Abstract
mRNA translation in many ciliates utilizes variant genetic codes where stop codons are reassigned to specify amino acids. To characterize the repertoire of ciliate genetic codes, we analyzed ciliate transcriptomes from marine environments. Using codon substitution frequencies in ciliate protein-coding genes and their orthologs, we inferred the genetic codes of 24 ciliate species. Nine did not match genetic code tables currently assigned by NCBI. Surprisingly, we identified a novel genetic code where all three standard stop codons (TAA, TAG, and TGA) specify amino acids in Condylostoma magnum. We provide evidence suggesting that the functions of these codons in C. magnum depend on their location within mRNA. They are decoded as amino acids at internal positions, but specify translation termination when in close proximity to an mRNA 3′ end. The frequency of stop codons in protein coding sequences of closely related Climacostomum virens suggests that it may represent a transitory state.
Keywords: the genetic code, ciliates, translation termination, stop codon reassignment, alternative genetic decoding
Introduction
The standard genetic code contains 61 amino acid specifying codons and 3 codons that specify translation termination. It was long considered to be unchangeable and its origin was described as a “frozen accident” (Crick 1968). Since then a number of variant genetic codes have been discovered, and the National Center for Biotechnology Information (NCBI) currently reports 18 additional genetic codes alongside the standard one (http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi). The majority of them have been found in mitochondrial and bacterial genomes. The rise of variant genetic codes is due to a change in codon meaning which is referred to as codon reassignment. This phenomenon can occur due to alterations in the components of translation machinery (tRNAs, aminoacyl-tRNA synthetases or release factors), see Baranov et al. (2015) for a review.
Stop codon reassignments are a particularly common feature of mRNA translation in ciliates (Knight et al. 2001; Lozupone et al. 2001). Species belonging to the genera Paramecium, Tetrahymena and Oxytricha are known to translate TAA and TAG as glutamine (Q) and only recognize TGA as a signal for termination (Horowitz and Gorovsky 1985), whereas Blepharisma translates TGA as tryptophan (W) and recognizes TAA and TAG as signals for translation termination (Liang and Heckmann 1993). In Euplotes, TGA is reassigned to cysteine (C) (Meyer et al. 1991) and high frequency of +1 frameshifting during mRNA translation occurs at TAA and TAG codons (Klobutcher and Farabaugh 2002; Wang et al. 2016). It has been conjectured recently that Euplotes species use additional mechanisms to discriminate between TAA/TAG codons specifying ribosomal frameshifting and termination of translation (Lobanov AV, Heaphy SM, Turanov AA, Gerashchenko MV, Pucciarelli S, Devaraj RR, Xie F, Petyuk VA, Smith RD, Klobutcher LA, Atkins JF, Miceli C, Hatfield DL, Baranov PV, Gladyshev VN, unpublished data).
To obtain a more detailed picture of stop codon reassignment events in ciliates, we took advantage of recent advances in large scale sequencing projects. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) (Keeling et al. 2014) provides transcriptomic data for over 650 different marine microbes including ciliates. We obtained RNA-seq from 18 different ciliate genera from the MMETSP. In addition, transcriptomics for four additional genera were obtained from (Feng et al. 2015), one from (Kodama et al. 2014) and one from (Lobanov AV, Heaphy SM, Turanov AA, Gerashchenko MV, Pucciarelli S, Devaraj RR, Xie F, Petyuk VA, Smith RD, Klobutcher LA, Atkins JF, Miceli C, Hatfield DL, Baranov PV, Gladyshev VN, unpublished data). We assembled each transcriptome de novo using Trinity (Grabherr et al. 2011), see Methods section.
Using BLAST (Altschul et al. 1997), we searched each transcript against the NCBI Reference Sequence (RefSeq) protein database with an e-value of 10− 30 as a threshold for significant sequence similarity for individual transcript hits. Table 1 summarizes characteristics of each transcriptome composition and provides information on the number of transcripts with statistically significant similarity hits.
Table 1.
Summary of 24 Species Analyzed; Including the Assembled Transcriptome Size and The Number of Significant Alignment Hits.
| Genus and Species | Assembled | Transcripts | TAA | TAG | TGA | Source |
|---|---|---|---|---|---|---|
| Transcripts | E=10−30 | NCBI/Here | NCBI/Here | NCBI/Here | ||
| Anophryoides haemophila | 14,853 | 2,189 | Q/Q | Q/Q | −/- | Keeling et al. (2014) |
| Aristerostoma sp. ATCC | 30,326 | 3,950 | Q/Q | Q/Q | −/- | Keeling et al. (2014) |
| Blepharisma japonicum | 32,295 | 6,392 | −/- | −/- | W/W | Keeling et al. (2014) |
| Campanella umbellaria | 171,018 | 16,384 | Q/E | Q/E | −/- | Feng et al. (2015) |
| Carchesium polypinum | 87,362 | 8,610 | Q/E | Q/E | −/- | Feng et al. (2015) |
| Climacostomum virens | 23,177 | 5,718 | −/? | −/? | C/? | Keeling et al. (2014) |
| Colpoda aspera | 87,297 | 9,079 | −/- | −/- | C/- | Feng et al. (2015) |
| Condylostoma magnum | 29,437 | 4,510 | Q/Q | Q/Q | −/W | Keeling et al. (2014) |
| Euplotes focardii | 34,984 | 3,939 | −/? | −/? | C/C | Keeling et al. (2014) |
| Euplotes crassus | 33,701 | 3,619 | −/- | −/- | C/C | Lobanov et al. (under review) |
| Fabrea salina | 15,706 | 4,340 | −/- | −/- | C/- | Keeling et al. (2014) |
| Favella ehrenbergii | 31,448 | 3,387 | Q/Q | Q/Q | −/- | Keeling et al. (2014) |
| Litonotus pictus | 30,341 | 2,692 | −/- | −/- | −/- | Keeling et al. (2014) |
| Mesodinium pulex | 84,288 | 7,615 | −/? | −/Y | −/- | Keeling et al. (2014) |
| Myrionecta rubra | 40,881 | 3,579 | −/Y | −/Y | −/- | Keeling et al. (2014) |
| Paralembus digitiformis | 108,308 | 5,579 | Q/Q | Q/Q | −/- | Feng et al. (2015) |
| Paramecium bursaria | 128,693 | 13,341 | Q/Q | Q/Q | −/- | Kodama et al. (2014) |
| Platyophrya macrostoma | 46,111 | 7,407 | Q/- | Q/- | −/- | Keeling et al. (2014) |
| Protocruzia adherens | 42,999 | 4,835 | Q/- | Q/- | −/- | Keeling et al. (2014) |
| Pseudokeronopsis sp.OXSA | 32,771 | 3,919 | Q/Q | Q/Q | −/- | Keeling et al. (2014) |
| Strombidinopsis acuminata | 66,812 | 7,693 | Q/Q | Q/Q | −/- | Keeling et al. (2014) |
| Strombidium inclinatum | 38,510 | 3,545 | Q/Q | Q/Q | −/- | Keeling et al. (2014) |
| Tiarina fusus | 77,484 | 6,261 | Q/Q | Q/? | −/- | Keeling et al. (2014) |
| Uronema sp.Bbcil | 14,501 | 2,843 | Q/Q | Q/Q | −/- | Keeling et al. (2014) |
Note.—Comparison between NCBI genetic codes and the genetic codes inferred in this study (separated with/). - refers to no reassignment and “?” shows that the function of the codon cannot be classified based on threshold used in this study. Organisms with the genetic codes different from the NCBI classification are highlighted in gray.
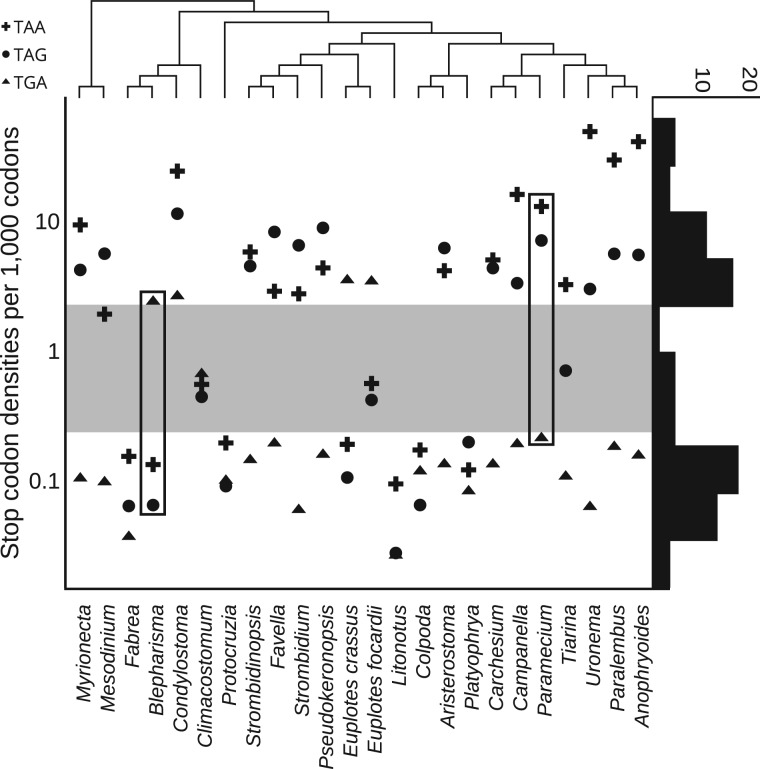
To infer stop codon reassignment events, we first calculated the density of stop codons in pairwise alignments of conceptually translated ciliate mRNAs (with stop codons translated as an unknown amino acid) for each data set. Figure 1 shows the densities of each stop codon (see Methods section for the description of the pipeline). Blepharisma and Paramecium were used as reference organisms for determining a threshold for discrimination between stop codons that were reassigned to code for amino acids and stop codons that function as signals for termination. The threshold is shown as a gray-shaded area in figure 1. It can be seen that the distribution of stop codon1\ frequencies is bimodal with a clear separation between two classes. The few stop codons falling into the gray area may represent very recent stop codon reassignments, transitory states, or may correspond to organisms with a large number of pseudogenes in their genomes or frequent utilization of recoding mechanisms in translation of their transcriptomes. Most organisms have either 1 or 2 stop codons reassigned to amino acids. It is also clear that evolution of TAA and TAG codon meanings is coupled, i.e., if one of these codons is reassigned the other codon is also reassigned. This is most likely because these two codons differ at the wobble position and could be recognized by the same tRNA. A few exceptions where one of these two codons occur in the gray area could be due to inability of the threshold used to provide a clear discrimination (see Discussion section below). Most striking, however, is that all three stop codons in Condylostoma magnum show frequencies indicative of reassignment to sense codons.
Fig. 1.
Classification of ciliate stop codons. Stop codon densities (axis y) in protein coding sequences are indicated for each species (bottom). Rectangles specify stop codons of the organisms used for defining a threshold (gray area) for discriminating reassigned codons (above gray area) from those that retained their function as signals for termination (below gray area). The phylogenetic tree constructed with 18S rRNA sequences above indicates the relatedness of each species. The histogram on the right shows distribution of stop codon densities.
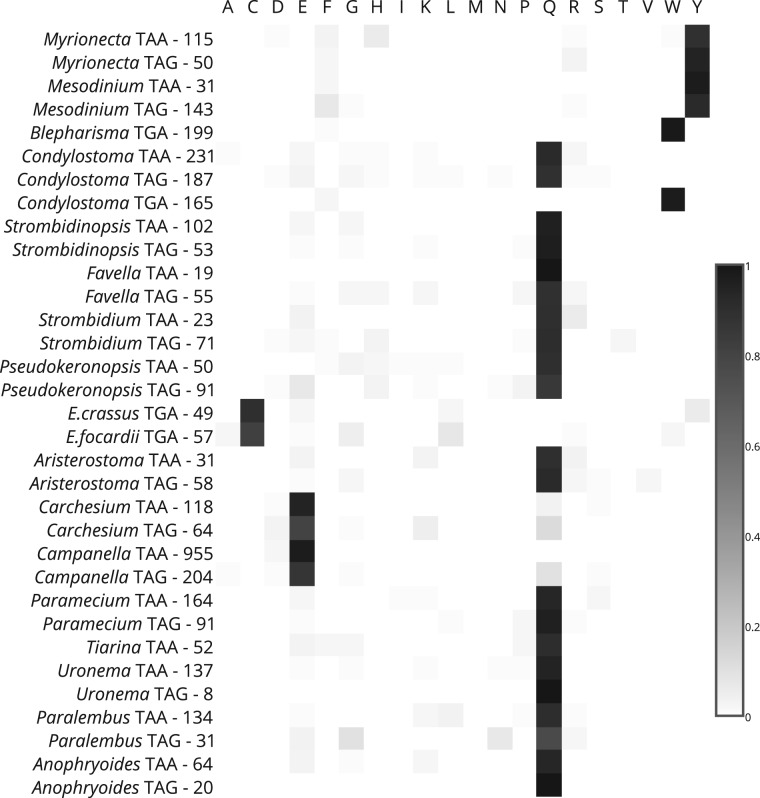
To determine the meaning of reassigned stop codons, we evaluated the frequency of amino acid substitutions in pairwise alignments of translated mRNAs and their close homologs from other species. Occasional matching of a ciliate stop codon (functioning as a terminator) to a sense amino acid in a homolog may occur close to N- or C-termini if a ciliate homolog is shorter, in case of transcribed pseudogenes containing nonsense mutations, when a ciliate transcript contains a sequencing error or when a specific stop codon is recoded to an amino acid in the context of a specific mRNA. However, if a stop codon reassignment took place, it is expected that the reassigned stop codon would frequently match the specific amino acid to which it was reassigned. We provide the total substitution values of all three stop codons for each ciliate in supplementary tables S1–S3, Supplementary Material online. Supplementary figure S1, Supplementary Material online, shows z-scores of amino acid substitution frequencies for each likely reassigned stop codon. It can be seen that for each reassigned stop codon there is only a single amino acid with exceptionally high Z-score. An even clearer picture is obtained when substitution frequencies are calculated only for amino acid residues evolving under strong stabilizing selection (fig. 2 and supplementary table S4, Supplementary Material online).
Fig. 2.
Identification of amino acid specifications of the reassigned codons. Each row corresponds to a single reassigned codon. The organism, the codon identity and the total number of occurrences at highly conserved positions of aligned sequences are indicated on the left. The normalized frequencies of amino acid substitutions are shown as heatmaps.
For Paramecium, we observe that Q is the most frequently substituted amino acid for both TAA and TAG, and for Blepharisma and both Euplotes species tryptophan (W) and cysteine (C) are the most frequently substituted amino acids for TGA, respectively. With the same frequency as Blepharisma, we can clearly see that TGA in Condylostoma is likely reassigned to W along with TAA and TAG also reassigned to Q. The specificity of substitutions in Condylostoma further supports the notion that all three codons are reassigned in this organism. In addition, we report novel stop codon reassignments in Campanella and Carchesium where TAA and TAG appear to code for glutamic acid (E) and in Mesodinium and Myrionecta where TAA and TAG appear to code for tyrosine (Y). In total, we provide evidence in support of redefining the genetic codes of nine ciliates. Table 1 compares the genetic code of each ciliate species analyzed with the NCBI assigned code.
The unclassified, gray-shaded region of figure 1 requires additional attention. It is likely that Mesodinium TAA is reassigned to Y. It is very close to the threshold and such reassignment would be consistent with the function of TAG in Mesodinium. Climacostomum is closely related to Condylostoma and may represent a transitory state that potentially could provide an answer to how Condylostoma emerged as an organism with the genetic code composed of 64 sense codons. Recently, we carried out ribosome profiling analysis of E. crassus translatome and mass-spectrometry analysis of its proteome (Lobanov AV, Heaphy SM, Turanov AA, Gerashchenko MV, Pucciarelli S, Devaraj RR, Xie F, Petyuk VA, Smith RD, Klobutcher LA, Atkins JF, Miceli C, Hatfield DL, Baranov PV, Gladyshev VN, unpublished data). While the analysis revealed thousands of ribosomal frameshifting occurrences at TAA/TAG codons, it revealed no cases of stop codon readthrough that preserved the frame. As can be seen in figure 1, the density of TAA/TAG codons is much higher in E. focardii than in E. crassus and this could be due to potential utilization of stop codon readthrough in addition to ribosomal frameshifting.
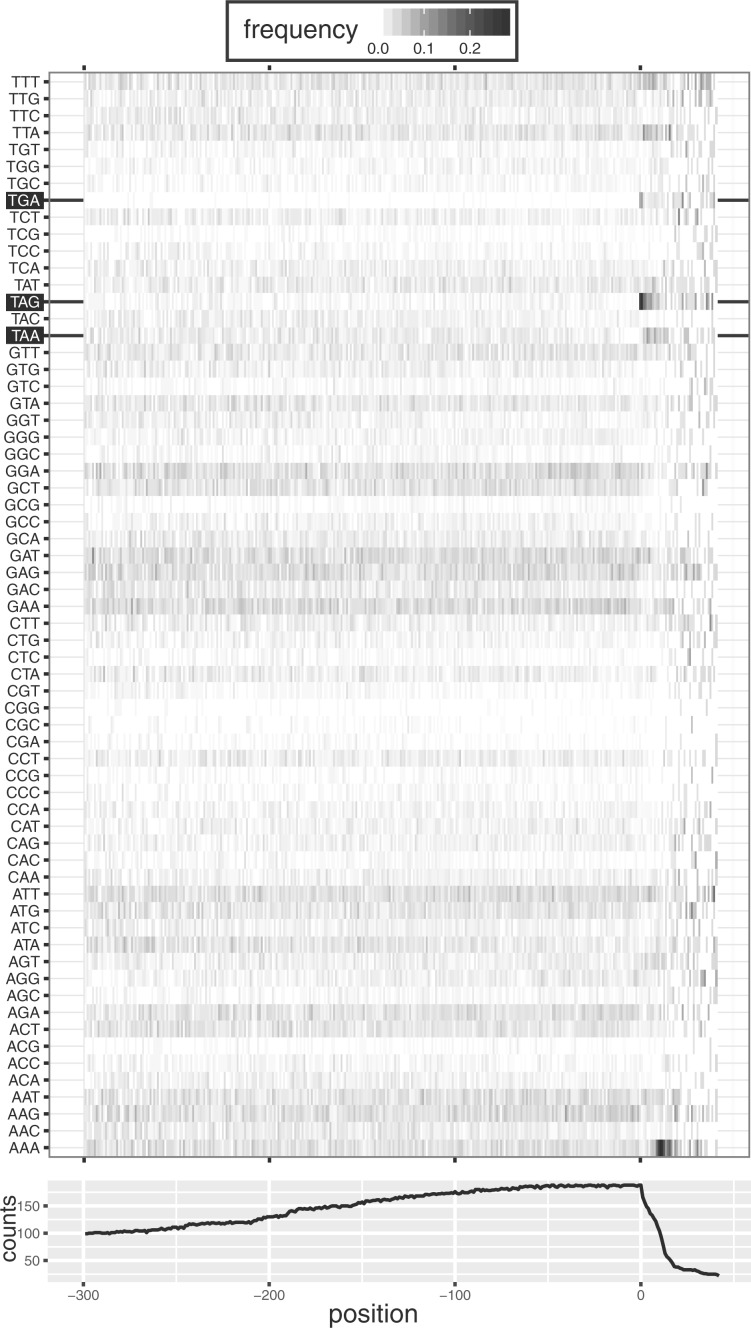
Identification of an organism with all stop codons reassigned to sense codons poses a question of how translation termination is accomplished in Condylostoma. A theoretical possibility is a regulated termination where stop codon function would depend on specific ligands whose expression is regulated by a specific condition. Such a situation has been observed previously in Acetohalobium arabaticum, where the function of the TAG codon as signal for termination or as a codon for pyrrolysine depends on the energy source used by these bacteria (Prat et al. 2012). This, however, seems an unlikely possibility because of very high frequency of stop codons in protein coding genes and tremendous impact of such switches on the whole proteome. An alternative possibility is that the function of a stop codon depends on its position within mRNA. Based on our recent characterization of E. crassus translatome and proteome (Lobanov AV, Heaphy SM, Turanov AA, Gerashchenko MV, Pucciarelli S, Devaraj RR, Xie F, Petyuk VA, Smith RD, Klobutcher LA, Atkins JF, Miceli C, Hatfield DL, Baranov PV, Gladyshev VN, unpublished data) we proposed that the translational machinery of E. crassus is able to discriminate stop codons in internal positions of mRNAs from those at the ends and use only the latter for termination of translation. Such a mechanism could also explain the enigmatic reassignment of all three stop codons in Condylostoma. To address this possibility, we analyzed codon frequencies relative to the expected ends of protein coding sequences (CDS). For this purpose, Condylostoma transcriptome was aligned to the most conserved eukaryotic proteins using eukaryotic orthologous groups (KOGs) (Tatusov et al. 2003). The positions in pairwise alignments matching the stop codons of homologous sequences were considered as expected locations of stop codons in the corresponding Condylostoma sequence. Figure 3 shows frequencies of all 64 codons relative to expected CDS ends. It can be seen that stop codons (TAG and TGA in particular) are overrepresented at the expected locations of CDS ends. Importantly, it also can be seen that ∼15 nt downstream of expected termination locations there is overrepresentation of AAAs which probably reflects locations of mRNA polyA tails. This suggests that 3′-UTRs in Condylostoma are very short and conserved and may be implicated in recognition of stop codons signalling for termination of translation. Consistent with this hypothesis, a depletion of stop codons is observed within ∼30 nt upstream of expected locations of termination sites, which probably is due to selection to avoid premature termination due to close location of stop codons to the polyA tails.
Fig. 3.
Frequency of codons relative to expected positions (zero on axis x) of translation termination in Condylostoma. Top panel—frequency of each out of 64 codons (stop codons are highlighted). Bottom panel—total number of codons found at corresponding location. The total number differs due to variance in transcript and CDS lengths and also due to presence of ambiguous nucleotides (codons with ambiguous nucleotides were ignored).
Since the strength of stop codons as signals for termination is highly dependent on the identity of the nucleotide adjacent at the 3′ end (McCaughan et al. 1995; Poole et al. 1995) we explored the possibility of a particular context preference at internal (reassigned) or terminal positions of coding regions. We observed that in both cases A and T occur more frequently than G and C consistent with AT richness of Candylostoma genome (supplementary fig. S2, Supplementary Material online). However, Ts downstream of TAGs and TGAs are more frequent than As at the sites of termination, but not at the internal positions.
Given that Euplotes and Condylostoma are distant relatives within the ciliophora phylum, it is possible that a polyA distance mechanism of translation termination has emerged in the course of convergent evolution; however, it is also conceivable that the mechanism evolved earlier in the evolution and is common to all ciliates. If the latter is true, it could explain the high propensity of ciliates to stop codon reassignment. The difference in genetic codes among ciliates would depend primarily on the availability of specific tRNAs for recognition of stop codons when those occur in internal positions. Emergence of such tRNAs is not an unlikely event in the light of a recent discovery of substantial variability in identity of codons recoded as selenocysteine in bacteria (Mukai et al. 2016). Sequence analysis of ciliate tRNAs and future experimental studies may shed a light on this intriguing possibility and disclose the molecular machinery used by ciliates to discriminate between stop codons at different positions. Potential possibilities include interactions between polyA biding proteins (PABP) and ribosome complexes with release factors, it has been shown recently that PABPs enhance termination in a mammalian system in vitro (Ivanov et al. 2016). It is also conceivable that the first ribosome reading through all stop codons could stall in the beginning of polyA tails and serve as a barrier for trailing ribosomes favouring termination of translation when the trailing ribosomes are located at stop codons shortly upstream of polyA tails. Ribosome stalling at the beginnings of polyA tails have been observed in a yeast strain lacking ribosome rescue factor Dom34 (Guydosh and Green 2014).
Methods
Data Sources and Assembly
We obtained RNA-seq data for 19 of the MMETSP ciliate species from iMicrobe (http://data.imicrobe.us/), along with four sequence read archive (SRA) files from (Feng et al. 2015) and one SRA file from (Kodama et al. 2014). SRA files were converted to fastq with FASTQ-DUMP. We used RNA-seq forward strand reads to assemble a transcriptome de novo using Trinity version r20140413p1 (Grabherr et al. 2011) for each species. A summary of the assemblies is tabulated in table 1.
Stop Codon Densities and Substitution Frequencies
We performed pairwise alignments of conceptually translated ciliate mRNAs using standalone BLASTX 2.2.31 for each transcriptome against NCBI Reference Sequence (RefSeq) protein sequences database with an e-value of 10−30 as a threshold for significant sequence similarity for individual transcript hits. In order to indicate each stop codon individually, we performed pseudo reassignments of two stop codons to amino acids with the one remaining stop codon translated as an unknown amino acid, denoted by “*”. In total, we carried out three alignments for each of the species analyzed, one per stop codon. The alignments were output in format option 2 “query-anchored no identities”. We removed alignments where hits were originating from mitochondrial and bacterial species to reduce contamination from unintended assembled transcripts. From this output, we were able to calculate the density of stop codons in each query sequence, based on the frequency in the pairwise alignments and the length of the alignment size, as illustrated in figure 1.
Using a custom Python script, we calculated the frequency of amino acid substitutions (20 standard amino acids) in pairwise alignments for each stop codon classified as reassigned (fig. 1). For each amino acid substitution, we calculated corresponding z-scores which are displayed as a heatmap in supplementary figure S1, Supplementary Material online. For figure 2, the analysis was limited only to substitutions at the positions evolving under strong stabilizing selection, i.e., the positions that are at least 95% identical across 100 closest homologs found in RefSeq database. The absolute substitution counts among conserved positions is summarized in supplementary table S4, Supplementary Material online.
Position Specific Codon Frequencies in Condylostoma
Individual transcripts from MMETSP0210 Condylostoma magnum, strain COL2 from iMicrobe (http://data.imicrobe.us/) were searched using a collection of eukaryotic orthologous groups, KOGs (Tatusov et al. 2003). One “profile” alignment was built for each KOG and the pipeline Selenoprofiles (Mariotti and Guigo 2010) was used to perform protein-to-RNA alignments. The hits were filtered with a BLAST e-value threshold 10−10 and a minimum profile coverage of 90% (i.e., the predicted Condylostoma protein sequence was required to span at least 90% of the input KOG alignment). When multiple transcripts matched the same KOGs family, only the best scoring sequence was chosen for further analysis. The sequences at the 3′ of the homologous regions identified in this way in the Condylostoma transcriptome were treated as expected locations of translation termination. The frequency of each of the 64 codons was counted at each position relative to the expected location of termination (fig. 3). The analysis of nucleotide context at the 3′ ends of stop codons was performed in the same way, except that quadruplets (stop codon and adjacent 3′ end nucleotide) were used instead of codons.
Supplementary Material
Supplementary tables S1–S4 and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work has been supported by the Science Foundation Ireland grants number (12/IA/1335 to P.V.B. and 13/1A/1853 to J.F.A.) and National Institute of Health grant number (GM061603 to V.N.G).
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Atkins JF, Yordanova MM. 2015. Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nat Rev Genet. 16:517–529. [DOI] [PubMed] [Google Scholar]
- Crick FH. 1968. The origin of the genetic code. J Mol Biol. 38:367–379. [DOI] [PubMed] [Google Scholar]
- Feng JM, Jiang CQ, Warren A, Tian M, Cheng J, Liu GL, Xiong J, Miao W. 2015. Phylogenomic analyses reveal subclass Scuticociliatia as the sister group of subclass Hymenostomatia within class Oligohymenophorea. Mol Phylogenet Evol. 90:104–111. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R. 2014. Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S, Gorovsky MA. 1985. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci USA. 82:2452–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Mikhailova T, Eliseev B, Yeramala L, Sokolova E, Susorov D, Shuvalov A, Schaffitzel C, Alkalaeva E. 2016. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. doi: 10.1093/nar/gkw635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Farabaugh PJ. 2002. Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell 111:763–766. [DOI] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. 2001. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2:49–58. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Suzuki H, Dohra H, Sugii M, Kitazume T, Yamaguchi K, Shigenobu S, Fujishima M. 2014. Comparison of gene expression of Paramecium bursaria with and without Chlorella variabilis symbionts. BMC Genomics 15:183.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang A, Heckmann K. 1993. Blepharisma uses UAA as a termination codon. Naturwissenschaften 80:225–226. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight RD, Landweber LF. 2001. The molecular basis of nuclear genetic code change in ciliates. Curr Biol. 11:65–74. [DOI] [PubMed] [Google Scholar]
- Mariotti M, Guigo R. 2010. Selenoprofiles: profile-based scanning of eukaryotic genome sequences for selenoprotein genes. Bioinformatics 26:2656–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. 1995. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci USA. 92:5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Schmidt HJ, Plumper E, Hasilik A, Mersmann G, Meyer HE, Engstrom A, Heckmann K. 1991. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc Natl Acad Sci USA. 88:3758–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Englert M, Tripp HJ, Miller C, Ivanova NN, Rubin EM, Kyrpides NC, Soll D. 2016. Facile recoding of Selenocysteine in nature. Angew Chem Int Ed Engl. 55:5337–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole ES, Brown CM, Tate WP. 1995. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 14:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat L, Heinemann IU, Aerni HR, Rinehart J, O’Donoghue P, Soll D. 2012. Carbon source-dependent expansion of the genetic code in bacteria. Proc Natl Acad Sci USA. 109:21070–21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xiong J, Wang W, Miao W, Liang A. 2016. High frequency of + 1 programmed ribosomal frameshifting in Euplotes octocarinatus. Sci Rep. 6:21139.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.