Staphylococcus aureus is a versatile human pathogen that produces an array of virulence factors, including several proteases. Of these, six proteases called the Spls are the least characterized. Previous evidence suggests that the Spls are expressed during human infection; however, their function is unknown. Our study shows that the Spls are required for S. aureus to cause disseminated lung damage during pneumonia. Further, we present the first example of a human protein cut by an Spl protease. Although the Spls were predicted not to cut staphylococcal proteins, we also show that an spl mutant has altered abundance of both secreted and surface-associated proteins. This work provides novel insight into the function of Spls during infection and their potential ability to degrade both staphylococcal and human proteins.
KEYWORDS: Staphylococcus aureus, mucin, pneumonia, proteases, Spl, virulence factors
ABSTRACT
The Spl proteases are a group of six serine proteases that are encoded on the νSaβ pathogenicity island and are unique to Staphylococcus aureus. Despite their interesting biochemistry, their biological substrates and functions in virulence have been difficult to elucidate. We found that an spl operon mutant of the community-associated methicillin-resistant S. aureus USA300 strain LAC induced localized lung damage in a rabbit model of pneumonia, characterized by bronchopneumonia observed histologically. Disease in the mutant-infected rabbits was restricted in distribution compared to that in wild-type USA300-infected rabbits. We also found that SplA is able to cleave the mucin 16 glycoprotein from the surface of the CalU-3 lung cell line, suggesting a possible mechanism for wild-type USA300 spreading pneumonia to both lungs. Investigation of the secreted and surface proteomes of wild-type USA300 and the spl mutant revealed multiple alterations in metabolic proteins and virulence factors. This study demonstrates that the Spls modulate S. aureus physiology and virulence, identifies a human target of SplA, and suggests potential S. aureus targets of the Spl proteases.
IMPORTANCE Staphylococcus aureus is a versatile human pathogen that produces an array of virulence factors, including several proteases. Of these, six proteases called the Spls are the least characterized. Previous evidence suggests that the Spls are expressed during human infection; however, their function is unknown. Our study shows that the Spls are required for S. aureus to cause disseminated lung damage during pneumonia. Further, we present the first example of a human protein cut by an Spl protease. Although the Spls were predicted not to cut staphylococcal proteins, we also show that an spl mutant has altered abundance of both secreted and surface-associated proteins. This work provides novel insight into the function of Spls during infection and their potential ability to degrade both staphylococcal and human proteins.
INTRODUCTION
Staphylococcus aureus is a Gram-positive opportunistic pathogen that is a significant cause of both health care- and community-associated infectious disease and is responsible for nearly 500,000 hospitalizations per year in the United States (1). This versatile organism uses a wide array of virulence factors to cause many types of infections, including cutaneous lesions, pneumonia, osteomyelitis, and toxic shock syndrome (2). Although many virulence factors have been characterized, S. aureus produces other putative virulence factors that are poorly studied. One example is the spl (serine protease-like) operon, which is found on the νSaβ pathogenicity island and carries six serine protease genes (splA, splB, splC, splD, splE, and splF) (3, 4). The spl operon is not found in the other staphylococci but is present in most strains of S. aureus, although some strains do not have the full operon (3, 5). The SplD and SplF amino acid sequences are 94.6% identical (3). The natural substrates and virulence roles of the Spl proteases are unknown; however, there is evidence that they are involved in colonization or infection of the host.
The spl locus was first identified in a study of anti-S. aureus antibodies generated in patients with invasive S. aureus infections. In a screen of S. aureus proteins, one that had strong reactivity with the patients’ antisera was characterized. Sequence analysis of the open reading frame (ORF) identified revealed that it was a putative serine protease and appeared to be within an operon (6). The ORF identified was later found to be SplC (3). Since then, the Spls have been demonstrated to be immunogenic in individuals with S. aureus infections, as well as healthy individuals colonized by S. aureus (5, 7). A recent study also identified the Spls as immunogenic in the airway, particularly in patients with severe asthma (8). Strikingly, the same study also discovered peptide fragments of SplD and SplF in human nasal polyp tissue. Therefore, it is well established that Spl proteases are secreted in vivo and are potentially involved in S. aureus-host interaction.
The goal of this study was to investigate the role of the Spl proteases in infection and identify possible Spl cleavage targets. Using a community-associated methicillin-resistant S. aureus (CA-MRSA) USA300 strain, we found that an spl deletion mutant was not attenuated in lethality in a rabbit pneumonia model but was able to induce severe damage in only one lung. In contrast, the USA300 wild-type (WT) strain induced more diffuse disease, affecting both lungs equally. We also demonstrated that SplA is able to cleave mucin 16 from the human lung cell line CalU-3, which is the first identification of a host protein as a substrate for an Spl protease. Proteomic studies show that the spl mutant has an altered abundance of many proteins both on the cell surface and secreted, suggesting that Spls may target S. aureus proteins as well.
RESULTS
spl mutant produces localized pneumonia in rabbit model of infection.
To study the function of the Spls during host infection, we compared the USA300 WT strain LAC and an allelic replacement spl operon mutant (Δspl::erm) in a rabbit pneumonia model of infection. LAC is a CA-MRSA USA300 strain (here USA300 WT), and this USA300 lineage has become the most frequent cause of CA-MRSA infections in North America and is increasingly reported both overseas and in hospital-associated MRSA (HA-MRSA) infections (9, 10). USA300 strains have been isolated from both CA-MRSA and HA-MRSA pneumonia patients (11, 12). Animal models have demonstrated that the S. aureus sae regulatory system and various sae-regulated factors are important in S. aureus pneumonia (13–17). Since the Spls are directly regulated by sae (13), we predicted that pneumonia would be a relevant model to investigate their role in virulence.
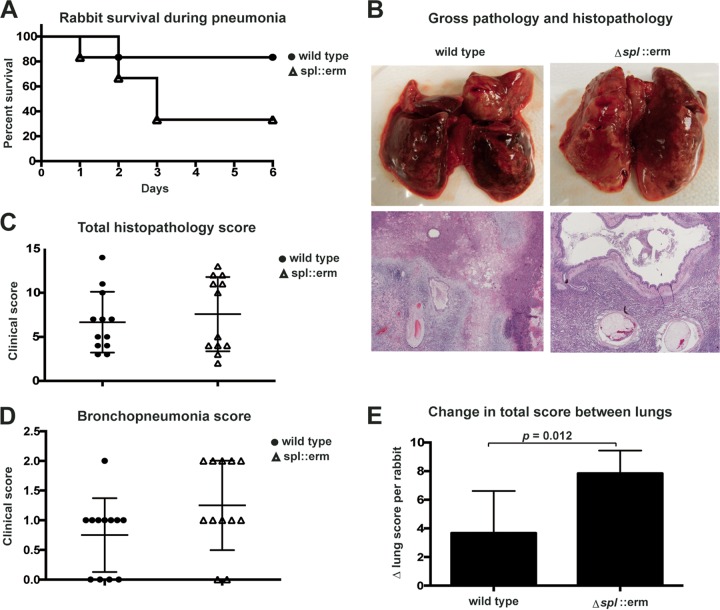
The rabbit pneumonia model was performed as previously described (18). Rabbits were infected with either USA300 WT or Δspl::erm mutant bacteria at a dose of 2 × 109 CFU delivered directly to the lung through a catheter that was inserted at the trachea. After 6 days of infection, surviving rabbits were sacrificed. In a total of two experiments (n = 6 rabbits per group), 83% of the USA300 WT-infected rabbits (5 of 6) survived to day 6, while 33.3% of the spl mutant-infected rabbits (2 of 6) survived to day 6 (P = 0.11, Fig. 1A). Gross pathology and histopathology revealed significant lung damage in both groups. Gross pathology of a USA300 WT-infected rabbit that was euthanized on day 6 showed hemorrhage in both the left and right lungs (Fig. 1B, upper left). In a Δspl::erm mutant-infected animal that also was euthanized on day 6, the left lung also showed a large area of hemorrhage, while the right lung appeared more intact, suggesting a more constrained distribution of lesions (Fig. 1B, upper right). Histopathology also revealed edema, inflammatory infiltrates, and pleuritis in infected lungs from both groups of animals. Hematoxylin and eosin (H&E) straining of the left lungs of USA300 WT-infected (Fig. 1B, lower left) and Δspl::erm mutant-infected (Fig. 1B, lower right) rabbits euthanized on day 6 demonstrated these findings in both animals.
FIG 1 .
USA300 spl mutant causes unilateral pneumonia. (A) Survival of rabbits infected with the USA300 WT or spl mutant strain (n = 6). Two experiments were performed, with three rabbits per group in each experiment. The P value (0.11) was calculated with a log rank (Mantel-Cox) test. (B) Gross pathology (top) and histopathology (bottom) of USA300 WT and spl mutant strain-infected rabbit lungs euthanized on day 6. Histopathology of H&E-stained infected lung tissue is shown. (C) Total histopathology score of each lung, combined from six scoring categories. n = 12 lungs per group. (D) Bronchopneumonia score of each lung. n = 12 lungs per group. P = 0.09 calculated with an unpaired, two-tailed t test. (E) Difference between the left and right lung scores of each rabbit. For each rabbit, the value of the lung with the lower total clinical score was subtracted from the value of the lung with the higher clinical score. n = 6 rabbits per group. The P value was calculated with an unpaired, two-tailed t test.
Histopathology scoring of the lungs of all of the infected rabbits in both groups was also performed, regardless of the time of death. The lungs were scored on the presence of pleuritis, edema, bronchopneumonia, necrosis, lymphoid cell/heterophil infiltration, and Gram-positive bacteria. Each of the six categories was given a score of 0 (within normal limits) to 3 (severe and extensive). The average total histopathology scores were nearly identical in WT strain-infected rabbits and Δspl::erm mutant-infected rabbits (Fig. 1C), and rabbits in both groups exhibited bronchopneumonia (Fig. 1D). Interestingly, the Δspl::erm mutant-infected rabbits displayed a pattern of one high-scoring lung and one low-scoring lung, while lung scores appeared more evenly distributed in the WT strain-infected rabbits (Fig. 1C and E). A D’Agostino-Pearson normality test determined that the WT strain-infected lung scores follow a normal distribution (P = 0.27), while mutant-infected lung scores do not (P = 0.01), suggesting that the mutant-infected lungs represent two distinct populations. When the difference in score between the two lungs was calculated (Δ lung score) for each rabbit, the spl mutant-infected rabbits had a significantly greater difference between their lungs than WT strain-infected rabbits did (Fig. 1E, P = 0.012). On the whole, these findings indicate that although no significant difference in the lethality of the Δspl::erm mutant was observed, this mutant also produced a clear phenotype of lung damage that has a constrained localization within the lungs.
SplA induces mucin 16 release from CalU-3 cells.
Mucin 16 is an ~25-MDa, heavily glycosylated cell surface protein that is found on multiple epithelial tissues in the body, including the ocular surface and airway epithelia (19–21). Like other mucins, mucin 16 provides lubrication of epithelia, as well as a protective barrier against pathogens. Two studies have reported that RNA interference-mediated knockdown of mucin 16 in human corneal-limbal epithelial cells renders these cells more susceptible to S. aureus adherence (22, 23). Additionally, the Streptococcus pneumoniae metalloprotease ZmpC has been shown to cleave mucin 16 from a variety of human epithelial cell types, allowing the bacteria to increasingly invade these cells in vitro (24). An adenovirus capable of causing keratoconjunctivitis also was recently shown to induce mucin 16 release from ocular epithelial cells, facilitating infection (25). These findings demonstrate that S. aureus and other pathogens encounter mucin 16 as a barrier to the host epithelium and are able to disrupt this barrier in order to colonize or infect the host.
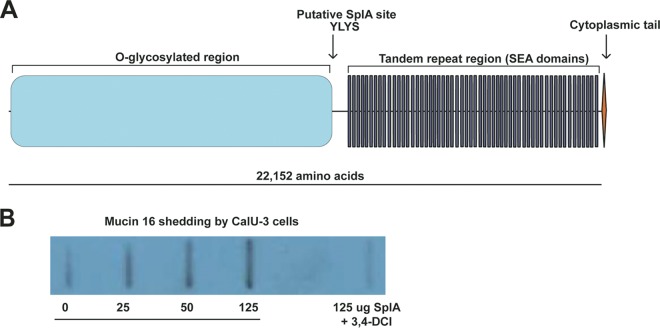
A study describing the crystal structure and cleavage site preference of SplA found that this enzyme has highly specific substrate requirements. In an experiment with a cellular library of peptide substrates, SplA was found to cleave exclusively substrates containing the Trp/Tyr-Leu-Tyr-Thr/Ser (W/Y-L-Y-T/S) motif (26). That study also noted that mucin 16 contains a Y-L-Y-S sequence and could be a likely target for SplA cleavage (Fig. 2A) (26). On the basis of this evidence, as well as our observation that the Δspl::erm mutant exhibited more localized disease in the rabbit pneumonia model, we hypothesized that SplA cleaves mucin 16 from lung cells. To test this, confluent monolayers of the lung epithelial cell line CalU-3 were treated with purified, His6-tagged SplA. Because of the large size of mucin 16 (Fig. 2A), CalU-3 medium was tested for mucin 16 with a slot blot apparatus rather than by gel electrophoresis. Probing with a mucin 16 antibody revealed that mucin 16 is endogenously released from CalU-3 cells (Fig. 2B). This is in accordance with previous reports that mucin 16 is naturally shed from epithelia (19, 27, 28). Incubation with SplA increased this shedding in a dose-dependent manner. When SplA was treated with the serine protease inhibitor 3,4-dichloroisocoumarin (3,4-DCI) before it was added to the cells, mucin 16 shedding was decreased to a level similar to that in untreated cells (Fig. 2B). This suggests that SplA activity is required for mucin 16 removal, by either cleaving it at the Y-L-Y-S site or inducing its shedding indirectly by degradation of another substrate. This activity may allow S. aureus to perturb the airway epithelial barrier and cause more disseminated disease.
FIG 2 .
SplA cleaves human mucin 16. (A) Diagram of human mucin 16. Mucin 16 is exposed on the cell surface and contains one putative SplA cleavage site. The N-terminal portion is heavily glycosylated, and the tandem repeat region contains several SEA (sea urchin sperm protein, enterokinase, and agrin) domains, which are present in various mucins. (B) Anti-mucin 16 slot blot assay of CalU-3 medium following treatment with SplA. One representative blot is shown.
The Δspl::erm mutant retains viability in whole blood, hemolysis activity, and protease production.
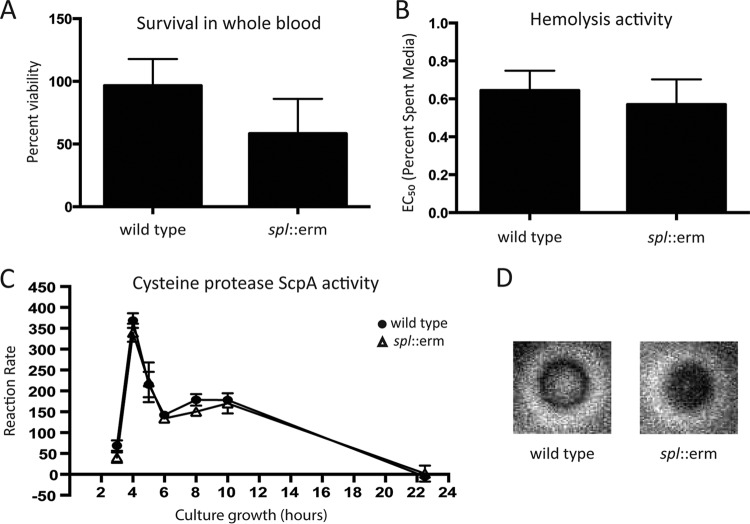
S. aureus produces a multitude of virulence factors that function by thwarting the innate immune defenses of the infected host (29). Based on the in vivo phenotype of the Δspl::erm mutant, we hypothesized that it may have an altered capacity to survive these immune assaults. To test this, we compared the survival of USA300 WT and Δspl::erm mutant cells in whole human blood. Whole human blood contains cellular and complement-mediated innate immunity components and thus is used as a general assay of bacterial survival under these conditions. When log-phase bacteria were inoculated into whole human blood and incubated for 3 h, 60% of USA300 WT bacteria remained viable. However, 40% of Δspl::erm mutant bacteria remained viable (Fig. 3A), although this difference was not statistically significant.
FIG 3 .
Virulence factor production is unchanged in a USA300 spl mutant. (A) Survival of the USA300 WT and Δspl::erm mutant strains in whole human blood. Bacteria were incubated with whole blood for 3 h. (B) Hemolysis activities of the USA300 WT and Δspl::erm mutant strains. A dilution series of spent medium was incubated with rabbit erythrocytes. The graph shows the percentage of medium needed to lyse 50% of the erythrocytes (EC50). (C) ScpA activity of the USA300 WT and Δspl::erm mutant strains measured over the course of culture growth. At each time point, spent medium was incubated with a FRET substrate for ScpA activity. Activity is shown as the rate at which fluorescence was generated by FRET substrate cleavage. (D) Milk plate proteolysis activities of the USA300 WT and Δspl::erm mutant strains. Overnight cultures were spotted onto an agar plate containing 5% milk, and the halo around the colony corresponds to clearing of the milk.
Alternatively, we hypothesized that the Δspl::erm mutant had altered production of one or more secreted virulence factors whose levels could be modulated by proteolytic degradation by the Spl proteases in S. aureus. To investigate this, the activity of the secreted alpha toxin (Hla) and the secreted proteases staphopain A (ScpA) and aureolysin (Aur) were compared in USA300 WT and Δspl::erm mutant bacteria. Hla is a potent cytolysin and immunomodulatory virulence factor that has been shown to contribute to pathogenesis in a number of S. aureus infection models, including a murine model of pneumonia (30, 31). In an Hla activity assay (32), the Δspl::erm mutant did not exhibit a change in Hla activity from the USA300 WT strain (Fig. 3B), indicating that hemolysis is unaffected by the Spls.
Next, we tested the activity levels of the S. aureus secreted proteases staphopain A (ScpA) and aureolysin (Aur). Both of these proteases inhibit the classical and alternative pathways of complement activation (33). ScpA also cleaves the neutrophil chemotaxis receptor CXCR2, blocking neutrophil recruitment (34), and exhibits activity against the extracellular matrix protein collagen, whose degradation could promote host damage during infection (35). Aur is able to inhibit complement activation by cleaving the complement protein C3 (36). ScpA activity in cell-free spent medium was tested with a FRET substrate containing the ScpA cleavage site from human CXCR2 (34, 37). Over the course of growth, the Δspl::erm mutant displayed the same ScpA activity as the USA300 WT strain, indicating that the Spls do not affect ScpA activity (Fig. 3C). A milk plate assay to detect Aur activity showed no decrease in the Δspl::erm mutant (Fig. 3D). On the whole, these results suggest that the Spls do not modulate ScpA or Aur activity.
Proteomic studies reveal differences between USA300 WT and Δspl::erm mutant strain secreted and surface proteins.
Finally, the surface and secreted proteomes of the USA300 WT and Δspl::erm mutant strains were analyzed. The objective of these experiments was to identify changes in protein levels that might explain the altered virulence phenotype of the Δspl::erm mutant and suggest potential S. aureus proteins that are degraded by the Spls. The proteomic data were analyzed with the Scaffold program using Fisher’s exact test, and all hits with a P value of ≤0.010 were considered significant. The surface proteomics identified 63 proteins that were detected with increasing abundance in the Δspl::erm mutant relative to USA300 WT (Table 1) and 21 proteins detected with decreasing abundance in the Δspl::erm mutant (Table 2). A few genes involved in virulence and immune evasion were increased in the Δspl::erm mutant, including those encoding IsdA and SpA. IsdA promotes S. aureus adherence to desquamated nasal epithelial cells (38), fibrinogen, and fibronectin (39) and enhances S. aureus survival of bactericidal fatty acids and peptides in human skin (40). SpA binds to IgG, which protects S. aureus from opsonophagocytosis and dysregulates the host adaptive immune response (41, 42). The cell wall-associated protein EbpS was also increased on the surface of spl mutant bacteria. Although EbpS was once thought to act as an adhesin by binding to elastin, recent studies have found that its contribution to bacterial adhesion is minimal (43). However, EbpS does mediate Zn2+-dependent growth and biofilm formation (44). Further, an investigation of the differential expression of S. aureus genes in murine models of colonization and pathogenesis found that ebpS was significantly upregulated in the blood relative to the nares (45). Therefore, this protein may play a role during infection. Increased levels of these proteins could enhance S. aureus immunomodulation and virulence.
TABLE 1 .
Proteins increased on spl mutant surface
| GenBank (UniProt) accession no. | Name | Biological function(s) | Avg no. of assigned spectra |
P valuea | |
|---|---|---|---|---|---|
| WT | Δspl::erm | ||||
| Q2FGW1 (EBPS_STAA3) | Elastin-binding protein EbpS | Adhesion, biofilm formation | 24.00 | 47.67 | 0.00000021 |
| Q2FHV1 (ISDA_STAA3) | Iron-regulated surface determinant protein A IsdA | Adhesion, immune evasion | 113.33 | 136.00 | 0.0018 |
| Q2FDT8 (ISAA_STAA3) | Transglycosylase IsaA | Autolysis | 37.67 | 60.33 | 0.000013 |
| Q2FGW9 (Q2FGW9_STAA3) | DNA-binding protein HU/Hup | Housekeeping, DNA structure | 40.67 | 68.67 | 0.00000046 |
| Q2FF94 (CH10_STAA3) | 10-kDa chaperonin GroS | Housekeeping, protein folding | 5.67 | 12.67 | 0.0022 |
| Q2FDV8 (CLPL_STAA3) | ATP-dependent protease ClpL | Housekeeping, protein turnover | 2.33 | 13.00 | 0.00000054 |
| Q2FGB6 (GREA_STAA3) | Transcription elongation factor GreA | Housekeeping, transcription | 115.33 | 153.67 | 0.0000033 |
| Q2FJA3 (RL11_STAA3) | 50S ribosomal protein R11 RplK | Housekeeping, translation | 250.00 | 320.67 | 2.4E−09 |
| Q2FFJ4 (GATC_STAA3) | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C GATC | Housekeeping, translation | 41.67 | 57.00 | 0.0019 |
| Q2FER1 (IF1_STAA3) | Translation initiation factor IF-1 InfA | Housekeeping, translation | 15.33 | 26.33 | 0.0011 |
| Q2FEQ4 (RL6_STAA3) | 50S ribosomal protein L6 RplF | Housekeeping, translation | 8.33 | 14.67 | 0.01 |
| Q2FFJ6 (GATB_STAA3) | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B GatB | Housekeeping, translation | 0.00 | 6.00 | 0.0002 |
| Q2FF08 (RL31B_STAA3) | 50S ribosomal protein L31 type B RpmE2 | Housekeeping, translation | 2.67 | 7.00 | 0.0096 |
| Q2FEQ7 (RL30_STAA3) | 50S ribosomal protein L30 RpmD | Housekeeping, translation | 0.00 | 2.33 | 0.007 |
| Q2FJU4 (Q2FJU4_STAA3) | Triacylglycerol lipase GehB/SAL2 | Lipase | 8.67 | 32.33 | 1.2E−11 |
| Q2FH26 (ODO2_STAA3) | Dihydrolipoyllysine residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex OdhB | Metabolism, amino acid degradation | 0.00 | 2.33 | 0.007 |
| Q2FHD0 (Q2FHD0_STAA3) | Glutamine synthetase GlnA | Metabolism, amino acid metabolism, glutamine synthesis | 26.33 | 47.33 | 0.0000045 |
| Q2FDV3 (Roca_STAA3) | 1-Pyrroline-5-carboxylate dehydrogenase RocA | Metabolism, amino acid metabolism, proline catabolism | 0.00 | 31.67 | 5E−30 |
| Q2FJC8 (Q2FJC8_STAA3) | Cysteine synthase CysK | Metabolism, amino acid synthesis | 82.33 | 133.00 | 7.4E−11 |
| Q2FH38 (Q2FH38_STAA3) | Diaminopimelate decarboxylase LysA | Metabolism, amino acid synthesis | 0.00 | 2.33 | 0.007 |
| Q2FF22 (ATPA_STAA3) | ATP synthase subunit alpha AtpA | Metabolism, ATP synthesis (general metabolism) | 22.67 | 32.00 | 0.01 |
| Q2FKD2 (Q2FKD2_STAA3) | Acetoin (diacetyl) reductase SAUSA300_0129 | Metabolism, carbohydrate metabolism | 6.00 | 12.33 | 0.0052 |
| Q2FK29 (LDH1_STAA3) | l-Lactate dehydrogenase 1 Ldh1 | Metabolism, carbohydrate metabolism | 0.00 | 6.00 | 0.0000028 |
| Q2FFV5 (PCKA_STAA3) | Phosphoenolpyruvate carboxykinase PckA | Metabolism, carbohydrate metabolism, gluconeogenesis | 12.67 | 35.00 | 0.000000003 |
| Q2FG39 (Q2FG39_STAA3) | 6-Phosphofructokinase PfkA | Metabolism, carbohydrate metabolism, glycolysis | 18.00 | 36.67 | 0.0000028 |
| Q2FDQ4 (ALF1_STAA3) | Fructose-bisphosphate aldolase class 1 Fda | Metabolism, carbohydrate metabolism, glycolysis | 0.00 | 6.67 | 0.00000069 |
| Q2FGM3 (Q2FGM3_STAA3) | 6-Phosphogluconate dehydrogenase, decarboxylating Gnd | Metabolism, carbohydrate metabolism, pentose phosphate pathway | 139.00 | 166.33 | 0.00072 |
| Q2FFW0 (Q2FFW0_STAA3) | Transaldolase SAUSA300_1725 | Metabolism, carbohydrate metabolism, pentose phosphate pathway | 40.67 | 66.33 | 0.0000026 |
| Q2FEZ2 (Q2FEZ2_STAA3) (+1) | Deoxyribose-phosphate aldolase DeoC | Metabolism, carbohydrate metabolism, pentose phosphate pathway | 4.00 | 10.67 | 0.0013 |
| Q2FHZ7 (Q2FHZ7_STAA3) | Phosphocarrier protein HPr/PtsH | Metabolism, carbohydrate metabolism, sugar PTS b | 22.67 | 50.67 | 1.7E−09 |
| Q2FE56 (Q2FE56_STAA3) | NAD-dependent epimerase/dehydratase family protein SAUSA300_2387 | Metabolism, cofactors and vitamins | 7.67 | 14.33 | 0.0065 |
| Q2FJM5 (GUAA_STAA3) | GMP synthase GuaA | Metabolism, nucleotide metabolism | 0.00 | 8.00 | 0.00000004 |
| Q2FI07 (PUR5_STAA3) | Phosphoribosylformylglycinamidine cyclo-ligase PurM | Metabolism, nucleotidemetabolism | 0.00 | 2.67 | 0.0034 |
| Q2FI11 (Q2FI11_STAA3) | Phosphoribosylformylglycinamidine synthase PurS | Metabolism, nucleotide metabolism | 0.00 | 2.67 | 0.0034 |
| Q2FI15 (FOLD_STAA3) | Bifunctional protein FolD | Metabolism, one-carbon metabolism, tetrahydrofolate interconversion | 317.33 | 350.00 | 0.0017 |
| Q2FFV8 (Q2FFV8_STAA3) | Oxidoreductase SAUSA300_1728 | Oxidoreductase | 99.67 | 123.33 | 0.00084 |
| Q2FIB9 (Q2FIB9_STAA3) | NADH-dependent flavin oxidoreductase SAUSA300_0859 | Oxidoreductase/FMNc binding | 0.00 | 3.00 | 0.0017 |
| Q2FEB6 (Q2FEB6_STAA3) | Putative uncharacterized protein SAUSA300_2327 | Putative FMN binding and pyridoxamine-phosphate oxidase (general metabolism/oxidation) | 30.67 | 52.67 | 0.0000055 |
| Q2FFZ4 (Q2FFZ4_STAA3) | Putative thioredoxin SAUSA300_1690 | Putative housekeeping, redox homeostasis | 246.67 | 285.67 | 0.00016 |
| Q2FEY0 (Q2FEY0_STAA3) | Haloacid dehalogenase-like hydrolase SAUSA300_2102 | Putative hydrolase activity | 0.00 | 2.67 | 0.0034 |
| Q2FFH4 (Y1902_STAA3) | Conserved hypothetical protein, putative lactonase SAUSA300_1902 | Putative lactonase activity | 9.33 | 22.33 | 0.00002 |
| Q2FGJ2 (Q2FGJ2_STAA3) | Proline dipeptidase SAUSA300_1491 | Putative peptidase activity (aminopeptidase) | 3.67 | 10.00 | 0.0016 |
| Q2FFL6 (Q2FFL6_STAA3) | Aminopeptidase PepS | Putative peptidase activity (aminopeptidase) | 0.00 | 4.00 | 0.0002 |
| Q2FG30 (Y1654_STAA3) | Uncharacterized peptidase SAUSA300_1654 | Putative peptidase activity (dipeptidase) | 1.00 | 19.67 | 3.4E−15 |
| Q2FFM0 (Q2FFM0_STAA3) | Uncharacterized putative glycosyl hydrolase SAUSA300_1856 | Putative peptidase activity (endopeptidase) | 0.00 | 16.00 | 1.6E−15 |
| Q2FI96 (Q2FI96_STAA3) | Conserved hypothetical protein SAUSA300_0882 | Putative phospholipid-binding protein | 0.00 | 2.33 | 0.007 |
| Q2FIJ2 (OHRL_STAA3) | Organic hydroperoxide resistance protein-like protein SAUSA300_0786 | Putative stress response, oxidative stress (putative) | 0.00 | 2.33 | 0.007 |
| Q2FJY6 (Q2FJY6_STAA3) | ESAT-6-like protein SAUSA300_0278 | Secretion | 22.33 | 33.00 | 0.0044 |
| Q2FDH3 (DRP35_STAA3) | Lactonase Drp35 | Stress response, cell envelope stress | 735.67 | 768.33 | 0.0044 |
| Q2FEV0 (ASP23_STAA3) | Alkaline shock protein 23 Asp23 | Stress response, cell envelope stress/alkaline shock protein | 6.00 | 25.67 | 1.3E−10 |
| Q2FGH0 (SODM1_STAA3) | Superoxide dismutase (Mn/Fe) 1 SodA | Stress response, oxidative stress | 2.00 | 14.00 | 0.000000028 |
| Q2FHD4 (Q2FHD4_STAA3) | Glutathione peroxidase SAUSA300_1197 | Stress response, oxidative stress | 17.67 | 26.33 | 0.0089 |
| Q2FIW1 (Q2FIW1_STAA3) | Conserved hypothetical protein SAUSA300_0664 | Unknown | 72.33 | 113.67 | 0.000000009 |
| Q2FKC5 (Q2FKC5_STAA3) | Cell wall surface anchor family protein SAUSA300_0136 | Unknown | 51.67 | 77.33 | 0.000013 |
| Q2FEJ7 (Q2FEJ7_STAA3) | Conserved hypothetical protein SAUSA300_2246 | Unknown | 50.33 | 75.00 | 0.000021 |
| Q2FDZ7 (Q2FDZ7_STAA3) | Conserved hypothetical protein SAUSA300_2447 | Unknown | 53.00 | 73.67 | 0.00027 |
| Q2FDH4 (Y2620_STAA3) | UPF0312 protein SAUSA300_2620 | Unknown | 7.67 | 26.33 | 4.4E−09 |
| Q2FHB6 (Q2FHB6_STAA3) | Conserved hypothetical protein SAUSA300_1215 | Unknown | 4.00 | 19.67 | 3.1E−09 |
| Q2FIT2 (Q2FIT2_STAA3) | Putative lipoprotein SAUSA300_0693 | Unknown | 18.00 | 33.00 | 0.000078 |
| Q2FG19 (Q2FG19_STAA3) | GAF domain-containing protein SAUSA300_1665 | Unknown | 10.00 | 19.33 | 0.0011 |
| Q2FKE8 (Q2FKE8_STAA3) | IgG-binding protein SpA | Virulence factor, immunomodulator | 470.00 | 563.67 | 5.9E−10 |
| Q2FHR3 (Q2FHR3_STAA3) | Antibacterial protein PSM-β2 | Virulence factor, immunomodulator | 31.00 | 42.33 | 0.0067 |
| Q2FES8 (Q2FES8_STAA3) | MHCd class II analog protein Map-w SAUSA300_2164 | Virulence factor, immunomodulator | 11.67 | 19.67 | 0.0056 |
Fisher’s exact test.
PTS, phosphotransferase system.
FMN, flavin mononucleotide.
MHC, major histocompatibility complex.
TABLE 2 .
Proteins decreased on spl mutant surface
| GenBank (UniProt) accession no. | Name | Biological function(s) | Avg no. of assigned spectra |
P valuea | |
|---|---|---|---|---|---|
| WT | Δspl::erm | ||||
| Q2FJ78 (SDRD_STAA3) | Serine-aspartate repeat-containing protein D SdrD | Adhesion, biofilm formation | 2.33 | 0.00 | 0.0087 |
| Q2FJ77 (SDRE_STAA3) | Serine-aspartate repeat-containing protein E SdrE | Adhesion, biofilm formation | 34.33 | 22.67 | 0.0082 |
| Q2FJ79 (SDRC_STAA3) | Serine-aspartate repeat-containing protein C SdrC | Adhesion, biofilm formation | 317.67 | 32.67 | 0 |
| Q2FHT6 (THIO_STAA3) | Thioredoxin TrxA | Housekeeping, cell redox homeostasis | 7.67 | 0.67 | 0.000014 |
| Q2FJA0 (RL7_STAA3) | 50S ribosomal protein L7/L12 RpiL | Housekeeping, translation | 81.00 | 45.67 | 0.00000015 |
| Q2FKP7 (SYS_STAA3) | Seryl-tRNA synthetase SerS | Housekeeping, translation, aminoacyl tRNA biosynthesis | 38.00 | 13.00 | 1.6E−09 |
| Q2FH16 (Q2FH16_STAA3) | Glucose-specific IIA component crr | Metabolism, carbohydrate metabolism, sugar PTSb | 163.67 | 108.33 | 0.000000038 |
| Q2FJ55 (Q2FJ55_STAA3) | Phosphate acetyltransferase Pta | Metabolism, carbohydrate metabolism | 14.67 | 6.67 | 0.0027 |
| Q2FIU1 (Q2FIU1_STAA3) | Fructose 1-phosphate kinase FruB | Metabolism, carbohydrate metabolism | 30.67 | 8.33 | 4.7E−10 |
| Q2FIB3 (G6PI_STAA3) | Glucose-6-phosphate isomerase Pgi | Metabolism, carbohydrate metabolism | 149.33 | 114.67 | 0.00061 |
| Q2FEL8 (MOAC_STAA3) | Molybdenum cofactor biosynthesis protein MoaC | Metabolism, cofactors and vitamins | 2.33 | 0.00 | 0.0087 |
| Q2FJD1 (Q2FJD1_STAA3) | Hypoxanthine phosphoribosyltransferase Hpt | Metabolism, purine metabolism | 538.00 | 36.00 | 0 |
| Q2FHN7 (Q2FHN7_STAA3) | Dihydroorotase PyrC | Metabolism, pyrimidine metabolism | 16.67 | 1.33 | 3.9E−11 |
| Q2FHR1 (Q2FHR1_STAA3) | Uncharacterized N-acetyltransferase | Putative acetyltransferase | 6.33 | 0.00 | 0 |
| Q2FIC1 (PPI1_STAA3) | Putative peptidyl-prolyl cis-trans isomerase SAUSA300_0857 | Putative housekeeping, protein folding | 98.00 | 40.67 | 1.1E−16 |
| Q2FG31 (Y1653_STAA3) | UPF0173 metal-dependent hydrolase SAUSA300_1653 | Putative hydrolase activity | 183.67 | 126.00 | 0.000000094 |
| Q2FEC8 (Y2315_STAA3) | Conserved hypothetical protein SAUSA300_2315 | Putative lipoprotein | 16.67 | 5.67 | 0.000057 |
| Q2FH36 (CSPA_STAA3) | Cold shock protein CspA | Regulation, transcription factor | 12.33 | 5.00 | 0.0023 |
| Q2FFR1 (TRAP_STAA3) | Signal transduction protein TRAP | Stress response, oxidative stress | 143.33 | 100.67 | 0.0000086 |
| Q2FEZ0 (Q2FEZ0_STAA3) | General stress protein 20U Dps | Stress response, starvation-inducible | 74.67 | 53.00 | 0.0015 |
| Q2FDL5 (Q2FDL5_STAA3) | N-Acetylmuramoyl-l-alanine amidase domain protein SAUSA300_2579 | Unknown | 81.67 | 47.33 | 0.00000044 |
Fisher’s exact test.
PTS, phosphotransferase system.
Proteins with decreased levels on the surface included the cell wall-anchored adhesins SdrC, SdrD, and SdrE. These proteins are members of the Clf-Sdr MSCRAMM family, and all have been demonstrated to promote S. aureus adhesion or immune evasion (46–48). SdrC self-associates to promote biofilm formation (49), while both SdrC and SdrD promote S. aureus adherence to desquamated nasal epithelial cells (38). SdrE inhibits classical (50) and alternative (51) complement activation. The decrease of these proteins in the spl mutant suggests that one or more Spl proteases indirectly affect their abundance, rather than directly degrading them.
Secreted proteomics identified 48 proteins increased (Table 3) and 28 proteins decreased (Table 4) in the spl mutant relative to the USA300 WT strain. As in the surface proteomics, IsdA was increased in the Δspl::erm mutant secretome, suggesting that it is more abundant both on the cell surface and in shedding from the cell due to cleavage or cell lysis. Multiple virulence factors were also increased, including LukD, Sbi, Nuc, and Efb. LukD is part of the LukED bicomponent pore-forming toxin, which is cytolytic to several cell types, including monocytes and polymorphonuclear leukocytes (52, 53). LukED has also been reported to promote S. aureus growth in human blood by facilitating iron scavenging (54). Like SpA, Sbi binds IgG, which prevents opsonophagocytosis of S. aureus (55). Nuc is a potent secreted DNase that is able to degrade neutrophil extracellular traps (NETs), promoting S. aureus survival in the lung and host killing in a murine model of respiratory infection (56). NET degradation by Nuc also induces macrophage death (57). Efb is a secreted protein that binds fibrinogen and complement C3 to form a protective shield around S. aureus and inhibit its phagocytic clearance (58, 59). The increased production of these proteins in the Δspl::erm mutant suggests that it may have enhanced immunomodulatory properties that contribute to its increased lethality in the rabbit model of pneumonia.
TABLE 3 .
Proteins increased in spl mutant spent medium
| GenBank (UniProt) accession no. | Name | Biological function(s) | Avg no. of assigned spectra |
P valuea | |
|---|---|---|---|---|---|
| WT | Δspl::erm | ||||
| Q2FJV7 (Q2FJV7_STAA3) | 5′-Nucleotidase, lipoprotein e(P4) family SAUSA300_0307 | Acid phosphatase | 80.00 | 128.67 | 2.7E−09 |
| Q2FHV1 (ISDA_STAA3) | Iron-regulated surface determinant protein A IsdA | Adhesion, immune evasion | 7.67 | 23.67 | 0.00000036 |
| Q2FF95 (CH60_STAA3) | 60-kDa chaperonin GroL | Housekeeping, protein folding | 12.67 | 32.67 | 0.00000014 |
| Q2FF94 (CH10_STAA3) | 10-kDa chaperonin GroS | Housekeeping, protein folding | 0.00 | 2.33 | 0.0078 |
| Q2FIM5 (CLPP_STAA3) | ATP-dependent Clp protease proteolytic subunit ClpP | Housekeeping, protein turnover | 5.33 | 11.67 | 0.0055 |
| Q2FJ98 (RPOB_STAA3) | DNA-directed RNA polymerase subunit beta RpoB | Housekeeping, transcription | 0.00 | 2.67 | 0.0039 |
| Q2FHI1 (EFTS_STAA3) | Elongation factor Ts/Tsf | Housekeeping, translation | 80.33 | 122.00 | 0.00000021 |
| Q2FES2 (RS9_STAA3) | 30S ribosomal protein S9 RpsI | Housekeeping, translation | 3.00 | 19.00 | 5.9E−10 |
| Q2FG80 (RL21_STAA3) | 50S ribosomal protein L21 RplU | Housekeeping, translation | 5.67 | 14.67 | 0.00037 |
| Q2FFJ4 (GATC_STAA3) | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C GATC | Housekeeping, translation | 0.00 | 2.67 | 0.0039 |
| Q2FEX8 (Q2FEX8_STAA3) | Glucosamine-fructose-6-phosphate aminotransferase (isomerizing) GlmS | Metabolism, amino acid metabolism | 0.00 | 5.67 | 0.0000077 |
| Q2FJ90 (Q2FJ90_STAA3) | Putative pyridoxal phosphate-dependent acyltransferase SAUSA300_0535 | Metabolism, amino acid metabolism | 0.00 | 2.33 | 0.0078 |
| Q2FDQ7 (LDH2_STAA3) | l-Lactate dehydrogenase 2 Ldh2 | Metabolism, carbohydrate metabolism, glycolysis | 25.00 | 55.00 | 0.000000003 |
| Q2FHY7 (Q2FHY7_STAA3) | Pyruvate dehydrogenase E1 component, alpha subunit PdhA | Metabolism, carbohydrate metabolism | 12.33 | 26.67 | 0.000044 |
| Q2FHY5 (Q2FHY5_STAA3) | Dihydrolipoamide acetyltransferase SAUSA300_0995 | Metabolism, carbohydrate metabolism | 5.33 | 14.33 | 0.00029 |
| Q2FKD2 (Q2FKD2_STAA3) | Acetoin (diacetyl) reductase SAUSA300_0129 | Metabolism, carbohydrate metabolism | 0.00 | 2.33 | 0.0078 |
| Q2FIB3 (G6PI_STAA3) | Glucose-6-phosphate isomerase Pgi | Metabolism, carbohydrate metabolism, glycolysis, gluconeogenesis | 45.33 | 71.67 | 0.000014 |
| Q2FIL9 (TPIS_STAA3) | Triosephosphate isomerase TpiA | Metabolism, carbohydrate metabolism, glycolysis, gluconeogenesis | 27.00 | 52.00 | 0.00000063 |
| Q2FE81 (GPMA_STAA3) | 2,3-Bisphosphoglycerate-dependent phosphoglycerate mutase GmpA | Metabolism, carbohydrate metabolism, glycolysis, gluconeogenesis | 13.33 | 33.67 | 0.00000014 |
| Q2FGM3 (Q2FGM3_STAA3) | 6-Phosphogluconate dehydrogenase, decarboxylating Gnd | Metabolism, carbohydrate metabolism, pentose phosphate pathway | 29.33 | 44.33 | 0.0015 |
| Q2FHZ7 (Q2FHZ7_STAA3) | Phosphocarrier protein HPr/PtsH | Metabolism, carbohydrate metabolism, sugar PTSb | 41.00 | 105.50 | 1.4E−14 |
| Q2FIQ8 (Q2FIQ8_STAA3) | Ribonucleoside-diphosphate reductase, beta subunit SAUSA300_0717 | Metabolism, nucleotide metabolism | 3.33 | 8.67 | 0.0057 |
| Q2FI15 (FOLD_STAA3) | Bifunctional protein FolD | Metabolism, one-carbon metabolism, tetrahydrofolate interconversion | 2.67 | 7.33 | 0.0081 |
| Q2FJ56 (Y569_STAA3) | UPF0447 protein SAUSA300_0569 | Putative heme-dependent oxidoreductase | 0.00 | 9.00 | 7.5E−09 |
| Q2FJP2 (Q2FJP2_STAA3) | Conserved hypothetical protein SAUSA300_0372 | Putative lipoprotein | 92.33 | 122.33 | 0.00022 |
| Q2FKJ0 (Q2FKJ0_STAA3) | Putative lysophospholipase SAUSA300_0070 | Putative lysophospholipase | 0.00 | 5.00 | 0.000031 |
| Q2FE21 (Y2422_STAA3) | Uncharacterized oxidoreductase SAUSA300_2422 | Putative oxidoreductase | 0.67 | 4.00 | 0.0065 |
| Q2FEG8 (Q2FEG8_STAA3) | Oxidoreductase, short-chain dehydrogenase/reductase family GN=SAUSA300_2275 | Putative oxidoreductase | 0.00 | 3.00 | 0.002 |
| Q2FFM0 (Q2FFM0_STAA3) | Uncharacterized putative glycosyl hydrolase SAUSA300_1856 | Putative peptidase activity (endopeptidase) | 11.33 | 22.67 | 0.0005 |
| Q2FG28 (Y1656_STAA3) | Putative universal stress protein SAUSA300_1656 | Putative stress response protein | 3.33 | 12.33 | 0.00005 |
| Q2FH36 (CSPA_STAA3) | Cold shock protein CspA | Regulation, transcription factor | 5.33 | 27.33 | 3.7E−12 |
| Q2FJY5 (Q2FJY5_STAA3) | Type VII secretion system protein EsaA SAUSA300_0279 | Secretion | 0.00 | 2.33 | 0.0078 |
| Q2FEV0 (ASP23_STAA3) | Alkaline shock protein 23 Asp23 | Stress response, cell envelope stress/alkaline shock protein | 120.00 | 173.67 | 0.000000029 |
| Q2FGH0 (SODM1_STAA3) | Superoxide dismutase [Mn/Fe] 1 SodA | Stress response, oxidative stress | 17.33 | 35.00 | 0.000014 |
| Q2FDV8 (CLPL_STAA3) | ATP-dependent Clp protease ClpL | Stress response, thermotolerance | 19.33 | 42.33 | 0.00000021 |
| Q2FJK5 (Q2FJK5_STAA3) | Conserved hypothetical protein SAUSA300_0409 | Unknown | 2.33 | 12.00 | 0.0000045 |
| Q2FGB1 (Y1572_STAA3) | Conserved hypothetical protein UPF0473 protein SAUSA300_1572 | Unknown | 0.00 | 6.00 | 0.0000038 |
| Q2FJN1 (Q2FJN1_STAA3) | Conserved hypothetical protein SAUSA300_0383 | Unknown | 0.00 | 4.00 | 0.00025 |
| Q2FFJ2 (Q2FFJ2_STAA3) | CamS sex pheromone cAM373 SAUSA300_1884 | Unknown | 1.67 | 5.67 | 0.0085 |
| Q2FKC5 (Q2FKC5_STAA3) | Cell wall surface anchor family protein SAUSA300_0136 | Unknown | 0.00 | 3.33 | 0.00098 |
| Q2FG32 (Q2FG32_STAA3) | Universal stress protein family protein SAUSA300_1652 | Unknown | 0.00 | 3.33 | 0.00098 |
| Q2FEV8 (Q2FEV8_STAA3) | Transporter gate domain protein SAUSA300_2133 | Unknown | 0.00 | 2.67 | 0.0039 |
| Q2FIK2 (Q2FIK2_STAA3) | Thermonuclease Nuc | Virulence factor | 323.00 | 369.67 | 0.0011 |
| Q2FIH7 (Q2FIH7_STAA3) | Staphylococcal enterotoxin Q Seq/SelQ | Virulence factor | 21.00 | 56.00 | 1.6E−12 |
| Q2FFA2 (LUKL2_STAA3) | Uncharacterized leukocidin-like protein 2 SAUSA300_1975 | Virulence factor, cytolysin | 202.33 | 230.33 | 0.01 |
| Q2FFR9 (Q2FFR9_STAA3) | Leukotoxin LukD | Virulence factor, cytolysin | 2.00 | 2.33 | 0.00000021 |
| Q2FE79 (SBI_STAA3) | Immunoglobulin-binding protein Sbi | Virulence factor, immunomodulator | 54.67 | 90.00 | 0.0000002 |
| Q2FHS5 (Q2FHS5_STAA3) | Fibrinogen-binding protein Efb | Virulence factor, immunomodulator | 0.00 | 5.00 | 0.000031 |
Fisher’s exact test.
PTS, phosphotransferase system.
TABLE 4 .
Proteins decreased in spl mutant spent medium
| GenBank (UniProt) accession no. | Name | Biological function | Avg no. of assigned spectra |
P valuea | |
|---|---|---|---|---|---|
| WT | Δspl::erm | ||||
| Q2FE04 (Q2FE04_STAA3) | Fibronectin-binding protein B FnBPB | Adhesion | 5.67 | 1.00 | 0.0013 |
| Q2FE08 (Q2FE08_STAA3) | Putative cell wall surface anchor family protein surface protein G SAUSA300_2436 | Adhesion, biofilm formation (nonfunctional in SAUSA300) | 193.33 | 126.67 | 4E−11 |
| Q2FI25 (Q2FI25_STAA3) | Autolysin Atl | Autolysis | 657.67 | 605.33 | 0.0043 |
| Q2FJH7 (SLE1_STAA3) | N-Acetylmuramoyl-l-alanine amidase Sle1 | Cell wall turnover | 72.67 | 54.33 | 0.0027 |
| Q2FDT8 (ISAA_STAA3) | Transglycosylase IsaA | Cell wall turnover | 140.00 | 99.33 | 0.0000026 |
| Q2FEP2 (RL2_STAA3) | 50S ribosomal protein L2 RplB | Housekeeping, translation | 6.00 | 0.67 | 0.0002 |
| Q2FJP8 (RS6_STAA3) | 30S ribosomal protein s6 RpsF | Housekeeping, translation | 11.67 | 1.00 | 0.000000033 |
| Q2FE84 (Q2FE84_STAA3) | Amino acid ABC transporter, amino acid-binding protein SAUSA300_2359 | Membrane transporter | 33.33 | 22.00 | 0.005 |
| Q2FIL7 (ENO_STAA3) | Enolase Eno | Metabolism, carbohydrate metabolism, glycolysis | 354.67 | 297.00 | 0.000037 |
| Q2FJM5 (GUAA_STAA3) | GMP synthase (glutamine-hydrolyzing) GuaA | Metabolism, nucleotide metabolism, purine metabolism | 18.00 | 9.67 | 0.004 |
| Q2FFI6 (Q2FFI6_STAA3) | Staphopain A ScpA | Protease, cysteine protease | 102.33 | 69.00 | 0.0000053 |
| Q2FFT4 (SPLF_STAA3) | Serine protease SplF | Protease, serine protease | 3.67 | 0.00 | 0.00000024 |
| Q2FFT0 (SPLB_STAA3) | Serine protease SplB | Protease, serine protease | 9.67 | 0.00 | 3.3E−18 |
| Q2FFT1 (SPLC_STAA3) | Serine protease SplC | Protease, serine protease | 15.00 | 0.00 | 2.8E−14 |
| Q2FFS8 (Q2FFS8_STAA3) | Conserved hypothetical protein SAUSA300_1759 | Putative β-lactamase | 68.33 | 53.33 | 0.01 |
| Q2FJK6 (Q2FJK6_STAA3) | Conserved hypothetical protein SAUSA300_0408 | Putative surface protein | 58.00 | 43.33 | 0.0065 |
| Q2FHI3 (Cody_STAA3) | GTP-sensing transcriptional pleiotropic repressor CodY | Regulation, transcription factor | 5.67 | 1.33 | 0.0036 |
| Q2FI16 (Q2FI16_STAA3) | Chitinase-related protein SAUSA300_0964 | Unknown | 7.67 | 2.33 | 0.0026 |
| Q2FEJ0 (Q2FEJ0_STAA3) | Secretory antigen precursor SsaA | Unknown | 18.67 | 8.33 | 0.00037 |
| Q2FI95 (Q2FI95_STAA3) | Putative surface protein similar to Map-w SAUSA300_0883 | Unknown | 279.00 | 238.67 | 0.00098 |
| Q2FE76 (Q2FE76_STAA3) | Gamma-hemolysin component B HlgB | Virulence factor, cytolysin | 190.67 | 153.33 | 0.0002 |
| Q2FFA3 (LUKL1_STAA3) | Uncharacterized leukocidin-like protein 1 SAUSA300_1974 | Virulence factor, cytolysin | 153.00 | 105.33 | 0.00000013 |
| Q2FF89 (Q2FF89_STAA3) | Delta-hemolysin Hld | Virulence factor, cytolysin, immunomodulator | 15.33 | 2.67 | 0.000000067 |
| Q2FHR4 (Q2FHR4_STAA3) | Antibacterial protein PSM-β1 | Virulence factor, immunomodulator | 459.33 | 383.00 | 0.0000017 |
| Q2FFF8 (SCIN_STAA3) | SCIN | Virulence factor, immunomodulator | 326.67 | 250.33 | 0.000000013 |
| Q2FES8 (Q2FES8_STAA3) | MHCb class II analog protein Map-w SAUSA300_2164 | Virulence factor, immunomodulator | 578.67 | 500.67 | 0.000012 |
| Q2FHR3 (Q2FHR3_STAA3) | Antibacterial protein PSM-β2 | Virulence factor, immunomodulator | 210.00 | 86.67 | 6E−37 |
| Q2FIH8 (Q2FIH8_STAA3) | Staphylococcal enterotoxin K Sek/SelK | Virulence factor, toxin | 32.00 | 19.33 | 0.0013 |
Fisher’s exact test.
MHC, major histocompatibility complex.
In contrast, multiple secreted virulence factors are decreased in abundance in the spl mutant, including the Υ hemolysin component HlgB, the staphylococcal complement inhibitor (SCIN), and the δ hemolysin Hld (Table 4). HlgB is the F subunit of the Υ hemolysin pore-forming toxins (60–62). Hlg toxins have been shown to contribute to virulence in a murine model of septic arthritis (63) and to induce eye injury when directly injected intraocularly into rabbits (64), as well as promote S. aureus virulence in rabbit endophthalmitis infection models (65–67). SCIN is a secreted protein that inhibits complement activation by binding to C3 convertases (68, 69). The Hld cytolysin is translated from RNAIII, the regulatory RNA that is the effector of the staphylococcal agr system (70). Decreased RNAIII would indicate lower agr activation in the spl mutant; however, our finding that Hla and ScpA activities are not decreased in the spl mutant suggests that this is not the case (Fig. 3B and C). Altered Hld levels therefore may be due to a posttranslational modification.
DISCUSSION
The Spl proteases are an intriguing topic because, despite evidence that they contribute to S. aureus-host interaction, their specific functions have not been identified. Structural and activity analyses of SplA, SplB, and SplD have revealed that these enzymes are likely highly specific, given that they have cleavage preferences at subsites beyond the P1 residue of cleavage (26, 71–73). Further, the Spls are not produced as zymogens but, in some cases, appear to require the binding of a preferred substrate in order to take on an enzymatically active conformation (72), as has been demonstrated by characterization of SplB activity (71). The goals of this study were to investigate the function of the spl operon in virulence and to identify possible substrates of the Spl proteases.
The virulence phenotype of the spl mutant is complex, since the mutant-infected animals had a statistically insignificant change in lethality but more localized, less disseminated lung damage. We tested SplA cleavage of predicted substrate mucin 16 (26) and found that it induces the shedding of this protein from CalU-3 cells. SplA may therefore promote S. aureus invasion and spreading by removing mucin 16 from epithelial cells. Since mucin 16 is found on ocular, airway, and female reproductive tract epithelial cells (19), this function could facilitate infection at multiple body sites. This finding is also the first report of an Spl enzyme potentially cleaving a host target.
Proteomic studies of the secreted and surface proteins present on the USA300 WT and spl mutant strains revealed many proteins altered in abundance. The fact that intracellular and housekeeping genes were identified is one caveat of this experiment, suggesting that cell lysis may have occurred during sample preparation. However, none of the proteins identified contain the consensus motifs that have been identified for substrates of the Spls. These are W/Y-L-Y-S/T for SplA (26), W-E-L-Q for SplB (71), and R-Y/W-P/L-T/L/I/V-S for SplD (73). We therefore cannot predict any specific hits that are directly cleaved. However, it is possible that the Spls are able to cleave substrates with some deviation from these consensus motifs. The abundance of some proteins may also be indirectly affected by the Spls. Alternatively, SplC or SplE could be responsible for some effects, since only SplA, SplB, and SplD cleavage preferences have been published. Considering that the amino acid sequences of SplD and SplF are 94.6% identical (3), their activities are likely identical.
The proteomic results show virulence factors in both the increased and decreased protein groups, so it is difficult to predict specific virulence factors that are responsible for the virulence phenotype observed. A few of the proteomic hits also do not match our in vitro studies. For example, ScpA was found to be decreased in the spl mutant (Table 4) but unaltered when we tested its activity with a specific ScpA substrate (Fig. 3C). The decrease in Hld also could correspond to lower RNAIII, indicating lower agr activation, but this was also not observed in our Hla and protease studies (Fig. 3B and C). Hld translation could therefore be altered, or it may be degraded posttranslationally. Hld is a member of the phenol-soluble modulin (PSM) family of peptides, which are known to be protease labile (74, 75). Thus, the lower Hld levels in the spl mutant could be due to degradation by an unknown protease.
Our findings have some similarities to a previous study that tested the virulence and proteomics of a total protease knockout of S. aureus (74). In a murine model of sepsis, the protease null mutant had increased lethality in mice but reduced invasion of organs. Proteomics also identified an increase in the abundance of agr-regulated virulence factors such as Hla and other leukocidins. Although we did not observe an increase in Hla, some of our proteomic results mirrored those of the previously reported study. For example, the previous paper reported increased abundance of Sbi, LukE, Seq, Geh, Efb, IsdA, IsaA, Spa, and EbpS in the protease null mutant, all of which were increased in our spl mutant as well (Tables 2 and 4). This suggests that the Spl proteases contributed to the previously reported phenotypes and corroborates the model in which secreted proteases modulate virulence factor production.
A recent study found that the Spls are important mediators of the host allergic airway response to S. aureus (8). In the serum of five healthy S. aureus carriers, strong IgG4 antibody binding to the Spls was observed. Purified SplD induced a Th2-type response in a tissue explant model with tissue from human nasal polyps. In a mouse allergy model, inhalation of SplD also induced eosinophil infiltration and IgE antibodies to SplD (8). IgG4, IgE, and other Th2-mediated responses are involved in allergic airway disease (75). Interestingly, many allergens that induce allergic responses in the lungs are proteases, including proteases produced by Aspergillus fumigatus (76) and mites and cockroaches (77). These findings indicate that, in addition to SplD, the other Spls may cleave substrates in airway cells and contribute to allergic airway disease and other immune responses in the airway. Our finding that SplA induces mucin 16 shedding adds to this model, as mucin 16 removal could facilitate the ability of the other proteases to act on host cell targets.
The Spls are clearly highly specific proteases, and their natural targets have been difficult to identify. However, mounting evidence suggests that they are expressed in vivo and affect host immune responses. Future work will tease apart the roles of individual Spls in interaction with the host and unravel their host effects, hopefully by identification of their cleavage substrates in the host.
MATERIALS AND METHODS
Strains and plasmids.
LAC is a USA300 CA-MRSA strain (78), and the erythromycin-sensitive version called USA300 WT (79) was used in this study. The LAC Δspl::erm mutant strain was generated by phage transduction from RN6390 Δspl::erm (3) into LAC with φ80α. Strains were cultured on tryptic soy agar (TSA) plates or in tryptic soy broth (TSB) at 37°C, with shaking at 200 rpm for broth culture.
The SplA purification plasmid pSK236/splA6xHis was kindly provided by Suzan Rooijakkers of the University Medical Center Utrecht. This plasmid contains C-terminally His6-tagged splA that is under control of the S. aureus scn (SCIN) promoter. The plasmid was moved by electroporation into S. aureus Newman to maximize SplA production, since scn is positively regulated by the sae system (80) and sae expression is enhanced in the Newman strain (81).
Rabbit pneumonia model.
The rabbit pneumonia model was prepared in accordance with reference 18. The USA300 WT and Δspl::erm mutant strains were grown overnight in TSB, washed once in sterile TSB, and resuspended at 1 × 1010 CFU/ml. Adult Dutch belted rabbits (Bakkom) were anesthetized with ketamine (25 mg/kg) and xylazine (20 mg/kg) administered subcutaneously. A ventral midline incision was made through the skin and into the trachea; a catheter was then inserted and fed into the lungs. Approximately 2 × 109 CFU of washed bacteria in 200 µl of TSB was delivered through the catheter to each rabbit. Inocula were plated to confirm the dose given. The actual dose was 2 × 109 to 3 × 109 CFU. The incisions were closed, and the rabbits were monitored for 6 days. Over the course of the experiment, the rabbits were treated for pain with buprenorphine (0.05 mg/kg twice daily) through intramuscular injection. Rabbits were euthanized by intravenous injection of 1 ml of a mixture of the sodium salts of phenytoin and pentobarbital (Beuthanasia-D) either on day 6 or if they were found during the course of the experiment to be unable to right themselves or exhibit escape behavior. At the time of death, rabbit lungs were photographed; tissues were fixed (10% neutral buffered formalin), processed, and embedded; and tissue sections were stained with H&E and histopathologically scored in accordance with standard principles (82).
SplA purification.
S. aureus Newman expressing pSK236/splA6xHis was grown overnight in 5 ml of TSB plus 10 µg/ml chloramphenicol, and then the entire culture was added to 200 ml of TSB and grown overnight. The spent medium was harvested by centrifugation and filtration, and 121 g of ammonium sulfate was slowly dissolved in the spent medium to achieve approximately 85% saturation. The sample was centrifuged at 30,000 × g at 4°C for 30 min to collect the pellet, which was dissolved in 10 ml of PBS and dialyzed into 50 mM Na2HPO4 with 0.3 M NaCl at pH 8.0 and then into the same buffer with 10 mM imidazole for loading onto the His-Select nickel affinity gel (Sigma). Batch purification with the His-Select nickel affinity gel was performed according to the manufacturer’s protocol. The final protein was dialyzed into PBS and stored at −80°C.
Mucin 16 slot blot assay.
CalU-3 cells were seeded at 500,000/well into 24-well plates and cultured with 1 ml/well minimum essential medium (Gibco) containing 10% heat-inactivated fetal bovine serum (Gibco) and 1× nonessential amino acids (Gibco) at 37°C in 5% CO2 for 2 days. For the SplA-induced shedding experiment, purified SplA was added to fresh medium without serum at 500 µl/well to avoid serum cross-reactivity with the mucin 16 antibody. For inhibition of SplA, 3,4-DCI was added at a final concentration of 10 µM. The cells were incubated with SplA at 37°C in 5% CO2 for 2 h, and the medium was removed for Western blotting. For the slot blot assay, samples were loaded onto polyvinylidene difluoride (PVDF) with a vacuum and a slot blot apparatus. The PVDF was first equilibrated in methanol for 15 s, in distilled H2O (dH2O) for 2 min, and in TBS plus 0.5% Tween 20 for 5 min. It was then placed into the slot blot apparatus on top of two pieces of filter paper wetted with TBS plus 0.5% Tween, and the vacuum was turned on low to drain excess buffer. A 100-µl volume of each sample was loaded into the slots, and then the vacuum was turned on until the liquid was absorbed. TBS plus 0.5% Tween 20 at 100 µl/well was then loaded to wash any unbound sample from the wells. The membrane was dipped into methanol and then dH2O and then blocked for 2 h in 5% BSA in TBS plus 0.5% Tween 20. A primary mouse anti-CA 125 antibody was diluted 1:200 in blocking buffer and incubated for 1 h, and a horseradish peroxidase-conjugated goat anti-mouse secondary antibody was diluted 1:5,000 in blocking buffer and incubated for 1 h. The membrane was washed with TBS plus 0.5% Tween 20 between steps. The membrane was incubated in chemiluminescent substrate and exposed to film.
Blood survival assay.
Blood survival of the USA300 WT and Δspl::erm mutant strains was measured as previously described (83). Strains were subcultured and grown to mid-log phase (optical density at 600 nm [OD600] of 0.8) and centrifuged at 8,000 × g for 5 min. Bacteria were washed twice in TSB and resuspended to 1 ml. A 100-µl volume was added to 1 ml of heparinized human blood, and the culture was placed on a tumbler at 37°C for 3 h. Serial dilutions were plated on TSA and counted.
Hemolysis activity assay.
An Hla activity assay was performed as previously described (84). Briefly, overnight cultures of the USA300 WT and Δspl::erm mutant strains were subcultured at 1:500 in TSB and grown for 24 h. Spent medium from these cultures was serially diluted 2-fold across a 96-well plate. Defibrinated rabbit blood (HemoStat Laboratories) was centrifuged to pellet rabbit erythrocytes, which were washed three times in 1.1× PBS and resuspended to a final concentration of 3%. A 30-µl volume of each spent medium dilution was mixed with 70 µl of the erythrocyte solution in the 96-well plate and then incubated statically at room temperature for 1 h, after which the OD630 was measured. The OD630 was plotted versus the percent spent medium on a four-parameter logistic curve with the Prism program, and the midpoint of the curve (the percentage of medium needed to lyse 50% of the erythrocytes) was used as an indicator of activity.
ScpA activity assay.
Overnight cultures of the USA300 WT and Δspl::erm mutant strains were diluted to an OD600 of 0.1 in 25 ml of TSB and grown for 24 h. At selected time points, spent medium was collected and incubated with a fluorescent resonance energy transfer (FRET) substrate that is based on the CXCR2 ScpA cleavage site (34). The FRET assay was performed in accordance with reference 85. The substrate was resuspended to a concentration of 50 µM in 20 mM Tris-HCl, pH 7.4. Spent medium was also buffered to 20 mM Tris-HCl, pH 7.4. The substrate (25 µl) was mixed with 175 µl of buffered spent medium, and fluorescence (excitation, 490 nm; emission, 520 nm) measurements were taken every 2 min for 30 min with a Tecan plate reader. The activity at each time point was expressed as the slope (fluorescence/time).
Aur and hemolysis plate activity assays.
Overnight cultures of the USA300 WT and Δspl::erm mutant strains were spotted in 5-µl amounts onto blood agar (5% [vol/vol] rabbit blood and 3% Bacto agar) or milk agar (5% nonfat dry milk and 3% Bacto agar) plates. The plates were incubated overnight at 37°C, and pictures of colonies with zones of clearing were taken.
Proteomics.
Overnight cultures of the USA300 WT and Δspl::erm mutant strains were grown in TSB and harvested for secreted and surface proteomic profiling. Secreted proteome profiling was carried out in accordance with reference 86, and surface proteomic analysis was carried out in accordance with reference 87.
ACKNOWLEDGMENTS
We thank Les Shaw (University of South Florida, Tampa, FL) for assistance with proteomic studies and Suzan Rooijakkers (Universiteit Utrecht, Utrecht, the Netherlands) for kindly providing the SplA purification plasmid.
A.E.P. was funded by American Heart Association predoctoral fellowship award 14PRE19910005. Studies in the laboratory of A.R.H. are supported by Project 3 of NIH grant AI083211. W.S.-P. and P.M.S. were supported by startup funds from the University of Iowa Carver College of Medicine.
REFERENCES
- 1.Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis 13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Reed SB, Wesson CA, Liou LE, Trumble WR, Schlievert PM, Bohach GA, Bayles KW. 2001. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect Immun 69:1521–1527. doi: 10.1128/IAI.69.3.1521-1527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zdzalik M, Karim AY, Wolski K, Buda P, Wojcik K, Brueggemann S, Wojciechowski P, Eick S, Calander A-M, Jonsson I-M, Kubica M, Polakowska K, Miedzobrodzki J, Wladyka B, Potempa J, Dubin G. 2012. Prevalence of genes encoding extracellular proteases in Staphylococcus aureus—important targets for triggering immune response in vivo. FEMS Immunol Med Microbiol 66:220–229. doi: 10.1111/j.1574-695X.2012.01005.x. [DOI] [PubMed] [Google Scholar]
- 6.Rieneck K, Renneberg J, Diamant M, Gutschik E, Bendtzen K. 1997. Molecular cloning and expression of a novel Staphylococcus aureus antigen. Biochim Biophys Acta 1350:128–132. doi: 10.1016/S0167-4781(96)00216-3. [DOI] [PubMed] [Google Scholar]
- 7.Holtfreter S, Nguyen TT, Wertheim H, Steil L, Kusch H, Truong QP, Engelmann S, Hecker M, Völker U, van Belkum A, Bröker BM. 2009. Human immune proteome in experimental colonization with Staphylococcus aureus. Clin Vaccine Immunol 16:1607–1614. doi: 10.1128/CVI.00263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stentzel S, Teufelberger A, Nordengrün M, Kolata J, Schmidt F, van Crombruggen K, Michalik S, Kumpfmüller J, Tischer S, Schweder T, Hecker M, Engelmann S, Völker U, Krysko O, Bachert C, Bröker BM 10 May 2016. Spls are pacemakers of allergic airway reactions to Staphylococcus aureus. J Allergy Clin Immunol. doi: 10.1016/j.jaci.2016.03.045. [DOI] [PubMed] [Google Scholar]
- 9.Thurlow LR, Joshi GS, Richardson AR. 2012. Virulence strategies of the dominant USA300 lineage of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). FEMS Immunol Med Microbiol 65:5–22. doi: 10.1111/j.1574-695X.2012.00937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. 2012. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr Opin Microbiol 15:588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, Albrecht V, Limbago B, Talan DA, EMERGEncy ID NET Study Group . 2012. Prevalence of methicillin-resistant Staphylococcus aureus as an etiology of community-acquired pneumonia. Clin Infect Dis 54:1126–1133. doi: 10.1093/cid/cis022. [DOI] [PubMed] [Google Scholar]
- 12.Pasquale TR, Jabrocki B, Salstrom SJ, Wiemken TL, Peyrani P, Haque NZ, Scerpella EG, Ford KD, Zervos MJ, Ramirez JA, File TM Jr., IMPACT-HAP Study Group . 2013. Emergence of methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of late-onset nosocomial pneumonia in intensive care patients in the USA. Int J Infect Dis 17:e398–e403 doi: 10.1016/j.ijid.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. 2010. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis 201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diep BA, Afasizheva A, Le HN, Kajikawa O, Matute-Bello G, Tkaczyk C, Sellman B, Badiou C, Lina G, Chambers HF. 2013. Effects of linezolid on suppressing in vivo production of staphylococcal toxins and improving survival outcomes in a rabbit model of methicillin-resistant Staphylococcus aureus necrotizing pneumonia. J Infect Dis 208:75–82. doi: 10.1093/infdis/jit129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashida A, Bartlett AH, Foster TJ, Park PW. 2009. Staphylococcus aureus beta-toxin induces lung injury through syndecan-1. Am J Pathol 174:509–518. doi: 10.2353/ajpath.2009.080394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hongo I, Baba T, Oishi K, Morimoto Y, Ito T, Hiramatsu K. 2009. Phenol-soluble modulin alpha 3 enhances the human neutrophil lysis mediated by Panton-Valentine leukocidin. J Infect Dis 200:715–723. doi: 10.1086/605332. [DOI] [PubMed] [Google Scholar]
- 17.Schlievert P. 2009. Cytolysins, superantigens, and pneumonia due to community-associated methicillin-resistant Staphylococcus aureus. J Infect Dis 200:676–678. doi: 10.1086/605333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson GJ, Seo KS, Cartwright RA, Connelley T, Chuang-Smith ON, Merriman JA, Guinane CM, Park JY, Bohach GA, Schlievert PM, Morrison WI, Fitzgerald JR. 2011. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog 7:e1002271. doi: 10.1371/journal.ppat.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haridas D, Ponnusamy MP, Chugh S, Lakshmanan I, Seshacharyulu P, Batra SK. 2014. MUC16: molecular analysis and its functional implications in benign and malignant conditions. FASEB J 28:4183–4199. doi: 10.1096/fj.14-257352. [DOI] [PubMed] [Google Scholar]
- 20.O’Brien TJ, Beard JB, Underwood LJ, Shigemasa K. 2002. The CA 125 gene: a newly discovered extension of the glycosylated N-terminal domain doubles the size of this extracellular superstructure. Tumour Biol 23:154–169. doi: 10.1159/64032. [DOI] [PubMed] [Google Scholar]
- 21.Govindarajan B, Gipson IK. 2010. Membrane-tethered mucins have multiple functions on the ocular surface. Exp Eye Res 90:655–663. doi: 10.1016/j.exer.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blalock TD, Spurr-Michaud SJ, Tisdale AS, Heimer SR, Gilmore MS, Ramesh V, Gipson IK. 2007. Functions of MUC16 in corneal epithelial cells. Invest Ophthalmol Vis Sci 48:4509–4518. doi: 10.1167/iovs.07-0430. [DOI] [PubMed] [Google Scholar]
- 23.Gipson IK, Spurr-Michaud S, Tisdale A, Menon BB. 2014. Comparison of the transmembrane mucins MUC1 and MUC16 in epithelial barrier function. PLoS One 9:e100393. doi: 10.1371/journal.pone.0100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govindarajan B, Menon BB, Spurr-Michaud S, Rastogi K, Gilmore MS, Argüeso P, Gipson IK. 2012. A metalloproteinase secreted by Streptococcus pneumoniae removes membrane mucin MUC16 from the epithelial glycocalyx barrier. PLoS One 7:e32418. doi: 10.1371/journal.pone.0032418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menon BB, Zhou X, Spurr-Michaud S, Rajaiya J, Chodosh J, Gipson IK. 2016. Epidemic keratoconjunctivitis-causing adenoviruses induce MUC16 ectodomain release to infect ocular surface epithelial cells. mSphere 1:e00112-15. doi: 10.1128/mSphere.00112-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stec-Niemczyk J, Pustelny K, Kisielewska M, Bista M, Boulware KT, Stennicke HR, Thogersen IB, Daugherty PS, Enghild JJ, Baczynski K, Popowicz GM, Dubin A, Potempa J, Dubin G. 2009. Structural and functional characterization of SplA, an exclusively specific protease of Staphylococcus aureus. Biochem J 419:555–564. doi: 10.1042/BJ20081351. [DOI] [PubMed] [Google Scholar]
- 27.Davies JR, Kirkham S, Svitacheva N, Thornton DJ, Carlstedt I. 2007. MUC16 is produced in tracheal surface epithelium and submucosal glands and is present in secretions from normal human airway and cultured bronchial epithelial cells. Int J Biochem Cell Biol 39:1943–1954. doi: 10.1016/j.biocel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Blalock TD, Spurr-Michaud SJ, Tisdale AS, Gipson IK. 2008. Release of membrane-associated mucins from ocular surface epithelia. Invest Ophthalmol Vis Sci 49:1864–1871. doi: 10.1167/iovs.07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thammavongsa V, Kim HK, Missiakas D, Schneewind O. 2015. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 13:529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berube BJ, Bubeck Wardenburg J. 2013. Staphylococcus aureus alpha-toxin: nearly a century of intrigue. Toxins 5:1140–1166. doi: 10.3390/toxins5061140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kebaier C, Chamberland RR, Allen IC, Gao X, Broglie PM, Hall JD, Jania C, Doerschuk CM, Tilley SL, Duncan JA. 2012. Staphylococcus aureus alpha-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J Infect Dis 205:807–817. doi: 10.1093/infdis/jir846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. 2010. agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun 2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jusko M, Potempa J, Kantyka T, Bielecka E, Miller HK, Kalinska M, Dubin G, Garred P, Shaw LN, Blom AM. 2014. Staphylococcal proteases aid in evasion of the human complement system. J Innate Immun 6:31–46. doi: 10.1159/000351458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laarman AJ, Mijnheer G, Mootz JM, van Rooijen WJ, Ruyken M, Malone CL, Heezius EC, Ward R, Milligan G, van Strijp JA, de Haas CJ, Horswill AR, van Kessel KP, Rooijakkers SH. 2012. Staphylococcus aureus staphopain A inhibits CXCR2-dependent neutrophil activation and chemotaxis. EMBO J 31:3607–3619. doi: 10.1038/emboj.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohbayashi T, Irie A, Murakami Y, Nowak M, Potempa J, Nishimura Y, Shinohara M, Imamura T. 2011. Degradation of fibrinogen and collagen by staphopains, cysteine proteases released from Staphylococcus aureus. Microbiology 157:786–792. doi: 10.1099/mic.0.044503-0. [DOI] [PubMed] [Google Scholar]
- 36.Laarman AJ, Ruyken M, Malone CL, van Strijp JA, Horswill AR, Rooijakkers SH. 2011. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol 186:6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 37.Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Büttner H, Dunman PM, Rohde H, Cech NB, Fey PD, Horswill AR. 2014. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol 196:3482–3493. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corrigan RM, Miajlovic H, Foster TJ. 2009. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol 9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarke SR, Wiltshire MD, Foster SJ. 2004. IsdA of Staphylococcus aureus is a broad spectrum, iron-regulated adhesin. Mol Microbiol 51:1509–1519. doi: 10.1111/j.1365-2958.2003.03938.x. [DOI] [PubMed] [Google Scholar]
- 40.Clarke SR, Mohamed R, Bian L, Routh AF, Kokai-Kun JF, Mond JJ, Tarkowski A, Foster SJ. 2007. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 1:199–212. doi: 10.1016/j.chom.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Forsgren A, Quie PG. 1974. Effects of staphylococcal protein A on heat labile opsonins. J Immunol 112:1177–1180. [PubMed] [Google Scholar]
- 42.Falugi F, Kim HK, Missiakas DM, Schneewind O. 2013. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio 4:e00575-13. doi: 10.1128/mBio.00575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roche FM, Downer R, Keane F, Speziale P, Park PW, Foster TJ. 2004. The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J Biol Chem 279:38433–38440. doi: 10.1074/jbc.M402122200. [DOI] [PubMed] [Google Scholar]
- 44.Nakakido M, Aikawa C, Nakagawa I, Tsumoto K. 2014. The staphylococcal elation-binding protein regulates zinc-dependent growth/biofilm formation. J Biochem 156:155–162. doi: 10.1093/jb/mvu027. [DOI] [PubMed] [Google Scholar]
- 45.Jenkins A, Diep BA, Mai TT, Vo NH, Warrener P, Suzich J, Stover CK, Sellman BR. 2015. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. mBio 6:e02272-14. doi: 10.1128/mBio.02272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paharik AE, Horswill AR. 2016. The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol Spectr 4. doi: 10.1128/microbiolspec.VMBF-0022-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Josefsson E, McCrea KW, Ní Eidhin D, O’Connell D, Cox J, Höök M, Foster TJ. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144:3387–3395. doi: 10.1099/00221287-144-12-3387. [DOI] [PubMed] [Google Scholar]
- 49.Barbu EM, Mackenzie C, Foster TJ, Höök M. 2014. SdrC induces staphylococcal biofilm formation through a homophilic interaction. Mol Microbiol 94:172–185. doi: 10.1111/mmi.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hair PS, Foley CK, Krishna NK, Nyalwidhe JO, Geoghegan JA, Foster TJ, Cunnion KM. 2013. Complement regulator C4BP binds to Staphylococcus aureus surface proteins SdrE and Bbp inhibiting bacterial opsonization and killing. Results Immunol 3:114–121. doi: 10.1016/j.rinim.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharp JA, Echague CG, Hair PS, Ward MD, Nyalwidhe JO, Geoghegan JA, Foster TJ, Cunnion KM. 2012. Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS One 7:e38407. doi: 10.1371/journal.pone.0038407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyes-Robles T, Alonzo F III, Kozhaya L, Lacy DB, Unutmaz D, Torres VJ. 2013. Staphylococcus aureus leukotoxin ED targets the chemokine receptors CXCR1 and CXCR2 to kill leukocytes and promote infection. Cell Host Microbe 14:453–459. doi: 10.1016/j.chom.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alonzo F III, Kozhaya L, Rawlings SA, Reyes-Robles T, DuMont AL, Myszka DG, Landau NR, Unutmaz D, Torres VJ. 2013. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493:51–55. doi: 10.1038/nature11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoong P, Torres VJ. 2015. Counter inhibition between leukotoxins attenuates Staphylococcus aureus virulence. Nat Commun 6:8125. doi: 10.1038/ncomms9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gonzalez CD, Ledo C, Giai C, Garófalo A, Gómez MI. 2015. The Sbi protein contributes to Staphylococcus aureus inflammatory response during systemic infection. PLoS One 10:e0131879. doi: 10.1371/journal.pone.0131879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Köckritz-Blickwede M. 2010. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun 2:576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thammavongsa V, Missiakas DM, Schneewind O. 2013. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342:863–866. doi: 10.1126/science.1242255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ko YP, Kuipers A, Freitag CM, Jongerius I, Medina E, van Rooijen WJ, Spaan AN, van Kessel KP, Höök M, Rooijakkers SH. 2013. Phagocytosis escape by a Staphylococcus aureus protein that connects complement and coagulation proteins at the bacterial surface. PLoS Pathog 9:e1003816. doi: 10.1371/journal.ppat.1003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ko YP, Kang M, Ganesh VK, Ravirajan D, Li B, Höök M. 2016. Coagulase and Efb of Staphylococcus aureus have a common fibrinogen binding motif. mBio 7:e01885-15. doi: 10.1128/mBio.01885-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dalla Serra M, Coraiola M, Viero G, Comai M, Potrich C, Ferreras M, Baba-Moussa L, Colin DA, Menestrina G, Bhakdi S, Prévost G. 2005. Staphylococcus aureus bicomponent gamma-hemolysins, HlgA, HlgB, and HlgC, can form mixed pores containing all components. J Chem Inf Model 45:1539–1545. doi: 10.1021/ci050175y. [DOI] [PubMed] [Google Scholar]
- 61.Alonzo F III, Torres VJ. 2014. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol Mol Biol Rev 78:199–230. doi: 10.1128/MMBR.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prévost G, Cribier B, Couppié P, Petiau P, Supersac G, Finck-Barbançon V, Monteil H, Piemont Y. 1995. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect Immun 63:4121–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nilsson IM, Hartford O, Foster T, Tarkowski A. 1999. Alpha-toxin and gamma-toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect Immun 67:1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siqueira JA, Speeg-Schatz C, Freitas FI, Sahel J, Monteil H, Prévost G. 1997. Channel-forming leucotoxins from Staphylococcus aureus cause severe inflammatory reactions in a rabbit eye model. J Med Microbiol 46:486–494. doi: 10.1099/00222615-46-6-486. [DOI] [PubMed] [Google Scholar]
- 65.Dajcs JJ, Austin MS, Sloop GD, Moreau JM, Hume EB, Thompson HW, McAleese FM, Foster TJ, O’Callaghan RJ. 2002. Corneal pathogenesis of Staphylococcus aureus strain Newman. Invest Ophthalmol Vis Sci 43:1109–1115. [PubMed] [Google Scholar]
- 66.Dajcs JJ, Thibodeaux BA, Girgis DO, O’Callaghan RJ. 2002. Corneal virulence of Staphylococcus aureus in an experimental model of keratitis. DNA Cell Biol 21:375–382. doi: 10.1089/10445490260099656. [DOI] [PubMed] [Google Scholar]
- 67.Supersac G, Piémont Y, Kubina M, Prévost G, Foster TJ. 1998. Assessment of the role of gamma-toxin in experimental endophthalmitis using a hlg-deficient mutant of Staphylococcus aureus. Microb Pathog 24:241–251. doi: 10.1006/mpat.1997.0192. [DOI] [PubMed] [Google Scholar]
- 68.Rooijakkers SH, Ruyken M, Roos A, Daha MR, Presanis JS, Sim RB, van Wamel WJ, van Kessel KP, van Strijp JA. 2005. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat Immunol 6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 69.Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJ, van Strijp JA, Gros P. 2009. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol 10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem Rev 111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dubin G, Stec-Niemczyk J, Kisielewska M, Pustelny K, Popowicz GM, Bista M, Kantyka T, Boulware KT, Stennicke HR, Czarna A, Phopaisarn M, Daugherty PS, Thøgersen IB, Enghild JJ, Thornberry N, Dubin A, Potempa J. 2008. Enzymatic activity of the Staphylococcus aureus SplB serine protease is induced by substrates containing the sequence Trp–Glu–Leu–Gln. J Mol Biol 379:343–356. doi: 10.1016/j.jmb.2008.03.059. [DOI] [PubMed] [Google Scholar]
- 72.Popowicz GM, Dubin G, Stec-Niemczyk J, Czarny A, Dubin A, Potempa J, Holak TA. 2006. Functional and structural characterization of Spl proteases from Staphylococcus aureus. J Mol Biol 358:270–279. doi: 10.1016/j.jmb.2006.01.098. [DOI] [PubMed] [Google Scholar]
- 73.Zdzalik M, Kalinska M, Wysocka M, Stec-Niemczyk J, Cichon P, Stach N, Gruba N, Stennicke HR, Jabaiah A, Markiewicz M, Kedracka-Krok S, Wladyka B, Daugherty PS, Lesner A, Rolka K, Dubin A, Potempa J, Dubin G. 2013. Biochemical and structural characterization of SplD protease from Staphylococcus aureus. PLoS One 8:e76812. doi: 10.1371/journal.pone.0076812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kolar SL, Ibarra JA, Rivera FE, Mootz JM, Davenport JE, Stevens SM, Horswill AR, Shaw LN. 2013. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2:18–34. doi: 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berger A. 2000. Th1 and Th2 responses: what are they? BMJ 321:424. doi: 10.1136/bmj.321.7258.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Namvar S, Warn P, Farnell E, Bromley M, Fraczek M, Bowyer P, Herrick S. 2015. Aspergillus fumigatus proteases, Asp f 5 and Asp f 13, are essential for airway inflammation and remodelling in a murine inhalation model. Clin Exp Allergy 45:982–993. doi: 10.1111/cea.12426. [DOI] [PubMed] [Google Scholar]
- 77.Jeong SK, Kim HJ, Youm JK, Ahn SK, Choi EH, Sohn MH, Kim KE, Hong JH, Shin DM, Lee SH. 2008. Mite and cockroach allergens activate protease-activated receptor 2 and delay epidermal permeability barrier recovery. J Invest Dermatol 128:1930–1939. doi: 10.1038/jid.2008.13. [DOI] [PubMed] [Google Scholar]
- 78.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Saïd-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol 175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 79.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rooijakkers SH, Ruyken M, van Roon J, van Kessel KP, van Strijp JA, van Wamel WJ. 2006. Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell Microbiol 8:1282–1293. doi: 10.1111/j.1462-5822.2006.00709.x. [DOI] [PubMed] [Google Scholar]
- 81.Steinhuber A, Goerke C, Bayer MG, Döring G, Wolz C. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J Bacteriol 185:6278–6286. doi: 10.1128/JB.185.21.6278-6286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gibson-Corley KN, Olivier AK, Meyerholz DK. 2013. Principles for valid histopathologic scoring in research. Vet Pathol 50:1007–1015. doi: 10.1177/0300985813485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White MJ, Boyd JM, Horswill AR, Nauseef WM. 2014. Phosphatidylinositol-specific phospholipase C contributes to survival of Staphylococcus aureus USA300 in human blood and neutrophils. Infect Immun 82:1559–1571. doi: 10.1128/IAI.01168-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flack CE, Zurek OW, Meishery DD, Pallister KB, Malone CL, Horswill AR, Voyich JM. 2014. Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. Proc Natl Acad Sci U S A 111:E2037–E2045. doi: 10.1073/pnas.1322125111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mootz JM, Malone CL, Shaw LN, Horswill AR. 2013. Staphopains modulate Staphylococcus aureus biofilm integrity. Infect Immun 81:3227–3238. doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rivera FE, Miller HK, Kolar SL, Stevens SM Jr., Shaw LN. 2012. The impact of CodY on virulence determinant production in community-associated methicillin-resistant Staphylococcus aureus. Proteomics 12:263–268. doi: 10.1002/pmic.201100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gatlin CL, Pieper R, Huang ST, Mongodin E, Gebregeorgis E, Parmar PP, Clark DJ, Alami H, Papazisi L, Fleischmann RD, Gill SR, Peterson SN. 2006. Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus. Proteomics 6:1530–1549. doi: 10.1002/pmic.200500253. [DOI] [PubMed] [Google Scholar]