Abstract
Allopolyploidization combines parental genomes and often confers broader species distribution. However, little is known about parentally transmitted gene expression underlying quantitative traits following allopolyploidization because of the complexity of polyploid genomes. The allopolyploid species Arabidopsis kamchatica is a natural hybrid of the zinc hyperaccumulator Arabidopsis halleri and of the nonaccumulator Arabidopsis lyrata. We found that A. kamchatica retained the ability to hyperaccumulate zinc from A. halleri and grows in soils with both low and high metal content. Hyperaccumulation of zinc by A. kamchatica was reduced to about half of A. halleri, but is 10-fold greater than A. lyrata. Homeologs derived from A. halleri had significantly higher levels of expression of genes such as HEAVY METAL ATPASE4 (HMA4), METAL TRANSPORTER PROTEIN1 and other metal ion transporters than those derived from A. lyrata, which suggests cis-regulatory differences. A. kamchatica has on average about half the expression of these genes compared with A. halleri due to fixed heterozygosity inherent in allopolyploids. Zinc treatment significantly changed the ratios of expression of 1% of homeologous pairs, including genes putatively involved in metal homeostasis. Resequencing data showed a significant reduction in genetic diversity over a large genomic region (290 kb) surrounding the HMA4 locus derived from the A. halleri parent compared with the syntenic A. lyrata-derived region, which suggests different evolutionary histories. We also estimated that three A. halleri-derived HMA4 copies are present in A. kamchatica. Our findings support a transcriptomic model in which environment-related transcriptional patterns of both parents are conserved but attenuated in the allopolyploids.
Keywords: allopolyploid, homeolog, hyperaccumulation, heavy metal, selective sweep, transcriptomics
Introduction
The divergence of phenotypes plays a primary role in speciation and is often driven by ecological adaptations (Coyne and Orr 2004; Rundle and Nosil 2005). Once isolated, species with highly divergent ecological adaptations become less likely to come into contact, and if they do, viable offspring are unlikely because of genetic incompatibilities that accumulate during parental divergence (Wu and Ting 2004). Therefore, it is unclear how allopolyploid hybrids not only survive, but also are able to inhabit broad ecological habitats compared with parental species (Stebbins 1971; Ehrendorfer 1980; Levin 2000). Allotetraploids inherit chromosomes from two parental species through hybridization and genome duplication, and thus represent a fixed heterozygote state of two parental genomes (reviewed in Comai 2005). In allopolyploids, differential gene expression can result from vertically transmitted bias in parental expression (Yoo et al. 2014) or “parental legacy” (Buggs et al. 2014), which results in one homeolog being preferentially expressed over the other. Studies in Arabidopsis polyploids have demonstrated that divergent parental expression can be transmitted to an allopolyploid (Wang et al. 2006) and parental (allelic) expression patterns can persist over multiple generations (Shi et al. 2012).
Comparative transcriptomics studies in natural and synthesized allopolyploids have become a powerful tool for detecting homeolog-specific expression and estimating additive and nonadditive gene regulation due to cis and trans effects (Wang et al. 2006; Shi et al. 2012; Yoo et al. 2013). Studies have often identified genes enriched for gene ontologies corresponding to a particular treatment condition that affects homeolog expression (Akhunova et al. 2010; Combes et al. 2013; Akama et al. 2014; Shimizu-Inatsugi et al. 2016), but comparisons of both expression levels and quantitative phenotypes have shown a relationship between quantitative traits and transcriptomics in allopolyploids and diploid parents (Bertrand et al. 2015). To gain an empirical perspective into the possible advantages or disadvantages of polyploidy that affect species distributions, it is first necessary to identify phenotypes that were inherited from diploid ancestors, which can be measured precisely in both parents and hybrids (Takumi et al. 2009; McCarthy et al. 2015), along with the knowledge of the corresponding genetic loci underlying such phenotypes. We propose that heavy metal hyperaccumulation represents a highly tractable and inducible quantitative trait that has tremendous potential for detailed analyses of gene regulation between diploid and polyploid relatives. Next-generation sequencing can now be used to enhance both genome-wide studies of hyperaccumulation (Verbruggen et al. 2013) and allopolyploid evolution (Buggs et al. 2012).
Hyperaccumulator species transport toxic heavy metals from roots to shoots and can grow on soils with high cadmium (Cd) and zinc (Zn) contamination. Only a limited number of hyperaccumulator species have evolved independently among the angiosperms (Krämer 2010) and are often distributed in toxic ecological niches with heavy metal contamination. Hyperaccumulation of Cd and Zn has been studied extensively in Brassicaceae at the physiological and molecular level in Arabidopsis halleri and Noccaea caerulescens (formerly Thlaspi caerulescens), and many candidate genes responsible for metal tolerance have been identified (Hanikenne et al. 2008; Milner and Kochian 2008, Shahzad et al. 2010; Ó Lochlainn et al. 2011). Both genera have closely related hyperaccumulator and nonaccumulator species enabling comparative genomic and phenotyping studies (Becher et al. 2004; Talke et al. 2006; reviewed in Verbruggen et al. 2009; Krämer 2010).
Metal homeostasis involves multicellular transport of ions and intricate detoxification of cells, presenting considerable challenges for finding loci with large effects. Nevertheless, quantitative trait locus (QTL) studies using a backcross family (BC1) of A. halleri and Arabidopsis lyrata have demonstrated two main loci involved in hyperaccumulation and tolerance to both Cd and Zn: HEAVY METAL ATPASE4 (HMA4) coding for an ATPase transporter protein and METAL TRANSPORTER PROTEIN1 (MTP1, also called ATCDF1 or ZAT1) coding for a cation diffusion facilitator (CDF) protein (Dräger et al. 2004; Desbrosses-Fonrouge et al. 2005; Filatov et al. 2006; Courbot et al. 2007; Willems et al. 2007). These two loci are constitutively expressed even under low metal conditions (Talke et al. 2006), and both have known paralogous duplications in A. halleri (Hanikenne et al. 2008; Shahzad et al. 2010) giving them enhanced cis-regulation. The discovery in A. halleri of the tandemly triplicated HMA4 as the major locus responsible for transporting heavy metals from roots to shoots was confirmed by yeast complementation tests (Talke et al. 2006) and RNA silencing (RNAi, Hanikenne et al. 2008). Silencing of HMA4 in A. halleri effectively inhibited Zn translocation to leaves, but transformation of the functional A. halleri HMA4 (AhHMA4) gene into a nontolerant Arabidopsis thaliana background conferred the ability to transport Zn to the xylem, although it was not sufficient to confer the ability to hyperaccumulate Zn into leaves. This indicates that transport is tightly integrated with cellular detoxification mediated by other factors involving vacuolar sequestration of toxic ions, such as MTP1 (Kobae et al. 2004; Filatov et al. 2006; Willems et al. 2007) and possibly HMA3 (Becher et al. 2004; Morel et al. 2008). In short, the triplication as well as enhanced promoter activity of each copy contributed to high HMA4 expression (Hanikenne et al. 2008), which is presumably responsible for the constitutive hyperaccumulation ability in the entire species (Krämer 2010).
The time of evolutionary divergence of A. halleri and A. lyrata has been estimated to coincide with the duplication of HMA4 (Roux et al. 2011), which presumably confers the ability to hyperaccumulate heavy metals to A. halleri, but not A. lyrata. This suggests a mechanism of ecological speciation by enhanced tolerance to toxic soils. Despite that habitats of European A. halleri encompass both metalliferous and nonmetalliferous soils, A. halleri appears to be constitutively metal tolerant regardless of low to intermediate heavy metal concentrations in most populations (Bert et al. 2002; Pauwels et al. 2006; Meyer et al. 2010). The HMA4 diversity analysis of A. halleri by Hanikenne et al. (2013) estimated a hard selective sweep surrounding the HMA4 locus (Hanikenne et al. 2013), which further suggests that hyperaccumulation in A. halleri is adaptive. Quantitative PCR (qPCR) showed three HMA4 copies in numerous A. halleri genotypes, which indicates that the triplication is widespread in the species. The elemental defense hypothesis may support the species-wide ability to hyperaccumulate metals where even small amounts of Cd or Zn accumulation in leaves deter herbivores (Coleman et al. 2005; Boyd 2007). Elemental defense was recently supported by Kazemi-Dinan et al. (2014), who discovered a significant reduction in herbivory of A. halleri as a result of elevated Cd and Zn concentrations in leaves. Metal concentrations from above 1,000 μg Zn g−1 dry weight (DW) and 18 μg Cd g−1 DW reduced feeding of A. halleri by three specialist insects (Kazemi-Dinan et al. 2014).
The allopolyploid species Arabidopsis kamchatica is a natural hybrid of diploid A. halleri and A. lyrata without chromosome reduction. Phylogenetic analysis showed that the nearest ancestors of A. kamchatica are A. halleri subsp. gemmifera, native to East Asia, and A. lyrata subsp. petraea (also called A. petraea subsp. umbrosa), native to Siberia (Shimizu et al. 2005; Shimizu-Inatsugi et al. 2009; Schmickl et al. 2010). The diploid progenitors of A. kamchatica are highly divergent in both hyperaccumulation and gene expression (Filatov et al. 2006; Willems et al. 2007), which provides the opportunity to study a quantitative phenotype in a hybrid species whose diploid parents possess opposite phenotypes. The divergence of A. kamchatica from its parental diploid species was estimated to have occurred 20,417 years ago (95% confidence interval 0–75,460 years) using mutation rates estimated by Koch et al. (2000) and 245,070 years (95% confidence interval 37,385–532,953 years) using mutation rates estimated by Ossowski et al. (2010) (Tsuchimatsu et al. 2012). The possible divergence of A. halleri and A. lyrata due to ecological speciation (Roux et al. 2011) followed by recent hybridization to form the allopolyploid A. kamchatica can provide insights into genome-wide transcriptional changes maintaining potentially adaptive phenotypes in these Arabidopsis species.
To test whether A. kamchatica retained Zn hyperaccumulation following allopolyploid hybridization between hyperaccumulating and nonaccumulating parental species, we grew plants hydroponically under Zn stress to compare hyperaccumulation in both diploid parental species. We then characterized transcriptional changes among homeologous gene copies using algorithms to assign RNA-seq reads to their ancestral genomes (Akama et al. 2014) and validated expression ratios using pyrosequencing. We identified several genes previously reported as having roles in metal tolerance and ion transport that show strong bias toward the hyperaccumulating A. halleri (H-origin) parental genome and significant regulatory changes in both H-origin and A. lyrata (L-origin)-derived homeologs following Zn treatment. We examined whether the three HMA4 copies of A. halleri are also present in A. kamchatica and whether HMA4 showed high homeolog-specific expression. Using polymorphism data flanking the HMA4 locus in A. kamchatica, we detected significantly different levels of diversity between H-origin and L-origin homeologs surrounding HMA4 over a large genomic region. The combination of the Zn accumulation phenotype, transcriptomics data, and polymorphism analysis suggests that despite allopolyploid hybridization with a nontolerant A. lyrata-like ancestor, the allopolyploid A. kamchatica has retained the hyperaccumulation phenotype from the A. halleri parent.
Results
Zinc Accumulation
We expected that the concentration of Zn in A. kamchatica would be high in leaves and low in roots if hyperaccumulation was inherited from the A. halleri parent. Using hydroponic growth chambers, after 1 week of exposure to 500 μM Zn, we first compared Zn accumulation between diploid A. halleri, A. lyrata, and four A. kamchatica genotypes (two Japanese and two Alaskan accessions) (Experiment 1). In leaf tissues, the hyperaccumulator A. halleri had the highest Zn accumulation (mean = 8,180 μg g − 1 DW, standard deviation (SD) = 3,533) while A. lyrata accumulated less than one-tenth the concentration of Zn (mean = 614.6 μg g − 1, sd = 262.9; fig. 1A, supplementary table S1, Supplementary Material online) as A. halleri. Zinc levels in leaves of the Tada mine A. halleri genotype showed elevated levels similar to those in previous reports of European A. halleri collected in the field (Bert et al. 2002) and grown under controlled Zn treatments (Chiang et al. 2006; Talke et al. 2006; Kashem et al. 2010). In A. kamchatica, the mean accumulation of Zn in leaf tissues was about half of A. halleri. The average Zn accumulation of each genotype (with replication) varied from 3,200 to 4,100 μg g − 1 DW (the maximum accumulation in the PAK genotype was 6,720 μg g − 1 DW). Statistical comparisons confirmed that all A. kamchatica genotypes showed significantly lower Zn accumulation in leaves than A. halleri (0.003 < P < 0.02), while both A. halleri and A. kamchatica showed the same magnitude of Zn accumulation, and both accumulated significantly higher concentrations than A. lyrata (all P values <1.20e−05, supplementary table S1B, Supplementary Material online).
Fig. 1.
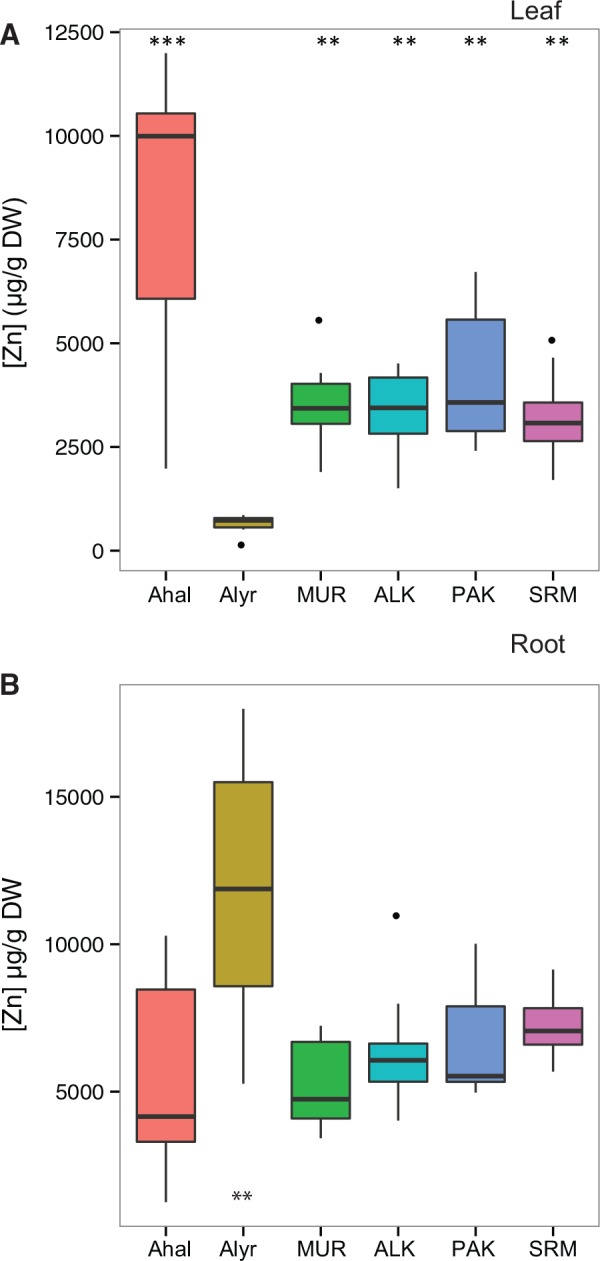
Zinc (Zn) accumulation in leaf (A) and root tissue (B) in Arabidopsis halleri subsp. gemmifera (Ahal), Arabidopsis lyrata subsp. petraea (Alyr), and Arabidopsis kamchatica (MUR, ALK, PAK, and SRM genotypes) after 7 days of 500 μM Zn exposure estimated as μg per gram of dry weight (DW). Box plots indicate the first quartile, median, and third quartile, whiskers indicate the minimum and maximum interquartile range (no more than 1.5 times), and dots show outlier values. Asterisks in (A) show significant pairwise differences relative to mean A. lyrata leaf Zn concentration (P < 10−5; supplementary table S1, Supplementary Material online). For roots (B), asterisks indicate that A. lyrata is significantly different from both A. halleri and all four A. kamchatica genotypes (P < 0.008). Pairwise differences were estimated using a Wilcoxon signed rank-sum test to determine significance.
In root tissues, the Zn concentration of A. lyrata (mean = 13,754 μg g−1) was significantly higher than both A. halleri and A. kamchatica (fig. 1B), which indicated a lack of Zn transport to shoots. Zinc quantities in A. kamchatica (5,300–6,600 μg g − 1) and A. halleri (mean concentration of 6,756 μg g − 1) roots were not significantly different (supplementary table S1D, Supplementary Material online). The level of Zn in A. lyrata roots was significantly higher than in A. halleri and all four A. kamchatica genotypes (supplementary table S1D, Supplementary Material online). Among A. kamchatica, the SRM genotype had the highest level of Zn in roots, and the difference between the MUR and SRM genotypes in roots was significant. Strong hyperaccumulators typically show leaf-to-root ratios of Zn accumulation greater than one (Krämer 2010). Ratios of shoot-to-root concentrations of Zn from Experiment 1 were 1.21 for A. halleri and 0.04 for A. lyrata. Shoot-to-root Zn ratios for each A. kamchatica genotype (0.52–0.66) were about half that of A. halleri, but were >10 times higher than that of A. lyrata. Two additional experiments that varied Zn treatments (250–1,000 μM) showed proportional increases in Zn accumulation indicating highly responsive heavy metal transport (supplementary figs. S1 and S2, Supplementary Material online) in A. kamchatica.
Genome-Wide Patterns of Homeolog Expression and Hyperaccumulation-Related Genes
To quantify the expression patterns of homeologs, we conducted RNA-seq analysis on 48 cDNA libraries (4 genotypes × 2 tissues × 2 treatments × 3 replicates) using leaf and root tissues of two A. kamchatica genotypes (MUR from Murodo, Japan and PAK from Potter, Alaska), one A. halleri genotype, and one A. lyrata genotype. Tissues were collected during Experiment 1 before and after 48 h of 500 μM Zn treatment to determine the early transcriptional response. Reads were sorted using the HomeoRoq pipeline described in Akama et al. (2014) and mapping statistics can be found in supplementary file 1, Supplementary Material online. We estimated similar proportions of reads sorted as H-origin and L-origin with a slight excess of the halleri-derived reads.
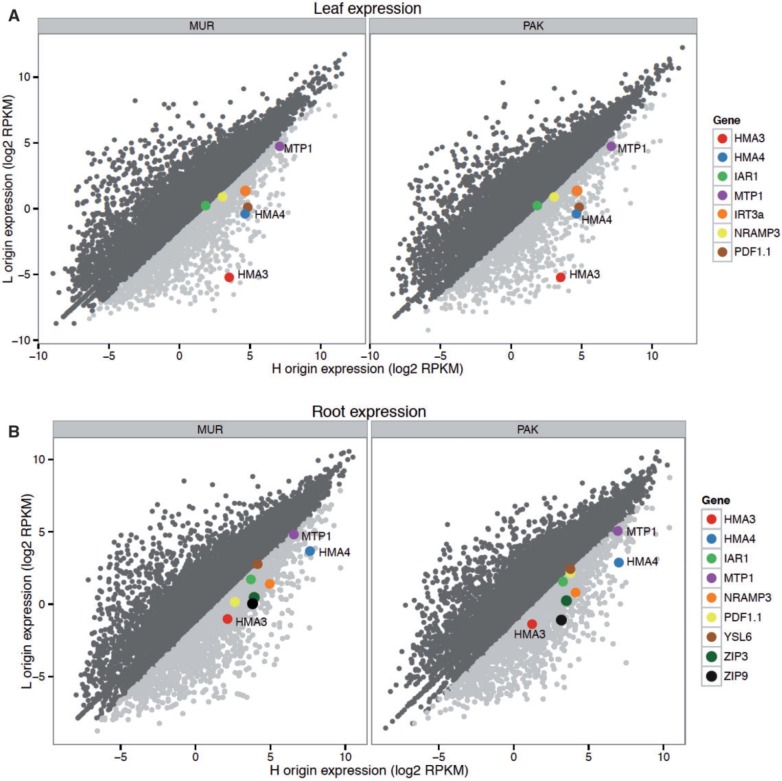
We expected genes affecting hyperaccumulation in A. kamchatica would be inherited from A. halleri and would show significantly higher expression in the H-origin compared to the L-origin homeologs. Comparing homeolog expression levels before Zn treatment that were ≥3-fold (read per kilobase per million reads (RPKM)) higher in H-origin than in L-origin copies in leaves and root tissues, we found that the Japanese genotype (MUR) showed 1,410 (leaf) and 1,208 (root) genes with ≥3-fold higher expression in H-origin than in L-origin homeologs. The Alaska genotype (PAK) showed 1,041 genes in leaves and 1,281 in roots with ≥3-fold higher H-origin expression (fig. 2). Among these genes, we focused on those identified in previous differential expression studies between the hyperaccumulator A. halleri and nonaccumulators A. thaliana (Becher et al. 2004; Talke et al. 2006; Chiang et al. 2006, Shanmugam et al. 2011) or A. lyrata (Filatov et al. 2006). We found that many of the candidates for hyperaccumulation, metal tolerance, or metal ion transport (hereafter we refer to these genes as HM genes) previously detected in differential expression studies are above the 3-fold threshold of H-origin over L-origin homeolog expression (fig. 2). Among these HM genes for leaves, the top four genes with >5-fold H-origin expression bias were HMA3, HMA4, PDF1.1, and MTP1, followed by IRT3, NRAMP3, and IAR1, which show 3- to 5-fold H-origin bias in at least one of the samples (supplementary file 2, Supplementary Material online). For roots, we found (again using ≥3-fold bias as a cutoff) higher H-origin expression in HMA3, HMA4, HMA5, IAR1, MTP1, NRAMP3, PDF1.1, ZIP3, and ZIP9. Nearly all of the differences in homeolog-specific expression were statistically significant using edgeR (FDR < 0.05; supplementary file 2, Supplementary Material online), suggesting cis-regulatory differences (He et al. 2012; Shi et al. 2012).
Fig. 2.
Genome-wide expression patterns in the leaves (A) and roots (B) of H-origin and L-origin homeologs before Zn treatment in the Murodo, Japan (MUR) and Potter, Alaska (PAK) Arabidopsis kamchatica genotypes. Genes with ≥3-fold higher expression (RPKM) in H-origin over L-origin homeologs are shaded light gray and those with ≤3-fold higher expression are shown as dark-gray points. Colored points represent candidate orthologs in A. kamchatica previously shown to be differentially expressed by Talke et al. (2006) and Filatov et al. (2006) or with annotated metal transport function in TAIR10 (Lamesh et al. 2012). Expression is log2 of mean RPKM among three replicates. Genome-wide correlations between homeologs are r2 = 0.79–0.84.
Compared with A. kamchatica, we found similar tendencies for the diploid parents for most of these same HM genes, but often stronger A. halleri bias because of higher expression of many metal-tolerance-related genes compared with A. kamchatica (supplementary file 2, Supplementary Material online). For diploids, the top five genes showing the strongest A. halleri bias in leaves were HMA3, PDF1.1, MTP1, HMA4, and IRT3. In particular, both MTP1 and PDF1.1 had dramatically higher expression and stronger bias in A. halleri than in A. lyrata compared with the H-origin homeolog of these genes in A. kamchatica. The Zn transporter HMA4 had greater expression in A. halleri, but a lower fold increase over A. lyrata following Zn treatment compared with A. kamchatica HMA4 homeologs (supplementary file 2, Supplementary Material online). Using the procedure and the genome assembly here, when there are duplicated copies within a diploid species (e.g., duplicated copies of HMA4 and MTP1 in A. halleri and consequently in halleri-homeologs), the sum of their expression would be detected (see the section HMA4 copy number by pyrosequencing and 3′ amplification).
To compare the levels of expression between diploid and allopolyploid species better, we calculated the sum of the expression of homeologs for sets of hyperaccumulator-related genes. The sum of the expression levels of the homeologs of hyperaccumulation genes is potentially relevant, because no differences in protein function have been reported between orthologs derived from hyperaccumulator and nonaccumulator species (Krämer 2010). Because the expression level in A. halleri is much higher than A. lyrata for heavy metal-related genes, and because the allotetraploid represents a fixed heterozygous condition, the total expression level in A. kamchatica of most of these genes would be expected to be much higher than A. lyrata, but still reduced (to about half) compared with A. halleri. This can be described as follows: supposing that the levels of expression in A. halleri (denoted by H) is x times higher than A. lyrata (denoted by L), H = xL. In a simple case with cis-regulation only, the total expression level of A. kamchatica (denoted by K, the sum of a pair of homeologs) is the average of two parents because allotetraploids represent a fixed heterozygous condition, that is, K = (L + xL)/2 = (1 + x)L/2. In an average situation with the same expression level of both homeologs (x = 1), the ratio K/H = (1 + x)/2x) is 1. In contrast, when x is large, K/H approaches 1/2 and K/L becomes large. We confirmed these predictions using our expression data, in which H and L were diploid expression levels, and K was measured as the sum of the pair of homeologs for each gene. The majority of the gene pairs show a similar expression level and consistently the genome-wide median of K/H in both A. kamchatica genotypes was close to 1 (supplementary fig. S3, Supplementary Material online). In contrast, from our HM gene list (supplementary file 2, Supplementary Material online), the ratio of K/H was less than 1 (mean 0.34–0.67, median 0.19–0.45), while the ratio of both homeologs divided by the A. lyrata expression (K/L) was higher than 1 (mean 6.33–48.81, median 1.8–8.07). The variation between genes also suggests the contribution of both cis- and trans-regulation. This finding also conforms well with the Zn accumulation phenotype, where on average A. kamchatica exhibits about one-half the Zn concentration in leaves compared with A. halleri, but more than ten times greater concentration than that in A. lyrata.
Differential Expression Following Zn Treatment
To quantify the regulatory changes affected by Zn treatment in both the allopolyploid and diploid relatives, we examined differential expression before and after 48 h of Zn treatment in leaf and root tissues. Although several genes directly involved in hyperaccumulation are known to be expressed in A. halleri in a broad range of Zn concentrations (“constitutive expression”), a large number of differentially regulated genes and homeologs (both up- and down-regulated) were found after the Zn treatment (table 1). In A. halleri, leaves showed over twice the number of differentially regulated genes compared with roots (1,992/846). A. lyrata showed the opposite pattern, whereby roots had nearly 20 times more differentially regulated genes than leaves (table 1A). This pattern is consistent with the distribution of Zn with high concentration in roots of A. lyrata and leaves of A. halleri because of root-to-shoot transport and also indicative of a strong stress response in A. lyrata roots.
Table 1.
Numbers of Significantly Up or Down Regulated Genes Before and After 48 h Zinc Treatment in Leaf and Root Tissues.
| Leaf | Root | |||
|---|---|---|---|---|
| (A) Diploids | ||||
| A. halleri | A. lyrata | A. halleri | A. lyrata | |
| Upregulated | 889 | 355 | 232 | 5,095 |
| Downregulated | 1,103 | 187 | 614 | 5,458 |
| Total | 1,992 | 542 | 846 | 10,553 |
| (B) A. kamchatica | ||||
| MUR (Murodo, JP) | H-origin | L-origin | H-origin | L-origin |
| Upregulated | 313 | 297 | 1,030 | 1,006 |
| Downregulated | 384 | 358 | 1,345 | 1,332 |
| Total | 697 | 655 | 2,375 | 2,338 |
| (C) A. kamchatica | ||||
| PAK (Potter, AK) | H-origin | L-origin | H-origin | L-origin |
| Upregulated | 220 | 226 | 1,527 | 1,441 |
| Downregulated | 696 | 661 | 1,480 | 1,353 |
| Total | 916 | 887 | 3,007 | 2,794 |
Note.—(A) Diploids A. halleri subsp. gemmifera genotype (Tada mine) and A. lyrata subsp. petraea genotype w1178. Allopolyploid genes significantly up- or down-regulated in A. halleri-derived (H-origin) and A. lyrata-derived (L-origin) homeologs for A. kamchatica genotypes (B) MUR (Murodo, Japan) and (C) PAK (Potter, Alaska). The significance threshold is set to a FDR-adjusted P < 0.05 and expression between control to zinc-treated conditions must add up to RPKM ≥ 0.2.
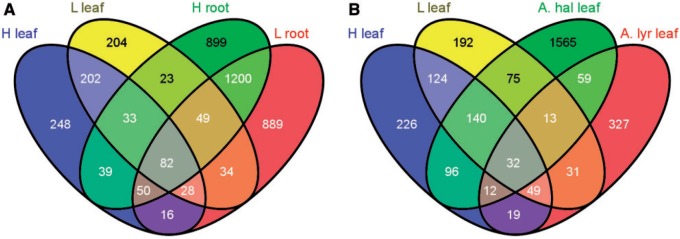
Compared with the diploids, A. kamchatica showed an intermediate pattern with 2.5–4 times as many differentially regulated homeologs in roots compared with leaf tissues and similar numbers of total H-origin and L-origin homeologs showing regulatory changes in each tissue (table 1B and C). Similar numbers of differentially regulated homeologs are also consistent with the highly correlated homeolog expression for our genome-wide dataset, which we found for both A. kamchatica genotypes (r2 = 0.79–0.83). However, despite the similar numbers of homeologs showing regulatory changes in each tissue type, the overlap among differentially regulated homeologs was about half the total number for both tissues (fig. 3A). The largest number of unique genes showing differential regulation was found for A. halleri leaves (fig. 3B), which indicated the regulation of many genes that are likely necessary for complete detoxification to achieve high Zn accumulation in A. halleri.
Fig. 3.
Venn diagram of overlapping significant differentially regulated genes in the Arabidopsis kamchatica Japan (MUR) genotype (A) for H-origin and L-origin homeologs in both leaf and root tissues. (B) Overlap between the number of differentially regulated genes for H-origin and L-origin homeologs with Arabidopsis halleri and Arabidopsis lyrata for leaf tissue.
We performed a gene ontology (GO) enrichment test to determine whether differentially regulated genes are enriched for molecular functions (MFs) that have ontologies related to metal homeostasis in leaf tissues. For GO categories with >30 query genes, A. halleri showed enrichment for transporter activity (GO:0005215), transmembrane transporter activity (GO:0022857), and ion transmembrane transporter activity (GO:0015075) (supplementary table S2, Supplementary Material online). A. lyrata showed no enrichment for any transporter activity or metal ion-related GO. For A. kamchatica homeologs, we found enrichment for the same categories as A. halleri, and in addition, several additional categories showed enrichment including substrate-specific transporter activity (GO:0022892), ion binding (GO:0043167), cation binding (GO:0043169), and metal ion binding (GO:0046872). The complete GO results using TAIR gene IDs can be found in supplementary file 3, Supplementary Material online.
We found differential regulation of many genes related to hyperaccumulation and tolerance as reported in previous studies (Becher et al. 2004; Filatov et al. 2006; Talke et al. 2006; supplementary table S2, Supplementary Material online). These studies showed the effect of increased Zn stress on constitutively expressed genes related to hyperaccumulation and tolerance in A. halleri often results in the downregulation of known ion transporters because the plant’s unusually high demand for Zn is attained (Talke et al. 2006; Hanikenne et al. 2008). For the main Zn transporter gene HMA4, both A. halleri and A. kamchatica H-origin homeologs show high expression before Zn treatment, then following Zn stress, the gene was downregulated in both shoots and roots, characteristic of a released deficiency response (supplementary table S2, Supplementary Material online). Downregulation was also detected in A. halleri and H-origin homeologs of PDF1.1, IRT3, NRAMP3, ZIP3, and ZIP9. For MTP1, both the A. halleri and the A. kamchatica H-origin homeologs of MTP1 show an approximately 1.5-fold increase in expression following Zn treatment. This suggests that after the initiation of Zn stress and transport to leaves, upregulation of MTP1 in leaves is required for detoxification of excess Zn in cells following transport from roots and shoots.
Ratio Change Test of Differential Regulation Among Homeologs
As an interaction of the response to Zn treatment and the ratio of homeolog expression, we tested for significant changes in homeolog ratios following Zn treatment using a statistical test controlling for overdispersion (Akama et al. 2014). We found that approximately 1% (0.3–1.5%) of the homeolog pairs showed significant ratio changes in both leaves and roots of A. kamchatica (table 2), which is similar to the results reported using a synthesized A. kamchatica hybrid and cold treatments (Akama et al. 2014) where 1.11% of homeologs showed ratio changes following cold stress. The relatively small proportion of homeologous pairs showing significant ratio changes can be explained by highly stable genome-wide H-origin to L-origin ratios across treatment conditions (supplementary fig. S4, Supplementary Material online), and also by the high correlation of expression ratios before and after the Zn treatment (r2 = 0.89–0.96; table 2).
Table 2.
Number of Genes Showing Significant Ratio Changes in Leaf and Root for Each Genotype.
| A. kamchatica | Significant genes | % total | H ratio (ctl) | H ratio (zinc) | H ratio r2 |
|---|---|---|---|---|---|
| MUR-leaf | 68 | 0.3 | 53.8% | 53.7% | 0.94 |
| MUR-root | 309 | 1.5 | 53.6% | 53.6% | 0.93 |
| PAK-leaf | 174 | 0.9 | 52.3% | 52.3% | 0.89 |
| PAK-root | 293 | 1.5 | 52.0% | 52.3% | 0.90 |
Note.—Percent total is the number of genes showing significant ratio changes out of our total gene dataset (N = 19820 genes). H ratio ctl (before treatment) and H ratio zinc (after 48 h zinc treatment) indicates the median Arabidopsis halleri-origin expression over the Arabidopsis lyrata-origin copies for all genes in the dataset. Correlation coefficients (r2) indicate the correlation of genome wide expression ratios before and after zinc treatment. Significance for the ratio change statistic based on FDR-adjusted P < 0.05
Although many genes responsible for tolerance and hyperaccumulation are expressed constitutively, we found significant ratio changes in several genes, suggesting that both constitutive and induced expression changes are important. Among the genes showing significant ratio changes (table 2) in leaves, only 8 were common between the Japanese and Alaskan genotypes, while 51 were common between genotypes in roots. Among the genes showing ratio changes in leaves, two had annotated roles in putative metal homeostasis, ZIP3 (AT2G32270.1) and a Yippee family putative Zn-binding protein (AT4G27740.1). Among the significant genes identified in roots, several were found with previously implicated roles in metal tolerance and hyperaccumulation such as MTP1 (AT2G46800.1), MTP3 (AT3G58810.1), NAS4 (AT1G56430.1), HIPP25 (AT4G35060.1), SAM2 (AT4G01850.1), ZIP9 (AT4G33020.1), and ZIP10 (AT1G31260.1), and an unnamed gene annotated as a “heavy metal transport protein” (AT5G26690.1).
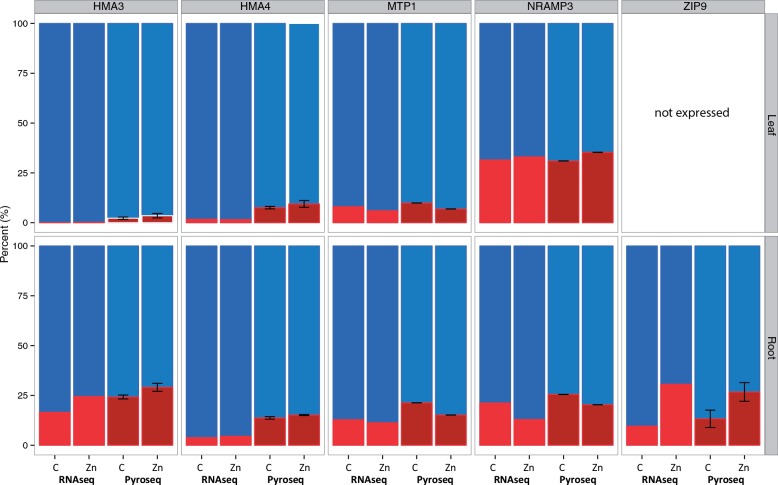
Pyrosequencing Validation of Homeolog Expression and HMA4 Alignments
We validated the homeolog expression ratios using pyrosequencing assays for known genes in both leaf and root tissues using cDNA from before and after the Zn treatment. Because pyrosequencing uses diverged SNPs in the parental genomes (and homeologs) to design sequencing assays, this method can be considered an allele-specific test of expression ratios. Pyrosequencing confirmed that our homeolog read assignment and expression ratio quantifications were accurate for these genes and we estimated a strong correlation (r2 = 0.96) between homeolog ratios using both RNA-seq and pyrosequencing (fig. 4). Pyrosequencing validated the strong expression bias in the H-origin homeologs HMA3, HMA4, MTP1, and NRAMP3 and a significant ratio change in expression between homeologs from control to Zn treatment for ZIP9 in root tissues. For an example of pyrosequencing assay positions, see supplementary fig. S5, Supplementary Material online for an alignment that includes both parental HMA4 alleles, and four halleri- and lyrata-origin homeologs with target positions of diverged SNPs between homeologs that were sequenced by PyroMark. Similar to RNA-seq reads mapping to single-copy homeologs, putatively duplicated genes in the HMA4 and MTP1 A. halleri-derived homeolog would result in amplified PCR products being the sum of duplicated H-origin (i.e., the sum of 3 copies of HMA4) gene expression.
Fig. 4.
Ratio of H-origin (blue) and L-origin (red) relative expression by RNA-seq and PyroMark pyrosequencing of the MUR genotype. Correlation between RNAseq and pyrosequencing is r2 = 0.94. Error bars are SDs between three replicates in PyroMark assays.
HMA4 Copy Number by Pyrosequencing and 3′ Amplification
Recently duplicated genes with nearly identical sequences are difficult to assemble separately using short-read NGS libraries. Our reference genome assembly (v. 1.0) of A. halleri (Tada mine) contains only a single copy of HMA4, despite evidence that the Langelsheim A. halleri accession contains three tandemly duplicated HMA4 copies (Hanikenne et al. 2008). The HMA4 diversity analysis by Hanikenne et al. (2013) indicates that duplicated copies of HMA4 are widespread in A. halleri (including subsp. gemmifera) based on successful sequencing of PCR fragments of each copy among 18 genotypes and qPCR of genomic DNA amplification of HMA4. The multiple copies contribute to high HMA4 expression and widespread hyperaccumulation ability in the species. To estimate the copy number of A. halleri-derived HMA4 in A. kamchatica, we used pyrosequencing assays with the same experimental setup as for cDNA, but using genomic DNA as templates. If A. kamchatica inherited three copies of HMA4 from A. halleri and one copy from A. lyrata, the assay should show a 3:1 ratio. Pyrosequencing results showed 67–76% halleri-origin SNPs and 25–31% lyrata-origin SNPs in five A. kamchatica genotypes (supplementary table S3 and fig. S6, Supplementary Material online), which indicates a 3:1 ratio for HMA4. Because the five genotypes come from distant locations, including two Japanese accessions (MUR lowland, SRM, highland), one subspecies A. kamchatica subsp. kawasakiana (HMK), one Taiwanese sample (TWN), and one Alaskan accession (PAK), it appears the triplicated halleri-origin HMA4 is widespread in A. kamchatica. Finally, because our A. halleri HMA4 reference contig was truncated at amino acid position 983, we performed 3′ RACE to complete the C-terminus, which appears similar to known A. halleri (translated from GenBank DQ221101.1) amino acid sequences (supplementary fig. S7, Supplementary Material online).
Genetic Diversity of the HMA4 Locus
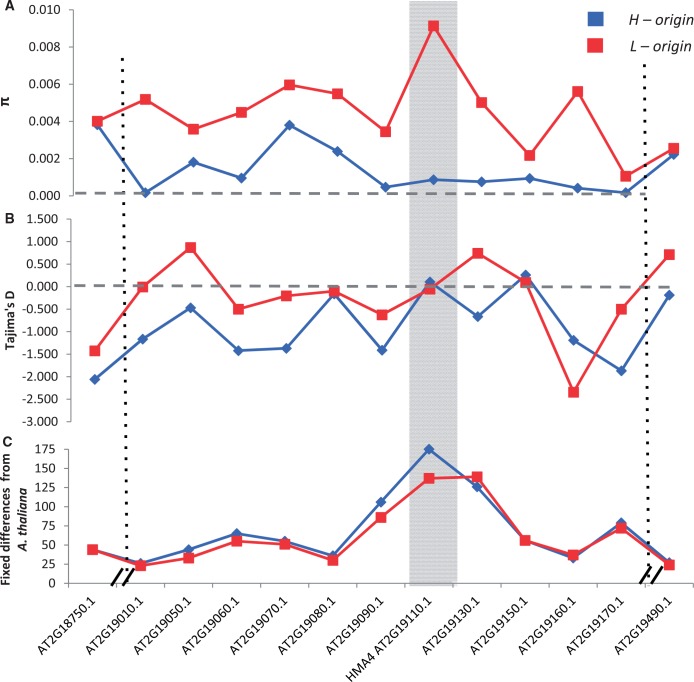
The recently estimated signature of a hard selective sweep in the HMA4 region of A. halleri Hanikenne et al. (2013) led us to examine the patterns of genetic diversity of the HMA4 regions of both A. kamchatica homeologous regions. Using resequencing data from 20 A. kamchatica genotypes, we created sequence alignments for 11 genes surrounding and including the HMA4 locus plus two reference genes that are located distally on the each side of the genomic region to determine whether there are significant differences in H-origin and L-origin “background” genetic diversity (S1 and S13 in Hanikenne et al. 2013). The 11 homeologs were aligned to the Langelsheim A. halleri HMA4 BAC scaffold (EU382072.1 and EU382073.1; supplementary fig. S8A, Supplementary Material online) and also to the syntenic A. lyrata region of LG3 (Hu et al. 2011; supplementary fig. S8B, Supplementary Material online). We first compared the average pairwise diversity (π) among the 11 coding sequences and found a significant reduction in diversity in all of the H-origin homeologs compared with L-origin diversity (fig. 5A), while the two background genes on either side show highly similar levels of diversity between H-origin and L-origin homeologs (S1: AT2G18750: πhalleri = 0.0038, πlyrata = 0.004; S13: AT2G19490: πhalleri = 0.0022, πlyrata = 0.0026; fig. 5A, supplementary file 4, Supplementary Material online). The mean π of the H-origin homeologs of the 11 genes (mean πhalleri = 0.0012) was about four times lower than that of L-origin homeologs (mean πlyrata = 0.0046, P < 0.001; supplementary file 4, Supplementary Material online). Although pyrosequencing showed three H-origin HMA4 copies in A. kamchatica, a single copy H-origin HMA4 from our reference genome assembly was used to estimate diversity for the HMA4 coding sequence. This copy can be aligned to any of the three HMA4 copies in the Langelsheim BAC assembly due to 99% homology among the three copies. We also assume that all three copies in A. kamchatica should have high homology, similar to A. halleri (Hanikenne et al. 2008). This assumption is also supported by the phylogeny, in which the HMA4 homeolog sequences clustered with diploid parental alleles and showed little divergence from the parental sequences (see supplementary fig. S9, Supplementary Material online for a phylogeny of diploid alleles and A. kamchatica homeologs, including the three A. halleri subsp. halleri Langelsheim HMA4 copies). This procedure could combine polymorphisms at the three duplicated copies and could result in a bias for higher diversity in halleri-origin HMA4, but the estimated diversity at the H-origin HMA4 was very low and supported the low diversity in the surrounding H-origin homeologs. To be conservative, we calculated the difference using the surrounding ten genes excluding HMA4, and the difference was still highly significant for both nonsynonymous π, and synonymous π (supplementary fig. S10A and B, Supplementary Material online). Similarly, we found significant differences between both homeologs in the total number of segregating sites (supplementary fig. S10D, Supplementary Material online), although the difference in Tajima’s D was not significant (fig. 5B, supplementary fig. S10C, Supplementary Material online).
Fig. 5.
Average pairwise nucleotide diversity at 13 genes (A) around the HMA4 locus (gray shaded area; BAC accessions EU382072.1 and EU382073.1 comprising approximately 290 kb) of H-origin (blue) and L-origin (red) homeologs. Dotted vertical lines marking genes AT2G18750.1 and AT2G19490.1 that are outside of the BAC sequence and considered unlinked to the HMA4 locus show nearly identical diversity. Diversity at the 11 genes that align to the BAC: mean πhalleri = 0.0012, mean πlyrata = 0.0046, P < 0.001. Dashed horizontal gray line for (A) and (B) mark zero for diversity and Tajima’s D, respectively. Tajima’s D estimates (B) for each of the genes show that L-origin copies have on average higher D, but are overall not significantly different (dashed gray lines, D = 0 representing neutrality). Fixed differences (C) (both nonsynonymous and synonymous combined) between H-origin and L-origin homeologs and Arabidopsis thaliana show that both homeologs have the greatest number of fixed differences at the HMA4 gene itself. The x-axis represents the physical orientation of each gene alone, not the actual physical distance. The HMA4 coding sequence is represented by a single contig in each of our Arabidopsis halleri subsp. gemmifera (C1829917.g40478) and Arabidopsis lyrata subsp. petraea (scaffold16994.g19792) reference genomes and does not contain duplicated CDS regions.
Divergence from a related species can be used to test neutrality because polymorphism within focal species can increase or decrease expected ratios of divergence to polymorphism (Hudson et al. 1987). We included A. thaliana orthologs in each H-origin and L-origin alignment as a related outgroup species, and used a HKA test to compare a pair of homeologs (Small et al. 1999) using the sum of all segregating sites for the 11 genes in each of the HMA4 region homeologs. Under neutrality, the number of diverged substitutions between a homeolog relative to an outgroup species (e.g., H-origin homeologs relative to A. thaliana orthologs, or L-origin homeologs relative to A. thaliana orthologs) is proportional to the time since the split of A. thaliana and the mutation rate. For within-species polymorphism, the number of segregating sites is proportional to the effective population size and the mutation rate. Therefore, it would be expected that the ratio of divergence and polymorphism (e.g., Sdiv/Spoly) in neutral mutations should be roughly equivalent between different genes (here between a pair of homeologs), and significant differences in Sdiv/Spoly between species or homeologous genes suggests nonneutral evolution (Hudson et al. 1987). Divergence estimates from A. thaliana (the number of fixed differences with A. kamchatica homeologs) were highly similar for each gene, with the HMA4 gene itself showing similarly elevated divergence for both H-origin and L-origin homeologs (fig. 5C, supplementary fig. S10E, Supplementary Material online). This similar divergence from A. thaliana resulted in significantly different Sdiv/Spoly ratios between H-origin and L-origin homeologs (P = 0.002, supplementary fig. S10F, Supplementary Material online), indicating that there are significantly fewer polymorphic sites in the H-origin genes despite both homeologs having similar divergence from a common outgroup (fig. 5C). This was also significant both when segregating sites in each homeolog were summed and compared using the HKA test (supplementary fig. S4, Supplementary Material online). We also found that the low diversity in the HMA4 region of H-origin, but not that of L-origin in A. kamchatica, appears generally similar to that reported for A. halleri.
Quantification of Metal Ions in Soil
We measured soil concentrations of metal ions for several of our sample locations of A. halleri and A. kamchatica. The A. halleri subsp. gemmifera accession used in this experiment comes from a mine site in Japan known to be contaminated with metals (Tada mine site) with soil estimates for Zn of 724.7–2,461.5 mg kg − 1 and Cd of 4.7–12.8 mg kg − 1 (supplementary table S5, Supplementary Material online). While there is no international consensus on the threshold value for metal contamination, Bert et al. (2002) proposed that soil with more than 300 mg kg−1 Zn, 100 mg kg−1 Pb, or 2 mg kg−1 Cd be classified as metalliferous (or metal-contaminated) soil according to the French agricultural norm. Soil concentrations from two of the three A. kamchatica sample locations show levels consistent with previously described nonmetalliferous sites (Meyer et al. 2010; Hanikenne et al. 2013) for A. halleri (Zn < 160 mg kg−1 and Cd < 0.42 mg kg−1). By the above standard, the soil at Mt. Shirouma is metalliferous (supplementary table S5, Supplementary Material online; Zn > 300 mg kg−1), with high Zn (mean 800 mg kg−1, up to 1,164.3 mg kg−1), along with high Mg (average 74,000 mg kg−1) and Ni (mean 600 mg kg−1), which are consistent with reported serpentine soil conditions with a mixture of other soil types at this site (Schlüchter and Heuberger 1981; Hatano and Matsuzawa 2008).
Discussion
Zinc Accumulation in A. kamchatica is High in Leaves and Low in Roots
We have demonstrated that the allopolyploid species A. kamchatica possesses high Zn accumulation ability in leaves, similar to the order expected of known hyperaccumulators (Bert et al. 2002; Talke et al. 2006). Estimates of Zn accumulation among the three species showed that A. kamchatica accumulates about half of the amount of Zn in its leaves compared with A. halleri and about ten times the concentration accumulated by the A. lyrata parent. This result is similar to that shown by Willems et al. (2007) using a F1 cross of diploid A. halleri and A. lyrata, which showed Zn tolerance was still very high, but was reduced from A. halleri, as expected for a stable hybrid. Despite the lower Zn accumulation in the leaves of A. kamchatica compared with A. halleri, there were no significant differences in Zn accumulation in root tissues between the two species. Both A. halleri and A. kamchatica also showed significantly lower Zn accumulation in roots compared with A. lyrata, which indicates that A. kamchatica still transports substantial amounts of Zn from roots to shoots. Moreover, while the leaf-to-shoot ratios for A. kamchatica were slightly less than half the shoot-to-root ratio of A. halleri, this ratio was an order of magnitude higher than in A. lyrata. Despite lower levels of Zn accumulation in leaves of A. kamchatica compared with A. halleri when treated with 500 μM Zn, we found that exposing A. kamchatica to increasing Zn concentrations resulted in proportional increases in Zn accumulation, which demonstrates that the allopolyploid is highly responsive to Zn treatments over short time periods. These experiments indicated up to 10,000 μg g−1 leaf accumulation can be achieved in A. kamchatica when treated with 1,000 μM of Zn over 1 or 2 week time periods (supplementary figs. S1 and S2, Supplementary Material online), which is consistent with levels found for both European and Asian A. halleri collected in heavy metal-contaminated sites (Bert et al. 2000; Bert et al. 2002; A. halleri subsp. gemmifera called Arabis gemmifera by Kubota and Takenaka 2003). While we did not test tolerance directly, these high levels of hyperaccumulation of levels also suggest increased tolerance to heavy metals because these traits are tightly linked.
Genes Regulating Metal Homeostasis by Hyperaccumulation and Detoxification Show Significantly Higher Expression in A. halleri-Derived Copies
As expected, if high Zn accumulation is maintained in A. kamchatica, constitutively expressed genes for hyperaccumulation and detoxification should show a strong expression bias in the H-origin over the L-origin homeolog. Significantly higher expression in Arabidopsis diploid and polyploid hybrids suggests cis-regulatory divergence (He et al. 2012; Shi et al. 2012) and our data indicates that conserved cis-regulatory differences between A. halleri and A. lyrata HM orthologs were retained among homeologs (supplementary file 2, Supplementary Material online). Although the expression patterns of these genes alone do not unequivocally implicate their function, it is known that high expression of genes such as the Zn transporter, HMA4, and detoxification of tissues by MTP1 in A. halleri, is essential for hyperaccumulation (Willems et al. 2007; Hanikenne et al. 2008; Shahzad et al. 2010). The patterns of differences in homeolog expression and regulatory changes for genes such as HMA3, HMA4, MTP1, PDF1.1, NRAMP3, ZIP3, and ZIP9 (and others listed in supplementary file 2, Supplementary Material online) are consistent with studies of differential expression comparing A. halleri with A. lyrata or A. thaliana (Chiang et al. 2006; Filatov et al. 2006; Talke et al. 2006), which provides a connection to their conserved role in A. kamchatica.
For the four HM genes showing the largest H-origin-derived expression bias over the L-origin homeolog, several studies have also demonstrated their roles in hyperaccumulation and tolerance. In A. thaliana, HMA3 is responsible for vacuolar sequestration of both Cd and Zn based on cellular localization using fluorescent probes (Morel et al. 2008). Differential expression analyses between A. halleri and A. thaliana show significantly greater AhHMA3 expression (Filatov et al. 2006, Talke et al. 2006) and AhHMA3 confers greater Zn detoxification in complementation tests using Saccharomyces cerevisiae (Becher et al. 2004) supporting its putative function in vacuolar sequestration of Zn. In A. halleri, the ATPase transporter HMA4 has a direct role for both Cd and Zn root to shoot transport through xylem tissue based on gene silencing in planta (RNAi, Hanikenne et al. 2008). Studies of differential expression of HMA4 between A. halleri and nontolerant A. lyrata and A. thaliana show patterns of ortholog-specific bias similar to our homeolog-biased expression results in A. kamchatica (Chiang et al. 2006, Filatov et al. 2006). Our study also shows that A. kamchatica has similarly greater expression in the H-origin homeolog of HMA4 in roots compared with leaves, consistent with the results of Talke et al. (2006). Also, like AhHMA4, the H-origin copy of HMA4 in A. kamchatica exhibits significant downregulation after 500 μM Zn treatment. This indicates that under low-Zn or Zn-deficient conditions (before Zn stress), A. halleri and A. kamchatica express HMA4 highly to maximize Zn transport, a mechanism that would also provide benefit to the plants under the elemental defense hypothesis, even if a small amount of Zn is present in the environment (Coleman et al. 2005).
Along with HMA4, AhMTP1 shows the strongest QTL for the Zn-tolerance phenotype in the A. halleri × A. lyrata BC1 family (Willems et al. 2007), which indicates that these are large-effect loci, and the duplicated MTP1 copies (3–5 copies) found in A. halleri strongly enhance tolerance (Shahzad et al. 2010). Like HMA4, MTP1 shows significantly higher gene expression in A. halleri over A. thaliana or A. lyrata (Weber et al. 2004, Talke et al. 2006). MTP1 has been demonstrated to be involved in cytoplasmic detoxification of Zn (Dräger et al. 2004; Desbrosses-Fonrouge et al. 2005), which would be consistent with its extremely high expression bias in leaves in our study. This would also explain the upregulation of the H-origin MTP1 homeolog following the 500 μM Zn treatment where increased cellular Zn induces a detoxification response.
Plant defensins, such as PDF1s in A. halleri, have recently been demonstrated to play a role in both fungal resistance and heavy metal tolerance in plants (Mirouze et al. 2006; Shahzad et al. 2013). The gene PDF1.1 (AT1G75830) showed >100-fold expression in A. halleri over A. lyrata orthologs before Zn treatment in our study, which is consistent with previous studies comparing A. halleri with nontolerant species of Arabidopsis. Shahzad et al. (2013) showed that AhPDF1.1 could confer greater Zn tolerance in yeast complementation tests, while Mirouze et al. (2006) also showed that AhPDF1.1 resulted in greater Zn tolerance when transformed in A. thaliana. The strong bias in the A. kamchatica H-origin homeolog of PDF1.1 and significant downregulation following Zn treatment suggests that plant defensins may also play an important role in Zn accumulation in the allopolyploid.
Homeolog and Diploid Transcriptional Patterns are Consistent with Leaf and Root Accumulation Phenotypes of Each Species
Comparisons of tissue-specific transcriptional patterns between the two diploids and the allopolyploid species show remarkably different levels of gene regulation in leaves and roots that reflect the levels of Zn accumulation in these tissues. A. halleri shows by far the greatest number of regulatory changes in the leaves and the highest Zn accumulation compared with leaves of both A. kamchatica and A. lyrata. The patterns for both A. kamchatica and A. lyrata indicate that direct exposure to Zn stress induces significant regulatory changes in roots, while the highly efficient metal transport from roots to leaves in A. halleri (shoot-to-root ratio > 1) suggests a more complex transcriptional response for heavy metal detoxification. Efficient heavy metal transport to leaves driven by transcriptional responses inherently protects roots from metal toxicity, which may be indicated by the far fewer regulatory changes in A. halleri roots than in A. lyrata and A. kamchatica.
Another interesting pattern of the regulatory changes in A. kamchatica is the similar numbers of H-origin and L-origin homeologs showing responses to Zn treatment in both tissues (table 1). This pattern could be explained by coregulation of homeologs by trans-acting factors, which have known influences on gene expression in other Arabidopsis allopolyploids (Shi et al. 2012; Chen 2013). However, only about half of these homeologs are shared in each tissue (fig. 3A), which would not support trans-regulation of equal numbers of homeologs. It is more likely that the high correlation in homeolog gene expression in both control and Zn-treated samples (r2 = 0.79–0.84) results in correlated expression changes among homeologs (table 2).
We applied a new statistical method that considers overdispersion to exclude pseudopositives (Akama et al. 2014) and found the majority of homeolog pairs in the genome show stable expression ratios across stress treatments (supplementary fig. S4, Supplementary Material online), with about 1% of homeologs showing significant ratio changes (table 2). For most HM genes, we found greater constitutive expression of H-origin homeologs, which indicates strong cis-regulation. However, the ratio changes were not always predictable toward either homeolog driving them following Zn treatment. An interesting example is the gene MTP3, where the strong upregulation of the L-origin copy results in a significant ratio change following Zn treatment, which could potentially influence the Zn accumulation phenotype. Because overexpression of this gene is thought to result in vacuolar sequestration in roots, preventing toxic metal transport to leaves in nontolerant A. thaliana (Arrivault et al. 2006), the strong L-origin expression response in roots could indicate a candidate gene responsible for longer root retention and lower Zn transport to leaves. Finally, while metal hyperaccumulation clearly has a genetic basis, it is unclear how other factors, such as larger cell size or structural differences common in tetraploid relative to diploid cytology, may also contribute at least partly to increased quantities of metal ion accumulation in tissues (Levin 2002; Comai 2005).
Triplicated HMA4 from A. halleri is Common in the Allopolyploid A. kamchatica
The tandem triplication of the HMA4 gene in A. halleri was shown to have contributed to its ability to hyperaccumulate by constitutive expression together with the enhanced promoter activity of each copy. Hyperaccumulation is a species-wide trait that explains the signature of a hard selective sweep in A. halleri (Hanikenne et al. 2013). Based on pyrosequencing of HMA4 using genomic DNA, we found a 3:1 ratio of H-origin to L-origin SNPs, which indicated that three H-origin copies of HMA4 were present in all tested A. kamchatica individuals that were estimated to represent three independent polyploid origins (Shimizu-Inatsugi et al. 2009). RNA-seq data and pyrosequencing showed that the HMA4 homeologs derived from A. halleri were preferentially expressed (fig. 4). Therefore, it is very unlikely that the triplication occurred independently in A. halleri and in multiple origins of A. kamchatica, but rather this strongly suggests that the HMA4 triplication of the A. halleri founder individuals was inherited during the hybridization with A. lyrata and genome duplication, and its high expression contributed to hyperaccumulation in A. kamchatica.
Until recently, the search for genetic loci having differential contributions with large phenotypic effects arising from either diploid ancestor has eluded researchers studying polyploidy in the genus Arabidopsis (Chen 2013; Bomblies and Madlung 2014). We found significantly different patterns of homeolog diversity surrounding the HMA4 locus in A. kamchatica, which bears a potential signature of a species-wide selective sweep in the A. halleri-derived HMA4 region. In A. kamchatica, the HMA4 locus and surrounding genes derived from the A. halleri ancestor has approximately three times fewer nonsynonymous and synonymous substitutions, despite showing nearly equal diversity in both homeologs for the two genes unlinked to the A. halleri BAC sequence (AT2G18750 and AT2G19490.1 in fig. 5). Previous estimates of nuclear and chloroplast haplotype diversity and population structure analyses (Shimizu-Inatsugi et al. 2009; Tsuchimatsu et al. 2012) indicated that allopolyploid hybridization occurred at least three times in the ancestors of our collection of A. kamchatica genotypes. The most conservative and likely scenario is that current diversity at the HMA4 locus is the result of very low diversity in the ancestral A. halleri parents in this genomic region compared with the ancestral A. lyrata parents. These data support that the triplication and hyperaccumulation evolved once in A. halleri, and three copies were transmitted to A. kamchatica during the hybridization with A. lyrata.
Ecological Relevance of Hyperaccumulation
In an attempt to assess the ecological relevance of hyperaccumulation, we estimated soil concentrations in A. halleri and A. kamchatica sites for genotypes used in this study. The habitats of European A. halleri encompass both metalliferous (>300 up to 35,000 mg kg−1 of Zn) and nonmetalliferous soils (Bert et al. 2002). The Tada mine site of the Japanese A. halleri subsp. gemmifera accession (Tada mine) had high concentrations of both Zn and Cd characteristic of a highly metalliferous site as the result of mining in the area. We found that the habitats of A. kamchatica encompass both nonmetalliferous and metalliferous soils (supplementary table S5, Supplementary Material online). Native soil conditions in the sites at Murodo, Japan and Potter, Alaska show negligible levels of heavy metals and no A. kamchatica has yet been collected from any known mining areas. The Mt. Shirouma population in Japan showed ion concentrations typical of serpentine soils (high in Ni and Mg) (Hatano and Matsuzawa 2008) and soils with elevated Zn (>800 mg kg−1), but low Cd (supplementary table S5, Supplementary Material online).
Ecological studies and Zn accumulation experiments using many European A. halleri populations show that native soils are often not remarkable for heavy metals, but plants collected from these sites still possess tolerance and general hyperaccumulation ability (Bert et al. 2000; Pauwels et al. 2006). Elemental defense against insect herbivores could explain the constitutive ability of A. halleri to hyperaccumulate metals where a significant reduction in herbivory by specialist and generalist insects occurs when leaves contain high levels of Cd and Zn. The recent study by Kazemi-Dinan et al. (2014) showed that Zn concentrations from above 1,000 μg g−1 in A. halleri leaves were sufficient to reduce herbivory. We have shown that A. kamchatica can accumulate four times this level of Zn. It is yet to be shown whether native soils are sufficient to provide these concentrations, or whether insects would be deterred by similar concentrations in A. kamchatica.
A Model of Gene Expression following Allopolyploidization: Transcriptional Patterns of Both Parents Are Conserved but Attenuated
A long-standing question in the study of polyploids is why some species have a broader environmental tolerance and habitat distribution than their diploid parents (Stebbins 1950; Stebbins 1971; te Beest et al. 2012). Based on our quantitative estimates of Zn hyperaccumulation and transcriptional patterns, we propose a model for an allopolyploid (A. kamchatica) to combine the transcriptional patterns of both parental species by merging their regulatory networks. First, the ancestral diploid species diverged and a relatively small number of genes evolved species-specific expression patterns, such as induced or constitutively high expression levels of hyperaccumulation-related genes in A. halleri. After the gene networks of the two parental species were merged by allopolyploidization, despite the redundancy of most homeologous pairs, parent-specific transcriptional patterns can be maintained in the allopolyploid by cis-regulatory divergence. We found that 99% of the homeologous pairs did not show significant changes in ratios after Zn treatment. Importantly, in many hyperaccumulation-related genes, high expression of halleri-derived homeologs was maintained in the allopolyploid by cis-regulatory divergence, which strongly suggests that the high expression level of these genes contributed to hyperaccumulation in A. kamchatica. A similar pattern was also observed in cold-induced transcriptomes of A. kamchatica, where about 99% of the homeologous pairs did not show significant changes in ratios (Akama et al. 2014), and the genes that showed significant ratio changes were enriched in the GO categories of stress responses. Among them, lyrata-derived homeologs were much more induced than A. halleri-derived homeologs in well-known cold response genes such as RD29B and COR15A that are known to confer cold tolerance in A. thaliana (Yamaguchi-Shinozaki and Shinozaki 1993; Thomashow 1999; Nakayama et al. 2007). Our data indicates that the allopolyploid A. kamchatica can encompass expression patterns of Zn hyperaccumulation characteristic of A. halleri and that of cold response genes characteristic of A. lyrata (fig. 6).
Fig. 6.
A model showing that the transcriptional patterns underlying divergent traits of both diploid parents can be combined in the allopolyploid. Candidate genes for heavy metal hyperaccumulation show strong Arabidopsis halleri-derived patterns of constitutive expression and response to Zn treatments (this study), while cold treatments show Arabidopsis lyrata-derived expression responses detected by Akama et al (2014).
Despite that the high expression of hyperaccumulation-related genes was transmitted from A. halleri to A. kamchatica, our data also indicates that the absolute expression level in the allopolyploid may be attenuated (or diluted) compared with parental species (supplementary fig. S3, Supplementary Material online). Due to the fixed heterozygosity inherent in allopolyploids, the total level of gene expression (sum of both homeologs) with dominant cis-regulation would be the average of the expression levels of the parental species. When the expression in a parent is much higher than that of another, the total expression level will be reduced to nearly half that of the former parent (see the Results section on K/H ratio). Such attenuated expression would result in a quantitatively reduced phenotype. We found that the total level of expression (the sum of homeologs in the allopolyploid) of hyperaccumulation-related genes of A. kamchatica is generally about half of that in the hyperaccumulator A. halleri, and at least two times higher in A. kamchatica than that in the nonhyperaccumulator A. lyrata for these genes. Consistent with the expression data, the degree of hyperaccumulation of the shoot-to-root Zn concentration of A. kamchatica is about half that of A. halleri, but >10 times greater than that of A. lyrata. Depending on the genetic architecture in other phenotypic traits, the reduction may not necessarily be half, but it is expected that the trait cannot maintain the same level as the specialist diploid. It is also possible that the homeologs from another parent have an inhibitory function, that is, the A. lyrata-derived metal ion transporters network may reduce the efficiency of Zn transport in A. kamchatica, perhaps by slower uptake from the environment or by longer retention of Zn in the roots.
The ability to use the environmental responses of both parents can provide a generalist strategy for allopolyploid species to respond to the broader environmental conditions. Although transcriptional data and Zn hyperaccumulation does not directly show tolerance and fitness in natural environments, the broad distribution range of A. kamchatica suggests that it has a generalist property. The study of the climatic niches of Arabidopsis species (Hoffmann 2005) showed that A. kamchatica (called A. lyrata subsp. kamchatica by Hoffmann 2005) has one of the broadest temperature and precipitation gradients in the genus, spanning a large latitudinal range (from Taiwan to Alaska) and highly variable altitudes (from 0 to 3,000 m) (Kenta et al. 2011). The diploid parents, A. lyrata subsp. petraea and A. halleri subsp. gemmifera, are distributed in colder and warmer regions, respectively, which indicates that allopolyploid hybridization resulted in greater environmental plasticity. Natural A. kamchatica is tolerant to cold temperatures (Armstrong et al. 2015). Hyperaccumulation/metal tolerance may provide additional benefits to high variability in soil types across the broad distribution of A. kamchatica (Kenta et al. 2011). This model of transcriptome network merging could provide a molecular basis for the “general purpose genotypes,” which Stebbins (1971) defined as polyploid genotypes that are able to tolerate a wide range of environmental conditions. Because there are few allopolyploid systems that have been used as empirical models to address species distributions, the combination of temperature and soil adaptations derived from both diploid parents suggests that A. kamchatica holds considerable promise in this area.
Materials and Methods
Plant Cultivation
All accessions of A. kamchatica, A. halleri subsp. gemmifera, and A. lyrata subsp. petraea used in this study are cultivars of strains that have been repeatedly self-fertilized or grown clonally in the laboratory. For hydroponic experiments, we used five genotypes of A. kamchatica: MUR (seed stock w1880; Murodo, Japan, near the population of kamD19 described by Shimizu-Inatsugi et al. 2009), ALK (seed stock w1661; Darling Creek, Alaska, from the same population as kamH46), PAK (seed stock w1889; Potter, Alaska, also kamH43), SRM (seed stock w2347; Mt. Shirouma, Japan, from the same population as kamD17), HMK (seed stock w2075; Hamakurosaki, Toyama, Japan, from the same population as kamkwsB10). The last strain belongs to the subspecies kawasakiana, while the others belong to the subspecies kamchatica. Chloroplast, ribosome, and low-copy nuclear data obtained by Shimizu-Inatsugi et al. (2009) suggested at least three independent polyploid origins (each represented by MUR, HMK, and PAK/ALK), and SRM represents another unique genotype. For Experiments 1 and 2 (see details below), seeds were germinated on fine quartz sand in separate wells in 12-well Greiner plates for ca. 2 weeks at room temperature and kept moist with deionized water. Far East A. lyrata subsp. petraea (also named A. petraea subsp. umbrosa) was collected from the banks of the Suharnaya River, alluvium of Kolyma, Yakutia (Sakha Republic), Far East Russia (named lyrpet4 by Shimizu-Inatsugi et al. 2009) and the seeds (w1178 in our stock number) were similarly germinated on fine quartz sand. Our accession of A. halleri subsp. gemmifera is a clone of halgem2 used by Shimizu-Inatsugi et al. (2009) and of w302 used by Tsuchimatsu et al. (2010), which was treated with five rounds of self-fertilization by bud pollination in the lab to reduce heterozygosity, and can be propagated clonally because of inflorescence reversion.
After successful germination, seedlings were then transferred to Oil-Dri US-special substrate (Damolin, www.damolin.fr) and placed in a growth chamber under a 16 h light and 8 h dark regime at 20°C for 1.5 weeks to allow establishment of primary roots. The Oil-Dri gravel contains no soil or additional nutrients and allows for easy removal from roots. The seedlings were then transferred to 5-L hydroponic containers, with 12 plants randomized per pot. Hydroponic pots were covered and aerated to limit algal growth and provide oxygen to roots. The third experiment (Experiment 3) used a more compact hydroponic chamber design using 0.5 mL thermo PCR tubes containing phytoagar for seed germination and 1,000 μL (700 mL volume) pipette tip boxes used as the hydroponic containers. The 0.5 mL tubes were then cut at the bottom to allow roots to penetrate the phytoagar into the hydroponic medium.
The hydroponic nutrient recipe is as follows: for 5 L final volume we added each nutrient at the following concentrations: 4 mM KNO3, 1.2 mM Ca(NO3)2, 0.8 mM MgSO4, 0.8 mM KH2PO4, 0.8 mM NH4Cl, and 5 μM Fe(III)EDTA. A separate 1 L stock of oligoelements was made with the following elements and 16.25 mL of oligonutrients was added to the final 5 L solution: 0.2 mM KCL, 0.12 mM H3BO3, 0.04 mM MnSO4, 4 μM CuSO4, 3 μM ZnSO4, and 1 μM (NH4)6Mo7O2. Ten-liter batches were mixed in one container and then dispensed to individual hydroponic containers. The final pH was adjusted to 5.6–5.8 and the solution was changed weekly. Zn supplements were added after about 4.5 weeks of plant growth (with slight variability among genotypes because of seed germination).
Zn Treatment Experiments
Because our goal was to induce a strong Zn stress response at both the phenotypic and transcriptomic levels, our Zn treatment concentrations were selected based on pilot experiments and other studies of A. halleri and A. lyrata (Filatov et al. 2006; Willems et al. 2007) that used 100 μM, 500 μM, and 1,000 μM Zn treatments. The study of Willems et al. (2007) tested tolerance in A. halleri × A. lyrata F1s and BC1 up to 3,000 μM Zn and found high tolerance in BC1 plants at 250–1,000 μM Zn. Other studies have shown hyperaccumulation in A. halleri under Zn stress up to 2,000 μM Zn in hydroponic conditions (Kashem et al. 2010) and exceptionally high leaf accumulation even in plants collected from sites contaminated with low levels of heavy metals.
Three independent experiments were conducted, with ionomics analyses performed in two separate labs, thereby ensuring repeatability. The first experiment (Experiment 1) compared Zn accumulation among four A. kamchatica genotypes (MUR, ALK, PAK, and SRM), A. halleri subsp. gemmifera, and A. lyrata subsp. petraea using a Zn supplement of 500 μM added after about 4.5 weeks of plant growth. Leaf and root tissue samples for metal analysis were harvested after 7 days exposure to 500 μM Zn. Each genotype was represented by 8–11 replicates for leaf tissue and 6–13 replicates for roots. Tissues from Experiment 1 were also harvested before Zn was added (T = 0) and after 48 h (T = 48) for RNA-seq analyses.
The second experiment (Experiment 2) used three A. kamchatica genotypes (MUR, PAK, and HMK), and Zn was added at 250, 750, and 1,000 μM to the hydroponic medium in 5 L hydroponic containers after about 4.5 weeks of plant growth using three replicates of each genotype. Tissues were then harvested 2 weeks after Zn treatment. A third experiment (Experiment 3) tested three A. kamchatica genotypes (MUR, SRM, and PAK) at both 500 and 1,000 μM Zn accumulation after 7 days of added Zn with phytoagar germination and 700 mL hydroponic containers. This experiment was represented by 10–12 replicates of each plant genotype for the 500 μM Zn treatment and 6–8 replicates for the 1,000 μM Zn treatment.
Elemental Analysis in Plant Tissues
Measurements of elemental concentrations in leaves were performed as described by Lahner et al. (2003) in the Salt Lab, University of Aberdeen (Experiments 1 and 2), and the Schulin Lab in the Institute of Terrestrial Ecosystems at ETH Zurich (Experiments 1 and 3). Leaves and roots (approximately 2–5 mg DW) were harvested from plants grown for 5.5 or 6.5 weeks following Zn treatment as described above. Leaf tissue was dried at room temperature and then at 50 °C for 24 h. Root tissue was first rinsed in deionized water, and then washed in 5 mm CaCl2 followed by 1 mM MES-KOH to remove residual ions and rinsed again in double-distilled H2O. Tissues were rinsed again with 18 MΩ water and placed into Pyrex digestion tubes. Samples were placed into an oven at 92 °C to dry for 20 h. After cooling, seven reference samples from each planted block were weighed. Samples were digested in a microwave oven with 2 mL concentrated nitric acid (HNO3, ACS reagent; Sigma-Aldrich) and 30% hydrogen peroxide (Normapur; VWR Prolabo) and diluted to 10 mL with 18 MΩ water. Analytical blanks and standard reference material (WEPAL IPE 980) were digested together with plant samples in the same manner. After samples and controls were prepared, elemental analysis was performed using inductively coupled mass spectrometry (ICP-MS) for Zn. An internal standard with yttrium and indium were added to the samples to correct for instrumental drift. All samples were normalized to calculated weights, as determined with a heuristic algorithm using the best-measured elements, the weights of the samples, and the elemental solution concentrations. We used ANOVA to detect differences in Zn accumulation between species and genotypes (linear model: μg per gram DW approximately equal to the genotype + tissue + species + species × tissue). All statistical analyses were conducted using R.
Elemental Analysis of Soils
Soil samples from the Tada mine site where our A. halleri reference accession and from three A. kamchatica populations were collected (supplementary table S5, Supplementary Material online). Soil samples were weighed and ground, and then dried at 40 °C. Soils were digested using the DigiPREP MS digestion system (SCP Science, Canada) and 2 M HNO3 for 90 min at 120 °C. The samples were then cooled and diluted with up to 50 mL nanopure water. The digested soils were then filtered using 41 Whatman filter paper into 50 mL centrifuge tubes. Samples were then diluted for measurement using ICP-OES at ETH Zurich.
RNA Extraction and cDNA Synthesis
For transcriptomics experiments, leaf and root tissues were harvested from three replicates at time zero (control), and then at 48 h following the replacement of the hydroponic solution containing an additional 500 μM of Zn from Experiment 1 described above. Leaf and root tissues collected before and after Zn treatment were flash-frozen in liquid nitrogen and stored at –80 °C. RNA was extracted with TRIzol (Invitrogen) and further purified with the RNeasy Mini Kit (Qiagen). RNA concentrations were measured using a Qubit (Invitrogen). The RNA sample (1 μg each) library synthesis was done using TruSeq RNA Sample Prep Kit, version 2 (Illumina), polyA-unstranded, at the Functional Genomics Center Zurich (http://www.fgcz.ch). Samples were sequenced using Illumina HiSeq 2000 to generate 2× paired-end 100 bp reads.
Reference Genome Assembly
The diploid parental species A. halleri and A. lyrata each possess eight chromosomes (n = 8, 2n = 2x = 16) and the allopolyploid hybrid has 2n = 4x = 32 chromosomes. Because A. kamchatica is a self-fertilizing species (Shimizu & Tsuchimatsu 2015), the A. halleri- and A. lyrata-derived chromosomes are generally homozygous. This effectively allows us to treat the eight A. halleri- and eight A. lyrata-derived chromosomes as haploid genomes. We created homeologous A. kamchatica (A. halleri-derived = H-origin) and (A. lyrata-derived = L-origin) diploid-guided reference genomes for a Japanese (MUR) and an Alaskan (PAK) accession using de novo assemblies of A. halleri subsp. gemmifera and A. lyrata subsp. petraea from Akama et al. (2014). Genomic DNA from both accessions of A. kamchatica (accessions MUR: Murodo, Japan and PAK: Potter, Alaska) was extracted from leaf tissue using the DNeasy Plant Kit (Qiagen) and the total DNA was sequenced using Illumina HiSeq 2000 with insert size of 200 bp. DNA reads from A. kamchatica were mapped using BWA-MEM version 0.7.5a on the two diploid genomes independently. We classified the reads to each parental (H-origin or L-origin) using HomeoRoq (http://seselab.org/homeoroq, last accessed July 14, 2016). In this method, reads are first mapped to both parental genomes, and then classified as H-origin, L-origin, common, or unclassified (see fig. 1 in Akama et al. 2014 for schematic diagram). If a read was mapped to only one of the parental genomes, it was discarded. After mapping to the A. halleri genome, we detected A. kamchatica halleri-origin (H-origin) reads and identified single-nucleotide polymorphisms (SNPs) and short insertions and deletions using Samtools 0.1.18. Then, the nucleotides were replaced on the detected variant position with the alternative nucleotides if the positions were covered by at least five reads. At least 80% of the reads were required to have the alternative nucleotide from the reference to be called a SNP. The A. kamchatica lyrata-origin (L-origin) genome was updated in a similar manner. These processes, mapping, read classification, and reference modification, were repeated ten times. We used only origin reads the first five times and both origin and common reads the last five times for the reference modification. Then, using the updated genomes, we performed the SNP replacement iteration three more times. For the final reference of each A. kamchatica accession, we assumed that both imputed references derived from parental species were homeologous genomes. The de novo diploid genome assemblies and the polyploid SNP replacement strategy decreases mapping bias that would come from mapping only to a single common genome, which improves our estimates of homeolog expression levels for RNA-seq experiments.
RNA-Seq Read Classification and Homeolog Assignment
We extracted total RNA using RNeasy (Qiagen) from three replicates of both leaf and root tissues from two A. kamchatica (MUR: Murodo, Japan and PAK: Potter, Alaska), A. halleri subsp. gemmifera (Tada mine), and A. lyrata subsp. petraea (Siberia) sampled at time zero (no Zn treatment) and 48 h after 500 μM Zn treatment. In total, 48 cDNA libraries were constructed at the Functional Genomics Center, University of Zurich, and then sequenced using Illumina HiSeq 2000 generating 2× paired-end 100 bp reads. Low-quality regions of the sequenced reads were trimmed using Trimmomatic-0.30 (Bolger et al. 2014) before mapping. We used STAR-2.3.0 (Dobin et al. 2013) to map RNA-seq reads in A. kamchatica with default parameter settings to map reads and the HomeoRoq pipeline to classify reads to H-origin or L-origin genomes. Mapping and read classification were done using the reference genomes with SNP replacement for two A. kamchatica accessions as described above for the genome assembly step. Therefore, the reference genomes of each A. kamchatica genotype match those used in the RNA-seq experiments. If a read was mapped to multiple positions, the aligned position with a “primary” flag (the position with the best alignment score) was adopted to quantify expression levels to avoid ambiguous results caused by multiply mapped reads. Reads that only mapped to one parental genome were discarded. Using this type of read to quantify homeolog expression levels may cause a biased estimation of expression levels attributed to the difference in quality of the assembled genomes of two parental species, even though part of them would truly represent species-specific gene fragments. We mapped A. halleri and A. lyrata RNA-seq reads directly using STAR-2.3.0 without the read classification step.
We identified 25,509 homeologous contigs and scaffolds in the two A. kamchatica genotypes using a reciprocal blast hit (best-to-best) strategy using a reciprocal best hit (best-to-best) based on BLAST E values of 10−15 for each A. kamchatica genotype. Among these, 19,820 are orthologous with A. thaliana also using a reciprocal blast hit strategy. We also detected orthologous genes between the predicted genes on diploid A. halleri and A. lyrata contigs with A. thaliana genes using TAIR 10 gene annotations (Lamesch et al. 2012) using reciprocal best hits based on BLAST E values of 10−15. We limited our dataset by TAIR gene IDs that are common to both A. halleri- and A. lyrata-derived copies in both diploids and polyploid genomes to compare gene expression between homeologs and between species. The use of reciprocal best hits and limiting the gene dataset to items containing A. thaliana orthologous gene models is somewhat conservative, but we are primarily concerned with homologous gene pairs with annotated function. We also used one-way blast of A. lyrata-derived scaffolds as the query against the A. halleri-derived reference genome to confirm that our reciprocal blast results blasted to the same contig if there were multiple hits in the one-way blast. In addition, when gene duplications occur in a particular species, the sum of the expression of those duplicates can drive the function underlying the phenotype of interest (e.g., HMA4 and hyperaccumulation). The highly similar duplicates are likely to be assembled as a single copy in the current assembly (see the section regarding pyrosequencing validation of homolog expression) because of their short overall contig length.
Differential Expression Analysis
Differentially regulated (up- and down-regulated) homeologs in control and Zn treatments were estimated using edgeR (Robinson et al. 2010) with only H-origin or L-origin read count data. We first performed differential expression analyses for each homeolog (H-origin or L-origin), tissue (leaf or root), and genotype (Alaskan or Japanese) separately. Diploid gene regulation was estimated similarly using orthologous read count data to detect up- and downregulation from control to Zn-treated samples. Significance was determined using a false discovery rate (FDR; BH test) cutoff of <0.05. If the RPKM was <0.1 in both control and Zn-treated samples, the gene was considered unexpressed and was not counted as differentially regulated if significant. We conducted GO analysis on differentially regulated genes using agriGO (Du et al. 2010) and GO categories with ≥30 query genes for MF and a FDR P value threshold of <0.05. Changes in homeolog ratios from control to Zn-treated conditions were estimated using the statistical test described by Akama et al. (2014) to control for overdispersion and reduce false-positive ratio-change estimates.
RPKM reported as total levels of gene expression were estimated in the allopolyploids including both origin and common reads counted using HTSeq 0.5.4 in union mode after the read classification was bundled in the HomeoRoq pipeline as described in the following equation (1):
| (1) |
where is the number of reads that are mapped on gene in species , is the length of gene in the A. halleri genome, and is the total number of mapped reads. In our procedure, there are three types of count values produced after read classification: 1) : the number of reads mapped to A. halleri-derived homeolog i, 2) : the number of reads mapped to A. lyrata-derived homeolog i, and 3) : the number of reads that cannot be classified in the parental species because the reads are identical to the genomes of both species (i.e., common reads). The reads were allocated to one of the species at a rate proportional to the H-origin and L-origin ratio of each homeolog for total RPKM comparisons. That is, two RPKM values were estimated corresponding to each gene in the A. kamchatica genome, one for the A. halleri-derived homeolog (H-origin), and one for the A. lyrata-derived homeolog (L-origin). In the diploid, RPKM values were calculated normally after counting mapped reads to the reference without the read classification. The RPKM values of tetraploids and diploids would be comparable despite minor methodological differences.
Complete datasets of A. kamchatica (MUR and PAK genotype leaf and root) transcriptomics can be found in supplementary files 5 and 6, Supplementary Material online, and the A. halleri and A. lyrata transcriptomics datasets can be found in supplementary file 7, Supplementary Material online. Supplementary file 2, Supplementary Material online shows candidate gene expression ratios and for both polyploid genotypes and parental diploid species.
Pyrosequencing
We used pyrosequencing to validate the expression ratios between H-origin and L-origin homeologs in A. kamchatica. The RNA samples of three replicates per genotype were reverse transcribed to cDNA using a High Capacity RNA-to-cDNA kit (Invitrogen) separately for leaf and root tissues for replicates of each polyploid genotype before and after Zn treatments. We designed pyrosequencing assays using diverged SNPs in homeologous gene copies for the following genes orthologous to A. thaliana: HMA3 (AT4G30120), HMA4 (AT2G19110), MTP1 (AT2G46800), NRAMP3 (AT2G23150), and ZIP9 (AT4G33020). Each set of gene-specific PCR primers and sequencing primers was designed using PyroMark Assay Design software (v. 2.0, Qiagen) at the Genomic Diversity Center, ETH, Zurich (supplementary table S6, Supplementary Material online). The gene-specific amplification primers were designed at conserved regions between H- and L-homeologs so that they include SNP positions inside the amplified region. The sequencing primer was designed at an adjacent conserved region to targeted SNP positions between H- and L-homeologs (see supplementary fig. S5, Supplementary Material online for example assay design using HMA4 homeologs).
The amplified PCR fragments were sequenced using the PyroMark ID system (Qiagen) at the ETH Genetic Diversity Center, Zurich. The amplification peaks were analyzed using the PyroMark system in allele quantification (AQ) mode to determine the SNP amplification ratio. The multiple SNP ratios obtained for each gene fragment were averaged to estimate the H-origin and L-origin homeolog ratios. Genes such as HMA4 and MTP1 that presumably have multiple copies in the A. halleri genome are represented in our assembly on short contigs in both A. halleri and A. lyrata and have only a single copy. Therefore, pyrosequencing assays identify target SNPs between A. halleri- and A. lyrata-derived homeologs that are only represented by these single-copy contigs. Pyrosequencing expression ratios between H-origin and L-origin are potentially the sum of duplicated A. halleri-derived copies if the duplicates also possess these target SNPs. To determine whether A. kamchatica shares the tandemly duplicated copies also found in A. halleri, we used genomic DNA from A. kamchatica accessions for the template for PCR amplification. Because we designed assays in the same manner using SNPs that are divergent among H-origin and L-origin homeologs (supplementary fig. S5, Supplementary Material online), these assays can detect whether there are three A. halleri-derived copies, compared with a single copy from the A. lyrata parent by quantifying the A. halleri-derived SNPs. Pyrosequencing primers can be found in supplementary table S5, Supplementary Material online. We also used 3′ rapid amplification of cDNA ends (3′ RACE) to amplify the C-terminus end of HMA4 using the Invitrogen RACE kit. PCR products were subcloned and amplified using M13 forward and reverse primers, and then Sanger sequenced.
HMA4 Region Resequencing and Diversity Analysis
Using the approach described above for diploid-guided reference assembly for 20 A. kamchatica genotypes, we generated consensus contigs for H-origin and L-origin genes for each genotype that align to the BAC sequences from A. halleri subsp. halleri (GenBank BAC accession numbers: EU382072.1 and EU382073.1) containing the HMA4 locus (Hanikenne et al. 2008; Hanikenne et al. 2013). Coding sequences from the 20 A. kamchatica H-origin and L-origin HMA4 regions were aligned using Muscle and Geneious (v. 6.06). We also aligned our H-origin and L-origin contigs to a corresponding region on JGI’s A. lyrata (Hu et al. 2011) chromosome 3 (Alyr_JGI_scaffold_3.23427106-23532642) containing the orthologous HMA4 region. This showed the same physical order of 11 H-origin and L-origin contigs surrounding and including the HMA4 coding sequences on both A. halleri (BAC) and A. lyrata (JGI) genomes (supplementary fig. S8 A and B, supplementary material online), and we assume the same orientation and synteny in A. kamchatica for our diversity comparisons.
The A. halleri and A. lyrata genomes have some important structural differences surrounding the HMA4 locus, where the published A. halleri BAC sequence is nearly 2.5 times longer (including intergenic space and introns) than that of the corresponding region containing these 11 genes on chromosome 3 in the JGI A. lyrata assembly. This is largely because of the triplication of the HMA4 gene on the BAC assembly. Our de novo assemblies of A. halleri and A. lyrata each contain only a single contig with the HMA4 gene (homologous to A. thaliana AT2G19110.1) identified as homeologs by both reciprocal blast hit and one-way blast of the A. lyrata genome to the A. halleri genome. Moreover, many of the genes identified as surrounding the HMA4 locus by aligning to the BAC sequence in A. halleri are also on separate contigs in our diploid reference assemblies described in supplementary file 3, Supplementary Material online.
The 11 gene alignments are orthologous to A. thaliana TAIR10 annotated gene models: AT2G19010.1, AT2G19045.1, AT2G19050.1, AT2G19060.1, AT2G19070.1, AT2G19080.1, AT2G19090.1, AT2G19110.1, AT2G19130.1, AT2G19150.1, AT2G19160.1, and AT2G19170.1. We also included two contigs corresponding to two genes outside of the A. halleri BAC sequence and A. lyrata scaffold corresponding to the S1 and S13 genes used by Hanikenne et al. (2013) to estimate background diversity between homeologs (supplementary file 4, Supplementary Material online). The genes outside of the BAC sequence are: AT2G18750.1 and AT2G19490.1. Eight of these genes correspond to the gene regions S1, S2, S3, S4 (including HMA4-1, AT2G19110.1), S11, S12, and S13, with S1 (AT2G18750.1) and S13 (AT2G19490.1) not linked (“background genes”) and outside of the BAC sequence. We then aligned the 13 H-origin and L-origin coding sequences from 20 A. kamchatica genotypes to A. thaliana orthologs to use as a common outgroup for each homeolog. Summary and diversity statistics, including divergence from A. thaliana, were estimated using libsequence (Thornton 2003) packages and custom R and Perl shell scripts. The HKA test was performed using DNASP (Librado and Rozas 2009).
Data Accessibility
RNA-seq reads were sumitted to DDBJ. Submission: DRA004363 (masa-0001_Submission). BioProject: PRJDB4054 (PSUB004762). BioSample: SAMD00035297 (SSUB004394). Experiment: DRX049283 (masa-0001_Experiment_0001). Run: DRR054434 (masa-0001_Run_0001). Illumina data for the MUR and PAK accessions. DDBJ: DRA003243 and DRA004363. HMA4 homoeolog alignments and Supplementary Files 5–7 can be downloaded from at http://dx.doi.org/10.5061/dryad.tr85d.
Supplementary Material
Supplementary figures S1–S10, tables S1–S6, and files 1–4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Author Contributions
TP, RSI, and KKS conceived the experiments. TP and TC conducted the hydroponic experiments. TP and MH assembled and analyzed transcriptomics datasets. TP conducted polymorphism analysis. JS assembled reference genomes. KT, YO, RIS, and KKS contributed plant and soil samples. TP, RSI, and KKS wrote the paper, with editorial contributions by all coauthors.
Supplementary Material
Acknowledgments
We thank Enrico Martinoia at UZH Plant Biology for material contributions to hydroponics experiments and useful discussions about experimental design. David Salt and John Danku at the University of Aberdeen for ion estimates. Rainer Schulin and Bjoern Studer at ETH Zurich for metal analysis of plants and soils as well as technical support and useful discussions. Lucas Mohn for PyroMark sequencing and the Genetic Diversity Center at ETH Zurich for PyroMark software and equipment. We also thank the Functional Genomic Center Zurich for technical support. The study was supported by Swiss National Science Foundation, the University Research Priority Program of Evolution in Action of the University of Zurich to K.K.S, MEXT KAKENHI Grant Numbers 16H06469 and 26113709, Young Investigator Award of Human Frontier Science Program to K.K.S and J.S., EU cofund Plant Fellow to T.P. and Marie-Heim Hoegtlin grant by Swiss National Science Foundation to R.S.I., ISCB (Indo-Swiss Collaboration in Biotechnology) to K.K.S. and M.H., the Special Coordination Funds for Promoting Science and Technology from MEXT Japan, an Inamori Foundation research grant, a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (Young Researchers B, 2277023), and Research and Education Funding for Japanese Alps Inter-Universities Cooperative Project, MEXT, Japan to KT.
References
- Akama S, Shimizu-Inatsugi R, Shimizu KK, Sese J. 2014. Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis. Nucleic Acids Res. 42:e46–ee4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunova AR, Matniyazov RT, Liang H, Akhunov ED. 2010. Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11:505.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JJ, Takebayashi N, Sformo T, Wolf DE. 2015. Cold tolerance in Arabidopsis kamchatica. Am J Bot. 102:439–448. [DOI] [PubMed] [Google Scholar]
- Arrivault S, Senger T, Krämer U. 2006. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46:861–879. [DOI] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U. 2004. Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri: transcript profiling in shoots of A. halleri. Plant J. 37:251–268. [DOI] [PubMed] [Google Scholar]
- Bert V, Bonnin I, Saumitou-Laprade P, de Laguerie P, Petit D. 2002. Do Arabidopsis halleri from nonmetallicolous populations accumulate zinc and cadmium more effectively than those from metallicolous populations? New Phytol. 155:47–57. [DOI] [PubMed] [Google Scholar]
- Bert V, Macnair MR, De Laguerie P, Saumitou-Laprade P, Petit D. 2000. Zinc tolerance and accumulation in metallicolous and nonmetallicolous populations of Arabidopsis halleri (Brassicaceae). New Phytol. 146:225–233. [DOI] [PubMed] [Google Scholar]
- Bertrand B, Bardil A, Baraille H, Dussert S, Doulbeau S, Dubois E, Severac D, Dereeper A, Etienne H. 2015. The greater phenotypic homeostasis of the allopolyploid Coffea arabica improved the transcriptional Homeostasis over that of both diploid parents. Plant Cell Physiol. 56:2035–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Madlung A. 2014. Polyploidy in the Arabidopsis genus. Chromosome Res. 22:117–134. [DOI] [PubMed] [Google Scholar]
- Boyd RS. 2007. The defense hypothesis of elemental hyperaccumulation: status, challenges and new directions. Plant Soil 293:153–176. [Google Scholar]
- Buggs RJA, Renny-Byfield S, Chester M, Jordon-Thaden IE, Viccini LF, Chamala S, Leitch AR, Schnable PS, Barbazuk WB, Soltis PS, et al. 2012. Next-generation sequencing and genome evolution in allopolyploids. Am J Bot. 99:372–382. [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Wendel JF, Doyle JJ, Soltis DE, Soltis PS, Coate JE. 2014. The legacy of diploid progenitors in allopolyploid gene expression patterns. Philos Trans R Soc B Biol Sci. 369:20130354–20130354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. 2013. Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet. 14:471–482. [DOI] [PubMed] [Google Scholar]
- Chiang H-C, Lo J-C, Yeh K-C. 2006. Genes associated with heavy metal tolerance and accumulation in zn/cd hyperaccumulator Arabidopsis halleri: a genomic survey with cDNA microarray. Environ Sci Technol. 40:6792–6798. [DOI] [PubMed] [Google Scholar]
- Coleman CM, Boyd RS, Eubanks MD. 2005. Extending the elemental defense hypothesis: dietary metal concentrations below hyperaccumulator levels could harm herbivores. J Chem Ecol. 31:1669–1681. [DOI] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nat Rev Genet. 6:836–846. [DOI] [PubMed] [Google Scholar]
- Combes M-C, Dereeper A, Severac D, Bertrand B, Lashermes P. 2013. Contribution of subgenomes to the transcriptome and their intertwined regulation in the allopolyploid Coffea arabica grown at contrasted temperatures. New Phytol 200:251–260. [DOI] [PubMed] [Google Scholar]
- Courbot M, Willems G, Motte P, Arvidsson S, Roosens N, Saumitou-Laprade P, Verbruggen N. 2007. A major quantitative trait locus for cadmium tolerance in Arabidopsis halleri. Colocalizes with HMA4, a gene encoding a heavy metal atpase. Plant Physiol. 144:1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. Sunderland (MA): Sinauer Associates [Google Scholar]
- Desbrosses-Fonrouge A-G, Voigt K, Schröder A, Arrivault S, Thomine S, Krämer U. 2005. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 579:4165–4174. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger DB, Desbrosses-Fonrouge A-G, Krach C, Chardonnens AN, Meyer RC, Saumitou-Laprade P, Krämer U. 2004. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J. 39:425–439. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis 90 toolkit for the agricultural community. Nucleic Acids Res. 38:W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrendorfer F. 1980. Polyploidy and distribution In: Lewis WH, editor. Polyploidy, biological relevance. New York: Plenum Press; p. 45–60. [Google Scholar]
- Filatov V, Dowdle J, Smirnoff N, Ford-Lloyd B, Newbury HJ, Macnair MR. 2006. Comparison of gene expression in segregating families identifies genes and genomic regions involved in a novel adaptation, zinc hyperaccumulation: gene expression in segregating gene families. Mol Ecol. 15:3045–3059. [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U. 2008. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453:391–395. [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Kroymann J, Trampczynska A, Bernal M, Motte P, Clemens S, Krämer U. 2013. Hard selective sweep and ectopic gene conversion in a gene cluster affording environmental adaptation. PLoS Genet. 9:e1003707.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H, Matsuzawa T. 2008. Serpentine soil environment and distribution of alpine plants in the Shirouma mountain range. JPN J Ecol. 58:199–204. [Google Scholar]
- He F, Zhang X, Hu J, Turck F, Dong X, Goebel U, Borevitz J, de Meaux J. 2012. Genome-wide analysis of cis-regulatory divergence between species in the Arabidopsis genus. Mol Biol Evol. 29:3385–3395. [DOI] [PubMed] [Google Scholar]
- Hoffmann MH. 2005. Evolution of the realized climatic niche in the genus: Arabidopsis (BRASSICACEAE). Evolution 59:1425–1436. [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 43:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashem MA, Singh BR, Kubota H, Sugawara R, Kitajima N, Kondo T, Kawai S. 2010. Zinc tolerance and uptake by Arabidopsis halleri ssp. gemmifera grown in nutrient solution. Environ Sci Pollut Res. 17:1174–1176. [DOI] [PubMed] [Google Scholar]
- Kazemi-Dinan A, Thomaschky S, Stein RJ, Krämer U, Müller C. 2014. Zinc and cadmium hyperaccumulation act as deterrents towards specialist herbivores and impede the performance of a generalist herbivore. New Phytol 202:628–639. [DOI] [PubMed] [Google Scholar]
- Kenta T, Yamada A, Onda Y. 2011. Clinal variation in flowering time and vernalisation requirement across a 3000-m altitudinal range in perennial Arabidopsis kamchatica ssp. kamchatica and annual lowland subspecies kawasakiana. J Ecosys Ecograph S6:001. [Google Scholar]
- Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, Maeshima M. 2004. Zinc transporter of arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 45:1749–1758. [DOI] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol. 17:1483–1498. [DOI] [PubMed] [Google Scholar]
- Krämer U. 2010. Metal hyperaccumulation in plants. Ann Rev Plant Biol. 61:517–534. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L, et al. 2003. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nature Biotechnol. 21:1215–1221. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- McCarthy EW, Arnold SEJ, Chittka L, Le Comber SC, Verity R, Dodsworth S, Knapp S, Kelly LJ, Chase MW, Baldwin IT, et al. 2015. The effect of polyploidy and hybridization on the evolution of floral colour in Nicotiana (Solanaceae). Ann Bot. 115:1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C-L, Kostecka AA, Saumitou-Laprade P, Créach A, Castric V, Pauwels M, Frérot H. 2010. Variability of zinc tolerance among and within populations of the pseudometallophyte species Arabidopsis halleri and possible role of directional selection. New Phytol. 185:130–142. [DOI] [PubMed] [Google Scholar]
- Milner MJ, Kochian LV. 2008. Investigating heavy-metal hyperaccumulation using Thlaspi caerulescens as a model system. Ann Bot. 102:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Sels J, Richard O, Czernic P, Loubet S, Jacquier A, François IEJA, Cammue BPA, Lebrun M, Berthomieu P, et al. 2006. A putative novel role for plant defensins: a defensin from the zinc hyper-accumulating plant, Arabidopsis halleri, confers zinc tolerance. Plant J. 47:329–342. [DOI] [PubMed] [Google Scholar]
- Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. 2008. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 149:894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Okawa K, Kakizaki T, Honma T, Itoh H, Inaba T. 2007. Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol. 144:513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó Lochlainn S, Bowen HC, Fray RG, Hammond JP, King GJ, White PJ, Graham NS, Broadley MR. 2011. Tandem quadruplication of HMA4 in the Zinc (Zn) and Cadmium (Cd) hyperaccumulator Noccaea caerulescens. PLoS One 6:e17814.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels M, FréRot H, Bonnin I, Saumitou-Laprade P. 2006. A broad-scale analysis of population differentiation for Zn tolerance in an emerging model species for tolerance study: Arabidopsis halleri (Brassicaceae). J Evol Biol. 19:1838–1850. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Castric V, Pauwels M, Wright SI, Saumitou-Laprade P, Vekemans X. 2011. Does speciation between Arabidopsis halleri and Arabidopsis lyrata Coincide with major changes in a molecular target of adaptation? PLoS One 6:e26872.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle HD, Nosil P. 2005. Ecological speciation. Ecol Lett. 8:336–352. [Google Scholar]
- Schmickl R, Jørgensen MH, Brysting AK, Koch MA. 2010. The evolutionary history of the Arabidopsis lyrata complex: a hybrid in the amphi-Beringian area closes a large distribution gap and builds up a genetic barrier. BMC Evol Biol. 10:98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüchter C, Heuberger H, Horie S. 1981. New evidence for multiglaciation in the high mountains of Japan. I. New observations in Hakuba (Shirouma)-dake. Proc JPN Acad Ser B Phys Biol Sci. 57:296–299. [Google Scholar]
- Shahzad Z, Gosti F, Frérot H, Lacombe E, Roosens N, Saumitou-Laprade P, Berthomieu P. 2010. The five AhMTP1 zinc transporters undergo different evolutionary fates towards adaptive evolution to zinc tolerance in Arabidopsis halleri. PLoS Genet. 6:e1000911.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad Z, Ranwez V, Fizames C, Marquès L, Le Martret B, et al. 2013. Plant defensin type 1 (PDF1): protein promiscuity and expression variation within the Arabidopsis genus shed light on zinc tolerance acquisition in Arabidopsis halleri. New Phytol. 200:820–833. [DOI] [PubMed] [Google Scholar]
- Shanmugam V, Lo J-C, Wu C-L, Wang S-L, Lai C-C, Connolly EL, Huang J-L, Yeh K-C. 2011. Differential expression and regulation of iron-regulated metal transporters in Arabidopsis halleri and Arabidopsis thaliana—the role in zinc tolerance. New Phytol. 190:125–137. [DOI] [PubMed] [Google Scholar]
- Shi X, Ng DW-K, Zhang C, Comai L, Ye W, Jeffrey Chen Z. 2012. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun. 3:950.. [DOI] [PubMed] [Google Scholar]
- Shimizu KK, Fujii S, Marhold K, Watanabe K, Kudoh H. 2005. Arabidopsis kamchatica (Fisch. ex DC.) K. Shimizu & Kudoh and A. kamchatica subsp. kawasakiana (Makino) K. Shimizu & Kudoh, new combinations. Acta Phyt Geobot. 56:165–174. [Google Scholar]
- Shimizu KK, Tsuchimatsu T. 2015. Evolution of selfing: recurrent patterns in molecular adaptation. Ann Rev Ecol Evol Syst. 46:593–622. [Google Scholar]
- Shimizu-Inatsugi R, Lihová J, Iwanaga H, Kudoh H, Marhold K, Savolainen O, Watanabe K, Yakubov VV, Shimizu KK. 2009. The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri. Mol Ecol. 18:4024–4048. [DOI] [PubMed] [Google Scholar]
- Shimizu-Inatsugi R, Terada A, Hirose K, Kudoh H, Sese J, Shimizu KK. 2016. Plant adaptive radiation mediated by polyploid plasticity in transcriptomes. Mol Ecol. DOI: 10.1111/mec.13738. [DOI] [PubMed] [Google Scholar]
- Small RL, Ryburn JA, Wendel JF. 1999. Low levels of nucleotide diversity at homoeologous Adh loci in allotetraploid cotton (Gossypium L.). Mol Biol Evol. 16:491–501. [DOI] [PubMed] [Google Scholar]
- Stebbins GJ. 1950. Variation and evolution in plants. New York: Columbia University Press. [Google Scholar]
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold. [Google Scholar]
- Talke IN, Hanikenne M, Kramer U. 2006. Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol. 142:148–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumi S, Naka Y, Morihiro H, Matsuoka Y. 2009. Expression of morphological and flowering time variation through allopolyploidization: an empirical study with 27 wheat synthetics and their parental Aegilops tauschii accessions. Plant Breed. 128:585–590. [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesova M, Pysek P. 2012. The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot. 109:19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Ann Rev Plant Physiol Plant Mol Biol. 50:571–599. [DOI] [PubMed] [Google Scholar]
- Thornton K. 2003. libsequence: a C ++ class library for evolutionary genetic analysis. Bioinformatics 19:2325–2327. [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T, Suwabe K, Shimizu-Inatsugi R, Isokawa S, Pavlidis P, Städler T, Suzuki G, Takayama S, Watanabe M, Shimizu KK. 2010. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464:1342–1346. [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T, Kaiser P, Yew C-L, Bachelier JB, Shimizu KK. 2012. Recent loss of self-incompatibility by degradation of the male component in allotetraploid Arabidopsis kamchatica. PLoS Genet. 8:e1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hanikenne M, Clemens S. 2013. A more complete picture of metal hyperaccumulation through next-generation sequencing technologies. Front Plant Sci. 4:388.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. 2009. Molecular mechanisms of metal hyperaccumulation in plants: tansley review. New Phytol. 181:759–776. [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, et al. 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172:507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Harada E, Vess C, Roepenack-Lahaye E. v, Clemens S. 2004. Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors: A. halleri metal hyperaccumulation factors. Plant J. 37:269–281. [DOI] [PubMed] [Google Scholar]
- Willems G, Drager DB, Courbot M, Gode C, Verbruggen N, Saumitou-Laprade P. 2007. The genetic basis of zinc tolerance in the metallophyte Arabidopsis halleri ssp. halleri (Brassicaceae): an analysis of quantitative trait loci. Genetics 176:659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I, Ting C-T. 2004. Genes and speciation. Nat Rev Genet. 5:114–122. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1993. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet. 236-236:331–340. [DOI] [PubMed] [Google Scholar]
- Yoo M-J, Szadkowski E, Wendel JF. 2013. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M-J, Liu X, Pires JC, Soltis PS, Soltis DE. 2014. Nonadditive gene expression in polyploids. Ann Rev Genet. 48:485–517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq reads were sumitted to DDBJ. Submission: DRA004363 (masa-0001_Submission). BioProject: PRJDB4054 (PSUB004762). BioSample: SAMD00035297 (SSUB004394). Experiment: DRX049283 (masa-0001_Experiment_0001). Run: DRR054434 (masa-0001_Run_0001). Illumina data for the MUR and PAK accessions. DDBJ: DRA003243 and DRA004363. HMA4 homoeolog alignments and Supplementary Files 5–7 can be downloaded from at http://dx.doi.org/10.5061/dryad.tr85d.