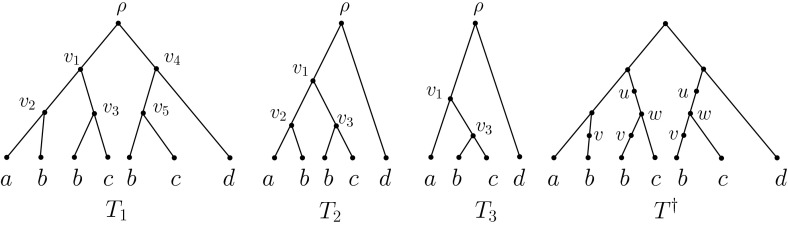
We now describe the folding operation F for constructing an X-network F(T) from a MUL-tree T introduced in (Huber and Moulton 2006, p. 628). This operation can be thought of intuitively as the reverse of the unfolding operation U, and it works by repeatedly finding a maximal inextendible subMULtree, subdividing the incoming arcs of the roots of the subMUL-trees that are isomorphic with it, and finally identifying the subdivision vertices and removing all but one copy of that subMUL-tree. This continues until an X-network is obtained or, equivalently, no further maximal inextendible subMUL-tree can be found. Formally, a pseudo MUL-tree is constructed from T to guide this process. To do this we need to define a sequence of MUL-trees. Suppose is such that we have already constructed tree . Then we obtain as follows: If there is no inextendible subMUL-tree of , we declare to be the last tree in . Otherwise, we take a maximal inextendible subMUL-tree of . Let v be the root of this tree and let be the subset of vertices w of with isomorphic to . Then, to obtain , for each we remove the subtree and the arc with head w from . If this has rendered the root of a vertex with out-degree one then we collapse the remaining arc with tail . Otherwise, we suppress the resulting vertex with in-degree and out-degree one. For the MUL-tree T in Fig. 1, an example of such a MUL-tree sequence is depicted in Fig. 2.
Now, to obtain , we consider each tree in other than T in turn. Let and assume that is such that is constructed from . Then we subdivide all of those arcs a in T for which and T(v) are isomorphic (as pseudo MUL-trees). The pseudo MUL-tree obtained once the last element in has been processed is . Note that although the pseudo MUL-tree sequences are not necessary unique, they all result in the same intermediate tree .
Finally, to obtain F(T), we define an equivalence relation on that identifies all pairs of vertices v, w in with isomorphic with (as pseudo-MUL-trees), and let F(T) be the X-network obtained by taking the quotient of by . More precisely, let G(T) denote the DAG with vertex set and (multi)-set of arcs obtained by joining any two vertices for which holds by arcs if and only if for one (and hence for all) the size of is m. The X-network obtained from G(T) by suppressing all vertices of indegree one and outdegree one and defining the leaf labels in the natural way is F(T).
For example, consider the MUL-tree T in Fig. 1. Then the pseudo MUL-tree is depicted in Fig. 2. The two vertices labelled u in make up and the vertex in F(T), representing is labelled u. Similarly, the two vertices labelled w in make up which we again represent in F(T) in terms of w. Clearly and holds for all . Hence, there is precisely one arc in F(T) from u to w.
Note that any MUL-tree T is isomorphic with U(F(T)) (as MUL-trees) (Huber and Moulton 2006). Thus, if there is no risk of confusion we will sometimes identify T and U(F(T)). Also, note that if T is binary, then F(T) is semi-resolved, that is, every tree vertex in F(T) has out-degree 2. Moreover, in Huber and Moulton (2006), Proposition 3 it is shown that if F(T) is semi-resolved, then F(T) has the minimum number of reticulation vertices amongst all phylogenetic networks that exhibit T.
In general, the folding of an arbitrary MUL-tree on X need not be a phylogenetic network. For example, consider the MUL-tree U(N) in Fig. 3(ii); then its fold up F(U(N)) is not a phylogenetic network as it contains parallel arcs. We now characterize those MUL-trees T for which F(T) is a phylogenetic network.
Proposition 1
Suppose T is a binary MUL-tree on X. Then F(T) is a phylogenetic network if and only if there is no pair of distinct vertices v, w in T which share a parent in T and are such that T(v) and T(w) are isomorphic.
Proof
We prove the claim that if F(T) is a phylogenetic network then there is no pair of distinct vertices in T with the stated property by establishing the contrapositive. Suppose T is a binary MUL-tree on X that contains two distinct vertices v and w which share a parent in T and are such that T(v) and T(w) are isomorphic. Without loss of generality, we may assume that v and w are such that there exist no two vertices and of T on the directed paths from the root of T to v and w, respectively, such that and are isomorphic and the parent of is also the parent of . Thus, T(v) is inextendible. If T(v) is maximal inextendible then F(T) contains a parallel arc and so F(T) is not a phylogenetic network, as required. So, assume that T(v) is not maximal inextendible. Then there must exist some vertex in T distinct from v and w such that is maximal inextendible and T(v) is a subMUL-tree of .
Let , , denote the vertices on the directed path from to v such that is inextendible and is rendered maximal inextendible during the folding of T. Then must hold as every MUL-tree , , must contain both T(v) and T(w) as a subMUL-tree. Thus, T(v) is rendered maximal inextendible at some stage in the construction of F(T). Applying the operation F to T(v) introduces a parallel arc into F(T) and thus F(T) is not a phylogenetic network, as required.
Conversely, suppose that T is a binary MUL-tree on X such that there exist no two vertices v, w in T which share a parent in T such that T(v) and T(w) are inextendible. Assume for contradiction that F(T) is not a phylogenetic network. Then F(T) must contain parallel arcs a and . Put and . Then v is a tree vertex and w is a reticulation vertex of F(T). Let z denote the unique child of w in F(T). Note that since the folding operation implies that F(T) cannot contain an arc both of whose end vertices are reticulation vertices, z must in fact be a tree vertex in F(T).
Now, let denote two directed paths from the root of F(T) to z which contain arcs a and , respectively, and which differ only on those arcs. Denoting the MUL-tree U(F(T)) on X by , the subMUL-trees and of are isomorphic. But this is impossible, since there is a directed path from to v such that is the parent of both and in which is isomorphic to T. Thus, F(T) must be a phylogenetic network.
As mentioned above, the folding operation F can be considered as the reverse of the operation U. However, there exist phylogenetic networks N such that F(U(N)) is not isomorphic to N (see e.g. Fig. 3). Therefore, it is of interest to understand those networks N for which F(U(N)) and N are isomorphic.
Stable networks
In this section, we shall give a characterization of phylogenetic networks N for which F(U(N)) is isomorphic to N. We call such networks stable.
We start by recalling the definition of an irreducible network (Huber and Moulton 2006). Suppose that N is a phylogenetic network on X. We call two distinct tree vertices v and w in N identifiable if there exist directed paths from the root of N to v and from to w such that the subMUL-trees and of U(N) are isomorphic. In addition, we say that N is irreducible if it does not contain an identifiable pair of tree vertices. To illustrate, the network N depicted in Fig. 3(i) is not irreducible, since the two vertices v and w are identifiable.
If N is a phylogenetic network then let Ret(N) denote the set of reticulation vertices of N. We call N compressed if the child of each vertex in Ret(N) is a tree vertex. Note that in Cardona et al. (2009), this property is taken as part of the definition of a phylogenetic network, the rationale being that we cannot expect to reconstruct the order in which hybridization events occur.
Theorem 1
Suppose that N is a semi-resolved phylogenetic network. Then the following statements are equivalent.
-
(i)
N is stable.
-
(ii)
N is compressed and irreducible.
-
(iii)
N is compressed and there does not exist a pair of distinct tree vertices v, w in N such that .
Proof
(ii) (iii): Suppose that (ii) holds and assume for contradiction that there exists a pair of distinct tree vertices v, w in N such that . Then . Since N is semi-resolved we have . Let . Since N is compressed the children and of a and b, respectively, are tree-vertices of N. Let and denote the directed paths from the root of N to that cross v and w, respectively. Similarly, let and denote the directed paths in N from to that cross v and w, respectively. Then the subMUL-trees and of U(N) are isomorphic and so are the subMUL-trees and . Let denote the subpath obtained from by terminating at v. Similarly, let denote the subpath obtained from by terminating at w. Then the MUL-tree obtained from and by adding the vertex labelled and the arcs and is . Similarly, the MUL-tree obtained from and by adding the vertex labelled and the arcs and is . Since and are clearly isomorphic it follows that v, w is an identifiable pair in N. Hence N is not irreducible which provides the required contradiction.
(iii) (ii): Suppose that (iii) holds and assume for contradiction that N is not irreducible. Then N contains an identifiable pair of vertices v, w. Without loss of generality, we may assume that v and w are such that there are no vertices and below v and w, respectively, that also form an identifiable pair.
To obtain the required contradiction, we first claim that ch(v) and ch(w) are contained in Ret(N). Suppose that . For all non-root vertices u of N let denote a directed path from the root of N to u. If s is a leaf of N then, since v and w are an identifiable pair, the MUL-trees and are isomorphic and the underlying bijection is the identity on X. Hence, holds too and, so, which is impossible as s is a leaf of N.
If s is a non-leaf tree-vertex of N then, since and are isomorphic and every tree vertex z of N gives rise to a subset of vertices in the MUL-tree U(N), it follows that there exists a non-leaf tree vertex below w such that and are isomorphic. By the choice of v and w, we cannot have that s and form an identifiable pair and so must hold. Hence, , which is impossible as s is assumed to be a tree vertex of N. Since every non-root vertex of N is either a tree-vertex or a reticulation vertex of N, it follows that . Similar arguments imply that also holds which completes the proof of the claim.
To complete the proof, assume for contradiction that there exists some . Then, , by the previous claim. Since N is compressed, the child of s must be a tree-vertex of N. Since and are isomorphic it follows that there exists a tree vertex r in N below w such that and are isomorphic. Note that as otherwise must be a reticulation vertex of N which is impossible. Hence, and r form an identifiable pair in N with below v and r below w which is impossible in view of the choice of v and w. Thus, . Similar arguments imply that and so must hold, as required. But this is impossible in view of (iii).
(i) (ii): This follows by Huber and Moulton (2006), Theorem 3.
(ii) (i): Suppose that N is compressed and irreducible. Let and denote some binary resolution of N and F(U(N)), respectively. Since N is irreducible so is , and since N exhibits U(N) so does . Hence, by applying Huber and Moulton (2006), Corollary 2 to and and using the assumption that N is compressed, it follows that N is stable.
As an immediate corollary (Corollary 1) of this last theorem, we see that the collection of binary, stable phylogenetic networks contains a well-known class of phylogenetic networks. More specifically, suppose that N is a phylogenetic network. A vertex w of N distinct from some vertex v of N is a sibling of v if v and w share the same parent, and a sibling that is a tree vertex is called a tree-sibling vertex. In addition, N is called a tree-child network if every non-leaf vertex of N has a child that is a tree vertex of N (Cardona et al. 2008), and N is called a tree-sibling network if every reticulation vertex of N has a tree-sibling (Cardona et al. 2009). Note that a tree-child network is a tree-sibling network.
Corollary 1
Suppose N is a binary, compressed, tree-sibling network. Then N is stable.
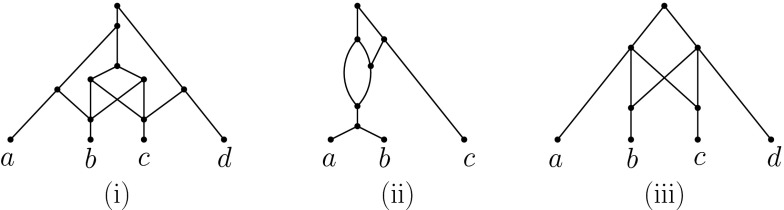
By Corollary 1, it follows that the fold up of an unfolded binary, compressed, tree-sibling network is the network itself and that it cannot contain a pair of distinct tree vertices that have the same set of children. Note that there exist semi-resolved, compressed, tree-sibling networks that are not stable (Fig. 4(i)), binary, stable phylogenetic networks that are not tree-sibling (Fig. 4(ii)), and non-binary, tree-child networks that are not stable (Fig. 4(iii)).
Fig. 4.
(i) The network on is semi-resolved, compressed tree-sibling but not stable. (ii) The network on is binary, stable but not tree-sibling. (iii) The network on is non-binary, tree-child but not stable
Folding maps
In this section, we explore a relationship between the folding/unfolding operations and graph fibrations. For simplicity, we shall follow the presentation of the latter topic in Boldi and Vigna (2002). Results from this section will be used to establish a main result in Sect. 7.
Recall that the head of an arc a in an X-network N is denoted by and its tail by . Now, suppose that is a pseudo MUL-tree on X and that N is a phylogenetic network on X. An X -morphism is a pair of functions , such that (i) for all , we have and , and (ii) if with , , then . An X-morphism f is called a rooted X -morphism if also holds. In case the context is clear, we denote both and by f. We call an X-morphism a folding map 1 if both maps are surjective, and for each arc and such that there is a unique arc (the lifting of the arc a at v) such that and . Note that a folding map is necessarily a rooted X-morphism. We call the inverse image , , the fibre over v. Informally, the fibre over v is the subset of V(T) that is mapped to v under f. For example, for the tree and the phylogenetic network N depicted in Fig. 2 and Fig. 1, respectively, the fibers of the vertices u, v and w in N are given by the vertices of labelled with the same letters.
We begin by stating a result which illustrates how folding maps naturally arise from the unfolding U(N) of a network N. This result is an analogue of Boldi and Vigna (2002), Theorem 15; the proof is quite similar and straight-forward and so we omit it.
Theorem 2
Let N be a phylogenetic network on X. Then the map that takes each vertex in to its last vertex, and each arc of to the corresponding arc in N is a folding map.
As we shall now show, the folding F(T) of a MUL-tree T can also give rise to a folding map. In analogy with Boldi and Vigna (2002), p. 25, we say that an equivalence relation on the vertex set V(T) of a pseudo MUL-tree T satisfies the local out-isomorphism property (LOIP) if the following holds for all .
LOIP: If then there is a bijection from the set of arcs in T with tail v to the set of arcs in T with tail w such that , for all arcs a in T with tail v.
We now use the LOIP-property to characterize when equivalence relations on MUL-trees give rise to folding maps (cf. Boldi and Vigna 2002, Theorem 2). To aid clarity of presentation, we denote the parent of a non-root vertex v in a rooted directed tree by pa(v).
Theorem 3
Suppose that is a pseudo MUL-tree on X, and is an equivalence relation on V(T). Then the equivalence classes are the fibres of a folding map (for some phylogenetic network N on X) if and only if satisfies the following five properties:
-
(i)
LOIP,
-
(ii)
for all with in-degree and out-degree 1, , and for some ,
-
(iii)
for all with in-degree 1 and out-degree not equal to 1, for all ,
-
(iv)
for all and , , and
-
(v)
for all , for all distinct from v.
Proof
Assume first that N is a phylogenetic network on X and is a folding map such that the equivalence classes of are the fibres of f. For each with , define a map from the set of arcs a in T with tail v to the set of arcs in T with tail w by putting equal to . Then . Hence , and so satisfies (i). Moreover, as N is a phylogenetic network, it is straight-forward to check that (ii) must hold as no vertex in N can have in-degree and out-degree 1, (iii) must hold as every vertex of N that is not the root of N is either a reticulation vertex or a tree vertex (but not both), and that (iv) must hold as all elements in must be mapped by f to a vertex labeled by x which has in-degree 1. Finally, (v) follows from the fact that N does not contain parallel arcs.
Conversely, assume that is an equivalence relations on V(T) that satisfies properties (i)–(v). To simplify notation, put for all vertices u in V(T). Let be the network obtained by taking the quotient of T by (as described in Sect. 3). In particular, is a rooted DAG with vertex set , and ([u], [v]) an arc in for if and only if for some and (note that this definition is independent of the choice of and ). In addition, we identify each leaf [u] in with the necessarily unique element x in X with whose existence follows from property (iv). It is straight-forward to check that properties (i)–(v) ensure that is a phylogenetic network on X.
Now, define to be the X-morphism that maps each vertex u in V(T) to its equivalence class [u], and each arc (u, v) in A(T) to the arc ([u], [v]). It is straight-forward to check that f is a folding map as properties (i) and (iv) imply that f yields a well-defined surjective X-morphism from T to that satisfies the aforementioned arc lifting property.
Given a MUL-tree T, consider the equivalence relation on the vertex set of the pseudo MUL-tree defined in Sect. 3. Since satisfies properties (i)–(v) of the last theorem it follows that, in case F(T) is a phylogenetic network, we obtain a folding map whose fibres are the equivalence classes of .
We now state a result that provides additional insight into unfoldings of networks, and that will also be useful in the last section. It can be regarded as a phylogenetic analogue of path lifting in topology (cf. also Boldi and Vigna 2002, Theorem 13 and Corollary 14).
Theorem 4
Suppose that T and are pseudo MUL-trees on X, that N is a phylogenetic network on X and that is an X-morphism. If is a folding map, then there exists an X-morphism such that . Moreover, if g is a rooted X-morphism, then so is , and is necessarily unique.
Proof
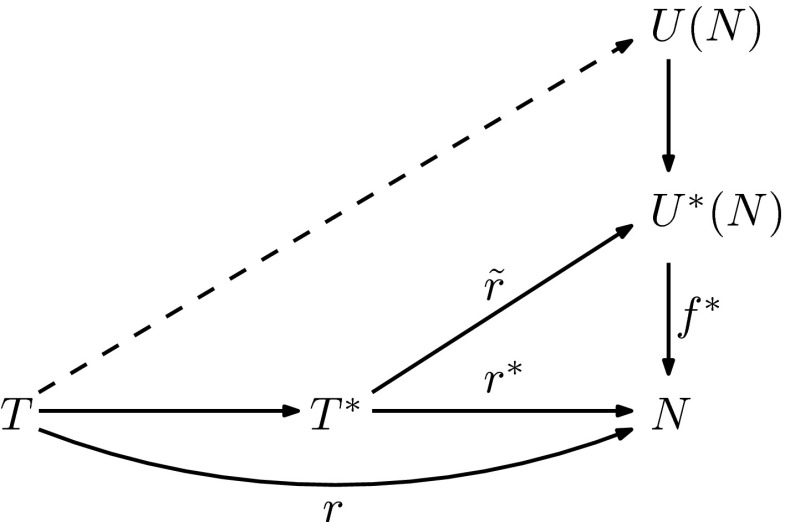
Using a top-down approach, we define recursively as follows. Since f is a folding map, there exists a vertex u in . We set . Now, if the map has been defined on the parent of some as well as the arcs and vertices on the directed path from to , and , then we define , and to be the head of this arc in T. It is straight-forward to check that the mapping that we obtain in this way yields an X-morphism with the desired property. Moreover, if g is a rooted X-morphism, then and hence . This implies that is a rooted X-morphism, and that is the only such map.
As a corollary of this result, we now see that the pseudo MUL-tree can be regarded as a phylogenetic analogue of the universal total graph of N (at ), a graph theoretical variant of the universal cover of a topological space (cf. Boldi and Vigna 2002, Section 3.1).
Corollary 2
Suppose that is a pseudo MUL-tree and N is a phylogenetic network, both on X, and that is a folding map. Then is isomorphic to .
Proof
Applying Theorem 4 with and , it follows that there exists a unique rooted X-morphism with . Since g is a folding map, it follows that is also a folding map, and hence an isomorphism, as required.
Using again the notation for a guidetree for the operation F, we now use this last result to provide an alternative characterisation for stable networks.
Corollary 3
Suppose that N is a phylogenetic network. Then N is stable if and only if is isomorphic to .
Proof
Suppose N is stable, that is, N is isomorphic to F(U(N)). By the comment following Theorem 3, there exists a folding map from the pseudo MUL-tree to F(U(N)). As N is isomorphic to F(U(N)), there also exists a folding map from to N. By Corollary 2, it follows that is isomorphic to .
Conversely, suppose is isomorphic to and write rather than . By Theorem 2 we have a folding map . Hence, by Theorem 3, there exists an equivalence relation on such that N is isomorphic to . Moreover, in if and only if the pseudo MUL-trees and are isomorphic.
Now, F(U(N)) is isomorphic to , where in if and only if is isomorphic to . Therefore, the two equivalence relations and are equal (up to the isomorphism between and ), and hence N is isomorphic to F(U(N)), as required.
Note that our definition for folding maps can be extended to obtain folding maps between X-networks in general. We will not pursue this possibility further here, but it could be of interest to understand categorical properties of such maps (cf. Boldi and Vigna 2002, Section 6).
Displaying trees in stable networks
Following Iersel et al. (2010), we say that a phylogenetic network N on X displays a phylogenetic tree T on X if there is a subgraph of N that is a subdivision of T (i. e. can be obtained from T by replacing arcs (u, v), by directed paths from u to v). We illustrate this concept in Fig. 5.
Fig. 5.
The phylogenetic tree in (i) is displayed by the network in (iii), but the tree in (ii) is not
In Kanj et al. (2008) it is shown that it is NP-complete to decide whether or not a given phylogenetic tree is displayed by a given phylogenetic network. On the other hand, in Iersel et al. (2010) it is shown that there are polynomial algorithms for this problem for certain classes of networks e. g. binary tree-child networks. Thus it is of interest to know the complexity of this question for stable phylogenetic networks. We show that the following decision problem is NP-complete.
TreeDisplaying
Instance: A binary stable phylogenetic network on X and a binary phylogenetic tree on X.
Question: Is T displayed by N?
To establish this fact, we show that this problem is NP-complete when restricted to compressed, binary tree-sibling networks and apply Corollary 1. In the proof, we shall use the following operation, which is a modification of an operation with the same name defined in Iersel et al. (2010). Suppose that N is a binary phylogenetic network on X and that R is a binary phylogenetic tree on X. Let denote the root of N, let denote the root of R, and let . Assume that are pairwise distinct vertices not already contained in N and that , , and are also not contained in R. Then the operation HangLeaves( v ) adds the vertices to N as well as the arcs , , , , and . In addition, it adds the vertices to R as well as the arcs , , , and .
Theorem 5
TreeDisplaying is NP-complete, even when restricted to the class of binary, compressed tree-sibling networks.
Proof
By Corollary 1, it suffices to restrict attention to the class of binary, compressed tree-sibling networks. Let T be a binary phylogenetic tree on X and let N be a binary phylogenetic network on X. We will (in polynomial time) modify N to a binary, compressed tree-sibling network on some leaf set that contains X and, simultaneously, modify T to a binary phylogenetic tree on . For and we then show that T is displayed by N if and only if is displayed by . The result then follows as it has been shown in Kanj et al. (2008) that it is NP-complete to decide whether or not a binary phylogenetic tree is displayed by a binary phylogenetic network.
The construction of is in two steps. In the first step, we repeatedly apply the operation HangLeaves to transform N into a compressed tree-sibling network on some yet to be specified leaf set and T to a binary phylogenetic tree on . To do this we associate to N a phylogenetic network in which every reticulation vertex has a unique child and that child is a tree-vertex. This is achieved by carrying out the following operation. For each arc e in N whose head is a reticulation vertex of N we subdivide e by a new vertex and then apply HangLeaves to . We denote the resulting rooted DAG by . Note that is clearly a binary phylogenetic network on X, every reticulation vertex of has a unique child, and that child is a tree-vertex. Furthermore, every reticulation vertex of that is also a reticulation vertex of N has two siblings in both of which are reticulation vertices.
Next, we follow the proof of Iersel et al. (2010), Theorem 3 and choose for every reticulation vertex v of that is also a reticulation vertex in N one of its two siblings. Let s denote that sibling. Let denote the joint parent of s and v in . Then we subdivide the arc of by a new vertex and apply HangLeaves to . We denote the resulting DAG by . Note that is a tree-sibling of v in , and that is a tree-sibling of in . Let denote the union of X and all of the leaves added to N this way. Then it is easy to check that the resulting DAG is a binary, compressed tree-sibling network on . Moreover, the phylogenetic tree constructed in concert with is clearly binary and has leaf set .
We now establish our claim that T is displayed by N if and only if is displayed by . To do so, we first show that T is displayed by N if and only if is displayed by where and are a phylogenetic network and a phylogenetic tree on , respectively, that are the result of a single application of operation HangLeaves, to a vertex v of N.
Assume first that T is displayed by N. To see that is displayed by note first that there exists a subgraph of N that is a subdivision of T. Combined with the fact that the subgraph of with vertex set and arc set , , , and is a subdivision of the subtree of of whose vertex set is and whose arc set is , , , and , it is easy to see that gives rise to a subgraph of that is a subdivision of . Thus, is displayed by .
Conversely, assume that is displayed by , that is, there exists a subgraph of that is a subdivision of . Clearly, the restriction of to is a subgraph of N that is a subdivision of restricted to , that is T. Thus, T is displayed by N which completes the proof of the claim. A repeated application of the last claim implies that T is displayed by N if and only if is displayed by .
Weakly displaying trees
Given a phylogenetic tree T and a network N on X, we say that T is weakly displayed by N if it is displayed by U(N) (that is, there exists a subgraph of U(N) that is a subdivision of T). For example, both of the trees in Fig. 5 are weakly displayed by the phylogenetic network N, but the tree in (ii) is not displayed by N. As we shall see, this concept is closely related to the problem of reconciling gene trees with species networks. In Sect. 6, we studied the problem of displaying trees in networks, in particular showing that it is NP-complete to decide whether or not a binary phylogenetic tree T is displayed by a phylogenetic network N even if it is stable. In this section, we show that, in contrast, one can decide in polynomial time whether or not a tree is weakly displayed by any given phylogenetic network.
Before presenting our algorithm, we first derive a characterization for when a tree is weakly displayed by a phylogenetic network in terms of so-called tree reconciliations. Given a phylogenetic network N on X, let be the set consisting of the tree vertices in V(N) together with the root on N. Following Zhang et al. (2011), a reconciliation map between a phylogenetic tree T on X and a phylogenetic network N is a map such that for all , holds for all , and every arc (u, v) in A(T) is associated with a directed path in N with initial vertex r(u) and terminal vertex r(v).
We now give the aforementioned characterization for when a tree is weakly displayed by a phylogenetic network. We call a reconciliation r between T and N locally separated if for each pair of vertices and in T that have the same parent v, both and contain at least one arc, and the initial arc in is distinct from the initial arc in .
Theorem 6
Suppose that N is a phylogenetic network on X. Then a phylogenetic tree T on X is weakly displayed by N if and only if there exists a locally separated reconciliation between T and N.
Proof
We first prove that if there is a locally separated reconciliation r between T and N, then T is weakly displayed by N. We illustrate the main idea of the proof in Fig. 6—essentially, the map r induces an X-morphism from a subdivision of T into N, and so, using Theorem 4, we obtain an X-morphism from to , from which we can then deduce that T is displayed by U(N).
Fig. 6.
The relationship between the maps described in the proof of Theorem 6
More specifically, suppose that r is a locally separated reconciliation between T and N. Since each arc in T is associated with a directed path in N which contains at least one arc, it follows that r induces an X-morphism from a subdivision of T to N. By Theorem 4, let be an X-morphism from to such that . Since r is locally separated, it follows that the map is injective, and hence is isomorphic to a subgraph of .
Now, consider the set that is the pre-image of the in-degree one and out-degree one vertices in under . Then, since is an X-morphism, each vertex in has in-degree one and out-degree one. Let be the tree obtained from by suppressing all vertices in . Then is a subdivision of T. Since U(N) does not contain any in-degree one and out-degree one vertices and is a subdivision of U(N) it follows that is isomorphic to a subdivision of . Thus, T is displayed by U(N), and so T is weakly displayed by N, as required.
Conversely, suppose that T is weakly displayed by N. Then there exists a subdivision of T such that is isomorphic to a subgraph of U(N). Since is a subdivision of U(N), there exists a subdivision of (and hence also a subdivision of T) such that is isomorphic to a subgraph of . Denote the X-morphism from to induced by this isomorphism by and let be the folding map from to N given by Theorem 2. Then the X-morphism from to N induces a map r from V(T) to V(N) defined by putting , for all . Clearly, r(v) exists because is contained in and thus in V(U(N)) as is an X-morphism. Moreover, since and are X-morphisms it follows that r(v) is a tree vertex of N.
Now, for every arc (u, v) in T, denote the subdivision of (u, v) in by (that is, is the necessarily unique path from u to v in ) and let be the image of under , a directed path from r(u) to r(v) in N which contains at least one arc. Then it follows that r is a reconciliation between T and N. Moreover, to see that r is locally separated, consider an arbitrary pair of distinct vertices and in V(T) that have the same parent v. Denote the initial arcs of the two (necessarily distinct) directed paths and in by and , respectively. Furthermore, for any arc a of U(N) put and . Since is induced by an isomorphism between and a certain subgraph of , we obtain . Combined with and Property (v) in Theorem 3 it follows that . Therefore and contain distinct initial arcs, from which it follows that r is locally separated, as required.
In light of the last result, deciding whether or not a phylogenetic tree is weakly displayed by a phylogenetic network is equivalent to the following decision problem:
Locally separated reconciliation
Instance: A phylogenetic network N on X and a binary phylogenetic tree T on X.
Question: Does there exist a locally separated reconciliation between T and N?
We now present a dynamic programming algorithm to solve this problem. Let N be a phylogenetic network on X and let T be a binary phylogenetic tree on X. Then for every tree vertex v in N we denote by N(v) the phylogenetic network obtained from by N by first restricting N to v and all the vertices of N below v and then suppressing any resulting vertices with in-degree one and out-degree one. In addition, we define a function as follows. If v is not a leaf in V(T), then we set if and only if there exists some such that (i) or is below u in N, and (ii) there exists a locally separated reconciliation between T(v) and . If v is a leaf with label x, then we set if and only if u is a leaf in N labeled with x or x is a leaf in N below u. We remark that implies that holds for all such that u is below in N.
By definition, there exists a locally separated reconciliation between T and N if and only if . In order to compute the value of , we will use the following result concerning the function .
Proposition 2
Let T be a binary phylogenetic tree on X, and N a phylogenetic network on X. Suppose that v is an interior vertex in T with two children and and . Then if and only if u is an interior vertex in N with for a child of u, or there exist two distinct children of u in N such that and .
Proof
We begin by establishing the ‘if’ direction. Note that if holds for a child of u then, by the previous remark, follows. Therefore we may assume that u is an interior vertex in N with two children in N such that and . This implies that there exist two (not necessarily distinct) vertices and in N such that for , there exists a locally separated reconciliation between and . Fix a directed path in N obtained by combining the arc and an arbitrary path from to . Since the paths and both contain at least one arc and their respective first arcs are distinct.
Now consider the map defined, for all , by if , if is contained in , and otherwise. Since and are the two children of v and holds for and holds for each arc in it follows that f is a reconciliation between T(v) and N(u). Combined with the fact that and are locally separated, it follows that f is also locally separated. Hence, , as required.
Conversely, suppose that for v an interior vertex in T and . We may further assume that for each child of u as otherwise the proposition clearly follows. Under this assumption, it follows that there exists a locally separated reconciliation f between T(v) and N(u) with .
Now, let for (where is not necessarily distinct from ). Since is a child of v and f is a locally separated reconciliation, it follows that is below u and that . Considering the two directed paths which have the same starting vertex v but distinct initial arcs, it follows that there exist two distinct children and of u such that is contained in for . Together with , this implies , as required.
Proposition 2 forms the basis of a dynamic programming algorithm for computing in polynomial time, which we now briefly describe.
Let , , and let k be the maximum number of children that any vertex in N may have. Note first that a topological ordering of V(T) (that is, a linear ordering of V(T) such that is below in T implies ), can be computed in O(m) time. Similarly, we can compute a topological ordering of V(N) in O(kn) time. Now, noting that and , consider the matrix whose (i, j)th entry is . Then by Proposition 2, it takes O(mnk) time to fill this matrix and, therefore, to compute . Since a binary phylogenetic tree T on X has vertices (Semple and Steel 2003, Proposition 1.2.3), we summarize this last discussion in the following corollary.
Corollary 4
Suppose that T is a binary phylogenetic tree on X, and that N is a phylogenetic network on X. Then, using a dynamic programming algorithm, it can be decided in
time whether or not T is weakly displayed by N.
Acknowledgments
VM and MS thank O. Gascuel for organizing the “Mathematical and Computational Evolutionary Biology” meeting, June 2012, Hameau de l’Etoile, France, where first ideas for the paper were conceived. All authors would also like to thank the two anonymous referees for their helpful and constructive comments on a previous version of this paper.
Footnotes
A folding map is analogous to an “opfibration” for digraphs (cf. Boldi and Vigna 2002, Definition 4).
Contributor Information
Katharina T. Huber, Email: katharina.huber@cmp.uea.ac.uk
Vincent Moulton, Email: vincent.moulton@cmp.uea.ac.uk.
Mike Steel, Email: mike.steel@math.canterbury.ac.nz.
Taoyang Wu, Email: taoyang.wu@gmail.com, Email: taoyang.wu@uea.ac.uk.
References
- Boldi P, Vigna S. Fibrations of graphs. Discrete Math. 2002;243(1):21–66. [Google Scholar]
- Cardona G, Llabrés M, Rosselló F, Valiente G. A distance metric for a class of tree-sibling phylogenetic networks. Bioinformatics. 2008;24(13):1481–1488. doi: 10.1093/bioinformatics/btn231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona G, Rossello F, Valiente G. Comparison of tree-child phylogenetic networks. IEEE/ACM Trans Comput Biol Bioinf (TCBB) 2009;6(4):552–569. doi: 10.1109/TCBB.2007.70270. [DOI] [PubMed] [Google Scholar]
- Gusfield D. ReCombinatorics: the algorithmics of ancestral recombination graphs and explicit phylogenetic networks. Cambridge: MIT Press; 2014. [Google Scholar]
- Huber K, Moulton V. Phylogenetic networks from multi-labelled trees. J Math Biol. 2006;52(5):613–632. doi: 10.1007/s00285-005-0365-z. [DOI] [PubMed] [Google Scholar]
- Huber KT, Moulton V, Spillner A, Storandt S, Suchecki R (2012) Computing a consensus of multilabeled trees. In: Proceedings of the meeting on algorithm engineering & experiments (ALENEX 12), SIAM, pp 84–92
- Huson DH, Rupp R, Scornavacca C. Phylogenetic networks: concepts, algorithms and applications. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- Kanj I, Nakhleh L, Than C, Xia G. Seeing the trees and their branches in the network is hard. Theor Comput Sci. 2008;401(1):153–164. [Google Scholar]
- Lott M, Spillner A, Huber KT, Petri A, Oxelman B, Moulton V. Inferring polyploid phylogenies from multiply-labeled gene trees. BMC Evol Biol. 2009;9(1):216. doi: 10.1186/1471-2148-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T, Heier L, Brysting AK, Oxelman B, Jakobsen KS. From gene trees to a dated allopolyploid network: insights from the angiosperm genus viola (violaceae) Syst Biol. 2015;64(1):84–101. doi: 10.1093/sysbio/syu071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi F, Scornavacca C. Reconstructible phylogenetic networks: do not distinguish the indistinguishable. PLoS Comput Biol. 2015;11(4):e1004,135. doi: 10.1371/journal.pcbi.1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple C, Steel M. Phylogenetics. Oxford: Oxford University Press; 2003. [Google Scholar]
- van Iersel L, Semple C, Steel M. Locating a tree in a phylogenetic network. Inf Process Lett. 2010;110:1037–1043. [Google Scholar]
- Willson S. CSD homomorphisms between phylogenetic networks. IEEE/ACM Trans Comput Biol Bioinf (TCBB) 2012;9(4):1128–1138. doi: 10.1109/TCBB.2012.52. [DOI] [PubMed] [Google Scholar]
- Wu T, Zhang L. Structural properties of the reconciliation space and their applications in enumerating nearly-optimal reconciliations between a gene tree and a species tree. BMC Bioinf. 2011;12(Suppl 9):S7. doi: 10.1186/1471-2105-12-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ng YK, Wu T, Zheng Y (2011) Network model and efficient method for detecting relative duplications or horizontal gene transfers. In: 2011 IEEE 1st international conference on computational advances in bio and medical sciences (ICCABS). IEEE, pp 214–219