Abstract
Identifying phylogenetically distinct lineages and understanding the evolutionary processes by which they have arisen are important goals of phylogeography. This information can also help define conservation units in endangered species. Such analyses are being transformed by the availability of genomic-scale data sets and novel analytical approaches for statistically comparing different historical scenarios as causes of phylogeographic patterns. Here, we use genomic-scale restriction-site-associated DNA sequencing (RADseq) data to test for distinct lineages in the endangered Eastern Massasauga Rattlesnake (Sistrurus catenatus). We then use coalescent-based modeling techniques to identify the evolutionary mechanisms responsible for the origin of the lineages in this species. We find equivocal evidence for distinct phylogenetic lineages within S. catenatus east of the Mississippi River, but strong support for a previously unrecognized lineage on the western edge of the range of this snake, represented by populations from Iowa, USA. Snakes from these populations show patterns of genetic admixture with a nearby non-threatened sister species (Sistrurus tergeminus). Tests of historical demographic models support the hypothesis that the genetic distinctiveness of Iowa snakes is due to a combination of isolation and historical introgression between S. catenatus and S. tergeminus. Our work provides an example of how model-based analysis of genomic-scale data can help identify conservation units in rare species.
Introduction
The capacity to produce genomic-scale data sets from nonmodel organisms has transformed our ability to characterize patterns of genetic diversity within species and understand the historical processes that generate such patterns (Ekblom and Galindo, 2011). High-throughput sequencing technologies, novel cost-effective methods for constructing reduced representation libraries (that is, restriction site-associated DNA sequencing (RADseq); Davey and Blaxter, 2011; Davey et al., 2011) and the development of statistical approaches that allow historical demographic analyses of these data (that is, Gutenkunst et al., 2009; Excoffier et al., 2013; Leaché et al., 2014) are important factors contributing to this transformation. These novel genomic-scale data sets can reveal cryptic genetic diversity within recognized taxa, estimate demographic parameters characterizing populations and identify important recent and historical events that have shaped existing genetic diversity, such as hybridization between lineages (Luikart et al., 2003; Ellegren, 2013). Such information can inform decisions about how to designate conservation units in rare taxa and, as a result, genomic-scale analyses now play an important role in conservation genetic research (Avise, 2010; Funk et al., 2012; Narum et al., 2013).
The Eastern Massasauga Rattlesnake (Sistrurus catenatus) is a snake of conservation concern that occurs in small, isolated populations across eastern North America. It is designated as threatened or endangered in every US state and Canadian province in which it occurs (Szymanski, 1998), and is currently being considered for listing as a federally endangered species under the Endangered Species Act in the United States (United States Fish and Wildlife Service, 2015). Genetic studies to date have drawn different conclusions as to whether distinct phylogenetic lineages that would represent evolutionary significant units under the Endangered Species Act occur in this species. For example, Kubatko et al. (2011) found no evidence for phylogeographic structure based on a multilocus analysis of 20 nuclear DNA markers, although the number and geographic range of samples analyzed was limited. In contrast, Ray et al. (2013) analyzed a much larger and more widespread set of samples with a single mitochondrial DNA (mtDNA) locus and identified three geographically separate mtDNA lineages (Western, Central and Eastern). These designations are currently being considered for use in a captive breeding program for this species (Ray et al., 2013). However, given that this inference is based on a single mtDNA locus, it is unclear whether this pattern represents variation across the genome as a whole, and hence whether it provides an accurate representation of the phylogenetic history of this species.
Both of the studies highlighted above were focused on inferring patterns of phylogenetic diversity within S. catenatus. However, recent analytical advances that use the site frequency spectrum (SFS) generated from genomic-scale data sets provide opportunities to also assess the evolutionary and ecological mechanisms that generate observed phylogeographic patterns (Gutenkunst et al., 2009; Excoffier et al., 2013). In this study, we use a genomic-scale single-nucleotide polymorphism (SNP) data set that includes samples collected from across the range of S. catenatus to reexamine whether important phylogenetic lineages are present within this species. We do this by first using clustering algorithms to identify major genetic groups within S. catenatus, and then use species tree-based phylogenetic methods to test for significant phylogenetic differentiation between these groups. We then apply recently described likelihood-based methods that use the SFS to test various historical demographic scenarios as the cause of the observed patterns of diversity, and to estimate associated demographic parameters. We discuss the implications of our results in terms of how they contribute to the designation of conservation units within S. catenatus.
Materials and methods
Samples and sequencing methods
We obtained blood or tissue samples for S. catenatus individuals from 35 populations spanning the geographic range of this species (N=1–13 per population), and for an additional eight Sistrurus tergeminus individuals (Figure 1 and Table 1). We also obtained one Sistrurus miliarius sample that was used to polarize SNPs when generating unfolded SFSs. Genomic DNA was extracted using either DNA Blood and Tissue Kits (Qiagen, Valencia, CA, USA) or phenol–chloroform. Double-digest RADseq libraries (Peterson et al., 2012) were generated with EcoRI and SbfI restriction enzymes (New England Biolabs, Ipswich, MA, USA) and a modified version of the RADseq protocol described in DaCosta and Sorenson (2014). Details of the library prep methods are included in the Supplementary Information. Pooled libraries of up to 36 individuals were run in single-end 50 bp runs on either entire lanes of an Illumina GAII (San Diego, CA, USA) sequencer or as partial lanes (ranging from 10 to 25%, depending on the number of individuals in the specific library) on an Illumina HiSeq 2500 sequencer.
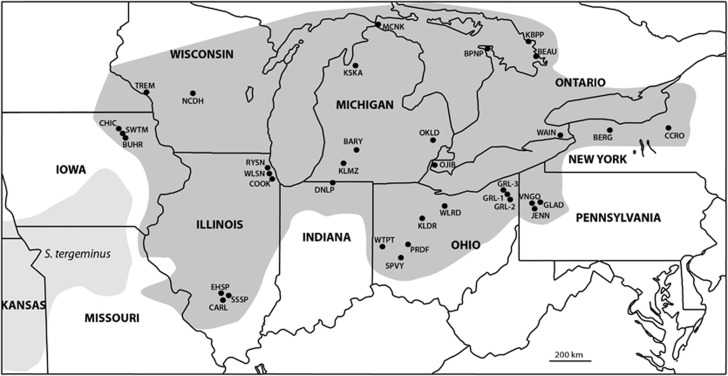
Figure 1.
Map of sampling locations for S. catenatus samples (darker shading) and S. tergeminus samples (lighter shading). Populations are indicated with smaller bold font, and state or province names are indicated with larger bold font. See Table 1 for population abbreviations and sample sizes.
Table 1. Sampling locations with location codes for Sistrurus catenatus and Sistrurus tergeminus included in the study.
| Location | N, total | N, clustering | N, fastsimcoal |
|---|---|---|---|
| Illinois | |||
| Carlyle Lake (CARL) | 3 | 3 | |
| Cook County (COOK) | 1 | 1 | |
| Eldon Hazlet State Park (EHSP) | 2 | 2 | |
| Ryerson (RYSN) | 2 | 2 | |
| South Shore State Park (SSSP) | 3 | 3 | |
| Willow Sanders (WLSN) | 2 | 2 | |
| Indiana | |||
| Dunlap (DNLP) | 3 | 3 | |
| Iowa | |||
| Buhr Access (BUHR) | 13 | 3 | 13 |
| Chickasaw (CKSW) | 2 | 2 | |
| Sweet Marsh (SWTM) | 1 | 1 | |
| Kansas | |||
| Ellsworth County (TERG) | 4 | 2 | 4 |
| Russell County (TERG) | 1 | 1 | |
| Michigan | |||
| Barry (BARY) | 2 | 2 | |
| Kalamazoo (KMZO) | 1 | 1 | |
| Kalkaska (KSKA) | 2 | 2 | |
| Mackinac Island (MKNC) | 1 | 1 | |
| Oakland (OKLD) | 2 | 2 | |
| Missouri | |||
| Pershing State Park (TERG) | 1 | 1 | |
| Squaw Creek NWR (TERG) | 2 | 2 | |
| New York | |||
| Bergen (BERG) | 3 | 3 | |
| Cicero (CCRO) | 3 | 3 | |
| Ohio | |||
| Grand River Lowlands Site 1 (GRL-1) | 3 | 3 | |
| Grand River Lowlands Site 2 (GRL-2) | 3 | 3 | |
| Grand River Lowlands Site 3 (GRL-3) | 3 | 3 | |
| Killdeer Plains (KLDR) | 13 | 3 | 13 |
| Prairie Road Fen (PRDF) | 3 | 3 | |
| Spring Valley (SPVY) | 3 | 3 | |
| Willard (WLRD) | 3 | 3 | |
| Wright Patterson AFB (WTPT) | 2 | 2 | |
| Ontario | |||
| Beausoleil Island (BEAU) | 3 | 3 | |
| Bruce Peninsula (BPNP) | 3 | 3 | |
| Killbear Provincial Park (KBPP) | 3 | 3 | |
| Ojibway (OJIB) | 2 | 2 | |
| Wainfleet Bog (WAIN) | 3 | 3 | |
| Pennsylvania | |||
| State Game Lands 95 (GLAD) | 3 | 3 | |
| Jennings (JENN) | 3 | 3 | |
| Venango (VNGO) | 3 | 3 | |
| Wisconsin | |||
| Necedah (NCDH) | 1 | 1 | |
| Trempelau (TREM) | 2 | 2 | |
Total sample sizes from each location, along with sample sizes for the clustering and fastsimcoal analyses, are included. Data were also generated for one additional S. miliarus sample that served as an outgroup to generate an unfolded site frequency spectrum for fastsimcoal analyses.
Bioinformatic methods
De novo locus assembly, SNP identification and genotyping were carried out on the raw fastq data using AftrRAD version 4.1 (Sovic et al., 2015) that efficiently assembles RADseq loci while accounting for indel variation. Two separate AftrRAD analyses were performed. The first analysis generated data that were used for the clustering and species tree analyses, and included a maximum of three individuals per population (see Table 1). Based on results from these analyses, we performed a second AftrRAD run to generate a separate data set for demographic modeling analyses. This run only included S. catenatus samples from Buhr Access, Iowa and Killdeer Plains, Ohio (N=13 each), samples from S. tergeminus and one S. miliarius individual that was used to polarize SNPs when building an unfolded SFS. Details on the parameter settings for these AftrRAD runs are provided in the Supplementary Information.
Genetic clustering analyses
We performed clustering analyses on the genetic data using two programs: Structure (Pritchard et al., 2000) and adegenet (Jombart and Ahmed, 2011). The purpose of these analyses was to identify major genetic groups that could be subsequently tested for distinctiveness with phylogenetic methods. In both analyses we used a maximum of three samples per population to reduce possible effects of unequal sample sizes on clustering results (Kalinowski, 2011). Structure uses a Bayesian approach to cluster samples into groups that most closely meet Hardy–Weinberg equilibrium expectations, and runs were performed for K values ranging from 1 to 6. Three independent runs were completed for each K value, with 2 × 105 Markov chain Monte Carlo iterations, plus a 10% burn-in period, per run. Optimal K values were identified using the ΔK method of Evanno et al. (2005) as calculated using Structure Harvester (Earl and Vonholdt, 2012).
We also identified genetic clusters using the find.clusters function in adegenet 1.3–9.2 (Jombart and Ahmed, 2011) that uses a model-free K-means clustering approach to identify distinct groups and, thus, unlike Structure, does not rely on assumptions of Hardy–Weinberg equilibrium and linkage equilibrium within clusters. As for the Structure runs, optimal clustering solutions were generated for K values ranging from 1 to 6, with clustering solutions at each k value evaluated based on Bayesian information criterion scores. Finally, to summarize how variation is partitioned among the identified groups, we performed a discriminant analysis of principal components (Jombart et al., 2010) based on the optimal clustering solution identified in adegenet, and used Arlequin v3.5 (Excoffier and Lischer, 2010) to calculate Fst values between each identified cluster.
Species tree analyses
Given the results from the clustering analyses described above, we used two approaches to test for significant phylogenetic differentiation among the major genetic groups identified within S. catenatus. First, we performed species tree phylogenetic analyses using SNAPP (Bryant et al., 2012). Because the computational demands of SNAPP depend on the number of individuals included in the analysis (Bryant et al., 2012), we used a subsampling approach similar to that used by Harvey and Brumfield (2015) in which we randomly selected three individuals (six chromosomes) to represent each of the groups identified in the clustering analyses (see Results below), and used a script available in AftrRAD to generate a SNAPP data set that consisted of unlinked biallelic SNPs for these samples. We repeated this 10 times to capture any variation associated with subsampling individuals, and performed SNAPP runs on each of the 10 data sets. The sister species S. tergeminus was included as an outgroup in all runs to root the trees. For all subsampled data sets, we generated maximum clade credibility consensus trees with TreeAnnotator (http://beast.bio.ed.ac.uk/treeannotator), and from these we obtained clade support values and 95% highest probability densities for all branch lengths separating taxa. Mutation rates u and v were sampled as part of the analysis, and default priors as set in Beauti 2 were used for the remaining run parameters. All SNAPP runs were carried out for a minimum of 5 × 106 Markov chain Monte Carlo iterations, with the first 1 × 106 discarded as burn-in, and samples collected every 1000 trees. Convergence of runs was evaluated with Tracer (Drummond and Rambaut, 2007; http://tree.bio.ed.ac.uk/software/tracer/), and only runs for which all ESS values exceeded 100 and no trends were observed in traces for individual parameters were retained for analyses.
In addition to the SNAPP runs described above, we performed a species delimitation analysis as implemented in BFD* (Leaché et al., 2014) to further test for phylogenetic differentiation among genetic clusters. Because BFD* relies on SNAPP, this analysis was subject to similar computational demands as those described above. Therefore, we again chose three individuals to represent each group identified in our clustering analyses (S. tergeminus, Iowa, West and East—see clustering results below), and also included three samples to represent the Central group proposed by Ray et al. (2013) based on previous mtDNA analyses (these ‘Central' samples were included as part of the Eastern group in the current analyses). These samples were used to test alternative models that ranged from a fully differentiated five-lineage model to a two-lineage model in which S. tergeminus and S. catenatus were included as the only two recognized taxa. Five independent runs of BFD* were performed, each with a different subset of individuals to represent each lineage. For each run, models were ranked based on their marginal likelihood estimates, and Bayes Factors were calculated and interpreted in the context of the recommendations provided by Kass and Raferty (1995).
Demographic model testing
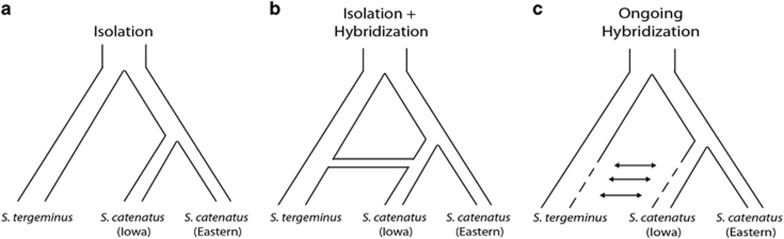
Our analyses (see below) detected a phylogenetically distinct lineage within S. catenatus (from Iowa, USA) that, based on clustering analyses, demonstrated a pattern of genetic admixture with S. tergeminus. To evaluate the historical processes that may have given rise to the pattern of admixture, we tested a set of models (Figure 2) that differed based on the occurrence and timing of hybridization between S. tergeminus and the S. catenatus lineage represented by our samples from Iowa. The three models include: (1) a model in which the Iowa lineage was formed because of isolation in allopatry (‘Isolation', Figure 2a); (2) a model in which the Iowa lineage initially diverges in allopatry, with a subsequent hybridization event between this lineage and S. tergeminus (‘Isolation+Hybridization', Figure 2b); and (3) a model in which hybridization currently occurs between the Iowa lineage and S. tergeminus, and continues back in time for some duration that is bounded by the divergence time of the Iowa lineage (‘Ongoing Hybridization', Figure 2c). For all models, we used the population from Killdeer Plains, Ohio, as representative of S. catenatus east of the Mississippi River. This population represents one of the largest and most stable populations of S. catenatus (Chiucchi and Gibbs, 2010) and therefore likely better represents the genetic diversity characteristic of this group than would smaller populations more strongly affected by genetic drift. Each model is described in more detail in the Supplementary Information.
Figure 2.
Representation of the three models (Isolation, Isolation+Hybridization, and Ongoing Hybridization, a–c respectively) compared in fastsimcoal to evaluate the historical process giving rise to the pattern of admixture observed between S. catenatus samples from Iowa and the sister taxon S. tergeminus. These models differ in the occurrence and/or timing of hybridization between these lineages. Nearly all of the model weight from Akaike information criterion (AIC) analysis was attributed to the Isolation+Hybridization model (b).
A multidimensional, unfolded SFS was generated with AftrRAD for model testing and parameter estimation, and included three groups: S. tergeminus from Ellsworth County, Kansas (N=4), S. catenatus from Buhr Access, Iowa (N=13) and S. catenatus from Killdeer Plains, Ohio (N=13). A maximum of one SNP was included in the SFS from each locus (unlinked-1 flag in AftrRAD). We used the coalescent-based modeling package fastsimcoal version 2.5.2 (Excoffier et al., 2013) to evaluate each model based on this observed SFS. Fastsimcoal offers advantages over methods such as approximate Bayesian computation (Beaumont et al., 2002) by efficiently generating likelihoods for genomic data sets, and relying on the SFS, eliminating the need to choose specific summary statistics upon which to base inferences. For each model, we performed 100 independent runs of fastsimcoal (30 expectation conditional maximization cycles and 5 × 104 simulations per run), and the run with the highest likelihood for each model was chosen to perform model selection with Akaike information criterion. Maximum likelihood estimates for parameters such as ancestral and current effective population sizes, times of historical events and levels of gene flow were obtained from the optimal model/run and used to generate confidence intervals by parametric bootstrapping. Specifically, we simulated 100 independent SFSs for the optimal model and parameter estimates, treated each of these 100 data sets as observed data and performed parameter estimation for each in a similar way as above (50 runs, 30 expectation conditional maximization cycles and 5 × 104 simulations per run). We identified the maximum likelihood parameter estimates for each of the 100 simulated data sets, and we report the confidence intervals as the minimum and maximum values of these sets of parameter estimates. We are not aware of any direct estimates for genome-wide mutation rates in snakes and hence we used a rate of 2.5 × 10−8, as estimated for humans (Nachman and Crowell, 2000). Finally, we evaluated how well the chosen model explains the data with a G-test statistic as used in Excoffier et al. (2013).
We followed the approach of Lande et al. (2003) to convert estimates of time from generations to years. Specifically, we assumed an age of first maturity of 3 years (Siegel, 1986) and an annual adult survival rate of 0.67 (Jones et al., 2012), and estimated a generation time of 5.03 years using the equation T=α+[s/(1−s)], where T is the generation time, α is the age at first maturity and s is the annual adult survival rate.
Results
Sequencing and de novo SNP identification
For the AftrRAD run associated with the clustering and phylogenetic analyses, a mean of 8.2 × 105 sequence reads were assigned to each of the 91 samples (range 122 001–1 756 507) after quality filtering. The mean read depth per locus was 107.6 reads, and the median read depth was 65. A total of 19 827 nonparalogous loci were identified for the data set. Of these loci, 16 804 were monomorphic, with the remaining 3023 containing at least one polymorphic site. Of the 3023 polymorphic loci, 1466 (48.5%) were scored in at least 95% of the samples in the data set. When only S. catenatus was included (no S. tergeminus samples), a total of 19 141 loci were identified, with 17 068 of these monomorphic, 2073 polymorphic and 1019 (49.2%) of the polymorphic loci scored in at least 95% of the samples. The decreased number of polymorphic loci after S. tergeminus was removed is expected, as many loci that are monomorphic in S. catenatus samples alone become polymorphic when S. tergeminus is included. For the second AftrRAD run in which data were generated for the fastsimcoal analyses, a mean of 6.6 × 105 reads were assigned to each of the 31 samples (range 103 469–1 596 353). The mean read depth was 85.5, and the median was 52. A total of 22 829 loci were identified. Of these, 3281 were polymorphic and 1099 were polymorphic and scored in all samples. This set of 1099 loci was used to construct the SFS for analyses in fastsimcoal.
Clustering analyses
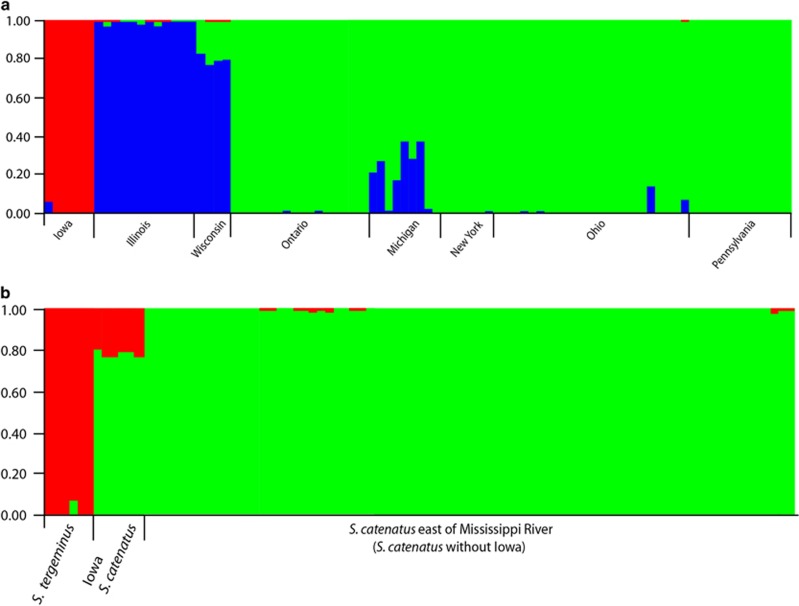
When only S. catenatus samples were analyzed, Structure identified K=3 as optimal (maximum ΔK), with groups generally consisting of samples from (1) Iowa, (2) Illinois and Wisconsin and (3) all remaining samples to the north and east (Figure 3a). Some moderate levels of genetic variation were shared between the Wisconsin and Illinois samples from the second group and samples from Michigan in the third group. When S. tergeminus samples were included in the data set, Structure identified K=2 as optimal, with the major division being between S. catenatus and S. tergeminus, as expected. However, S. catenatus samples from Iowa showed a pattern of admixture at K=2, in which they are assigned with ~20% probability (confidence interval (CI) 17.2–26.0%) to S. tergeminus, suggesting possible hybridization between these groups (Figure 3b).
Figure 3.
Results from clustering analyses in Structure with S. catenatus samples only (a), and with the outgroup S. tergeminus included (b). Each plot represents individual assignment proportions at the optimal ΔK value (K=3 for (a) and K=2 for (b)). When S. tergeminus is included, S. catenatus samples from Iowa demonstrate a pattern of genetic admixture between the two groups (b).
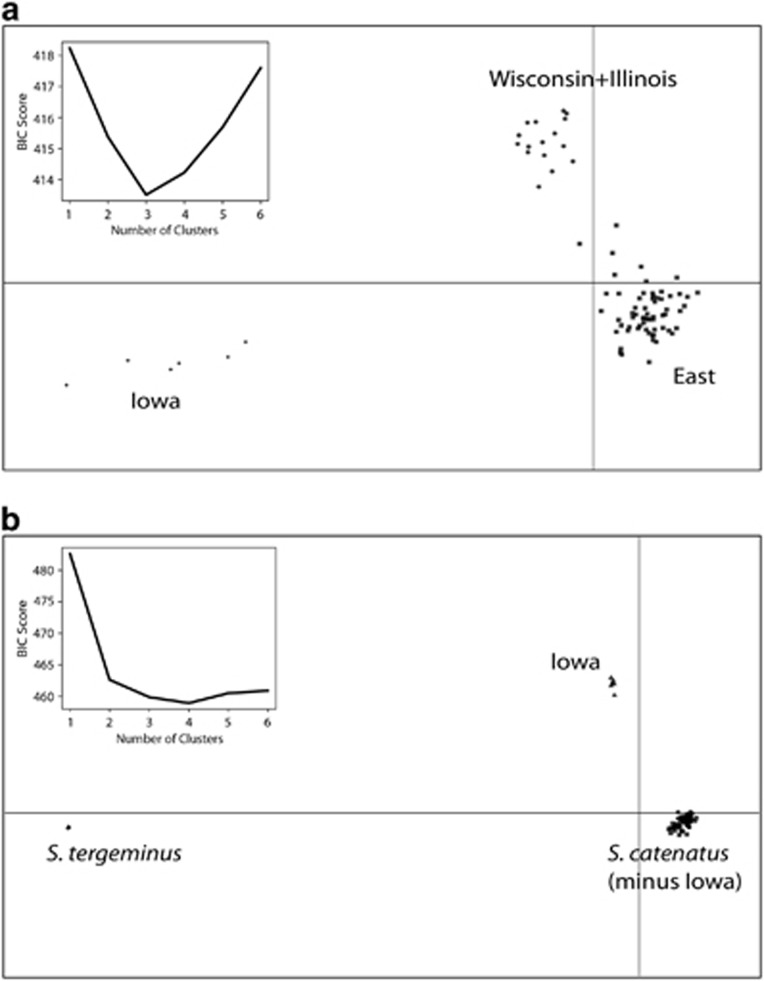
Comparisons of Bayesian information criterion values in adegenet showed that three and four clusters were optimal for the ingroup only data set and the data set including S. tergeminus, respectively (Figures 4a and b). Samples in the adegenet analyses were assigned to clusters in a similar way as in the Structure analyses: S. catenatus individuals were separated into clusters of samples from (1) Iowa, (2) Illinois and Wisconsin and (3) all remaining samples to the east and north. All individual samples were assigned with >99% probability to these respective clusters, and when plotted along the first two discriminant function axes, the greatest differentiation between S. catenatus clusters was associated with the Iowa populations (Figure 4).
Figure 4.
Results from clustering analyses in adegenet with S. catenatus samples only (a), and with the outgroup S. tergeminus included (b). Main plots show results of discriminant analysis of principal components for the optimal K value inferred from Bayesian information criterion (BIC; top left insets). Within S. catenatus, the greatest separation in discriminant function space was observed between the Iowa samples and all the remaining S. catenatus samples from east of the Mississippi River.
Finally, we used Fst values to estimate the magnitude of genetic divergence between the identified clusters of samples. Within S. catenatus, levels of divergence were lowest between the Illinois+Wisconsin group and the cluster consisting of the remaining samples to the north and east. (Fst=0.13). These Fst values were ∼2–3 times higher when the Iowa cluster was compared with either of the other two S. catenatus clusters (0.29 and 0.32, for the two other groups, respectively). All these values were substantially less than the Fst observed between samples of S. catenatus and S. tergeminus (0.65).
SNAPP phylogenetic analyses
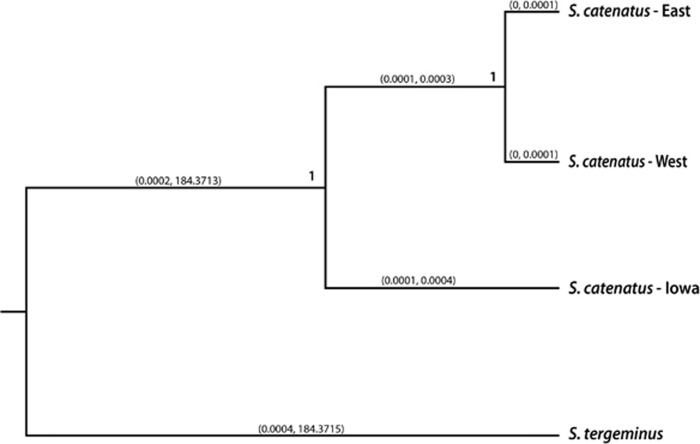
As expected from Kubatko et al. (2011), ingroup S. catenatus samples formed a monophyletic clade, clustering together to the exclusion of S. tergeminus, with high support (1.0 posterior probability) in all SNAPP runs (Figure 5). Within S. catenatus, Iowa samples formed a phylogenetically distinct lineage basal to the remaining ingroup samples with high support (1.0 posterior probability) in all experimental runs. In addition, the 95% highest probability density interval for branch lengths separating the Iowa group from the remaining S. catenatus did not include zero for any of the 10 subsampled runs (Figure 5). In contrast, the analyses did not provide support for differentiation between the Illinois+Wisconsin cluster and the cluster consisting of the remaining samples to the north and east, as the highest probability density intervals for branch length between these groups included zero as a lower bound. Together, these results suggest that Iowa lineage represents a distinct phylogenetic lineage within S. catenatus, but that there is not strong support for the presence of phylogenetically distinct lineages among S. catenatus populations east of the Mississippi River.
Figure 5.
Tree summarizing results from the 10 SNAPP runs that each included different subsets of samples. Values at nodes represent posterior support for clades after the first 20% of trees were removed as burn-in (topologies and clade support values were the same across all 10 runs). Values on the branches represent the minimum and maximum values observed in the set of 95% highest probability density (HPD) values for branch length across the 10 runs. Lower bounds of the branch lengths separating Iowa from the remaining S. catenatus do not include zero, supporting the presence of significant phylogenetic divergence between Iowa and other S. catenatus populations.
BFD* phylogenetic analyses
Results from the independent runs of BFD* varied in terms of the specific delimitation model that was best supported. However, across all runs the best supported model was one in which samples from the Iowa group represented a distinct lineage compared with models in which Iowa samples were combined with other groups (Supplementary Table S1).
Demographic modeling
Among the demographic models in Figure 2, most of the model weight from Akaike information criterion was associated with the Isolation+Hybridization model (Table 2). This suggests that differentiation of the Iowa lineage from other S. catenatus involved a combination of divergence through isolation and a hybridization event that resulted in introgression between S. tergeminus and the Iowa lineage. Based on the values we used for the average mutation rate and generation time in this system, maximum likelihood parameters under this model (Table 3) estimate that divergence of the Iowa lineage from other S. catenatus (TCAT) occurred 6756 generations bp, corresponding to ~34 000 ybp (CI 19 401–46 739 ybp), and the introgression event occurred 2242 generations bp, corresponding to ~11 000 ybp (CI 2429–29 602 ybp). The difference in these values represents the amount of time that the Iowa lineage evolved in allopatry before the hybridization/introgression event, and is estimated to be 4514 generations or ~23 000 years before present (CI 4502–36 095 years). A G-test to evaluate the model's fit to the data (Excoffier et al., 2013) was significant (P=0.01), suggesting that the specified models do not provide a complete explanation of the processes generating the observed patterns of genetic variation.
Table 2. Akaike information criterion (AIC) model selection results for fastsimcoal analyses.
| Model | No. of parameters | Ln likelihood | AIC | Akaike weight |
|---|---|---|---|---|
| Isolation+Hybridization | 9 | −6000.05 | 27 649.25 | 1 |
| Ongoing Hybridization | 10 | −6014.22 | 27 716.51 | 2.49e−15 |
| Isolation | 7 | −6015.61 | 27 716.89 | 2.05e−15 |
Three historical demographic models differing in the presence and/or timing of hybridization between S. tergeminus and the Iowa lineage of S. catenatus were compared, and are depicted in Figure 2. Models are ordered based on relative model weights.
Table 3. Maximum likelihood estimates for demographic parameters estimated in the fastsimcoal analysis for the best-supported Isolation+Hybridization model (Figure 2b).
| Parameter | Point estimate | Confidence interval |
|---|---|---|
| TSIS | 30 894 (155 397) | 24 287–34 721 (122 164–174 647) |
| TCAT | 6756 (33 988) | 4953–9289 (24 914–46 723) |
| THYB | 2242 (11 277) | 687–4781 (3456–24 048) |
| PTERG | 0.066 | 0.021–0.17 |
| NSIS | 34 142 | 26 285–40 824 |
| NCAT_ANCES | 32 730 | 20 359–44 632 |
| NTERG | 90 988 | 61 200–95 533 |
| NIOWA | 7590 | 4344–9610 |
| NKLDR | 4987 | 3911–9127 |
Times are given in number of generations, with the number of years included in parentheses based on the estimated generation time. Confidence intervals for parameters were generated from parametric bootstrapping, as described in the Materials and methods. Parameters are described in Supplementary Information.
The modeling analyses generated additional population parameter estimates of conservation interest (Table 3). In particular, current effective population sizes were smaller in both S. catenatus populations than in the S. tergeminus (a nonthreatened species) population: Specifically, for S. catenatus, the genetically effective population size (Ne) for the Killdeer Plains population was estimated at 4987 (CI 2696–9127) and the Iowa population was 7590 (CI 4344–9610), whereas the S. tergeminus was much larger at 90 988 (CI 54 764–96 718). In terms of historical changes in population sizes, the S. tergeminus population was best modeled as a growing population, whereas the two S. catenatus populations were best modeled as declining populations since their divergence from each other (Table 3).
Discussion
Our main results are that (1) phylogeographic analyses identify S. catenatus in Iowa as phylogenetically distinct from all other populations, whereas there is equivocal evidence for significant phylogenetically distinct lineages among remaining S. catenatus populations east of the Mississippi River; and (2) historical demographic modeling suggests that isolation and hybridization between S. tergeminus and Iowa S. catenatus has contributed to the genetic distinctiveness of Iowa snakes. We discuss the evolutionary and conservation implications of these results below.
Identifying significant phylogenetic lineages
Our results suggest that individuals from three populations located within 30 km of each other in northeastern Iowa may represent a phylogenetically distinct lineage within S. catenatus. This conclusion is based on a two-step analytical approach for identifying distinct lineages in a taxon that has been used in some recent species delimitation studies (that is, Leaché and Fujita, 2010; Satler et al., 2013; Hedin et al., 2015). The first step (discovery) involves assigning samples to putative lineages, and is followed by a second (validation) step in which the putative lineages are tested for phylogenetic distinctiveness.
For the first (discovery) step in the analysis, we used Structure (Pritchard et al., 2000) and adegenet (Jombart and Ahmed, 2011) to identify genetic clusters in S. catenatus. These two programs differ in their approach to clustering and in the assumptions made about the data. Using the two separate approaches allowed us to evaluate the sensitivity of the genetic clusters identified to alternative clustering algorithms. Both clustering methods grouped similar sets of geographically related samples together, suggesting that the identified clusters are robust to the clustering algorithm used. However, because clusters are inferred without reference to the history of population diversification, they may not directly reflect phylogenetic branching patterns (Kalinowski, 2011). Therefore, the second (validation) step in our approach utilized the species tree-based programs SNAPP and BFD* to evaluate the phylogenetic distinctiveness of each identified cluster. Results from these phylogenetic analyses strongly support Iowa as a distinct lineage, but provide equivocal support for significant phylogenetic differentiation between genetic clusters east of the Mississippi River, with some analyses supporting distinctiveness whereas others do not. We conclude that these clusters east of the Mississippi River have not been isolated for a sufficient time following the post-glacial colonization of their present range to show evidence of being evolutionarily independent lineages.
These conclusions differ from those of Ray et al. (2013) in terms of the number and geographic location of potential conservation units within S. catenatus. That study was limited to a single mtDNA locus, whereas our analyses used >1000 nuclear DNA loci. Although mtDNA alone can reveal phylogeographic patterns that are consistent with genome-wide patterns (Barrowclough and Zink, 2009), analyses based on multiple nuclear DNA markers often provide a more reliable estimate of the true phylogeographic history of a species (Edwards and Bensch, 2009). In addition, Ray et al. (2013) used a clade support metric (the gsi statistic; Cummings et al., 2008) to define conservation units that is less stringent than our more conservative approach in which we assessed and required differentiation across a series of metrics including clade support values, branch lengths and BFD* analyses.
Hey (2009) discussed the arbitrary nature of delineating formal biological groupings such as species or conservation units, and we acknowledge that judgment of what constitutes distinct phylogenetic lineages in our study depends on the specific criteria applied. As shown by Hedin et al. (2015), groups characterized by strong population structure present some of the most challenging systems for delimitation, especially when inferences are limited to genetic data. In such cases, methods for delineating formal groups (that is, species, conservation units) may ‘over-split' major genetic groups, and factors such as sample size can play an important role in the inference of whether such groups are recognized or not (see Niemiller et al., 2012 for an empirical example). As demonstrated by Chiucchi and Gibbs (2010), S. catenatus exhibits characteristics such as strong population structure and small population sizes that may make these snakes susceptible to potential ‘over-splitting' in lineage delimitation studies. Factors such as these led Carstens et al. (2013a) to highlight the importance of using conservative criteria for delimiting evolutionary groups, and Hedrick (2001) has also emphasized the need to be conservative when using neutral genetic markers to identify conservation units such as evolutionary significant units. Given these points, we emphasize that our main result, that the greatest degree of genetic divergence within S. catenatus occurs between lineages east and west of the Mississippi River, is not affected by the specific choice of criteria. However, even this ‘large' divergence within S. catenatus is relatively small (that is, branch lengths and Fst values are considerably smaller) compared with the divergence between S. catenatus and its sister taxon S. tergeminus. For these reasons, we suggest that the differentiation observed between the Iowa groups and the remaining S. catenatus reflects intraspecific rather than interspecific diversity.
Previous analyses failed to detect Iowa populations of S. catenatus as distinct. We see several potential reasons for this. Most simply, samples from Iowa were not included in Kubatko et al. (2011), and hence their distinctness at the SNP-based nuclear DNA loci used in that study could not be evaluated. Ray et al. (2013) included five samples from the Iowa populations surveyed here (three of these five samples were included in the present study) and found that these samples fell within their Western S. catenatus mtDNA clade that also included samples from Wisconsin and Illinois. Given our conclusion that past hybridization occurred between Iowa snakes and S. tergeminus, the fact that no S. tergeminus mtDNA was observed in these samples could be explained in a number of ways. These include loss of rare S. tergeminus mitochondrial genomes because of drift, sampling effects or the possibility that introgression was primarily a result of successful hybridization between male S. tergeminus and female S. catenatus (see Toews and Brelsford, 2012 for a review on discordant patterns between mtDNA and nuclear DNA). At a minimum, the mtDNA results from Ray et al. (2013) demonstrate that the Iowa lineage has not been isolated for a sufficient period to achieve monophyly at the sampled mtDNA locus, constituting one criterion previously proposed and applied for designating evolutionary significant units for conservation purposes (Moritz, 1994). Regardless, our study demonstrates the value in combining genomic-scale data sets with broad geographic sampling for identifying potentially important cryptic genetic diversity within taxa.
Origin of the Iowa lineage
Historical demographic modeling has been used to identify the evolutionary mechanisms responsible for observed patterns of lineage diversity within species (Carstens et al., 2013b). Analysis using Structure showed a pattern consistent with genetic admixture in the Iowa snakes that could reflect the effects of hybridization, incomplete lineage sorting or both. Our modeling approach incorporated in the program fastsimcoal provided strong support for a two-step process in which the lineage represented by the Iowa samples initially diverged in allopatry, and then became genetically admixed thousands of years later because of introgression with S. tergeminus. Thus, the distinctiveness of the Iowa snakes is because of the joint effects of divergence in allopatry before and after a hybridization event, and the introgression of novel genetic material via a historically limited hybridization event.
In terms of isolation, two mechanisms could contribute to the initial divergence between S. catenatus samples from Iowa and the remaining populations to the east: refugial isolation or large-scale vicariance. One possibility is that the group from Iowa represents a refugial population from the ‘Driftless Area' that remained unglaciated during the Wisconsinan glaciation (Holliday et al., 2002). Phylogeographic structure associated with the Driftless Area has been documented in a number of taxa (Rowe et al., 2004; Lee-Yaw et al., 2008). However, our Wisconsin samples are also likely from the Driftless Area, and yet do not show a similar pattern of divergence as the Iowa samples, as expected under this hypothesis.
Vicariance effects due to the influence of the Mississippi River as a long-term isolating barrier could also potentially explain the differentiation. Similar patterns of divergence coinciding with the Mississippi River have been demonstrated in other snakes including Diadophis spp. (Fontanella et al., 2008) and Lampropeltis spp. (Pyron and Burbrink, 2009), supporting this hypothesis. To our knowledge, the known distribution of S. catenatus populations west of the Mississippi River is currently restricted to the area represented by our samples. Additional sampling is needed to confirm the restricted range of this lineage. If populations were identified from other geographic areas, these samples could be used to further test vicariance because of the Mississippi River as a diversifying mechanism that could have potentially influenced other S. catenatus populations on the western extreme of the species distribution.
Hybridization has been previously reported between related species of rattlesnakes (Murphy and Crabtree, 1988) including these species of Sistrurus (Evans and Gloyd, 1948, but see Gibbs et al., 2011). Our modeling approach allowed us to estimate parameters related to historical demographic events such as the timing of divergences and admixture events. We note that these estimates are dependent on a number of factors that include the assumed mutation rate for our markers and generation times for these snakes, the appropriateness of the model under which the values were estimated and the degree to which any linkage among markers may not have been accounted for. For such reasons specific values should be interpreted with caution. With this in mind, our parameter estimates date the divergence of the Iowa lineage at ~34 000 ybp and the time at which introgression occurred at ~11 000 ybp. These dates roughly correspond with the late Pleistocene and early Holocene eras, respectively. This time was characterized by retreat of the Wisconsin glacial ice sheet, altered habitats and associated distribution shifts for many species (Schmidt, 1938; King, 1981; Webb, 1981) that could have led to secondary contact between the Iowa lineage and S. tergeminus. These dates suggest that introgression occurred on the order of 1000s of years before any significant human impacts on the landscape that in this region of North America resulted from modern agriculture and development in the nineteenth and twentieth centuries. Thus, the introgression appears to have occurred as a natural process, as opposed to being caused by human activities, and hence the observed pattern of admixture is not likely to represent recent genetic swamping such as has been observed in other threatened taxa (Allendorf et al., 2001). In contrast, human impacts on habitat may have had the opposite impact: habitat conversion to agriculture and resulting fragmentation now severely limit the distributions of Sistrurus populations in Iowa, and there are currently no sympatric populations of S. tergeminus and S. catenatus in the state (VanDeWalle and Christiansen, 2002), eliminating the potential for any gene exchange on contemporary timescales.
Conservation status of Iowa snakes
A final question is whether the genetically distinct Iowa snakes warrant special conservation status. Our results suggest that Iowa snakes diverged from other S. catenatus as a result of both isolation and historical hybridization over evolutionary timescales. Isolation can help promote local adaptation through selection acting in different environments, and this is the primary mechanism suggested to be responsible for adaptive differences between evolutionary significant units (Crandall et al., 2000). In terms of hybridization, we conclude that this lineage represents an example of either a Type 1 (Natural Hybrid Taxon) or Type 2 (Natural Hybridization) scenario under the scheme of Allendorf et al. (2001) for characterizing hybridization in threatened species. Allendorf et al. (2001) argue that rare hybrid groups originating through these scenarios should be considered for protection because hybridization can be an important evolutionary process capable of creating novel genetic diversity. The fact that Iowa snakes also appear to be stabilized, introgressed individuals that have persisted over evolutionary timescales suggests that they may contain a unique set of adaptive variation within the species as a whole. However, the presence or absence of adaptive variation cannot be inferred given the (presumably) neutral markers used in this study. Additional studies of divergence at functional loci (Funk et al., 2012), phenotypic comparisons of Iowa snakes with other S. catenatus populations or ecological studies including Environmental Niche Modeling (Wooten and Gibbs, 2012) are needed to establish whether Iowa snakes contain unique adaptive variation, as little is currently know in this regard. Until this work is completed, we feel our analyses suggest that a conservative conclusion is that Iowa snakes represent a distinct portion of the evolutionary history of S. catenatus as a whole, and hence deserve consideration as a Distinct Population Segment under the US Endangered Species Act because of their discreteness, significance and population status within the species. We emphasize that additional considerations independent of genetics such as local abundance and ecological importance are also important in establishing the most appropriate conservation status of these snakes.
Data archiving
RADseq data sets are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.nt65q. Historical demographic models used in the Fastsimcoal analyses are available at https://github.com/mikesovic.
Acknowledgments
We thank all those individuals (listed in Chiucchi and Gibbs, 2010) who have provided samples or assisted with collections across the range of S. catenatus over the past 20 years—this work would not be possible without their help. For this study in particular, we thank Rich King and Terry VanDeWalle for generously providing key samples. We also thank Laura Kubatko, Adam Leaché, Jordan Satler, Elizabeth Murray, Oleksandr Zinenko, Pearlly Yan, Matt Holding, other members of the Gibbs Lab and especially Bryan Carstens for advice and comments on the manuscript, Jose Diaz for help with lab work, Crisley de Camargo for assistance with bioinformatics and Carolyn Caldwell for her long-time support of our conservation genetic work on endangered snakes. Illumina sequencing was performed at the OSUCCC Genomics Shared Resource (OSU Cancer Center (CORE) Support Grant 5P30CA016058-12), and analyses were performed on the Ohio Biodiversity Conservation Partnership Computing Cluster. This work was supported by the State Wildlife Grants Program, administered jointly by the US Fish and Wildlife Service and the Ohio Division of Wildlife, with funds provided by the Ohio Biodiversity Conservation Partnership between Ohio State University and the Ohio Division of Wildlife.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. (2001). The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16: 613–622. [Google Scholar]
- Avise JC. (2010). Perspective: conservation genetics enters the genomics era. Conserv Genet 11: 665–669. [Google Scholar]
- Barrowclough GF, Zink RM. (2009). Funds enough, and time: mtDNA, nuDNA and the discovery of divergence. Mol Ecol 18: 2934–2936. [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. (2002). Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. (2012). Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol Biol Evol 29: 1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens BC, Brennan RS, Chua V, Duffie CV, Harvey MG, Koch RA et al. (2013. b). Model selection as a tool for phylogeographic inference: an example from the willow Salix melanopsis. Mol Ecol 22: 4014–4028. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Pelletier TA, Reid NM, Satler JD. (2013. a). How to fail at species delimitation. Mol Ecol 22: 4369–4383. [DOI] [PubMed] [Google Scholar]
- Chiucchi JE, Gibbs HL. (2010). Similarity of contemporary and historical gene flow among highly fragmented populations of an endangered rattlesnake. Mol Ecol 19: 5345–5358. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. (2000). Considering evolutionary processes in conservation biology. Trends Ecol Evol 15: 290–295. [DOI] [PubMed] [Google Scholar]
- Cummings MP, Neel MC, Shaw KL. (2008). A genealogical approach to quantifying lineage divergence. Evolution 62: 2411–2422. [DOI] [PubMed] [Google Scholar]
- DaCosta JM, Sorenson MD. (2014). Amplification biases and consistent recovery of loci in a double-digest RAD-seq protocol. PLoS One 9: e106713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JL, Blaxter MW. (2011). RADSeq: next-generation population genetics. Brief Funct Genomics 9: 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. (2011). Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12: 499–510. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, Vonholdt BM. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361. [Google Scholar]
- Edwards S, Bensch S. (2009). Looking forwards or looking backwards in avian phylogeography? A comment on Zink and Barrowclough 2008. Mol Ecol 18: 2930–2933. [DOI] [PubMed] [Google Scholar]
- Ekblom R, Galindo J. (2011). Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. (2013). Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol 29: 51–63. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Evans PD, Gloyd HK. (1948). The subspecies of the massasauga, Sistrurus catenatus, in Missouri. Bull Chicago Acad Sci 8: 225–232. [Google Scholar]
- Excoffier L, Lischer HEL. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sanchez E, Sousa VC, Foll M. (2013). Robust demographic inference from genomic and SNP data. PLOS Genet 9: e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanella FM, Feldman CR, Siddall ME, Burbrink FT. (2008). Phylogeography of Diadophis punctatus: extensive lineage diversity and repeated patterns of historical demography in a trans-continental snake. Mol Phylogenet Evol 46: 1049–1070. [DOI] [PubMed] [Google Scholar]
- Funk WC, McKay JK, Hohenlohe PA, Allendorf FW. (2012). Harnessing genomics for delineating conservation units. Trends Ecol Evol 27: 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs HL, Murphy M, Chiucchi JE. (2011). Genetic identity of endangered massasauga rattlesnakes (Sistrurus sp.) in Missouri. Conserv Genet 12: 433–439. [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. (2009). Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5: e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey MG, Brumfield RT. (2015). Genomic variation in a widespread Neotropical bird (Xenops minutus reveals divergnce, population expansion, and gene flow. Mol Phylogenet Evol 83: 305–316. [DOI] [PubMed] [Google Scholar]
- Hedin M, Carlson D, Coyle F. (2015). Sky island diversification meets the multispecies coalescent - divergence in the spruce-fir moss spider (Microhexura montivaga, Araneae, Mygalomorphae) on the highest peaks of southern Appalachia. Mol Ecol 24: 3467–3484. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. (2001). Conservation genetics: where are we now? Trends Ecol Evol 16: 629–636. [Google Scholar]
- Hey J. (2009). On the arbitrary identification of real species. In: Butlin RK, Bridle JR, Schluter D (eds). Speciation and Patterns of Diversity. Cambridge University Press: Cambridge, UK, pp 15–28. [Google Scholar]
- Holliday VT, Knox JC, Running IV, Mandel RD, Ferring CR. (2002). The central lowlands In: Orme AR (ed). The Physical Geography of North America. Oxford University Press: New York, pp 335–362. [Google Scholar]
- Jombart T, Devillard S, Balloux F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. (2011). adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PC, King RB, Bailey RL, Bieser ND, Bissell K, Campa H III et al. (2012). Range-wide analysis of eastern massasauga survivorship. J Wildl Manag 76: 1576–1586. [Google Scholar]
- Kalinowski ST. (2011). The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human populations structure. Heredity 106: 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass RE, Raferty AE. (1995). Bayes factors and model uncertainty. J Am Stat Assoc 90: 773–795. [Google Scholar]
- King JE. (1981). Late Quaternary vegetational history of Illinois. Ecol Monogr 51: 43–62. [Google Scholar]
- Kubatko LS, Gibbs HL, Bloomquist EW. (2011). Inferring species-level phylogenies and taxonomic distinctiveness using multilocus data in Sistrurus rattlesnakes. Syst Biol 60: 393–409. [DOI] [PubMed] [Google Scholar]
- Lande R, Engen S, Saether BE. (2003) Stochastic Population Dynamics in Ecology and Conservation. Oxford University Press: Oxford, UK. [Google Scholar]
- Leaché AD, Fujita MK. (2010). Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus. Proc R Soc B 277: 3071–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché AD, Fujita MK, Minin VN, Bouckaert RR. (2014). Species delimitation using genome-wide SNP data. Syst Biol 63: 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Yaw JA, Irwin JT, Green DM. (2008). Postglacial range expansion from northern refugia by the wood frog, Rana sylvatica. Mol Ecol 17: 867–884. [DOI] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. (2003). The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4: 981–994. [DOI] [PubMed] [Google Scholar]
- Moritz C. (1994). Defining 'Evolutionarily Significant Units' for conservation. Trends Ecol Evol 9: 373–375. [DOI] [PubMed] [Google Scholar]
- Murphy RW, Crabtree CB. (1988). Genetic identification of a natural hybrid rattlesnake: Crotalus scutulatus scutulatus x C. viridis viridis. Herpetologica 44: 119–123. [Google Scholar]
- Nachman MW, Crowell SL. (2000). Estimate of the mutation rate per nucleotide in humans. Genetics 16: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA. (2013). Genotyping-by-sequencing in ecological and conservation genomics. Mol Ecol 22: 2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemiller ML, Near TJ, Fitzpatrick BM. (2012). Delimiting species using multilocus data: diagnosing cryptic diversity in the southern cavefish, Typhlichthys subterraneus (Teleostei: Amblyopsidae). Evolution 66: 846–866. [DOI] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. (2012). Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PloS One 7: e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA, Burbrink FT. (2009). Systematics of the Common Kingsnake (Lampropeltis getula; Serpentes: Colubridae) and the burden of heritage in taxonomy. Zootaxa 32: 22–32. [Google Scholar]
- Ray JW, King RB, Duvall MR, Robinson JW, Jaeger CP, Dreslik MJ et al. (2013). Genetic analysis and captive breeding program design for the eastern massasauga Sistrurus catenatus catenatus. J Fish Wildl Manag 4: 104–113. [Google Scholar]
- Rowe KC, Heske EJ, Brown PW, Paige KN. (2004). Surviving the ice: Northern refugia and post-glacial colonization. Proc Natl Acad Sci USA 101: 10355–10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satler JD, Carstens BC, Hedin M. (2013). Multilocus species delimitation in a complex of morphologically conserved trapdoor spiders (mygalomorphae, antrodiaetidae aliatypus). Syst Biol 62: 805–823. [DOI] [PubMed] [Google Scholar]
- Schmidt KP. (1938). Herpetological evidence for the postglacial eastward extension of the steppe in North America. Ecology 19: 396–407. [Google Scholar]
- Siegel RA. (1986). Ecology and conservation of an endangered rattlesnake, Sistrurus catenatus, in Missouri, USA. Biol Conserv 35: 333–346. [Google Scholar]
- Sovic MG, Fries AC, Gibbs HL. (2015). AftrRAD: a pipeline for accurate and efficient de novo assembly of RADseq data. Mol Ecol Resour 15: 1163–1171. [DOI] [PubMed] [Google Scholar]
- Szymanski J. (1998) Sistrurus c. catenatus Rangewide Status Assessment. U.S. Fish and Wildlife Service: Fort Snelling, Minnesota. [Google Scholar]
- Toews DPL, Brelsford A. (2012). The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 12: 3907–3930. [DOI] [PubMed] [Google Scholar]
- United States Fish and Wildlife Service. (2015) Endangered and threatened wildlife and plants; threatened species status for the Eastern Massasauga Rattlesnake. Fed Regis 80: 58688–58701. [Google Scholar]
- VanDeWalle TJ, Christiansen JL. (2002) Survey of Eastern Massasauga Rattlesnake (Sistrurus catenatus catenatus) Populations in Iowa. Iowa Department of Natural Resources: Des Moines, IA. [Google Scholar]
- Webb T. (1981). The past 11,000 years of vegetational change in eastern North America. Bioscience 31: 501–506. [Google Scholar]
- Wooten JA, Gibbs HL. (2012). Niche divergence and lineage diversification among closely related Sistrurus rattlesnakes. J Evol Biol 25: 317–328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.