Abstract
Wild boar (Sus scrofa), one of the most widespread wildlife species, has entered a stage of continuous growth in Europe, and could even be considered a pest species. We analysed microsatellite variability in 723 wild boars from across Europe, including the northern Dinaric Balkans. Our aims were: (1) to define the population structure of wild boars in the Balkans and its relation with other European populations; (2) to estimate effective populations sizes, levels of intra- and inter-population diversity, inbreeding migration and gene flow patterns; (3) to test subpopulations for bottlenecks; (4) to interpret these results in light of current knowledge about the demographic history of wild boars in Europe; and (5) to discuss the relevance of these findings for management and conservation. Strong population structuring was observed and 14 subpopulations were revealed. High genetic diversity was found, and besides the well-known identity of the Italian populations of Sardinia and Castelporziano, we bring new insights into other potential relevant, refugial populations such as Littoral Slovenia, South Portugal, North-western Iberia and an entire cluster in the Balkans. There was evidence of gene flow going from these refugial subpopulations towards less peripheral and more admixed subpopulations. Recent population bottlenecks and expansions were detected, mostly in the peninsular refuge subpopulations. The results are consistent with the fluctuations of wild boar numbers in Europe since the beginning of the twentieth century. These results should be taken into account in future conservation and management plans for wild boar populations in Europe.
Introduction
The wild boar (Sus scrofa) is one of the most important and widespread wildlife species in the world. The geographical distribution and present-day genetic diversity of wild boars across Europe has been shaped by Quaternary glacial/interglacial cycles (Scandura et al., 2008; Alexandri et al., 2012; Vilaça et al., 2014; Veličković et al., 2015), and also by human activities in the past centuries (Apollonio et al., 2010). During the nineteenth and early twentieth centuries, the distribution and numbers of wild boar populations in Europe were reduced, mainly because of human overexploitation of both their habitat and the species itself (Apollonio et al., 1988; Massei and Genov, 2000; Apollonio et al., 2010; Linnel and Zachos, 2011). In the past four decades, a remarkable increase in the European wild boar population has been recorded that is mainly related to its high reproductive rate, lack of large predators, reforestation, climate change, supplementary feeding and decreased hunting across Europe (Saez-Royuela and Telleria, 1986; Fonseca, 2004; Ferreira et al., 2009; Apollonio et al., 2010; Scandura et al., 2011a; Massei et al., 2015). European wild boar populations appear to have entered a stage of continuous growth. Population expansions have affected human societies through the spread of diseases, crop damage and vehicle collisions (Massei et al., 2015), causing human–wildlife conflicts (Frank et al., 2015). In some cases, the wild boar is even considered a pest species.
Management strategies for wild boar populations should be based on an appropriate understanding of the factors affecting current population trends and consideration for its adaptation to a wide range of environmental conditions (Scandura et al., 2011a; Massei et al., 2015). Detailed information on the genetic diversity of wild boars is of great importance, as wild boar populations are also a reservoir of genetic variation of suid species and its conservation is important not only for preserving the species itself, but also for preserving more diversity for better breeding for future generations, as local European domestic pig breeds are currently threatened (Frantz et al., 2016). Genetic data can support population management and conservation by: defining population structure (and thus management units); informing about relevant demographic parameters such as levels of inbreeding, diversity and effective population sizes; measuring gene flow between subpopulations; and identifying potential risks associated with demographic changes and inbreeding (Shafer et al., 2015). Of particular importance is to understand whether the current patterns of structure and diversity among wild boar populations are mostly influenced by human activities or whether they actually retain the signature of past events such as Quaternary climatic fluctuations and, consequently, are unaffected by human-mediated demographic fluctuations and gene flow.
All these major concerns, together with the development of molecular genetic tools for wildlife management and conservation, have stimulated a number of studies on the genetic diversity and phylogeography of wild boar populations in Europe (Vernesi et al., 2003; Ferreira et al., 2006, 2009; Scandura et al., 2008, 2011a; Alves et al., 2010; Alexandri et al., 2012; Kusza et al., 2014; Vilaça et al., 2014; Iacolina et al., 2016; Sprem et al., 2016). One of the most recent study included data from large areas in the northern Dinaric Balkans, providing evidence that the Balkans represent an important genetic reservoir of European wild boar and that the present-day genetic structure and geographical distribution of wild boars across Europe arose from ancient processes during the last glaciations, indicating that similar phylogeographic patterns emerged in all southern European peninsulas and that all three peninsulas played a similar role in the post-glacial recolonization of Europe by wild boar (Veličković et al., 2015). On the other hand, Frantz et al. (2016) pointed out that in Western Europe, many local populations had gone extinct and restocked with other wild boar populations. This process resulted in loss of genetic diversity of wild boar populations in Europe compared with that of domestic breeds and wild boars in Asia.
This study was designed in order to provide a more detailed insight into the patterns of genetic variation and structure of wild boars across Europe, especially in the Balkans, to infer the processes that gave rise to the present-day genetic diversity and structure and to discuss their relevance in terms of the future management of wild boar in Europe. Based on microsatellite analyses, we specifically aimed: (1) to define the population structure of wild boars in the Balkans and its relation with other European populations; (2) to estimate effective populations sizes, levels of intra- and inter-population diversity, inbreeding, migration and gene flow patterns; (3) to test subpopulations for bottlenecks; (4) to interpret these results in light of current knowledge about the demographic history of wild boars in Europe and the possible influence of human interventions, besides the already well-documented influence from Quaternary climatic fluctuations; and (5) to discuss the relevance of these findings for management and conservation, providing new insights through the determination of management units.
Materials and methods
Material collection and DNA extraction
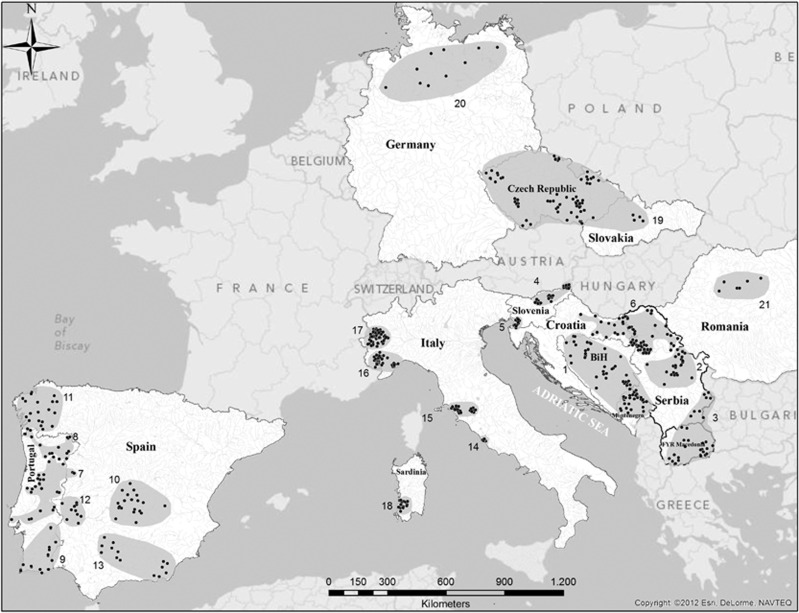
A total of 581 wild boar samples were collected from different localities across Serbia, FYR Macedonia, Montenegro, Bosnia and Herzegovina, Croatia, Slovenia, Italy, Portugal, Spain, Czech Republic, Germany, Slovakia and Romania during regular hunting seasons (Figure 1). The analysis also included 142 wild boars from Northern Italy, for which we retrieved complete genotypes from the study of Caratti et al. (2010) and these sampling sites are also included in Figure 1. In addition, 35 domestic pig samples from two Iberian breeds (Bisaro and Malhado de Alcobaça) were included. Total DNA was extracted using Proteinase K digestion, followed by standard phenol–chloroform–isoamyl alcohol extraction (Sambrook and Russell, 2001) and saline extraction procedures (Bruford et al., 1992).
Figure 1.
Map of Europe showing the distribution of wild boar sampling sites. Samples were divided into 21 subpopulations according to their geographic proximity, the biogeographical features of the sampling areas and, when available, prior information on local genetic structure: 1, Dinaric Balkans; 2, Peridinaric Balkans; 3, South Central Balkans; 4, Continental Slovenia; 5, Littoral Slovenia; 6, South Pannonia; 7, Central-western Iberia; 8, North Portugal; 9, South Portugal; 10, Castilla la Mancha; 11, Galicia; 12, Extremadura; 13, Andalusia; 14, Castelporziano; 15, Central-western Italy; 16, North-western Italy; 17, Alps; 18, Sardinia; 19, Czech Republic and Slovakia; 20, Germany; and 21, Romania.
Microsatellite genotyping
Multiplex PCR amplification of 11 tetranucleotide microsatellites was carried out using the Animaltype Pig PCR Amplification Kit (Biotype AG, Dresden, Germany) following the manufacturer's recommendations. Typing was done by capillary electrophoresis on an ABI3730 × l Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The PCR product fragments of these 11 microsatellites were analysed using Peak Scanner v1.0 (Applied Biosystems) and Gene Marker (Softgenetics, State College, PA, USA) software against an allelic ladder and a control DNA sample (DL157) of known genotype provided by the kit manufacturer.
Statistical analysis
All wild boar samples were divided into 21 a priori subpopulations according to their geographic proximity, biogeographical features of the sampling areas and, when available, prior information on local genetic structure: (1) Dinaric Balkans (68 samples); (2) Peridinaric Balkans (26); (3) South Central Balkans (33); (4) Continental Slovenia (36); (5) Littoral Slovenia (17); (6) South Pannonia (79); (7) Central-western Iberia (26); (8) North Portugal (22); (9) South Portugal (19); (10) Castilla la Mancha (20); (11) Galicia (20); (12) Extremadura (10); (13) Andalusia (14); (14) Castelporziano (19); (15) Central-western Italy (28); (16) North-western Italy (57); (17) Alps (120); (18) Sardinia (16); (19) Czech Republic and Slovakia (77); (20) Germany (10); and (21) Romania (6) (Figure 1). The two domestic pig breeds were treated as two different a priori subpopulations. In order to verify whether stutter bands, large allele dropout and null alleles have influenced microsatellite allele frequencies and heterozygosity calculations, the programme MICRO-CHECKER (Van Oosterhout et al., 2004) was used to test our data set for these potential error sources. Quality check was performed separately for each a priori subpopulation in order to minimize the effect of structure on the observed level of homozygosity. The number of different alleles, observed and expected heterozygosity values, deviations from Hardy–Weinberg equilibrium and inbreeding coefficients with confidence intervals were estimated using the divbasic function of diveRsity package (Keenan et al., 2013) in R software (R Core Team, 2015). Deviations from linkage equilibrium, analysis of molecular variance (AMOVA) and pairwise genetic distances among the presumed groups (FST) were calculated using ARLEQUIN 3.5.1.2 (Excoffier and Lischer, 2010). The significance level for multiple comparisons was modified using Bonferroni correction (Rice, 1989). The rarefied private alleles were calculated using ADZE (Szpiech et al., 2008) by setting rarefaction indexed to the minimum sample size.
Genetic structure of populations throughout a species' distribution range might reflect past demographic and migratory events, rather than current geographic distances among individuals or random-mating population units. Therefore, we performed Bayesian cluster analyses implemented in STRUCTURE 2.3.4 (Pritchard et al., 2000; Falush et al., 2003, 2007; Hubisz et al., 2009) in order to infer the number of distinct genetic clusters represented in our sample of wild boars and domestic pigs in Europe. Three independent sets of STRUCTURE runs were performed assuming different models: for the first set of runs, we adopted an admixture model with correlated allele frequencies; for the second, we assumed no admixture between K groups and no correlation between allele frequencies; and for the third, we used a no admixture model with correlated allele frequencies. Initially, all three analyses were done with a burn-in length of 100 000 followed by 1 000 000 Markov chain Monte Carlo iterations. Independent runs from the first two sets of analyses were convergent using this number of iterations, but the third analysis (that is, no admixture model with correlated allele frequencies) had to be repeated with a burn-in of 200 000 followed by 2 000 000 Markov chain Monte Carlo iterations to achieve convergence. For all three analyses, we ran 3 replicates for each K ranging between 1 and 17.
We estimated the most likely number of clusters through the ΔK method (Evanno et al., 2005), using CLUMPAK (Kopelman et al., 2015) and STRUCTURE HARVESTER (Earl and vonHoldt, 2012). CLUMPAK was also used for detailed inspection of convergence between independent runs for each K and graphical interpretation of the results. For the selected K-value, we evaluated the population membership coefficient (Qpop) of the inferred clusters. For all three models, the most probable number of genetic clusters always included K=3 and K=7. However, it has been shown that the ΔK method detects the uppermost level of population structure when several hierarchical levels exist (Evanno et al., 2005). Therefore, we have reason to assume that, with this first round of analyses using the total sample set (758 individuals), we detected the first two levels of structuring. In order to fully understand the genetic structuring of wild boars in the Balkans and Europe, we performed additional STRUCTURE analyses for some of the K groups inferred in the previous step (see Results for more details).
The correlation between genetic distances among a posteriori-defined subpopulations and geographical distances was tested using Mantel's nonparametric test on pairwise distance matrices (Mantel, 1967) using ARLEQUIN. In addition, in order to better understand spatial distribution and possible geographic barriers for wild boars in Europe, we performed spatial analyses of molecular variance (SAMOVA) using SAMOVA 2.1 (Dupanloup et al., 2002). We performed analyses for K = 2 to 20 to identify the most likely number of groups, with the 21 a priori subpopulations. The best K was determined when Fct value (degree of differentiation among groups) reached plateau.
The basic genetic indices for a posteriori-defined subpopulations were calculated in the same way as described for a priori subpopulations. In addition, for a posteriori subpopulations, we estimated the GST (Nei, 1973) and D (Jost, 2008) differentiation statistics and the relative migration network using the functions diffCalc and divMigrate of diveRsity R package, respectively. The latter implements the method described by Sundqvist et al. (2016). The networks were estimated using the statistics GST, D and Nm (Slatkin, 1993, estimated according to Alcala et al. (2014). Significant migration flows were estimated based on a bootstrap procedure with 49999 replicates. This method allows for the estimation of asymmetric migration among individual populations, assuming a finite island model (or one where migration has an unconstrained dispersal kernel) with infinite number of alleles and low mutation rates (Sundqvist et al., 2016). Mutation rates for porcine microsatellites average 7.52 × 10−5 per locus per generation (Yue et al., 2002), and can thus be considered small (μ2<<μ) in this context. Microsatellite loci are considered neutral markers, but can be influenced by closely related adaptative genes (Nei and Tajima, 1981; Ford, 2002). This effect should however be diluted by using a set of independent markers. Effective population size (Ne) based on linkage disequilibrium was inferred using LDNE 1.31 (Waples and Do, 2008). The 95% confidence intervals were obtained via a jackknife method and estimates excluded all alleles with a frequency of <0.05 in order to correct for known biases from rare alleles.
In order to gain an insight into the demographic history of wild boars in the Balkans and Europe, each subpopulation was tested for recent bottleneck events. We used two approaches, suggested as being particularly promising (Williamson-Natesan, 2005). The first approach was based on the detection of heterozygosity excess relative to the number of alleles, across all loci, under a two-phased mutation model, as implemented in BOTTLENECK (Cornuet and Luikart, 1996). The second approach was based on the analysis of the M ratio described by Garza and Williamson (2001), using the executable files M_P_VAL.exe and critical_M.exe, developed by the same authors. The approach of Cornuet and Luikart (1996) is based on the heterozygosity excess (relative to the number of alleles) that is expected to build up in a population that has recently undergone a bottleneck. The Garza and Williamson (2001) method is based on the ratio M between the number k of observed alleles of a given locus and the range r of the distribution of allele sizes for that locus. According to the authors, the number of alleles will be more strongly affected by a bottleneck than the range of allele size distribution. The first approach should be more efficient for the detection of more recent and short-term bottlenecks, whereas the second would allow the detection of older and prolonged bottlenecks (Williamson-Natesan, 2005).
Results
Data quality and marker polymorphism
The majority of genotypes were successfully scored and missing data amounted to 0.17% (0 to 0.9% per locus). We did not find any significant evidence caused by allele dropout and only very occasionally detected significant evidence for genotyping errors due to stutter. This occurred only for four subpopulations, at one marker per subpopulation (SBH13 or SBH22). However, after carefully going through the electophoretograms, this possibility could be ruled out. There was few or no evidence for the presence of null alleles for most of the markers (up to two subpopulations, out of 23, per marker). Only in two markers (SBH10 and SBH22) we found evidence of null alleles in six or more subpopulations. Most of the evidence for null alleles came from five subpopulations (Peridinaric Balkans, SC Balkans, CW Iberia, Alps and Czech Republic and Slovakia) that were later proven to include individuals from different gene pools. The detection of null alleles is based on the expected proportions of homozygotes that can also be affected by the presence of internal population structure or admixture. After carefully going through the electrophoretograms, we found no reason to suspect that these nonsystematic deviations from the expected level of homozygosity should be consequence of the presence of null alleles, rather than other biologically meaningful processes causing population structure or admixture. All loci showed high levels of polymorphism and we found a total of 209 alleles across all loci. The number of observed alleles per locus varied from 9 at locus SBH19 to 29 at locus SBH20, with an average of 19 alleles per locus.
Genetic diversity and differentiation on a priori subpopulations
The total number of alleles per a priori-defined subpopulation varied from 44 in Castelporziano to 144 in the Alps, whereas rarefied allelic richness varied from 2.9 alleles in Castelporziano to 5.5 in South Central Balkans (Supplementary Table S1). The lowest private allelic richness was observed in Castelporziano subpopulation. The expected heterozygosity varied from 0.46 in Castelporziano to 0.79 in the South Central Balkans, Alps and Castilla La Mancha. The lowest observed heterozygosity was 0.44 in Castelporziano, whereas the highest value of HO was found in Dinaric Balkans (HO=0.74). The inbreeding coefficients ranged from −0.037 to 0.172, and in 11 out of 21 a priori subpopulations the FIS values were highly significant. Genetic distances (FST) between pairs of a priori-defined subpopulations were highly significant for 250 out of 253 possible pairwise combinations among subpopulations (P<0.001). Most estimated values corresponded to moderate to high values of genetic differentiation (Wright, 1978), and only 29 out of 253 combinations showed low levels of genetic differentiation (Supplementary Figure S2). For the AMOVA, 21 a priori-defined subpopulations were merged into 4 geographical/regional groups of wild boars (Balkans, Italy, Iberia and Central-eastern Europe) and 1 group consisting of domestic pigs. The hierarchical AMOVA showed the highest variability within subpopulations (90.9%), whereas variability among groups was 1.9% and variation among subpopulations within groups was 7.2% (P<0.001).
Genetic structure of wild boar in Europe
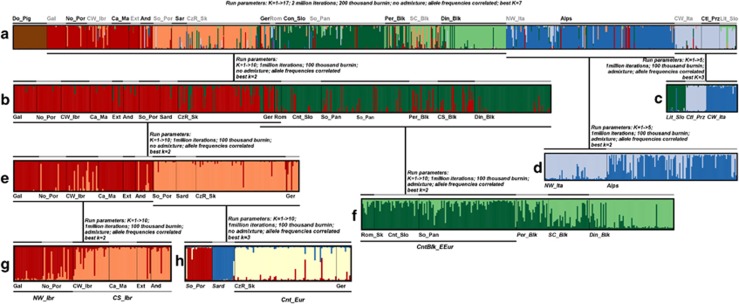
For each of three sets of STRUCTURE runs including all samples, K=3 and K=7 were among the best K numbers of genetic clusters (Table 1). At K=3, regardless of the model, three genetic clusters were found, each mainly associated with one of the southern European peninsulas. There was consistent agreement among the results obtained with the three models at K=7, despite differences in the proportions of genomes attributed to each cluster, for the same subpopulations. As the ΔK method detects the uppermost level of population structure, we conclude that K=3 is the first level of structuring present in wild boars across Europe, suggesting the existence of three main gene pools in Europe At K=3, domestic pigs are clustered with Iberian wild boars, and at K=7 (Figure 2a) domestic pigs are clearly separated from all wild boars in all three models, and also at this level of structure there were some well-defined wild boar clusters. As for biological conservation it is very important to understand the demographic history of a species and regional levels of structuring, we also examined lower levels of population structuring by running STRUCTURE on the clusters defined in the previous analysis (Figures 2b–h).
Table 1. Best K number of clusters and a priori subpopulations assigned to each cluster for the three models tested in STRUCTURE.
| Model and simulated K | Best K (Evanno's method) | Clusters at K=3 | Clusters at K=7 |
|---|---|---|---|
| Admixture+allele frequencies correlated (K=1 to K=17) | K=13 (ΔK=117.9) K=3 (ΔK=111.7) K=7 (ΔK=93.3) | (1) Balkans+Ctl_Prz+Rom (57%)+CW_Ita (55%)+Ger and CzR_Sk (26%)+Sard (19%) (2) NW_Ita+Alps+CW_Ita (31%)+Rom (26%)+Sard (20%)+Ger and CzR_Sk (19%) (3) Iberia+Domestic Pig+Sard (61%)+Ger and CzR_Sk (55%)+Rom (17%)+CW_Ita (14%) | (1) Din_Blk+SC_Blk (49%)+Per_Blk (22%)+So_Pan (10%) (2) Cnt_Slo+So_Pan (69%)+Per_Blk (49%)+SC_Blk (16%)+Ger (10%)+CzR_Sk (15%)+Rom (41%) (3) Ctl_Prz+CW_Ita+Lit Slo+Rom (15%) (4) NW_Ita+Alps+Rom (21%)+Ger (11%) (5) No_Por+Gal+Ca_Ma+Ext+CW_Ibr (68%)+And (61%)+Per_Blk (10%)+Ger (14%) (6) So_Por+Sard+Ger (55%)+CzR_Sk (64%)+CW_Ibr (18%)+And (16%)+SC_Blk (16%) (7) Domestic Pig |
| No admixture+allele frequencies not correlated (K=1 to K=17) | K=2 (ΔK=2085.0) K=3 (ΔK=760.6) K=7 (ΔK=139.2) | (1) Balkans+Sard+Ger and CzR_Sk (72%)+Rom (50%) (2) Italy+Rom (34%)+Ger and CzR_Sk (16%) (3) Iberia+Domestic Pig+Rom (16%)+Ger and CzR_Sk (12%) | (1) Din_Blk+SC_Blk (46%)+Lit_Slo (53%)+Per_Blk (23%) (2) Cnt_Slo+So_Pan+Per_Blk (58%)+SC_Blk (23%)+Rom (47%) (3) Ctl_Prz+CW_Ita+Lit_Slo (44%) (4) NW_Ita+Alps+Ger (10%)+Rom (22%) (5) No_Por+Gal+Ca_Ma+Ext+CW_Ibr (69%)+And (68%)+Ger (13%) (6) So_Por+Sard+CzR_Sk+Ger (65%)+Rom (25%)+SC_Blk (22%)+CW_Ibr (26%)+And (17%) (7) Domestic Pig |
| No admixture+allele frequencies correlated (K=1 to K=17) | K=7 (ΔK=271.5) K=3 (ΔK=235.6) K=2 (ΔK=163.8) | (1) Balkans+Ctl_Prz+Rom (59%)+CW_Ita (52%)+Ger and CzR_Sk (30%)+Sard (23%) (2) NW_Ita+Alps+CW_Ita (37%)+Rom (25%)+Ger and CzR_Sk (15%)+Sard (13%) (3) Iberia+Domestic Pig+Sard (64%)+Ger and CzR_Sk (55%)+ Rom (16%)+CW_Ita (11%) | (1) Din_Blk+SC_Blk (50%)+Per_Blk (25%) (2) So_Pan+Cnt_Slo+Per_Blk (56%)+Rom (50%)+ SC_Blk (15%) (3) Ctl_Prz+CW_Ita+Lit Slo+Rom (19%) (4) NW_Ita+Alps+Rom (22%) (5) Ibr except So_Por+Ger (15%) (6) So_Por+Sard+CzR_Sk+Ger (67%)+SC_Blk (24%) (7) Domestic Pig |
Abbreviations: Balkans: Cnt_Slo, Continental Slovenia; Din_Blk, Dinaric Balkans; Lit_Slo, Littoral Slovenia; Per_Blk, Peridinaric Balkans; SC_Blk, South and Central Balkans; So_Pan, South Pannonia; Central Europe: CzR_Sk, Czech Republic and Slovakia; Ger, Germany; Rom, Romania; Iberia: And, Andalusia; Ca_Ma, Castilla La Mancha; CW_Ibr, Central-western Iberia; Ext, Extremadura; Gal, Galiza; No_Por, North Portugal; So_Por, South Portugal; Domestic Pig, Iberian domestic pig (Bísaro and Malhado de Alcobaça); Italy: Alps, Alps; Ctl_Prz, Castelporziano; CW_Ita, Central-western Italy; NW_Ita, North-western Italy; Sard, Sardinia.
A priori subpopulations (or groups of populations) established a priori with <75% proportion of genome attributed to a single cluster (K=3 and K=7) are not in bold (if percentage of assignment to that cluster is >10%).
Figure 2.
Hierarchical distribution of a priori wild boar subpopulations into the genetic clusters inferred using STRUCTURE. Results are presented for consecutively less inclusive runs (a–h). Each individual is represented by a vertical line; a thin black line separates genotypes from different subpopulations. The parameters of each STRUCTURE run are given above the representative STRUCTURE plot. DinBlk, Dinaric Balkans; PerBlk, Peridinaric Balkans; SC_Blk, South Central Balkans; Cnt_Slo, Continental Slovenia; Lit_Slo, Littoral Slovenia; So_Pan, South Pannonia; CW_Ibe, Central-western Iberia; No_Por, North Portugal; So_Por, South Portugal; Ca_LMa, Castilla la Mancha; Gal, Galicia; Ext, Extremadura; And, Andalusia; Ctl_Prz, Castelporziano; CW_Ita, Central-western Italy; NW_Ita, North-western Italy; Alps, Alps; Sard, Sardinia; CzR, Czech Republic; Ger, Germany; Rom, Romania; Rom_Sk, Romania and Slovakia; NW_Ibr, North-western Iberia; CS_Ibr, Central South Iberia; Cnt_Eur, Central Europe; CntBlk_EEur, Continental Balkans and Eastern Europe.
For all tested models, the a priori-defined subpopulations of North-western Italy and Alps represented one genetic cluster, but as we had five sampling localities in that area we investigated genetic structuring within this cluster. We found that NW Italy represents a distinct genetic cluster, whereas the Alps represent an admixed cluster (Figure 2). In all three models, Castelporziano+Central-western Italy+Littoral Slovenia represented a consistent cluster and further STRUCTURE analyses supported K=3, echoing the division into the three sampling localities (Figure 2c). In all tested models, wild boar samples from Central-eastern Europe and Sardinia were clustered with both the Balkan and Iberian peninsulas. Therefore, we ran STRUCTURE with samples from the Balkans (excluding Littoral Slovenia), Iberia, Sardinia and Central-eastern Europe (Figure 2b). In this analysis, two clusters were revealed: (1) Balkans+samples from Romania and Slovakia and (2) Iberia+samples from Sardinia, Czech Republic and Germany. Further analyses (Figures 2e–h) of these two clusters revealed seven clearly distinct genetic clusters: (1) Dinaric Balkans, (2) Continental Balkans+Eastern Europe (Continental Slovenia, South Pannonia, Romania and Slovakia), (3) Sardinia, (4) North-western Iberia (North Portugal and Galicia), (5) South Portugal, (6) Central South Iberia (Central-western Iberia, Extremadura, Castilla La Mancha and Andalusia) and (7) Central Europe (Czech Republic and Germany). These analyses also evidenced two admixed clusters: Peridinaric Balkans and South Central Balkans.
The SAMOVA corroborated results obtained by STRUCTURE. The revealed best K was 14, and 11 defined groups matched a posteriori subpopulations. Results from SAMOVA group Peridinaric Balkans together with Continental Balkans+Eastern Europe. Subopulation Central South Iberia was divided in two groups in SAMOVA analyses: (1) Central-western Iberia+Andalusia and (2) Extremadura+Castilla la Mancha. Mantel's nonparametric test performed for a posteriori-defined groups showed no significant correlation between genetic and geographical distances of a posteriori subpopulations (r=−0.000003).
Genetic diversity and differentiation on a posteriori subpopulations
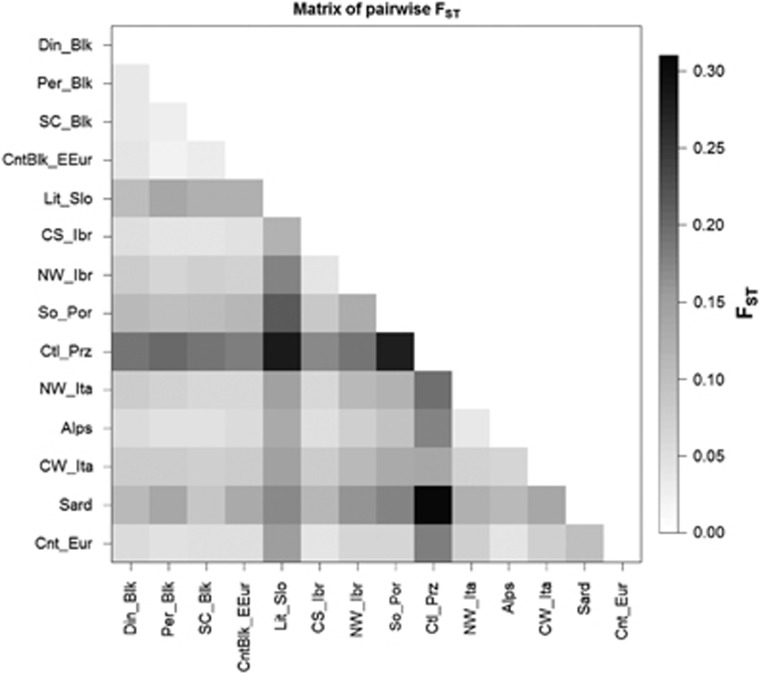
The basic parameters of genetic diversity were also calculated for each a posteriori subpopulation. The lowest number of alleles, allelic richness and private allelic richness (44, 3.5 and 0.028, respectively) were found in Castelporziano subpopulation (Table 2). The highest number of alleles and the highest private allelic richness were observed in Alps, whereas the highest allelic richness was found in South and Central Iberia. Observed heterozygosity for all loci varied between 0.44 and 0.72, with a mean of 0.65, whereas expected heterozygosity ranged from 0.46 to 0.79, with a mean of 0.71. In 12 of the 14 a posteriori-defined subpopulations, 1 to 5 loci showed significant deviations from Hardy–Weinberg equilibrium and in these subpopulations significantly positive inbreeding coefficients were observed. In 13 subpopulations, 1 to 14 pairs of loci (out of 55 combinations) were in linkage disequilibrium (Table 2). The highest Hardy–Weinberg equilibrium and linkage equilibrium deviations were observed in the Central South Iberia and Alps subpopulations, also suggesting that these subpopulations are highly admixed. The mean effective population size was 77.8, and ranged from 22 in Sardinia to 223.1 in Central Europe (Table 2). In one of 14 a posteriori subpopulations (Alps) we observed that Ne is smaller than mean harmonic sample size. The AMOVA indicated higher intrapopulation (92.6%) than interpopulation (7.4%) variability (P<0.001). Genetic distances between pairs of subpopulations were always highly significant (P<0.001). Most estimated values corresponded to moderate to high values of genetic differentiation (Wright, 1978). Only 17 out of 91 combinations showed low levels of genetic differentiation (Figure 3).
Table 2. General genetic diversity indices for 14 a posteriori wild boar subpopulations, based on 11 microsatellite markes.
| Population | 2n | Ne | Allele number | Allele richness (rarefied) | Private alleles (rarefied) | Ho | He | FIS | Loci in HWD (out of 11) | LD (pair loci/55) |
|---|---|---|---|---|---|---|---|---|---|---|
| Dinaric Balkans | 136 | 85.2 | 115 | 7.2 | 0.342 | 0.740 | 0.780 | 0.045 | 2 | 5 |
| Peridinaric Balkans | 52 | 48.4 | 97 | 7.2 | 0.221 | 0.720 | 0.770 | 0.065 | 2 | 2 |
| Cont. Balkans and East. Europe | 250 | 129.3 | 135 | 7.6 | 0.083 | 0.690 | 0.760 | 0.089 | 3 | 5 |
| South Central Balkans | 66 | 57.4 | 107 | 7.7 | 0.362 | 0.720 | 0.790 | 0.092 | 2 | 3 |
| Littoral Slovenia | 34 | 25.5 | 53 | 4.2 | 0.219 | 0.500 | 0.580 | 0.136 | 1 | 1 |
| Castel Porziano | 38 | 23.6 | 44 | 3.5 | 0.028 | 0.440 | 0.460 | 0.041 | 0 | 2 |
| Central-western Italy | 56 | 52.4 | 87 | 6.4 | 0.295 | 0.640 | 0.720 | 0.121 | 3 | 2 |
| North-western Italy | 114 | 86.5 | 119 | 7.2 | 0.383 | 0.710 | 0.740 | 0.044 | 1 | 3 |
| Alps | 240 | 99.2 | 144 | 7.7 | 0.437 | 0.700 | 0.790 | 0.113 | 5 | 14 |
| Sardinia | 32 | 22.0 | 53 | 4.4 | 0.155 | 0.660 | 0.650 | −0.015 | 0 | 1 |
| North-western Iberia | 84 | 93.1 | 89 | 6.5 | 0.309 | 0.710 | 0.750 | 0.053 | 3 | 2 |
| Central South Iberia | 140 | 93.8 | 123 | 7.9 | 0.320 | 0.650 | 0.790 | 0.174 | 2 | 13 |
| South Portugal | 38 | 49.4 | 65 | 5.0 | 0.231 | 0.560 | 0.630 | 0.098 | 1 | 0 |
| Central Europe | 166 | 223.1 | 124 | 7.7 | 0.234 | 0.690 | 0.750 | 0.082 | 2 | 6 |
Abbreviations: FIS, inbreeding coefficient; HE, expected heterozygosity; HO, observed heterozygosity; HWD, Hardy–Weinberg disequilibrium; LD, linkage disequilibrium; 2n, number of gene copies; Ne, effective population size.
Statistically significant values (P<0.01) are in bold.
Figure 3.
Matrix of genetic differentiation (pairwise FST) for 14 a posteriori-defined wild boar subpopulations in Europe. Colour gradient represents the degree of genetic differentiation: low for FST<0.05, moderate for 0.05<FST<0.15, high for 0.15<FST<0.25 and very high for FST>0.25, according to the criterion for genetic differentiation by Wright (1978). DinBlk, Dinaric Balkans; PerBlk, Peridinaric Balkans; SC_Blk, South Central Balkans; CntBlk_EEur, Continental Balkans and Eastern Europe; Lit_Slo, Littoral Slovenia; CS_Ibr, Central South Iberia; NW_Ibr, North-western Iberia; So_Por, South Portugal; Ctl_Prz, Castelporziano; CW_Ita, Central-western Italy; NW_Ita, North-western Italy; Alps, Alps; Sard, Sardinia; Cnt_Eur, Central Europe.
Evidences for bottlenecks and directional migration flow
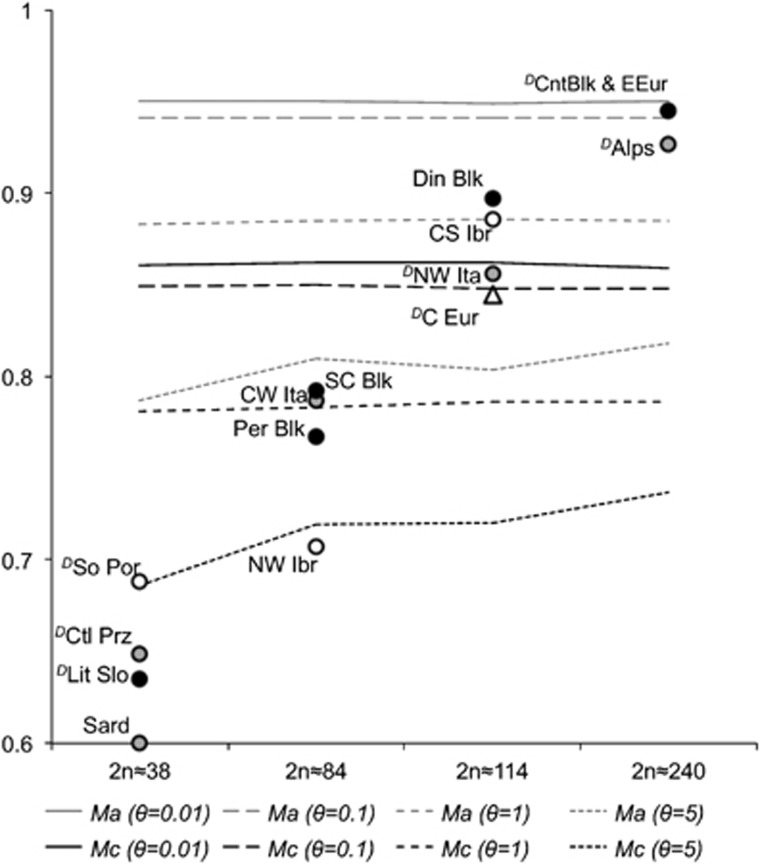
For theta values ranging from 0.01 to 5, significant evidence of a bottleneck (M value for sample below critical Mc value) was found for the following subpopulations: Littoral Slovenia, Sardinia, Castelporziano, South Portugal and North-western Iberia (Figure 4). We also found significant evidence of a bottleneck for theta from 0.01 to 1 for Peridinaric Balkans. For this subpopulation and theta=5, sample M value was below the average Ma value for populations at equilibrium, but was above the critical Mc value. The significant excess of heterozygosity that is expected in bottlenecked populations (Cornuet and Luikart, 1996) was not observed in any of the a posteriori subpopulations. However, we observed heterozygote deficiency in seven wild boar subpopulations (Figure 4).
Figure 4.
M ratio values for all a posteriori-defined subpopulations. Critical values (Mc), below which populations are considered to show significant evidence of a bottleneck, and average (Ma) values for equilibrium populations are represented for different theta values. Subopulations of different regions are represented by increasing order of sample size and by different symbols: Iberia (white circles); Italy (grey circles); Balkans (black circles); and Central Europe (grey triangles). The symbol ‘D' indicates a significant heterozygote deficiency detected using the BOTTLENECK software. DinBlk, Dinaric Balkans; PerBlk, Peridinaric Balkans; SC_Blk, South Central Balkans; CntBlk_EEur, Continental Balkans and Eastern Europe; Lit_Slo, Littoral Slovenia; CS_Ibr, Central South Iberia; NW_Ibr, North-western Iberia; So_Por, South Portugal; Ctl_Prz, Castelporziano; CW_Ita, Central-western Italy; NW_Ita, North-western Italy; Alps, Alps; Sard, Sardinia; Cnt_Eur, Central Europe.
On average, differentiation estimates based on GST (average: 0.060; range: 0.011–0.181) and D (average: 0.264; range: 0.051–0.560) were low (Supplementary Figures S3 and S4) in the context of the estimation of demographic parameters (Alcala et al., 2014). The analysis of migration dynamics revealed the following patterns, regardless of the differentiation statistic: (1) intense relative migration flows among wild boar subpopulations (Supplementary Figures S5–S7), mostly within more admixed and less peripheral subpopulations (for example, Czech Republic and Germany, Continental Balkans and Eastern Europe, Central South Iberia, Alps); (2) significant relative migration flows (Figure 5 and Supplementary Figures S8–S10), with some subpopulations functioning systematically as source of migration flows (Littoral Slovenia, Castelporziano, Sardinia, South Portugal) whereas others functioning systematically as sinks (Peridinaric Balkans, South Central Balkans, Alps or Central South Iberia); and (3) no evidence of significant relative migration flow between Iberian domestic pig breeds and wild boar subpopulations (Nm) or significant only from wild boar to domestic pig (GST and D).
Figure 5.
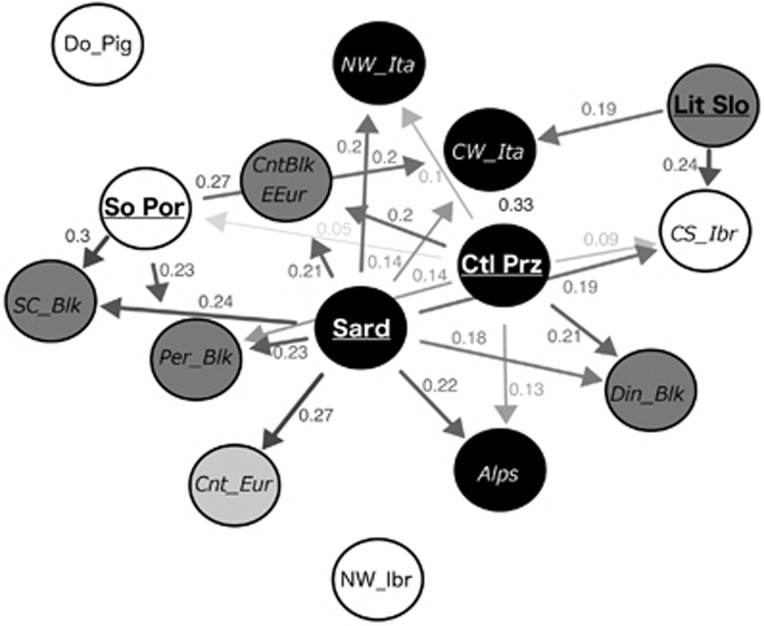
Network of relative migration levels between subpopulations, using the Nm parameter. Only significant (after 49 999 bootstrap replicates) relative migration levels were plotted in the network. Arrows indicate the direction of gene flow. The numbers on arrows indicate relative migration coefficients. Subopulations are coded by colours: Balkans, dark grey; Iberia, white; Italy, black; Continental Europe, light grey. Codes for source populations are written in bold and underlined, and for sink populations are written in italics. DinBlk, Dinaric Balkans; PerBlk, Peridinaric Balkans; SC_Blk, South Central Balkans; Cnt_Slo, Continental Slovenia; Lit_Slo, Littoral Slovenia; So_Pan, South Pannonia; CW_Ibe, Central-western Iberia; No_Por, North Portugal; So_Por, South Portugal; Ca_LMa, Castilla la Mancha; Gal, Galicia; Ext, Extremadura; And, Andalusia; Ctl_Prz, Castelporziano; CW_Ita, Central-western Italy; NW_Ita, North-western Italy; Alps, Alps; Sard, Sardinia; CzR, Czech Republic; Ger, Germany; Rom, Romania; Rom_Sk, Romania and Slovakia; NW_Ibr, North-western Iberia; CS_Ibr, Central South Iberia; Cnt_Eur, Central Europe; CntBlk_EEur, Continental Balkans and Eastern Europe; Do_Pigs, domestic pig breeds.
Discussion
Genetic structure and migration patterns of wild boars in the Balkans and Europe
Previous genetic studies of wild boars in Europe have shown that populations are rarely homogenous and usually genetically differentiated into subpopulations (Ferreira et al., 2006, 2009; Scandura et al., 2008, 2011a; Nikolov et al., 2009; Veličković et al., 2012, 2015; Iacolina et al., 2016). Thus, the first step in our comprehensive analysis was an assessment of genetic structure. The existence of three gene pools in Europe was revealed. The geographical distribution of the three clusters appears to be consistent with genetic differentiation among the three southern European peninsulas (Balkans, Italy and Iberia), whereas Central-eastern Europe evidences the admixture of the three gene pools. These findings provide support to the hypothesis stated by Veličković et al. (2015) that all three refugees could have had a role in the postglacial recolonization of Europe. Furthermore, hierarchical AMOVA with a priori subpopulations showed more differentiation among populations within peninsulas than among peninsulas, suggesting that there are distinct gene pools in the peninsulas, but there are common elements across all Europe. It may be concluded that some subpopulations establish the bridge among European regions, consistent with a scenario of ‘refugial populations' and ‘source populations to the recolonization of Europe' in the same regions, and fitting ‘leading edge hypothesis' proposed by Alexandri et al. (2012) and Veličković et al. (2015). In detail, structure analysis allowed us to define 14 wild boar subpopulations, among which we found both isolated, genetically distinct subpopulations and admixed subpopulations (for example, Alps, Central South Iberia, Peridinaric Balkans). A possible explanation for the admixed nature of Italian Alps subpopulation can be the historical records reporting the recolonization of North-western Italy by wild boars coming from France since 1919 (De Beaux and Festa, 1927).
We identified subpopulations that exhibited gene flow with Central-eastern European subpopulations, whereas others are exclusively peninsular and nonadmixed. This is the case of some Italian subpopulations, such as Castelporziano and Sardinia, that are well known for their distinctive genetic assembly, such as the presence of E2 mitochondrial DNA clade haplotypes (Scandura et al., 2008; Vilaça et al., 2014). Similar pattern was revealed in our study for two subpopulations on the Balkans (Littoral Slovenia and Dinaric Balkans) and two on Iberia (South Portugal and North-western Iberia) that had not before been identified as distinct genetic clusters.
These patterns are also consistent with the detected migration flows. As expected, according to the leading edge hypothesis, more intense relative migration flows were detected within less peripheral, more admixed, peninsular and central European subpopulations. However, there were no significant relative migration flows within these subpopulations. On the other hand, subpopulations like Littoral Slovenia, Castelporziano, Sardinia or South Portugal were systematically identified as source populations for less peripheral subpopulations, although these latter subpopulations, more closely related with continental and other peninsular subpopulations, appear to function as sink populations. This source–sink dynamics occurred frequently among regions, and more rarely within peninsulas.
These results appear counterintuitive, if we assume that the net gene flow coming from the refugial subpopulations is contemporary. Namely, Sardinia is an island and Castelporziano is a fenced subpopulation. However, we suspect that this pattern probably reflects historical contributions related with the Last Glacial Maximum (LGM) and is consistent with the leading edge hypothesis of wild boar recolonization of Europe (Alexandri et al., 2012; Veličković et al., 2015). No significant directional migration flows were detected among continental and nonrefugial peninsular subpopulations because migration, within these subpopulations, should have been essentially symmetric. This should be true, even if the flows were asynchronous: a first migration flow from north to south, towards the peninsulas, and then backwards, towards continental Europe, as suggested by Veličković et al. (2015). If the hypothesis advanced by these authors is correct, more peripheral (refugial) peninsular subpopulations should have had a minor role in these migratory exchanges. Nevertheless, the results here presented suggest that even these refugial subpopulations were net contributors to the contemporary gene pool of other peninsular and continental wild boar subpopulations. The paradigm of postglacial colonization patterns has been challenged in various species based on wider sampling across refugial areas, and this is particularly the case in Iberia and the Balkans. Instead of regarding them as continuous and homogenous refugial areas, each of those peninsulas seems to contain genetically diverse groups of populations, leading to a more recent ‘refugia within refugia' concept (Gomez and Lunt, 2007; Feliner, 2011).
A genetic snapshot of wild boar subpopulations in the Balkans and Europe
Because of the presence of distinct subpopulations, the Balkans and other southern European peninsulas seem to harbour a greater extent of wild boar genetic diversity in Europe. However, these subpopulations are not necessarily more diverse per se. In fact, these relict subpopulations are characterized by lower numbers of alleles, allelic richness, private allelic richness and levels of expected heterozygosity compared with less peripheral subpopulations. This is probably a consequence of the admixture process itself, as admixed subpopulations are influenced by different gene pools. As expected, levels of linkage disequilibrium and Hardy–Weinberg deviations were also higher in central and eastern European subpopulations and peninsular subpopulations of admixed origin. We did not find similar patterns for the levels of inbreeding that varied almost 10-fold among southern peninsular subpopulations. The significantly high FIS values may also indicate the excess of homozygosity, not only related to inbreeding, but also as a result of admixture of different gene pools that is shown for Alps and Central South Iberia subpopulations. The estimated effective population size was higher than harmonic mean sample size for all subpopulations, except Alps that can be consequence of deviation from linkage disequilibrium observed in this subpopulation.
Evidence of bottlenecks was biased towards southern subpopulations, with Sardinia, Littoral Slovenia, Castelporziano, South Portugal and North-western Iberia presenting significant evidence of bottlenecks. This evidence came solely from the Garza and Williamson (2001) approach, suggesting that we might be looking at older and longer bottlenecks (Williamson-Natesan, 2005). We could be tempted to interpret these results as an evidence of bottlenecks occurring during the LGM (Veličković et al., 2015). However, two arguments challenge this interpretation: (1) in this case, we should expect the opposite pattern: peninsular subpopulations would be less affected by LGM population bottlenecks than subpopulations from higher latitudes; (2) M ratio should recover almost completely before 500 generations had passed after the bottleneck, and hence this method would not be able to detect bottlenecks >1000 years old (Garza and Williamson, 2001). A different interpretation is that the observed patterns are a consequence of more recent bottlenecks, like those resulting from generalized overhunting across Europe during the nineteenth and early twentieth centuries (Fonseca, 2004; Ferreira et al., 2009; Apollonio et al., 2010; Scandura et al., 2011a). Indeed, the effects of these bottlenecks should be more evident in smaller populations, occupying smaller geographic ranges, that would be more vulnerable to stochastic events and genetic drift than larger and more widespread populations.
The significant evidence of heterozygosity deficiency found in several subpopulations (Figure 4) might be an evidence of recent population expansions (Cornuet and Luikart, 1996). This decline followed by expansion is actually documented for Castelporziano (Imperio et al., 2010) and South Portugal (Vingada et al., 2010) subpopulations. Despite prior evidence of a global wild boar population expansion across Europe after the LGM (Veličković et al., 2015), we suspect that the signal for population expansions detected here is linked to much more recent events, such as the expansion occurring across Europe in recent decades (Massei et al., 2015). It is therefore possible that wild boar in Europe has gone through two large and widespread bottlenecks: an historical one at the LGM (Veličković et al., 2015), and a more recent one probably related with generalized overhunt in Europe, as evidenced here, through the use of fast-evolving microsatellite markers.
Management units and cross-border management of wild boars in Europe
This study highlights two important issues about wild boar conservation and management: (1) European wild boars are divided into several subpopulations; (2) the distribution range of several subpopulations crosses national borders, particularly in the Balkans where most of the subpopulations are distributed across borders.
The identification of different biological populations is highly relevant for defining conservation or management units. Population or disease monitoring and control and culling plans by local hunting associations or forestry services have better chances of success if applied to biological rather than administrative units. As an example, the three genetic clusters previously identified in Portugal (Ferreira et al., 2009) show very good agreement with the division of wild boar into three distinct management units based on ecological differences (that is, phenology) (Fonseca, 2004). According to these authors, more effective management would be achieved if culling plans and hunting seasons were defined at regional level rather than at a national level.
Integrating national approaches into cross-border management is one of the major current conservation challenges (Fonseca et al., 2014). We provide a first insight into potential cross-border wild boar's management units across Europe. For what concern the Balkans, we found five genetic clusters, one of which (Continental Balkans and Eastern Europe) includes territory in five countries: Serbia, Croatia, Slovenia, Romania and Slovakia. The ranges of another two Balkan subpopulations also span different countries: (1) the Dinaric Balkans subpopulation includes wild boars from Bosnia and Herzegovina and Montenegro; (2) the South Central Balkans subpopulation includes wild boars from the southern part of Serbia and from FYR Macedonia. Thus, the specific emphasis should be put on transboundary management in this region and should involve the authorities of different countries. The Littoral Slovenia is clearly divergent from other wild boars collected in Slovenia and, at the higher level of structuring, it is more related to Italian subpopulations (Castelporziano and North-western Italy; Table 1 and Figure 2). The wild boars from this region are also phenotypically different. These boars have been characterized as having ‘small boars' because of low body weight during the entire lifecycle, and accordingly investigations were probably introduced into this part of Slovenia from Italy (Sila and Koren, 2010). In addition, there are evidences of natural colonization of Western Italian Alps from wild boars coming from Slovenia in the 1950s and 1960s of the past century (Monaco et al., 2007). Thus, it should be important that future management of Slovenian and Italian subpopulations include authorities of both countries.
Can humans manage wild boars or do we need another glaciation?
Recent studies are unanimous in stating that human actions might have a strong impact on the demography but a limited impact on the genetic make-up of the European wild boar (Scandura et al., 2008, 2011b; Vilaça et al., 2014; Veličković et al., 2015), despite the existing evidence of introgression of domestic gene pool into wild boar populations (Koutsogiannouli et al., 2010; Goedbloed et al., 2013) and from wild boar into domestic stocks (Frantz et al., 2015). There is a growing body of evidence that patterns of genetic diversity and structure of wild boar in Europe have mostly been shaped by Quaternary climate fluctuations (Scandura et al., 2008; Vilaça et al., 2014; Veličković et al., 2015). This does not mean that human actions, such as long-distance translocations, overhunting or hybridization with domestic pig breeds, do not interfere with the gene pools of European wild boar subpopulations. However, these actions seem to be relevant mostly on a local scale. On the other hand, the evidence of recent bottleneck reported here is probably related with human-mediated and widespread population decline across Europe, in the beginning of the past century. However, the structure and migration patterns of wild boar populations are consistent with a ‘refugia within refugia' theory (Gomez and Lunt, 2007; Feliner, 2011) and, this way, preserve the genetic signature of more ancient events.
The population genetic structure and genetic diversity of wild boars in Europe we detail here provides unique information for the development of management strategies aimed to maintain the highest possible level of genetic variation across the species distribution. Particular attention should be paid to locally isolated subpopulations such as Castelporziano, Sardinia, Littoral Slovenia, North-western Iberia and South Portugal. Smaller, less diverse and peripheral subpopulations are probably more susceptible to the effect of maladaptative hybridization with domestic stocks (Lowe et al., 2015). In conclusion, each defined subpopulation should be managed in an integrative way, embedded on the concept of cross-border management plans. Despite the limited overall impact confirmed by the present study, wild boar management in Europe should avoid translocations, because of not only genetic concerns, but also sanitary concerns as wild boar is a well-known tuberculosis vector and carrier of other infectious diseases.
Data archiving
The data used in the present study available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.jm090. The deposit file contains microsatellite genotypes for each sample.
Acknowledgments
This work was financially supported by the Ministry of Education, Science and Technological Development, Republic of Serbia (Grant No. 43002), the Provincial Secretariat for Science and Technological Development (Grant No. 114-457-2173/2011-01) and the Provincial Secretariat for Agriculture, Water Management and Forestry Project ‘The assessment of genetic potential of wild boar populations in hunting areas in Vojvodina'. The study was also conducted under Project No. TA02031259 of the Technology Agency of the Czech Republic. We thank all those who helped us in collecting samples: Mladen Gagrčin, Zoran Rašić, Dragan Rogožarski, the public company ‘Vojvodinašume' Novi Sad (Serbia); Vladimir Maletić (FYR Macedonia); Matija Stergar (Slovenia); Ranko Savić, Darko Cvijović, Veterinary Ambulance Nikšić (Montenegro); Albert Marinculić (Croatia); Ivona Panić (Bosnia and Herzegovina); Aderbal Cavalcante-Neto (Brasil); Juan Carranza (Spain); Luca Aicardo, Barbara Franzetti, Aurelio Perrone, Andrea Marsan, Massimo Bellavita, Luca Cimino (Italy); and all the Portuguese and Balkan biologists, veterinarians, hunters and hunter associations that collected samples. We thank Carlo Robino for providing us with full genotypes for wild boar populations from North-western Italy. Nevena Veličković was supported by a FEBS Collaborative Experimental Scholarship for Central and Eastern Europe. Eduardo Ferreira was supported by the Fundação para a Ciência e Tecnologia, POPH and European Union (Grant SFRH/BPD/72895/2010). We thank University of Aveiro (Department of Biology) and FCT/MEC for the financial support to CESAM RU (UID/AMB/50017) through national funds co-financed by the FEDER, within the PT2020 Partnership Agreement. We thank John O'Brien for proofreading the manuscript.
Author contributions
NV, MD, DOV, EF and CF devised the sampling strategy; NV, CF, AM and ME performed the sampling; NV, MD, EF and CF conceived and designed the study; NV and EF performed laboratory work; NV, MD and EF performed data analysis; NV, MD, EF, AM and CF contributed to discussions during the preparation of the manuscript; NV, MD and EF wrote the manuscript. All authors read, commented on and approved the final version of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Alcala A, Goudet J, Vuilleumier S. (2014). On the transition of genetic differentiation from isolation to panmixia: what we can learn from GST and D. Theor Pop Biol 93: 75–84. [DOI] [PubMed] [Google Scholar]
- Alexandri P, Triantafyllidis A, Papakostas S, Chatzinikos E, Platis P, Papageorgiou N et al. (2012). The Balkans and the colonization of Europe: the post-glacial range expansion of the wild boar, Sus scrofa. J Biogeogr 39: 713–723. [Google Scholar]
- Alves PC, Pinheiro I, Godinho R, Vicente J, Gortázar C, Scandura M. (2010). Genetic diversity of wild boar populations and domestic pig breeds (Sus scrofa in Southwestern Europe. Biol J Linn Soc 101: 797–822. [Google Scholar]
- Apollonio M, Andersen R, Putman R. (2010) European Ungulates and Their Management in the 21st Century. Cambridge University Press: Cambridge, UK. [Google Scholar]
- Apollonio M, Randi E, Toso S. (1988). The systematics of the wild boar (Sus scrofa L.) in Italy. Boll Zool 55: 213–221. [Google Scholar]
- Bruford MW, Hanotte O, Brookfield JFY, Burke T (1992). Single-locus and multilocus DNA fingerprint. In: Hoelzel AR (ed). Molecular Genetic Analysis of Populations, a Practical Approach. IRL Press: Oxford. pp 225–270. [Google Scholar]
- Caratti S, Rossi L, Sona B, Origlia S, Viara S, Martano G et al. (2010). Analysis of 11 tetrameric STRs in wild boars for forensic purposes. Forensic Sci Int Genet 4: 339–342. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G. (1996). Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beaux O, Festa E. (1927). La ricomparsa del cinghiale nell'Italia settentrionale occidentale. Mem Soc It Sc Nat Mus Civ S.N. Milano 9: 265–342. [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. (2002). A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11: 2571–2581. [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliner GN. (2011). Southern European glacial refugia: a tale of tales. Taxon 60: 365–372. [Google Scholar]
- Ferreira E, Souto L, Soares AMVM, Fonseca C. (2006). Genetic structure of the wild boar (Sus scrofa L.) population in Portugal. Wildl Biol Pract 2: 17–25. [Google Scholar]
- Ferreira E, Souto L, Soares AMVM, Fonseca C. (2009). Genetic structure of the wild boar population in Portugal: evidence of a recent bottleneck. Mamm Biol 74: 274–285. [Google Scholar]
- Fonseca C. (2004) Population dynamics and management of wild boar (Sus scrofa L.) in Central Portugal and Southeastern Poland. PhD thesis University of Aveiro: Portugal. [Google Scholar]
- Fonseca C, Torres RT, Santos JPV, Vingada J, Apollonio M (2014). Challenges in the management of cross-border populations of ungulates. In: Putman R, Apollonio M (eds). Behaviour and Management of European Ungulates. Whittles Publishing: Scotland, UK. [Google Scholar]
- Ford MJ. (2002). Applications of selective neutrality tests to molecular ecology. Mol Ecol 11: 1245–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank BF, Monaco A, Bath AJ. (2015). Beyond standard wildlife management: a pathway to encompass human dimension findings in wild boar management. Eur J Wildl Res 61: 723–730. [Google Scholar]
- Frantz L, Meijaard E, Gongora J, Haile J, Groenen MAM, Larson G. (2016). The evolution of suidae. Annu Rev Anim Biosci 4: 61–85. [DOI] [PubMed] [Google Scholar]
- Frantz LAF, Schraiber JG, Madsen O, Megens H-J, Cagan A, Bosse M et al. (2015). Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet 47: 1141–1148. [DOI] [PubMed] [Google Scholar]
- Garza JC, Williamson EG. (2001). Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10: 305–318. [DOI] [PubMed] [Google Scholar]
- Goedbloed DJ, Megens HJ, Van Hooft P, Herrero-Medrano JM, Lutz W, Alexandri P et al. (2013). Genome-wide single nucleotide polymorphism analysis reveals recent genetic introgression from domestic pigs into Northwest European wild boar populations. Mol Ecol 22: 856–866. [DOI] [PubMed] [Google Scholar]
- Gomez A, Lunt DH (2007). Refugia within refugia: patterns of phylogeographic concordance in the Iberian Peninsula. In: Weiss S, Ferrand N (eds). Phylogeography of Southern European Refugia. Springer: Dordrecht. pp 155–188. [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. (2009). Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacolina L, Scandura M, Goedbloed DJ, Alexandri P, Crooijmans RPMA, Larson G et al. (2016). Genomic diversity and differentiation of a managed island wild boar population. Heredity 116: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperio S, Ferrante M, Grignetti A, Santini G, Focardi S. (2010). Investigating population dynamics in ungulates: do hunting statistics make up a good index of population abundance? Wildlife Biol 16: 205–214. [Google Scholar]
- Jost L. (2008). GST and its relatives do not measure differentiation. Mol Ecol 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA. (2013). diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Eco Evol 4: 782–788. [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. (2015). Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15: 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsogiannouli EA, Moutou KA, Sarafidou T, Stamatis C, Mamuris Z. (2010). Detection of hybrids between wild boars (Sus scrofa scrofa and domestic pigs (Sus scrofa f. domestica in Greece, using the PCR-RFLP method on melanocortin-1 receptor (MC1R) mutations. Mamm Biol 75: 69–73. [Google Scholar]
- Kusza S, Podgórski T, Scandura M, Borowik T, Jávor A, Sidorovich VE et al. (2014). Contemporary genetic structure, phylogeography and past demographic processes of wild boar Sus scrofa population in Central and Eastern Europe. PLoS One 9: e91401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnel J, Zachos F (2007). Status and distribution patterns of European ungulates: genetics, population history and conservation. In: Putman R, Apollonio M, Andersen R (eds). Ungulate Management in Europe: Problems and Practices. Cambridge University Press: Cambridge, UK. pp 12–53. [Google Scholar]
- Lowe WH, Muhlfeld CC, Allendorf FW. (2015). Spatial sorting promotes the spread of maladaptive hybridization. Trends Ecol Evol 30: 456–462. [DOI] [PubMed] [Google Scholar]
- Mantel N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220. [PubMed] [Google Scholar]
- Massei G, Genov P. (2000) Il Cinghiale. Calderini Edagricole: Bologna. [Google Scholar]
- Massei G, Kindberg J, Licoppe A, Gačić D, Šprem N, Kamler J et al. (2015). Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Manag Sci 71: 492–500. [DOI] [PubMed] [Google Scholar]
- Monaco A, Carnevali L, Riga F, Toso S (2007). Il cinghiale sull'arco alpino: status e gestione delle popolazioni. In: Hauffe HC, Crestanello B, Monaco A (eds). Il Cinghiale Sull'arco Alpino: Status e Gestione. Report no. 38 del Centro di Ecologia Alpina: Torino, Italy. pp 5–24. [Google Scholar]
- Nei M. (1973). Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Tajima F. (1981). Genetic drift and estimation of effective population size. Genetics 98: 625–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov IS, Gum B, Markov G, Kuehn R. (2009). Population genetic structure of wild boar Sus scrofa in Bulgaria as revealed by microsatellite analysis. Acta Theriol 54: 193–205. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2007). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available at http://www.R-project.org/. [Google Scholar]
- Rice WR. (1989). Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Saez-Royuela C, Telleria JL. (1986). The increased population of the wild boar (Sus scrofa L.) in Europe. Mamm Rev 16: 97–101. [Google Scholar]
- Sambrook JF, Russell DW. (2001) Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY. [Google Scholar]
- Scandura M, Iacolina L, Apollonio M. (2011. a). Genetic diversity in the European wild boar Sus scrofa: phylogeography, population structure and wild x domestic hybridization. Mamm Rev 41: 125–137. [Google Scholar]
- Scandura M, Iacolina L, Cossu A, Apollonio M. (2011. b). Effects of human perturbation on the genetic make-up of an island population: the case of the Sardinian wild boar. Heredity 106: 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandura M, Iacolina L, Crestanello B, Pecchioli E, di Benedetto MF, Russo V et al. (2008). Ancient vs. recent processes as factors shaping genetic variation of the European wild boar: are the effects of the last glaciation still detectable? Mol Ecol 17: 1745–1762. [DOI] [PubMed] [Google Scholar]
- Shafer ABA, Wolf JBW, Alves PC, Bergström L, Bruford MW et al. (2015). Genomics and the challenging translation into conservation practice. Trends Ecol Evol 30: 78–87. [DOI] [PubMed] [Google Scholar]
- Sila A, Koren I. (2010) Wild boar on the western border - the problems of the population and managing it. 2nd Slovenian-Croatian consultations on the management of wild animals: wild boar: Velenje, Slovenia. 17–18 September 2010. pp 72–76. [Google Scholar]
- Slatkin M. (1993). Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47: 264–279. [DOI] [PubMed] [Google Scholar]
- Sprem N, Safner T, Treer T, Florijancic T, Jurić J, Cubric-Curic V et al. (2016). Are the dinaric mountains a boundary between continental and mediterranean wild boar populations in Croatia? Eur J Wildl Res 62: 167–177. [Google Scholar]
- Sundqvist L, Keenan K, Zackrisson M, Prodöhl P, Kleinhans D. (2016). Directional genetic differentiation and relative migration. Ecol Evol 6: 3461–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. (2008). ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. (2004). Micro-checker: a software for identifying and correcting genotyping errors in microsatellite data. Mol Eco Notes 4: 535–538. [Google Scholar]
- Veličković N, Djan M, Ferreira E, Stergar M, Obreht D, Maletić V et al. (2015). From north to south and back: the role of the Balkans and other southern peninsulas in the recolonization of Europe by wild boar. J Biogeogr 42: 716–728. [Google Scholar]
- Veličković N, Djan M, Obreht D, Vapa L. (2012). Population genetic structure of wild boars in the West Balkan region. Russ J Genet 48: 859–863. [PubMed] [Google Scholar]
- Vernesi C, Crestanello B, Pecchioli E, Tartari D, Caramelli D, Hauffe H et al. (2003). The genetic impact of demographic decline and reintroduction in the wild boar (Sus scrofa: a microsatellite analysis. Mol Ecol 12: 585–595. [DOI] [PubMed] [Google Scholar]
- Vilaça ST, Biosa D, Zachos F, Iacolina L, Kirschning J, Alves PC et al. (2014). Mitochondrial phylogeography of the European wild boar: the effect of climate on genetic diversity and spatial lineage sorting across Europe. J Biogeogr 41: 987–998. [Google Scholar]
- Vingada J, Fonseca C, Cancela J, Ferreira J, Eira C (2007). Ungulates and their management in Portugal. In: Apollonio M, Andersen R, Putman RJ (eds). European Ungulates and Their Management in the 21st Century. Cambridge University Press: Cambridge, UK. pp 392–418. [Google Scholar]
- Waples RS, Do C. (2008). LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Eco Res 8: 753–756. [DOI] [PubMed] [Google Scholar]
- Williamson-Natesan EG. (2005). Comparison of methods for detecting bottlenecks from microsatellite loci. Conserv Genet 6: 551–562. [Google Scholar]
- Wright S (2007). Evolution and the Genetics of Populations. Vol. 4Variability Within and Among Natural Populations. University of Chicago Press: Chicago. [Google Scholar]
- Yue GH, Beeckmann P, Geldermann H. (2002). Mutation rates at swine microsatellite loci. Genetica 114: 113–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.