Abstract
Background:
Pancreatic ductal adenocarcinoma (PDAC) remains a devastating disease due to the lack of specific early diagnostic markers. To improve the outcomes, proteomic approaches are being developed for the discovery of novel biomarkers of PDAC.
Methods:
Using tandem mass tag labelling and LC-MS/MS, we performed comparative analyses of pre- and postoperative sera from PDAC patients to identify specific serum biomarkers for PDAC. In validation studies, we evaluated the discriminatory power of candidate proteins.
Results:
Among the 302 proteins analysed, 20 were identified as potential biomarkers, with C4b-binding protein α-chain (C4BPA) and polymeric immunoglobulin receptor (PIGR) being selected for further analysis. The sera levels of C4BPA and PIGR were significantly higher in the preoperative PDAC patients than in the postoperative ones (P<0.008, P<0.036, respectively). In addition, serum C4BPA levels, but not PIGR, in patients with PDAC were significantly higher than those in healthy controls as well as in patients with pancreatitis and other malignancies including biliary tract cancers (BTC) (P<0.001). The respective area under the receiver operator characteristics (ROC) curve (AUC) was 0.860 for C4BPA, 0.846 for CA19-9 and 0.930 for the combination of C4BPA and CA19-9 in PDAC vs non-cancer individuals. The respective AUC was 0.912 for C4BPA, 0.737 for CA19-9 in Stages I and II of PDAC, 0.854 for C4BPA and 0.264 for CA19-9 in PDAC vs BTC.
Conclusions:
We have demonstrated that C4BPA is a novel serum biomarker for detecting early stage PDAC, as well as for distinguishing PDAC from other gastroenterological cancers. Further analysis in large cohort studies will warrant C4BPA as a promising biomarker of PDAC in clinical use.
Keywords: pancreatic cancer, biomarkers, serum, TMT, early detection, proteomics, C4BPA, PIGR
Pancreatic ductal adenocarcinoma (PDAC) accounts for 85–90% of all pancreatic tumours. The median survival of all PDAC patients is less than 6 months, and the 5-year-survival rate is 3–5% (Jemal et al, 2010). One of the most crucial reasons for the poor prognosis is the lack of early diagnostic markers for PDAC. To overcome this and improve the outcomes of patients with PDAC, there is an urgent need to identify highly sensitive and specific markers for early detection. The widely used serum-circulating markers for PDAC, carbohydrate tumour–associated antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA), are not sufficiently accurate to be used as early diagnostic markers. Specifically, CA19-9 is elevated in only ∼65% of individuals with resectable PDAC; in addition, it is frequently elevated in patients with various benign pancreaticobiliary disorders, notably, cholestasis and chronic pancreatitis (PT) (Goggins, 2005). Moreover, it cannot discriminate the PDAC from other diseases such as pancreatitis and major gastroenterological cancers including biliary tract cancer (BTC). Therefore, CA19-9 is not recommended for diagnostic use (Locker et al, 2006).
Glycosylation is one of the most common post-translational modifications of a secreted protein (Rudd and Dwek, 1997). Glycoproteins are generally modified by the attachment and processing of various glycans at each glycosylation site. Thus, each glycoprotein is a collection of glycoforms. The glycoform population of a glycoprotein is cell- and protein-specific and highly reproducible in a given physiological state; however, the glycoform signature can be significantly altered in disease. Alterations to cellular pathways in cancer affect glycan processing, and these changes can be monitored by analysing the oligosaccharides attached to individual glycoproteins (Arnold et al, 2008). Thus, analysis of the serum glycome and specific serum glycoproteins could provide useful tumour biomarkers (Arnold et al, 2008).
Tandem mass tags (TMTs) are chemical labels used for MS-based identification and quantification of biological molecules. TMTs has an important role in comparative proteomics in various samples such as cells, tissues, and body fluids using gel-free proteomic approaches (Baeumlisberger et al, 2010; Arntzen et al, 2011; Dayon et al, 2011; Giron et al, 2011). In terms of biomarker discovery for PDAC, Britton et al. and O'Brien et al. have recently showed that TMT-labelling of proteins in tissues and sera is a useful technique (Britton et al, 2014; O'Brien and Timms, 2014).
In the present study, we employed the TMT method to generate comparative protein profiles of sera samples obtained from pre- and postoperative PDAC patients. Furthermore, we compared the serum levels of candidate proteins to evaluate the ability to discriminate between PDAC and healthy controls. Serum CA19-9 cannot discriminate the PDAC from other diseases such as pancreatitis and major gastroenterological cancers including biliary tract cancer (BTC). Furthermore, we compared the serum levels of candidate proteins to evaluate the ability to discriminate between PDAC and healthy controls as well as pancreatitis and other malignancies including BTC. Here, we identify novel serum biomarkers that are potentially superior to classical markers for early and specific detection of patients with PDAC.
Materials and methods
Patient serum samples
The test-set consisted of serum samples collected pre- and postoperative (3–4 weeks after surgery, when the serum levels of C-reactive protein returned to the normal range) from three PDAC patients who underwent curative surgery. To confirm results, we measured the levels of candidate proteins in the sera of 14 pre- and postoperative PDAC patients, including three pairs of samples from the test-set in validation set 1. For the second validation study, blood samples were obtained from 52 patients with PDAC, 20 patients with PT and age/gender-matched 40 healthy volunteers (HVs) as controls. Blood samples were obtained from all patients in the Department of General Surgery, Chiba University Hospital, who were histologically diagnosed with PDAC and PT from December 2006 to September 2012. Samples were also obtained from HVs in the Kashiwado Hospital (Table 1).
Table 1. Clinical characteristics of participants.
|
Sex |
Tumour stage (UICC stagea) |
||||||
|---|---|---|---|---|---|---|---|
| Age (Mean±s.d.) | M | F | I | II | III | IV | |
|
Test set: TMT reagents experiments | |||||||
| Pre- and postoperative sera of pancreatic cancer (n=3) | 66±5 | 1 | 2 | 0 | 1 | 1 | 1 |
|
Validation set 1 | |||||||
| Pre- and postoperative sera of pancreatic cancer (n=14) | 68±7 | 7 | 7 | 0 | 8 | 1 | 5 |
|
Validation set 2 | |||||||
| Healthy volunteers (HVs) (n=40) | 67±8 | 20 | 20 | — | — | — | — |
| Pancreatitis (PT) (n=20) | 56±12 | 10 | 10 | — | — | — | — |
| Pancreatic cancer (PDAC) (n=52) | 67±9 | 26 | 26 | 5 | 5 | 15 | 27 |
|
Validation set 3 | |||||||
| Cancers | |||||||
| Pancreas (n=66: set 1+set 2) | 67±10 | 33 | 33 | 5 | 13 | 16 | 32 |
| Stomach (n=20) | 70±8 | 10 | 10 | 8 | 7 | 3 | 2 |
| Colorectal (n=20) | 68±6 | 10 | 10 | 7 | 6 | 5 | 2 |
| Liver (n=20) | 70±9 | 10 | 10 | 5 | 5 | 5 | 5 |
| Biliary tract (n=20) | 69±8 | 10 | 10 | 5 | 5 | 5 | 5 |
Abbreviations: HVs=healthy volunteers; PDAC=pancreatic ductal adenocarcinoma; PT=pancreatitis; TMT=tandem mass tag.
Union for International Cancer Control; TNM Classification of malignant tumours.
In another validation study (validation set 3), 80 samples from patients with other major gastroenterological cancers were analysed, including 20 gastric, 20 colorectal, 20 liver and 20 biliary tract cancers (Table 1). Blood samples were obtained from all patients in the Department of Medicine and Clinical Oncology, Chiba University Hospital, who were diagnosed with the respective malignancies from January 2008 to December 2010.
All blood samples were processed according to a standardised protocol, and serum sample aliquots were frozen until subsequent analysis. None of the patients underwent any therapeutic measures, such as radiation, chemotherapy or surgery until serum samples were collected. The ethics committee for each institute approved the protocol. Written informed consent was obtained from all patients and HVs.
Magnetic bead sample preparation using the Glycoprotein Isolation Kit
Paramagnetic nonporous particles coupled with a concanavalin A lectin ligand (Con A; Bruker Daltonics, Germany) were used to process the serum samples. Binding, washing and elution solutions were prepared according to the manufacturer's instructions. MB–LCA Con A binding solution (10 μl) and serum (5 μl) were transferred to a 0.2 ml thin-walled PCR-tube (ABgene, UK). A homogenous magnetic-particle solution (10 μl) was added, mixed, and incubated for 5 min. The tubes were placed in an 8 × 12-well magnetic-bead separator (MBS) (Bruker Daltonics, Germany) for 30 s for magnetic fixation of the MB-LCA Con A particles. The supernatant was aspirated, and the tubes were removed from the MBS device. Washing solution (100 μl) was added and carefully mixed with the magnetic beads. The tube was replaced into the MBS device and moved back and forth sequentially between adjacent wells on each side of the device's magnetic bar. After fixation of the magnetic beads for 30 s, the supernatant was aspirated. This washing procedure was repeated three times. After the final washing step, the bound molecules were eluted by incubation with 10 μl MB-LCA Con A elution solution for 1 min, before the eluate was collected using the MBS device.
TMT labelling
TMTsixplex Isobaric Label Reagent Set (Pierce, Idaho, ID, USA) was used according to the manufacturer's instructions. Tubes containing the different isobaric chemical tags (0.8 mg each) were added with 41 μl of anhydrous ACN, and dissolved for 5 min with occasional vortexing at room temperature. TMT solution (20 μl) was added to each tube and allowed to react at room temperature for 60 min. Each sample received 4 μl of 5% hydroxylamine, and the mixture was allowed to incubate for 15 min to quench the reaction (Tsuchida et al, 2013). Samples were finally pooled and lyophilised.
We analysed two different samples for each PDAC patient (n=3), pre- and postoperative; 20 μl of serum samples from three preoperative subjects were labelled with TMT6-126, 127 and 128, and 20 μl of serum samples from three postoperative subjects were labelled with TMT6-129, 130 and 131, respectively.
SDS-PAGE analysis
The lyophilised sample was dissolved in PAGE sample buffer (pH 6.8; 50 mM Tris–HCl, 50 mM dithiothreitol, 0.5% SDS, and 10% glycerol). The solution was then analysed using SDS–PAGE (Perfect NT Gel System W, 10– 20% acrylamide, 20 wells; DRC Co. Ltd., Tokyo, Japan) according to the manufacturer's protocol. The gel was stained with Coomassie brilliant blue (CBB) (PhastGel Blue R; GE Healthcare, Little Chalfont, UK) (Sogawa et al, 2011). The protein bands were excised from the gel. In-gel tryptic digestion was performed to identify the protein, as previously described (Sogawa et al, 2011).
Protein identification and quantification by LC-MS/MS analysis
Each fractionated peptide was injected into a trap column (C18, 0.3 × 5 mm; DIONEX, Sunnyvale, CA, USA) and an analytical column (C18, 0.075 × 120 mm; Nikkyo Technos, Tokyo, Japan), which was attached to the Ultimate 3000 (DIONEX). The flow rate of the mobile phase was 300 nl min−1. The solvent composition of the mobile phase was programmed to change in 120-min cycles, with varying mixing ratios of solvent A (2% v/v CH3CN and 0.1% v/v HCOOH) to solvent B (90% v/v CH3CN and 0.1% v/v HCOOH) as follows: 5–10% B, 5 min; 10–13.5% B, 35 min; 13.5–35% B, 65 min; 35–90% B, 4 min; 90% B, 0.5 min; 90–5% B, 0.5 min; and 5% B, 10 min. Purified peptides were introduced from HPLC to the LTQ-Orbitrap XL (Thermo Scientific, San Jose, CA, USA), a hybrid ion-trap Fourier transform mass spectrometer. One full scan cycle was applied (350–1200 m/z, resolution 60 000) followed by top three data-dependent collision induced dissociation (CID): higher-energy collision-activated dissociation (HCD) dual MS/MS scans acquired for TMT reporter ions. CID scans were acquired in LTQ, and full scans and HCD scans in Orbitrap. The normalised collision energy was 35% in CID and 45% in HCD. Activation time was 30 min for CID and 75 min for HCD. Dynamic exclusion was applied: 1 repeat count, 30 min repeat duration, 500 exclusion list sizes, and 180 s exclusion duration.
The database search engine (Proteome discoverer; Thermo Scientific) was used to identify and quantify proteins from the mass, tandem mass and reporter ion spectra of peptides. Peptide mass data were matched by searching the Human International Protein Index database (IPI, July 2008, 72 079 entries, European Bioinformatics Institute). The database search parameters employed were, peptide mass tolerance=2 p.p.m. and fragment tolerance=0.6 Da. Furthermore, the enzyme parameter was set to trypsin, allowing up to one missed cleavage variable modifications, methionine oxidation, and cysteine alkylation. The minimum criteria of protein identification were filtered with Xcorr vs charge state and set as false discovery rate (FDR)<1%. The FDR was estimated by searching against a randomised decoy database created by the Proteome Discoverer 1.2 program supplied by Thermo Scientific (Tsuchida et al, 2013).
Enzyme-linked immunosorbent assay
Serum levels of C4b-Binding Protein Alpha (C4BPA) and polymeric immunoglobulin receptor (PIGR) were determined by enzyme-linked immunosorbent assays according to the manufacturer's instructions (ELISA Kits for C4BPA and PIGR, Cloud-Clone Corp. Houston, TX, USA). Optical density was measured at 450 nm using a microplate reader (iMark Microplate Reader S/N 10288).
Statistical analyses
Numerical data are presented as the mean±s.d. Data were analysed using SPSS version 19.0 statistical package (SPSS, Chicago, IL, USA). Data were evaluated by linear regression analysis and correlations were assessed using Pearson correlation coefficients. For non-parametric data, the differences between two groups were analysed using the Mann–Whitney U-test. P-values<0.05 were considered statistically significant.
Results
Identification of candidate proteins by comparing the sera of pre- and postoperative PDAC patients using the TMT method
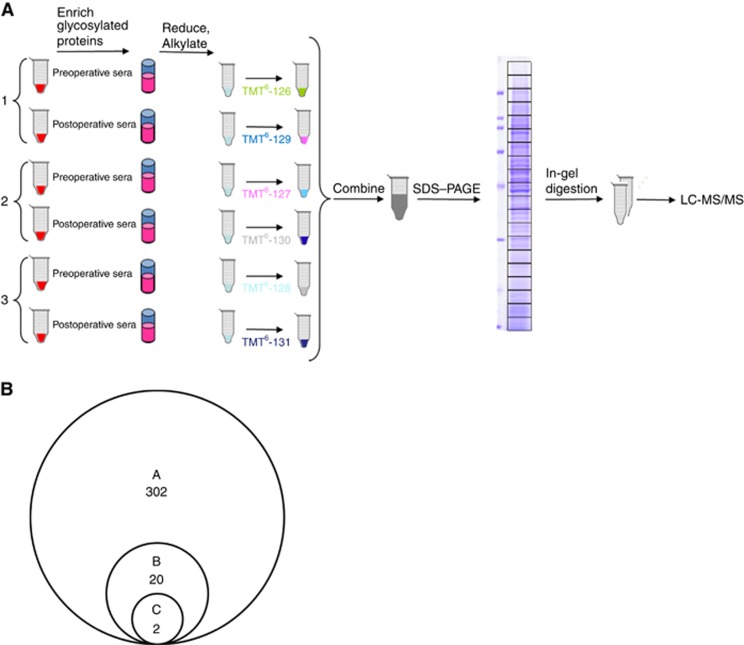
To identify new serum biomarkers for PDAC, six serum samples (three pairs of sera from pre- and postoperative PDAC patients) were alkylated with iodoacetamide, digested with trypsin, differentially labelled with TMT, pooled and subjected to analysis using LC-MS/MS (Figure 1A). Of these, three preoperative patient samples were labelled with TMT with reporter ions at m/z=126 127 128, and three postoperative patient samples were labelled with TMT with reporter-ions at m/z=129 130 131. After comparing the profiles of proteins by LC-MS/MS using an LTQ Orbitrap XL mass spectrometer, 302 proteins with unique peptide sequences were found (Figure 1B).
Figure 1.
Identification of candidate proteins using TMT labelling and LC-MS/MS analyses. (A) TMT six-plex approach. A total of six serum samples (three pairs of sera from pre- and postoperative PDAC patients) were differentially labelled with TMT, pooled, and subjected to analysis using LC-MS/MS. (B) A: Pre- and postoperative sera of three PDAC patients were analysed by LC-MS/MS using an LTQ Orbitrap XL mass spectrometer. Proteomic analysis revealed 302 proteins with unique peptide sequences. B: Twenty proteins had serum levels increased by twofold in at least one preoperative patient compared with the postoperative levels. C: The serum levels of two candidate proteins, C4BPA and PIGR were elevated in all three preoperative patients by greater than twofold compared with those in postoperative patients.
Among these, we identified 20 proteins whose serum levels were elevated more than twofold before and after the surgery, and they are listed in Supplementary Table S1. In the 20 identified proteins, C4b-binding protein alpha chain (C4BPA) and polymeric immunoglobulin receptor (PIGR) serum expression levels were elevated in all three preoperative patients by greater than twofold compared with those in postoperative patients (Figure 1B). Therefore, we selected these two proteins as candidates for further analysis.
C4BPA and PIGR levels are increased in the sera of patients with PDAC
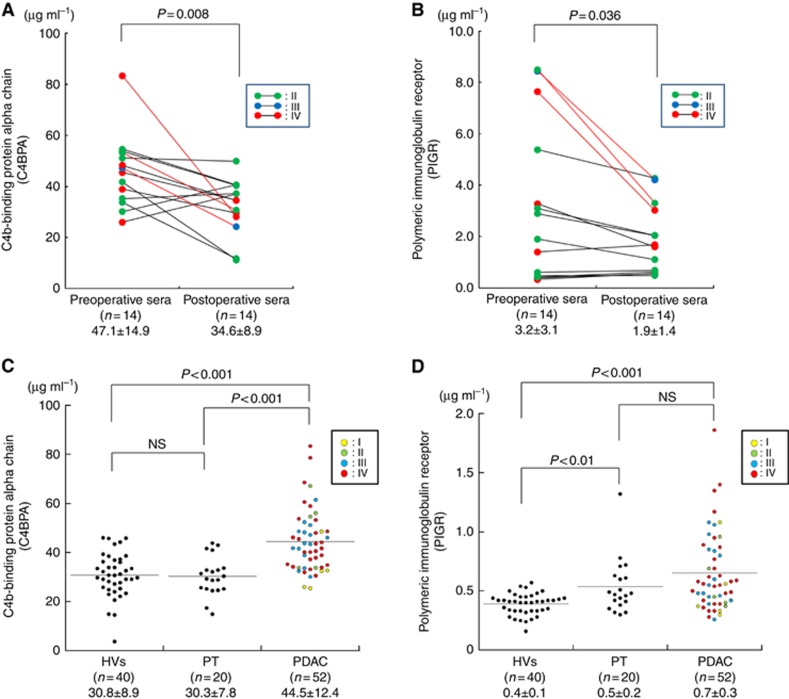
To validate these results, we added 11 pairs of pre- and postoperative sera to three former test-set samples to measure both C4BPA and PIGR levels by ELISA assay. The mean C4BPA levels in the preoperative sera (47.1±14.9 μg ml−1) were significantly higher than in the postoperative sera (34.6±8.9 μg ml−1) in all 14 PDAC patients (P<0.008; Student's paired t-test; Figure 2A). The mean PIGR levels in the preoperative sera (3.2±3.1) were also significantly higher than in the postoperative sera (1.9±1.4) in the same cohort (P<0.036; Student's paired t-test; Figure 2B). These results confirmed that C4BPA and PIGR, identified using TMT method, are significantly decreased in the sera of PDAC patients after surgery. Furthermore, we evaluated the correlations between the tumour resectability and the rate of change after surgery in both serum C4BP and PIGR levels. Comparing with non-R0 resection group (R1/R2/RX resection group), R0 resection group showed not significant but a decrease in the percentage of change of the postoperative levels to the preoperative levels in both C4BPA and PIGR (Supplementary Figures 1A and 1B).
Figure 2.
The serum C4BPA and PIGR levels are increased in PDAC patients. (A) Comparison of serum C4BPA levels in pre- and postoperative sera of PDAC patients (P=0.008; Mann–Whitney U-test). Samples of test set are indicated as red lines. (B) Comparison of serum PIGR levels in pre- and postoperative sera of PDAC patients (P=0.036; Mann–Whitney U-test). Samples of test set are indicated as red lines. (C) Comparison of serum C4BPA levels in healthy volunteers (HVs) and patients with pancreatitis (PT) and PDAC using ELISA. The serum C4BPA levels in PDAC patients were significantly greater than in the HVs and PT patients (PDAC vs HVs: P<0.001, PDAC vs PT: P<0.001; Mann–Whitney U-test). (D) Comparison of serum PIGR levels in HVs, PT and PDAC using ELISA. The serum PIGR levels in PDAC patients were not significantly higher than in PT patients but in HVs (P<0.001; Mann–Whitney U-test).
Next, to assess the discriminatory power of serum expression levels of these two candidate proteins, we compared serum C4BPA and PIGR levels among 52 PDAC patients and age-matched benign controls including 40 HVs and 20 PT patients in validation set 2 (Table 1). As shown in Figure 2C, the serum C4BPA levels in PDAC patients (44.5±12.4 μg ml−1) were significantly greater than in the HVs (30.8±8.9 μg ml) and PT patients (30.3±7.8 μg ml−1) (PDAC vs HVs: P<0.001, PDAC vs PT: P<0.001; Mann–Whitney U-test). Importantly, there was no significant difference in serum C4BPA levels between HVs and PT patients. The serum PIGR levels in PDAC patients (0.7±0.3 μg ml−1) were not significantly higher than in PT patients (0.5±0.2 μg ml−1) but in HVs (0.4±0.1 μg ml−1) (P<0.001; Mann–Whitney U-test; Figure 2D). These results imply that C4BPA is a potential sensitive and specific serum marker for PDAC.
C4BPA is superior to CA19-9 in the early detection of PDAC
The serum levels of C4BPA, CA19-9, and CEA in 66 PDAC patients (validation sets 1 and 2) are summarised according to UICC Stage (Stage I+II: n=18, Stage III: n=16, Stage IV: n=32) as well as HVs and PT patients in Table 2.
Table 2. Summary of the levels of C4BPA, CA19-9 and CEA in sera of participants.
|
Non-cancer |
Pancreatic cancer (validation set 1 and 2; n=66) |
||||
|---|---|---|---|---|---|
| HVs (n=40) | PT (n=20) | Stage I+II (n=18) | Stage III (n=16) | Stage IV (n=32) | |
| C4BPA (μg ml−1) | 30.8±8.9 | 30.3±7.8 | 42.5±12.4 | 44.3±8.0 | 46.4±14.8 |
| CA19-9 (U ml−1) | 12.1±8.7 | 18.4±16.5 | 117.1±268.0 | 232.1±383.8 | 25709.6±134054.2 |
| CEA (ng ml−1) | 2.5±1.3 | 2.7±1.7 | 6.8±10.9 | 7.0±10.1 | 8.8±14.4 |
Abbreviations: HVs=healthy volunteers; PT=pancreatitis.The levels are indicated as mean±s.d.
The cutoff values of the three markers were set at levels that gave 95.4% specificity compared with those for HVs and PT: 38.4 μg ml−1, 47.7 U ml−1 and 5.5 ng ml−1 for C4BPA, CA19-9, and CEA, respectively. Among the 66 PDAC patients, the sensitivities of C4BPA, CA19-9, and CEA at these cutoff levels were 67.3%, 71.2% and 34.6%, respectively. C4BPA levels were above the cutoff in 10 (52.6%) of 19 PDAC patients in whom the serum levels of CA19-9 were below their respective cutoff values. Together with this, the sensitivity and the specificity of a combination assay for C4BPA and CA19-9 were 86.4% and 80.0%, respectively.
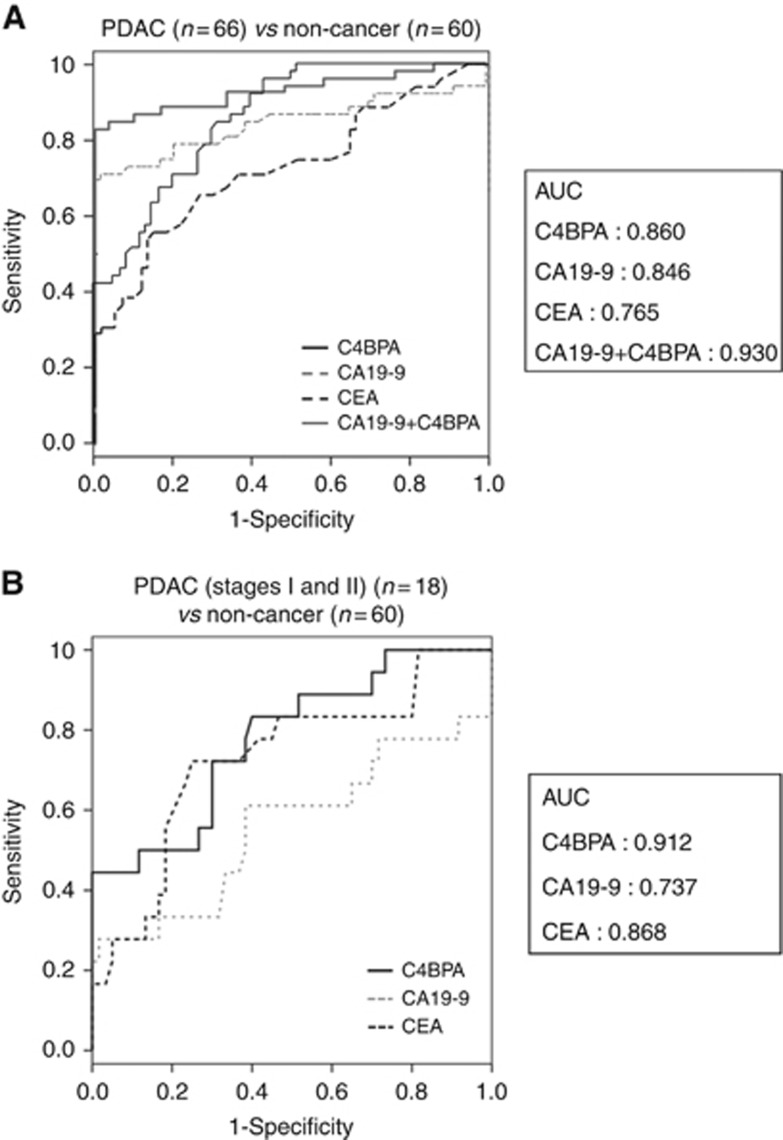
Receiver operator characteristic (ROC) curves were conducted to evaluate the value of C4BPA, CA19-9, CEA, and the combination of C4BPA and CA19-9 levels for distinguishing PDAC patients from HVs and PT patients. For distinguishing between cancer and non-cancer (HVs and PT) patients, the respective area under the ROC curve (AUC) was 0.860 for C4BPA, 0.846 for CA19-9, 0.765 for CEA, and 0.930 for the combination of C4BPA and CA19-9. These results suggest that serum C4BPA expression is potentially a better diagnostic PDAC biomarker than serum CA19-9 and CEA expressions (Figure 3A).
Figure 3.
C4BPA excels CA19-9 and CEA in the early detection of PDAC. (A) The ROC analyses were performed for the serum levels of C4BPA, CA19-9 and CEA between PDAC and non-cancer (HVs and PT) populations. The respective AUC was 0.860 for C4BPA, 0.846 for CA19-9, 0.765 for CEA, and 0.930 for the combination of C4BPA and CA19-9. (B) The ROC analyses were performed for the serum levels of C4BPA, CA19-9, and CEA between the early stage (stages I and II) of PDAC patients and non-cancer (HVs and PT) populations. The respective AUC was 0.912 for C4BPA, 0.737 for CA19-9, and 0.868 for CEA.
To evaluate whether C4BPA is useful for early detection of PDAC, serum C4BPA, CA19-9 and CEA levels in patients with early stage of PDAC (stages I and II) were compared with those in non-cancer individuals (HVs and PT patients). In 18 early stage PDAC patients, the sensitivities of C4BPA, CA19-9 and CEA at cutoff levels that gave 95.4% specificity were 50.0%, 22.2% and 22.2%, respectively. ROC curves were also conducted to estimate the value of C4BPA, CA19-9 and CEA levels for distinguishing PDAC patients with early stage from HVs and PT patients. For distinguishing between cancer and non-cancer groups, the respective AUC was 0.912 for C4BPA, 0.737 for CA19-9 and 0.868 for CEA. Taken together, these results predominantly indicate that C4BPA is superior to CA19-9 and CEA as an early diagnostic biomarker for resectable PDAC (Figure 3B).
C4BPA is a potentially new specific serum biomarker for PDAC
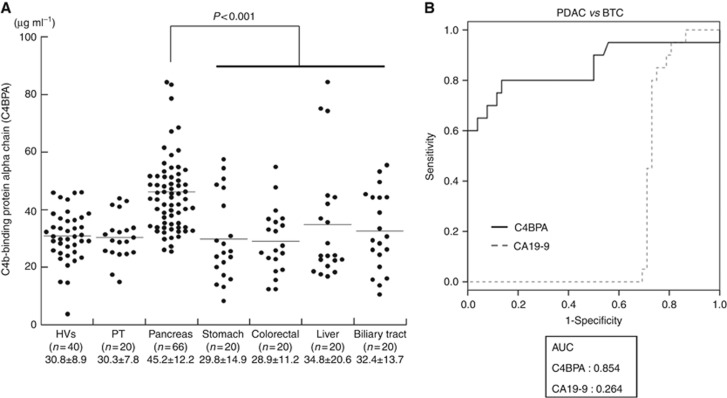
To assess the discriminatory ability of a specific serum marker for PDAC, serum C4BPA levels were measured in major gastroenterological malignancies such as stomach, colorectal, liver and biliary tract cancers. Serum C4BPA levels of these four cancer groups were significantly lower than those of the PDAC group (P<0.001; Mann–Whitney U-test; Figure 4A). PDAC and BTC are virtually indistinguishable by conventional clinical examination, diagnostic imaging and histopathology. Therefore, to evaluate the discriminatory power of C4BPA compared with CA19-9 ROC curves were plotted for distinguishing PDAC patients from BTC patients. Interestingly, the AUC for C4BPA was much higher than that for CA19-9 (C4BPA: 0.854, CA19-9: 0.264, Figure 4B). These results suggest that C4BPA is a specific biomarker for PDAC.
Figure 4.
C4BPA is a potentially new specific serum biomarker for PDAC. (A) Serum C4BPA levels in HVs, and in patients with PT, PDAC and other gastroenterological cancers. Serum C4BPA levels in patients with PDAC were significantly higher than those in HVs and in patients with PT and other gastroenterological cancers (P<0.001; Mann–Whitney U-test). (B) The ROC analyses were performed for the serum levels of C4BPA and CA19-9 between patients with PDAC and biliary tract cancers (BTC). The respective AUC was 0.854 for C4BPA and 0.264 for CA19-9.
Discussion
In this study, we have demonstrated that 302 proteins with unique peptide sequences were revealed, and C4b-binding protein α-chain (C4BPA) and Polymeric immunoglobulin receptor (PIGR) were validated with additional samples. To the end, we have described that serum C4BPA level is a potential serum biomarker to distinguish PDAC from chronic pancreatitis and major gastroenterological cancers, including BTC.
PIGR is a member of the immunoglobulin superfamily. It binds polymeric immunoglobulin molecules and presents them at the mucosal lining of the gastrointestinal tract and exocrine glands (Phalipon and Corthesy, 2003). The finding that high PIGR expression is associated with more favourable clinic-pathological characteristics and its loss with an adverse clinical outcome is in line with the vast majority of published studies in a number of cancers, for example, pancreatic, gastro-oesophageal, ovarian, bladder, colon and non-small cell lung cancer (Fristedt et al, 2014). In this study, the serum PIGR levels in PDAC patients were significantly greater than in HVs. The mechanistic basis underlying the potential tumour-suppressing role for PIGR in PDAC, as well as in several other cancer forms, remains to be elucidated in further analyses.
C4BPA has binding sites for many ligands, such as the CD40 molecule, C-reactive protein, and heparin, which are key molecules involved in inflammatory and coagulation pathways (Antoniades et al, 2009). Specifically, C4BPA binds directly to CD40 on human B cells at a site that differs from CD40 ligand; therefore C4BPA does not compete with CD40 ligand for binding to CD40 (Brodeur et al, 2003). Subsequently, C4BPA induces B cell proliferation and CD40 activation, which can reverse immune suppression and drive antitumour T cell responses. Beatty et al. suggested that systemic CD40 activation with an agonist CD40 monoclonal antibody was sufficient to circumvent tumour-induced immune suppression and invoke productive macrophage and T-cell-dependent antitumour immunity in PDAC (Beatty et al, 2011). Furthermore, a recent paper has also described that CD40, a binding partner of C4BPA, mediates clonal T-cell expansion with decreased regulatory T cells, therefore CD40 functions as a non-redundant mechanism to convert tumour microenvironment immunologically in PDAC (Byrne and Vonderheide, 2016). These immunological findings are implicated that C4BPA is produced by host immune responses against the tumour in the sera of PDAC patients. Although this immune reaction might not be specific for PDAC, it is speculated to be more sensitive in PDAC compared with other gastrointestinal cancers. In our study, the serum level of C4BPA protein is upregulated in the early stage of PDAC, and the response of PDAC patients might be one of the reasons why the levels of C4BPA increase even in the early stage of PDAC. Further analyses are needed to elucidate the underling mechanism for this observation.
To evaluate these two PDAC biomarkers for the possibility of clinical application, we have analysed the correlations between the efficacy of treatment for the patients and both C4BPA and PIGR serum levels based on the clinical data of patients in validation set 1. Interestingly, the change rate of these two biomarkers after surgical treatment showed a decrease in R0 curative resection group compared with non-R0 resection group. However a large number of samples are needed to show a significant difference, the serum levels of these two biomarkers might be influenced by the effect of treatment for PDAC patients.
In this study, we have demonstrated that serum C4BPA is a potential biomarker for PDAC. Various attempts have been made to identify new serum biomarkers, and some molecular targets have been found attractive as diagnostic or prognostic markers for PDAC (Takano et al, 2008). Wang et al (2014) reported on macrophage inhibitory cytokine 1 (MIC-1) as a novel diagnostic serum biomarker in PDAC. In terms of the comparison of diagnostic power of patients with PDAC from healthy controls in each biomarker, the respective sensitivities were 65.8% for serum MIC-1 (the specificity was 96.4%) and 67.3% for serum C4BPA (the specificity was 95.4%). Furthermore, we have previously discovered the phosphorylated extracellular signal-regulated kinase 1/2 (ERK1/2) as a novel diagnostic serum biomarker for PDAC using an immunoaffinity approach (Takano et al, 2010). As for distinguishing between PDAC and non-cancer (HVs and PT) patients, the AUC of C4BPA is superior to that of p-ERK1/2 (0.86 for C4BPA and 0.75 for p-ERK1/2).
In this study, we have also revealed that C4BPA can discriminate PDAC from BTC, which is an undistinguishable disease with the elevation of CA19-9 in some clinical cases. Recent studies show that C4BPA is highly expressed in non-small cell lung cancer (Liu et al, 2012). Furthermore, fully-sialylated C4BPA, A2160, is a newly identified serum biomarker for ovarian cancer, and interestingly, A2160 levels in patient sera are particularly useful for distinguishing early-stage clear cell carcinoma from endometrioma (Mikami et al, 2015).
In conclusion, we have used TMT labelling and proteomics to identified C4BPA protein as a novel serum biomarker. It is possible to distinguish PDAC from benign tumours of the pancreas as well as from other gastroenterological cancers including BTC, using this serum biomarker. One of the critical limitations is that this study has been analysed retrospectively. Moreover, the candidate proteins were identified by single analysis of LC-MS/MS, and the diagnostic window of C4BPA is narrow in all stages of patients with PDAC. Further validation in an independent and prospective large cohort will be needed to establish C4BPA as a new diagnostic marker for PDAC in a clinical setting.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research ‘KAKENHI' C: 16K08979 (KS, MS, FN), B: 15H04925 (ST, HY, MM), Challenge Exploratory Research: 16K15607 (ST, MS, HY, MM), B: 26293299 (MM, ST, HY), C: 15K08610 (MS).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Antoniades C, Bakogiannis C, Tousoulis D, Antonopoulos AS, Stefanadis C (2009) The CD40/CD40 ligand system: linking inflammation with atherothrombosis. J Am Coll Cardiol 54: 669–677. [DOI] [PubMed] [Google Scholar]
- Arnold JN, Saldova R, Hamid UM, Rudd PM (2008) Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Proteomics 8: 3284–3293. [DOI] [PubMed] [Google Scholar]
- Arntzen MØ, Koehler CJ, Barsnes H, Berven FS, Treumann A, Thiede B (2011) IsobariQ: software for isobaric quantitative proteomics using IPTL, iTRAQ, and TMT. J Proteome Res 10: 913–920. [DOI] [PubMed] [Google Scholar]
- Baeumlisberger D, Arrey TN, Rietschel B, Rohmer M, Papasotiriou DG, Mueller B, Beckhaus T, Karas M (2010) Labeling elastase digests with TMT: informational gain by identification of poorly detectable peptides with MALDI-TOF/TOF mass spectrometry. Proteomics 10: 3905–3909. [DOI] [PubMed] [Google Scholar]
- Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, Huhn RD, Song W, Li D, Sharp LL, Torigian DA, O'Dwyer PJ, Vonderheide RH (2011) CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science 331: 1612–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton D, Zen Y, Quaglia A, Selzer S, Mitra V, Löβner C, Jung S, Böhm G, Schmid P, Prefot P, Hoehle C, Koncarevic S, Gee J, Nicholson R, Ward M, Castellano L, Stebbing J, Zucht HD, Sarker D, Heaton N, Pike I (2014) Quantification of pancreatic cancer proteome and phosphorylome: indicates molecular events likely contributing to cancer and activity of drug targets. PLoS One 9: e90948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur SR, Angelini F, Bacharier LB, Blom AM, Mizoguchi E, Fujiwara H, Plebani A, Notarangelo LD, Dahlback B, Tsitsikov E, Geha RS (2003) C4b-binding protein (C4BP) activates B cells through the CD40 receptor. Immunity 18: 837–848. [DOI] [PubMed] [Google Scholar]
- Byrne KT, Vonderheide RH (2016) CD40 stimulation obviates innate sensors and drives T cell immunity in cancer. Cell Rep 15: 2719–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayon L, Turck N, Garcí-Berrocoso T, Walter N, Burkhard PR, Vilalta A, Sahuquillo J, Montaner J, Sanchez JC (2011) Brain extracellular fluid protein changes in acute stroke patients. J Proteome Res 10: 1043–1051. [DOI] [PubMed] [Google Scholar]
- Fristedt R, Elebro J, Gaber A, Jonsson L, Heby M, Yudina Y, Nodin B, Uhlén M, Eberhard J, Jirström K (2014) Reduced expression of the polymeric immunoglobulin receptor in pancreatic and periampullary adenocarcinoma signifies tumour progression and poor prognosis. PLoS One 9: e112728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giron P, Dayon L, Turck N, Hoogland C, Sanchez JC (2011) Quantitative analysis of human cerebrospinal fluid proteins using a combination of cysteine tagging and amine-reactive isobaric labeling. J Proteome Res 10: 249–258. [DOI] [PubMed] [Google Scholar]
- Goggins M (2005) Molecular markers of early pancreatic cancer. J Clin Oncol 23: 4524–4531. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics. CA Cancer J Clin 60: 277–300. [DOI] [PubMed] [Google Scholar]
- Liu YS, Luo XY, Li QR, Li H, Li C, Ni H, Li RX, Wang R, Hu HC, Pan YJ, Chen HQ, Zeng R (2012) Shotgun and targeted proteomics reveal that pre-surgery serum levels of LRG1, SAA, and C4BP may refine prognosis of resected squamous cell lung cancer. J Mol Cell Biol 4: 344–347. [DOI] [PubMed] [Google Scholar]
- Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr ASCO (2006) ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol 24: 5313–5327. [DOI] [PubMed] [Google Scholar]
- Mikami M, Tanabe K, Matsuo K, Miyazaki Y, Miyazawa M, Hayashi M, Asai S, Ikeda M, Shida M, Hirasawa T, Kojima N, Sho R, Iijima S (2015) Fully-sialylated alpha-chain of complement 4-binding protein: diagnostic utility for ovarian clear cell carcinoma. Gynecol Oncol 139: 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien DP, Timms JF (2014) Employing TMT quantification in a shotgun-MS platform. Methods Mol Biol 1156: 187–199. [DOI] [PubMed] [Google Scholar]
- Phalipon A, Corthesy B (2003) Novel functions of the polymeric Ig receptor: well beyond transport of immunoglobulins. Trends Immunol 24: 55–58. [DOI] [PubMed] [Google Scholar]
- Rudd PM, Dwek RA (1997) Glycosylation: heterogeneity and the 3D structure of proteins. Crit Rev Biochem Mol Biol 32: 1–100. [DOI] [PubMed] [Google Scholar]
- Sogawa K, Kodera Y, Satoh M, Kawashima Y, Umemura H, Maruyama K, Takizawa H, Yokosuka O, Nomura F (2011) Increased serum levels of pigment epithelium-derived factor by excessive alcohol consumption-detection and identification by a three-step serum proteome analysis. Alcohol Clin Exp Res 35: 211–217. [DOI] [PubMed] [Google Scholar]
- Takano S, Yoshitomi H, Togawa A, Sogawa K, Shida T, Kimura F, Shimizu H, Tomonaga T, Nomura F, Miyazaki M (2008) Apolipoprotein C-1 maintains cell survival by preventing from apoptosis in pancreatic cancer cells. Oncogene 27: 2810–2822. [DOI] [PubMed] [Google Scholar]
- Takano S, Sogawa K, Yoshitomi H, Shida T, Mogushi K, Kimura F, Shimizu H, Yoshidome H, Ohtsuka M, Kato A, Ishihara T, Tanaka H, Yokosuka O, Nomura F, Miyazaki M (2010) Increased circulating cell signalling phosphoproteins in sera are useful for the detection of pancreatic cancer. Br J Cancer 103: 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida S, Satoh M, Kawashima Y, Sogawa K, Kado S, Sawai S, Nishimura M, Ogita M, Takeuchi Y, Kobyashi H, Aoki A, Kodera Y, Matsushita K, Izumi Y, Nomura F (2013) Application of quantitative proteomic analysis using tandem mass tags for discovery and identification of novel biomarkers in periodontal disease. Proteomics 13: 2339–2350. [DOI] [PubMed] [Google Scholar]
- Wang X, Li Y, Tian H, Qi J, Li M, Fu C, Wu F, Wang Y, Cheng D, Zhao W, Zhang C, Wang T, Rao J, Zhang W (2014) Macrophage inhibitory cytokine 1 (MIC-1/GDF15) as a novel diagnostic serum biomarker in pancreatic ductal adenocarcinoma. BMC Cancer 14: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.