Abstract
Computerized evaluation of histological preparations of prostate tissues involves identification of tissue components such as stroma (ST), benign/normal epithelium (BN) and prostate cancer (PCa). Image classification approaches have been developed to identify and classify glandular regions in digital images of prostate tissues; however their success has been limited by difficulties in cellular segmentation and tissue heterogeneity. We hypothesized that utilizing image pixels to generate intensity histograms of hematoxylin (H) and eosin (E) stains deconvoluted from H&E images numerically captures the architectural difference between glands and stroma. In addition, we postulated that joint histograms of local binary patterns and local variance (LBPxVAR) can be used as sensitive textural features to differentiate benign/normal tissue from cancer. Here we utilized a machine learning approach comprising of a support vector machine (SVM) followed by a random forest (RF) classifier to digitally stratify prostate tissue into ST, BN and PCa areas. Two pathologists manually annotated 210 images of low- and high-grade tumors from slides that were selected from 20 radical prostatectomies and digitized at high-resolution. The 210 images were split into the training (n = 19) and test (n = 191) sets. Local intensity histograms of H and E were used to train a SVM classifier to separate ST from epithelium (BN + PCa). The performance of SVM prediction was evaluated by measuring the accuracy of delineating epithelial areas. The Jaccard J = 59.5 ± 14.6 and Rand Ri = 62.0 ± 7.5 indices reported a significantly better prediction when compared to a reference method (Chen et al., Clinical Proteomics 2013, 10:18) based on the averaged values from the test set. To distinguish BN from PCa we trained a RF classifier with LBPxVAR and local intensity histograms and obtained separate performance values for BN and PCa: JBN = 35.2 ± 24.9, OBN = 49.6 ± 32, JPCa = 49.5 ± 18.5, OPCa = 72.7 ± 14.8 and Ri = 60.6 ± 7.6 in the test set. Our pixel-based classification does not rely on the detection of lumens, which is prone to errors and has limitations in high-grade cancers and has the potential to aid in clinical studies in which the quantification of tumor content is necessary to prognosticate the course of the disease. The image data set with ground truth annotation is available for public use to stimulate further research in this area.
Keywords: Machine learning, Image analysis, Prostate cancer, Tissue classification, Tissue quantification
1. Introduction
Prostate cancer (PCa) remains the most commonly diagnosed cancer in men in developed countries. Fortunately, cancer deaths are steadily declining despite a fairly steady rate of new incidences per year [1]. Microscopic evaluation of prostate needle biopsies is the gold standard for PCa diagnosis and criteria have been established to manage patients based on histopathologic observations in the biopsy and radical prostatectomies. While normal glands are organized into ducts and acini and well separated by stroma, as PCa develops, the malignant acinar structures undergo excessive branching morphogenesis. This is the reason for the histological appearance of small and tightly packed glands with little or no intervening stroma that has become a diagnostic hallmark of low-grade PCa. The architecture in high-grade cancer is different. Cancer cells form glands within glands (Gleason grade 4 (G4) cribriform) loose their ability to form glands that possess a lumen (G4 non-cribriform) or grow in sheets (G4 or G5) [2]. The association between the severity and growth pattern of the prostate cancer provides the basis for the Gleason grading scheme [2,3], which is used clinically. Accurate grading by pathologists requires extensive experience and is occasionally associated with disagreement about low- versus high-grade diagnostic interpretation. In fact, in the early days of the Gleason grading scheme, the inter-observer reproducibility to distinguish low-grade (Gleason grade 3 (G3)) from high-grade tumor growth patterns (G4) ranged between 25% and 47% depending on the grade distribution in the study cohort [4–6].
One way to potentially improve the reproducibility and accuracy of tumor grading is through a computer-assisted approach. Tools for recognition and quantification of morphological characteristics, which correlate with individual Gleason grades have been under intense development by computational pathology researchers. The vast majority of software algorithms for image analysis employs context-based gland quantification to distinguish benign/normal tissue from low- and high-grade areas of cancer [7–12]. As a starting point for image analysis, a typical scenario involves the generation of image tiles with several areas of tumor cells, which receive a grade annotation by an experienced pathologist. A set of descriptors that reflect the cellular organization, inflammation or various secretions is first extracted from the image tiles and then classified according to the cancer grade annotation of the image. Typically, the image content of the entire tile is predefined as stroma (ST), benign/normal (BN), low-grade (G3) or high-grade (G4) cancer [13,14].
Numerous approaches have been developed to capture the growth pattern of prostate cancers. Existing techniques involve various kinds of image features to capture growth patterns that are related to color, texture and nuclear topology [7–11,14].
Yet, the performance of these classification methods varies greatly. The uniformity of the image content, ideally with only one tissue component in each tile, has a major impact on the accuracy of the tissue classification. For images with heterogeneous content, such as those from cancer tissues, which contain admixtures of benign structures and cancer, the performances of the classifiers decline. While the speed of evaluating the entire slide constitutes a major advantage of the tile-based analysis, the approach has several shortcomings. The impact of the tile size, which varies among studies on the performance of the classifier, is unknown. Moreover, the predictive power of tissue descriptors and classifiers can be artificially high, if training and validation is performed on small sets of tiles (usually <100). This problem is particularly grave in prostate cancer since large tiles with homogenous tissue content cannot be generated in sufficient quantity.
To overcome the problems that are caused by tissue heterogeneity, tissue classes can be manually delineated by a pathologist for algorithm training and validation [13,15–17]. The training set consists of similar manually annotated tissue regions. While this approach is more laborious, it provides additional opportunities for computational image classification [15–17]. Low-power image magnification (<20×) is often employed for prostate cancer image analysis mainly, because it is the most efficient way to manually grade prostate cancer [7,8,10] [15]. However, a recent study shows the improved performance of high-resolution imaging. In the work published by Kwak et al. [16], 21 intensity and 42 texture features (including local binary patterns) were utilized to segment stroma from the epithelium and the analysis was conducted on 4 different resolution scales. The training was performed manually by a pathologist and ROC curves showed high concordance rates with final algorithm output. However, since the robustness of image analysis is a complex product of the quality of manual ground truth, image resolution and tissue heterogeneity, it is important to determine the effect of each component on the accuracy of segmentation
Recently, machine learning approaches have become popular to quantify tumor areas in histopathological preparations. When expression of protein biomarkers in breast cancer specimens was visualized by staining with antibodies and quantified by image analysis, human and software-derived annotations showed strong agreement in the classification of cancer areas. Furthermore, software developed by academic or commercial groups efficiently separated cancer from stroma within image tiles on a sub-tile resolution [18,19]. However, quantitative analysis of specimens stained with hematoxylin and eosin (H&E), which is routinely used for histopathologic evaluations, is much more challenging and the development of software for analysis of H&E stained slides is the main determinant of the pace at which the image analysis field advances.
Since benign prostate glands and G3 and G4 cancer areas are morphologically distinct from stroma in H&E stained tissue sections, the differences can be numerically captured by image analysis. These slide preparations provide an ideal starting point to demonstrate the power of machine learning tools for classification of H&E images on a sub-tile resolution. In other words, instead of classifying the whole tile content into one class, a machine learning tool can classify individual pixels and deliver pixel-based tissue quantifications. Towards the development of such tools, our team designed three separate classifiers to identify and quantify areas of stroma in images with benign glands, or G3, or G4 prostate cancer with excellent performance [17]. In contrast to other methods, the histograms composed of pixels from H&E intensity measurements that we utilized to describe and classify images were superior to histograms of oriented gradients and provided the highest tissue classification rates. Encouraged by the preliminary results, we continued to improve the approach through the employment of intensity features combined with a more complex texture features for the capture of patterns within areas of glandular architecture. The tissue segmentation results were compared to manual annotations by a pathologist in a large set of high-resolution images of radical prostatectomies. Overall, this approach combines improvements in classification performance and speed and is ready for preliminary testing in prognostic and predictive biomarker studies.
2. Materials
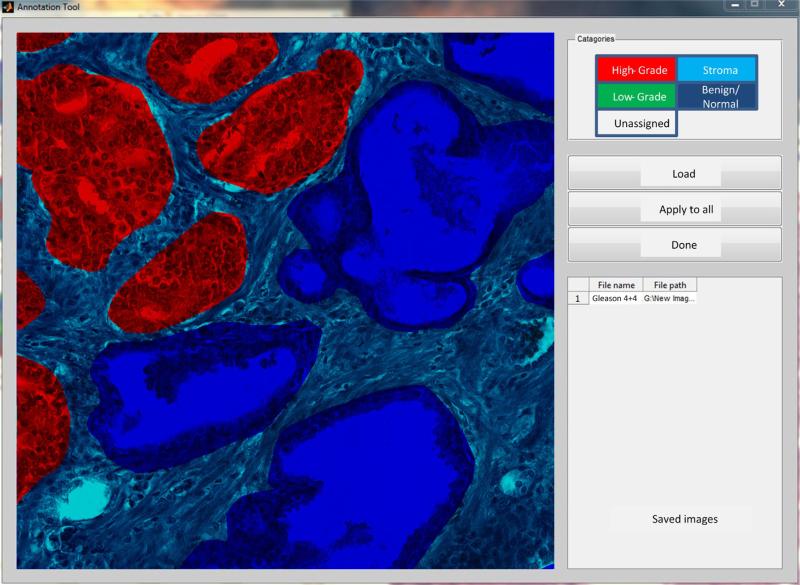
Radical prostatectomy specimens from 20 patients with a diagnosis of G3 or G4 prostate cancer according to the contemporary grading criteria [2,3] were retrieved from archives in the Pathology Department at our institution under an Institutional Review Board approval no. Pro00029960. Slides were digitized by a high resolution whole slide scanner SCN400F (Leica Biosystems, Buffalo Grove, IL) dedicated for pathology research. The scanning objective was set to 20× and the focusing was automatically adjusted by the scanner. The output was a color RGB image with the pixel size of 0.5 μm × 0.5 μm and 8 bit intensity depth for each color channel. We utilized freely available libraries from OpenSlide.org [20] to import Leica (.scn) images, select histopathologically important fields of view (FOV) that were converted to TIFFs for methods development. Furthermore, the FOVs were split into tiles of 1200 × 1200 pixels for image analysis. Of the total 5000 tiles, a subset of 210 images was selected by pathologists (SM, SB) who identified ST, BN glands, G3 cancer and G4 cancer containing cribriform and non-cribriform growth patterns. Depending on their content, tiles were categorized into groups (Table 1), and then annotated manually using a custom graphical user interface (GUI) which we specifically developed for this task (Fig. 1). The GUI facilitated: (a) import–export of images in common formats (tiff, jpg, png, etc), (b) free-hand contouring with the capability of an intuitive contour closing and space filling of the contour, (c) tissue labeling by colors: high-grade tumor (red), low-grade tumor (yellow), benign/normal glands (blue) and stroma (cyan), (d) easy correction of wrongly delineated areas, and (e) delineation of stroma by using a semi-automatic image flood-filling procedure after all glandular annotations are finished. At the end of this procedure all annotated image tiles were returned to pathologists for cross-evaluation.
Table 1.
Tissue images utilized in our study. Tissue components in each image were delineated by a pathologist for training and validation for the proposed tissue classification algorithms.
| Epithelial components | BN | G3 | G4 | BN + G3 | BN + G4 | G3 + G4 | BN + G3 + G4 | Total |
|---|---|---|---|---|---|---|---|---|
| No. of images | 33 | 20 | 54 | 23 | 15 | 44 | 21 | 210 |
Fig. 1.
Example PCa image with overlaid manual annotation by pathologist who used the dedicated graphical user interface (GUI) that we developed. Color coding and image transparency facilitated the delineation of tissue components: red color was used for G4, blue for BN and cyan for ST.
3. Methods
3.1. Overview of the image analysis strategy
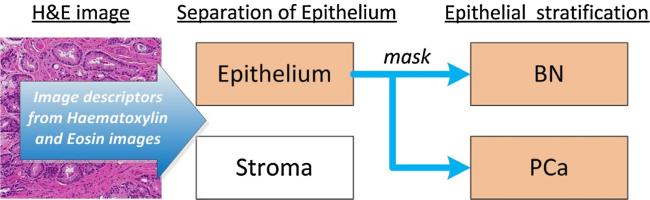
Tissue classification based on H&E images is a challenging and computationally expensive process [13,19]. Our approach involves the stratification of prostate tissue in two sequential steps: (1) separating stroma (ST) from the epithelium (EP) and (2) differentiating of benign/normal glands (BN) from prostate cancer (PCa) (Fig. 2). In each step only two tissue classes are analyzed at once. In step 1, a mask covering epithelial areas is generated to facilitate recognition of BN and PCa tissues in the next step. This strategy is significantly different from other published approaches [8–10,13], because it does not involve the per se glandular segmentation and extraction of glandular features. Many gland segmentation approaches known to date [9,21–24] are based a priori on the segmentation of nuclei, and use the glandular lumen to recognize the presence of glands. However, nuclear segmentations often fail in dense, overstained or understained tissue areas and glandular lumens can be occluded by corpora amylacia or secretions. Furthermore, empty spaces caused by tissue retraction artifacts or blood vessels can falsely be recognized as glandular lumens. In areas of high-grade cancer lumens are absent or difficult to detect. To overcome the limitations of existing methods, we formulate the problem as pixel-wise semantic segmentation based on intensity and texture descriptors that are extracted from images after color deconvolution. We apply novel and robust descriptors to distinguish BN from PCa. Due to the complexity of the cellular architecture in prostate tissue, different classifiers and descriptors are utilized at each step (Fig. 2). Since non-epithelial tissues such as ST are morphologically less heterogeneous than BN glands and PCa, the computational expense can be decreased by removing ST before analyzing glandular structures.
Fig. 2.
Image analysis workflow for prostate cancer tissue quantification. In the preprocessing steps, areas of stroma are first separated from the epithelium. Subsequently, areas of epithelium are stratified into benign/normal glands (BN) and prostate cancer (PCa).
Our methodology was developed based on the following premises: (a) the density of epithelial nuclei is higher than the density of stromal nuclei, (b) the intensity of eosin is different in ST versus BN glands and areas of PCa and (c) the texture of hematoxylin and eosin in glands differs from the texture in ST. Hence, we utilize the difference in pixel intensity characteristics of eosin and hematoxylin to classify areas of prostate tissue. The classifiers are able to distinguish the three tissue classes based solely on the descriptors (intensity and/or texture histograms) that are extracted from hematoxylin (H) and eosin (E) image stains after color deconvolution [25]. The color deconvolution algorithm developed by Ruifrok and Johnston, was used to process RGB color images for digital separation of immunohistochemical dyes in a tissue image and performs well even if the dyes have overlapping absorption spectra or co-localize in same cellular compartment. Using optical density vectors of the pure dyes and an ortho-normal transformation of image intensities the algorithm deconvolves optical density of dyes for all image pixels. As a result, blue hematoxylin and pink eosin images are obtained as single channel intensity images. For details regarding color deconvolution the reader is referred to [25].
Histogram-based features are extracted by sliding a window W over H and E images with the interval of W/4 in horizontal and vertical directions. Descriptors are calculated for W and assigned to its central pixel. Descriptors for the remaining pixels are obtained via bicubic interpolation and then classified. Descriptors obtained this way form a three dimensional descriptor matrix with x and y dimensions that are same as in the input image and the z-dimension determined by the descriptor length. W slides from the top left corner to the bottom right corner vertically and horizontally throughout the image and stops systematically at W/4 intervals (defined by x and y) to calculate the descriptor (histogram). Subsequently, the descriptor is inserted to the three dimensional matrix and at x and y positions defined by W/4 intervals. Descriptors at image borders are calculated after mirroring pixels that are up to W/2 away from the image border. The mirroring provides a sufficient number of pixels to calculate a descriptor at pixels for which W positioned at the upper left corner and along the borders of the original image. A bicubic interpolation is then applied to the three dimensional descriptor matrix to obtain values at positions not visited by W. It is applied to each z-plane separately using a third degree polynomial and values from a 4 × 4 grid of descriptor components at which W stopped.
Prior to color deconvolution, images are color-normalized [26] to account for variability in staining intensity. Bright areas (background and lumens) are removed by thresholding of gray-level images obtained from color-normalized H&E images. This intensity threshold was manually optimized and set to the value of 210. The resulting binary mask of the background is cleaned up by removing isolated blobs with areas smaller than 50 pixels followed by a morphological closing with a flat disk-like structuring element with the radius of 7. Pixels under the background mask are excluded from analysis.
3.1.1. Intensity and texture histograms of hematoxylin and eosin stained images
Prediction of tissue content requires a set of descriptors–numerical features extracted from images that have the capacity to capture differences in tissue morphology. Tissue descriptors utilized in our study are divided into two groups: intensity histograms and joint distributions of the uniform rotation-invariant local binary patterns and rotation invariant local variance. The first type of histograms represents a distribution of pixel intensities, whereas the second histogram represents the spatial relationships (mostly differences) between intensities of pixels, which have a close proximity. Thus, the second type is an image texture descriptor.
Our approach utilizes texture and intensity based features that are extracted from hematoxylin and eosin images after color deconvolution (Fig. 3). For each image pixel the descriptors: (a) intensity histograms: Hist(H) and Hist(E) and (b) joint distributions of the uniform rotation-invariant local binary patterns (LBP) [27] and rotation invariant local variance (VAR) [27] denoted as (H) and (E) respectively for hematoxylin and eosin. P and R denote neighborhood size and radius for LBP and VAR. riu2 stands for the uniform rotation-invariant type of LBP. To obtain the descriptor the intensity and contrast insensitive histogram is combined with a single value representing image contrast VARP,R. The resulting descriptor is a one-dimensional histogram with length k × n, where: k is the number of quantizing levels of VARP,R and n is the length of the histogram. Since each image pixel can contribute one and VARP,R it makes sense to calculate for pixels in a window W. For instance, for a 10 bin long histogram and VARP=8,R=1 quantized as 1 the bins of histogram are added to the first 10 bins of . If VARP=8,R=1 is quantized as 3, then the bins of are added to the third tenth of bins in . We quantized VARP,R into 8 bins. After calculating all histograms in W and sorting them according to values of VARP=8,R=1, the joint histogram will be 8 × 10 = 80 bin long. Source codes for calculating LBP and VAR can be found here [28].
Fig. 3.
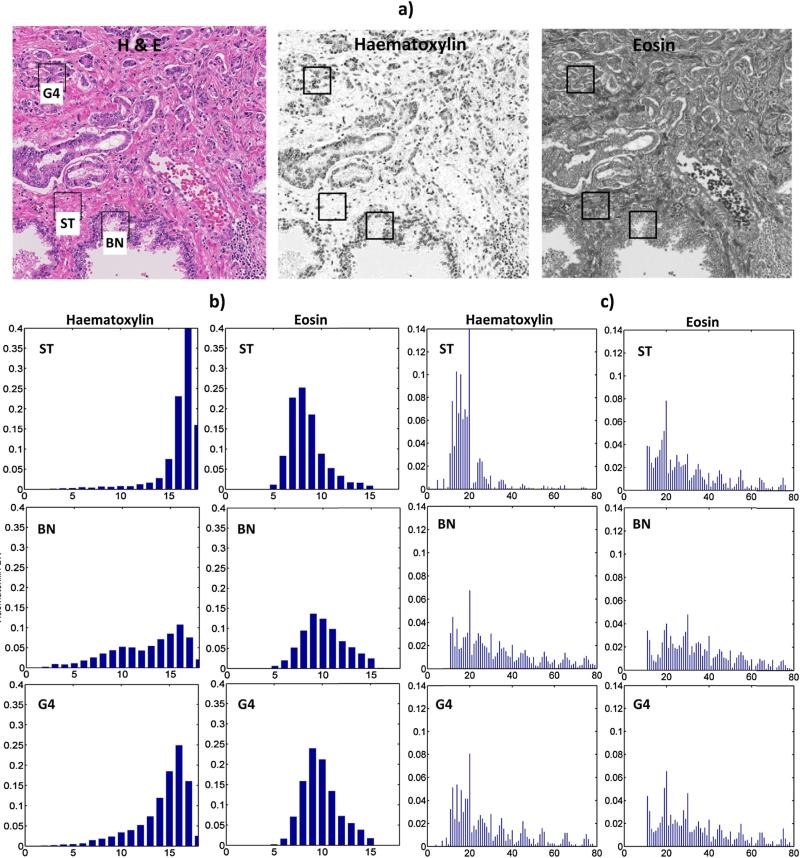
Examples of intensity and texture pixel descriptors from deconvoluted H&E images of prostate cancer tissue: (a) original image with deconvoluted components, (b) pixel intensity histograms and (c) × VARP=8,R=1 histograms, representing the image texture. Boxes in (a) indicate locations of windows W (size of window: N = 64 pixels) from which the descriptors were extracted. Histograms from windows in areas of stroma (ST), benign/normal epithelium (BN) and cancer Gleason pattern 4 (G4) are shown in (b) and (c). Note, that background or luminal pixels found in H&E image are masked out and do not contribute to the histogram calculation. Histograms in (b) have 18 bins to map 0–255 gray level intensity range (x-axes), whereas in (c) 80 bins are used to represent the distribution of local binary patterns (LBP) from low (bins 0–40) and high (bins 41–80) contrast image regions. Y-axes are the normalized density of pixels in each histogram bin.
The Hist(H), Hist(E) and descriptors are derived from a N × N pixel window W centered at each image pixel. Intensity histograms Hist(.) are sorted into 18, and each into (P + 2) × 8 equally spaced bins. Paired values of P = {8,24} and R = {1,3}, are tested for best classification performances. For deconvoluted H and E images the histograms: Hist(H), Hist(E), (H) and (E) are concatenated, and then used as descriptors for tissue classification.
The window size N depends on scanner resolution and needs to be chosen to provide enough data points to derive meaningful descriptors. According to [17] several G3 glands fit W with N = 64 and may be sufficient to obtain spatial precision in predicting ST and EP. Since its impact on epithelial stratification is unknown, we tested performance of descriptors obtained for N = {64, 128, 256}. The prediction of ST and EP regions is conducted using full size images (highest resolution). Prior to extracting the descriptors for the epithelial stratification (Fig. 2) the image was downsized by 50% with bicubic interpolation to reduce computation and storage costs.
4. Classifiers training
4.1.1. Stromal and epithelial descriptors
The first step in the proposed tissue prediction workflow (Fig. 2) is to train an algorithm to separate stroma from the epithelium. Training descriptors are extracted from windows W, which were manually and independently placed over tissue areas by two pathologists (SM, SB) (Fig. 4). Using a graphical user interface, the pathologists placed W over regions with homogenous tissues patterns of stroma (ST category) or epithelium (EP category). Stromal windows were placed mostly over fibroblasts between glands. Some windows were placed over blood vessels, erythrocytes, muscles and immune cells. To generate a training set with different epithelial content: BN, G3 and G4 areas were used.
Fig. 4.
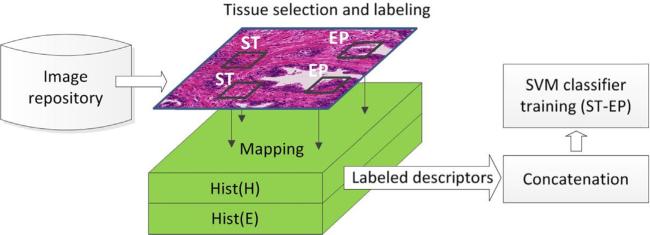
Formation of pixel descriptors to train a SVM classifier for differentiating stroma (ST) from epithelium (EP). Training descriptors are extracted from matrices containing local intensity histograms of hematoxylin and eosin images. After a window is placed over a selected area, the respective descriptor is extracted by mapping the window location onto the descriptor matrix Hist(H), Hist(E). The descriptors are labeled based on their tissue origin that was assigned by a pathologist as stroma (ST) or epithelium (EP).
After a window was placed, the interface automatically loaded up corresponding file with pre-calculated descriptor matrix and extracted a sample associated with the selected window. The sample was then labeled as either ST or EP by a pathologist. The two pathologists worked independently and selected respectively 92 and 60 windows with equal number of ST and EP components. Up to 4 ST and up 4 EP windows within a single H&E image were picked, and only 19 H&E images from the entire collection of 210 images were used. Two training sets: ST-EP1 and ST-EP2 were formed from samples extracted by the first and the second pathologist. A third training set: ST-EP3 was generated by concatenating ST-EP1 with ST-EP2. The training sets contained Hist(H), Hist(E) descriptors that together were 36 bins long (b). Hist(H) and Hist(E) were normalized by dividing each bin by the total number of pixels in W. The training samples were z-transformed before training the classifier. Using ST-EP3 a test involving non-interpolated Hist(H), Hist(E) descriptor matrices was ran to compare tissue classification performances obtained for the interpolated Hist(H), Hist(E) descriptor matrices. Classification results were validated using the remaining 191 images (test set) annotated by pathologists.
4.1.2. Benign/normal and PCa descriptors
BN and PCa training samples for RF classifier training were selected in a semi-supervised manner. Annotations provided by pathologists were mapped onto matrices with pre-calculated histograms to find those specific for BN and PCa (Fig. 5), and the mapping was continued for multiple images. Training samples found in this way were automatically labeled as either BN or PCa. We used the same set of 19 training images and extracted samples for all pixels covered by pathologist annotations. This process yielded a very large (6th magnitude order) training set with redundant and highly correlated samples. To reduce their number we randomly selected only 25% of the samples for each tissue type. We tested the effect of descriptor sampling variability on the tissue prediction performance (see Section 7).
Fig. 5.
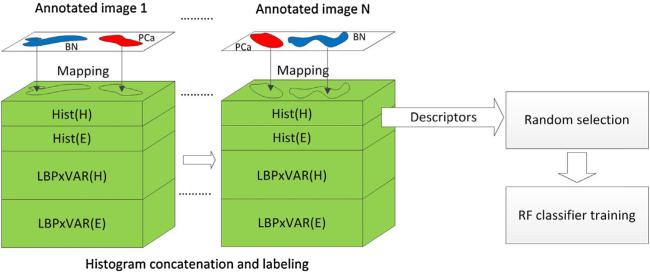
Formation of pixel-based descriptors and training set for benign/normal glands (BN) and prostate cancer (PCa) classification. Tissue annotations from multiple images [1. . .N] are mapped onto combinations of histogram matrices detailed in Table 2, to extract and label samples for BN and PCa. A number of descriptors reduced by random selection (to 25%) is used to train the random forest (RF) classifier.
Hist(H), Hist(E), (H) and (E) histograms were used to configure 10 different types of descriptors (Table 2) and train RF classifiers to test their predictive power. After the 25% reduction the remaining 561,562 samples were used to train the RF classifiers. The proportions of BN and PCa used for training constituted respectively 45% and 55% of the total number of samples.
Table 2.
Types of descriptors. Intensity, texture and combined intensity and texture histograms were used as descriptors for training machine learning models.
| Descriptor | Descriptor number |
|---|---|
| Hist(H), Hist(E) | 1 |
| Hist(H) | 2 |
| Hist(H), | 3 |
| Hist (H), Hist (E), , | 4 |
| , | 5 |
| , | 6 |
| Hist(H), | 7 |
| Hist(H), Hist(E), , | 8 |
| , | 9 |
| , | 10 |
5. Classifiers
We trained two different types of classifiers to predict prostate tissue components. For stromal and epithelial tissues a support vector machine (SVM) [29] was chosen. For each of the training sets, ST-EP1, ST-EP2 and ST-EP3, a separate SVM classifier with Gaussian radial basis function with width of σ = 15 was trained. The output of the SVM classifier was a binary image with “1” masking epithelial and “0” masking stromal pixels respectively.
To stratify epithelial pixels into BN and PCa categories a random forest (RF) classifier with 20 trees was trained with each of the ten descriptor types from Table 2. The RF provided labeled binary output with “2” representing BN and “3” representing PCa. All image analysis tasks including classification were performed in MATLAB R2013b (Mathworks, Natick, MA).
6. Validation
To assess classification performance of the proposed system several measures of agreement that are frequently applied when a computed result (C) is compared to a manual ground truth (G) were computed. The measure include the area overlap (O), Jaccard similarity coefficient (J) and Rand index (Ri) [30] and are defined respectively as: , and where, a is the number of pairs with elements that are in the same set in G and in the same set in C, b is the number of pairs with elements that are in different sets in G and in different sets in C and is the total number of possible unordered pairs in the dataset were used. The Rand index is a widely accepted method for the evaluation of classification and clustering results. An O, J and Ri yield 1 for a perfect agreement. A score of 0 shows a complete disagreement between G and C. For this work the range [0 1] was linearly scaled to [0 100] to facilitate comparison with published results. Similarly to Salman et al. [17], the background areas including lumens were removed from G and C before the validation.
7. Results
7.1.1. Stroma–epithelium separation
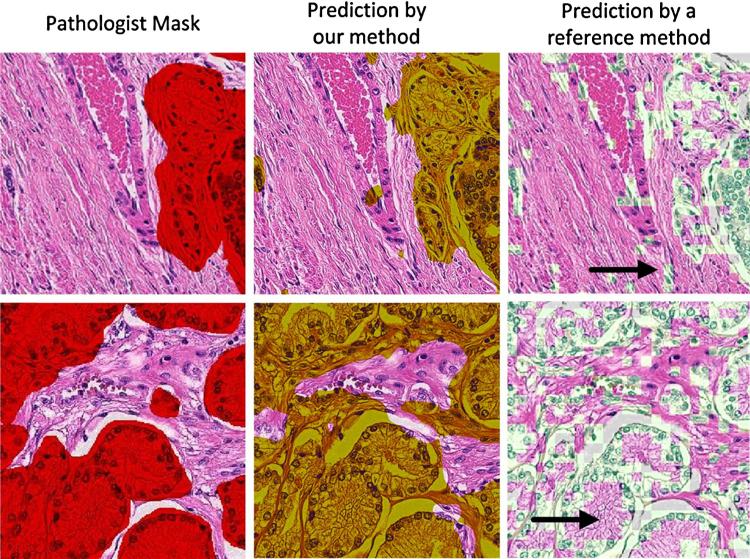
Utilizing data from clinical images and the presented framework, we first tested the performance of our approach in the separation of stroma from the epithelium. Three training sets: ST-EP1, ST-EP2 and ST-EP3 containing respectively 50% of stromal and 50% epithelial samples with Hist(H), Hist(E) descriptors (descriptor 1 in Table 2) were used to train three separate SVM classifiers: SVM(ST-EP1), SVM(ST-EP2) and SVM(ST-EP3). Their classification performance was compared to a method published by Chen et al. [31] that predicts ST and EP based on distances of color component ratios from centroids derived from k-means clustering. Chen's method was retrained on our data set. Furthermore, the training set, similar to ours, involves areas that are manually selected by a user. Results in Table 3 were obtained by evaluating the significance of the differences between groups with the Tukey's Studentized range test with a Bonferroni correction of α = 0.056. Differences in JST for all classification methods were not significant. JEP from all SVM-based results was higher from JEP obtained by Chen's method (p < 10−4). OST from Chen's method was not significantly higher than OST obtained for SVM(ST-EP1). Yet, SVM (ST-EP1) and Chen's classification methods yielded higher values (p < 10−4) comparing to SVM(ST-EP2) and SVM(ST-EP3). On the other hand, SVM(ST-EP2) and SVM(ST-EP3) yielded highest OEP than the other two methods (p < 10−4). Ri value for Chen's method was much lower than Ri of any of the SVM based methods (p < 10−4). Fig. 6 shows classification results of Chen's and our method. Chen's method often misclassified blood vessels as glands. In addition, it misclassified the eosinophilic cytoplasm of glandular luminal cells as areas of stroma. Therefore, the overall misclassification rate was higher than with the proposed framework. For this comparison, Chen's method was trained using the same pathologist delineated images used in training of our method.
Table 3.
Comparison of epithelial tissue classification performances. Training sets ST-EP1.3 were established using 60.152 windows from 19 images. Trained SVM classifiers were applied to 191 testing images to separate epitehelium from stroma. Algorithms were evaluated by Jaccard, area overlap (O) and Rand (Ri) indices to determine the concordance of the algorithm-based prediction with pathologist manual annotations. J and O indices were calculated separately for concordance of ST and EP areas, whereas Ri was calculated for ST and EP together. Respective columns contain mean value ± standard deviation.
| J ST | J EP | O ST | O EP | Ri | |
|---|---|---|---|---|---|
| Chen et al. [31] | 49.9 ± 19.1 | 48.9 ± 13.0 | 75.0 ± 13.5 | 63.2 ± 17.7 | 57.6 ± 7.4 |
| SVM(ST-EP1) | 53.2 ± 18.8 | 55.9 ± 16.8 | 72.0 ± 14.2 | 72.8 ± 19.6 | 62.1 ± 8.2 |
| SVM(ST-EP2) | 49.6 ± 19.2 | 58.9 ± 14.8 | 61.9 ± 15.9 | 81.4 ± 12.2 | 61.5 ± 7.7 |
| SVM(ST-EP3) | 50.8 ± 18.2 | 59.5 ± 14.6 | 63.7 ± 14.1 | 81.5 ± 12.1 | 62.0 ± 7.5 |
Fig. 6.
Prediction of areas of stroma and epithelium by different methods. Tissue areas in the images in the first column were delineated by a pathologist using the graphical user interface (GUI) to generate a mask with the red color indicating epithelial areas, in this example PCa. The second column demonstrates our classification method applied to the same images with epithelial areas marked in yellow, whereas the third column demonstrates the application of Chen's method (reference method, white areas) [31]. Arrows point to areas where the two algorithms differ the most. In row 1, the arrow points to misclassified cells in or around blood vessels and in row 2, the arrow points to the cytoplasm of luminal cells that was not included in the glandular areas. Note that our approach is pixel-based whereas Chen's involves small image blocks. Luminal and background areas were removed prior calculating the performance of the predictor.
A SVM(ST-EP3) classifier that was separately trained using non-interpolated matrices of Hist(H), Hist(E) descriptors yielded the following rates: JST = 58.9 ± 20.32, JEP = 58.73 ± 16.74, OST = 81.13 ± 9.84, OEP = 72.36 ± 20.64 and Ri = 65.25. The calculation of the descriptor matrix without interpolation was 180 s versus 2 s for the calculation of an interpolated descriptor matrix.
7.1.2. Prediction of benign/normal and PCa
The identification of areas with epithelial glands was tested individually using 10 different descriptor s (Table 2), random forest (RF) classifiers and sliding windows with three different sizes of 64, 128 and 256 pixels. The values for pixel-based descriptors were extracted and then classified to predict the tissue content. RF classifiers were trained as shown in Fig. 4. The prediction performance of BN and PCa was first evaluated in the training set containing 19 images (Table 4) and then in the test set with 191 images (Table 5). First, the training error of the RF models was assessed on the 19 training images. Since only 25% of the samples were used in the training phase, the idea of measuring the predictive performance for all the descriptors on the training images could serve as an indirect assessment of prediction accuracy. Table 4 shows average performances of best and worst performing descriptors, whereas Table 5 shows results for all descriptors and all three window sizes. Fig. 7 shows one of the images as an example, after applying our method.
Table 4.
Average tissue prediction performance in the training set using 25% of the total number of samples. Values reflect best and worst performances of descriptors from Table 2.
| J BN | O BN | J PCa | O PCa | Ri | |
|---|---|---|---|---|---|
| Worst average prediction | 60.3 | 76.6 | 55.6 | 76.3 | 67.6 |
| Best average prediction | 68.4 | 81.42 | 56.8 | 77.6 | 68.9 |
Table 5.
Comparison of BN and PCa tissue prediction by different descriptors and three different scanning window sizes. Respective columns contain mean value ±SD of performance indices. Best three results of each index are bolded.
| Scanning window size N = 64 | |||||
|---|---|---|---|---|---|
| Descriptor | J BN | O BN | J PCa | O PCa | Ri |
| 1 | 34.3 ± 24.1 | 51.7 ± 28.9 | 45.5 ± 20.7 | 64.4 ± 19.5 | 60.4 ± 7.5 |
| 2 | 30.4 ± 22.5 | 45.6 ± 28.1 | 45.6 ± 20.3 | 65.5 ± 20.0 | 60.0 ± 7.4 |
| 3 | 31.4 ± 21.2 | 43.4 ± 26.7 | 47.2 ± 18.7 | 69.0 ± 15.1 | 60.2 ± 7.1 |
| 4 | 33.0 ± 23.5 | 46.5 ± 29.6 | 47.4 ± 19.1 | 69.0 ± 15.9 | 60.3 ± 7.4 |
| 5 | 28.0 ± 18.2 | 40.8 ± 19.9 | 44.4 ± 17.1 | 64.4 ± 14.2 | 59.0 ± 7.3 |
| 6 | 30.9 ± 23.4 | 41.8 ± 29.5 | 47.8 ± 18.4 | 71.0 ± 14.8 | 60.2 ± 7.4 |
| 7 | 33.8 ± 21.6 | 45.6 ± 26.7 | 48.0 ± 18.1 | 70.5 ± 13.9 | 60.3 ± 7.2 |
| 8 | 35.4 ± 22.7 | 48.6 ± 28.7 | 48.4 ± 17.9 | 71.3 ± 14.2 | 60.4 ± 7.4 |
| 9 | 27.0 ± 15.4 | 37.0 ± 17.2 | 46.7 ± 17.0 | 68.6 ± 12.5 | 59.3 ± 7.0 |
| 10 | 33.2 ± 22.0 | 43.8 ± 26.9 | 48.7 ± 17.7 | 72.6 ± 14.4 | 60.2 ± 7.3 |
| Scanning window size N = 128 | |||||
|---|---|---|---|---|---|
| Descriptor | J BN | O BN | J PCa | O PCa | Ri |
| 1 | 32.1 ± 24.9 | 47.4 ± 32.0 | 46.3 ± 20.2 | 67.1 ± 20.1 | 60.3 ± 7.5 |
| 2 | 30.5 ± 23.4 | 45.3 ± 29.8 | 46.0 ± 20.9 | 66.6 ± 22.3 | 60.2 ± 7.5 |
| 3 | 31.7 ± 23.3 | 44.5 ± 29.9 | 47.6 ± 19.4 | 69.4 ± 17.2 | 60.5 ± 7.3 |
| 4 | 31.7 ± 25.6 | 46.0 ± 33.5 | 48.1 ± 19.5 | 70.0 ± 16.8 | 60.5 ± 7.7 |
| 5 | 29.4 ± 20.6 | 42.0 ± 26.0 | 47.1 ± 18.8 | 68.6 ± 16.5 | 60.1 ± 7.2 |
| 6 | 32.1 ± 25.6 | 46.2 ± 33.1 | 48.3 ± 19.0 | 70.7 ± 16.3 | 60.4 ± 7.6 |
| 7 | 31.6 ± 23.7 | 41.4 ± 29.7 | 49.3 ± 18.0 | 73.2 ± 13.3 | 60.5 ± 7.2 |
| 8 | 33.4 ± 25.4 | 46.2 ± 33.0 | 49.1 ± 18.6 | 72.3 ± 15.4 | 60.6 ± 7.4 |
| 9 | 27.3 ± 18.4 | 38.7 ± 21.4 | 46.6 ± 17.4 | 68.0 ± 14.5 | 59.5 ± 7.2 |
| 10 | 32.9 ± 24.9 | 45.3 ± 32.4 | 49.0 ± 18.3 | 72.4 ± 15.3 | 60.4 ± 7.5 |
| Scanning window size N = 256 | |||||
|---|---|---|---|---|---|
| Descriptor | J BN | O BN | J PCa | O PCa | Ri |
| 1 | 30.7 ± 24.8 | 44.7 ± 31.2 | 46.3 ± 21.0 | 66.5 ± 20.8 | 60.2 ± 7.8 |
| 2 | 30.4 ± 25.1 | 45.1 ± 33.0 | 46.4 ± 20.7 | 67.1 ± 21.3 | 60.3 ± 7.6 |
| 3 | 21.0 ± 15.0 | 46.2 ± 26.6 | 26.4 ± 19.1 | 36.9 ± 26.9 | 58.5 ± 8.2 |
| 4 | 35.8 ± 24.3 | 50.9 ± 31.2 | 48.5 ± 19.6 | 71.3 ± 17.9 | 60.6 ± 7.6 |
| 5 | 30.2 ± 21.2 | 44.2 ± 27.3 | 47.2 ± 20.2 | 67.4 ± 18.7 | 60.3 ± 7.4 |
| 6 | 33.3 ± 25.8 | 48.2 ± 32.7 | 48.4 ± 19.6 | 70.2 ± 17.9 | 60.6 ± 7.9 |
| 7 | 33.5 ± 22.9 | 46.4 ± 28.8 | 48.9 ± 18.9 | 71.1 ± 15.7 | 60.7 ± 7.2 |
| 8 | 35.2 ± 24.9 | 49.6 ± 32.0 | 49.5 ± 18.5 | 72.7 ± 14.8 | 60.6 ± 7.6 |
| 9 | 31.3 ± 21.1 | 44.5 ± 26.1 | 46.9 ± 18.2 | 68.0 ± 15.8 | 59.9 ± 7.4 |
| 10 | 34.3 ± 25.9 | 48.5 ± 33.2 | 49.1 ± 19.1 | 72.2 ± 16.4 | 60.6 ± 7.8 |
Fig. 7.
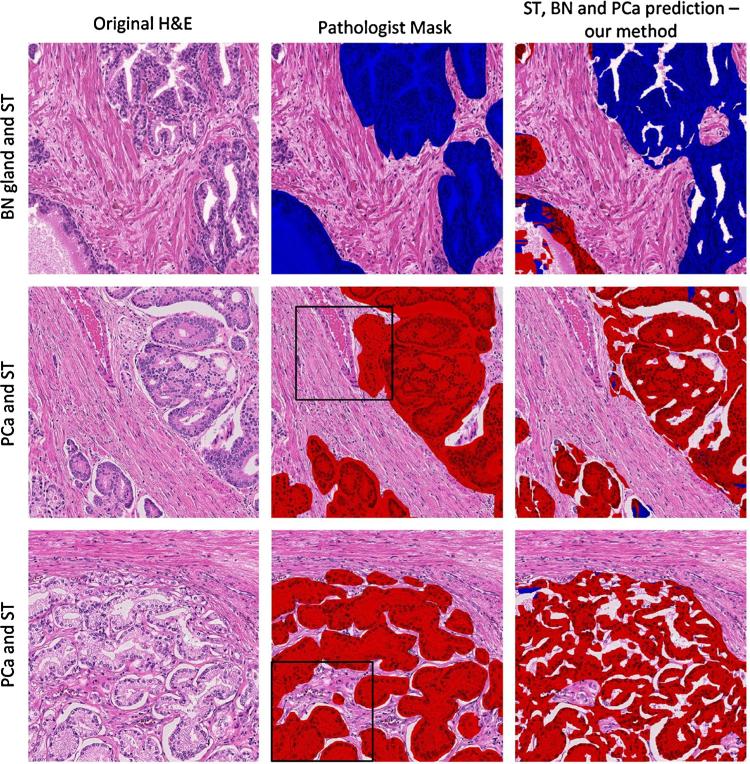
Classification of glandular areas from benign/normal glands and prostate cancer using the random forest (RF) classifier. Original H + E images in column 1 of a representative area of benign/normal glands (BN) and stroma (ST) (row 1) and 2 different areas of prostate cancer PCa Gleason pattern 4 (rows 2 and 3). The annotation of the pathologist in column 2 is compared to the annotation with the RF classifier in column 3. The areas marked with black squares are identical to the ones analyzed in Fig. 6. The epithelium was classified as BN or PCa using a descriptor formed by Hist(H), Hist(E), (H), (E) with P = 24 and R = 3.
7.1.3. Effects of sampling variability
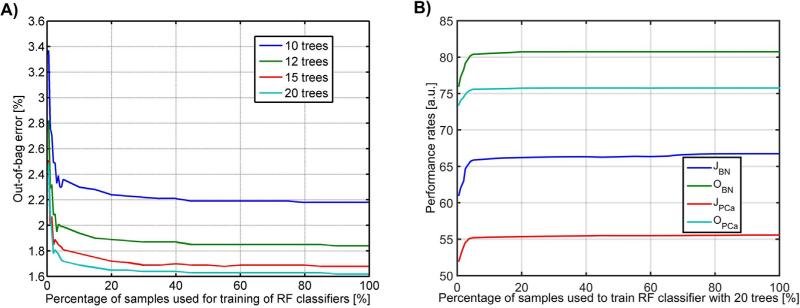
Building a RF classifier that involves multiple descriptors can be computationally expensive. To reduce the computational burden we determined the fraction of pixels that is needed for adequate performance of the classifier. To this point we conducted two experiments in which we randomly sub-sampled the training set and measured the classifier performance. In the first experiment, we gradually decreased the percentage of the samples used for training (100–5% range with 5% interval and 4.5–0.5% with 0.5% interval, respectively) and observed the performance of an RF classifier in the training set of 19 images by measuring the out-of-bag error (OOB) and the Jaccard index for BN and PCa areas (Fig. 8). OOB is the unbiased classification error estimated during the RF classifier training involving approximately 1/3 of cases that were not used to construct trees [32]. The performance measured for 100% of the training samples was used as a baseline. For 20 trees the OOB error was 1.62% and it increased to 1.65% for the sampling rate in the range of 20–100% (Fig. 8a). In this range JPCa and JBN increased respectively from 55.39 to 55.58 and from 66.25 to 66.72 (Fig. 8b). A decrease of the sampling rate below 25% resulted in an increase of OOB and in a decrease of Jaccard indices. Second, we trained 10 RF classifiers with 10 different sets of sub-samples, each one of only 25% of the original size. The performance was measured using the Jaccard index for BN and PCa. Since no statistically significant difference in results was noted between results from the 10 versus 20 RF classifiers (paired t-test yielded p < 0.05) we collectively assumed that the random selection of 25% of descriptors can be used to reduce the computational burden of the classifier training without influencing the results of the performance of tissue classification in the test set.
Fig. 8.
RF classification performance for different sampling rates of pixels in the training set: (a) the out-of-bag error estimate calculated for the RF classifier trained with a different number of trees and (b) JBN, JPCa, OBN and OPCa performance measures for the RF classifier with 20 trees.
8. Discussion
The main goal of this study was to develop, implement and validate a machine learning approach for computer-assisted classification of images from histopathologic preparations of prostate tissues. For this purpose, we implemented a workflow with 210 images, divided into training and test sets and annotated by pathologists to stratify pixels in image tiles into stroma, benign/normal glands and prostate cancer using binary SVM and RF classifiers. The results from our classifications were evaluated by comparison to the manual delineation of tissue structures and the performance of our classifier reported by calculating the overlap between the computational approach and the pathologist.
8.1.1. Prediction of stromal and epithelial areas
The proposed framework for predicting areas of epithelium in digital images from sections of prostate tissue differs from previous approaches in [7,8,10] in terms of image acquisition gold standard data (i.e., the annotation of images by a pathologist), and methodology design. First, studies reported in [7,8,10] utilized 40, 62 and 44 images that were acquired with 5×, 1.25× and 4× objective magnification respectively. In contrast, we utilized a 20× objective in this work. Second, the main goal of previous studies was to segment and then classify glands based on identification of opened glandular lumens. For example, in a study by Nguyen et al. [8] glands without opened lumens were not included in their evaluation. Another approach [7] utilized luminescence image channel thereby ignoring the color information in the images, which resulted in gland segmentation that was solely based on the presence of bright pixels in opened lumens. Likewise, k-means based clustering of principal components of image intensity applied to detect open lumens of benign and PCa glands proposed in Yahui et al. [10] scored poorly, because 84% of low- and high- grade PCa glands segmented by this method were not accepted by pathologist. While the glandular lumens are prominent in the majority of BN glands and G3 cancer, they do not exist in cancer areas of non-cribriform G4 growth patterns. Thus, as the three previously published gland-segmentation methods rely on opened glandular lumens, they cannot be applied for analysis of prostate cancer with non-cribriform G4 or G5 growth patterns. However, since only glands with lumens were used in the gold standard, the JEP = 66 reported by Nguyen et al. [8] is higher than the one calculated with our best method (JEP = 59.5) or with the one from Monaco et al. (JEP = 31) [7] as shown by Nguyen et al. [8]. For these reasons, a direct comparison of our approach with other methods that rely on the detection of glandular lumens may not be fully justified. For such a comparison the testing and training of all the methods should be performed on the same dataset.
To the best of our knowledge the only published method that does not depend on glandular lumens was proposed by Chen and colleagues [31]. Similarly to our approach, Chen's method solely requires manual training and is not affected by the scanning magnification. Although this method can predict stroma with OST = 75.0, the performance of classifying epithelial areas, as assessed by JEP, OEP, Ri, is significantly lower than the performances of our SVM-based classifier when applied to same set of 191 images from our collection. The selections of 92 and 60 windows were performed independently by two observers. Both sets of windows selected for training were placed away from ST-EP borders to reduce the chance of including descriptors from neighboring tissues of another type. Essentially, training windows labeled as ST did not contain EP pixels and vice versa. The performance rates obtained from all three sets of training windows (ST-EP1, ST-EP2 and ST-EP3) were generally superior to those from the reference method Chen et al. [31]. Tissue predictions by our as well as Chen's method are less accurate in areas that consist of heterogeneous tissue types or that contain glands with a small number of nuclei due to the plane of sectioning. Errors of this kind can also be seen in Fig. 2 of [16]. Since only the epithelial classification is critical for diagnostic purposes, it is unlikely that further refinements of the Chen's method will provide adequate assistance for histological diagnoses.
In addition Kwak and colleagues [16] have shown very promising concordance rates based on ROC curves between manual and computer generated segmentations of stroma and epithelium in whole slide analysis using a multiresolution approach. However, it is unclear how this algorithm would perform in normal and cancerous glands. It is also not clear how the ground truth was generated and compared to the final computer-based result since 4 different resolution scales were involved in the image processing. One has to be aware that the way annotations are used for evaluation (pixels versus patches) has a significant impact on the performance. Figure 13 in the work by Dolye et al. [15] shows that the performance rates derived from ROC curves calculated for a pixel-based approach are significantly lower comparing to those derived from patch-based analyses in the same set of images. Thus, considering Kwak's study as a reference, our methodological advances seem to be a good alternative for the research community because: (a) our method uses the highest image resolution (20×) for training and validation, (b) very good performance rates were obtained for separation of stroma from benign/normal, low and high-grade cancer and (c) we only use intensity features (2 intensity histograms with 36 bins total per each image pixel), which is computationally less expensive and faster in comparison to Kwak's approach, which uses 63 features per pixel.
The proposed method was tested and developed on interpolated descriptor matrices. Performance tests involving non-interpolated descriptor matrices indicated that they are generally superior to those obtained from interpolated ones. However, we think that their high computational cost (90 times longer calculation time) outweighs benefits. Interestingly, the newly obtained performances demonstrate the limitation of the proposed method.
8.1.2. Prediction of benign/normal and cancer
Numbers reported in Table 4 demonstrate the overall performance of the RF classifier, when applied to all image tiles from the training set and without the level of detail shown in Table 5. After the training of 30 different RF classifiers (10 for each of the three window sizes) we observed high predictive power for BN and good predictive rates for PCa. Although there were discrepancies in the performance originating from different descriptors, the average Jaccard index for the worst average prediction in Table 4 was above 60, indicating an excellent concordance between the two raters. Furthermore, the O and Ri indices were consistently above 76.3 and 67.7 indicating excellent tissue prediction (either BN or PCa) regardless of the chosen descriptor and the scanning window size. One should also note that the RF classifiers were trained using samples derived from 25% of all epithelial pixels delineated in the training set. Results in Table 4 reflect averaged performances from all epithelial descriptors including those selected for training and the remaining 75% that were not. Thus, the high prediction rates in Table 4 confirmed that descriptors and samples chosen for the training were suitable and therefore allowed us to test them in images uninvolved in the test set.
When compared to the performance in the training set, performance rates were uniformly lower in the 191 images from the test set. To determine the best performing descriptor type amongst the 10 descriptors from Table 2, we ranked the values that were obtained with each descriptor in each of 3 window sizes and for each of the 5 performance measures for test set images. Ranking of descriptors was conducted irrespectively of the window size and ranking of windows irrespectively of the descriptor category, with top three results taken into consideration each time. The descriptor 8 formed as a concatenation of Hist(H), Hist(E), (H) and (E) was the most frequent one (6/15 times) amongst descriptors with top three JBN, JPCa, OPCa, OBN and Ri indices. In addition, the largest window with N = 256 was selected 9/15 times. Surprisingly, performances of descriptors 5 and 9 - (H) – which quantify the texture in the hematoxylin image, were collectively worse than performances of the descriptor 8. Similarly, under the same criteria descriptors 1 and 2 that utilized solely the pixel intensities, Hist(H) or Hist(H) combined with Hist(E), were overall inferior to the performances of descriptor 8. These results demonstrate that descriptor types utilizing both the intensity and texture of hematoxylin and eosin images are better suited for classification than descriptors based solely on a single stain image or one that quantifies either texture or signal intensity.
When we ranked the descriptors according to their performance, descriptor 8 ranked in the top 10% of results 6/15 times and therefore it is considered the most promising one. Since the evaluation of descriptor performances using established statistical methods was not feasible because of the absence of published statistical methods for this type of image analysis data, it is uncertain which descriptor performs best in a statistical sense. In order to develop a statistical approach for a more stringent evaluation of descriptors that takes advantage of the Jaccard, Overlap and Rand indices one needs to take into account the data structure and relationship between the indices, which include: (a) different effects of image tiles with a single epithelial component (n = 33 for BN and n = 118 for PCa) compared to tiles with data from the 2 types of epithelial components (n = 59), (b) OBN/PCa is undefined and JBN/PCa is 0 for image tiles that do not contain a BN or PCa component, (c) the positive correlations between Jaccard and Overlap indices for the same tissue component and (d) the nonlinearity, difference in dynamic ranges and large spread of J, O and Ri measurements. Based on the unique challenges that image analysis data pose, it will be necessary to develop appropriate statistical methods for analysis in the future.
In general, we observe a better classification performance for PCa than for BN glands (third column of Fig. 7). In addition, BN glands adjacent to cancer were more frequently misclassified as PCa compared to those farther away from PCa. Misclassification also occurred in small tangential sections of BN glands. On the contrary, PCa areas were consistently predicted with higher accuracy and lower variability than areas of BN glands. The large variability in the prediction of BN glands is the main reason for the large standard deviations in the performance measurements in Table 5. The best results that were obtained with descriptor 8 and N = 256 revealed an average OBN = 49.6 and JBN = 35.2 for BN glands. The latter is 10% higher than the Jaccard index from a paper demonstrating performances of an early gland segmentation technique [7] published four years ago. For PCa, the respective averages are higher than for BN glands. The calculated indices of OPCa = 71.1 and JPCa = 48.9 install confidence that the proposed method is suited for classification of low-grade and high-grade PCa, including high grade cribriform and non-cribriform tumor growth patterns. To further improve the distinction between BN glands and PCa and reach J indices above 60, a more powerful descriptor or a window covering a larger area needs to be employed. A less subjective method for selecting regions to train classifiers should also be sought.
8.2. Implementation
The overall computational expense with the approach was much higher than calculations utilizing Hist(H) and Hist(E). The complexity of calculations with particularly for P = 24 and R = 3 results requires a high consumption of RAM, storage and CPU power. For instance, the descriptor 8 amounts to a length of 2*(18 + 208) = 452 and the size of the related single precision descriptor matrix is image_width × image_height × descriptor_length. Consequently, the RF classifier training demands a large amount of CPUs and RAM. On a PC-based workstation with 2 CPUs, each consisting of 12 cores with 2.67 GHz clock and 24 GB of RAM, the average duration to complete the calculation of the descriptor 8 matrix was 40 min, the RF training time was 15 min and the tissue classification time was less than 40 s per image. For comparison, the average classification time by SVM took 15 s.
8.3. Concluding remarks
Our method to separate stroma from benign/normal glands and cancer in tissue sections of the prostate involved a training set and a SVM classifier. The classifier was tested on images that included high-grade PCa and were able to distinguish low-grade and high-grade cancer growth patterns from stroma, even in the absence of lumen formation in areas of high-grade cancer. A major advance of our technology is to first employ a pixel-based classification strategy to the sub-stratification of glands into benign/normal and cancer with good accuracy. Based on the performance measures, we conclude that a machine learning approach that is designed to classify individual pixels in H&E stained tissues bears promise for evaluation of images from prostate cancer tissues.
To facilitate and encourage further advancements in this field, the full set of 210 original images with the gold standard annotations used in our study is made available from the corresponding author upon request.
9. Conclusions
We have developed and evaluated two machine learning techniques and applied them to identify and classify benign/normal and malignant prostate glands. The performance of the proposed framework was thoroughly evaluated in independent training and test sets and constitutes an automated and consistent approach for quantification of disease related histopathological parameters in microscopic images. Our method has the potential to improve the measurement of parameters in tissue sections that are needed for diagnosis and prognostication of patients with prostate cancer.
Acknowledgments
This work was performed in part with the support from the Departments of Surgery, Pathology and Biomedical Sciences at Cedars-Sinai Medical Center. Institutional support was provided from the Department of Surgery to AG and from the Department of Biomedical Sciences to BSK. The authors would like to thank Elena Chang MD for selecting cases and marking tumor areas on glass slides before slide digitization and Dr Yuan Xiaopu for verification of ground truth annotations.
Footnotes
10. Conflict of interest statement
All the data, results as well as the methodology developed are the authors’ own work and research effort. The authors declare that they have no conflict of interest.
References
- 1.Committee ACPRW. Sawyers CL, Abate-Shen C, Anderson KC, Barker A, Baselga J, Berger NA, et al. AACR Cancer Progress Report 2013. Clin Can Res: J Am Assoc Can Res. 2013;19(20 Suppl):S4–98. doi: 10.1158/1078-0432.CCR-13-2107. [DOI] [PubMed] [Google Scholar]
- 2.Fine SW, Amin MB, Berney DM, Bjartell A, Egevad L, Epstein JI, et al. A contemporary update on pathology reporting for prostate cancer: biopsy and radical prostatectomy specimens. Eur Urol. 2012;62(1):20–39. doi: 10.1016/j.eururo.2012.02.055. [DOI] [PubMed] [Google Scholar]
- 3.Brimo F, Montironi R, Egevad L, Erbersdobler A, Lin DW, Nelson JB, et al. Contemporary grading for prostate cancer: implications for patient care. Eur Urol. 2013;63(5):892–901. doi: 10.1016/j.eururo.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Oyama T, Allsbrook WC, Jr, Kurokawa K, Matsuda H, Segawa A, Sano T, et al. A comparison of interobserver reproducibility of Gleason grading of prostatic carcinoma in Japan and the United States. Arch Pathol Lab Med. 2005;129(8):1004–10. doi: 10.5858/2005-129-1004-ACOIRO. [DOI] [PubMed] [Google Scholar]
- 5.Allsbrook WC, Jr, Mangold KA, Johnson MH, Lane RB, Lane CG, Epstein JI. Inter-observer reproducibility of Gleason grading of prostatic carcinoma: general pathologist. Human Pathol. 2001;32(1):81–8. doi: 10.1053/hupa.2001.21135. [DOI] [PubMed] [Google Scholar]
- 6.Allsbrook WC, Jr, Mangold KA, Johnson MH, Lane RB, Lane CG, Amin MB, et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: uro-logic pathologists. Human Pathol. 2001;32(1):74–80. doi: 10.1053/hupa.2001.21134. [DOI] [PubMed] [Google Scholar]
- 7.Monaco JP, Tomaszewski JE, Feldman MD, Hagemann I, Moradi M, Mousavi P, et al. High-throughput detection of prostate cancer in histological sections using probabilistic pairwise Markov models. Med Image Anal. 2010;14(4):617–29. doi: 10.1016/j.media.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen K, Sarkar A, Jain AK. Structure and context in prostatic gland segmentation and classification. Medical image computing and computer-assisted intervention: MICCAI International Conference on Medical Image Computing and Computer-Assiste. Intervention. 2012;15(Pt 1):115–23. doi: 10.1007/978-3-642-33415-3_15. [DOI] [PubMed] [Google Scholar]
- 9.Tabesh A, Teverovskiy M, Pang HY, Kumar VP, Verbel D, Kotsianti A, et al. Multifeature prostate cancer diagnosis and Gleason grading of histological images. IEEE Trans Med Imag. 2007;26(10):1366–78. doi: 10.1109/TMI.2007.898536. [DOI] [PubMed] [Google Scholar]
- 10.Yahui P, Yulei J, Eisengart L, Healy MA, Straus FH, Yang XJ. Segmentation of prostatic glands in histology images.. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on; March 30 2011–April 2 2011.2011. pp. 2091–2094. [Google Scholar]
- 11.Yu E, Monaco JP, Tomaszewski J, Shih N, Feldman M, Madabhushi A. Detection of prostate cancer on histopathology using color fractals and Probabilistic Pairwise Markov models.. Engineering in Medicine and Biology Society, EMBC, 2011 Annual International Conference of the IEEE; Aug. 30 2011–Sept. 3 2011; 2011. pp. 3427–3430. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs TJ, Buhmann JM. Computational pathology: challenges and promises for tissue analysis. Comput Med Imag Graph: J Comput Med Imag Soc. 2011;35(7–8):515–30. doi: 10.1016/j.compmedimag.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Doyle S, Feldman MD, Shih N, Tomaszewski J, Madabhushi A. Cascaded discrimination of normal, abnormal, and confounder classes in histopathology: Gleason grading of prostate cancer. BMC Bioinform. 2012;13:282. doi: 10.1186/1471-2105-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorelick L, Veksler O, Gaed M, Gomez JA, Moussa M, Bauman G, et al. Prostate histopathology: learning tissue component histograms for cancer detection and classification. IEEE Trans Med Imag. 2013;32(10):1804–18. doi: 10.1109/TMI.2013.2265334. [DOI] [PubMed] [Google Scholar]
- 15.Doyle S, Feldman M, Tomaszewski J, Madabhushi A. A boosted Bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans Bio-med Eng. 2012;59(5):1205–18. doi: 10.1109/TBME.2010.2053540. [DOI] [PubMed] [Google Scholar]
- 16.Kwak JT, Xu S, Pinto PA, Turkbey B, Bernardo M, Choyke PL, Wood BJ. A multiview boosting approach to tissue segmentation. 2014:90410R–90410R-90417. [Google Scholar]
- 17.Salman S, Ma Z, Mohtanty S, Bhele S, Chu Y-T, Knudsen B, et al. A machine learning approach to identify prostate cancer areas in complex histological images. In: Piȩtka E, Kawa J, Wieclawek W, editors. Information Technologies in Biomedicine, Volume 3, 283. Springer International Publishing; Cham, Heidelberg, New York, Dordrecht, London: 2014. pp. 295–306. [Google Scholar]
- 18.Rizzardi AE, Johnson AT, Vogel RI, Pambuccian SE, Henriksen J, Skubitz AP, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol. 2012;7:42. doi: 10.1186/1746-1596-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linder N, Konsti J, Turkki R, Rahtu E, Lundin M, Nordling S, et al. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn Pathol. 2012;7:22. doi: 10.1186/1746-1596-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goode A, Gilbert B, Harkes J, Jukic D, Satyanarayanan M. OpenSlide: a vendor-neutral software foundation for digital pathology. J Pathol Inform. 2013;4:27. doi: 10.4103/2153-3539.119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Sparks R, Janowczyk A, Tomaszewski J, Feldman M, Madabhushi A. In: High-throughput prostate cancer gland detection, segmentation, and classification from digitized needle core biopsies. Prostate cancer imaging computer-aided diagnosis, prognosis, and intervention. Madabhushi A, Dowling J, Yan P, Fenster A, Abolmaesumi P, Hata N, editors. Springer; 6367. Berlin Heidelberg: 2010. pp. 77–88. [Google Scholar]
- 22.Naik S, Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology.. Biomedical Imaging: From Nano to Macro, 2008 ISBI 2008 5th IEEE International Symposium on; 14–17 May 2008.pp. 284–287. [Google Scholar]
- 23.Vidal J, Bueno G, Galeotti J, García-Rojo M, Relea F, Déniz O. A fully automated approach to prostate biopsy segmentation based on level-set and mean filtering. 2011:2. doi: 10.4103/2153-3539.92032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Janowczyk A, Chandran S, Madabhushi A. A high-throughput active contour scheme for segmentation of histopathological imagery. Medical image analysis. 15(6):851–862. doi: 10.1016/j.media.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Analyt Quantitat Cytol and Histol: Int Acad Cytol Am Soc Cytol. 2001;23(4):291–9. [PubMed] [Google Scholar]
- 26.Reinhard E, Ashikhmin M, Gooch B, Shirley P. Color transfer between images. IEEE Comput Graph Appl. 2001;21(5):34–41. [Google Scholar]
- 27.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. Pattern Anal Mach Intell IEEE Trans. 2002;24(7):971–87. [Google Scholar]
- 28. http://www.cse.oulu.fi/MVG/Downloads.
- 29.Scholkopf B, Smola AJ. Learning with kernels: support vector machines, regularization, optimization, and beyond. MIT Press; Cambridge, MA, USA: 2001. [Google Scholar]
- 30.Rand WM. Objective criteria for the evaluation of clustering methods. J Am Stat Assoc. 1971;66(336):846–50. [Google Scholar]
- 31.Chen J, Toghi ES, Bova GS, Li QK, Li X, Zhang H. Epithelium percentage estimation facilitates epithelial quantitative protein measurement in tissue specimens. Clin Proteom. 2013;10(1):18. doi: 10.1186/1559-0275-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]