Abstract
As the largest European herbivore, the wisent (Bison bonasus) is emblematic of the continent wildlife but has unclear origins. Here, we infer its demographic and adaptive histories from two individual whole-genome sequences via a detailed comparative analysis with bovine genomes. We estimate that the wisent and bovine species diverged from 1.7 × 106 to 850,000 years before present (YBP) through a speciation process involving an extended period of limited gene flow. Our data further support the occurrence of more recent secondary contacts, posterior to the Bos taurus and Bos indicus divergence (∼150,000 YBP), between the wisent and (European) taurine cattle lineages. Although the wisent and bovine population sizes experienced a similar sharp decline since the Last Glacial Maximum, we find that the wisent demography remained more fluctuating during the Pleistocene. This is in agreement with a scenario in which wisents responded to successive glaciations by habitat fragmentation rather than southward and eastward migration as for the bovine ancestors. We finally detect 423 genes under positive selection between the wisent and bovine lineages, which shed a new light on the genome response to different living conditions (temperature, available food resource, and pathogen exposure) and on the key gene functions altered by the domestication process.
Keywords: European bison, cattle, genetics, evolution, demography, adaptation, systems biology, domestication
Introduction
The European bison, Bison bonasus (BBO), also known as the wisent, is an emblematic species of the European wildlife. The oldest fossil remains were found in Eastern Europe and trace back to the Late Pleistocene, approximately 12,000 years before present (YBP) (Benecke 2005). The wisent is closely related to the North American bison (Bison bison), both species assumed to be derived from the extinct long-horned steppe bison (Bison priscus), which was widespread across the northern hemisphere in the mid and upper Pleistocene (i.e., 781,000–11,700 YBP). However, several morphological and behavioral traits distinguish the wisent from its American relative. In particular, adults have generally a smaller body length (Krasińska and Krasiński 2002), display a less hairy body, and tend to be mixed feeders that combine browsing and grazing (Kowalczyk et al. 2011; Merceron et al. 2014), whereas American bisons are essentially grazer. These various characteristics might be viewed as adaptation to living constraints associated to forest habitat that has been considered as a refuge (Kerley et al. 2012; Bocherens et al. 2015).
During the mid and late Holocene, the wisent was distributed in central and eastern Europe with a longest survival in north-eastern Europe (Benecke 2005). Nevertheless, the progressive replacement of the open steppe by forest cover in early Holocene and the human population growth associated with the spread of farming during the middle Holocene lead to a reduction of its habitat (Kuemmerle et al. 2012; Bocherens et al. 2015). Hence, by the middle age, the wisent was only present in a few natural forests of North-Eastern Europe and finally faced extinction in the wild at the beginning of the 20th century (Pucek et al. 2004). A species restoration program relying on a few tens of specimens kept in European zoos and private breeding centers was early initiated in the 1920s (Pucek et al. 2004; Tokarska et al. 2011). Thanks to these efforts, the wisent world population now consists of over 5,000 animals (including about 3,500 free-living individuals) that are mostly maintained in forests of eastern Europe (Kerley et al. 2012; Raczyński 2015).
The wisent is among the few large mammalian terrestrial species that survived the massive megafauna extinction of the last glacial/interglacial transition period, approximately 25,000–10,000 YBP (Lister and Anthony 2008; Lorenzen et al. 2011). Characterizing the specific conditions of its survival may thus help understanding how organisms evolve in response to environmental change. In this study, we explore the wisent demographic and adaptive histories using individual whole-genome sequences from two males of the wisent lowland line (Tokarska et al. 2011) living in the Białowieża forest. To that end, we perform a detailed comparative analysis of these newly generated wisent genome sequences with individual genome sequences representative of different European cattle breeds. Domestic cattle, the wild ancestor of which, the aurochs (Bos primigenius) went extinct in the 17th century, is indeed among the closest wisent relatives (Verkaar et al. 2004) and benefits from abundant genomic resources (Liu et al. 2009). Our genome-wide comparison provides an accurate characterization of the divergence between the bovine and wisent lineages and an estimation of their population size histories during Pleistocene and Holocene. In addition, the identification and functional annotation of genes under positive selection between both lineages shed light into the biological functions and the key adaptive traits that were affected by environmental constraints.
Results and Discussion
Comparison of Wisent and Cattle Genomes Reveals Low Amount of Nucleotide Divergence
High-throughput sequencing data from two wisent bulls, namely BBO_3569 and BBO_3574, were each aligned onto the UMD 3.1 Bos taurus (BTA) genome assembly (Liu et al. 2009). The read mapping statistics showed overall good performances (supplementary table S1, Supplementary Material online) with noticeably 97.4% (respectively, 96.6%) of the reads being properly paired for the BBO_3569 (respectively, BBO_3574). Conversely, following the same read mapping procedure but considering as a reference the OviAr3 genome assembly (Jiang et al. 2014) from the more distantly related domestic sheep (Ovis aries, OAR) strongly altered the statistics (supplementary table S1, Supplementary Material online). Altogether, these mapping results provided support to consider the cattle genome as closely related enough to provide a good reference to assemble our wisent reads. For autosomes, an approximately 10X-coverage was achieved for both individuals (9.81X and 11.6X for BBO_3569 and BBO_3574, respectively) with more than 95% of the (bovine) reference sequence covered by at least one read (table 1). As expected for males, the bovine chromosome X was only slightly more than half covered (∼56.8% and 55.8% of the autosome coverage for BBO_3569 and BBO_3574, respectively) due to the inclusion of pseudo-autosomal regions (PAR) in the estimation. Conversely, the (bovine) mitochondrial genome was found highly covered (>300X) for both individuals.
Table 1.
Read Mapping Statistics from the Alignment of each BBO_3569 and BBO_3574 European Bison Genome Sequences onto the UMD 3.1 Cattle Genome Assembly (Liu et al. 2009).
| Chromosome Type (BTA) | Size (in bp) | Coverage (Percentage of Sequence Covered)a |
Average Nucleotide Divergence in % (Number of Sites Compared)b |
||
|---|---|---|---|---|---|
| BBO_3569 | BBO_3574 | BBO_3569 | BBO_3574 | ||
| Autosomes | 2,512,082,506 | 9.81 (95.5) | 11.6 (95.7) | 0.870 (2.27 × 109) | 0.882 (2.29 × 109) |
| X-chromosome (including PAR) | 148,823,899 | 5.57 (90.9) | 6.48 (91.2) | 0.636 (1.11 × 108) | 0.642 (1.17 × 108) |
| Mitochondria | 16,338 | 397 (97.7) | 302 (97.1) | 3.58 (1.18 × 104) | 3.41 (1.20 × 104) |
aFor each individual, the average read coverages (after alignment) over autosomes, the X-chromosome, and the mitochondria are given together with the overall percentages of sites from the corresponding reference sequences covered with at least one read.
bFor each type of chromosomes, the average nucleotide divergence between the cattle reference genome and each European bison individual consensus sequence is given (see Materials and Methods).
Interestingly, the estimated autosomal nucleotide divergence between the bovine genome and each wisent individual genome was found equal to 0.870% for BBO_3569 and 0.882% for BBO_3574 (table 1) supporting a low amount of genome divergence between the two species. The estimated nucleotide divergence was about 1.37 times smaller for the chromosome X than for autosomes, as already observed in the Chimpanzee versus Human genome sequence comparison (Mikkelsen et al. 2005). This is likely related to a higher mutation rate in the male compared to the female germ line in mammalian species (Li et al. 2002; Mikkelsen et al. 2005). Also, the nucleotide divergence increased in distal chromosomal regions in both autosomes and X-chromosome comparisons while read coverage remained mostly uniform (supplementary fig. S1, Supplementary Material online). Such a regional pattern, also reported in the aforementioned Chimpanzee versus Human genome sequence comparison (Mikkelsen et al. 2005), might similarly result from the physical properties of the corresponding distal regions (i.e., high local recombination rate, high gene density, and higher GC content). Indirectly, observing such a pattern for wisent sequences mapped onto the cattle genome assembly supports a low amount of chromosome rearrangements between the two species. Finally, the estimated nucleotide divergence for the mitochondrial genomes was about four times higher than for autosomes. This suggests an absence of recent maternal bovine introgression at least within the lineages of the two wisents we considered.
The Wisent Demography Has Been More Fluctuating than the Bovine One during the Whole Pleistocene
We further sought to characterize the genetic variability of the wisent population that has long been considered as a threatened species and is still classified as “vulnerable” by the International Union for Conservation of Nature red list (http://www.iucnredlist.org/). The estimated average (autosomal) heterozygosities, as approximated by the population mutation rate (, where represents the mutation rate per site and per generation and the effective population size) measured on whole individual genomes, were consistent when considering either the BBO_3569 () or the BBO_3574 () individual. Interestingly, both estimates were similar to the ones obtained on four individual cattle genomes representative of the Angus (AAN_0037), Jersey (JER_0009), Holstein (HOL_0101), and Simmental (SIM_0043) European cattle breeds which ranged from to (supplementary fig. S2 and table S2, Supplementary Material online). The small heterozygosity observed for wisent might be explained not only by the sharp decline of its effective population size during the last 20,000 years (see below) but also by its more recent history since wisent experienced a strong bottleneck at the end of the First World War (Wójcik et al. 2009; Tokarska et al. 2011). However, we noted that our wisent estimates remained within the range observed for the four different European cattle breeds. This suggests that the constitution of the Białowieża wisent herd from a small number of presumably poorly related founders and subsequent management (Pucek et al. 2004) allowed to recover a reasonable amount of variability for the current population.
More generally, the patterns of variability observed at a local individual genome scale are informative about the demographic history of the population (Li and Durbin 2011). From our two individual genomes, we estimated the wisent past Ne using the Pairwise Sequentially Markovian Coalescent model (PSMC) introduced by Li and Durbin (2011) and compare it to that estimated for the bovine species based on each of the four cattle individual genomes aforementioned. Estimates of the scaled effective population sizes (in units of ) backward in time measured in scaled units of (where is in YBP and represents the mutation rate per site and per year) are plotted in supplementary figure S3, Supplementary Material online, for each of the six individual genome analyses (see supplementary fig. S4, Supplementary Material online, for bootstrap confidence intervals of each history). Overall, a similar trend was observed within each species, irrespective of the individual considered (although the AAN_0037 profile was more dissimilar than the three other cattle ones probably as a result of an overall lower coverage for this individual). In contrast, marked differences were found when comparing the inferred wisent and bovine population size histories. In particular, from backward time (in scaled units) t = 10 − 3 to t = 5.10 − 5 (supplementary fig. S3, Supplementary Material online), wisent displayed more pronounced oscillations and from two to three times higher effective population sizes. Because the PSMC model relies on local density of heterozygous sites, using the bovine genome assembly as a reference to derive the analyzed individual wisent genomes (see Materials and Methods) could have led to some inferential biases (Nevado et al. 2014). Nevertheless, the inferred wisent population size histories remained similar when considering a BBO consensus genome as a reference to derive the individual genome sequences (supplementary fig. S5, Supplementary Material online). Similarly, given the close relatedness of the bovine and bison species, chromosomal rearrangements might remain negligible and should not have substantial effect on the overall inferred histories.
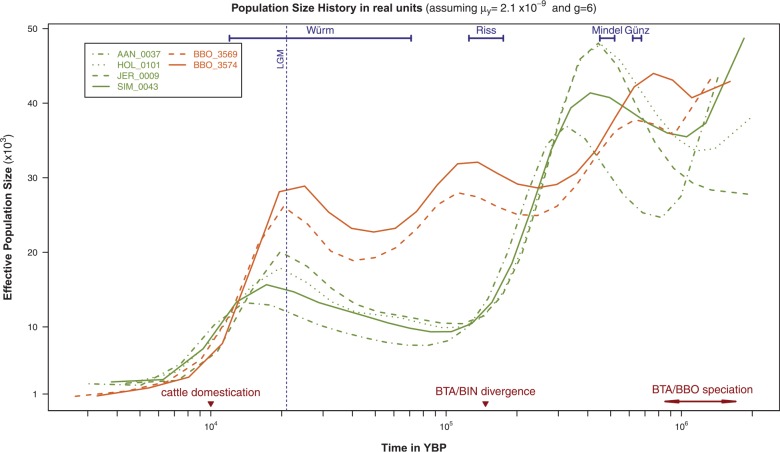
To facilitate biological interpretation of the observed demographic signals, we transformed the population size history profiles assuming (Liu et al. 2006) to translate the time scale in YBP. Further, to translate the effective population sizes (Ne) in real units, we assumed a generation time of g = 6 years (Keightley and Eyre-Walker 2000; Gautier et al. 2007), leading to . The resulting backward in time estimates of Ne are plotted in figure 1. For both the wisent and the bovine species, a sharp decline in Ne started at the end of the last glacial period, predating cattle domestication approximately 10,000 YBP as already suggested for the latter species by Macleod et al. (2013). This most presumably originates from a growing impact of human activities via hunting intensification and land anthropization especially after the development of agriculture (Soareset al. 2010). Interestingly, the estimated wisent population sizes clearly oscillated according to the succession of glacial Pleistocene periods to approximately 500,000 YBP (viewed backward in time) with interglacial periods coinciding with lower-population sizes. Conversely, from the Last Glacial Maximum (LGM) approximately from 20,000 (Clark et al. 2009) to 150,000 YBP, the estimated bovine population sizes only slightly decreased, remaining always between 10,000 and 20,000 individuals (more than two to three times lower than wisents, as mentioned above).
Fig. 1.
Population size histories inferred from the two wisent and the four bovine genomes under the PSMC model. Backward in time (in YBP) estimates of the effective population sizes derived from the psmc analyses of the BBO_3569 and BBO_3574 individual wisent genomes and the AAN_0037, HOL_0101, JER_0009, and SIM_0043 bull bovine genomes and assuming (Liu et al. 2006) and g = 6 (Keightley and Eyre-Walker 2000; Gautier et al. 2007). At the bottom of the figure, the timing of cattle domestication and the zebu (BIN) and taurine (BTA) divergence (Ho et al. 2008) are indicated by a dark red triangle while the timing interval of the bovine and wisent divergence (see the main text) is indicated by dark red arrows. Similarly, European ice ages (Würm, Riss, Mindel, and Günz glacial stages) according to the Penck and Bruckner work (Penck and Brückner 1901–1909) cited in Elias (2013) are indicated by blue intervals at the top of the figure. Finally, the LGM, approximately 20,000 YBP (Clark et al. 2009), is indicated by a vertical blue dotted line.
Hence, even if the fluctuating pattern observed for wisent might correspond to actual changes in the population sizes, the comparison with the bovine species might lead to an alternative scenario of population fragmentation. In such a scenario, the alternation of population isolation events due to fragmentation of the habitats during glacial periods leads to an increase of the overall estimated Ne, while a more continuously interbreeding between populations during interglacial periods leads to a decrease in the overall estimated Ne. Indeed, as recently evidenced by Mazet et al. (2015) when considering structured populations, changes in the gene flow patterns result in changes in the estimated Ne (even if the actual Ne remained constant) as PSMC assumes a panmictic model. As a result, one might expect an increase (respectively, a decrease) in the estimated Ne when gene flow and thus the amount of connectivity between (sub)populations decreases (respectively, increases). It is not possible from the PSMC analyses to distinguish between a scenario involving habitat fragmentation and a scenario of actual population size changes. However, it is tempting to speculate from the divergent patterns in the inferred population size across the wisent and bovine species that the ancient aurochs distribution range, possibly restrained to southern regions with more stable habitats, has only been partially overlapping the wisent one (hence the lower Ne).
In agreement with the “refugium theory” (Hofreiter and Stewart 2009), both species might have followed different strategies to survive during Pleistocene glaciations, the wisent remaining in small disconnected refugia while the aurochs migrated southward and possibly eastward (Mona et al. 2010). Conversely, during interglacial periods, wisent may have retrieved a larger distribution area with a reconnection of isolated populations, whereas the aurochs may have recolonized northern and central areas. Accordingly, the divergence of the wisent and the American bison which took place approximately 230,000 YBP (Hassanin et al. 2013) might result from an absence of reconnection between populations separated in Eurasian and North American refugia. Note also that the most recent common ancestor of all the Beringian bison lived from 164,000 to 111,000 YBP (Shapiro et al. 2004), a period that lies within the second peak observed in our estimated wisent population size history (fig. 1).
Finally, in agreement with Macleod et al. (2013) that used a different inference model (and a different time calibration procedure), we also observed a steep increase (viewed backward in time) starting approximately 150,000 YBP in the inferred bovine population sizes that might actually be explained by the divergence of the taurine (BTA) and zebu (Bos indicus [BIN]) lineages (see e.g., supplementary fig. S5 in Li and Durbin, 2011). Indeed, this divergence event is dated between 117,000 YBP and 275,000 YBP based on diversity of cattle mitochondrial sequences (Bradley et al. 1996) and at 147,000 YBP (with a 95% highest posterior density interval ranging from 84,000 to 219,000 YBP) using modern and radiocarbon-dated ancient mitochondrial DNA (Ho et al. 2008). A similar increase starting about 1 MYA could also be observed for the ancestral wisent population size which coincides with the timing of BTA/BBO speciation (see below).
The Wisent and Bovine Species Diverged in the Early Pleistocene and Experienced More Recent Secondary Contacts
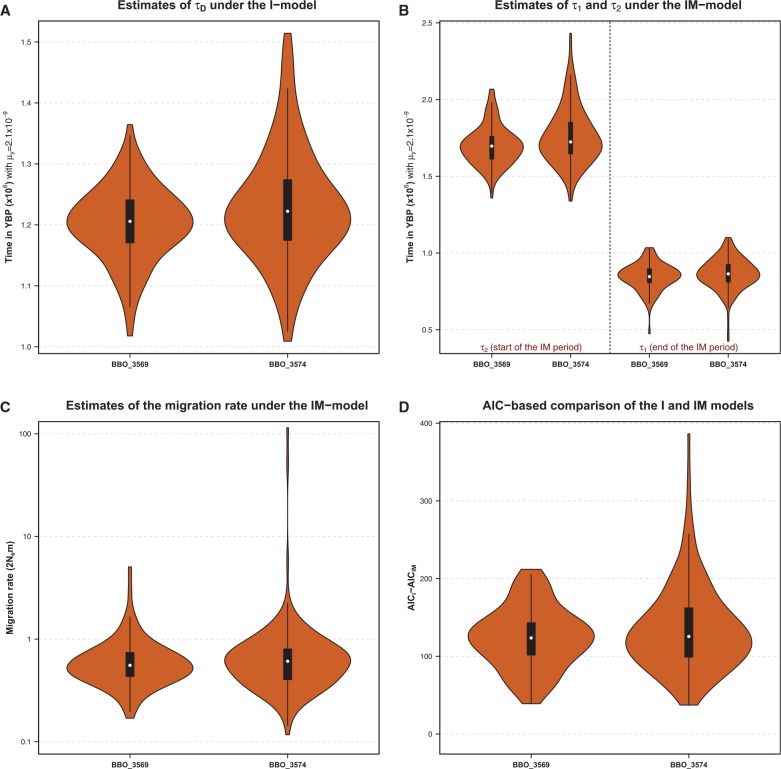
To characterize the divergence between the BBO and BTA lineages, we further relied on the coalescent hidden Markov models (CoalHMM) framework (Hobolth et al. 2007). Two models were considered corresponding to 1) an isolation (I) model that assumes that the ancestral population split into two populations at time τD into the past (Mailund et al. 2011) and 2) an isolation-with-migration (IM) model that assumes that the ancestral population started to split into two populations at time τ2 = τ1+τM and that these two populations exchanged gene with a migration rate M during a period τM until a later time τ1 when gene flow stopped (Mailund et al. 2012). Under the I-model and assuming as above , the median estimate of τD (over 10 Mb segments) was found equal to 1.20 YBP (ranging from 1.02 to 1.36) and 1.23 YBP (ranging from 1.01 to 1.51) when considering the alignments onto the UMD 3.1 bovine genome assembly of the BBO_3569 and BBO_3574 whole-genome sequences, respectively (fig. 2A). As expected, the τD estimates were always intermediate between the estimated start (τ2) and end (τ1) times of the divergence period obtained under the IM-model (fig. 2B). Hence, when considering the BBO_3569 (respectively, BBO_3574) sequence, the median estimates of τ1 and τ2 allowed to define a period ranging from 0.846 to 1.70 YBP (respectively, from 0.864 to 1.72 YBP) for the divergence of the BBO and BTA lineages. During the corresponding divergence periods, the estimated number of migrants (per generation) was found limited with median values of 0.56 and 0.61 for the BBO_3569 and BBO_3574 analyses, respectively (fig. 2C). It should also be noticed that the ancestral population sizes estimated under both the I-model (median values of 62,120 and 65,740 for the BBO_3569 and BBO_3574 analyses, respectively) and the IM-model (median values of 50,750 and 53,430 for the BBO_3569 and BBO_3574 analyses, respectively) were in agreement with those obtained above with the PSMC for the corresponding time period (fig. 1). Finally, as shown in figure 2D, the comparison of the I- and IM-models of speciation provided a clear support in favor of the IM-model. From these CoalHMM analyses, we thus conclude that the BBO and BTA lineages diverged in the early Pleistocene (from 1.7 to 850,000 YBP), in agreement with the previous studies that relied on alternative time calibrations (Bradley et al. 1996; Troy et al. 2001). Moreover, the divergence between these two lineages involved an extended period of limited gene flow similar to the speciation process previously reported in great apes (Mailund et al. 2012).
Fig. 2.
Characterization of the divergence between the BBO and BTA lineages under the isolation (I-) and the isolation-with-migration (IM-) models. (A) Estimation of the time of divergence between the BBO and BTA lineages under the I model (that assumes a clean split). The two violin plots show the distribution of estimates obtained for each 10 Mb nonoverlapping segments from the alignments of the BBO_3569 (n = 123 segments) and BBO_3574 (n = 177 segments) whole-genome sequence onto the UMD 3.1 bovine genome assembly. Time scale was translated into YBP assuming (Liu et al. 2006). (B) Estimation of starting () and ending () time estimates of the divergence between the BBO and BTA lineages under the IM model (that assumes the two ancestral lineages exchange migrants between and ). The violin plots show the distribution of and time estimates obtained for each 10 Mb nonoverlapping segments from the alignments of the BBO_3569 (n = 123 segments) and BBO_3574 (n = 177 segments) whole-genome sequence onto the UMD 3.1 bovine genome assembly. Time scale was translated into YBP assuming (Liu et al. 2006). (C) Estimation of the average number of migrant per generation () during the divergence of the BBO and BTA lineages under the IM model. The two violin plots show the distribution of the migration rate estimates obtained for each 10 Mb nonoverlapping segments from the alignments of the BBO_3569 (n = 123 segments) and BBO_3574 (n = 177 segments) whole-genome sequence onto the UMD 3.1 bovine genome assembly. (D) Model comparisons between the I and the IM models. The two violin plots show the distribution of the difference between the Akaike Information Criterion for the isolation (AICI) and for the IM (AICIM) obtained for each 10 Mb nonoverlapping segments from the alignments of the BBO_3569 (n = 123 segments) and BBO_3574 (n = 177 segments) whole-genome sequence onto the UMD 3.1 bovine genome assembly. Because the model with the smallest AIC should be preferred, the distributions provide strong support in favor of the IM-model (AICIM is always lower than AICI).
However, both the I- and IM-models were not designed to capture signals from more recent gene flow events associated to secondary contacts between the bovine and wisent species. To that end, we compared the relative abundance of ABBA and BABA site patterns (Green et al. 2010; Durand et al. 2011) defined across the following four taxons: 1) BBO; 2) BTA; 3) Bos indicus (BIN); and 4) OAR. Assuming a (((BTA;BIN);BBO);OAR) phylogeny (e.g., Buntjer et al. 2002; Ho et al. 2008; Jiang et al. 2014), ABBA (respectively, BABA) sites are those at which the derived allele (“B”) is shared between the nonsister BIN and BBO (respectively, BTA and BBO) lineages, whereas BTA (respectively, BIN) carries the ancestral allele (“A”), as defined by the OAR outgroup. In the absence of (recent) gene flow, both patterns may only result from incomplete lineage sorting and should be equally abundant in the genome while gene flow events between the BTA and BBO lineages would lead to a significant excess of BABA over ABBA sites (Durand et al. 2011). Whether considering the BBO_3569 or the BBO_3574 sequencing data to define the BBO reference (see Materials and Methods), we found a slight but significant excess of “BABA” (n = 49,337 and n = 58,032 for BBO_3569 and BBO_3574, respectively) over “ABBA” (n = 48,004 and n = 56,357 for BBO_3569 and BBO_3574, respectively). Accordingly, the D-statistic defined as the normalized difference in the counts of ABBA and BABA sites (Green et al. 2010; Durand et al. 2011) was found significantly negative for both the BBO_3569 and BBO_3574 derived consensus sequences with values equal to −1.25% (SD = 0.032%; Z = 39.0) and −1.37% (SD = 0.031%; Z = 44.4), respectively. The corresponding proportion of BBO ancestry into (European) cattle was found equal to (SD = 0.004%) which is actually a conservative minimum (Green et al. 2010). Assuming a constant population size, the bias is equal to ta/ts = 0.28/0.85 = 0.33 in the worst scenario since ta represents the timing of the BBO and BTA admixture (<275,000 YBP, the lower bound of the estimated divergence between the taurine and zebu lineages, see above) and ts the timing of the bovine/wisent speciation (>850,000 YBP, the upper bound found above under the IM-model). Even if the resulting corrected proportion () remains very small, the observed (significant) footprints of BBO ancestry into the genomes of European cattle supports the occurrence of secondary contacts between these two lineages that are posterior to the Bos taurus and BIN divergence (∼150,000 YBP, see above). Analyzing non-European (e.g., African) taurine genomes might allow to refine the timing of such secondary contacts by assessing whether they still occurred after (taurine) cattle domestication (∼10,000 YBP). To that respect also, sequencing individuals (or fossil remains) from extant (B. bison, most particularly) or extinct (e.g., aurochs and B. priscus) sister species would also be very informative. Conversely, it should be noticed that genomic data from Bison species would also allow to quantify the proportion of bovine ancestry (if any) in the BBO genome.
Identification and Functional Annotation of Genes under Positive Selection
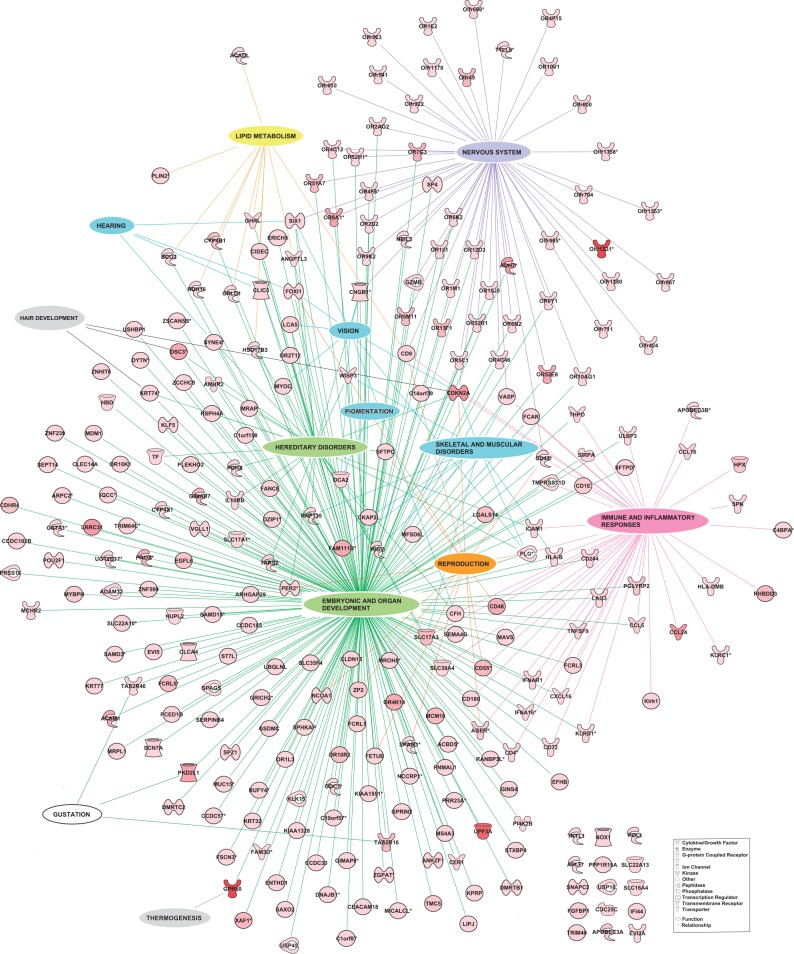
To identify genes under positive selection between the wisent and bovine lineages, we computed the KaKs ratio of nonsynonymous (Ka) and synonymous (Ks) substitution rates (Kimura 1983) for 17,073 protein sequence alignments between the BBO and BTA genomes. As expected from the effect of purifying selection (Hurst 2002), the average Ka/Ks was found equal to 0.273 (ranging from 0.00 to 17.2) with a distribution highly shifted toward 0 (supplementary fig. S6, Supplementary Material online). Transcripts with Ka/Ks > 1 were further considered under positive selection leading to the annotation of 425 genes ready to functional and gene network analyses (see Materials and Methods, supplementary table S3, Supplementary Material online). Overall, the most significant functions underlying these genes are related to 1) nervous system; 2) immune and inflammatory responses; 3) embryonic and organ development; 4) cellular morphology and organization; and 5) skeletal and muscular disorders (supplementary table S4, Supplementary Material online). Figure 3 illustrates the connection of the selected genes that were annotated with their key underlying functions and table 2 gives a more detailed list of functions and subfunctions. Interestingly, many genes are related to several functions (supplementary table S5, Supplementary Material online), suggesting a pleiotropic role and more strikingly, 85% of the genes participated to a global gene network which is defined by six significant networks connected by up to 11 common molecules (supplementary table S6 and supplementary fig. S7, Supplementary Material online). Note that very similar results are obtained when restricting the functional analysis to the 359 selected genes that are one to one orthologs between cattle and sheep (see table 2 and supplementary table S4, Supplementary Material online). Thanks to the method we used to define BBO/BTA gene sequence alignments, this suggests that our annotation remains somewhat robust to extra variation that could have been identified when mapping BBO reads in genic regions belonging to large gene families or due to copy number variants in the sequenced BBO individuals.
Fig. 3.
Representation of the genes with a Ka/Ks >1 connected to their key functions. Each global function and its links to the corresponding key genes are differently colored, that is, in purple for nervous system, blue for by-products of domestication, green for functions related to embryonic, organ development and hereditary disorders, pink for immune and inflammatory responses, orange for reproduction, gray for hair development and thermogenesis, yellow for lipid metabolism, and white for gustation. Global functions contain genes belonging to several Ingenuity Pathway Analysis functional categories. Gene symbols are colored in red and color intensity is correlated to the Ka/Ks value.
Table 2.
Main Genes under Selection Listed by Key Functions and by Presumed Adaptation.
| Key Function | Color in Figure 3 | Subfunction | Genes | Presumed adaptation |
|---|---|---|---|---|
| Hair development | Gray | Growth of hair follicules | CDKN2A | Adaptation to temperature conditions (cold/temperate) |
| Wooly hair | KRT74a | |||
| Hypotrichosis | DSC3a | |||
| Thermogenesis | Gray | GPR50a | ||
| Gustation | White | PKD2L1,a TAS1R1,a TAS2R16,a TAS2R46a | Adaptation to available food resources and vegetation diversity (forest/steppe habitat) | |
| Nervous system | Purple | Olfaction | ADH7, CNGB1,a Olfr1178,a Olfr1179, Olfr1231,a Olfr1280, Olfr1353, Olfr1358,a Olfr1535, Olfr424,a Olfr49, Olfr541, Olfr600,a Olfr606, Olfr610,a Olfr711, Olfr867,a Olfr905, Olfr922,a Olfr963,a OR10AG1, OR10J5,a OR10V1,a OR12D3,a OR13F1, OR1E2,a OR1J1,a OR1M1,a OR2AG2,a OR2D2,a OR2M5, OR4C12, OR4C46,a OR4F15,a OR4F6, OR51A7,a OR52E8,a OR52H1, OR52R1, OR5C1, OR5M11,a OR6K2,a OR6N2,a OR6Y1,a OR7G3, OR8A1, OR9K2 | |
| Neurogenesis, neurulation | CD44,a CD9,a CDKN2A, GZMB,a NEIL3,a OR8A, SIX1,a SP4,a TTLL8,a VASPa | Wildlife/Domestication | ||
| Hearing | Blue | CLIC5,a FOXL1, SIX1,a SYNE4a | Wildlife/Domestication | |
| Vision | Blue | CNGB1,a GPNMB, LCA5,a MYOC,a OCA2,a TMEM5 | Wildlife/Domestication | |
| Pigmentation | Blue | OCA2a | Wildlife/Domestication | |
| Sketetal and muscular development | Blue | CDKN2A, CKAP2L,a FAM111B,a FOXI1,a KLF5, NCOA1,a NDUFS6, PIGV,a PLG,a RNF135,a SLC17A3,a WISP3a | Wildlife/Domestication | |
| Reproduction | Orange | ACR, AGER,a CD44,a CD48, CD55,a CD9,a CDKN2A, FETUB,a FOXI1,a NCOA1,a PER2,a PLG,a SFTPC,a SPAM1,a ZP2a | Wildlife/Domestication | |
| Immune and inflammatory responses | Pink | ADAM8, AGER,a ANGPTL3,a APOBEC3B,a CCL16,a CCL24,a CCL5,a CD180,a CD1E,a CD244,a CD4,a CD44,a CD48, CD55,a CD72,a CD9,a CDKN2A, CFH, CKAP2L,a CXCL16,a EVI2A,a FCAR,a FCRL1,a FCRL3,a HBD,a HLA-B, HLA-DMB,a ICAM1,a IFI44,a IFNA16, IFNAR1,a KLRC1, KLRD1,a Klrk1,a LAG3,a MAVS,a OR12D3,a PGLYRP2,a PIGV,a PLG,a PPP1R15A,a PRSS16,a RHBDD3,a RTP4, SFTPD, SIRPA, SLC17A3,a SLC39A4,a SPN,a TF,a THPO,a TMPRSS11D,a TNFSF9,a TRIM40,a ULBP3, VASP,a XAF1a | Resistance/tolerance to pathogens (linked to wildlife/domestication) | |
| Metabolism | Yellow | Lipid metabolism | ACADL,a ANGPTL3,a APOF,a BCO2,a CCL5,a CD4,a CD9,a CIDEC,a CYP4B1,a GHRL,a GPLD1,a HSD17B3,a MOGAT3, PLIN2,a RDH16a | Adaptation to dietArtificial selection on dairy traits |
| Embryonic and organ development | Green | Mammary gland development | CD44,a CDN2A PLG,a | Artificial selection on dairy traits |
| Other | ACR, AGER,a CD44,a CD9,a CDKN2A, CKAP2L,a CLDN15,a FCAR,a FETUB,a FOXI1,a ICAM1,a IFNAR1,a KLF5, MAVS,a NCOA1,a ODC1, PLG,a POU2F1,a RNF135,a SFTPD, SIX1,a SPAM1,a TF,a THPO,a VASP,a VGLL1,a ZP2a |
aOne to one orthologs between cattle and sheep.
Among the genes under selection, some underlie obvious distinctive features between wisent and cattle providing in turn insights into the wisent adaptive history (table 2, fig. 3, and supplementary table S6, Supplementary Material online). First, KRT74 and DCS3 that are involved in wooly hair development and hypotrichosis, respectively (Ayub et al. 2009; Shimomura et al. 2010), and GPR50 that plays a role in thermogenesis (Bechtold et al. 2012) might be directly related to wisent adaptation to colder climatic conditions (table 2 and supplementary table S4, Supplementary Material online). Second, genes encoding olfactory and taste receptors are probably footprints of feeding behavior modifications, resulting, for example, from food resource differences in forest versus steppe habitat (trees–shrubs vs. grass). Third, the many genes related to immune and inflammatory responses (through their key role in activation, migration, binding, expansion, and modulation of a broad range of immune cells, supplementary table S5, Supplementary Material online) may sign adaptation of wisent and bovine to different pathogen exposures. Fourth, several genes were found involved in lipid metabolism or mammary gland development (supplementary table S5, Supplementary Material online), likely the result of selection in cattle for improved milk production performances.
More strikingly, the functional analysis of the genes under selection highlighted physiological functions associated to the domestication process (fig. 3, table 2, and supplementary table S6, Supplementary Material online), as expected from the close relatedness of wisent and domestic cattle. These functions underlie both key processes of nervous system (e.g., neurogenesis, neurulation, remodeling of dendrites, and differentiation of some neural cells) and by-product phenotypes of domestication (e.g., skeletal and muscular disorders, hair and skin properties, vision, hearing, and reproduction). Our analysis thus gives an empirical support to the unified explanation of the “domestication syndrome” in mammals (i.e., the general combination of observed traits in domestic mammals) formulated by Wilkins et al. (2014) following the pioneering work by Belyaev (1979) and Trut (2009). This explanation accords a central role to the neural crest through its developmental reduction as a result of the primary domestication pressure (i.e., the taming of animals). Furthermore, because neural crest cells are also cellular precursors of many different cells (e.g., osteocytes, chondrocytes, odontoblastes, and melanocytes), this reduction indirectly produces various secondary phenotypic changes (e.g., skeletal and craniofacial morphological modifications or coat-color changes). For instance, such an indirect relationship is well supported by the studies on coat coloration in wild and domesticated animals (Cieslak et al. 2011) and the description coat-color-associated mutations with pleiotropic effects (Reissmann and Ludwig 2013). Also, the numerous genes detected under selection and involved in immune response (table 2 and fig. 3) might be indirectly related to the domestication process. During this process and afterward, herding conditions may have incidentally lead to an increased pathogen exposure of domestic cattle due to proximity with individuals from the same or different species (Freeman et al. 2008).
Conclusions
To characterize the wisent demographic and adaptive histories, we carried out whole-genome sequencing of two males from the Białowieża lowland line. Although still considered as a vulnerable species, our results show that the conservation plan and subsequent management practices have been efficient to recover a reasonable amount of genetic variability that now compares to that observed in commercial cattle breeds. We further confirmed at the nucleotide level, the close relatedness of the wisent and cattle species. We estimated that the divergence between the bovine and wisent lineages occurred in the early Pleistocene through a speciation process involving limited gene flow, lasting from 1.7 × 10 − 6 to 850,000 YBP. We also found evidence for more recent secondary contacts, posterior to the BTA and BIN divergence (∼150,000 YBP), between the wisent and (European) taurine cattle lineages. Interestingly, whole individual genome-based demographic inference highlighted contrasting patterns in both species that might be reminiscent of different adaptive strategies (habitat fragmentation vs. migration) to survive Pleistocene glaciations. Our results are indeed in agreement with a scenario in which wisent survive in refugee pockets during glaciations (leading to habitat fragmentation), whereas aurochs migrate southward (and possibly eastward), where climate remained more temperate (Sommer and Nadachowski 2006). It is tempting to speculate that these two alternative strategies might have contributed to the survival of both species to the large mammal’s Quaternary extinction. Our results also show that wisent and aurochs display a similar trend toward extinction from the LGM, 20,000 years ago that is concomitant to human population growth (spread of farming and increased hunting pressure), to climate warming and to vegetation changes (e.g., replacement of open steppe by forests). For the wisent, in particular, the Holocene period was characterized by a tendency to shift toward forested habitats imposing a significant diet change (Kerley et al. 2012; Bocherens et al. 2015). These new constraints left footprints at the genomic level as illustrated by the genes that we found under selection when comparing the wisent and bovine lineages, that is, genes involved in feeding behavior and in adaptation to temperature conditions and to pathogen exposure. Conversely, wisent being the closest extant wild relative species of domestic cattle, several of the genes under selection could be related to the adaptive response to cattle domestication via their implication in nervous system development and in the expression of by-product domestication phenotypes. Strikingly, this result is in line with unified explanation of the domestication syndrome in mammals formulated by Wilkins et al. (2014) giving a shared developmental connection of these diverse traits via neural crest cells.
Materials and Methods
Sample Origin
Genomic DNA of two male wisents, namely BBO_3569 and BBO_3574, was extracted from blood samples collected in 1991 in the Białowieża forest. More precisely, BBO_3569 and BB0_3574 belong to the genetically isolated pure lowland line originates from seven founders kept in zoo and private breeding centers and used for the species restoration program at the beginning of the 1920s (Tokarska et al. 2009). In 1952, 40 of their descendants that were born in captivity were reintroduced to the wild in the Polish part of the Białowieża forest, the so-called lowland line now including more than 2,000 individuals (i.e., about half of the world wisent population).
High-Throughput Sequencing of Two Wisents
The Illumina TruSeq DNA sample preparation kit (FC-121-2001, Illumina Inc., San Diego, CA) was used according to the manufacturer’s protocol. Libraries were then validated on a DNA1000 chip on a Bioanalyzer (Agilent) to determine size and quantified by qPCR using the Kapa library quantification kit to determine concentration.
The cluster generation process was performed on cBot (Illumina Inc.) using the Illumina Paired-End DNA sample preparation kit (FC-102-1001, Illumina Inc.). Both BBO individuals were further paired-end sequenced on the HiSeq 2000 (Illumina Inc.) using the Sequence by Synthesis technique. Base calling was achieved by the RTA software (Illumina Inc.). Reads of 100 bp from both sides of the fragments were thus obtained after this step. Quality control of the sequences was checked using the FastQC software.
Mapping Wisent Sequencing Reads onto the Bovine Genome
In total, 331,975,598 and 407,585,788 reads paired in sequencing were available for BBO_3569 and BBO_3574, respectively, after Illumina Quality Check. After removal of sequencing adapters, these reads were mapped onto the UMD 3.1 cattle (Bos taurus) reference genome assembly (Liu et al. 2009) using default options of aln and sampe programs from the bwa (version 0.6.2) software package (Li and Durbin 2009). In total, 92.7% (respectively, 92.2%) of the reads from the BBO_3569 (respectively, BBO_3574) library were successfully mapped onto the bovine assembly, 94.5% (respectively, 93.6%) of which being properly paired. Read alignments with a mapping quality Phred-score < 20 and polymerase chain reaction duplicates were further removed using the view (option-q 20) and rmdup programs from the samtools (version 0.1.19) software package (Li and Durbin 2009). The resulting bam files are available for download from the Sequence Read Archive repository (http://www.ncbi.nlm.nih.gov/sra) under the accession number SRP070526. It should be noted that mapping the wisent reads onto the preliminary Y-chromosome (BTAY) sequence from the BosTau7 assembly (available at http://www.genome.ucsc.edu/) lead to a high proportion of reads improperly paired and only approximately 12% of the BTAY sequence was covered with an unexpected high read coverage (>60X). We thus decided not to consider the chromosome Y in further analyses.
Construction of Individual Wisent Consensus Genome Sequences
For the analyses of BBO/BTA cattle divergence, consensus genome sequences were built for each individual by first generating a mpileup file considering each European bison bam alignment file separately and using the mpileup program from the samtools (version 0.1.19) software package (Li and Durbin 2009) run with –C 50 and –d 5000 options. For each BBO individual and at each position (in the bovine reference assembly), the retained consensus base was randomly sampled among the aligned bases after discarding those showing a base alignment quality (BAQ) > 30 (allowing to account for uncertainty resulting from small indels). Such a procedure was aimed at limiting biases toward the bovine reference base at BBO heterozygous sites. Finally, positions with a read depth DP < 3 and DP > 30 (i.e., three times the average individual read coverage), or for which the retained base was supported by less than two reads (to limit sequencing error biases) were treated as N (not called) in the consensus sequences.
Whole-Genome Sequence of Four Individual Bovine Genomes
Four bovine individual whole-genome sequences were obtained from the 1,000 bull genome projects data (Daetwyler et al. 2014) stored in the NCBI sra archive website (http://www.ncbi.nlm.nih.gov/sra). More precisely, the four males selected were AAN_0037, HOL_0101, JER_0009, and SIM_0043 and belonged to the Angus, the Holstein, the Jersey, and the Simmental European taurine breeds, respectively, and were sequenced at a roughly similar coverage (8.2X, 9.6X, 11X, and 10X, respectively) than the European bisons. As for the European bison sequencing data, reads with a mapping quality Phred-score < 20 and polymerase chain reaction duplicates were filtered out from the downloaded bam files using the view (option-q 20) and the rmdup programs from the samtools (version 0.1.19) software package (Li and Durbin 2009).
Testing for Recent Gene Flow Events between the BBO and BTA Lineages Using the ABBA-BABA Statistics
To count the number of sites with ABBA and BABA patterns across the BTA, BIN, BBO, and OAR lineages, we first identified sites displaying a “BA” pattern by comparing the consensus sequences derived from each individual BBO to the OAR reference genome assembly. To that end and following the same procedure and program options as the ones described above for the mapping of reads onto the UMD 3.1 bovine genome assembly (see Results and supplementary table S1, Supplementary Material online), we mapped the sequencing reads originating from the BBO_3569 and BBO_3574 European bisons onto the OviAr3 OAR genome assembly (Jiang et al. 2014). Based on the resulting mpileup alignment files, we then defined for each BBO individual a consensus base as described above (see the section “Construction of Individual Wisent Consensus Genome Sequences”) except that the upper DP threshold was set to 20 (to account for the lower coverage when mapping reads onto the ovine genome as summarized in supplementary table S1, Supplementary Material online). The “BA” sites were those at which the consensus base differed from the OAR reference. To further identify whether the “A” or “B” allele was present in the BTA and BIN genomes, the OAR sequences surrounding each of these sites were extracted (60 nt upstream and 60 nt downstream) and aligned onto the BTA UMD 3.1 (Liu et al. 2009) and BIN genome (Canavez et al. 2012) assemblies using the blat (version v35 × 1) software with default options except for the minimum sequence identity that was set to 95% (Kent 2002). For a given comparison, sequences aligning to more than 10 positions were discarded from further analyses (otherwise, the alignment displaying the highest score was retained). Similarly, sites for which a base different from the “A” and “B” alleles was present in the BIN or BTA sequence or the two upstream and downstream flanking nucleotides did not perfectly match were although discarded. In total, 10,035,538 (respectively, 11,533,956) “BA” sites could be called in both the BIN and BTA assemblies based on the BBO_3564 (respectively, BBO_3574) genomic data. From the observed numbers of “ABBA” and “BABA,” we then computed the D-statistics and both their associated standard error and their corresponding Z-score (to assess whether D significantly differs from zero) using a Weighted Block Jacknife with 5 Mb blocks (Green et al. 2010; Durand et al. 2011). To further estimate the proportion of BBO ancestry in the BTA genome, we relied on the estimator proposed by Durand et al. (2011) here defined as , where S(P1,P2,P3,P4) represents the difference of the “ABBA” and “BABA” site numbers assuming a ((P1;P2);P3);P4) phylogeny and estimated following a procedure similar to the one described above. As for the D-statistic estimates, the standard deviation of was computed using a Weighted Block Jacknife with 5 Mb blocks.
Estimation of Genetic Heterozygosity from Individual Genomes
Genetic heterozygosities were estimated from the individual genome alignments onto the UMD 3.1 cattle assembly (i.e., based on the mpileup files described above) using mlrho version 2.8 (Haubold et al. 2010). Note that mlrho actually implements a maximum likelihood estimator of the population mutation rate (), which fairly approximates heterozygosity under an infinite sites model (and providing θ is small), while simultaneously estimating sequencing error rates and accounting for binomial sampling of parental alleles (Lynch 2008). For a given individual genome alignment, only sites covered by 3–30 reads (after discarding bases with a BAQ < 25) were retained in the computation (supplementary table S2, Supplementary Material online).
Population Size History Inference Using the PSMC
Effective population sizes in units of were estimated backward in time (in units of ) based on individual whole-genome sequences under the PSMC model using the program psmc (version 0.6.4) program (Li and Durbin 2011). Briefly, the analyzed individual whole-genome sequences were obtained from the above described mpileup files (aligned onto the UMD 3.1 cattle assembly) that were processed (individually) with bcftools (version 0.1.19) (Li and Durbin 2009) using the –c option. The resulting vcf files were further converted into fastq files using the vcf2fq utility of the vcfutils.pl program available in the samtools (version 0.1.19) suite (Li and Durbin 2009) discarding sites located less than five nucleotides from an indel (–l 5 option) and covered by less than 3 or more than 30 reads (-d 3 -D 30 options). As originally described, the psmc program was finally run with default options except for the pattern of atomic time intervals (-p option) that were set to “4 + 25*2 + 4 + 6” on a reduced version of the (autosomal) genomes (psmcfa files), with consecutive sites grouped into consecutive bins of 100 nucleotides marked as “K” (at least one heterozygote), “T” (no heterozygote sites), or “N” (less than 90 called sites).
Characterization of the Divergence between BBO and BTA Lineages under the coalHMM Framework
For each BBO individual consensus genome sequence, genome alignment onto the UMD 3.1 bovine reference assembly (see the section “Construction of individual consensus genome sequences” above) was divided into 10 Mb nonoverlapping segments. Segments mapping to the (bovine) X-chromosome or with more than 500,000 (5%) missing information (i.e., an uncalled base in either the BTA or BBO sequence) were further discarded leading to a total of 123 and 177 available segments for the BBO_3569 and BBO_3574 alignments, respectively. Maximum-likelihood inference was then carried out independently for each segment using the scripts isolation-model.py (version 1.1) for the I-model, and the initial-migration-model.py (version 1.2) for the IM-model, from the IMCoalHMM software package (https://github.com/mailund/IMCoalHMM). More precisely, the parameters underlying the I-model include the split time τD (in units of ) and also the ancestral effective population size (in scaled units of ) and the recombination rate ρ (in scaled units of ) that both parameterize the underlying coalescent process. The parameters underlying the IM-model include the times τ1 (completion of the split) and the IM period length τM that both are units of , the migration rate M (in number of migration per substitution) and θ and ρ (as for the I-model). Note that the product corresponds to the estimated number of migrant per generation (in the usual units of , where m is the net migration rate) over the IM period. Finally, for model comparison purposes, the Akaike Information Criterion (AIC) was computed as AICI = 2 × (3 − l) for the I-model and AICIM = 2 × (5 − l) for the IM-model, where l represents the estimated log likelihood of the corresponding model.
Detection of Genes under Selection
Based on the alignment of the BBO_3569 and BBO_3574 individual sequences onto the bovine UMD 3.1 reference genome assembly (mpileup file described above), a consensus BBO sequence was derived for all the n = 22,091 (bovine) ENSEMBL protein-coding sequences as defined in the UCSC genome database (http://genome.ucsc.edu/). At a given position, the retained consensus base corresponded to the most represented one among all the aligned reads from both the BBO_3569 and BBO_3574 individuals and after discarding bases with a BAQ < 30. Positions with a read depth DP < 3 and DP > 60 (three times the average coverage of BTA genome assembly by the combined BBO_3569 and BBO_3574 read sequencing data) were treated as N (not called) in the consensus sequences. BBO consensus protein sequences with more than 10% “N” were discarded from further analysis leading to a total of n = 19,372 remaining BBO/BTA alignments (with 1.11% “N” on average). For 1,306 (6.74%) of these, no nucleotide differences could be observed between the obtained BBO consensus sequence and the BTA one. Nonsynonymous (Ka) and synonymous (Ks) substitution rates were then computed using KaKs_calculator v1.2 (Zhang et al. 2006) assuming a modified Li-Wu-Luo (MLWL) model to estimate the number of synonymous and nonsynonymous sites (Zhang et al. 2006) allowing the estimation of the Ka/Ks ratio for a total of 17,073 protein coding sequences.
Functional Annotation of Candidate Genes under Positive Selection and Gene Network Analysis
Among the 17,073 Ensembl transcripts ID corresponding to the protein coding sequences for which a KaKs ratio was estimated, 873 transcripts with a KaKs ratio above one were considered under positive selection.
Functional annotation of these transcripts and networks analyses were carried out with the Ingenuity Pathway Analysis software (IngenuitySystems, www.ingenuity.com). Among the 873 transcripts with a KaKs ratio above one, 481 transcripts ID were mapped to the Ingenuity Pathway Knowledge Base (IPKB) and were representative of 425 individual genes ready for analysis (supplementary table S3, Supplementary Material online). A total of 405 of these transcripts were further identified as one to one orthologs between cattle and sheep, using Ensembl orthology information (http://www.ensembl.org/biomart/). The top significant functions and diseases (P value < 0.05) were obtained by comparing functions associated with the 425 (among which 359 were one to one orthologs between cattle and sheep) genes against functions associated with all genes in our reference set (14,465 transcripts mapped in IPKB from the 17,073 transcripts ID that were analyzed) using the right-tailed Fisher’s exact test. A total of 262 genes (among which 151 were one to one orthologs between cattle and sheep) participated to the significant functions thus determined (supplementary table S5, Supplementary Material online).
Among the 425 genes, 367 genes (among which 324 were one to one orthologs between cattle and sheep) were included in network analyses. For each network that contains at most 140 molecules, a score S was computed based on a right-tailed Fisher exact test for the overrepresentation of the genes with a KaKs ratio > 1 (S = −log[P value]). A network was considered as significant when S > 3.
Supplementary Material
Supplementary figures S1–S7 and tables S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors wish to thank Anna Madeyska-Lewandowska (Private Veterinary School, Warsaw, Poland) that provided the Bison bonasus samples in the early 1990s as part of a collaborative research project on the characterization of casein variants. They are also grateful to the four anonymous reviewers for their very helpful and constructive comments. This work was supported by an Institut National de la Recherche Agronomique (INRA)-Animal Genetics Department Grant (AFROSEQ project).
References
- Ayub M, Basit S, Jelani M, Ur Rehman F, Iqbal M, Yasinzai M, Ahmad W. 2009. A homozygous nonsense mutation in the human desmocollin-3 (DSC3) gene underlies hereditary hypotrichosis and recurrent skin vesicles. Am J Hum Genet. 85:515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold DA, Sidibe A, Saer BR, Li J, Hand LE, Ivanova EA, Darras VM, Dam J, Jockers R, Luckman SM, et al. 2012. A role for the melatonin-related receptor GPR50 in leptin signaling, adaptive thermogenesis, and torpor. Curr Biol. 22:70–77. [DOI] [PubMed] [Google Scholar]
- Belyaev DK. 1979. The Wilhelmine E. Key 1978 invitational lecture. Destabilizing selection as a factor in domestication. J Hered. 70:301–308. [DOI] [PubMed] [Google Scholar]
- Benecke N. 2005. The Holocene distribution of European bison-the archaeozoological record. Munibe (Antropologia-Arkeologia) 57:421–428. [Google Scholar]
- Bocherens H, Hofman-Kamińska E, Drucker DG, Schmolcke U, Kowalczyk R. 2015. European bison as a refugee species? Evidence from isotopic data on Early Holocene bison and other large herbivores in northern Europe. PLoS One 10:e0115090.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DG, MacHugh DE, Cunningham P, Loftus RT. 1996. Mitochondrial diversity and the origins of African and European cattle. Proc Natl Acad Sci U S A. 93:5131–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntjer JB, Otsen M, Nijman IJ, Kuiper MT, Lenstra JA. 2002. Phylogeny of bovine species based on AFLP fingerprinting. Heredity (Edinb) 88:46–51. [DOI] [PubMed] [Google Scholar]
- Canavez FC, Luche DD, Stothard P, Leite KR, Sousa-Canavez JM, Plastow G, Meidanis J, Souza MA, Feijao P, Moore SS, et al. 2012. Genome sequence and assembly of Bos indicus. J Hered. 103:342–348. [DOI] [PubMed] [Google Scholar]
- Cieslak M, Reissmann M, Hofreiter M, Ludwig A. 2011. Colours of domestication. Biol Rev Camb Philos Soc. 86:885–899. [DOI] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM. 2009. The last glacial maximum. Science 325:710–714. [DOI] [PubMed] [Google Scholar]
- Daetwyler HD, Capitan A, Pausch H, Stothard P, van Binsbergen R, Brondum RF, Liao X, Djari A, Rodriguez SC, Grohs C, et al. 2014. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 46:858–865. [DOI] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias SA. 2013. History of quaternary science In: Elias SA, editor. Encyclopedia of quartenary science, 2nd ed. Amsterdam: Elsevier; p. 10–18. [Google Scholar]
- Freeman AR, Lynn DJ, Murray C, Bradley DG. 2008. Detecting the effects of selection at the population level in six bovine immune genes. BMC Genet. 9:62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Faraut T, Moazami-Goudarzi K, Navratil V, Foglio M, Grohs C, Boland A, Garnier JG, Boichard D, Lathrop GM, et al. 2007. Genetic and haplotypic structure in 14 European and African cattle breeds. Genetics 177:1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanin A, An J, Ropiquet A, Nguyen TT, Couloux A. 2013. Combining multiple autosomal introns for studying shallow phylogeny and taxonomy of Laurasiatherian mammals: Application to the tribe Bovini (Cetartiodactyla, Bovidae). Mol Phylogenet Evol. 66:766–775. [DOI] [PubMed] [Google Scholar]
- Haubold B, Pfaffelhuber P, Lynch M. 2010. mlRho—a program for estimating the population mutation and recombination rates from shotgun-sequenced diploid genomes. Mol Ecol. 19(Suppl 1):277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SY, Larson G, Edwards CJ, Heupink TH, Lakin KE, Holland PW, Shapiro B. 2008. Correlating Bayesian date estimates with climatic events and domestication using a bovine case study. Biol Lett. 4:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolth A, Christensen OF, Mailund T, Schierup MH. 2007. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3:e7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Stewart J. 2009. Ecological change, range fluctuations and population dynamics during the Pleistocene. Curr Biol. 19:R584–R594. [DOI] [PubMed] [Google Scholar]
- Hurst LD. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486.. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, et al. 2014. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 344:1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. 2000. Deleterious mutations and the evolution of sex. Science 290:331–333. [DOI] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerley GIH, Kowalczyk R, Cromsigt JPGM. 2012. Conservation implications of the refugee species concept and the European bison: king of the forest or refugee in a marginal habitat? Ecography 35:519–529. [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Kowalczyk R, Taberlet P, Coissac E, Valentini A, Miquel C, Kamiński T, Wojcik JM. 2011. Influence of management practices on large herbivore diet—case of European bison in Białowieża Primeval Forest (Poland). Forest Ecol Manage. 261:821–828. [Google Scholar]
- Krasińska M, Krasiński ZA. 2002. Body mass and measurements of the European bison during postnatal development. Acta Theriologica 47:85–106. [Google Scholar]
- Kuemmerle T, Hickler T, Olofsson J, Schurgers G, Radeloff V. 2012. Reconstructing range dynamics and range fragmentation of European bison for the last 8000 years. Divers Distrib. 18:47–59. [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Yi S, Makova K. 2002. Male-driven evolution. Curr Opin Genet Dev. 12:650–656. [DOI] [PubMed] [Google Scholar]
- Lister AM, Anthony JS. 2008. The impact of climate change on large mammal distribution and extinction: evidence from the last glacial/interglacial transition. C R Geosci. 340:615–620. [Google Scholar]
- Liu GE, Matukumalli LK, Sonstegard TS, Shade LL, Van Tassell CP. 2006. Genomic divergences among cattle, dog and human estimated from large-scale alignments of genomic sequences. BMC Genomics 7:140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Qin X, Song XZ, Jiang H, Shen Y, Durbin KJ, Lien S, Kent MP, Sodeland M, Ren Y, et al. 2009. Bos taurus genome assembly. BMC Genomics 10:180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen ED, Nogues-Bravo D, Orlando L, Weinstock J, Binladen J, Marske KA, Ugan A, Borregaard MK, Gilbert MT, Nielsen R, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479:359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2008. Estimation of nucleotide diversity, disequilibrium coefficients, and mutation rates from high-coverage genome-sequencing projects. Mol Biol Evol. 25:2409–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod IM, Larkin DM, Lewin HA, Hayes BJ, Goddard ME. 2013. Inferring demography from runs of homozygosity in whole-genome sequence, with correction for sequence errors. Mol Biol Evol. 30:2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailund T, Dutheil JY, Hobolth A, Lunter G, Schierup MH. 2011. Estimating divergence time and ancestral effective population size of Bornean and Sumatran orangutan subspecies using a coalescent hidden Markov model. PLoS Genet. 7:e1001319.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailund T, Halager AE, Westergaard M, Dutheil JY, Munch K, Andersen LN, Lunter G, Prufer K, Scally A, Hobolth A, et al. 2012. A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species. PLoS Genet. 8:e1003125.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet O, Rodriguez W, Grusea S, Boitard S, Chikhi L. 2015. On the importance of being structured: instantaneous coalescence rates and human evolution-lessons for ancestral population size inference? Heredity (Edinb) 116:362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merceron G, Hofman-Kamińska E, Kowalczyk R. 2014. 3D dental microwear texture analysis of feeding habits of sympatric ruminants in the Białowieża Primeval Forest, Poland. Forest Ecol Manage. 328:262–269. [Google Scholar]
- Mikkelsen TS, Hillier LH, Eichler EE, Zody MC, Jaffe DB, Yang SP, Enard W, Hellmann I, Lindblad-Toh K, Altheide TK, et al. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Mona S, Catalano G, Lari M, Larson G, Boscato P, Casoli A, Sineo L, Di Patti C, Pecchioli E, Caramelli D, et al. 2010. Population dynamic of the extinct European aurochs: genetic evidence of a north-south differentiation pattern and no evidence of post-glacial expansion. BMC Evol Biol. 10:83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevado B, Ramos-Onsins SE, Perez-Enciso M. 2014. Resequencing studies of nonmodel organisms using closely related reference genomes: optimal experimental designs and bioinformatics approaches for population genomics. Mol Ecol. 23:1764–1779. [DOI] [PubMed] [Google Scholar]
- Penck A, Brückner E. 1901-1909. Die Alpen im Eiszeitalter. Leipzig: Tauchnitz. [Google Scholar]
- Pucek Z, Belousova Z, Krasińska M, Krasiński ZA, Olech W. 2004. IUCN/SSC Bison Specialist Group. European Bison. Status survey and conservation action plan. Gland, Switzerland and Cambridge, UK: IUCN-The World Conservation Union; p. 54. [Google Scholar]
- Raczyński J. 2015. European bison pedigree book 2014. Białowieża: Białowieża National Park.
- Reissmann M, Ludwig A. 2013. Pleiotropic effects of coat colour-associated mutations in humans, mice and other mammals. Semin Cell Dev Biol. 24:576–586. [DOI] [PubMed] [Google Scholar]
- Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, Sher AV, Pybus OG, Gilbert MT, Barnes I, Binladen J, et al. 2004. Rise and fall of the Beringian steppe bison. Science 306:1561–1565. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Wajid M, Petukhova L, Kurban M, Christiano AM. 2010. Autosomal-dominant woolly hair resulting from disruption of keratin 74 (KRT74), a potential determinant of human hair texture. Am J Hum Genet. 86:632–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Achilli A, Semino O, Davies W, Macaulay V, Bandelt HJ, Torroni A, Richards MB. 2010. The archaeogenetics of Europe. Curr Biol. 20:R174–R183. [DOI] [PubMed] [Google Scholar]
- Sommer RS, Nadachowski A. 2006. Glacial refugia of mammals in Europe:evidence from fossil records. Mammal Rev. 36:251–265. [Google Scholar]
- Tokarska M, Marshall T, Kowalczyk R, Wojcik JM, Pertoldi C, Kristensen TN, Loeschcke V, Gregersen VR, Bendixen C. 2009. Effectiveness of microsatellite and SNP markers for parentage and identity analysis in species with low genetic diversity: the case of European bison. Heredity (Edinb) 103:326–332. [DOI] [PubMed] [Google Scholar]
- Tokarska M, Pertoldi C, Kowalczyk R, Perzanowski K. 2011. Genetic status of the European bison Bison bonasus after extinction in the wild and subsequent recovery. Mammal Rev. 41:151–162. [Google Scholar]
- Troy CS, MacHugh DE, Bailey JF, Magee DA, Loftus RT, Cunningham P, Chamberlain AT, Sykes BC, Bradley DG. 2001. Genetic evidence for Near-Eastern origins of European cattle. Nature 410:1088–1091. [DOI] [PubMed] [Google Scholar]
- Trut L, Oskina I, Kharlamova A. 2009. Animal evolution during domestication: the domesticated fox as a model. Bioessays 31:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkaar EL, Nijman IJ, Beeke M, Hanekamp E, Lenstra JA. 2004. Maternal and paternal lineages in cross-breeding bovine species. Has wisent a hybrid origin? Mol Biol Evol. 21:1165–1170. [DOI] [PubMed] [Google Scholar]
- Wilkins AS, Wrangham RW, Fitch WT. 2014. The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 197:795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik JM, Kawałko A, Tokarska M, Jaarola M, Vallenback P, Pertoldi C. 2009. Post-bottleneck mtDNA diversity in a free-living population of European bison: implication for conservation. J Zool. 277:81–87. [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 4:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.